Summary
The Golgi-Associated Retrograde Protein (GARP) is a tethering complex involved in the fusion of endosome-derived transport vesicles to the trans-Golgi network through interaction with components of the Syntaxin 6/Syntaxin 16/Vti1a/VAMP4 SNARE complex. The mechanisms by which GARP and other tethering factors engage the SNARE fusion machinery are poorly understood. Herein we report the structural basis for the interaction of the human Ang2 subunit of GARP with Syntaxin 6 and the closely related Syntaxin 10. The crystal structure of Syntaxin 6 Habc domain in complex with a peptide from the N terminus of Ang2 shows a novel binding mode in which a di-tyrosine motif of Ang2 interacts with a highly conserved groove in Syntaxin 6. Structure-based mutational analyses validate the crystal structure and support the phylogenetic conservation of this interaction. The same binding determinants are found in other tethering proteins and syntaxins, suggesting a general interaction mechanism.
Introduction
Transport of macromolecular cargo between organelles of the endomembrane system occurs by budding of vesicles from a donor compartment followed by fusion of the vesicles with an acceptor compartment (Bonifacino and Glick, 2004). Budding most often involves protein coats that select specific cargos while promoting vesicle formation. On the other end, fusion is mediated by a protein ensemble comprising small GTPases of the Rab, Arl or Rho families, tethering factors, Soluble N-Ethylmaleimide-Sensitive Factor Attachment Protein Receptors (SNAREs) on both the vesicles (v-SNAREs) and the target organelle (t-SNAREs), and Sec1-Munc18 (SM) proteins (Sudhof and Rothman, 2009; Yu and Hughson, 2010). The specificity of different vesicular transport steps is determined by the use of distinct sets of budding and fusion proteins.
Tethering factors are long coiled-coil proteins or multi-subunit complexes that are recruited to membranes by small GTPases (Yu and Hughson, 2010). They have at least two functions: (i) capture of transport vesicles in the vicinity of the target organelle, and (ii) regulation of the fusion event through actions on the SNAREs. The Golgi-Associated Retrograde Protein (GARP) (also known as VFT) complex promotes fusion of retrograde transport vesicles derived from endosomes with the trans-Golgi network (TGN), a critical step for transmembrane proteins that cycle between these organelles (Bonifacino and Hierro, 2011). GARP is conserved from yeast to humans and consists of four subunits named Vps51 (Ang2 in humans), Vps52, Vps53 and Vps54 (Conibear and Stevens, 2000; Siniossoglou and Pelham, 2001; Siniossoglou and Pelham, 2002; Conibear et al., 2003; Liewen et al., 2005; Perez-Victoria et al., 2010b; Luo et al., 2011). The subunits of GARP are structurally related to each other as well as to subunits of other multi-subunit tethering complexes such as the exocyst, Conserved Oligomeric Golgi (COG) and Dsl1 complexes (Perez-Victoria et al., 2010a; Vasan et al., 2010). Most of these subunits share a structure consisting of tandem helical bundle domains, for which the complexes are collectively referred to as Complexes Associated with Tethering Containing Helical Rods (CATCHR) (Yu and Hughson, 2010).
GARP interacts with components of the Syntaxin 6/Syntaxin 16/Vti1a/VAMP4 SNARE complex involved in fusion of retrograde transport carriers with the TGN (Mallard et al., 2002; Perez-Victoria and Bonifacino, 2009; Perez-Victoria et al., 2010b). However, the mechanisms by which tethering complexes function in vesicle fusion are poorly understood, particularly with regard to their roles in SNARE complex assembly. Herein we present a biochemical and X-ray crystallographic analysis of the Ang2 interaction with Stx6 and the closely related Syntaxin 10 (Stx10). We show a novel binding mode significantly different from the yeast Vps51-Tlg1 interaction (Fridmann-Sirkis et al., 2006). Mutational analyses confirm the validity of this binding mode in vitro and in vivo. Importantly, the structural determinants of the interaction reported here are likely conserved in other tethers and syntaxins, suggesting a common interaction mechanism.
Results
A 50-amino-acid N-terminal segment from Ang2 interacts with the Habc domain from Stx6 and Stx10
In previous work, we found an interaction between the Ang2 subunit of human GARP and the TGN t-SNARE Stx6 (Perez-Victoria et al., 2010b) (Fig. 1A). To analyze further the specificity of this interaction, we performed yeast two-hybrid (Y2H) analysis of the binding of Ang2 to the cytosolic domains of several SNAREs that function at the TGN and endosomes. We found that full-length Ang2 interacted with the cytosolic domains of Stx6 (Stx6 1-238) and the paralogous (61% amino-acid sequence identity) Stx10 (Stx10 1-232), and more weakly with the cytoplasmic domain of the v-SNARE Vamp4 (Vamp4 1-114) (Fig. 1B). We did not detect interaction of Ang2 with the cytosolic domain of the more distantly related (31% amino-acid sequence identity) Stx8 (Stx8 1-218) (Fig. 1B). Further Y2H analysis showed that the N-terminal region of Ang2 (residues 1-299) (Fig. 1A), which mediates assembly with the other subunits of GARP (Perez-Victoria et al., 2010b), is sufficient for interaction with Stx6 and Stx10 (Fig. 1B).
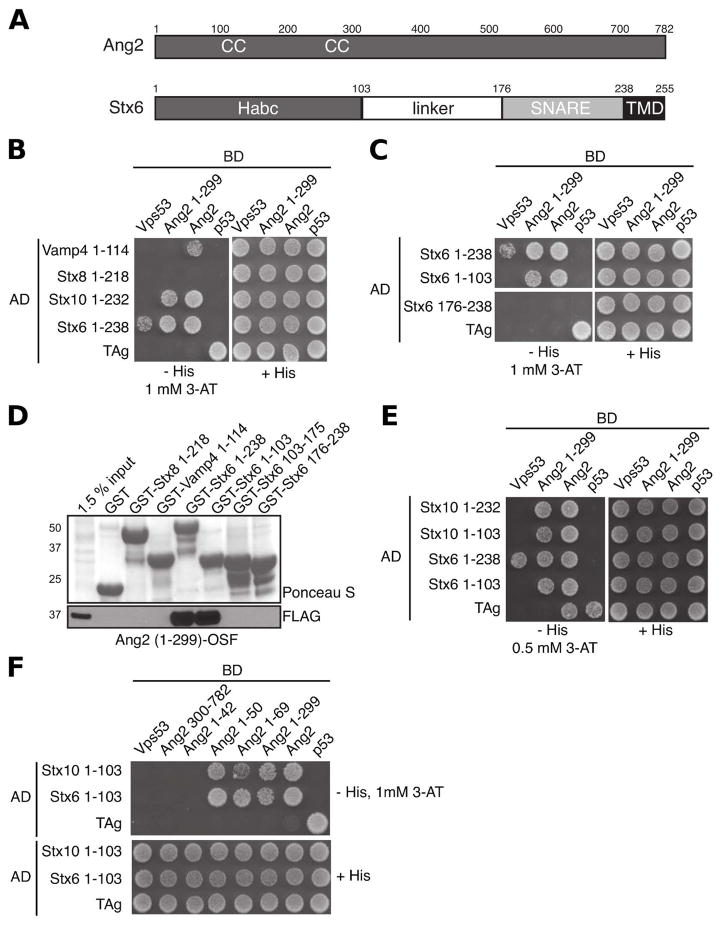
Figure 1.
The Habc domains of Stx6 and Stx10 bind a 50 amino-acid N-terminal segment from Ang2. (A) Schematic representation of human Ang2 and human Stx6. Amino acid numbers are indicated. Abbreviations used are CC (coiled coil), SNARE (SNARE domain) and TMD (transmembrane domain). (B) Y2H analysis of the interaction of SNARE cytosolic domains fused to Gal4-AD with Vps53, Ang2 and Ang2 (1-299) fused to Gal4-BD. The SV40 large T antigen (TAg) and p53 were used as controls. Growth in the absence of histidine (-His) is indicative of interactions. 3-amino-1,2,4-triazole (3-AT) was used to suppress self-activation. (C) Y2H analysis of the effect of truncations of the Stx6 cytosolic domain on interaction with Ang2 and Ang2 (1-299). (D) In vitro assay of Ang2 (1-299) binding to cytosolic portions of SNAREs. GST-tagged SNAREs or GST as a control were pre-bound to glutathione-Sepharose and incubated with detergent extracts from HeLa cells transiently transfected with a plasmid encoding Ang2 1-299 OSF. Bound proteins were subjected to SDS-PAGE and immunoblotting with an antibody to the FLAG epitope. To estimate the amounts of GST-protein fusions, immunoblots were stained with Ponceau S. A sample of the total extract was included as input control. The positions of molecular mass markers (in kDa) are indicated on the left. (E) Y2H analysis of the interaction of the Habc domains of Stx6 and Stx10 with Ang2 and Ang2 (1-299). (F) Y2H mapping of the Ang2 binding site to the first 50 residues of Ang2.
Both Stx6 and Stx10 belong to the Syntaxin family of Q-SNAREs (Fasshauer et al., 1998), and possess an N-terminal regulatory domain named Habc (Fernandez et al., 1998), followed by a linker region, a fusogenic SNARE domain and a C-terminal transmembrane anchor (Fig. 1A). Using the Y2H system, we found that the Stx6 Habc domain (Stx6 1-103) bound to both full-length Ang2 and Ang2 1-299, in contrast to the Stx6 SNARE domain (Stx6 176-238), which did not interact with either Ang2 construct (Fig. 1C). GST pull-down assays confirmed that both the Stx6 Habc (Stx6 1-103) and cytosolic (Stx6 1-238) domains bound the N-terminal region of Ang2 (Ang2 1-299) tagged with FLAG-One-STrEP (OSF) (Fig. 1D), whereas the Stx6 linker (Stx6 103-175) and SNARE domain (Stx6 176-238), the Stx8 cytosolic domain (Stx8 1-218) and Vamp4 cytosolic domain (Vamp4 1-114), did not bind (Fig. 1D). Y2H analysis also showed interaction of the Stx10 cytosolic (Stx10 1-232) and Habc (Stx10 1-103) domains with both full-length Ang2 and Ang2 1-299 (Fig. 1E). Further Y2H assays identified aminoacids 1-50 from Ang2 as the minimal segment capable of interacting with the Habc domains from both Stx6 (Stx6 1-103) and Stx10 (Stx10 1-103) (Fig. 1F). These experiments thus demonstrated a specific interaction between a 50-amino-acid N-terminal segment from Ang2 and the Habc domains from Stx6 and Stx10.
A di-tyrosine motif in the N-terminal region of Ang2 is required for binding to the Habc domain of Stx6 and Stx10
Inspection of the 50 N-terminal amino acids from human Ang2 in comparison to those from other species revealed a highly conserved tyrosine pair, Tyr40 and Tyr41 (Fig. 2A). Additional amino acids flanking the two aromatic amino acids also show a significant degree of conservation within an otherwise variable N-terminal segment (Fig. 2A). Indeed, using the Y2H system, we found that mutation of Tyr40 and Tyr41 to alanine (YY/AA mutant) completely abolished the interaction of a human Ang2 N-terminal fragment (Ang2 1-69) with both the cytosolic domains and Habc domains from Stx6 and Stx10 (Fig. 2B). In contrast, deletion of a variable segment comprising amino acids 2-23 (Fig. 2A) had no effect on the interactions (Fig. 2B). GST pull-down assays also showed a requirement of the di-tyrosine motif for interaction of Ang2 1-299-OSF with the cytosolic domains of Stx6 and Stx10 and the Habc domain of Stx6 in vitro (Fig. 2, C and D).
Figure 2.
A di-tyrosine motif in Ang2 is required for binding to the Habc domain of Stx6 and Stx10. (A) Alignment of N-terminal segments of Ang2/Vps51 from different species using ClustalW. (B) Y2H analysis of the interaction of Stx6 and Stx10 fragments with full-length Ang2, Ang2 1-69, Ang2 1-69 with a deletion of amino-acids 2-23 (Ang2 1-69 Δ2-23) and Ang2 with the conserved di-tyrosine motif mutated to alanine (Ang2 1-69 YY/AA). (C) In vitro binding of Ang2 1-299 OSF or (D) Ang2 1-299 OSF YY/AA expressed in HeLa cells to GST-SNAREs. The experiment was performed as described in the legend to Fig. 1D. (E) StrepTactin-pulldown assay of endogenous SNAREs with different Ang2 OSF constructs expressed by transfection in HeLa cells. Endogenous Stx6 and Vti1A SNAREs as well as clathrin heavy chain (Chc) and actin (loading controls) were detected by immunoblotting The positions of molecular mass markers (in kDa) are indicated on the left of panels C, D and E.
To determine if these interactions are required for association of Ang2 with endogenous SNAREs in vivo, we transfected HeLa cells with constructs encoding OSF-tagged Ang2 or Ang2 1-299 in both their wild-type and YY/AA mutant versions. We performed a Strep-Tactin pull-down, followed by immunoblotting for Stx6, Vti1a and several control proteins (Fig. 2E). We observed that wild-type Ang2 and Ang2 1-299 interacted with Stx6 and Vti1a, whereas the YY/AA mutant counterparts did not interact (Fig. 2E). The co-isolation of Vti1a with Stx6 suggested that Ang2 can bind via the di-tyrosine motif to Stx6 in complex with a cognate SNARE. As a control, we showed that mutation of the di-tyrosine motif had no effect on the incorporation of Ang2 into the GARP complex as evidenced by the co-isolation of the wild-type and mutant Ang2 species with endogenous Vps52 (Fig. 2E). From these experiments we concluded that a di-tyrosine motif within the 50 N-terminal amino acids from Ang2 is required for interaction with the Stx6 and Stx10 Habc domains in vitro and in vivo.
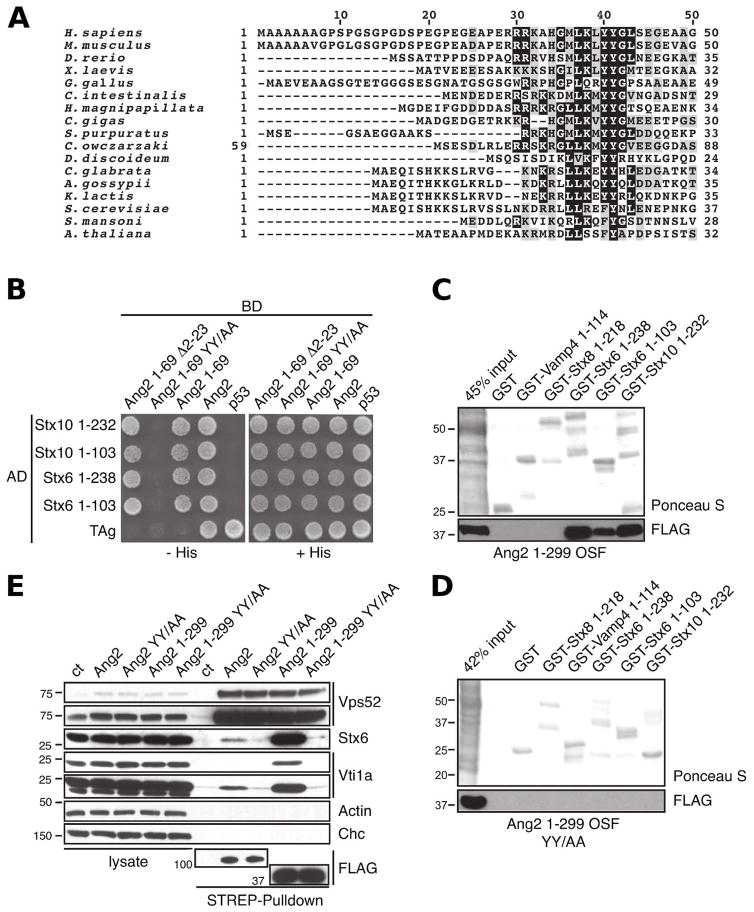
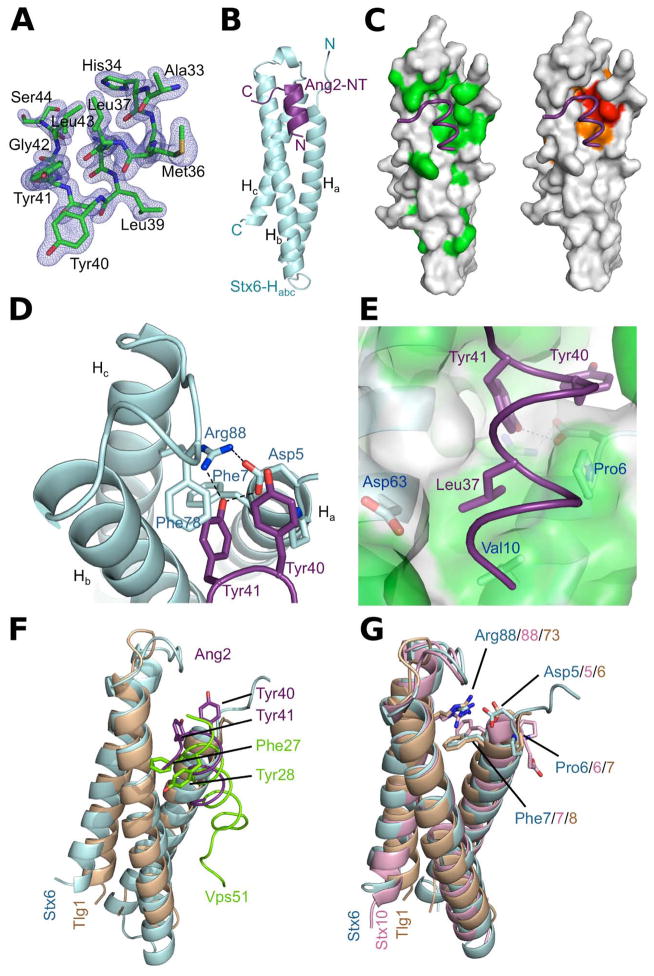
Structure of an Ang2 peptide bound to Stx6-Habc
To elucidate the structural basis for the Ang2-Stx6 interaction, we determined the crystal structure of the Ang2 peptide 33AHGMLKLYYGLSEGEAA49 in complex with the Stx6 Habc domain. The structure was solved by molecular replacement using the coordinates of Stx6-Habc (PDB 1LVF) (Misura et al., 2002) as the search model, and refined to 1.8 Å resolution with Rfactor and Rfree of 14.9 and 19.3, respectively (Table S1). The difference Fourier map showed clear electron density for a portion of the Ang2 peptide comprising residues 33AHGMLKLYYGLS44 (Fig. 3A). The remaining residues could not be modeled onto the electron density and were therefore deemed to be disordered. Overall, the Ang2 peptide forms a short α-helix that lies between α-helices a and b at the N-terminal side of Stx6 (Fig. 3B). The C-terminus of the Ang2 peptide, to which the rest of the GARP complex is bound, and the C-terminus of Stx6, which contains the membrane-anchoring domain, extend in opposite directions (Fig. 3B), consistent with a possible role of this interaction in tethering of apposed membranes. The contact region in the Stx6 Habc domain corresponds to a noticeably hydrophobic groove that is highly conserved as compared to other parts of the surface (Fig. 3C and Fig. S1). In agreement with the biochemical data described above, Ang2 is anchored to this groove by the burial of Tyr40 and Tyr41. The linchpin of this interaction is a network of hydrogen bonds between Tyr41Ang2, Asp5Stx6 and Arg88 Stx6 (Fig. 3D). The orientation of the Asp5Stx6 carboxyl group towards the hydrogen-bonding network is favored by the relative rigidity of the following Pro6Stx6. Similarly, Arg88Stx6 stacks against Phe7Stx6 holding its guanidinium group towards the hydrogen-bonding network (Fig. 3D). The proximal Phe78Stx6 further stabilizes the burial of Tyr40Ang2 and Tyr41Ang2. More peripherally, Leu37Ang2 is positioned in a shallow hydrophobic cavity formed by Pro6Stx6, Val10Stx6 and the beta carbon of Asp63Stx6 (Fig. 3E). This hydrophobic interaction, together with a hydrogen bond between His34Ang2 and Asp63Stx6, contribute to anchor the rest of the Ang2 α-helical segment to the Hab groove of Stx6. Thus, the high-resolution crystal structure identifies the most critical elements of the Stx6-Ang2 interaction.
Figure 3.
Structure of a human Ang2 peptide (amino acids 33-44) bound to the human Stx6 Habc domain. (A) 2Fo-Fc electron density map of Ang2 (amino acids 33-44) contoured at 1.1σ (blue mesh) with the final model superimposed. (B) Overall structure of human Stx6 Habc domain (cyan) in complex with the Ang2 peptide (purple) in ribbon cartoon. (C) Stx6 Habc domain showing the surface properties of the contact region with the Ang2 peptide in tube drawing. The molecular surface is colored by hydrophobicity (green) or by residue conservation (red and orange denote strictly and highly conserved residues, respectively). (D) Ang2 gets anchored through the burial of Tyr40 and Tyr41 (purple) into the conserved D5PF[…]R88 motif of Stx6 (cyan). (E) Stx6 residues (light blue) that contribute to the hydrophobic pocket shown under translucent surface in the vicinity of Leu37Ang2 (purple). (F) Structural comparison between the human Stx6-Ang2 complex (cyan Stx6 and purple Ang2 peptide) and the yeast Tlg1-Vps51 complex (PDB: 2C5K; brown Tlg1 and green Vps51 peptide). Note that a ~90º clockwise rotation along the helical axis of the Vps51 peptide would place Phe27Vps51 and Tyr28Vps51 over Tyr40Ang2 and Tyr41Ang2. (G) Superposition of the Habc domains of Stx6 (cyan), Stx10 (pdb id: 4DND; pink) and Tlg1 (pdb id: 2C5K; brown) in ribbon cartoon with the residues corresponding to the conserved DPF[...]R motif for each domain highlighted as stick model.
Stx6-Ang2 and Tlg1-Vps51 exhibit distinct crystallographic interactions
The closest homologue of human Stx6 in yeast is the t-SNARE Tlg1. Indeed, the crystal structure of the Tlg1 Habc domain in complex with a peptide derived from yeast Vps51 (residues 16-30) shows an association along the same Hab groove (Fig. 3F) and with the Vps51 peptide in a similar helical conformation (Fridmann-Sirkis et al., 2006). However, despite the apparent parallels between Stx6-Ang2 and Tlg1-Vps51 interactions, there are important differences in their crystallographic binding mode. In fact, the residues Phe27Vps51 and Tyr28Vps51, which are equivalent to Tyr40Ang2 and Tyr41Ang2, are positioned outside the Hab groove in the Tlg1-Vps51 structure (Fig. 2A and Fig. 3F). Furthermore, Tyr28Vps51, which is critical for binding in vivo (Fridmann-Sirkis et al., 2006), does not contact Tlg1. Instead it interacts with a symmetrically related molecule stabilizing the crystal lattice. Another difference concerns the strictly conserved 6DPF […]R73 residues of Tlg1, which do not participate in any hydrogen bonding network with Vps51 as the equivalent residues in the Stx6-Ang2 structure do (Fig. 3G and Fig. S1). Interestingly, a simple 90º rotation of the Vps51 peptide along its helical axis would place Phe27Vps51 and Tyr28Vps51 at positions similar to those of Tyr40Ang2 and Tyr41Ang2 (Fig 3F), raising the possibility that the crystallized yeast complex corresponds to an interaction intermediate.
Confirmation of the Stx6-Ang2 interface by mutational and binding analyses
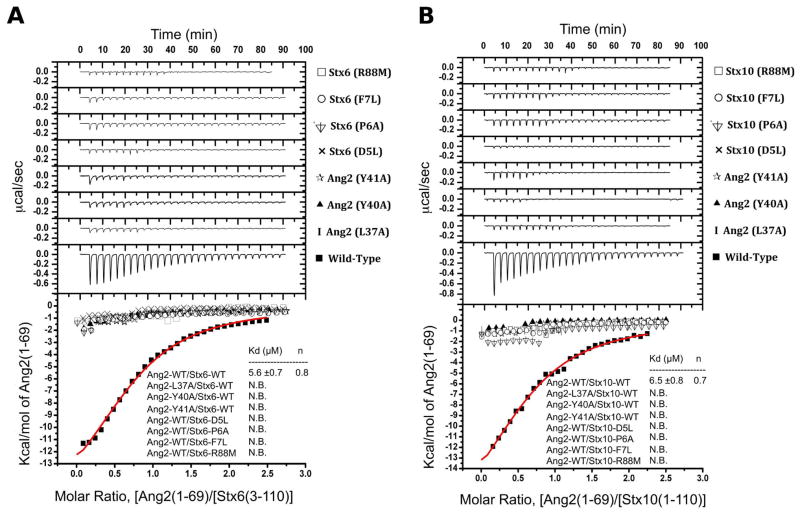
To confirm that the crystallographyc interaction between the 5DPF […]R88 signature sequence of Stx6 and the N-terminal 37LxxYY41 motif of Ang2 is preserved in solution, we performed isothermal titration calorimetry (ITC). We found that the Ang2 N-terminal region (residues 1-69) binds to the Stx6 Habc domain with Kd= 5.6 ± 0.7 μM and a best-fit stoichiometry of n = 0.8, which we interpret as a single binding site (Fig. 4A). The structure-based mutants D5LStx6 and R88MStx6 were designed to eliminate the central network of hydrogen bonds between Stx6 and Ang2, whereas the mutants P6AStx6 and F7LStx6 were designed to disrupt the orientation of Asp5Stx6 and the hydrophobic contact that stabilize Arg88Stx6, respectively. None of the mutations significantly altered Stx6 structure as measured by circular dichroism (CD) (Fig. S2). However, every single mutation rendered Stx6 Habc unable to bind Ang21-69, consistent with the central role of the 5DPF […]R88 binding motif in Stx6 (Fig. 4A). Similarly, the single mutants L37AAng2, Y40AAng2 or Y41A Ang2, designed to disrupt the anchoring of Ang2 to Stx6, completely abolished the binding (Fig. 4A), the latter two in agreement with the result of Y2H analyses (Fig. 2). These results confirmed that the crystallographic interaction and solution binding correspond to the same molecular association.
Figure 4.
Isothermal titration calorimetry of Ang2 interactions with Stx6 and Stx10. The top graphs represent the differential heat released during the titration of (A) Stx6 Habc with Ang2 (1-69) or (B) Stx10 Habc with Ang2 (1-69). The bottom graphs represent the fitted binding isotherms. All mutants within the conserved L37xxYY41 motif of Ang2 and the D5PF[…]R88 motif of Stx6 or Stx10 are annotated on the side of their respective titration curve. The Kd and binding stoichiometries (n values) are indicated as insets (N.B., no appreciable binding).
Ang2 displays the same binding mode for Stx6 and Stx10
The 5DPF […]R88 motif in Stx6 is also present at an equivalent position in Stx10 (Fig. 3G and S1). Furthermore, our Y2H and GST pull-down analyses showed that Ang2 interacts equally well with Stx6 and Stx10 (Figs. 1 and 2). Thus, to evaluate whether Ang2 employs the same binding mode for Stx6 and Stx10, we conducted an identical ITC analysis with Stx10. As expected, Ang21-69 exhibited a comparable affinity (Kd= 6.5 ± 0.8 μM) and stoichiometry (n = 0.7) for binding to the Stx10 Habc domain (Fig. 4B). In addition, the equivalent mutations in the 5DPF[…]R88 motif of Stx10 or in the 37LxxYY41 motif of Ang2 resulted in complete loss of interaction between both proteins (Fig. 4B). These results strongly support a common recognition mechanism for Ang2 and the t-SNARES Stx6 and Stx10. Furthermore, they imply that interaction of Ang2 with Stx6 and Stx10 cannot be simultaneous, suggesting that GARP may participate in more than one SNARE-mediated event at the TGN.
Discussion
Our biochemical and structural analyses reveal the molecular details of a highly specific interaction between a 37LxxYY41 motif in the N-terminal part of Ang2 and a 5DPF[…]R88 motif in the N-terminal Habc domain of Stx6 and Stx10. Notably, both motifs (Fig. S1) are conserved in most eukaryotic orthologs of these proteins, indicating that this interaction is evolutionarily conserved. Indeed, an analogous interaction has been reported for the S. cerevisiae orthologs Vps51 and Tlg1 (Fridmann-Sirkis et al., 2006). Also in this case, mutation of a di-aromatic motif (Phe27 and Tyr28) in a Vps51 peptide abolishes binding to the Tlg1 Habc domain in surface plasmon resonance and Y2H assays (Fridmann-Sirkis et al., 2006). However, in the published crystal structure Phe27Vps51 and Tyr28Vps51 do not contact the Tlg1 Habc domain to which the rest of the peptide is bound, but point outwards from the Hab groove (Fridmann-Sirkis et al., 2006). A structure like the one presented here would explain the requirement of Phe27Vps51 and Tyr28Vps51 for Vps51 binding to Tlg1 as well. We propose that the interaction details are conserved in most eukaryotes and that the published S. cerevisiae structure may correspond to a binding intermediate rather than the final state of the complex. Although disruption of the Vps51-Tlg1 interaction was not found to have any effect on GARP-dependent processes such as retrograde transport of the Sso1 and Snc1 SNAREs and maintenance of vacuolar integrity in S. cerevisiae (Fridmann-Sirkis et al., 2006), the evolutionary conservation of this interaction argues for an important role that remains to be determined.
A search for other proteins that have an LxxYY motif near the N-terminus using the ScanProsite tool (http://prosite.expasy.org/scanprosite/) identified the sequence L49ELYY53 within the vertebrate Cog2 subunit of the Conserved Oligomeric Golgi (COG) complex. Like GARP, COG is another CATCHR-type tethering complex (Yu and Hughson, 2010; Miller and Ungar, 2012) implicated in retrograde transport from endosomes to the TGN. This retrograde transport is partly dependent on interaction of the Cog6 subunit with the SNARE motif of Stx6 (Laufman et al., 2011). In light of our findings, it would be interesting to test whether Stx6 also interacts via its Habc domain with the N-terminal part of Cog2.
The DPF[…]R motif present in Stx6 and Stx10 also appears to be partially conserved in other syntaxins such Stx1, Stx2, Stx3, Stx4 and Stx8 (Fig. S3A). Indeed, the structure of Stx1A exhibits an arginine residue stacked against a phenylalanine residue at a position comparable to that on Stx6 and Stx10 (Fig. S3B). However, slight differences at the binding pocket may alter the specificity of recognition. For example, the lack of interaction of Stx8 with Ang2 (Fig. 1B) could be due to the stacking of arginine against tryptophan instead of phenylalanine, thus modifying the steric and electrostatic features of the site. The conservation of the LxxYY motif in Ang2 and possibly Cog2 and the presence of an analogous binding site in the Habc domain of several syntaxins suggest that the interaction described could be paradigmatic of tether-SNARE interactions at different subcellular locations.
The presence of an N-terminal Habc domain is a feature of all Syntaxin-type t-SNAREs (MacDonald et al., 2010). A well-known function of Habc domains is the regulation of SNARE-complex formation and vesicle fusion. For example, the Habc domain of mammalian Stx1 folds back onto the SNARE motif of the same protein to generate a closed conformation that is inactive for fusion (Zhou et al., 2012; Dulubova et al., 1999; Fiebig et al., 1999; Lerman et al., 2000). This interaction involves binding of α-helix H3 of the SNARE motif to a groove formed by α-helices b and c of the Habc domain. The SM protein Munc18-1 binds to the closed conformation of Stx1 also through interaction with the Stx1 Habc domain (Hata et al., 1993; Misura et al., 2000). Opening of the Stx1 structure is at least in part mediated by Munc13-1 (Ma et al., 2011), a large protein that is structurally homologous to CATCHR-type tethering factor subunits (Li et al., 2011). This conformational change depends on interactions of the CATCHR-fold MUN domain of Munc13-1 with both the Stx1 SNARE motif and Munc18-1 (Ma et al., 2011; Lerman et al., 2000). Several considerations suggest that this model may not apply to the Stx6-Ang2 interaction characterized in our study. First, the Stx6 Habc domain does not seem to interact with the SNARE motif (Misura et al., 2002). Second, although the Stx6 partner Syntaxin 16 binds the SM protein Vps45 (Dulubova et al., 2002), Stx6 itself is not known to interact with Vps45. Finally, the interaction of Stx6 with Ang2 does not involve the SNARE motif and CATCHR fold of these proteins (Fig. 1).
The Stx6-Ang2 interaction could instead function to tether endosome-derived retrograde vesicles bearing Stx6 to TGN sites containing associated GARP. It could also bring Stx6 into proximity of cognate SNAREs to facilitate SNARE-complex formation and vesicle fusion. In this regard it is worth noting that Ang2 pulls down via its di-tyrosine motif not only Stx6 but also Vti1a (Fig. 2E). Since Vti1a does not have a Habc domain, its pulldown by Ang2 is likely to depend on a different interaction or to be the indirect result of SNARE complex assembly. Finally, the Vps53 and Vps54 subunits of human GARP interact with the SNARE motifs of Syntaxin 6, Syntaxin 16 and VAMP4 (Perez-Victoria and Bonifacino, 2009). All of these interactions could work together in a molecular pathway of SNARE-complex assembly assisted by GARP.
Experimental Procedures
Recombinant DNAs, yeast two-hybrid and pull-down assays
A list of recombinant DNA constructs is shown in Supplementary Table 2. For constructs generated in this study, DNA sequences were PCR-amplified from human brain cDNA (Clontech, Mountain View, CA) and cloned into the corresponding plasmids. Site-directed mutagenesis was performed using the QuikChange kit (Stratagene, La Jolla, CA). Y2H experiments were performed as previously described (Ohno et al., 1995; Aguilar et al., 2001). GST-fusions to SNARE cytosolic domains were expressed in E. coli BL21 Rosetta (Novagen, Madison, WI) from pGEX-5X-1 and purified as described in Schindler and Spang, 2007. GST- and StrepTactin pull-downs were performed as previously described (Perez-Victoria and Bonifacino, 2009; Perez-Victoria et al., 2010b; Perez-Victoria et al., 2010a). HeLa cells (ATCC, Manassas, VA) were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were used 24 h after transfection. Antibodies used have been described in Perez-Victoria and Bonifacino, 2009. The antibody to clathrin heavy chain was from BD Biosciences (San Diego, CA).
Crystallization, data collection and model building
The synthetic peptide of Ang2 (aa 33-49) was obtained from GenScript USA Inc. (Piscataway, NJ). The peptide was resuspended in 6 M guanidine hydrochloride and mixed in a molar ratio 2:1 with Stx6 (3-110). The mixture was then dialyzed against 150 mM NaCl, 10 mM β-ME, 50 mM Tris-HCl pH 7.4 at 4°C during 48h. Crystallization trials were carried out in hanging drops with the protein-peptide mixture at 20 mg/ml. After 1–2 days large crystals appeared in 18% PEG 3350 and 100 mM ammonium dihydrogen phosphate. Crystals were flash-frozen under liquid nitrogen using paraffin oil as cryoprotectant. One data set at 1.8 Å resolution was collected at ESRF beamline ID14-1. Data reduction was carried with HKL2000 program (Otwinowski and Minor, 1997). The phases were calculated with PHASER (McCoy et al., 2007) using the Stx6 structure (PDB: 1LVF) as a search-model. Model building and refinement of the Ang2-Stx6 complex was performed using COOT (Emsley and Cowtan, 2004) and PHENIX (Adams et al., 2010). Data collection and refinement statistics are summarized in the Supplementary Table 1.
Isothermal titration calorimetry
Stx6 (3-110), Stx10 (1-110), Ang2 (1-69) and the corresponding mutant proteins were dialyzed overnight at 4°C against 150 mM NaCl, 50 mM Tris-HCl pH 7.4. For the ITC analysis, Ang2 (1-69) at 160mM was injected into the Stx6 (3-110) or Stx10 (1-110) solution at 13 mM in aliquots of 10 μl. Measurements were carried out at 25 °C on a VP-ITC Microcalorimeter (MicroCal/GE Healthcare). ITC data were processed using Origin software (OriginLab Corp., USA).
Supplementary Material
Table 1.
Data collection and structure refinement parameters
| Wavelength | 0.9334 |
| Images | 132 |
| Unit cell parameters | a = 41.64 Å, b = 45.66 Å, c=58.34 Å α=90º, β=90º, γ=90º |
| Space group | P21 |
| Resolution (Å) | 50.0–1.80 (1.86–1.80) |
| Rmerge (%) | 4.8 (35.8) |
| Completeness | 97.2 (91.0) |
| Multiplicity | 2.7 (2.1) |
| I/σ | 20.09 (2.15) |
| Alpha_twin | 0.49 |
| No. Reflections (observed) | 19877 |
| Structure refinement | |
| R-factor (%) | 14.95 |
| R-free (%) | 19.34 |
| No. Reflections (R-Free %) | 5% |
| No. water molecules | 510 |
| Rmsd values | |
| Bond lengths (A°) | 0.005 |
| Bond angles (deg.) | 0.835 |
| Ramachandran statistics | |
| Most favored (%) | 98.3 |
| Additional allowed (%) | 1.7 |
| Generously allowed (%) | 0 |
Acknowledgments
We thank A. Muga, University of the Basque Country, for help with ITC experiments; and A. Vidaurrazaga for technical assistance. This study made use of the European Synchrotron Radiation Facility (ESRF, Grenoble, France) under the Block Allocation Group MX1283/MX1420. This research was supported by the Carlos III Health Institute grant PI11/00121, the Basque Government grant PI2011-26 (to A.H.) and the intramural program of the National Institute of Child Health and Human Development (to J.S.B.). G.A-P has been supported by a fellowship from the Basque Government (BFI08.53).
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar RC, Boehm M, Gorshkova I, Crouch RJ, Tomita K, Saito T, Ohno H, Bonifacino JS. Signal-binding specificity of the mu4 subunit of the adaptor protein complex AP-4. J Biol Chem. 2001;276:13145–13152. doi: 10.1074/jbc.M010591200. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Hierro A. Transport according to GARP: receiving retrograde cargo at the trans-Golgi network. Trends Cell Biol. 2011;21:159–167. doi: 10.1016/j.tcb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Cleck JN, Stevens TH. Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol Biol Cell. 2003;14:1610–1623. doi: 10.1091/mbc.E02-10-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell. 2000;11:305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Sudhof TC, Rizo J. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Yamaguchi T, Gao Y, Min SW, Huryeva I, Sudhof TC, Rizo J. How Tlg2p/syntaxin 16 ‘snares’ Vps45. EMBO J. 2002;21:3620–3631. doi: 10.1093/emboj/cdf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci U S A. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez I, Ubach J, Dulubova I, Zhang X, Sudhof TC, Rizo J. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- Fiebig KM, Rice LM, Pollock E, Brunger AT. Folding intermediates of SNARE complex assembly. Nat Struct Biol. 1999;6:117–123. doi: 10.1038/5803. [DOI] [PubMed] [Google Scholar]
- Fridmann-Sirkis Y, Kent HM, Lewis MJ, Evans PR, Pelham HR. Structural analysis of the interaction between the SNARE Tlg1 and Vps51. Traffic. 2006;7:182–190. doi: 10.1111/j.1600-0854.2005.00374.x. [DOI] [PubMed] [Google Scholar]
- Hata Y, Slaughter CA, Sudhof TC. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- Laufman O, Hong W, Lev S. The COG complex interacts directly with Syntaxin 6 and positively regulates endosome-to-TGN retrograde transport. J Cell Biol. 2011;194:459–472. doi: 10.1083/jcb.201102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman JC, Robblee J, Fairman R, Hughson FM. Structural analysis of the neuronal SNARE protein syntaxin-1A. Biochemistry. 2000;39:8470–8479. doi: 10.1021/bi0003994. [DOI] [PubMed] [Google Scholar]
- Li W, Ma C, Guan R, Xu Y, Tomchick DR, Rizo J. The crystal structure of a Munc13 C-terminal module exhibits a remarkable similarity to vesicle tethering factors. Structure. 2011;19:1443–1455. doi: 10.1016/j.str.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liewen H, Meinhold-Heerlein I, Oliveira V, Schwarzenbacher R, Luo G, Wadle A, Jung M, Pfreundschuh M, Stenner-Liewen F. Characterization of the human GARP (Golgi associated retrograde protein) complex. Exp Cell Res. 2005;306:24–34. doi: 10.1016/j.yexcr.2005.01.022. [DOI] [PubMed] [Google Scholar]
- Luo L, Hannemann M, Koenig S, Hegermann J, Ailion M, Cho MK, Sasidharan N, Zweckstetter M, Rensing SA, Eimer S. The Caenorhabditis elegans GARP complex contains the conserved Vps51 subunit and is required to maintain lysosomal morphology. Mol Biol Cell. 2011;22:2564–2578. doi: 10.1091/mbc.E10-06-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Li W, Xu Y, Rizo J. Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat Struct Mol Biol. 2011;18:542–549. doi: 10.1038/nsmb.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C, Munson M, Bryant NJ. Autoinhibition of SNARE complex assembly by a conformational switch represents a conserved feature of syntaxins. Biochem Soc Trans. 2010;38:209–212. doi: 10.1042/BST0380209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard F, Tang BL, Galli T, Tenza D, Saint-Pol A, Yue X, Antony C, Hong W, Goud B, Johannes L. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J Cell Biol. 2002;156:653–664. doi: 10.1083/jcb.200110081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VJ, Ungar D. Re’COG’nition at the Golgi. Traffic. 2012;13:891–897. doi: 10.1111/j.1600-0854.2012.01338.x. [DOI] [PubMed] [Google Scholar]
- Misura KM, Bock JB, Gonzalez LCJ, Scheller RH, Weis WI. Three-dimensional structure of the amino-terminal domain of syntaxin 6, a SNAP-25 C homolog. Proc Natl Acad Sci U S A. 2002;99:9184–9189. doi: 10.1073/pnas.132274599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- Ohno H, Stewart J, Fournier MC, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino JS. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW, JaRMS, editors. Macromolecular Crystallography part A. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Perez-Victoria FJ, Abascal-Palacios G, Tascon I, Kajava A, Magadan JG, Pioro EP, Bonifacino JS, Hierro A. Structural basis for the wobbler mouse neurodegenerative disorder caused by mutation in the Vps54 subunit of the GARP complex. Proc Natl Acad Sci U S A. 2010a;107:12860–12865. doi: 10.1073/pnas.1004756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Victoria FJ, Bonifacino JS. Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-golgi network. Mol Cell Biol. 2009;29:5251–5263. doi: 10.1128/MCB.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Victoria FJ, Schindler C, Magadan JG, Mardones GA, Delevoye C, Romao M, Raposo G, Bonifacino JS. Ang2/Fat-free Is a Conserved Subunit of the Golgi-associated Retrograde Protein (GARP) Complex. Mol Biol Cell. 2010b doi: 10.1091/mbc.E10-05-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Spang A. Interaction of SNAREs with ArfGAPs precedes recruitment of Sec18p/NSF. Mol Biol Cell. 2007;18:2852–2863. doi: 10.1091/mbc.E06-08-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Pelham HR. An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J. 2001;20:5991–5998. doi: 10.1093/emboj/20.21.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Pelham HR. Vps51p links the VFT complex to the SNARE Tlg1p. J Biol Chem. 2002;277:48318–48324. doi: 10.1074/jbc.M209428200. [DOI] [PubMed] [Google Scholar]
- Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasan N, Hutagalung A, Novick P, Reinisch KM. Structure of a C-terminal fragment of its Vps53 subunit suggests similarity of Golgi-associated retrograde protein (GARP) complex to a family of tethering complexes. Proc Natl Acad Sci U S A. 2010;107:14176–14181. doi: 10.1073/pnas.1009419107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu IM, Hughson FM. Tethering Factors as Organizers of Intracellular Vesicular Traffic. Annu Rev Cell Dev Biol. 2010 doi: 10.1146/annurev.cellbio.042308.113327. [DOI] [PubMed] [Google Scholar]
- Zhou P, Pang ZP, Yang X, Zhang Y, Rosenmund C, Bacaj T, Sudhof TC. Syntaxin-1 N-peptide and H(abc)-domain perform distinct essential functions in synaptic vesicle fusion. EMBO J. 2012 doi: 10.1038/emboj.2012.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.