Abstract
In human cells ClpP and ClpX are imported into the mitochondrial matrix, where they interact to form the ATP-dependent protease ClpXP and play a role in the mitochondrial unfolded protein response. We find that reducing the levels of mitochondrial ClpP or ClpX renders human cancer cells more sensitive to cisplatin, a widely used anti-cancer drug. Conversely, overexpression of HClpP desensitizes cells to cisplatin. Overexpression of inactive HClpP-S97A had no effect. Cisplatin resistance correlated with decreased cellular accumulation of cisplatin and decreased levels of diguanosine-cisplatin adducts in both mitochondrial and genomic DNA. In contrast, higher levels of cisplatin-DNA adducts were found in cells in which HClpP had been depleted. Changes in the levels of ClpP had no effect on the levels of CTR1, a copper transporter that contributes to cisplatin uptake. However, the levels of ATP7A and ATP7B, copper efflux pumps that help eliminate cisplatin from cells, were increased when HClpP was overexpressed. HClpP levels were elevated in cervical carcinoma cells (KB-CP) and hepatoma cells (BEL-7404) independently selected for cisplatin resistance. The data indicate that robust HClpXP activity positively affects the ability of cells to efflux cisplatin and suggest that targeting HClpP or HClpX would offer a novel mechanism for sensitizing cancer cells to cisplatin.
Keywords: HClpP, HClpXP, mitochondria, ATP7A, copper transport, apoptosis
1. Introduction
The success of proteasome inhibitors in treating refractory cancers such as multiple myeloma has spurred interest in the development of inhibitors that target other components of the ubiquitin/proteasome system and other protein quality control enzymes in general [1, 2]. Recently inhibitors of p97/VCP used alone or in combination with proteasome inhibitors have been shown to effectively kill cancer cells in vitro [3, 4] and at least one major effort is underway to develop p97 inhibitors for treating cancer [5]. In addition to the cytosolic ubiquitin/ proteasome and protein quality control systems, mammalian cells have three other families of ATP-dependent proteases located within the mitochondria; Lon [6], FtsH [7, 8], and ClpXP [9–12]. These proteases are homologous to bacterial ATP-dependent proteases that perform regulatory protein degradation and maintain protein quality control [13–17]. While numerous targets and functions have now been identified for the eukaryotic Lon and FtsH homologs [18–23], relatively little has been known about the functions of human ClpXP (HClpXP). Recently mutations in human ClpP have been linked to Perrault’s syndrome, an autosomal-recessive condition accompanied by ovarian and neurosensory impairment [24], and knockout mice lacking ClpP have been shown to produce sick offspring with male and female infertility and auditory defects [25]. Interestingly, CLPP−/− mice have elevated levels (1.5-4-fold depending on the tissue) of mitochondrial DNA (MtDNA), although whether that effect underlies the developmental defects or is a compensatory mechanism to restore some degree of function is not yet known. Thus, mitochondrial ClpXP clearly performs important functions and is required for robust growth and development.
HClpXP is a bipartite chaperone/protease complex composed of two proteins, a homolog of the E. coli ATP-dependent protein unfoldase, ClpX, and a homolog of the self-compartmentalized protease, ClpP [11, 12, 26]. Human cells have a single copy of HCLPP located on chromosome 19p13.3 and a single copy of HCLPX located on chromosome 15q22.31 [9, 11]. Both proteins are imported to the mitochondrial matrix. Recombinant HClpX and HClpP are similar in structure and biochemical properties to their E. coli counterparts [12, 26, 27], although HClpP has a proline-rich 28-residue C-terminal extension that is absent in eClpP [26] suggesting that it might have an additional functional interaction either with HClpX or with another cellular factor.
Because the Clp proteolytic systems are involved in stress responses in bacterial cells, we investigated the effects of increased or decreased ClpP or ClpX expression on human cell cultures subjected to various stresses. We observed that changing the levels of HClpP in HeLa cells affected their sensitivity to cisplatin (cis-diaminodichloroplatinum II), a commonly used chemotherapeutic agent. Although cisplatin is highly reactive and targets many biological molecules, its effectiveness as a chemotherapeutic agent is undermined by the development of resistant tumors and by toxic side-effects inclduing hearing loss and infertility. Most cisplatin resistance is attributable to decreased uptake or increased efflux of cisplatin, thus increasing the therapeutic dose to levels that are prohibitively toxic to normal cells [28]. Cisplatin accumulates in mitochondria [29], which are thought to be the major target for cisplatin in cancer cells [30]. Among its many targets, cisplatin cross-links to DNA and interferes with transcription and replication. In human malignant melanoma cells, cisplatin preferentially binds to mitochondrial DNA and blocks synthesis of ATP [31]. Alterations in mitochondrial function have been implicated in cancer cell resistance to other chemotherapeutic agents [32, 33], and very recently studies have shown that lowering the copy number of mitochondrial DNA sensitizes cells to cisplatin (Mei et al 2015).
Here we report that elevated levels of HClpP result in lower sensitivity to cisplatin-induced apoptosis, and conversely lower levels of HClpP or HClpX lead to increased cisplatin sensitivity. Further, we find that HClpP is elevated in independently selected cisplatin-resistant cells derived from KB cervical adenocarcinoma (KB-CPr) and hepatoma BEL-7404-CPr cell lines. We propose that HClpP activity contributes to robust cell growth and protects cells by reducing accumulation of cisplatin thereby reducing damage to nuclear and mitochondrial DNA. While these studies were underway, Dan et al (2015) reported that inhibition of mitochondrial ClpP blocked oxidative phosphorylation and mitochondrial metabolism and kills human AML cells that require elevated levels of ClpP for viability. Our findings that cells with lower HClpP activity are more sensitive to cisplatin suggest that ClpP might be an even more broadly efficacious target for anticancer therapy.
2. Materials and methods
2.1 Reagents and antibodies
The sources for various reagents and kits were: Cisplatin (Sigma, St. Louis, MO); hygromycin B (Mediatech, Inc., Herndon, VA); ECL Western blotting detection reagents (GE Healthcare Bio-Sciences Corp., Piscataway, NJ); fetal bovine serum, DMEM, L-glutamine, penicillin, streptomycin, Lipofectamine Plus reagent, Gibco trypsin-EDTA reagent, PCR primers, and nitrocellulose membranes (Invitrogen, Carlsbad, CA); RIPA buffer (Thermo Scientific, Rockford, IL); Mitochondria Isolation Kit and BCA protein assay kit (Pierce, Rockford, IL); Protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN); TRIzol reagent and methionine-free DMEM (Life Technologies, Grand Island, NY); Wizard Genomic DNA purification kit (Promega, Madison, WI). Sources for antibodies were: anti-Hsp60, (Chemicon International, Temecula, CA); anti-PARP, anti-cleaved caspase 3 and cleaved caspase 7 (Cell Signaling, Beverly, MA); anti-cisplatin-modified DNA (Abcam, Cambridge, MA); anti-ATP7A, anti-ATP7B, and anti-Ctr1 (Santa Cruz Biotechnology Inc., Santa Cruz, CA). Anti-actin antibody was obtained from Sigma; the antibody recognizes an identical 11-amino acid peptide at the C-terminus of the three major forms, α-, β- and γ-actin. The proteins have identical mobility on SDS gels, and the band is labeled actin in all blots.
2.2 Growth of HeLa cells and stable transfectants
All transfectants used in this study were constructed in a parental HeLa cell line obtained from the American Type Culture Collection (Manassas, VA). For isolation of stable transfectants, the HeLa cells were transfected with empty vector pTRE2 or with pTRE2 into which coding regions expressing wild type or mutated HClpP proteins had been inserted (see below). Transfection was performed in the presence of Lipofectamine Plus reagent according to the manufacturer’s instructions. The single cell clones were maintained in DMEM containing 350 µg/ml of hygromycin. Two transfectants that overexpress HClpP was selected for study and are designated HP7 and HP23. Additional stable transfectants used in this study include: HP17, which expresses wild type HClpP with tandem C-terminal His and HA tags (protein referred to as HP-HA); HP5, which expresses the active site mutant HClpP-S97A with C-terminal His and HA tags; HP4, which expresses wild type HClpP from a doxycycline-inducible promoter.
2.3 Other cell cultures
KB-3-1, KB-CP.5, and KB-CP20 were from Dr. Michael Gottesman in the Laboratory of Cell Biology at the National Cancer Institute. KB-3-1 was originally derived from human KB cervical carcinoma cells (a variant of HeLa) after two subclonings from the parental cells. The cisplatin-resistant KB-CP.5 and KB-CP20 cells were selected for resistance to 0.5 µg/ml and 20 µg/ml cisplatin respectively [58]. KB-CP.5 cells were cultured in DMEM medium containing 0.5 µg/ml cisplatin. KB-CP20 cells were cultured in DMEM containing 5 µg/ml cisplatin.
2.4 Construction of plasmids expressing HClpP and HClpPS97A
A gene encoding HClpP was amplified by PCR from pVEX11HClpP [12] using the forward primer, 5’AGTCGGATCCACCACCATGTGGCCCGGAATATTG-3’ and the back primer 5’-AGTCACGCGTTCAGGTGCTAGCTGGGAC-3’ creating a BamHI site at the 5’ and an MluI site at the 3’ end of the coding region. The PCR fragment was digested and inserted between BamHI and MluI sites in the plasmid pTRE2 (BD-Biosciences, Clontech, Mountain View, CA), creating pTRE2-HClpP, which expresses HClpP under a mini CMV promoter. Additional clones expressing HClpP variants were constructed in pTRE2 in a similar manner using appropriate PCR products and engineered BamHI and MluI sites. pTRE2-HClpP-His encoding His-tagged wild type HClpP was amplified by PCR from pTRE2-HClpP using the above forward primer and the back primer 5’-AGTCACGCGTTCAATGGTGATGGTGATGGTGATGGTGGGTGCTAGCTGGGAC-3’ which added eight histidine residues to the C-terminus of HClpP. We created pTRE2-HClpP-HA expressing tandemly His and HA tagged HClpP by PCR from pTRE2-HClpP-His using the above forward primer and the back primer 5’-GCTAGCACGCGTTCATGCATAGTCTGGCACATCGTAAGGGTAGCTGCCATGGTGGGATGGTG GTGGTGGTGGGTGCTAGCTGG-3’. For catalytically inactive HClpP, we used primers 5’-AGTCGGATCCACCACCATGTGGCCCGGAATATTG-3’, 5’-CGGCGGCGAGAAGCAGGGAGCCCATGGCGGCGGCCTGGCCCACGCACCAGGTG-3’, 5’-CACCTGGTGCGTGGGCCAGGCCGCCGCCATGGGCTCCCTGCTTCTCGCCGCCG-3’, and 5’AGTCG GATCCACCACCATGTGGCCCGGAATATTG-3’ to change Ser97 to Ala by over lapping PCR. We used 5’-AGTCGGATCCACCACCATGTGGCCCGGAATATTG-3’ and 5’-AGT CACGCGTTCAATGGTGATGGTGATGGTGATGGTGGGTGCTAGCTGGGAC3’ to obtain pTRE2-HClpP-SA-His from pTRE2HClpP-SA and primers 5’-AGTCGGATCCACCACCATGTGGCCCGGAATATTG-3’ and 5’-GCTAGCACGCGTTCATGCATAGTCTGGCACATCGTAAGGGTAGCTGCCATGGTGGTGATGGT GGTGGTGGTGGTGCTAGCTGG-3’ to get pTRE2HClpP-SA-His-HA from pTRE2HClpP-SA-His. For all constructs, DNA sequencing confirmed the presence of the desired coding regions of HClpP variants including its native mitochondrial targeting sequence.
2.5 Human ClpP and ClpX depletion
Human CLPP and human CLPX siRNAs and non-silencing control siRNA (siGenome No-Targeting control #3) were purchased from Thermo Scientific (Lafayette, CO). To completely deplete HClpP, 100 nM of a mixture of four siRNAs or 50 nM individual siRNAs (HCLPP-siRNA#3 or #1) were used. Cultures were treated for 24 h with siRNAs introduced in the Dharmacon siRNA reagent, Fect1 (Thermo Scientific, Lafayette, CO), according to the manufacturer’s instructions. For partial depletion of HClpP, cells were treated with 10 nM HCLPP-siRNA#3 or #1 for 5–8 h and the medium was changed to remove unabsorbed siRNA. Cells were incubated in fresh medium without siRNA for 16–19 h prior to further treatment. Partial depletion of HClpX was obtained using 50 nM of a mixture of siRNAs for 24 h after which the siRNA was removed and replaced with fresh medium.
2.6 Western blotting and detection of proteins
After removing growth medium and washing the cells with phosphate buffered saline (PBS), cells were detached by treating with Gibco trypsin-EDTA reagent, collected by centrifugation, and washed three times with PBS. Cell lysates were prepared on ice with buffer containing 10 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.5% Nonidet P40, 100 mM NaF, 10 mM sodium pyrophosphate, 10 mM sodium orthovanadate, and a protease inhibitor cocktail (Cat No. 11873580001 Roche). The lysates were centrifuged at 700 × g for 30 min at 4 ° C, and the insoluble fraction was discarded. Extracts were mixed with an equal volume of 2× SDS sample buffer (100 mM Tris/HCl, pH 7.5, containing 2 mM EDTA, 4% SDS, 10% β-mercaptoethanol, and 30% glycerol) and heated at 95 °C except for detection of ATP7A, ATP7B, and Ctr1, in which case the samples in SDS were heated at 37 Ç. For 30 min. Protein concentrations were determined using the Pierce xxx assay and equal amounts of proteins (usually 25 or 50 µg) were loaded into the lanes and separated by SDS-PAGE. Proteins were electrophoretically transferred to a 0.2-µ nitrocellulose membrane, and the membranes were blocked with 5% non-fat dry milk in Tris-buffered saline containing 0.1 % Tween-20. The membranes were incubated at room temperature for 2 h or overnight at 4 °C in the same solution to which appropriate antibodies had been added. After incubation, the membrane was washed extensively, and immunoreactive proteins were detected by chemiluminescence after incubation with secondary antibodies conjugated to horseradish peroxidase. To reprobe blots for additional proteins, membranes were treated with Restore Western Blot Stripping Buffer (Pierce, Rockford, IL), blocked, and incubated with fresh antibodies as above.
Initially antibodies were added at the dilutions recommended by the supplier; the dilutions were adjusted as needed. A ladder of proteins was applied to lanes of all gels, and blots were aligned with the standards to verify that the sizes of the immunoreactive bands corresponded to the reported sizes of the protein species recognized by the antibodies.
2.7 Isolation and quantification of mRNA
Total RNA was isolated from cells using the TRIzol reagent (Invitrogen) according to the supplier’s protocol. For each sample, RNA (1 µg in 50 µl reaction) was converted to cDNA using the TaqMan reverse transcriptase from Applied Biosystems. After diluting the mixture to 100 µl, 5 µl-aliquots of the cDNA along with 15 nM of each appropriate primer were used for quantitative real-time PCR with the LightCycler RNA Amplification PCR kit with SYBR Green1 (Applied Biosystems, Foster City, CA) as recommended by the supplier. For HCLPP message, the sense primer was 5’-CAGCTCCTCTTCCTGCAATCC-3’ and the anti-sense primer was 5’-CACACCACCAGGGCTGTTG-3’. For HCLPX message, the sense primers 5’-GTCCTAAATGTGGCGACTTGTG-3’ and the anti-sense primer was 5’-CACTTGACAAAACGGGTGGAT-3’. Control GAPDH mRNA (1:100 dilution) was amplified with the primers, 5’-ATGGAATCCCATCACCATCTT-3’ and 5’-CCAGCATCGCCCCACTT-3’. To validate amplification across the GAPDH concentration range, a standard curve was constructed using the Cp values obtained with a serial dilution of cDNA amplified from 0.005–5 ng mRNA isolated from cells treated with 50 nM control si-RNA for 5 h. For HCLPP and HCLPX, standard curves were constructed amplification of serial dilutions of pTRE2-HClpP or pTRE2-HCLPX mRNA (0.001–10 pg). The relative amounts of mRNA in each sample were determined from the respective standard curves, and the ratio of HCLPP or HCLPX mRNA to GAPDH mRNA was calculated.
2.8 Cisplatin treatment
Cisplatin was dissolved in PBS, frozen rapidly, and stored in small aliquots at −80 °C. To test the sensitivity of cells to cisplatin, an aliquot of cisplatin was added directly to culture medium, and cell visibility was measured after 16–48 h. The concentration of cisplatin used and the length of time of exposure are indicated in the figure legends.
2.9 Measurement of apoptosis and cell killing
Several assays were used to assess cell killing or induction of apoptosis as a result of exposure to cisplatin. Cells were collected by trypsinization and stained with trypan blue. Cells were counted using a hemocytometer, and the percentage of non-viable cells was determined as the number of trypan blue-stained cells divided by the total number of cells. Cell viability was also measured using the Cell Counting Kit-8 (Dojindo Laboratories, Gaithersburg, MD) and following the procedure recommended by the manufacturer. About 5000 cells were seeded into each well of a 96-well plate, grown for one day, and treated in triplicate with different concentrations of cisplatin. The percentage of viable cells remaining was indicated by the remaining dehydrogenase activity determined by reduction of a tetrazolium dye. Lethal cell damage was also measured by monitoring the progress of apoptosis using the Annexin V-PE Apoptosis Detection Kit I (BD Biosciences, San Jose, CA). Briefly, after treating cells with cisplatin, both floating and adherent cells released by trypsin were collected and washed twice with cold PBS. Cells were suspended in binding buffer at a concentration of 1 × 106 cells/ml, and 1 × 105 cells were transferred to a 5 ml culture tube, treated with amounts of Annexin V-PE recommended by the manufacturer, and incubated at room temperature in the dark. After 15 min, 400 µl of binding buffer was added and samples were processed by flow cytometry. Data acquisition and analysis were performed using a Becton Dickinson FACScan flow cytometer using the CellQuest software (Becton Dickinson, Mountain View, CA).
2.10 Cell lysis and western blotting
Harvested cells were washed twice in PBS and suspended in RIPA buffer (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM sodium vanadate, 1 µg/ml leupeptin) containing a protease inhibitor cocktail to lyse the cells to give a protein concentration of 1–2 mg/ml. After 30 min on ice, the lysates were centrifuged for 30 min at 4 ° C, and the insoluble fraction was discarded. Extracts were mixed with an equal volume of 2× SDS sample buffer (100 mM Tris/HCl, pH 7.5, containing 2 mM EDTA, 4% SDS, 10% β-mercaptoethanol, and 30% glycerol) and heated at 95 °C. Proteins (25 or 50 µg) were separated by SDS-PAGE, and transferred to a 0.2-micron nitrocellulose membrane. The membranes were blocked with 5% non-fat dry milk in Tris-buffered saline containing with 0.1 % Tween-20 and then incubated overnight at 4 °C with antibody diluted in the same solution. The membrane was washed extensively, and immunoreactive proteins were detected by chemiluminescence after incubation with secondary antibodies conjugated to horseradish peroxidase. To reprobe blots for additional proteins, membranes were treated with Restore Western Blot Stripping Buffer (Pierce, Rockford, IL), blocked, and incubated with fresh antibodies as above.
2.11 Fractionation of mitochondria and cytosol
Mitochondria were fractionated according to the manufacturer’s instructions using a Mitochondria Isolation Kit (Pierce Cat. No. 89874). The protease inhibitor cocktail described above was added to all solutions. Briefly, 20 × 107 cells were collected by centrifugation, to which 800 µl of Mitochondria Isolation Reagent A was added. After 2 min of incubation on ice, 10 µl of Mitochondria Isolation Reagent B was added and incubation was continued on ice for 5 min followed by addition of 800 µl of Mitochondria Isolation Reagent C. Nuclei and other large particles were removed by centrifugation at 700g for 10 min at 4°C. The solution was then centrifuged at 12, 000 × g for 15 min at 4°C to separate the mitochondria in the pelleted fraction from the supernatant cytosolic fraction. Mitochondria were suspended in 500 µl Mitochondria Isolation Reagent C and collected again by centrifugation at 12,000 × g for 5 min at 4° C.
2.12 Platinum uptake and incorporation into DNA
Uptake and incorporation of platinum was measured by atomic absorption or by immunochemical detection with a specific antibody. For cisplatin uptake, the cells were incubated with different amounts of cisplatin for the indicated times, after which ~106 cells were suspended in 200 µl PBS and 400 µl HNO3 and placed in an ultrasonic bath for 5 min. Platinum was measured by flameless atomic absorption on an PerkinElmer 4100ZL atomic absorption spectrometer. To measure cisplatin uptake into mitochondria, cells were lysed and mitochondria were isolated as described above. Mitochondria were dissolved in 1.5 ml of 5 M nitric acid and incubated for 24 h at room temperature. Platinum was quantitated by inductively coupled plasma mass spectrometry. For measurement of platinum adducts in DNA, the cells were incubated with cisplatin for 5 and 16 h. Genomic DNA was extracted from the cells using the Wizard Genomic DNA Isolation kit as directed by the supplier. For mitochondrial DNA isolation, mitochondria were first prepared using the Mitochondria Isolation Kit described above and DNA was prepared from the mitochondria using the Wizard kit mentioned above. Equal aliquots of DNA were spotted onto nitrocellulose membranes and cross-linked by UV irradiation. Diguanosine-cisplatin adducts were detected immunochemically with a monoclonal antibody and HRP-conjugated secondary antibodies (Abcam).
2.13 Measurement of mitochondrial DNA
The mitochondrial DNA copy number was measured according to the procedure used by Dan et al (2015). HeLa cells were transfected with 10 nM HCLPP-siRNA for 5 hours, cells were transferred to fresh DMEM medium and grown for 16 h, after which they were treated with 2.5 µg/ml cisplatin for 0 or 24 h. The cells were harvested and total DNA was isolated using a Wizard Genomic DNA Purification Kit (Promega, San Luis Obispo, CA). The mitochondrial CYTB and the nuclear β-actin gene copy numbers were measured by quantitative PCR using the SYBR Green CFX96 Real-Time System (Bio-Rad Laboratories, Hercules, CA). Each reaction contained 100 ng DNA for CYTB and 250 ng DNA for actin. PCR amplification was performed under the following condition: denaturation at 95 C for 10 min, followed by 40 two-step cycles (95 C for 15 s and 60 C for 1min). The primers for CYTB were: upper 5’-ACTATCCGCCATCCCATAC-3’; Lower 5’-GCAAGAATAGGAGGTGGAG-3’. The primers for β-actin were: Upper 5’-ACCTTCTACAATGAGCTGCG-3’; Lower 5’-CCTGGATAGCAACGTACATGG-3’. Three independent cultures of each cell line were grown and duplicate DNA samples from each culture were analyzed. For each sample delta delta Cq measurements were used to obtain the ratio of the CYTB and ACTB copy numbers in the reaction. The average CYTB copy numbers per cell were calculated to obtain mitochondrial DNA copy number. Standard among the six samples from each cell line were <10% of the measured values; P was <0.01 for calculated values of the cellular copy numbers.
2.14 Statistical analysis
In most cases experimental data were obtained in triplicate, and data are expressed as the standard error of the mean of the measurements (SEM). Where data from single experiments are reported, the error bars represent the standard deviation (SD) of the data obtained in the three measurements. When statistical analysis was performed to compare two values, a two-tailed T-test was applied to calculate a P value at 95% confidence. In the graphs, P values ≤ 0.01 are indicated by double asterisks and P <0.1 by a single asterisk.
3. Results
3.1 Mitochondrial overexpression of HClpP protects HeLa cells from killing by cisplatin
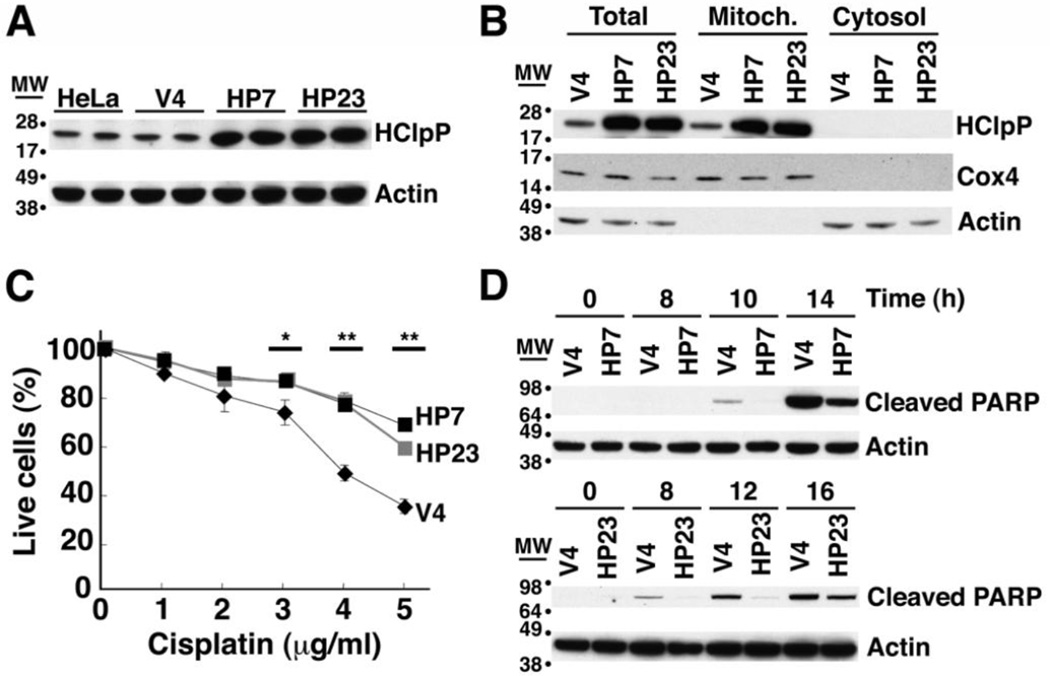
To investigate biological pathways affected by HClpXP, we monitored changes in growth rates and responses to various forms of stress in cultured human cells when HClpP expression was increased or decreased. HeLa cells were transfected with pTRE2-HCLPP1, which encodes full-length HClpP including its mitochondrial targeting sequence. Two stable hygromycin-resistant cell lines, HP7 and HP23, were selected for further study. Control cell lines, V3 and V4, which had been transfected with the unmodified pTRE2 vector, were also selected. HClpP levels determined by Western blotting were 6–8-fold higher in HP7 and HP23 than in non-transfected HeLa cells but were unchanged in control V3 and V4 cells (Fig. 1A and Supplementary Fig. S1A). Overexpressed HClpP was found only in the mitochondrial fraction (Fig. 1B). We then treated HP7, HP23, and V4 cells with different concentrations of cisplatin for 16 h and counted viable cells. More then 60% of V4 cells died after 16 h of exposure to 5 µg/ml cisplatin, whereas killing was decreased in cells with overexpressed HClpP (30% for HP7 and 32% for HP23 (P <0.01)) (Fig. 1C).
Fig. 1. Excess of HClpP protects HeLa cells from killing by cisplatin.
A. HClpP expression in stable transfectants. Proteins were extracted from stable transfectants of pTRE2-HClpP (HP7 and HP23) or the pTRE2 vector (V4) and separated by SDS-PAGE. HClpP was detected by western blotting with anti-HClpP antibodies. Actin was monitored as a loading control using an antibody against the C-terminal peptide common to α, β, and γ actins. B, Subcellular distribution of HClpP in stable transfectants. Mitochondrial and cytosolic fractions were prepared from HP7, HP23, and V4 cells. Protein aliquots (25 µg) from each fraction were separated by SDS-PAGE, and HClpP was measured by western blotting with antibodies to HClpP. Separation of mitochondria and cytosol was confirmed with antibodies to Cox4 as a mitochondrial marker or actin as a cytosolic marker. C, Sensitivity of transfectants to cisplatin. V4, HP7, and HP23 cells were treated with 0–5 µg/ml cisplatin (CP) for 16 h, and cell viability was measured by CCK-8 as described in "Materials and Methods". The viability of cells without cisplatin treatment was normalized to 100%. Results from a single experiment are shown. Error bars represent the standard error of measurements made in triplicate, and significant differences between overexpression and control cells determined by a two-tailed T-test are noted with asterisks. Similar results were obtained in at least three independent experiments. D, Delayed apoptotic response in HClpP-transfectants treated with cisplatin. Apoptotic cell death of cells treated with cisplatin was monitored by western blotting to detect the cleavage product of PARP, a marker for late stages of apoptosis. Aliquots (25 µg protein) from V4, HP7, and HP23 cells that had been treated with 5 µg/ml cisplatin were separated by SDS-PAGE and probed with an antibody specific for the proteolytically processed form of PARP. Actin served as the loading control.
Cisplatin damages DNA and induces an apoptotic response leading to cell death. The terminal stages of apoptosis can be monitored by the appearance of a truncated form of PARP, a 116-kDa nuclear poly (ADP-ribose) polymerase needed for DNA repair in response to environmental stress [34, 35]. In V4 cells, cleavage of PARP was obvious between 8 and 10 h after addition of cisplatin; PARP cleavage occurred much later in the HP7 and HP23 cell lines (Fig. 1D). This result further establishes that high levels of HClpP act to slow initiation or progression of apoptosis following exposure to cisplatin.
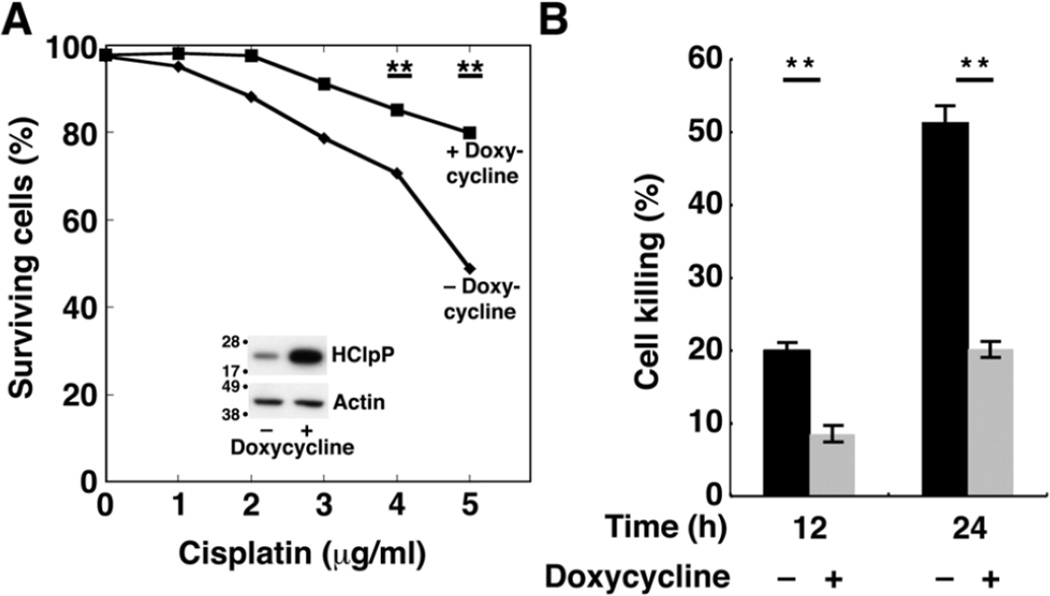
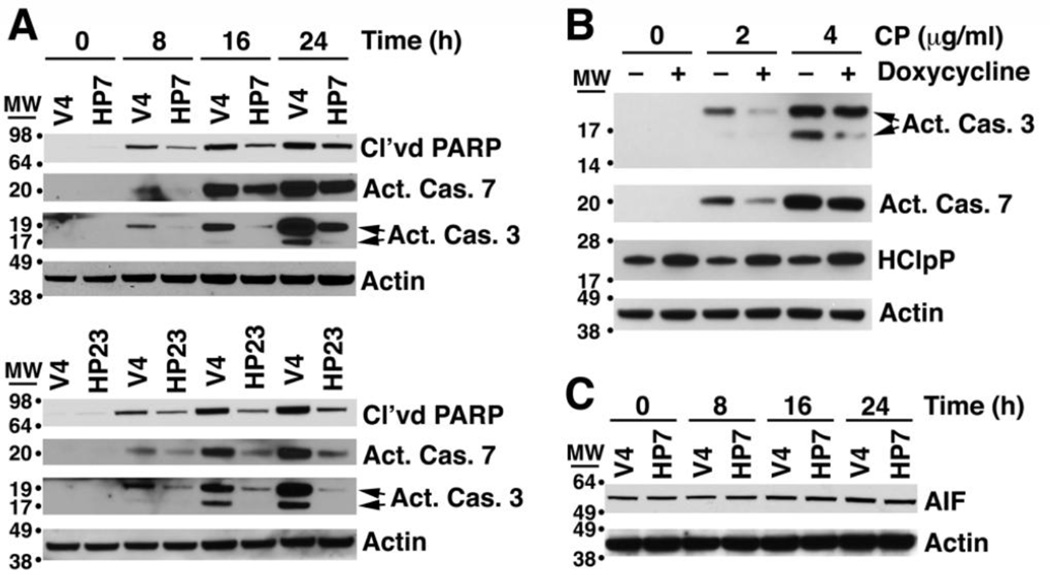
To rule out the possibility that the effects on cisplatin sensitivity were due to a host mutation picked up during selection of stable HCLPP transfectants, we obtained a stable transfectant of HCLPP (HP4) in which HClpP expression is under control of the tetracycline promoter and requires an inducer for high level expression of HClpP. HClpP levels were increased about 5–8-fold after 48 induction with doxycycline (Fig. 2A, inset). Uninduced HP4 cells were sensitive to cisplatin (Fig. 2A, diamonds) showing a dose response similar to that seen in Fig. 1C for V4 control cells. When HClpP was induced for 48 h prior to addition of cisplatin, HP4 cells showed increased resistance to cisplatin (Fig. 2A, squares). After 12 and 24 h in the presence of 5 µg/ml cisplatin (Fig. 2B), cultures in which HClpP had been induced had far fewer dead cells (20% without induction compared to 8% with induction at 12 h and 50 % without induction and 20 % after induction at 24 h (P < 0.01)).
Fig. 2. Increased resistance to cisplatin following induction of HClpP.
Cultures of HP4 cells, a stable transfectant with multiple copies of HCLPP under control of a tetracycline promoter, were grown in parallel. Doxycycline was added to one culture to induce expression of HClpP; DMSO was added to the other. A, Cisplatin dose response of cells with increased HClpP. After 48 h of continued growth, HClpP levels were monitored by western blotting following SDS-PAGE (inset). Induced (squares) and uninduced (diamonds) HP4 cells were treated with 0–5 µg/ml cisplatin for 24 h, and the percentage of cells that were not stained with Annexin V antibody was measured by fluorescence flow cytometry. B, Reduced rate of killing in cells with induced HClpP. HP4 cells were incubated with (gray bars) or without (black bars) doxycycline for 48 h, after which cells were treated with 5 µg/ml cisplatin for 12 or 24 h. Apoptotic cell death was monitored by Annexin V staining. Measurements were made in triplicate, and significance was estimated using a 2-tailed T-test. In A and B, results of single experiments are shown. Similar results were obtained in two independent experiments.
3.2 Sensitivity to cisplatin is increased in cells with lower levels of HClpP or HClpX
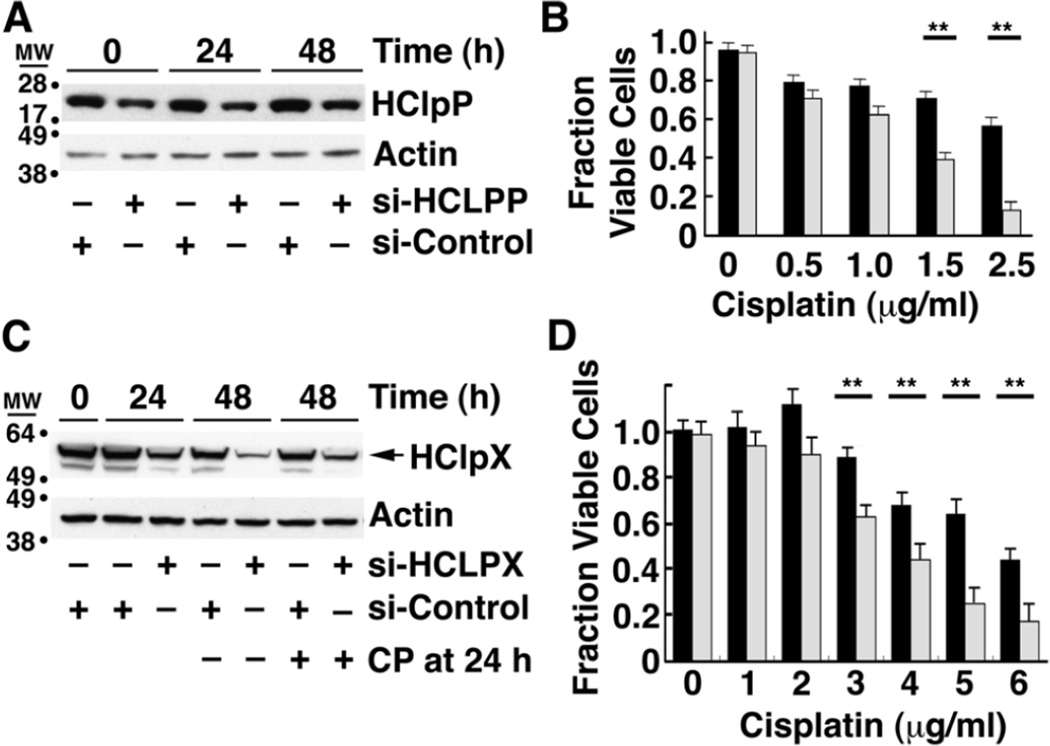
We next tested whether reducing HClpP had the opposite effect on cell killing by cisplatin. Treatment of HeLa cells with 50–100 nM siRNA specific for HCLPP results in a rapid depletion of HCLPP mRNA and a rapid reduction of HClpP protein levels (Supplementary Fig. S1B and C). Because depletion of HClpP by ≥90% for extended periods led to growth arrest and cell death (Y. Zhang and M. R. Maurizi, manuscript in preparation), we adopted a milder treatment to partially deplete HClpP. Transfection of HeLa cells with 10 nM HCLPP-siRNA#3 for 5 h followed by washing and suspension in fresh medium resulted in a 50–60% drop in HClpP levels (Fig. 3A) without loss of cell viability (Fig. 3B, no cisplatin). To assure that mitochondria integrity was maintained under these conditions, we monitored the levels of COX2, a subunit of the cytochrome C oxidase complex, as well as the α and β subunits of ATP synthase in cells treated with 10 nM HCLPP-siRNA. No changes were seen in any of these essential mitochondrial proteins in cells with partially depleted HClpP (Supplementary Fig. S2A and B). Cells with reduced HClpP were, however, were killed by lower concentrations of cisplatin than cells that had been treated with control siRNA (Fig. 3B). The cells with reduced HClpP also died much faster than control cells when both were treated with the same amount of cisplatin (Supplementary Fig. S3). Visual inspection of cells treated with cisplatin for 24 h revealed obvious shrinking, blebbing, and breaking of the cells with reduced HClpP, whereas the cells with normal HClpP levels had no abnormal morphology (data not shown). The above experiment was repeated with HClpP-siRNA#1, which targets a different region of the HCLPP mRNA, and the same results were obtained (Supplementary Table 1), making it highly unlikely that the acquired cisplatin sensitivity was due to off targeting by the HClpP siRNA. These data together with the data showing that increased expression of HClpP protect cells from killing by cisplatin indicate that HClpP performs a cellular function that helps cells avoid, repair, or resist the damage caused by cisplatin.
Fig. 3. Cells with decreased levels of HClpP or HClpX are more sensitive to cisplatin.
A, HClpP levels after siRNA transfection. HeLa cells were transfected with 10 nM HClpP siRNA or control siRNA for 5 h. The cells were washed to remove the siRNA and then incubated in DMEM medium for 16 h, after which they were treated with the indicated concentrations of cisplatin for an additional 24 or 48 h. The time shown is the time after addition of cisplatin. Equal aliquots of cell extracts were separated by SDS-PAGE, and HClpP was detected by western blotting with anti-HClpP antibodies. Actin was monitored as a loading control. B, Cisplatin sensitivity of cells with reduced levels of HClpP. Cells pre-treated as in panel A with control siRNA (black bars) or HCLPP-siRNA (white bars) were grown for 48 h in the presence of different amounts of cisplatin. The fraction of live cells is shown. C, HClpX levels after siRNA transfection. HeLa cells were transfected with 50 nM HClpX-siRNA or control siRNA for 24 h. The cells were washed to remove the siRNA and then incubated in DMEM medium with 2.5 µg/ml cisplatin (CP) for an additional 24 or 48 h. HClpX was detected by western blotting with anti-HClpX antibodies. Actin was monitored to confirm uniform protein loading. D, Cisplatin sensitivity of cells with reduced levels of HClpX. Cells pre-treated with control siRNA (black bars) or HCLPX-siRNA (white bars) were grown for 48 h in the presence of different amounts of cisplatin. In B and D, dead cells were counted after staining with trypan blue, and the percentage of live cells was calculated. The results of single experiments are shown. The error bars represent the standard error of measurements made in triplicate. Significance was estimated using a 2-tailed T-test. Similar results were obtained in several independent experiments.
We have not been able to establish stable transfectants expressing significantly higher levels of HClpX and thus have not investigated whether increased HClpX is also protective against cisplatin. However, treatment of HeLa cells with HCLPX-siRNA reduces HClpX levels in cells over a period of 24–48 h (Fig. 3C), and we asked whether cells with lower levels of HClpX are sensitive to cisplatin. HeLa cells were transfected with 50 nM HClpX siRNA (gray bars) or control siRNA (black bars) for 24 h and washed to remove the siRNA. Cells were then incubated in DMEM medium with different concentrations of cisplatin for an additional 48 h and trypan blue staining was used to monitor cell killing. Cells with partially depleted HClpX were killed by significantly lower concentrations of cisplatin (Fig. 3D). To ensure that cisplatin did not affect the level of HClpX, extracts of cells treated with 2.5 µg/ml cisplatin were run on an SDS gel and probed for HClpX by western blotting. HClpX levels were essentially the same with and without cisplatin addition for cells treated with either HCLPX-siRNA or control siRNA (Fig. 3C, four right lanes). These results indicate that increased sensitivity to cisplatin results from loss of a function or functions that are dependent on both ClpP and ClpX.
3.3 Overexpression of catalytically inactive HClpP (HClpP-S97A) does not protect HeLa cells from killing by cisplatin
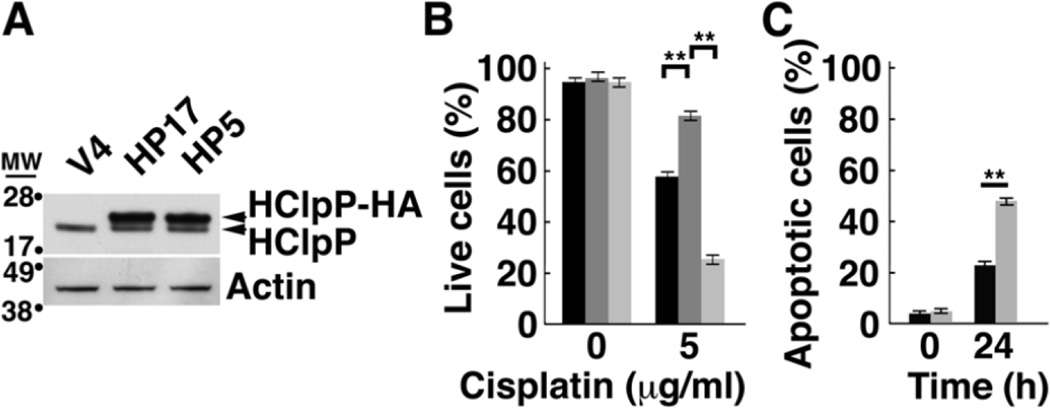
To confirm that the activity of HClpP is needed for protection from cisplatin, we selected a HeLa cell line (referred to as HP5) that overexpresses catalytically inactive HClpP-S97A-His10-HA, in which the catalytic serine is replaced by alanine; the protein also carries tandem C-terminal His10 and HA tags. We also selected HeLa cells that overexpress wild type HClpP with the same C-terminal tags (HP17). Western blots (Fig. 4A) showed that HClpP-His10-HA and HClpPS97A-His10-HA were expressed in similar amounts. We treated both stable transformants with 2.5 µg/ml cisplatin for 16 h and monitored cell death with trypan blue. As before, cells overexpressing active HClpP exhibited increased resistance to cisplatin, but HP5 cells with high levels of inactive HClpP did not exhibit resistance and appeared to be even more sensitive to cisplatin than vector control cells (Fig. 4B). To confirm that cells expressing inactive HClpP were more sensitive to cisplatin, we again treated HP5 cells with cisplatin and measured the appearance of apoptotic cells by Annexin V staining. After 24 h, there were twice as many Annexin V positive cells in the HP5 cultures as in control cell cultures (P >0.01) (Fig. 4C).
Fig. 4. Overexpression of the active site mutant HClpP-S97A does not protect cells from killing by cisplatin.
A, Levels of catalytically inactive HClpP-S97A. HP5 is a stable transfectant of HeLa that expresses a tagged form of the catalytically inactive mutant, HClpP-S97A. HP17 expresses an identically tagged form of active HClpP. Extracts (25 µg protein) from HP5, HP17, and control V4 cells were separated by SDS-PAGE. The wild type and mutated HClpP variants run slightly above endogenous HClpP in the gel and are labeled HClpP-HA. HClpP proteins were detected by Western blotting with anti-HClpP antibodies. Actin was monitored to confirm uniform loading. B, Inactive HClpP does not have protective effect against cisplatin. The number of V4 (black), HP17 (dark gray), and HP5 (light gray) transfectants killed following treatment with 5 µg/ml cisplatin for 16 h was determined by staining with trypan blue. The percentage of live cells is shown. Error bars represent the standard error of measurements made in triplicate. Significance was estimated using a 2-tailed T-test. The results were reproducible in at least three separate experiments. C, Cell killing by cisplatin is faster in cells overexpressing catalytically inactive HClpP. Control V4 cells (black bars) or the HP5 cells (gray bars) were treated with 2.5 µg/ml cisplatin for 24 h, and apoptotic cell death was monitored by Annexin V staining. Results of a single experiment are shown. Error bars represent the standard error of measurements made in triplicate.
3.4 HClpP acts upstream of caspases in protecting cells from killing by cisplatin
Cisplatin-induced apoptosis proceeds through a caspase-dependent pathway as well as a caspase-independent pathway involving release of the mitochondria-associated protein, AIF [36, 37]. To assess the stage at which HClpP was acting to delay killing, we treated cells with cisplatin and monitored activation of caspases 7 and 3 using antibodies specific for the cleaved activated forms of the caspases. Following addition of cisplatin to control V4 cells, activated caspase 3 accumulated in high amounts between 16 and 24 h; however, in HP7 and HP23 cells, activated caspase 3 accumulated more slowly and reached much lower levels (Fig. 5A). Activation of caspase 7 was also delayed in both HP7 and HP23 cells (Fig. 5A). To confirm that the slower rate of caspase activation was dependent on overexpressed HClpP and not a result of a second-site mutation in HP7 or HP23, we monitored caspase 3 and caspase 7 activation in cisplatin-treated HP4 with and without prior induction HClpP. Caspase 3 and caspase 7 were both activated to lesser extents in the HP4 cells with elevated levels of HClpP (Fig. 5B). AIF is a pro-apoptotic protein that is processed and released from mitochondria in response to DNA damage and other stress [38, 39]. We monitored the total cellular level of AIF and observed no change in the size or the amount of AIF following cisplatin addition to either HP7 or HP23 cells or to control V4 cells (Fig. 5C), suggesting that AIF does not play a role in cisplatin toxicity in HeLa cells under these conditions. These results suggested that HClpP acts upstream of permeabilization of the mitochondrial membrane and caspase activation, which prompted us to ask whether HClpP might exert its effect by reducing the rate or extent of DNA damage caused by cisplatin.
Fig. 5. HClpP acts upstream of caspase activation and apoptosis to protect cells from cisplatin.
A, Delayed activation of caspases 7 and 3 in HClpP overexpressing cells treated with cisplatin. HP7, HP23, and V4 control cells were treated with 5 µg/ml cisplatin. At the indicated times cells were collected, and proteins were separated by SDS-PAGE. Western blotting was performed using antibodies that recognize either the activated form of caspase 7 or the activated forms (19K and 17K) of caspase 3. The increase in apoptotic cells after cisplatin addition was monitored by the accumulation of cleaved PARP. Actin served as a loading control. Results were reproducible in three separate experiments. B, Delayed activation of caspase 3 and caspase 7 by cisplatin treatment of transfectants with induced levels of HClpP. HClpP was induced by doxycycline addition to HP4 cells for 48 h. HClpP levels increased 2–3-fold as indicated by western blotting. Cisplatin (CP) (0, 2, and 4 µg/ml) was added to the cultures for 24 h, and cells were collected for analysis of activated caspase 3 and 7 by western blotting as above. Uninduced HP4 cells were treated with cisplatin and processed in parallel. The experiment was repeated with similar results. C, Total AIF levels are not affected by overexpression of HClpP. The blot used for detection of caspases in HP7 and control cells was probed with antibody against AIF. The actin control is the same as that shown in panel A. The experiment has been repeated several times with similar results.
3.5 Cisplatin accumulation and DNA-bound platinum are reduced in HClpP-overexpressing cells and elevated in cells when HClpP is knocked down
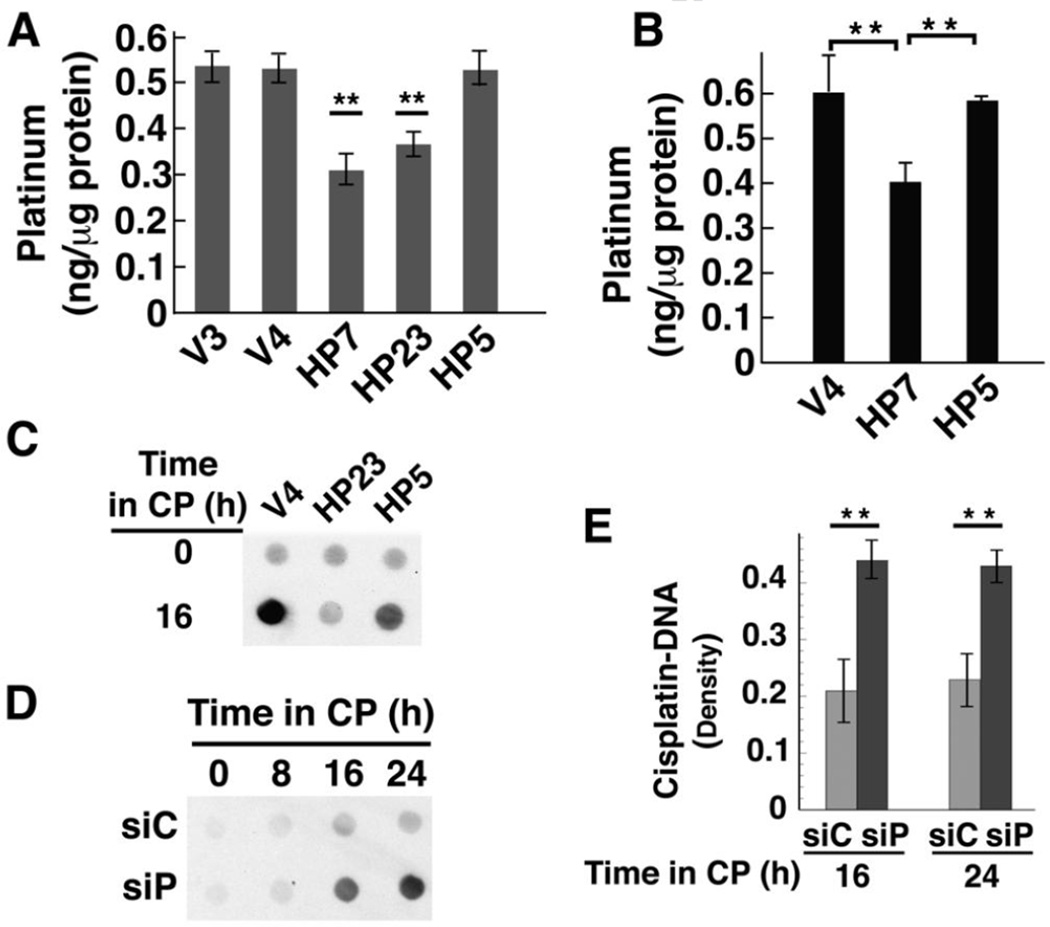
One mechanism by which tumors become resistant to cisplatin is through reduced cellular accumulation of cisplatin [40, 41]. Because HClpP appeared to act upstream of the cell death signaling pathway, we examined whether the levels of HClpP affected the uptake of cisplatin into whole cells and into mitochondria. Cells were treated with cisplatin for 16 h or 24 h and washed to remove excess cisplatin. Total platinum in whole cells or mitochondria was measured by atomic absorption (Fig. 6A) or by mass spectrometry (Fig. 6B). HP7 and HP23 cells overexpressing wild type HClpP showed significantly lower cellular accumulation of cisplatin than the control cells, V3 and V4. HP5 cells, which express catalytically inactive HClpP-S97A, accumulated cisplatin to levels similar to those in control cells. Isolated mitochondria from cells treated with cisplatin for 24 h followed a similar pattern. Mitochondria from control cells and cells expressing inactive HClpP had similar levels of platinum, whereas mitochondria from cells expressing wild type HClpP accumulated significantly lower levels. That reduced cisplatin accumulation was detectable at the level of total cellular uptake suggested that HClpP overexpression affected a major target of cisplatin or a fundamental process such as transport of cisplatin.
Fig. 6. Cisplatin accumulation and DNA-bound platinum are lower in cells over-expressing active HClpP and higher in cells with reduced HClpP.
A, Cellular accumulation of cisplatin. Cultures were treated with 3.5 µg/ml cisplatin for 16 h, after which the cells were washed and platinum levels in total cell lysates were measured in triplicate by atomic absorption. Error bars represent the SEM in two independent experiments. Platinum levels in HP7 and HP23 cells were between 50–60% of control V3 or V4 levels in two separate experiments (P < 0.01; noted with asterisks). Levels in HP5 were not significantly different from those in control cells (P >0.1). B, Mitochondrial accumulation of cisplatin. Cells were treated with 2.5 µg/ml cisplatin for 16 h, collected by centrifugation and washed. Mitochondria were isolated by differential centrifugation, washed, and dissolved in nitric acid. Platinum in the mitochondrial fraction was quantitated by ICP mass spectrometry. Error bars represent the SEM of the values obtained in two independent experiments. Levels in HP7 cells were 65 ± 2 of those in V4 cells (P < 0.01). C, V4, HP23, and HP5 cells were treated with 3.5 µg/ml cisplatin for 16 h. Total cellular DNA was isolated and 10 µg-aliquots of DNA were spotted and probed for platinum modified DNA using a monoclonal antibody that recognizes diguanosine-cisplatin-adducts. D and E, Accumulation of platinum in total cellular DNA after knockdown of HClpP. Parallel cultures of HeLa cells were transfected with 10 nM HClpP siRNA (siP) or control siRNA (siC) for 5 h, washed to remove the siRNA, and then incubated in DMEM medium for 16 h. Cisplatin (CP) (2.5 µg/ml) was added and individual cultures were harvested at different times. Total DNA was extracted from the cells and purified. D, Equal aliquots (12 µg) from each sample were spotted on a nitrocellulose membrane, and cisplatin adducts were detected as above. Spots contain < 0.1 ng MtDNA, and the signal reflects primarily modification of nuclear DNA. E, Serial dilutions of each DNA sample (12, 6, and 3 µg) were spotted and cisplatin adducts were detected immunochemically. The blots were scanned, and the densities for each sample were normalized for loading and averaged. The error bars represent the SEM of the normalized densities from the diluted samples.
As DNA is a major target of cisplatin, we asked whether DNA modification by cisplatin is affected by HClpP. Total cellular DNA was isolated and spotted onto nitrocellulose, and cisplatin in DNA was detected with an antibody specific for diguanosine-cisplatin adducts. Figure 6C shows that DNA from control cells gave a strong signal for cisplatin adducts, whereas DNA from HP23 cells gave a much weaker signal. HP5 cells also gave a strong signal, indicating that inactive HClpP does not reduce cisplatin uptake into DNA. Because knockdown of HClpP made cells more sensitive to cisplatin, we expected that cisplatin uptake into DNA would be higher after cells were treated with HCLPP-siRNA. Figure 6D shows that signal from cisplatin-DNA adducts was much stronger with DNA from cells in which HClpP had been knocked down. Densitometry was performed using spots from serial dilutions of DNA at each time point. The level of cisplatin incorporated into DNA was twice as high in HClpP-depleted cells as in control cells (P <0.01) (Fig. 6E). These data demonstrate that HClpP activity is important to reduce the extent of DNA damage caused by cisplatin.
3.6 Overexpressed HClpP reduces cisplatin uptake into MtDNA
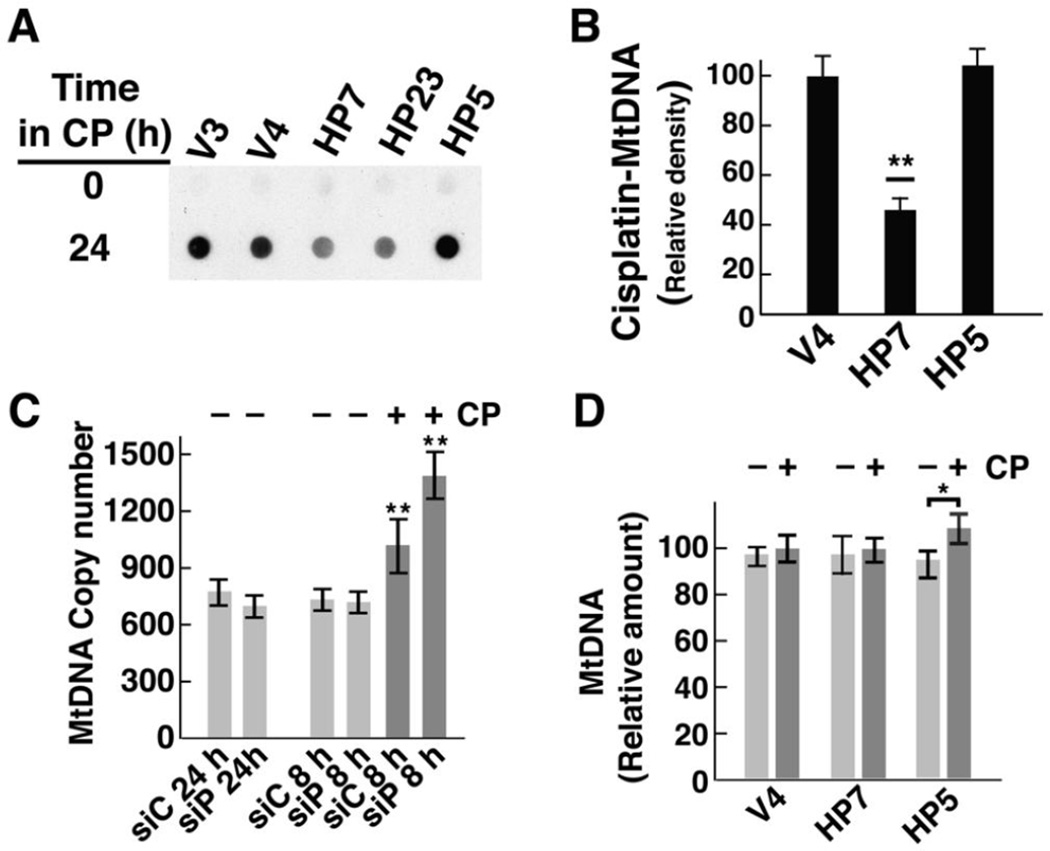
Cisplatin preferentially targets mitochondrial DNA, and MtDNA copy number influences the sensitivity of cells to cisplatin and other DNA damaging drugs (Mei H 2015). Dot blots with total cellular DNA (e. g., Fig. 5C and D) are dominated by nuclear DNA, which is 100–200 times more abundant than mitochondrial DNA. To determine if HClpP affected cisplatin incorporation into MtDNA, we blotted MtDNA and probed for cisplatin adducts. When equal amounts of MtDNA from control cells or HClpP-overexpressing cells were blotted, cisplatin adducts were readily detected in MtDNA from V4 and V3 cells but gave significantly lower signals in HP7 or HP23 cells (Fig. 7A). MtDNA from HP5 cells elicited a signal comparable to that from control cells (Fig. 7A). Densitometry of the dot blots showed that MtDNA from cells with overexpressed HClpP took up 40–70% as much cisplatin as MtDNA from control cells (Fig. 7B) (P <0.01). Cisplatin levels in HP5 MtDNA were not different from those found in control cells.
Fig. 7. Decreased cisplatin damage to MtDNA in cells overexpressing HClpP.
A, HeLa cells were treated with 3.5 µg/ml of cisplatin (CP) for 16 h. DNA was purified from isolated mitochondria, equal aliquots of DNA were spotted, and cisplatin incorporation was detected immunochemically using antibody against diguanosine-cisplatin adducts. Similar results were obtained in two other separate experiments. B, The blot was scanned and the densities from two control cultures and two HClpP overexpressing cultures were averaged. Error bars represent the SEM. HP7 levels were 40–50% of control levels (P <0.01). C, Mitochondrial DNA copy number in cells with reduced HClpP. DNA was purified from HeLa cells treated with control or HCLPP-siRNA. Cells treated for 24 h received 50 nM siRNA to deplete >90% of the HClpP. Cells treated for 8 h received 10 nM siRNA, which reduces HClpP by ~50% (e. g. see Fig. 3A). Cells partially depleted of HClpP were transferred to fresh medium and subsequently treated with 2.5 µg/ml cisplatin (CP) (dark gray bars) or left untreated (light gray bars) for 24 h. Mitochondrial CYTB copy number was measured relative to the nuclear ACTB gene encoding β-actin by Q-PCR as described in Materials and Methods. DNA was isolated from three independent cultures for each condition. Copy numbers were calculated from duplicate measurements for each sample. The averages of the copy numbers and the SEM are shown; values significantly different from those obtained with DNA from cells not treated with cisplatin are noted with asterisks. D, Mitochondrial DNA copy number in cells with elevated levels of HClpP. DNA was purified from stable transfectants (V4, HP7, and HP5) that had been left untreated (light gray bars) or had been treated with 2.5 µg/ml cisplatin (CP) for 24 h (dark gray bars). Copy numbers were calculated from duplicate measurements made with DNA isolated from nine independent cultures. To simplify the comparisons, the copy number for the untreated V4 cells was set to 100 and the copy numbers fore the other cells are expressed relative to that for V4. The error bars represent the SEM for the calculated copy numbers.
Studies with homozygous knockout mice lacking any functional mitochondrial ClpP revealed that surviving animals have significantly increased copy numbers of MtDNA, which, depending on the tissue source, can vary from 1.5- to 4-times that found in control animals [25]. To ask whether the differences in cisplatin uptake (or the sensitivity of cells to killing by cisplatin) could reflect differences in the copy number of mitochondrial DNA in cells when HClpP is decreased under our experimental conditions, we measured MtDNA after knocking down HClpP with either moderate or high levels of siRNA. MtDNA copy number was measured by Q-PCR using CYTB as a marker for MtDNA and compared to ACTB as the nuclear DNA marker. MtDNA copy numbers were within 8–10% of one another both in cells treated with high levels of HCLPP-siRNA for 24 h to reduce HClpP by ≥90% and in cells treated with lower levels of HCLPP-siRNA for 8 h to reduce HClpP ~50% (Fig. 7C). Thus the sensitivity and resistance to cisplatin observed under our conditions is not a function of changes in MtDNA copy numbers. Interestingly, cisplatin treatment alone produced a small increase in MtDNA, which was amplified when HClpP was partially depleted. Addition of 3.5 µg/ml cisplatin for 24 h to cells previously treated with control RNA resulted in 30–40% increase in MtDNA (P ≤0.01). Cisplatin addition to cells previously treated with HCLPP-siRNA to reduce HClpP by ~50% resulted in a 70–80% increase in MtDNA (Fig. 7C) (P <0.01). As the copy number of MtDNA was not increased by knockdown of HClpP alone, our interpretation of this result is that cisplatin damage results in up-regulation of MtDNA synthesis and that the increased damage caused by cisplatin when HClpP levels are low amplifies this response.
We also measured MtDNA levels in cells that overexpress either active or inactive HClpP. Figure 7D shows that there were no significant differences in MtDNA in V4, HP7, and HP5 cells. When these cells were treated with cisplatin for 24 h, neither control cells or HP7 cells showed a consistent different in MtDNA, however HP5 cells expressing inactive HClpP-S97A had slightly elevated levels of MtDNA (Fig. 7D). As HP5 cells are more sensitive to cisplatin than control or HP7 cells, the increase in MtDNA might reflect the effect of increased DNA damage as seen in
3.7 Levels of the copper efflux pump, ATP7A, are higher in cells that overexpress HClpP
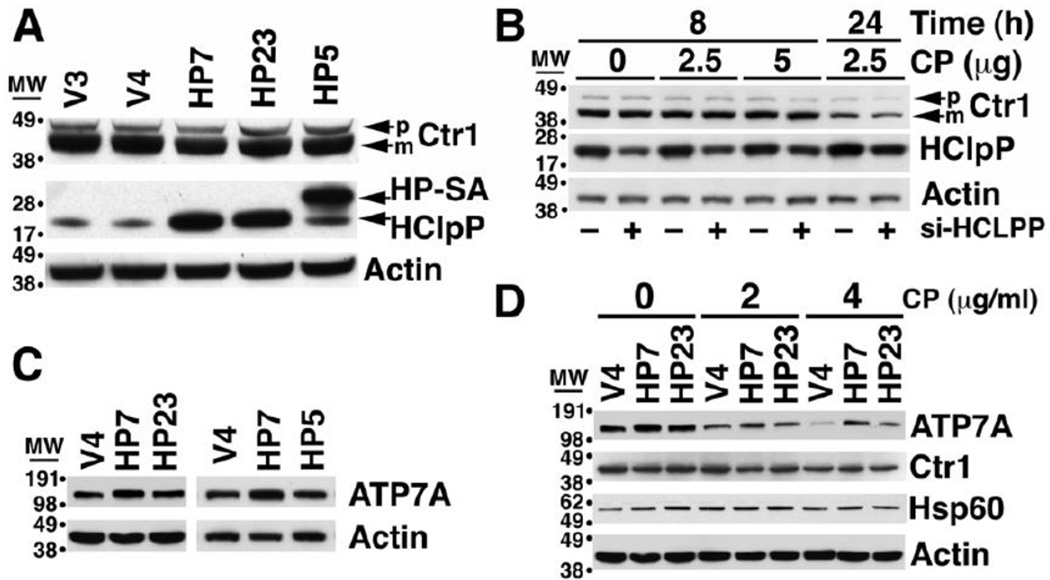
Changes in HClpP correlated with changes in cellular accumulation of platinum, and thus we asked whether HClpP might affect the ability of cells to take up or release cisplatin. Biological and biochemical data have established a tight association between cisplatin and copper transport pathways. For example, cisplatin-resistant ovarian carcinoma cell lines accumulate less copper and are cross resistant to copper [42], and copper-resistant cell lines are cross resistant to cisplatin [43]. The copper transporters, Ctr1 and Ctr2, have been shown to catalyze cisplatin uptake, and the copper efflux pumps, ATP7A and ATP7B, promote its release from cells. We first measured expression of Ctr1 in HeLa cells with either increased or reduced expression of HClpP. The levels of Ctr1 in the HClpP-overexpressing HP7 or HP23 cells were similar to those in V4 cells as well as in SA-HA5 cells that overexpress inactive HClpP-S97A (Fig. 8A). We next measured Ctr1 levels in cells with reduced amounts of HClpP. The levels were unchanged after partial knockdown of HClpP (Fig. 8B, no cisplatin). Ctr1 was shown to be degraded in cells after addition of cisplatin [44]. In our experiment, there was no change in Ctr1 levels during the first 8 h in cells treated with cisplatin (Fig. 8B), but the levels decreased after 24 h. The decrease was similar in cells treated with control or HCLPP-siRNA (Fig. 8B). These and other experiments in which HClpP was knocked down to very low levels (data not shown) establish that Ctr1 is not affected by changes in HClpP activity.
Fig. 8. The levels of the copper transporter Ctr1 and the copper efflux pump ATP7A in cells with increased or decreased HClpP.
A. Ctr1 levels are unchanged in cells overexpressing active or inactive HClpP. Ctr1 was detected by western blotting following SDS-PAGE separation of proteins. The major band is mature Ctr1, a multiple-pass integral membrane protein that undergoes N-terminal truncation. The minor band above mature Ctr1 is the unprocessed precursor. No consistent differences in intracellular levels Ctr1were seen in cells with endogenous levels of HClpP (V3 and V4), with >8-fold overexpression of HClpP (HP7 and HP23), or with >8-fold overexpression of the mutant HClpP-S97A (HP-SA). C-terminal HA and His tags cause HP-SA to run just above endogenous HClpP on SDS gels. B. Ctr1 levels are not affected by partial knock down of HClpP. HeLa cells were treated with HClpP-50 nM siRNA#3 or control siRNA for 5 h (see Materials and Methods) after which the cells were washed in PBS and transferred to fresh medium. Cisplatin (CP) was added as indicated prior to removing samples to measure the levels of Ctr1. C. Increased expression of HClpP is accompanied by an increase in ATP7A. ATP7A was detected by western blotting with anti-ATP7A antibodies following SDS-PAGE separation of proteins from HCLPP transfectants, HP7 and HP23, V4 control cells, and HP5 transfectant expressing catalytically inactive HClpP-SA. D. Following cisplatin treatment, ATP7A levels persist at higher levels in cells that overexpress HClpP. HP7, HP23, and V4 control cells were treated with 0, 2, or 4 µg/ml cisplatin (CP) for 24 h and the levels of ATP7A were measured by western blotting with anti-ATP7A antibodies.
We next assayed the level of the copper efflux pumps, ATP7A and ATP7B, in cells that overexpress HCLPP. ATP7A levels were higher in HP7 and HP23 than in the V4 control cells but were not elevated in HP5 cells that overexpress inactive HClpP-S97A (Fig. 8C). In multiple experiments the basal level of ATP7A has been found to be consistently higher by 30–70% in cells that overexpress active HClpP. A more pronounced difference was seen after treating cells with cisplatin. ATP7A levels declined following treatment with cisplatin, but the levels declined more slowly in cells with overexpressed HClpP (Fig. 8D). Consequently, after 24 h with 4 µg/ml cisplatin, the ATP7A levels in HP7 or HP23 cells were 4–5-fold higher than in control cells (Fig. 8D). To confirm that ATP7A levels were responsive to the change in HClpP and not to extraneous effects of transfection with siRNA, we treated induced and uninduced HP4 cells with cisplatin. ATP7A levels were higher in cells after induction of HClpP (Supplementary Fig. S3). When the cells were treated with cisplatin, the levels of ATP7A decreased in all the cells but declined significantly more slowly in the cells in which HClpP had been induced (Supplementary Fig. S3).
ATP7B levels were higher in cells that overexpress active HClpP but not in cells that overexpress inactive HClpP (Supplementary Fig. S4, lanes with no cisplatin addition). However, in contrast to ATP7A, ATP7B levels increased in cells treated with cisplatin and remained relatively more abundant in cells with overexpressed active HClpP (Supplementary Fig. S4). The increase in ATP7A and ATP7B in cells that overexpress HClpP is consistent with the decreased accumulation of cisplatin in those cells.
Because knockdown of HClpP made cells more sensitive to cisplatin we expected that ATP7A levels might be lower in cells treated with HCLPP-siRNA. However, we did not consistently observe lower levels of ATP7A under HClpP knockdown conditions. We conclude that overexpression of HClpP and knockdown of HClpP might not act on the same pathway to affect cisplatin sensitivity. Because higher levels of the HClpP protease led to higher levels of ATP7A it was clear that HClpP did not degrade ATP7A but more likely acted on an upstream pathway that affects expression or stability of ATP7A. To determine whether ATP7A is degraded in vivo, we added cycloheximide to growing cells to block new protein synthesis and measured the ATP7A remaining during the subsequent 24 h. There was no change in the intracellular levels of ATP7A after cycloheximide addition in control cells or in HClpP overexpressing cells (Supplementary Fig. S5), indicating that ATP7A is not subject to rapid degradation in the presence or absence of excess HClpP.
3.8 HClpP is elevated in spontaneous cisplatin-resistant cells
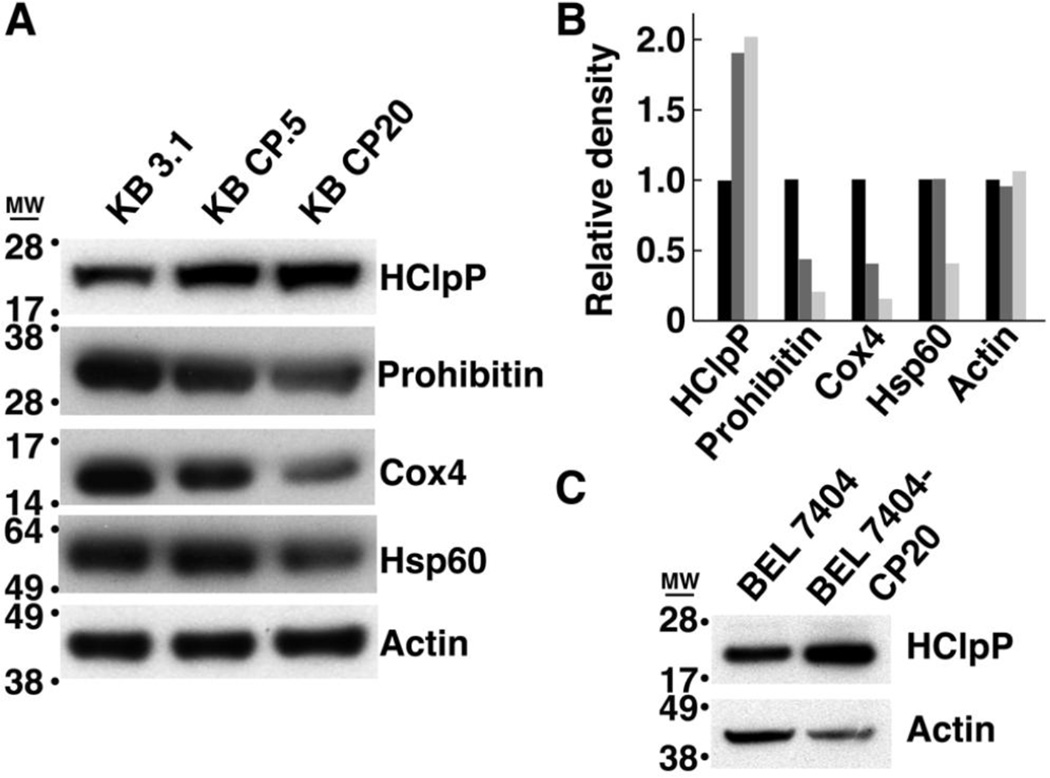
Cultured human cells selected for resistance to cisplatin have complex genotypes and display altered expression of genes associated with DNA damage repair, energy metabolism, apoptosis, and stress responses [45, 46]. Because our data showed a positive correlation between HClpP expression and cisplatin resistance, we measured the levels of HClpP in several cell lines selected for growth in either low or high levels of cisplatin. Compared to the levels in parental cell lines, HClpP levels were elevated in KB-CP0.5 cells previously selected for growth in 0.5 µg/ml cisplatin and HClpP levels were even higher in KB-CP20 cells selected for growth in 20 µg/ml cisplatin (Fig. 9A). As controls for changes in the number of mitochondria or expression of mitochondrial proteins, we also probed the same blot for Hsp60, a marker for the mitochondria matrix, Cox4, which resides in the outer mitochondrial membrane, and prohibitin, a protein that undergoes transient association with the mitochondrial membrane under stress conditions. Only HClpP was found to be higher in the cisplatin-resistant cell lines. Hsp60, Cox4, and prohibitin remained the same or were reduced in KB CP0.5 and KB CP20 cells (Fig. 9A). To further test the correlation between HClpP expression and cisplatin resistance we also examined BEL-7404 cells, another spontaneously cisplatin resistant cell line, for HClpP expression. HClpP expression was also elevated in BEL-7404 cells compared to the parental cisplatin-sensitive cell line (Fig. 9B).
Fig. 9. HClpP levels are elevated in cisplatin-resistant KB-CP cells.
A. Extracts were prepared from parental KB-3-1 cells and cisplatin-resistant KB-CP.5 and KB-CP20 cells, and aliquots (25 µg of protein) from each extract were separated by SDS-PAGE. Individual proteins were detected by western blotting with antibodies specific for mitochondrial proteins HClpP, Hsp60, Cox4, and prohibitin. The identical blot was used to probe each of the proteins to facilitate quantitative comparisons. Actin served as a protein loading control. B, The blots in panel A were scanned and analyzed by densitometry to estimate the protein levels in KB3.1 (black), KB CP0.5 (dark gray), and KB CP20 (light gray) cells. C, Protein (25 µg) from parental BEL-7404-CP0 cells and cisplatin-resistant BEL-7404-CP20 cells were separated on an SDS gel and probed by western blotting with antibodies specific for HClpP. Results of a single experiment are shown. A 2–5-fold increase in HClpP was observed in two separate experiments. Actin was used as a loading control.
3.9 Knockdown HClpP increases cisplatin sensitivity in cisplatin-resistant KB-CP20 cells
KB-20 cells grow well in DMEM containing 20 µg/ml cisplatin. To ask whether HClpP levels affected the ability of these cells to grow in the presence of cisplatin, we used siRNA to reduce the levels of HClpP, removed the siRNA, and continued growth of the cells with or without the addition of 10 or 20 µg/ml cisplatin. Cell viability was monitored by Annexin-V-PE staining. After 36 h of growth in the absence of cisplatin, 11–12% of the cells were Annexin-V positive with a mean of ~30 for Annexin-V fluorescence (Table 1). Cells treated with cisplatin showed increased Annexin-V staining and the increases were greater for cells with reduced HClpP. For cells treated with 10 µg/ml of cisplatin, HClpP depletion increased Annexin-V positive cells from 15 to 26 ± 2 % and mean fluorescence from 32 to 62 ± 4. (P < 0.01) For cells treated with 20 µg/ml of cisplatin, HClpP depletion increased Annexin-V positive cells from 17 to 28 ± 2 % and increased mean Annexin-V fluorescence from 37 to 61 ± 4 (Table 1). Thus, HClpP does not appear to directly impinge on the major pathway(s) conferring spontaneous resistance to cisplatin, but reducing HClpP in these cells shifts the sensitivity to cisplatin to a similar degree (2–4 fold) as observed in wild type cells. These results are consistent with a model in which HClpP reduces the dose of cisplatin felt by the cell.
Table 1. Knockdown of HClpP partially restores cisplatin sensitivity to KB CP20 cells.
KB CP20 cells were treated with si-HCLPP or control si-RNA for 8 h, placed in fresh medium for 16 h, after which they were incubated with 10 or 20 µg/ml cisplatin for 36 h. Apoptotic cell death was measured by flow cytometry of Annexin V-stained cells. Averages of the mean of the distribution of stained cells and the percentage of stained cells (apoptotic or dead) are reported. Significance was evaluated in Excel using the TTEST; siC vs. siP at 10 and 20 µg/ml CP): P <0.01; mean and % dead cells for 0 vs. 20 µg/ml CP): P <0.01.
| Annexin V staining | ||||
|---|---|---|---|---|
| Cisplatin Treatment* | si-Control | si-HCLPP | ||
| (µg/ml) | Mean staining |
% dead cells |
Mean staining |
% dead cells |
| 0 | 30 ± 2 | 11 ± 1 | 30 ± 2 | 12 ± 1 |
| 10 | 32 ± 2 | 15 ± 2 | 62 ± 4 | 26 ± 2 |
| 20 | 37 ± 3 | 17 ± 2 | 61 ± 4 | 28 ± 3 |
Cisplatin-treated cultures were harvested at 36 h. Cells without cisplatin were taken at the time of cisplatin addition to the other cultures.
4. Discussion
Our results establish a link between mitochondrial HClpP activity and the sensitivity of cells to cisplatin. We find that high levels of HClpP correlate with increased cisplatin resistance, whereas decreased levels of HClpP render cells more sensitive to cisplatin. Reduced levels of HClpX also confer sensitivity to cisplatin suggesting that the function reflected in the altered sensitivity to cisplatin requires the HClpXP complex. Our stable transfectants that constitutively overexpress HClpP produce 6–10-fold the endogenous levels of HClpP, but we see similar protective effects with just 2-fold higher levels obtained with the doxycycline-inducible HClpP transfectant. The continuum of responses to HClpP from partial knockdown to modest overexpression argues against the effects on cisplatin sensitivity representing an off-target activity or other artifact. Further support for this conclusion comes from the unexpected finding that three cell lines independently selected for cisplatin resistance all had elevated levels of HClpP and became more sensitive to cisplatin when the levels were reduced following knockdown with HCLPP-siRNA. Our results suggest that HClpP is limiting in cell lines grown in culture under our conditions and that HClpP plays an important role in maintenance of a pathway affecting cisplatin accumulation or metabolism in the cell.
Many mechanisms of cisplatin resistance have been described, the pathway is altered by changes in HClpP appears to involve the uptake or export of cisplatin. Total cellular accumulation of cisplatin and incorporation of cisplatin into both chromosomal and mitochondrial DNA varied inversely with the level of active HClpP, suggesting that HClpP directly or indirectly regulates the uptake or retention of cisplatin. Cells take up cisplatin by several mechanisms including import by the copper transporters, Ctr1 and Ctr2, and efflux by copper efflux pumps ATP7A and ATP7B. Mutations in many of the genes for components of the copper transport and utilization systems or changes in their expression levels have been shown to confer cisplatin resistance. We found that treatments that alter the level of active HClpP lead to changes in the level of the copper efflux pumps, ATP7A and ATP7B, but that levels of the copper transporter Ctr1 were largely unaffected. Although the changes in ATP7A we observe are rather modest, it is noteworthy that small changes in ATP7A expression in an ovarian cancer cell line were sufficient to confer >5-fold resistance to cisplatin [47]. Our finding that the cisplatin-resistant cell line BEL7404-CP20 remain responsive to changes in HClpP expression is consistent with our model that the effects of HClpP are mediated though effects on cisplatin efflux, because BEL7404-CP20 cells were reported to have reduced rates of cisplatin import but normal cisplatin efflux activity [48].
Cisplatin interacts with a multitude of cellular components, including DNA, RNA, proteins, lipids, and many small molecules. The ability of excess HClpP to reduce total cellular retention of cisplatin by as much as 70% is thus quite remarkable and suggests that elevated levels of ATP7A or ATP7B are only a part of the story. Moreover, the effect of HClpP on efflux itself is almost certainly indirect. Neither ATP7A not ATP7B is expected to be a direct substrate for HClpXP, which is localized to the mitochondrial matrix even under conditions in which it is overexpressed, whereas both pumps are distributed among the Golgi, small vesicular bodies, and the plasma membrane. Could HClpP affect other factors that control degradation of ATP7A? We observed that ATP7A was stable in HeLa cells for at least 24 h (Fig. 8D) with or without over expression of HClpP, indicating that homeostasis of ATP7A in the absence of stress or other perturbations does not occur at the level of protein turnover. However, ATP7A levels did decrease in response to cisplatin treatment, which suggests that the protein is degraded either in response to damage or through a regulatory pathway that is perturbed by cisplatin treatment. Overexpressing of HClpP affects the rate at which ATP7A declines in cisplatin-treated cells. One explanation for this effect could be that HClpP alters a regulatory response affecting ATP7A and ATP7B The copper efflux pumps are highly regulated, and their levels and/or subcellular distribution responds to changes in copper availability and utilization communicated by a plethora of copper chaperones and sensing proteins [49]. These factors and sensors are in turn regulated in response to cellular conditions, such as iron availability and redox potential, and extracellular conditions, such as nutrient sources and hormone signaling. HClpP could target a component of the copper utilization or signaling pathway or indirectly target a protein involved in redox regulation or metal ion homeostasis, both of which affect copper utilization and sensing. At least two mitochondria-associated proteins, PET191 and COX19, are known to affect the distribution of ATP7A between the endosomal vesicles and the plasma membrane [50, 51], however we did not detect any change in the levels of either of these proteins by western blotting (data not shown). An alternative explanation is that HClpP affects cisplatin uptake by a mechanism independent of the efflux pumps and that the effects on ATP7A or ATP7B are the result rather than a cause of altered cisplatin uptake. The slowed degradation of ATP7A observed when HClpP is overexpressed could reflect a specific mechanism, such as less direct damage to ATP7A and consequently slower degradation, or more general consequences, such as an reduced cellular stress response to cisplatin-induced damage, resulting in reduced levels of autophagy or other degradative pathways.
The effects of HClpP on cisplatin sensitivity should originate through its action on one or more mitochondrial proteins. No direct mitochondrial targets of ClpP have been identified, although recent data have linked loss of HClpP function to two human diseases, Perrault syndrome [24] and spastic paraplegia [52], and HClpP has been shown to undergo a progressive increase in a mouse heart model of Friedrich ataxia [53]. Interestingly, frataxin deficiency, a condition that underlies Friedrich ataxia, results in elevated HClpP and Mtcp1, a homolog of the copper-binding protein Cox23, providing another link between mitochondrial ClpP and copper sensing [54]. Human and other eukaryotic ClpPs have also been shown to play a role in protein quality control. ClpP is needed for a mitochondria-specific unfolded protein response [55], and unfolded proteins in the mitochondria up-regulate expression of ClpP [56]. In our studies, we find that down-regulation of HClpP induces Hsp60 and Lon protease, both stress-response proteins, and preliminary proteomics analysis of mitochondrial proteins show that the primary proteins affected by changes in ClpP levels are other stress response proteins (Y. Zhang, J. H. Kang and M. R. Maurizi, in preparation), suggesting that ClpP is needed to maintain protein quality control in mitochondria. Knockdown of HClpP in HEK2093T cells was reported to result in decreased oxygen consumption (A. W. Greene et al.), which implies that HClpP or HClpXP is needed for integrity of some component of the electron transport chain or ATP synthase complex. However, we saw no change in COX2 a component of the cytochrome C oxidase complex synthesized in the matrix nor in either the alpha or beta subunits of ATP synthase, the integrity of which is dependent on the activity of other mitochondrial ATP-dependent proteases (data not shown). Recently, HClpX was found to play a role in mitochondrial chromosome segregation, possibly by acting on the essential mitochondrial transcriptional regulator, TFAM [57]. Although HClpP was not required for the effects mediated by HClpX, the data suggest that HClpXP is required for stability or integrity of mitochondrial DNA, which could underlie the increased sensitivity of MtDNA to cisplatin when HClpXP levels are reduced. Mice with homozygous deletion of ClpP were found to have elevated MTDNA copy number in all tissues tested, although the fold-change varied between 4-fold and 1.5-fold depending on the tissue [25]. We find that the copy number of MtDNA is unchanged in HeLa cells with increased or decreased HClpP. One possibility is that short-term or modest decreases in HClpP affects other structural or functional properties of MtDNA. In our hands HeLa cells cannot be maintained in culture when HClpP is reduce >90%, and thus we are unable to reproduce the observations made in the knock-out mouse, which apparently has adapted to the loss of ClpP function in the mitochondria.
Because of its role in protein quality control and DNA maintenance, HClpP is needed for robust functioning of mitochondria and thus can influence many cellular functions that are affected by mitochondria. Mice with a homozygous knockout of ClpP survive but show severe loss of hearing as well as infertility [25]. ClpP thus appears to be dispensable for survival of most cells but plays a vital role in survival of specialized cells. In this light it is interesting that one of the major side effects of cisplatin chemotherapy is hearing loss, suggesting that some auditory cells are especially sensitive to cisplatin. One possibility is that such cells are more sensitive because they have reduced levels of ClpP. This question is under investigation. Conversely, HClpP can be overexpressed 5–25 times the endogenous levels without adverse effect; moreover, overexpression of HClpP conveys resistance to cisplatin. An implication of this finding is that endogenous levels of HClpP in cultured HeLa cells are suboptimal. These results are consistent with a dual role for HClpP as both a stress sensor and as an enzyme needed for a robust stress response in mitochondria. Thus it is possible that HClpP amplifies the response to mitochondrial protein damage caused by cisplatin. Because cisplatin has a broad range of targets and HClpP is likely to act in multiple pathways the effects of both will be pleiotropic. The pleiotropic effects of changing HClpP expression made it difficult to obtain consistent results when ATP7A levels were measured following knockdown of HCLPP. We cannot rule out that the increased sensitivity caused by HClpP depletion and the reduced sensitivity caused by HClpP overexpression operate by different mechanisms. Additional studies will be needed to identify in vivo targets of HClpXP, which should provide insight regarding the essential functions of HClpXP and the mechanism(s) by which the direct targets of HClpXP affect cisplatin sensitivity.
Supplementary Material
Highlights.
Acquired resistance to cisplatin is a major obstacle to effective treatment of many cancers.
Knockdown of HClpP and HClpX in human cervical carcinoma cells sensitizes them to cisplatin.
Formation of cisplatin-mitochondrial DNA adducts is correlated with the levels of active HClpP.
Targeting HClpP or HClpX will sensitize cancer cells to lower doses of cisplatin and might reduce the development of resistance.
Acknowledgments
This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD. The authors would like to thank Dr. Duck Yeon Lee, National Heart, Lung, and Blood Institute, Bethesda, MD and Dr. Kent Warnken of Thermo Fisher Scientific, Lanham, MD for, respectively, atomic absorption and ICP mass spectrometric measurements of platinum.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat. Rev. Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 2.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 3.Magnaghi P, D'Alessio R, Valsasina B, Avanzi N, Rizzi S, Asa D, Gasparri F, Cozzi L, Cucchi U, Orrenius C, Polucci P, Ballinari D, Perrera C, Leone A, Cervi G, Casale E, Xiao Y, Wong C, Anderson DJ, Galvani A, Donati D, O'Brien T, Jackson PK, Isacchi A. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat. Chem. Biol. 2013;9:548–556. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- 4.Auner HW, Moody AM, Ward TH, Kraus M, Milan E, May P, Chaidos A, Driessen C, Cenci S, Dazzi F, Rahemtulla A, Apperley JF, Karadimitris A, Dillon N. Combined inhibition of p97 and the proteasome causes lethal disruption of the secretory apparatus in multiple myeloma cells. PLoS One. 2013;8:e74415. doi: 10.1371/journal.pone.0074415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou TF, Li K, Nordin BE, Porubsky P, Frankowski K, Patricelli MP, Aube J, Schoenen FJ, Deshaies R. Selective, reversible inhibitors of the AAA ATPase p97. 2010 [PubMed] [Google Scholar]
- 6.Wang N, Gottesman S, Willingham MC, Gottesman MM, Maurizi MR. A human mitochondrial ATP-dependent protease that is highly homologous to bacterial Lon protease. Proc. Natl. Acad. Sci. USA. 1993;90:11247–11251. doi: 10.1073/pnas.90.23.11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, Filla A, Cocozza S, Marconi R, Durr A, Fontaine B, Ballabio A. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell. 1998;93:973–983. doi: 10.1016/s0092-8674(00)81203-9. [DOI] [PubMed] [Google Scholar]
- 8.Banfi S, Bassi MT, Andolfi G, Marchitiello A, Zanotta S, Ballabio A, Casari G, Franco B. Identification and characterization of AFG3L2, a novel paraplegin- related gene. Genomics. 1999;59:51–58. doi: 10.1006/geno.1999.5818. [DOI] [PubMed] [Google Scholar]
- 9.Corydon TJ, Bross P, Holst HU. A human homologue of Escherichia coli ClpP caseinolytic protease: recombinant expression, intracellular processing and subcellular localization. Biochem. J. 1998;331:309–316. doi: 10.1042/bj3310309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corydon TJ, Wilsbech M, Jespersgaard C, Andresen BS, Borglum AD, Pedersen S, Bolund L, Gregersen N, Bross P. Human and mouse mitochondrial orthologs of bacterial ClpX. Mamm. Genome. 2000;11:899–905. doi: 10.1007/s003350010173. [DOI] [PubMed] [Google Scholar]
- 11.Santagata S, Bhattacharyya D, Wang FH, Singha N, Hodtsev A, Spanopoulou E. Molecular cloning and characterization of a mouse homolog of bacterial ClpX, a novel mammalian class II member of the Hsp100/Clp chaperone family. J. Biol. Chem. 1999;274:16311–16319. doi: 10.1074/jbc.274.23.16311. [DOI] [PubMed] [Google Scholar]
- 12.Kang SG, Ortega J, Singh SK, Wang N, Huang NN, Steven AC, Maurizi MR. Functional proteolytic complexes of the human mitochondrial ATP-dependent protease, hClpXP. J. Biol. Chem. 2002;277:21095–21102. doi: 10.1074/jbc.M201642200. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman S, Maurizi MR. Regulation by Proteolysis: Energy-Dependent Proteases and Their Targets. Microbiol. Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottesman S, Maurizi MR, Wickner S. Regulatory subunits of energy-dependent proteases. Cell. 1997;91:435–438. doi: 10.1016/s0092-8674(00)80428-6. [DOI] [PubMed] [Google Scholar]
- 15.Gottesman S. Proteolysis in Bacterial Regulatory Circuits. Annu. Rev. Cell Dev. Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 17.Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, Neher SB, Oakes ES, Siddiqui SM, Wah DA, Baker TA. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venkatesh S, Lee J, Singh K, Lee I, Suzuki CK. Multitasking in the mitochondrion by the ATP-dependent Lon protease. Biochim. Biophys. Acta. 2012;1823:56–66. doi: 10.1016/j.bbamcr.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsushima Y, Kaguni LS. Matrix proteases in mitochondrial DNA function. Biochim. Biophys. Acta. 2012;1819:1080–1087. doi: 10.1016/j.bbagrm.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell. 2005;123:277–289. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Rugarli EI, Langer T. Translating m-AAA protease function in mitochondria to hereditary spastic paraplegia. Trends Mol. Med. 2006;12:262–269. doi: 10.1016/j.molmed.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Ugarte N, Petropoulos I, Friguet B. Oxidized Mitochondrial Protein Degradation and Repair in Aging and Oxidative Stress. Antioxid. Redox Signal. 2010;13:539–549. doi: 10.1089/ars.2009.2998. [DOI] [PubMed] [Google Scholar]
- 23.Anand R, Langer T, Baker MJ. Proteolytic control of mitochondrial function and morphogenesis. Biochim. Biophys. Acta. 2013;1833:195–204. doi: 10.1016/j.bbamcr.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 24.Jenkinson EM, Rehman AU, Walsh T, Clayton-Smith J, Lee K, Morell RJ, Drummond MC, Khan SN, Naeem MA, Rauf B, Billington N, Schultz JM, Urquhart JE, Lee MK, Berry A, Hanley NA, Mehta S, Cilliers D, Clayton PE, Kingston H, Smith MJ, Warner TT, Black GC, Trump D, Davis JR, Ahmad W, Leal SM, Riazuddin S, King MC, Friedman TB, Newman WG. Perrault syndrome is caused by recessive mutations in CLPP, encoding a mitochondrial ATP-dependent chambered protease. Am. J. Hum. Genet. 2013;92:605–613. doi: 10.1016/j.ajhg.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gispert S, Parganlija D, Klinkenberg M, Drose S, Wittig I, Mittelbronn M, Grzmil P, Koob S, Hamann A, Walter M, Buchel F, Adler T, Hrabe de Angelis M, Busch DH, Zell A, Reichert AS, Brandt U, Osiewacz HD, Jendrach M, Auburger G. Loss of mitochondrial peptidase Clpp leads to infertility, hearing loss plus growth retardation via accumulation of CLPX, mtDNA and inflammatory factors. Hum. Mol. Genet. 2013;22:4871–4887. doi: 10.1093/hmg/ddt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang SG, Dimitrova MN, Ortega J, Ginsburg A, Maurizi MR. Human mitochondrial ClpP is a stable heptamer that assembles into a tetradecamer in the presence of ClpX. J. Biol. Chem. 2005;280:35424–35432. doi: 10.1074/jbc.M507240200. [DOI] [PubMed] [Google Scholar]
- 27.Kang SG, Maurizi MR, Thompson M, Mueser T, Ahvazi B. Crystallography and mutagenesis point to an essential role for the N-terminus of human mitochondrial ClpP. J. Struct. Biol. 2004;148:338–352. doi: 10.1016/j.jsb.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Hall MD, Okabe M, Shen DW, Liang XJ, Gottesman MM. The role of cellular accumulation in determining sensitivity to platinum-based chemotherapy. Annu. Rev. Pharmacol. Toxicol. 2008;48:495–535. doi: 10.1146/annurev.pharmtox.48.080907.180426. [DOI] [PubMed] [Google Scholar]
- 29.Sharma RP, Edwards IR. cis-Platinum: subcellular distribution and binding to cytosolic ligands. Biochem. Pharmacol. 1983;32:2665–2669. doi: 10.1016/0006-2952(83)90073-4. [DOI] [PubMed] [Google Scholar]
- 30.Tacka KA, Dabrowiak JC, Goodisman J, Penefsky HS, Souid AK. Effects of cisplatin on mitochondrial function in Jurkat cells. Chem. Res. Toxicol. 2004;17:1102–1111. doi: 10.1021/tx0499564. [DOI] [PubMed] [Google Scholar]
- 31.Murata T, Hibasami H, Maekawa S, Tagawa T, Nakashima K. Preferential binding of cisplatin to mitochondrial DNA and suppression of ATP generation in human malignant melanoma cells. Biochem. Int. 1990;20:949–955. [PubMed] [Google Scholar]
- 32.Yadav N, Chandra D. Mitochondrial and postmitochondrial survival signaling in cancer. Mitochondrion. 2014;16C:18–25. doi: 10.1016/j.mito.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin DH, Choi YJ, Park JW. SIRT1 and AMPK mediate hypoxia-induced resistance of non-small cell lung cancers to cisplatin and doxorubicin. Cancer Res. 2014;74:298–308. doi: 10.1158/0008-5472.CAN-13-2620. [DOI] [PubMed] [Google Scholar]
- 34.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 35.Koh DW, Dawson TM, Dawson VL. Mediation of cell death by poly(ADP-ribose) polymerase-1. Pharmacol. Res. 2005;52:5–14. doi: 10.1016/j.phrs.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 36.Cepeda V, Fuertes MA, Castilla J, Alonso C, Quevedo C, Perez JM. Biochemical mechanisms of cisplatin cytotoxicity. Anticancer Agents Med. Chem. 2007;7:3–18. doi: 10.2174/187152007779314044. [DOI] [PubMed] [Google Scholar]
- 37.Susin SA, Daugas E, Ravagnan L, Samejima K, Zamzami N, Loeffler M, Costantini P, Ferri KF, Irinopoulou T, Prevost MC, Brothers G, Mak TW, Penninger J, Earnshaw WC, Kroemer G. Two distinct pathways leading to nuclear apoptosis. J. Exp. Med. 2000;192:571–580. doi: 10.1084/jem.192.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorenzo HK, Susin SA, Penninger J, Kroemer G. Apoptosis inducing factor (AIF): a phylogenetically old, caspase-independent effector of cell death. Cell Death Differ. 1999;6:516–524. doi: 10.1038/sj.cdd.4400527. [DOI] [PubMed] [Google Scholar]
- 39.Cande C, Vahsen N, Garrido C, Kroemer G. Apoptosis-inducing factor (AIF): caspase-independent after all. Cell Death Differ. 2004;11:591–595. doi: 10.1038/sj.cdd.4401400. [DOI] [PubMed] [Google Scholar]
- 40.Holford J, Sharp SY, Murrer BA, Abrams M, Kelland LR. In vitro circumvention of cisplatin resistance by the novel sterically hindered platinum complex AMD473. Br. J. Cancer. 1998;77:366–373. doi: 10.1038/bjc.1998.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang XJ, Shen DW, Gottesman MM. A pleiotropic defect reducing drug accumulation in cisplatin-resistant cells. J. Inorg. Biochem. 2004;98:1599–1606. doi: 10.1016/j.jinorgbio.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 42.Katano K, Kondo A, Safaei R, Holzer A, Samimi G, Mishima M, Kuo YM, Rochdi M, Howell SB. Acquisition of resistance to cisplatin is accompanied by changes in the cellular pharmacology of copper. Cancer Res. 2002;62:6559–6565. [PubMed] [Google Scholar]
- 43.Safaei R, Katano K, Samimi G, Naerdemann W, Stevenson JL, Rochdi M, Howell SB. Cross-resistance to cisplatin in cells with acquired resistance to copper. Cancer Chemother. Pharmacol. 2004;53:239–246. doi: 10.1007/s00280-003-0736-3. [DOI] [PubMed] [Google Scholar]
- 44.Holzer AK, Katano K, Klomp LW, Howell SB. Cisplatin rapidly down-regulates its own influx transporter hCTR1 in cultured human ovarian cancer cells. Clin. Can. Res. 2004;10:6744–6749. doi: 10.1158/1078-0432.CCR-04-0748. [DOI] [PubMed] [Google Scholar]
- 45.Gottesman MM. Mechanisms of cancer drug resistance. Annu. Rev. Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 46.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 47.Samimi G, Safaei R, Katano K, Holzer AK, Rochdi M, Tomioka M, Goodman M, Howell SB. Increased expression of the copper efflux transporter ATP7A mediates resistance to cisplatin, carboplatin, and oxaliplatin in ovarian cancer cells. Clin. Cancer Res. 2004;10:4661–4669. doi: 10.1158/1078-0432.CCR-04-0137. [DOI] [PubMed] [Google Scholar]
- 48.Johnson SW, Shen D, Pastan I, Gottesman MM, Hamilton TC. Cross-resistance, cisplatin accumulation, and platinum-DNA adduct formation and removal in cisplatin-sensitive and -resistant human hepatoma cell lines. Exp. Cell Res. 1996;226:133–139. doi: 10.1006/excr.1996.0211. [DOI] [PubMed] [Google Scholar]
- 49.Hasan NM, Lutsenko S. Regulation of copper transporters in human cells. Curr. Top. Membr. 2012;69:137–161. doi: 10.1016/B978-0-12-394390-3.00006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khalimonchuk O, Rigby K, Bestwick M, Pierrel F, Cobine PA, Winge DR. Pet191 is a cytochrome c oxidase assembly factor in Saccharomyces cerevisiae. Eukaryot. Cell. 2008;7:1427–1431. doi: 10.1128/EC.00132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leary SC. Redox regulation of SCO protein function: controlling copper at a mitochondrial crossroad. Antioxid. Redox Signal. 2010;13:1403–1416. doi: 10.1089/ars.2010.3116. [DOI] [PubMed] [Google Scholar]
- 52.Hansen J, Corydon TJ, Palmfeldt J, Durr A, Fontaine B, Nielsen MN, Christensen JH, Gregersen N, Bross P. Decreased expression of the mitochondrial matrix proteases Lon and ClpP in cells from a patient with hereditary spastic paraplegia (SPG13) Neuroscience. 2008;153:474–482. doi: 10.1016/j.neuroscience.2008.01.070. [DOI] [PubMed] [Google Scholar]
- 53.Guillon B, Bulteau AL, Wattenhofer-Donze M, Schmucker S, Friguet B, Puccio H, Drapier JC, Bouton C. Frataxin deficiency causes upregulation of mitochondrial Lon and ClpP proteases and severe loss of mitochondrial Fe-S proteins. FEBS J. 2009;276:1036–1047. doi: 10.1111/j.1742-4658.2008.06847.x. [DOI] [PubMed] [Google Scholar]
- 54.Schoenfeld RA, Napoli E, Wong A, Zhan S, Reutenauer L, Morin D, Buckpitt AR, Taroni F, Lonnerdal B, Ristow M, Puccio H, Cortopassi GA. Frataxin deficiency alters heme pathway transcripts and decreases mitochondrial heme metabolites in mammalian cells. Hum. Mol. Genet. 2005;14:3787–3799. doi: 10.1093/hmg/ddi393. [DOI] [PubMed] [Google Scholar]
- 55.Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol. Cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kasashima K, Sumitani M, Endo H. Maintenance of mitochondrial genome distribution by mitochondrial AAA+ protein ClpX. Exp. Cell Res. 2012;318:2335–2343. doi: 10.1016/j.yexcr.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 58.Shen DW, Akiyama S, Schoenlein P, Pastan I, Gottesman MM. Characterisation of high-level cisplatin-resistant cell lines established from a human hepatoma cell line and human KB adenocarcinoma cells: cross-resistance and protein changes. Br. J. Cancer. 1995;71:676–683. doi: 10.1038/bjc.1995.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.