Abstract
S-Adenosyl-L-methionine (AdoMet) is a ubiquitous methyl donor for a variety of biological methylation reactions catalyzed by methyltransferases (MTases). AdoMet analogs with extended propargylic chains replacing the sulfonium-bound methyl group can serve as surrogate cofactors for many DNA and RNA MTases enabling covalent deposition of these linear chains to their cognate targets sites in DNA or RNA. Here we describe synthetic procedures for the preparation of two representative examples of AdoMet analogs with a transferable hex-2-ynyl group carrying a terminal azide or amine functionality. Our approach is based on direct chemoselective alkylation of S-adenosyl-L-homocysteine at sulfur with corresponding nosylates under acidic conditions. We also describe synthetic routes to 6-substituted hex-2-yn-1-ols and their conversion to the corresponding nosylates. Using these protocols, synthetic AdoMet analogs can be prepared within one to two weeks.
Keywords: methyltransferase, cofactor engineering, synthetic AdoMet analogs, mTAG labeling, S-selective alkylation
INTRODUCTION
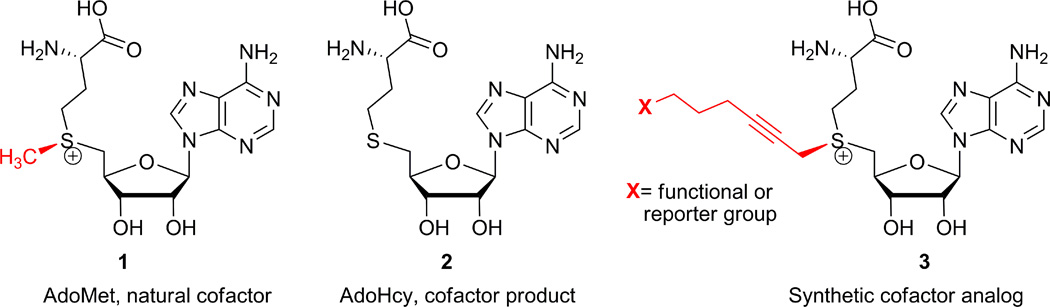
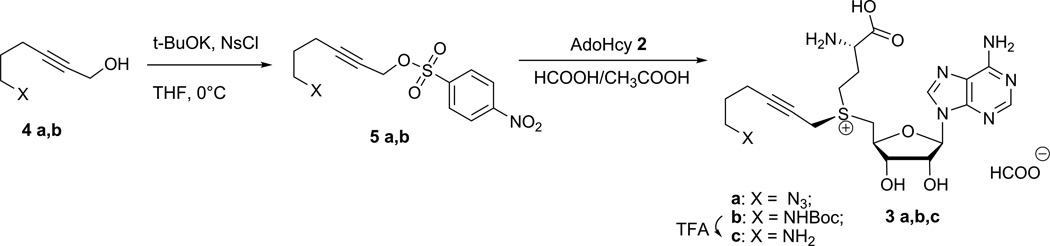
S-Adenosyl-L-methionine (AdoMet or SAM, 1) is the major methyl donor for biological methylations of various biomolecules such as DNA, RNA and proteins in the cell (Figure 1). These reactions are carried out by a vast plethora of methyltransferases (MTases) which catalyze a targeted transfer of the chemically activated sulfonium-bound methyl group from AdoMet 1 to nitrogen, oxygen, carbon or sulfur nucleophiles located on their target biomolecules (Klimašauskas and Lukinavičius, 2008; Grosjean, 2009). To expand the practical utility of this important enzymatic reaction, cofactors with activated sulfonium-bound side-chains have been produced and proved useful for the MTase-directed transfer of these extended side chains (Dalhoff et al., 2006a; Lukinavičius et al, 2007) (Figure 1) onto a variety of target sites in DNA, RNA and proteins (the approach named methyltransferase-directed Transfer of Activated Groups, mTAG). Here we present synthesis of hex-2-yn-1-yl moiety-based cofactor analogs (Figure 1) with a terminal azide 3a or amino 3b group suitable for targeted mTAG labeling of DNA or RNA in vitro and in bacterial cell lysates (Lukinavičius et al, 2012, 2013; Plotnikova et al., 2014). The synthetic strategy involves a direct chemoselective alkylation of S-adenosyl-L-homocysteine (AdoHcy, 2) under mild acidic conditions (Figure 2) (Dalhoff et al., 2006a,b). The chemical “recharging” of AdoHcy 2 yields AdoMet analogs as a 1:1 diastereomeric mixture at the sulfonium center. Since only the (S,S)-diastereoisomer of AdoMet 1 (and presumably its analogs) functions as cofactor for MTases, the protocol also describes chromatographic enrichment of the active diastereomer of cofactor 3a.
Figure 1.
Structure of the natural methyltransferase cofactor AdoMet 1, its synthetic analogs carrying extended propargylic (S-hex-2-yn-1-yl) side chains 3, and the methyltransferase reaction product AdoHcy 2.
Figure 2.
Synthetic route to AdoMet analogs carrying extended sulfonium bound chains with terminal azide (3a) and amine (3c) functional groups via S-selective alkylation of AdoHcy 2.
BASIC PROTOCOL 1
Synthesis of 6-substituted hex-2-yn-1-ols
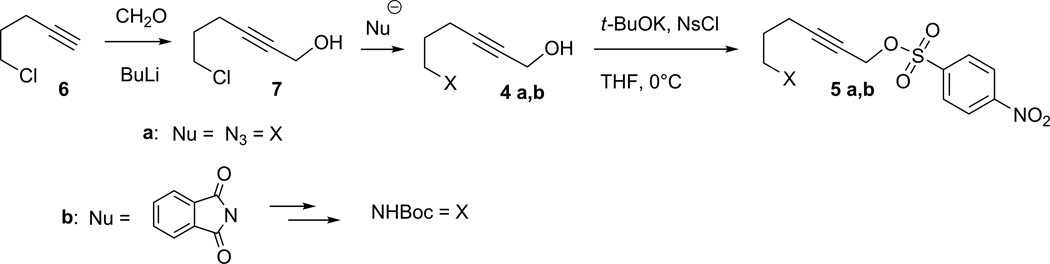
Target alcohols 4 can be synthesized from commercially available 5-chloropent-1-yne (6) by hydroxymethylation of the terminal alkyne group with paraformaldehyde (adapted from Brandsma, 2004) and subsequent nucleophilic substitution of the chlorine with a desired functionality (Figure 3). Here we provide examples of the synthesis of 6-substituted hex-2-yn-1-yl nosylates bearing azido and amino functional groups.
Figure 3.
Synthesis of 6-substituted hex-2-yn-1-yl nosylates.
Materials
5-Chloropent-1-yne (6)
Tetrahydrofuran, anhydrous (THF)
n-Butyllithium (2 M in hexane)
Paraformaldehyde
Ammonium chloride aqueous solution (saturated)
Diethyl ether
Sodium sulphate (Na2SO4)
Deuterated chloroform (CDCl3)
6-Chlorohex-2-yn-1-ol (7)
N,N-Dimethylformamide, anhydrous (DMF)
Silica gel (60 Ǻ, 230–400 mesh)
Methylene chloride (DCM)
Deuterated water (D2O)
Magnetic stir plate and stir bar
Double neck round-bottom flask (100-mL)
Freezing equipment or liquid nitrogen
50-mL Separatory funnel
Argon balloon
Syringe (50-mL)
Rotary evaporator equipped with a vacuum pump
Vacuum distillation apparatus
NMR tube
Round-bottom flask, oven dried
Sand bath
Chromatography column
Test tubes
Materials specific for Step 2A
Sodium azide (NaN3)
Tetra-n-butylammonium bromide (TBAB)
Benzene
Materials specific for Step 2B
Filtration system
Triethylamine (TEA)
Potassium phthalimide
Methanol
Hydrazine hydrate (N2H4, 50–60%)
Ethanol
Di-tert-butyl dicarbonate (Boc2O)
0.5 M Hydrochloric acid
Concd hydrochloric acid (HCl)
Brine (saturated solution of NaCl)
Chloroform
Ethyl acetate (EtOAc)
Step 1-Hydroxymethylation of 5-chloropent-1-yne (6)
Perform hydroxymethylation
-
1
Add 10 mL of 5-chloropent-1-yne (6) (93 mmol) into a double neck round-bottom flask.
-
2
Place a magnetic stir bar and pour anhydrous tetrahydrofuran (120 mL).
-
3
Cool to −78 °C using freezing equipment or liquid nitrogen.
-
4
Introduce an argon atmosphere through the one neck of a flask and maintain it throughout the reaction. Put a separatory funnel into other neck of the flask.
-
5
Transfer 48 mL of n-butyllithium (93 mmol, 1 eq) using a syringe from bottle to the separatory funnel.
-
6
Add n-butyllithium dropwise to the flask at such a rate that −78 °C is maintained and stir for another 30 min.
-
7
Add 3.36 g of paraformaldehyde (104 mmol, 1.1 eq) to the reaction vessel and stir for another 30 min.
-
8
Remove the cooling bath and leave the reaction vessel to warm to room temperature (approx. 1 h).
-
9
Quench the reaction with 100 mL of cold ammonium chloride.
-
10
Extract the product with diethyl ether (3 × 30 mL).
-
11
Dry combined organic layers over sodium sulphate.
-
12
Remove the solvent under reduced pressure using a rotary evaporator equipped with a vacuum pump.
-
13
Purify the product by distillation under vacuum (bp 55–57 °C (0.04 mmHg)). The resulting product, 6-chlorohex-2-yn-1-ol (7), is obtained in 58% yield (6.5 mL) as clear oil.
-
14
Take 10 mg of the product to an NMR tube and dissolve it in 0.65 mL of deuterated chloroform. Performed 1H NMR analysis gives following spectrum - 1H NMR (300 MHz, CDCl3): δ = 4.27 (t, 5J = 2.1 Hz, 2H, CH2OH), 3.67 (t, 3J = 6.3 Hz, 2H, CH2Cl), 2.44 (tt, 3J = 6.3 Hz, 5J = 2.1 Hz, 2H, C≡CCH2), 1.98 (m, 3H, CH2CH2CH2, OH).
Step 2 - Incorporation of a terminal functional group
A. Synthesis of 6-azidohex-2-yn-1-ol (4a)
Perform incorporation of a terminal azido functional group
-
15a
Add 0.2 g of 6-chlorohex-2-yn-1-ol (7) (1.5 mmol) in a round-bottom flask containing a magnetic stir bar.
-
16a
Pour anhydrous DMF (8 mL) and stir till the solution is clear.
-
17a
Add 0.294 g of sodium azide (4.5 mmol, 3 eq) and 0.048 g of tetra-n-butylammonium bromide (0.15 mmol, 0.1 eq) to the reaction vessel.
-
18a
Stir the reaction mixture for 24 h at 80 °C (sand bath).
-
19a
Add 5 mL of water to the reaction vessel.
-
20a
Extract the product with diethyl ether (3 × 15 mL), combine organic layers, dry over sodium sulphate and evaporate under reduced pressure using a rotary evaporator equipped with a vacuum pump.
-
21a
Purify the residue by column chromatography (elute impurities with benzene, Rf = 0.8, then elute the target product with methylene chloride Rf = 0.3). The resulting product, 6-azidohex-2-yn-1-ol (4a), is obtained in 70% yield (0.146 g, 1.05 mmol) as clear oil.
-
22a
Take 10 mg of the product to an NMR tube and dissolve it in 0.65 mL of deuterated chloroform. Performed 1H NMR analysis gives following spectrum - 1H NMR (300 MHz, CDCl3): δ = 4.28 (t, 5J = 2.4 Hz, 2H, CH2OH), 3.44 (t, 3J = 6.9 Hz, 2H, CH2N3), 2.37 (tt, 3J = 6.9 Hz, 5J = 2.4 Hz, 2H, C≡CCH2), 1.81 (m, 3H, CH2CH2CH2, OH).
B. Synthesis of 6-(N-Bocamino)hex-2-yn-1-ol (4b)
Perform incorporation of an amino group
-
15b
Add 0.2 g of 6-chlorohex-2-yn-1-ol (7) (1.5 mmol) in a round-bottom flask containing a magnetic stir bar.
-
16b
Pour anhydrous DMF and stir till the solution is clear.
-
17b
Add 0.315 g of potassium phthalimide (0.315 g, 1.7 mmol) to the reaction vessel.
-
18b
Stir the reaction mixture for 1 h at 80 °C (sand bath).
-
19b
Remove the solvent under reduced pressure using a rotary evaporator equipped with a vacuum pump. Formed 6-phthalimidohex-2-yn-1-ol is used in the following synthesis without further purification.
-
20b
Dissolve 0.85 g of the synthesized 6-phthalimidohex-2-yn-1-ol (3.5 mmol) in methanol (15 mL).
-
21b
Pour 0.35 mL of hydrazine hydrate (7 mmol) to the reaction vessel.
-
22b
Heat under reflux the reaction mixture for 2 h and then cool it to room temperature.
-
23b
Remove the solvent under reduced pressure using rotary evaporator equipped with a vacuum pump.
-
24b
Add a mixture of water and ethanol (10 mL, 1:1) and concd. hydrochloric acid (10 mL).
-
25b
Heat under reflux the mixture for 20 min.
-
26b
Remove the precipitate by filtration.
-
27b
Concentrate the filtrate under reduced pressure. The resulting product, 6-aminohex-2-yn-1-ol hydrochloride, is obtained in 70% yield (0.37 g, 2.45 mmol).
-
28b
Take 10 mg of the product to an NMR tube and dissolve it in 0.65 mL of deuterated water. Performed 1H NMR analysis gives following spectrum - 1H NMR (300 MHz, D2O): δ = 4.22 (t, 5J = 2.2 Hz, 2H, CH2OH), 3.13 (t, 3J = 7.0 Hz, 2H, CH2NH3), 2.39 (tt, 3J = 7.0 Hz, 5J = 2.2 Hz, 2H, C≡CCH2), 1.90-1.80 (m, 2H, CH2CH2CH2).
Perform Boc-protection of the amino group
-
29b
Add 0.37 g of 6-aminohex-2-yn-1-ol hydrochloride (2.48 mmol) in a round-bottom flask containing a magnetic stir bar.
-
30b
Pour methylene chloride (8 mL).
-
31b
Add 0.343 mL of triethylamine (2.46 mmol) to the suspension.
-
32b
When the solution is clear (after 5 min) add 0.49 g of di-tert-butyl dicarbonate (2.23 mmol).
-
33b
Stir the reaction mixture for 72 h at room temperature.
-
34b
Cool the reaction mixture to 0 °C using ice bath and acidify it with 5 mL of 0.5 M HCl (pH ~ 4).
-
35b
Separate the layers using a separatory funnel.
-
36b
Wash the organic layer with brine (2 × 5 mL), dry over sodium sulphate and remove the solvent under reduced pressure using rotary evaporator equipped with a vacuum pump.
-
37b
Purify the residue by column chromatography eluting with CHCl3/EtOAc (1:1, Rf = 0.5). The resulting product, 6-(N-Boc-amino)hex-2-yn-1-ol (4b), is obtained in 69% yield (0.36 g, 1.7 mmol) as yellowish oil.
-
38b
Take 10 mg of the product to an NMR tube and dissolve it in 0.65 mL of deuterated chloroform. Performed 1H NMR analysis gives following spectrum - 1H NMR (300 MHz, DMSO-d6): δ = 6.84 (br s, 1H, NH), 4.02 (t, 3J = 2.4 Hz, 2H, CH2OH), 2.96 (q, 5J = 6.9 Hz, 2H, CH2NH), 2.18 (tt, 3J = 6.9 Hz, 5J = 2.0 Hz, 2H, C≡CCH2), 1.59-1.51 (m, 2H, CH2CH2CH2), 1.38 (s, 9H, C(CH3)3).
BASIC PROTOCOL 2
Synthesis of 4-nitrobenzenesulfonates
Synthesis of nosylates could be achieved by reaction of a starting alcohol with nosyl chloride in the presence of pyridine (Lipshutz et al., 1990), or using sodium hydroxide as a general base (Lukinavičius et al., 2007), which in our hands proved either very slow, or not well reproducible, respectively. To achieve fast deprotonation of the alcohol and minimize hydrolysis of nosyl chloride we opted for a strong non-nucleophilic base – potassium tert-butoxide (Lukinavičius et al., 2013). This optimized approach furnishes the target nosylates 5a,b in high yields and in a matter of minutes.
Materials
Appropriate alcohol (e.g. 6-azidohex-2-yn-1-ol (4a))
Tetrahydrofuran, anhydrous (THF)
Potassium tert-butoxide (tert-BuOK)
4-Nitrobenzenesulfonyl chloride (NsCl)
Silica gel (60 Ǻ, 230–400 mesh)
Methylene chloride, anhydrous (DCM)
Magnetic stir plate and stir bar
5- and 50-mL Round-bottom flasks, oven dried
Drying tube with calcium chloride
Rotary evaporator equipped with a vacuum pump
Chromatography column
Test tubes
NMR tubes
Deuterated chloroform (CDCl3)
Perform nosylation of hydroxyl group
Add 0.073 g of 6-azidohex-2-yn-1-ol (4a) (0.52 mmol) in a round-bottom flask equipped with a drying tube (filled with calcium chloride).
Place a magnetic stir bar and cool to 0 °C in an ice bath.
Pour anhydrous THF (5 mL) and stir till the solution is clear.
- Add 0.055 g of potassium tert-butoxide (0.49 mmol) and stir till the solution is clear.If the solution remains turbid after 5 min, it is an indication of excess moisture in the reaction. Start over from Step 1 using freshly prepared anhydrous THF in Step 3.
- When solution is clear add 0.110 g of 4-nitrobenzenesulfonyl chloride (0.49 mmol).If the color of the solution changes to dark yellow or brown, the mole amount of added potassium tert-butoxide was probably larger than that of alcohol 4a, which leads to undesired side reactions. Start over from Step 1 using a lower amount of potassium tert-butoxide in Step 5.
Stir the reaction mixture for 20 min at 0 °C.
- Remove the solvent under reduced pressure using a rotary evaporator equipped with a vacuum pump and purify the residue by column chromatography eluting with methylene chloride (Rf = 0.5). The resulting product, 6-azidohex-2-yn-1-yl 4-nitrobenzenesulfonate (5a), is obtained in 42% yield (0.71 g, 2.19 mmol) as a yellowish powder.6-(N-Boc-amino)hex-2-yn-1-yl 4-nitrobenzenesulfonate (5b) is obtained in 79% yield (0.63 g, 1.58 mmol) from 0.43g of the starting alcohol 4b.
- Take 10 mg of the product to an NMR tube and dissolve it in 0.65 mL of deuterated chloroform. Performed 1H NMR analysis gives following spectra:For 6-azidohex-2-yn-1-yl 4-nitrobenzenesulfonate (5a) - 1H NMR (400 MHz, CDCl3): δ = 8.46 (d, 3J = 9.0 Hz, 2H, ArH), 8.19 (d, 3J = 9.0 Hz, 2H, ArH), 4.86 (t, 5J = 2.4 Hz, 2H, CH2O), 3.33 (t, 3J = 6.6 Hz, 2H, CH2N3), 2.23 (tt, 3J = 6.9 Hz, 5J = 2.4 Hz, 2H, C≡CCH2), 1.67 (tt, 3J = 6.9 Hz, 3J = 6.6 Hz, 2H, CH2CH2CH2).For 6-(N-Boc-amino)hex-2-yn-1-yl 4-nitrobenzenesulfonate (5b) - 1H NMR (300 MHz, CDCl3): δ = 8.43 (dd, 3J = 7.2 Hz, 4J = 2.1 Hz, 2H, ArH), 8.16 (dd, 3J = 7.2 Hz, 4J = 2.1 Hz, 2H, ArH), 4.84 (t, 5J = 2.1 Hz, 2H, CH2ONs), 4.61 (br s, 1H, NHBoc), 3.15-3.06 (m, 2H, CH2CH2CH2NHBoc), 2.13 (tt, 3J = 7.2 Hz, 5J = 2.1 Hz, 2H, CH2CH2CH2NHBoc), 1.63-1.53 (m, 2H CH2CH2CH2NHBoc), 1.45 (s, 9H, C(CH3)3).The prepared alkynyl nosylates are highly reactive and thus unstable compounds, and are best used in the next step without delay. If necessary, they can be stored at −20 °C for a short period of time (1–2 days, 5a; 1–2 weeks, 5b) before use.
BASIC PROTOCOL 3
S-alkylation of AdoHcy with nosylates
As mentioned above, selective S-alkylation of AdoHcy with corresponding nosylates is achieved in a mixture of formic and acetic acids, which confer transient protection of nucleophilic nitrogens in the adenine ring. The alkylation proceeds via an SN2 mechanism, thus the concentration of reactants is of utmost importance considering the reaction time (use larger excess of alkylating agent if a shorter reaction time is desired). It is advised to check reaction completion (disappearance of AdoHcy and appearance of a diastereomeric cofactor) using HPLC/MS.
Materials
S-Adenosyl-L-homocysteine (AdoHcy, 2)
Formic acid (HCO2H)
Acetic acid (CH3CO2H)
4-Nitrobenzenesulfonate (5)
Diethyl ether
Distilled water
Boc-protected cofactor (3b)
Trifluoroacetic acid (TFA)
Methylene chloride (DCM)
Magnetic stir plate and stir bar
5- and 50-ml Pear shaped flasks, oven dried
Ice bath
Argon balloon
50-mL Separatory funnel
Rotary evaporator equipped with a vacuum pump
Perform alkylation of AdoHcy (2)
-
1
Add 0.028 g of S-adenosyl-L-homocysteine (2) (0.072 mmol) into a pear shaped flask.
-
2
Place a magnetic stir bar and cool to 0 °C in an ice bath.
-
3
Introduce an argon atmosphere into the flask and maintain it throughout the reaction.
-
4
Add a mixture of formic and acetic acids (1:1, v/v, 1.5 mL) and stir till the solution is clear.
-
5
Add 0.141 g of 6-azidohex-2-yn-1-yl-4-nitrobenzenesulfonate (5a) (0.43 mmol) to the solution.
-
6After 1 h remove an ice bath and stir at room temperature for another 24 h.Reaction is monitored by HPLC-MS (for sample preparation see recipe).
-
7
Quench the reaction with 10 mL of cold water.
-
8
Wash the aqueous layer three times, each time with 5 mL of diethyl ether using a 50-mL separatory funnel and evaporate the aqueous residue to dryness giving colorless solids.
Deprotection of Boc-protected AdoMet analog
As mentioned above, nucleophilic functional groups in the extended side chain are protected during the sulfonation and S-alkylation steps. Selection of the protecting group is such that to permit its facile removal under mild acidic conditions because AdoMet and its analogs are unstable in neutral and, especially, in basic solutions (Hoffman, 1998; Lukinavičius et al., 2013). We therefore used Boc protection for the terminal amine, which is removed by brief treatment with trifluoroacetic acid.
Perform removal of the Boc group
-
9
Add solution of Boc-protected cofactor (3b) (all the material obtained in step 8) into a pear shaped flask.
-
10
Place a magnetic stir bar and cool to 0 °C in an ice bath.
-
11
Introduce an argon atmosphere into the flask and maintain it throughout the reaction.
-
12
Add a mixture of trifluoroacetic acid and methylene chloride (1:1, v/v, 2 mL) and stir for 1 h at 0 °C.
-
13
Remove the solvent under reduced pressure using a rotary evaporator equipped with a vacuum pump or evaporate using argon stream.
BASIC PROTOCOL 4
Purification and characterization of AdoMet analogs
Primary purification of AdoMet and its analogs can be performed using ion-exclusion chromatography, in which the positively charged cofactor elutes in the flow-through from an anion exchange column. This also leads to exchange of the remaining nosylate counterion, which otherwise interferes with photometric quantitation of the adenine chromophore (see below), with formate. Subsequently, reversed-phase preparative HPLC could be applied for producing higher purity of SAM analogues (e.g. complete removal of unreacted AdoHcy) and, in certain cases (3a), separation of the diastereomers (option).
Materials
Ammonium formate buffer (20 mmol/L, pH = 3.5)
Methanol (60%)
Formic acid solution (50%)
Sodium hydroxide (10 M)
Distilled water
Dowex 1×8, 50–100 mesh resin
5- and 50-ml Pear shaped flasks, oven dried
Filtration system
Rotary evaporator equipped with a vacuum pump
1.8 × 10.8-cm Chromatography column
1.5-mL Microcentrifuge tubes (e. g. Eppendorf)
TLC plate
UV lamp (254 nm)
Preparative reversed-phase HPLC (column - Agilent Prep-C18, dimensions: 30×150, 10 μm, PN 413910-302)
UV/Vis spectrophotometer
General procedure for purification of SAM analogues
Redissolve the crude product (all the material obtained in basic protocol 3 in step 13) in 1 mL of 20 mmol/L ammonium formate buffer.
Replace 4-nitrobenzenesulfonate counterion with formate using anion-exchange chromatography (formate-equilibrated column prepared from Dowex 1×8, 50–100 mesh, eluent – 20 mmol/L ammonium formate buffer (pH = 3.5), for resin preparation see recipe).
Collect fractions in microcentrifuge tubes manually and check them with UV light and by mass spectrometric analysis. Collect product-containing fractions and remove the solvent under reduced pressure.
Purify the residue using Agilent Prep-C18 reversed-phase HPLC column on an Agilent 1100 HPLC system. Elute the product using 20 mmol/L ammonium formate buffer (pH = 3.5) as eluent A and 60% methanol in water as eluent B at a flow rate of 30 mL/min (the eluent profile is shown in Table 1).
Collect the eluted peaks using a fraction collector and check fractions by mass spectrometric analysis. Collect product-containing fractions and remove the solvent under reduced pressure.
- Redissolve product in ~1 ml of distilled water and quantify the concentration by UV absorption of the adenosine chromophore (ε260 = 15400 L × mol−1 × cm−1).S-Adenosyl-S-(6-azidohex-2-yn-1-yl)-L-homocysteine formate (3a) is obtained in 12% yield (0.0044 g, 0.0087 mmol) of active diastereomer from 0.028g of AdoHcy 2. 1H NMR analysis of 3a gives following spectrum – 1H NMR (D2O, 300 MHz): δ = 8.18 (s, 1H, ArH), 8.17 (s, 1H, ArH), 6.01 (d, 3J = 5.7 Hz, 1H, C1’-H), 4.86 (dd, 3J = 5.7 Hz, 3J = 5.1 Hz, 1H, C2’-H), 4.49-4.41 (m, 1H, C4’-H), 4.21-4.18 (m, 2H, C≡CCH2S), 3.80-3.50 (m, 3H, SCH2CH, CHNH2), 3.48-3.30 (m, 2H, SCH2CH2CH), 3.15 (t, 3J = 6.3 Hz, 2H, CH2N3), 2.24-2.12 (m, 2H, SCH2CH2CH), 2.12-2.00 (m, 2H, C≡CCH2), 1.56-1.42 (m, 2H, CH2CH2CH2), signal of C3’-H in the furane ring overlaps with residual water signal in D2O.S-Adenosyl-S-(6-aminohex-2-yn-1-yl)-L-homocysteine formate (3c) is obtained in 57% yield (0.0146 g, 0.03 mmol) of diastereomeric mixture from 0.020 g of AdoHcy 2. 1H NMR analysis of 3c gives following spectrum – 1H NMR of S- and R- (D2O, 300 MHz): δ = 8.16-8.12 (m, 2H, ArHS/R), 5.99 (d, J = 2.8 Hz, 0.5H, H1’R), 5.96 (d, J = 3.8 Hz, 0.5H, H1’S), 4.84-4.78 (m, 1H, H2’S/R) 4.68-4.59 (m, 1H, H3’S/R) 4.47-4.37 (m, 1H, H4’S/R), 4.25-4.12 (m, 1.2H*, H1”R/S), 3.86-3.80 (m, 1H, H5’S), 3.76-3.45 (m, 3H, HαS/R, H5’R), 3.44-3.27 (m, 2H, HγS/R) 2.92 (t, J = 7.7 Hz, 1H, H6”S), 2.83 (t, J = 7.9 Hz, 1H, H6”R), 2.34-1.96 (m, 4H, H4”R/S, HβS/R), 1.80-1.57 (m, 2H, H5”R/S). * time-dependent loss of resonance in D2O due to exchange of H for D.Store the cofactors in acidic buffers (pH 2–3.5) at −80 °C.
Table 1.
Buffer composition for reversed-phase HPLC purification of AdoMet cofactor analogs
| T, min | 0 | 2 | 10 | 11 | 13 | 14 | 22 |
|---|---|---|---|---|---|---|---|
| A, % | 100 | 100 | 60 | 0 | 0 | 100 | 100 |
| B, % | 0 | 0 | 40 | 100 | 100 | 0 | 0 |
REAGENTS AND SOLUTIONS
Use distilled water for all recipes and protocol steps.
Sample preparation for HPLC analysis
Take 10 µL of reaction mixture and quench with 190 µL of water. Wash aqueous layer with diethyl ether (2 × 500 µL). Take 10 µL of aqueous layer and dilute to 100 µL with water.
200 mmol/L ammonium formate buffer
Dilute 7.55 mL of formic acid with approximately 980 mL of water. Add concentrated aqueous ammonia dropwise to pH = 3.5 (evaluated with a pH-meter). Dilute the solution to 1000 mL with water.
20 mmol/L ammonium formate buffer
100 mL 200 mmol/L ammonium formate
900 mL water
Anion resin preparation
Using filtration system, wash 20 g of the resin with 50 mL of 10 N NaOH.
Wash the resin with 150 ml of water (until the eluent become neutral).
Wash the resin with 30 mL of 50% formic acid solution.
Load the resin into a chromatography column.
Wash the resin with 50 mL of 20 mmol/L ammonium formate buffer (pH = 3.5).
COMMENTARY
Background Information
A vast plethora of methyltransferases (MTases) in the cell catalyze targeted transfers of the sulfonium-bound methyl group from AdoMet 1 to nitrogen, oxygen, carbon or sulfur nucleophiles located on their target biomolecules (Klimašauskas and Lukinavičius, 2008; Grosjean, 2009). To expand the practical utility of this highly specific enzymatic reaction, two major classes of synthetic AdoMet analogues have been developed (Klimašauskas and Weinhold, 2007). The first useful strategy exploited aziridine (Pignot et al., 1998; Pljevaljcic et al., 2003) or N-mustard (Weller and Rajski, 2006; Mai and Comstock, 2011) mimics of the sulfonium center whereby a whole cofactor molecule was transferred as the anchoring unit for attaching desired reporters to the target biomolecule (named sequence-specific methyltransferase-induced labeling, SMILing). Subsequently, cofactors with activated sulfonium-bound side-chains have been produced, which permit the transfer of these extended side chains alone (named methyltransferase-directed transfer of activated groups, mTAG) (Dalhoff et al., 2006a; Lukinavičius et al, 2007). The required activation of the extended side chain was achieved by a double or a triple bond imbedded next to the transferrable carbon atom (β position to the sulfonium center, Figure 1). A series of cofactor analogs have been designed and proved suitable for labeling a variety of biological targets in DNA, RNA and proteins.
The specificity of mTAG labeling is determined by a directing MTase. AdoMet-dependent methylation of DNA is performed by three classes of DNA MTases, which modify the N6 position of adenine, the N4 position or C5 position of cytosine within their specific target sequences ranging from two to eight base pairs. RNA methylation is much more diverse and includes enzymes that methylate a specific position in a particular RNA molecule (tRNA or rRNA) or in a specific class of RNA (miRNA or mRNA cap), or even perform RNA-directed sequence-specific methylation (which can be synthetically programmed in vitro to modify other sites). All of the mentioned MTases proved suitable for mTAG labeling either in native form or after simple steric engineering of their cofactor binding pocket (Motorin et al., 2011; Tomkuvienė et al., 2012; Sergeeva et al., 2012; Schultz et al., 2013; Plotnikova et al., 2014; Holstein et al., 2014). Examples of practical applications of sequence-specific mTAG labeling of DNA include single-molecule genotyping (Neely et al, 2010; Vranken et al., 2014) and epigenome profiling (Kriukienė et al., 2013).
We focused our attention on designing AdoMet analogs with triple bond-containing side chains carrying a terminal functional group, which proved particularly efficient with DNA C5-MTases. However, under certain circumstances the triple bond itself may become chemically active leading to undesired side reactions. For example, our previously described AdoMet analogues carrying sulfonium-bound 4-substituted but-2-yn-1-yl side chains (Lukinavičius et al, 2007), were ultimately found to exhibit poor stability in physiological buffers due to pH-dependent addition of a water molecule to the side chain. This side reaction was eradicated by increasing the separation between an electronegative functional group and the triple bond from one to three carbon units. The newly designed hex-2-yn-1-yl moiety-based cofactor analogs (Figure 1) with terminal amino, azide, or alkyne groups showed a markedly improved enzymatic transalkylation activity and proved well suitable for methyltransferase-directed sequence-specific labeling of DNA in vitro and in bacterial cell lysates (Lukinavičius et al, 2012, 2013).
The general synthetic strategy of AdoMet analogs involves a direct chemoselective alkylation of S-adenosyl-L-homocysteine (AdoHcy, 2) under mild acidic conditions (Figure 2) (Dalhoff et al., 2006a,b), which leads to protonation-mediated protection of all nucleophilic positions in AdoHcy 2 except the sulfur atom (De La Haba et al., 1959). An important element of this approach was the choice of a suitable nucleofuge (electron retaining leaving group) in the alkylating agent. Previously described examples of simple AdoMet analogs containing a prop-2-en-1-yl or a but-2-yn-1-yl group used commercially available 3-bromo-1-propene or triflate-activated but-2-yn-1-ol (Dalhoff et al., 2006b). However with larger and more complex linear groups, halogenides did not offer good conversions at suitable reaction times or reasonable excess amounts of an alkylating agent, whereas triflates often led to unwanted side reactions (unpublished observations). We therefore turned to arylsulfonates, which permit fine-tuning of their reactivity by selecting proper electron withdrawing substituents in the aryl moiety. The best results were achieved with 4-nitrobenzenesulfonates (nosylates) (Lukinavičius et al, 2013), which can be readily obtained from corresponding alcohols. Obviously, any functional group in the extended side chain more nucleophilic than a primary hydroxyl needs to be protected during the sulfonation and S-alkylation steps (Figure 2). Under these conditions, we used Boc protection for terminal amine, whereas no protection was required for azide.
Critical Parameters/Troubleshooting
Basic protocol 1: Reactions involving butyl lithium require strictly anhydrous conditions – glassware should be oven dried, anhydrous THF should be distilled from LiAlH4 before use, argon stream should be additionally dried over calcium chloride or other drying agent.
Basic protocol 2: If the formation of an ether product is observed (additional spot on TLC or/and additional 1H NMR signal upfield compared to the target product and not matching signal of starting alcohol) due to reaction of alcoholate 4 with the formed nosylate 5, lower the temperature to −50 °C. If the reaction solution is not clear after adding potassium tert-butoxide, THF could be wet. If the color of the reaction solution drastically changes after addition of nosyl chloride, the amount of potassium tert-butoxide was larger than required (prognosis of poor yield).
Basic Protocol 3: It is advised to check reaction completion (disappearance of AdoHcy and appearance of a double (diastereomeric) cofactor peak) using HPLC/MS. Note that the A260 signal of AdoHcy in HPLC chromatograms is often obscured by a broad UV peak of co-product nosylate, but AdoHcy is readily detectable by MS (M+H+ = 385.1289).
Basic Protocol 4: Checking fractions for target product via UV absorption by spotting on fluorescent silica gel TLC plates should be performed promptly (within 1 min) due to rapid loss of fluorescence in the acidic elution buffer (ammonium formate, pH=3.5). Crude preparative purification of synthetic cofactors by HPLC (without separation of diastereomers) could be performed collecting fractions based on UV response, while separation of diasteromers should be performed collecting smaller fractions based on time, then all said fractions should be checked on small scale HPLC/MS for diasteromeric purity.
Anticipated Results
The described protocols are suitable for preparation of synthetic AdoMet analogs at a scale of up to 100 mg. The prepared products could be used to perform hundreds of mTAG labeling reactions during the period of at least 2 years if stored below −78°C and not defrosted frequently (so it is advised to aliquot the final solution into smaller batches).
Time Considerations
The hydroxymethylation and nucleophilic substitution steps each take 2 days including purification. Nosylation can be performed in 1 day, and alkylation of AdoHcy will take another day. Purification of cofactors takes 1–3 days depending on whether purification by reverse phase HPLC is required. Therefore, full synthesis of 3a cofactor can be accomplished in 7–8 days, while synthesis of 3c takes 10–12 days due to additional protection/deprotection steps.
Acknowledgments
This work was supported by grants from the National Institutes of Health (HG004535 and HG007200).
LITERATURE CITED
- Brandsma L. Synthesis of acetylenes, allenes and cumulenes: Methods and Techniques. Amsterdam: Elsevier; 2004. Reactions with Aldehydes and Ketones; pp. 119–134. [Google Scholar]
- Dalhoff C, Lukinavičius G, Klimašauskas S, Weinhold E. Direct transfer of extended groups from synthetic cofactors by DNA methyltransferases. Nat. Chem. Biol. 2006a;2:31–32. doi: 10.1038/nchembio754. [DOI] [PubMed] [Google Scholar]
- Dalhoff C, Lukinavičius G, Klimašauskas S, Weinhold E. Synthesis of S-adenosyl-L-methionine analogs and their use for sequence-specific transalkylation of DNA by methyltransferases. Nat. Protoc. 2006b;1:1879–1886. doi: 10.1038/nprot.2006.253. [DOI] [PubMed] [Google Scholar]
- De La Haba G, Jamieson GA, Mudd SH, Richards HH. S-Adenosylmethionine: the relation of configuration at the sulfonium center to enzymatic reactivity. J. Am. Chem. Soc. 1959;81:3975–3980. [Google Scholar]
- Grosjean H. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin, TX: Landes Bioscience; 2009. [Google Scholar]
- Hoffman JL. Chromatographic analysis of the chiral and covalent instability of S-adenosyl-L-methionine. Biochemistry. 1986;25:4444–4449. doi: 10.1021/bi00363a041. [DOI] [PubMed] [Google Scholar]
- Holstein JM, Schulz D, Rentmeister A. Bioorthogonal site-specific labeling of the 5'-cap structure in eukaryotic mRNAs. Chem. Commun. 2014;50:4478–4481. doi: 10.1039/c4cc01549e. [DOI] [PubMed] [Google Scholar]
- Klimašauskas S, Lukinavičius G. Chemistry of AdoMet-dependent methyltransferases. In: Begley TP, editor. Wiley Encyclopedia of Chemical Biology. chapter 335. Weinheim: Wiley-VCH; 2008. [Google Scholar]
- Klimašauskas S, Weinhold E. A new tool for biotechnology: AdoMet-dependent methyltransferases. Trends Biotechnol. 2007;25:99–104. doi: 10.1016/j.tibtech.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Kriukienė E, Labrie V, Khare T, Urbanavičiūtė G, Lapinaitė A, Koncevičius K, Li D, Wang T, Pai S, Ptak C, Gordevičius J, Wang SC, Petronis A, Klimašauskas S. DNA unmethylome profiling by covalent capture of CpG sites. Nat. Commun. 2013;4:2190. doi: 10.1038/ncomms3190. [DOI] [PubMed] [Google Scholar]
- Mai V, Comstock LR. Synthesis of an Azide-Bearing N-Mustard Analogue of S-Adenosyl-l-methionine. J. Org. Chem. 2011;76:10319–10324. doi: 10.1021/jo2019637. [DOI] [PubMed] [Google Scholar]
- Lipshutz BH, Huff BE, McCarthy KE, Miller TA, Jaweed Mukarram SM, Siahaan TJ, Vaccaro WD, Webb H, Falick AM. Oxazolophanes as masked cyclopeptide alkaloid equivalents: cyclic peptide chemistry without peptide couplings. J. Am. Chem. Soc. 1990;112:7032–7041. [Google Scholar]
- Lukinavičius G, Lapienė V, Staševskij Z, Dalhoff C, Weinhold E, Klimašauskas S. Targeted labeling of DNA by methyltransferase-directed Transfer of Activated Groups (mTAG) J. Am. Chem. Soc. 2007;129:2758–2759. doi: 10.1021/ja0691876. [DOI] [PubMed] [Google Scholar]
- Lukinavičius G, Lapinaitė A, Urbanavičiūtė G, Gerasimaitė R, Klimašauskas S. Engineering the DNA cytosine-5 methyltransferase reaction for sequence-specific labeling of DNA. Nucleic Acids Res. 2012;40:11594–11602. doi: 10.1093/nar/gks914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavičius G, Tomkuvienė M, Masevičius V, Klimašauskas S. Enhanced chemical stability of AdoMet analogues for improved methyltransferase-directed labeling of DNA. ACS Chem. Biol. 2013;8:1134–1139. doi: 10.1021/cb300669x. [DOI] [PubMed] [Google Scholar]
- Motorin Y, Burhenne J, Teimer R, Koynov K, Willnow S, Weinhold E, Helm M. Expanding the chemical scope of RNA: methyltransferases to site-specific alkynylation of RNA for click labeling. Nucleic Acids Res. 2011;39:1943–1952. doi: 10.1093/nar/gkq825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely RK, Dedecker P, Hotta J, Urbanavičiūtė G, Klimašauskas S, Hofkens J. DNA fluorocode: A single molecule, optical map of DNA with nanometre resolution. Chem. Sci. 2010;1:453–460. [Google Scholar]
- Pljevaljcic G, Pignot M, Weinhold E. Design of a New Fluorescent Cofactor for DNA Methyltransferases and Sequence-Specific Labeling of DNA. J. Am. Chem. Soc. 2003;125:3486–3492. doi: 10.1021/ja021106s. [DOI] [PubMed] [Google Scholar]
- Pignot M, Siethoff C, Linscheid M, Weinhold E. Coupling of a nucleoside with DNA by a methyltransferase. Angew. Chem. Int. Ed. 1998;37:2888–2891. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2888::AID-ANIE2888>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Plotnikova A, Osipenko A, Masevičius V, Vilkaitis G, Klimašauskas S. Selective covalent labeling of miRNA and siRNA duplexes using HEN1 methyltransferase. J. Am. Chem. Soc. 2014;136:13550–13553. doi: 10.1021/ja507390s. [DOI] [PubMed] [Google Scholar]
- Sergeeva OV, Burakovsky DE, Sergiev PV, Zatsepin TS, Tomkuviene M, Klimasauskas S, Dontsova OA. Usage of rRNA-methyltransferase for site-specific fluorescent labeling. Moscow Univ. Chem. Bul. 2012;67:88–93. [Google Scholar]
- Schulz D, Holstein JM, Rentmeister A. A chemo-enzymatic approach for site-specific modification of the RNA cap. Angew. Chem. Int. Ed. 2013;52:7874–7878. doi: 10.1002/anie.201302874. [DOI] [PubMed] [Google Scholar]
- Tomkuvienė M, Clouet-d'Orval B, Černiauskas I, Weinhold E, Klimašauskas S. Programmable sequence-specific click-labeling of RNA using archaeal box C/D RNP methyltransferases. Nucleic Acids Res. 2012;40:6765–6773. doi: 10.1093/nar/gks381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken C, Deen J, Dirix L, Stakenborg T, Dehaen W, Leen V, Hofkens J, Neely RK. Super-resolution optical DNA Mapping via DNA methyltransferase-directed click chemistry. Nucleic Acids Res. 2014;42:e50. doi: 10.1093/nar/gkt1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller RL, Rajski SR. Design, synthesis and preliminary biological evaluation of a DNA methyltransferase-directed alkylating agent. Chem Bio Chem. 2006;7:243–245. doi: 10.1002/cbic.200500362. [DOI] [PubMed] [Google Scholar]
KEY REFERENCE
- Lukinavičius G, Tomkuvienė M, Masevičius V, Klimašauskas S. Enhanced chemical stability of AdoMet analogues for improved methyltransferase-directed labeling of DNA. ACS Chem. Biol. 2013;8:1134–1139. doi: 10.1021/cb300669x. Latest report on key synthetic and analytical procedures.