Abstract
The highly conserved Notch-signaling pathway mediates cell-to-cell communication and is pivotal for multiple developmental processes and tissue homeostasis in adult organisms. Notch receptors and their ligands are transmembrane proteins with multiple epidermal-growth-factor-like (EGF) repeats in their extracellular domains. In vitro the EGF repeats of mammalian ligands that are essential for Notch activation have been defined. However, in vivo the significance of the structural integrity of each EGF repeat in the ligand ectodomain for ligand function is still unclear. Here, we analyzed the mouse Notch ligand DLL1. We expressed DLL1 proteins with mutations disrupting disulfide bridges in each individual EGF repeat from single-copy transgenes in the HPRT locus of embryonic stem cells. In Notch transactivation assays all mutations impinged on DLL1 function and affected both NOTCH1 and NOTCH2 receptors similarly. An allelic series in mice that carried the same point mutations in endogenous Dll1, generated using a mini-gene strategy, showed that early developmental processes depending on DLL1-mediated NOTCH activation were differently sensitive to mutation of individual EGF repeats in DLL1. Notably, some mutations affected only somite patterning and resulted in vertebral column defects resembling spondylocostal dysostosis. In conclusion, the structural integrity of each individual EGF repeat in the extracellular domain of DLL1 is necessary for full DLL1 activity, and certain mutations in Dll1 might contribute to spondylocostal dysostosis in humans.
Keywords: Notch signaling, Notch-ligand interaction, Notch activation, mouse DLL1, targeted mutagenesis/allelic series
COMMUNICATION between adjacent cells mediated by the evolutionary conserved Notch-signaling pathway regulates multiple developmental processes in different tissues and species (reviewed in Andersson et al. 2011). Notch receptors and their ligands encode type 1 transmembrane proteins with multiple EGF-like repeats in their extracellular domains. The ligands contain an additional conserved extracellular cysteine-rich so-called DSL domain and a disulfide-bond stabilized module at the N terminus called MNNL (reviewed in Chillakuri et al. 2012). Mammalian genomes encode four Notch receptors, two Delta (DLL1 and DLL4), two Serrate-type ligands [called Jagged (JAG) 1 and 2] that activate Notch, and the untypical ligand DLL3 that can interact with, but does not activate, Notch (Ladi et al. 2005; Geffers et al. 2007; Chapman et al. 2011). Mutations in Notch pathway components underlie human diseases such as Alagille syndrome (ALGS) (mutations in JAG1 and NOTCH2) and CADASIL syndrome (mutations in NOTCH3) or spondylocostal dysostosis (mutations in DLL3, HES7, LFNG, and MESP2) (reviewed in Penton et al. 2012).
EGF repeats 11 and 12 constitute the major ligand binding site of Notch receptors, although additional repeats are essential for full Notch activation and function in vitro and in vivo (Shimizu et al. 1999; Hambleton et al. 2004; Xu et al. 2005; Cordle et al. 2008b; Andrawes et al. 2013; Luca et al. 2015). The MNNL and DSL domains and EGF2 of Drosophila Delta were shown to be required for Notch activation in vivo (Parks et al. 2006). In binding assays, the DSL domain of mouse Jag1 was essential for binding to mouse Notch2, and the presence of EGF1 and 2 enhanced this interaction (Shimizu et al. 1999). Likewise, a fragment of human Jag1 encompassing the DSL domain and first three EGF repeats was shown to bind to fragments of human Notch1 encompassing EGF repeats 10–13 (Cordle et al. 2008a), and deletion analyses of human DLL1 and DLL4 showed that the regions containing the MNNL to EGF3 were necessary and sufficient for full activation of Notch1 (Andrawes et al. 2013). Recently, the structure of a complex of EGF repeats 11–13 of Notch1 with the N-terminal portion of DLL4 up to and including EGF2 was published, showing that EGF repeats 11 and 12 of Notch interact with the DSL and MNNL domains of the ligand, respectively, placing EGF1 and -2 outside the essential Notch interaction surface (Luca et al. 2015).
While the first three EGF repeats of Notch ligands appear to be important for activation of Notch, the significance of other EGF repeats in the extracellular domains of ligands is less clear. Parks et al. (2006) identified cysteine missense mutations in EGF repeats 4 and 9 in Drosophila Delta that affect Notch signaling in some contexts, but were associated with aberrant subcellular localization and trafficking. Similarly, missense mutations in EGF repeats distant from the DSL domain in JAG1 of ALGS patients caused intracellular retention of the mutant protein (Morrissette et al. 2001; Bauer et al. 2010), preventing firm conclusions on how the region C-terminal to the interaction domain contributes to ligand function. Here, we focus on mouse DLL1, which has eight EGF-like repeats in its extracellular domain (Bettenhausen et al. 1995). To address the significance of all EGF repeats for DLL1 function, we disrupted the same two disulfide bridges individually in each EGF repeat. We introduced constructs expressing these protein variants as single-copy transgenes into the HPRT locus of embryonic stem (ES) cells and analyzed DLL1-mediated Notch activation in cell-based transactivation assays in vitro. In vivo we generated an allelic series introducing the same mutations into the endogenous Dll1 gene and analyzed somitogenesis, myogenesis, neurogenesis, and establishment of left–right asymmetry, developmental processes known to require Dll1 function. Our analyses show that disrupting disulfide bridges in any EGF repeat impairs ligand activity and reveals context-dependent different sensitivity of developmental processes to reduced DLL1-mediated Notch signaling, anterior–posterior patterning of somites being most sensitive. Mutations in Dll1 that specifically affected somitogenesis showed vertebral column malformations resembling spondylocostal dysostosis of varying severity, a human condition known to be caused by abnormal Notch signaling during somitogenesis (reviewed in Penton et al. 2012), but not yet associated with mutations in Dll1.
Materials and Methods
Ethics statement
Animal experiments were performed according to the German rules and regulations (German Animal Welfare Act Tierschutzgesetz) and approved by the ethics committee of Lower Saxony for care and use of laboratory animals (Lower Saxony State Office for Consumer Protection and Food Safety). Mice were housed in the central animal facility of Hannover Medical School Zentrales Tierlaboratorium (ZTL) and were maintained as approved by the responsible Veterinary Officer of the City of Hannover. Animal welfare was supervised and approved by the Institutional Animal Welfare Officer (Tierschutzbeauftragter).
Site-directed mutagenesis of EGF repeats
A 1.1-kb NotI/NdeI fragment of the Dll1 complementary DNA (cDNA) coding for the DSL domain and all eight EGF repeats was subcloned into pGem5zf. The fourth and fifth cysteine codons (TGT or TGC) of each individual EGF repeat were changed to glycine codons (GGT and GGC, respectively) using the Quick Change mutagenesis kit (Stratagene) according to the manufacturer’s instructions and the following primer pairs: EGF1 (CAAACCAGGGGAGGGCAAGGGCAGAGTTGGCTGG) and EGF1_R (CCAGCCAACTCTGCCCTTGCCCTCCCCTGGTTTG); EGF2 (CAGCAACCCTGGCAGGGTAACGGCCAGGAAGGC) and EGF2_R (GCCTTCCTGGCCGTTACCCTGCCAGGGTTGCTG); EGF3 (GGGGAGCTACACAGGTTCCGGCCGACCTGGG) and EGF3_R (CCCAGGTCGGCCGGAACCTGTGTAGCTCCCC); EGF4 (GGACAGCTTCTCTGGCACCGGCCCTCCCGGC) and EGF4_R (GCCGGGAGGGCCGGTGCCAGAGAAGCTGTCC); EGF5 (CGGAGGCTACACCGGCCATGGCCCCTTGGGC) and EGF5_R (GCCCAAGGGGCCATGGCCGGTGTAGCCTCCG); EGF6 (GCAACTCTTACCTGGGCCGGGGCCAGGCTGGC) and EGF6_R (GCCAGCCTGGCCCCGGCCCAGGTAAGAGTTGC); EGF7 (GAACGACTTCTCCGGTACCGGCCCACCTGGC) and EGF7_R (GCCAGGTGGGCCGGTACCGGAGAAGTCGTTC); and EGF8 (GCCAGCGCTACATGGGTGAGGGCGCCCAGGGCTATG) and EGF8_R (CATAGCCCTGGGCGCCCTCACCCATGTAGCGCTGGC).
All cDNA fragments were verified by sequencing.
Expression vectors
To generate expression constructs for DLL1 variants, the NotI/NdeI fragment of the wild-type cDNA in pTRACER was replaced with a mutated NotI/NdeI fragment generated by site-directed mutagenesis. For protein purification, the extracellular domains (ECDs) of DLL1 WT and EGF4m were fused to the Fc fragment of human IgG1 as the EcoRI/HindIII fragment in pCMV5. The plasmids were introduced in pTracerCMV as EcoRI/XbaI fragments. From a Notch1 cDNA containing the complete ORF the bases encoding the C-terminal 56 amino acids (PEST domain) were deleted and a C-terminal Flag tag was added using a fragment synthesized in vitro (MWG/Operon) and conventional cloning. The modified Notch1 cDNA (NOTCH1ΔC-Flag) was cloned into pcDNA3, resulting in pcDNA3-NOTCH1ΔC-Flag.
Constructs for introduction of Dll1 cDNAs into the HPRT locus
Flag-tagged Dll1 wild-type and mutant cDNAs were cloned as EcoRI/BamHI fragments into a shuttle vector containing MluI and SwaI sites and cloned into pMP8.CAG-stop (Singh et al. 2012) using these sites. The stop cassette was excised by Cre-mediated recombination in SW106 bacteria, bringing the cDNA expression under the control of the CAG promoter.
Construct for introduction of a Notch reporter into the HPRT locus
Four copies of a synthetic DNA fragment containing paired RBP-binding sites (TGAAAGTTACTGTGGGAAAGAAAGTTTGGGAAGTTTCACACGAGCCGTTCGCGTGCAGTCCCAGATATATATAGAGGCCGCCAGGGCCTGCGGATCACACAGGATCTGGAGCTGGTG) were cloned into pGa981-6 (Minoguchi et al. 1997), in front of the β-globin minimal promotor followed by the firefly luciferase gene and the SV40 polyadenylation signal generating RBP4xluc. The RBP4xluc cassette was cloned into a modified version of the HPRT-targeting vector pMP8 (referred to as pMP8-RBP4xluc) (Bronson et al. 1996; Alten et al. 2012).
Constructs for introduction of Notch1 or Notch2 and a Notch reporter into the HPRT locus
Notch1ΔC-Flag or Notch2-Flag cDNA linked to SV40pA and driven by the CAG promoter were introduced into pMP8-RBP4xluc. The chicken β-globin insulator (kind gift of Bernhard Herrmann) was cloned between the Notch cDNAs and RBP4xluc, resulting in H-Notch1ΔC-luc and H-Notch2-luc. Equal orientation of the insulator (3′-5′) was ascertained by PCR using primer pairs CGGATCTGATCAGCACGTGTTGAC and TCCTTTGCAACCCAGGCGTTC and CCACTGCAGCACCGCTCTTTG and GTTTAAACGAATTCGCCCTTATGTCG.
Constructs for introducing EGF mutations into the Dll1 locus
The strategy to modify endogenous Dll1 uses a Dll1 mini-gene build from a 2.2-kb EcoRI/NdeI cDNA fragment (encompassing exons 1 through part of exon 9) that was fused with a 1.6-kb genomic NdeI/EcoRI fragment that includes the remaining part of exon 9, intron 10, exon 10, intron 11, and exon 11 including the endogenous poly(A) signals. The 5′ homology region is a genomic 3.7-kb SalI (site derived from a phage vector)/EcoRI fragment. Three prime to the mini-gene the neo gene flanked by loxP sites (a 2-kb EcoRI/SalI fragment from pPNT lox2) was added in inverse orientation, followed by the 3′ homology region (a genomic 2.9-kb SalI/EcoRI fragment, in which NotI and NsiI sites were destroyed). On both sides a 1.1-kb Diphtheria toxin A cassette taken from pKO SelectDT was included. To generate the targeting vectors for the individual EGF repeat mutations, the NotI/NdeI fragment of the wild-type mini-gene was replaced with the fragments generated by site-directed mutagenesis. All vector constructs were generated by standard cloning techniques and were verified by sequencing.
ES cell culture
Embryonic stem cells were cultured in DMEM, supplemented with penicillin/streptomycin, glutamax, sodium pyruvat, essential amino acids, (Invitrogen), 15% fetal calf serum (Biochrom), β-mercaptoethanol, and leukemia inhibitory factor.
ES cells carrying Dll1 transgenes in the Hprt locus
Dll1 Hprt vectors were linearized with FseI and electroporated into HPRT-deficient E14tg2a ES cells (Hooper et al. 1987) that were re-derived from hybrid 129Sv/CD1 E14tg2a mice in our laboratory. Cells were selected with HAT in a concentration of 1:300 (Gibco). E14tg2a ES cells carry a deletion at the Hprt locus, allowing for efficient selection of single-copy transgene insertions into this locus using a targeting vector that restores HAT resistance (Hooper et al. 1987; Bronson et al. 1996; Redeker et al. 2013). Correct integration of HAT-resistant clones was verified by long-range PCR using the primers HPRT typ 5′ F3 GAT GGA CAA GGC CCT AAC TAG GTG AAC TG and HPRT typ 5′ F2 GGG AAC CTG TTA GAA AAA AAG AAA CTA TGA AGA AC and CAG rev GGC TAT GAA CTA ATG ACC CCG. Expression of DLL1 wild-type and mutant proteins was confirmed by Western blot analysis using anti-Flag-POD (SigmaA8592) diluted 1:4000. These cells are referred to as H-Dll1flag, H-Dll1EGF1mutflag, and H-Dll1EGF2mutflag, etc.
ES cells carrying RBP4xluc in the Hprt locus
pMP8-RBP4xluc was linearized with SacII, electroporated into E14Tg2a/CD1 ES cells, and selected as described before. HAT-resistant clones were verified for correct integration using the following primers: TGA GTG GGG GGG TTG ATA ATC TTG G and GTT TAA ACG AAT TCG CCC TTA TGT CG. These ES cells are referred to as H-RBPluc.
ES cells carrying Notch receptors and RBP4xluc in the Hprt locus
H-Notch1ΔC-RBPluc and H-Notch2-RBPluc constructs were linearized with SalI and electroporated into E14Tg2a/CD1 ES cells and selected as described before. Correctly targeted ES cells were identified by PCR using the primers described for ligand integrations. Expression of Notch1 and Notch2 was confirmed by Western blot analysis using anti-Flag-POD (SigmaA8592) diluted 1:4000. ES cells carrying these constructs are referred to as H-Notch1ΔCFlag-RBPluc and H-Notch2Flag-RBPluc.
H-RBPluc ES cells carrying randomly inserted Notch1ΔC
pcDNA3-N1ΔC-Flag was linearized with PvuI and electroporated into H-RBPluc ES cells. Clones carrying stable integrations were selected using 150 µg/ml G418. Notch1-expressing clones were identified by Western blot analysis using anti-Flag-POD (SigmaA8592) diluted 1:4000. ES cells are referred to as “Notch1ΔC-Flag/H-RBPluc.”
ES cells carrying targeted mutations in Dll1
Constructs for introducing mutations in individual EGF repeats were linearized with NsiI and electroporated into ES cells. ES cells were screened by long-range PCR amplifying a fragment spanning the 3′ homology region using primers EGF 3′#1 TGTCACGTCCTGCACGACG and EGF 3′#2 GGTATCGGATGCACTCATCGC and the Roche Expand High Fidelity PCR System. Homologous recombination was confirmed by Southern blot analyses. Probes were the following: a 5′ 320-bp AvaII/BamHI fragment derived from a PCR-amplified genomic fragment obtained with primers melta 121 GCGGAAAATGGACAGAAGGG and melta 122 AATGGGTGGATAGGGCAGACTC; 3′ a 500-bp genomic PCR fragment amplified with primers melta 124 CCTGTGAGACTTTCTACGTTGCTC and melta 125 CACAACCATGTCACCTTCTAGATTC cloned into pGemT Easy. These probes detect a 10-kb wild-type fragment. After homologous recombination the 5′ probe detected an 8-kb fragment and the 3′ probe a 6.5-kb fragment. Independently targeted ES cell clones obtained with each construct were used for chimera production.
Analysis of protein expression
Cell lysates were analyzed by Western blotting using anti-Flag antibodies. Expression levels were analyzed using ImageJ (Schneider et al. 2012) and compared using β-tubulin (detected by anti-tubulin antibodies; Sigma T7816 1:250000) as a reference. Chinese hamster ovary (CHO) cells transfected with expression vectors for DLL1 variants were analyzed by immunofluorescence using anti-Flag (M2, Sigma, 1:5000) antibodies and Alexa 488-coupled secondary antibodies (Invitrogen). Expression of ECD-Fc fusion proteins in stably transfected CHO cells was verified by Western blotting using an antibody against Fc (Dianova 209-005-088 1:1000) and as secondary antibody anti-mouse HRPOD (GE Healthcare NA031V 1:7000). For analysis under nonreducing conditions cell lysates were separated by PAGE in sample buffer without β-mercaptoethanol and probes were not boiled.
Purification of Fc fusion proteins
Six 150-mm dishes of CHO cells stably expressing DLL1-Fc fusion proteins were grown to confluency in medium containing fetal calf serum (FCS). Cells were extensively washed with PBS and DMEM/F12 and were grown for another 5 days in FCS free medium (ZAP, Invitria). Supernatants were briefly centrifuged to remove cell debris, concentrated with Pierce concentrators 20 ml/20 K (#89887; 4000 × g, 30 min), and incubated overnight with Sepharose G beads (GE Healthcare #17-0618-01) in the presence of protease inhibitors (Complete EDTA-free Roche #04693132001). Beads were washed several times with 50 mM sodium phosphate buffer, pH 7, containing 500 mM NaCl, 0.5% Triton, and protease inhibitor, followed by washes with 50 mM sodium phosphate buffer, pH 7, without Triton and Proteaseinhibitor. Protein was eluted with 0.1 M glycin/HCl, pH 2.0, and buffered with 0.5 M sodium phosphate, pH 8. Quality of the purified proteins was assessed by silver staining of 7.5% SDS-PAGE gels following standard procedures.
Circular dichroism
Circular dichroism (CD) spectra of Fc-fusion constructs of the entire extracellular DLL1 domain were measured at 25° in a buffer containing sodium phosphate (pH 8) and 0.1 M glycine. Melting temperatures of the constructs were determined by monitoring the temperature-dependent changes of ellipticity at 218 nm using a temperature-controlled π*-180 spectrometer equipped with a circular dichroism unit (Applied Photophysics, Leatherhead, UK), and a temperature gradient of 1°⋅min-1.
Surface biotinylation
Surface biotinylation and analysis of ES cells expressing DLL1 variants from the Hprt locus were performed essentially as described (Braune et al. 2014). Proteins were detected using anti-Flag antibodies (M2, Sigma, diluted 1:4000) and quantified using ImageJ (Schneider et al. 2012). Surface presentation was calculated as percentage of precipitated protein to the calculated total in the input. The surface presentation of DLL1 wild type was set to 1, and the surface presentations of DLL1 EGF mutants were normalized to the surface presentation of DLL1 wild-type protein.
Notch transactivation assay
To analyze Notch activation by DLL1EGF mutant proteins, Notch1ΔC-Flag/H-RBPluc or H-Notch2Flag-RBPluc ES cells were cocultered for 48 hr with ES cells expressing no, wild-type, or EGF mutant DLL1 proteins at a ratio of 1:12.5 in gelatinized 30-mm dishes (total cell number 1 × 106). Cells were harvested in 1× Cell Culture Lysis Reagent (Promega), and luciferase activity was measured using Luciferase Assay Reagent II (LARII Promega) in a TurnerBioSystems luminometer with Glomax software. Every lysate was measured three times. The activation potential of H-DLL1wtFlag ES cells compared to ES cells without DLL1 was set to 1, and the activation levels of the H-DLL1EGF mutants were normalized to the activation obtained by coculturing H-Dll1wt with Notch1deltaPest Flag/H-RBPluc ES cells or HPRT-Notch2Flag-insulator 3′-5′RBPluc ES cells. Proliferation of ES cells expressing DLL1 variants over the duration of the coculture was determined by seeding 1 × 106 cells and counting cells after 48 hr (three independent experiments, each experiment counted three times).
Generation of mice carrying targeted mutations in Dll1
Chimeric mice were generated as described (Alten et al. 2012).
Removal of the neo cassette
Germ-line chimeras were crossed to Zp3::Cre mice (de Vries et al. 2000) that had been backcrossed for 10 generations to 129SV/ImJ to excise the neo cassette in the female germ line. Dll1ki/+;ZP3::Cre double-heterozygous females were genotyped by PCR using the primers Cre1 TGATGAGGTTCGCAAGAACC and Cre2 CCATGAGTGAACGAACCTGG for Cre and the primers melta 38 ATCCCTGGGTCTTTGAAGAAG and melta 132 GGTTTTCTG TTGCGAGGTCATC for the Dll1ki alleles. Excision of the neo cassette in offspring of Dll1ki/+;ZP3-Cre females crossed to wild-type 129Sv/ImJ males was verified by PCR using primers EGF-neo FOR ATGGACAGCATTTCCTCCTGCCTC and EGF-neo REV GCCAGTCAGTTCCCAGTAAGAAGTC and Southern blot analysis with a 3′ probe. The presence of individual EGF mutations in each mouse line was reconfirmed by cloning and sequencing a 1423-bp genomic PCR fragment including the NotI/NdeI EGF cassette using the primers EGF-clone_FOR GCAACAGAAAACCCAGAAAGACTC and EGF-neo_REV GCCAGTCAGTTCCCAGTAAGAAGTC.
Mouse husbandry
Initially, all transgenic mouse lines were kept on the 129SV/ImJ genetic background (Dll1Dll1Ki, Dll1EGF5m, Dll1EGF6m, and Dll1EGF7m mice as homozygotes). Due to increasingly deteriorating breeding performance precluding the efficient collection of sufficient numbers of embryos, all lines were outcrossed to CD1 after the initial gross characterization and kept on a mixed 129Sv/ImJ/CD1 genetic background for further analyses.
Genotyping of mice and embryos
PCR typing was performed using genomic DNA isolated from tail biopsies or embryonic yolk sacs, respectively, using the allele-specific primer pairs listed in Table 1.
Table 1. Allele-specific primers used for genotyping.
| Repeat | Primer | Sequence |
|---|---|---|
| EGF1 | EGF1_mut_F1 | CCTGCACCTACGGCAGTGCTGTCACG |
| EGF1_mut_R3 | CAGCCAACTCTGCCCTTGCC | |
| EGF2 | EGF2-3_F1 | CAACCCCATCCGATTCCCC |
| EGF2_mut_R1 | CAGCCTTCCTGGCCGTTACC | |
| EGF3 | EGF3_mut_F1 | GACCACACAGAGGCACCTCACTGTG |
| EGF3_mut_R2 | ACCCAGGTCGGCCGGAACC | |
| EGF4 | EGF4_mut_F1 | TCTGCCGACCTCGGGATGACGCC |
| EGF4_mut_R1 | AAGCCGGGAGGGCCGGTGCC | |
| EGF5 | EGF5-6_F1 | CTGTCTGCCAGGGTGTGATGACCAAC |
| EGF5_mut_R1 | AGCCCAAGGGGCCATGGCC | |
| EGF6 | EGF5-6_F1 | CTGTCTGCCAGGGTGTGATGACCAAC |
| EGF6_mut_R1 | CCAGCCTGGCCCCGGCC | |
| EGF7 | EGF7_F1 | GGAGCCACCTGCACCAACACG |
| EGF7_mut_R2 | GCCAGGTGGGCCGGTACC | |
| EGF8 | EGF8_F1 | CCTAGCCCCTGCAAGAACGGAGC |
| EGF8_mut_R2 | CCC GGGCGCCCTCACC |
Whole-mount in situ hybridization
Whole-mount in situ hybridizations were done by standard procedures. Mutant and wild-type embryos were analyzed in parallel with a given probe under identical conditions. cDNAs for the generation of probes were originally obtained from M. Gessler (HeyL), M. Goulding (NeuroD, Neurogenin), A. Kispert (Uncx4.1, Tbx18, Pitx2, Tbx5), J. Rossant (Nodal), and T. Braun (Myogenin, MyoD). Probes were labeled with anti-digoxigen AP (Roche), and embryos were stained using BM purple. Skeletons of mouse fetuses were stained following standard procedures (Cordes et al. 2004). Pictures were taken with a MD628 microscope with a DFC 420 camera utilizing the Firecam software v3.4.1 (all Leica). All photos were processed equally using Adobe Photoshop.
Immunohistochemistry
Specimens were fixed in Bouin’s fixative for 24 hr, embedded in paraffin, and sectioned at 8 µM. Antibodies used were My32 (SigmaM4276) and NeuN (Millipore MAB377) at a dilution of 1:400. For NeuN, the MOM Kit from Vector Laboratories was used. Mouse biotinylated secondary antibody and the ABC Kit from Vector Laboratories were applied to sections incubated with My32. Detection of the signal was achieved with the DAB Kit (Vector Laboratories). Pictures were taken using a Leica DM5000B microscope and processed using Adobe Photoshop.
Statistical analyses
Statistical analyses were performed using Prism software (GraphPad). ImageJ quantifications and luciferase measurements were analyzed by one-way ANOVA, and compared using Bonferroni’s Multiple Comparison Test with a significance level of 0.05. Expression levels of NOTCH1 and NOTCH2 were analyzed using the Student’s t-test.
Data and reagent availability
Cell lines, constructs, and strains are available upon request to the corresponding author.
Results
Generation and characterization of cell lines expressing DLL1 variants
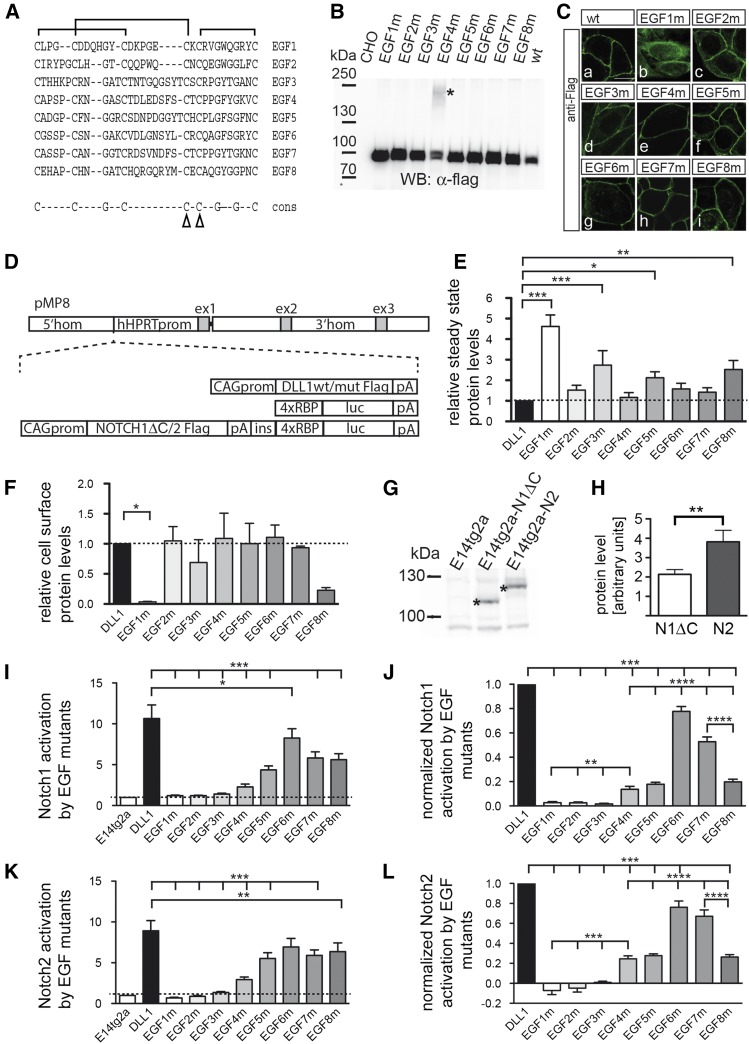
Since an EGF-like domain is thought to be an independently folding module (Downing et al. 1996), we reasoned that, by disrupting disulphide bridges in individual EGF repeats, the intrinsic structure of neighboring repeats should be only mildly affected, if at all (Suk et al. 2004; Mor-Cohen et al. 2012), and thus allows one to study the relevance of the structural integrity of individual EGF repeats in the context of full-length DLL1. We decided to exchange cysteine residues 4 and 5 of each individual EGF repeat by glycine, an amino acid that displays intrinsically high flexibility, to disrupt two disulfide bridges of each repeat (Figure 1A). To test how these mutations affect DLL1 function in cultured cells, we generated expression vectors for Flag-tagged DLL1 proteins carrying the Cys-Gly mutations in individual EGF repeats (from hereon referred to as “EGF#m”). Transiently transfected CHO cells expressed all mutant proteins, and all but EGF1m were predominantly detected at the cell membrane (Figure 1, B and C). For better comparability, we expressed the Flag-tagged DLL1 proteins from single-copy insertions into the Hprt locus of E14tg2a ES cells and used these cells for further analyses (Figure 1D and Materials and Methods). To obtain the first hints of whether the EGF mutants differ with respect to global structural disruptions or the formation of aberrant disulfide bridges, we compared their migration patterns during electrophoreses under nonreducing conditions. All mutant proteins except EGF1m showed a migration pattern similar to wild-type DLL1 (Supporting Information, Figure S1A), suggesting that these mutant proteins do not form mixed disulfides with other proteins during folding and transport to the cell surface. In addition, CD spectra of Fc-fusion proteins of the ECD of wild type or EGF4m (chosen as an example) were indistinguishable from each other (data not shown). Measuring the corresponding melting curves of these proteins revealed no significant differences in the melting temperatures, suggesting that no massive changes in the overall protein stability occur due to the introduction of the mutations (Figure S1, B and C). Quantification showed higher steady-state levels of EGF3m, EGF5m, EGF8m, and particularly EGF1m, compared to DLL1 wild type (P > 0.05; Figure 1E and Table S1). Surface biotinylation and quantification showed that all mutant proteins except EGF1m were present on the cell surface at similar relative levels (Figure 1F and Table S2). EGF8m consistently showed lower relative cell-surface levels that were, however, statistically not significant (P > 0.05 in one-way ANOVA).
Figure 1.
Analyses of EGF repeat mutant DLL1 proteins in cultured cells in vitro. (A) Alignment of amino acid sequences of DLL1 EGF repeats. The characteristic disulfide bridges are indicated by brackets above the sequence; arrowheads in the consensus sequence point to the mutated cysteine residues. (B) Western blot analysis of cell lysates of CHO cells transfected with expression vectors for DLL1 proteins as indicated at the top. CHO cells expressed all mutant proteins at the expected molecular weight. In the case of EGF4m, an additional slower-migrating protein species was observed (asterisk). This high-molecular-weight species was not shifted to a lower molecular weight by treatment with reducing agents (DTT and iodoacetamide) and thus is unlikely to be caused by aberrant disulfide bridges. When the extracellular domain of EGF4m was expressed as a soluble Fc fusion, only a protein of the expected size was observed (data not shown), suggesting that the intracellular domain or localization at the cell membrane leads to an as-yet-unknown modification of some EGF4m protein. For quantification, both protein species were taken into account. (C) Detection of DLL1 proteins in CHO cells by immunofluorescence. All DLL1 variants except EGF1m are at the cell surface. (D) Schematic representation of constructs used to generate single-copy transgene insertions in the Hprt locus. (E) Quantification of DLL1 proteins in E14tg2a ES cells expressing DLL1 variants from the Hprt locus (mean values and SEM; n = 4; Table S1). (F) Quantification of relative cell-surface levels of DLL1 variants normalized to DLL1 wild type (mean values and SEM; n = 4; Table S2). (G) Western blot analysis of cell lysates of E14tg2a H-RBPluc ES cells expressing NOTCH1ΔC-Flag from a random insertion (E14tg2a-N1) and NOTCH2-Flag from the Hprt locus (E14tg2a-N2). Asterisks indicate the S1 cleavage products of NOTCH1ΔC and NOTCH2. (H) Quantification of NOTCH1ΔC-Flag and NOTCH2-Flag stably expressed in E14tg2a H-RBPluc ES cells (mean values and SEM; n = 7; Table S3). (I) Activation of NOTCH1ΔC by DLL1 variants in coculture assays (mean values and SEM; n = 10; Table S4). (J) Normalized activation of NOTCH1ΔC by DLL1 variants in coculture assays. Values of EGF3m, -5m, and -8m were normalized to protein expression levels relative to DLL1 wild type (Table S5). (K) Activation of NOTCH2 by DLL1 variants in coculture assays (mean values and SEM; n = 9; Table S4). (L) Normalized activation of NOTCH2 by DLL1 variants in coculture assays. Values of EGF3m, -5m, and -8m were normalized to protein expression levels relative to DLL1 wild type (Table S5). ns = P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
We also generated ES cells that carry the RBP4xluc Notch reporter gene (Serth et al. 2015) in the Hprt locus of E14tg2a ES (referred to as H-RBPluc) cells. In addition, either NOTCH1 or NOTCH2 expression cassettes were introduced into these cells upstream of the reporter (Figure 1C) separated by the chicken β-globin insulator sequence (Vidigal et al. 2010). Since in our hands NOTCH1 is hard to detect by Western blot analysis, we deleted the C-terminal 56 amino acids composing the PEST domain (referred to as “NOTCH1ΔC”). For unknown reasons, only NOTCH2 expressed in the Hprt locus of ES cells (referred to as “H-Notch2Flag-RBPluc cells”) could be activated by DLL1 in coculture assays. Therefore, we introduced a Notch1ΔC expression construct randomly into E14tg2a H-RBPluc cells (referred to as Notch1ΔCFlag H-RBPluc cells). From NOTCH1ΔC-expressing clones, we selected the one that most closely matched NOTCH2 expression levels and (based on the S1-cleavage products) reached ∼56% of NOTCH2 levels (Figure 1, G and H, and Table S3).
Analysis of Notch activation by DLL1 variants
Notch1ΔCFlag H-RBPluc and H-Notch2Flag-RBPluc ES cells were cocultured with ES cells expressing wild-type or mutant DLL1. DLL1wt activated NOTCH1ΔC on average ∼11-fold and NOTCH2 ∼9-fold (Figure 1, I and K, and Table S4). As expected, EGF1m did not activate NOTCH1ΔC or NOTCH2 since it does not reach the cell surface and served as additional negative control. Likewise, EGF2m and EGF3m did not activate either receptor (Figure 1, I and K). All other DLL1 mutants activated NOTCH1ΔC and NOTCH2 although with different efficiencies ranging from ∼20 to 80% (Figure 1, I and K, un-normalized data). To compare the activity of EGF mutant proteins to DLL1wt, we also normalized their activity (setting DLL1 activity to 1; Figure 1, J and L) and corrected for expression levels [for EGF3m, EGF5m, and EGF8m, which showed significant (P > 0.05) differences in their steady-state levels compared to wild type; no correction was made for EGF1m because it was not present on the cell surface]. Both receptors showed a highly similar profile of activation by the EGF mutants. EGF4m, EGF5m, and EGF8m showed ∼15–20% activity of DLL1wt, EGF6m activated NOTCH1ΔC and NOTCH2 to ∼80% of wild-type levels, and EGF7 reached between 50 and 60% of activity (Figure 1, J and L, and Table S5). To exclude that different proliferation rates and thus different numbers of ES cells expressing DLL1 variants affect the results of the transactivation assays, we counted the number of ES cells after 48 hr of culture and did not find significant differences (Figure S1E). Thus, based on the in vitro Notch activation assays, disruption of the structural integrity of EGF repeats 2–8 impinges on DLL1-mediated Notch activation, and activation of both NOTCH1 and NOTCH2 is similarly affected. The significance of EGF1 for DLL1 ligand function cannot be assessed due to the intracellular retention of the mutated protein.
Generation and analysis of an allelic series of Dll1 mutations in mice
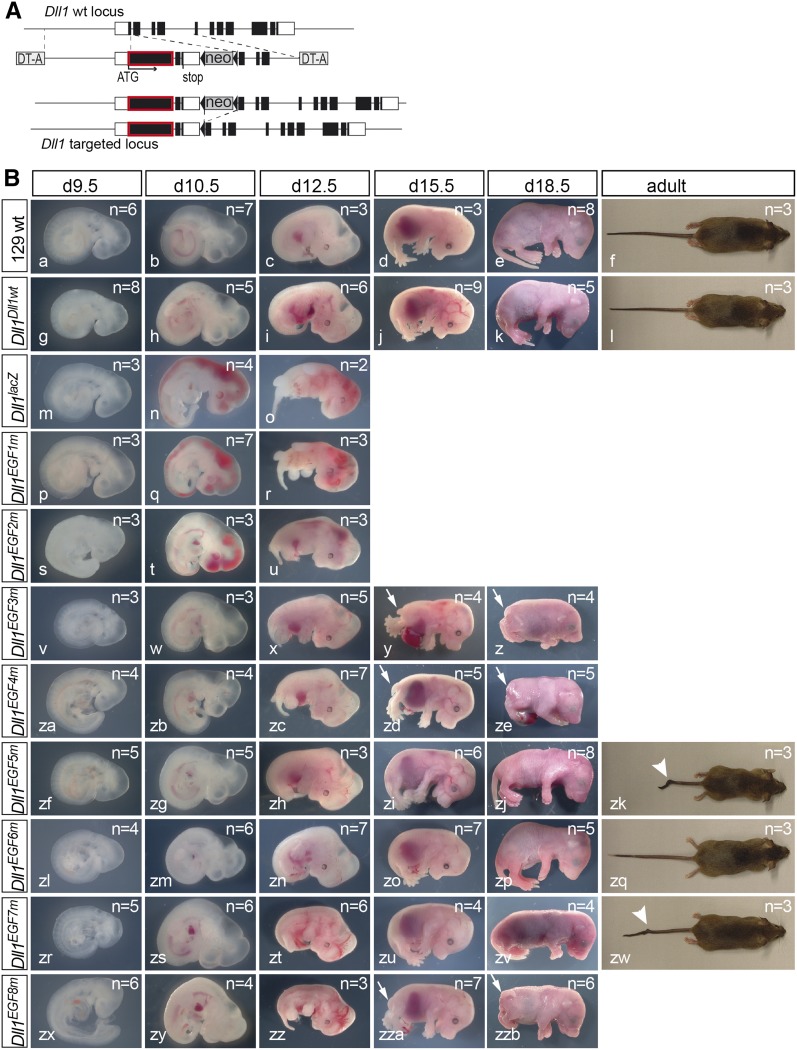
To assess the contribution of EGF2-8 to full DLL1 function under physiological conditions, we generated an allelic series of these Dll1 mutations in mice based on the knock-in of a Delta1 mini-gene (Figure 2A and Figure S1F). All heterozygous EGF repeat mutants were normal, indicating that one wild-type allele of Dll1 is sufficient for normal development and that the mutant proteins do not exert obvious dominant-negative effects on wild-type DLL1. Initially, homozygous mutants were analyzed on the isogenic 129Sv/ImJ genetic background. The Dll1Dll1wt allele carrying the Dll1 wild-type mini-gene was indistinguishable from wild-type mice (Figure 2B, a–f and g–l), indicating that the mini-gene compensates the disrupted endogenous gene. As previously described, mice lacking DLL1 (Dll1lacZ) were hemorrhagic from embryonic day 10.5 (E10.5) on and died around E12.5 (Figure 2B, m–o). Essentially the same phenotype was observed with Dll1EGF1m (Figure 2B, p–r) and Dll1EGF2m (Figure 2B, s–u) embryos. Dll1EGF3m embryos displayed a milder phenotype: embryos developed to E18.5, had only mild hemorrhages and a severely reduced body axis (Figure 2B, v–z), and were motionless. Dll1EGF4m (Figure 2B, za–ze) and Dll1EGF8m (Figure 2B, zx–zzb) embryos displayed a shortened body axis and did not survive after birth. Dll1EGF5m and Dll1EGF7m mutants were viable and fertile with shortened kinky tails, Dll1EGF5m being more strongly affected than Dll1EGF7m (Figure 2B, zf–zk and zr–zw). Dll1EGF6m embryos were indistinguishable from wild-type and Dll1Dll1wt, viable, and fertile (Figure 2B, zl–zq). Thus, the structural integrity of all but EGF repeat 6 appears to be important for DLL1 as a sufficiently active Notch ligand in vivo. For further comparative analyses of early developmental processes known to require DLL1, all alleles were outcrossed for three generations and analyzed on a mixed genetic background because the poor breeding performance of isogenic 129Sv/ImJ mice precluded the efficient collection of embryos. Mutants showed a virtually identical range of phenotypes on the mixed and the isogenic background (Figure S1H).
Figure 2.
Generation of an allelic series of EGF mutations in DLL1 and external phenotypes. (A) Structure of Dll1 before and after homologous recombination and of the targeting vector. Black boxes indicate coding and white boxes noncoding regions of Dll1. The cDNA portion is outlined in red. (B) Morphology of wild-type 129Sv/ImJ embryos and mice and of isogenic embryos and mice homozygous for the Dll1 alleles. Alleles are indicated at the left, developmental stages at the top. Arrows in y, z, zd, ze, zza and zzb point to shortened tails; arrowheads in zk and zw point to tail kinks.
Somite patterning
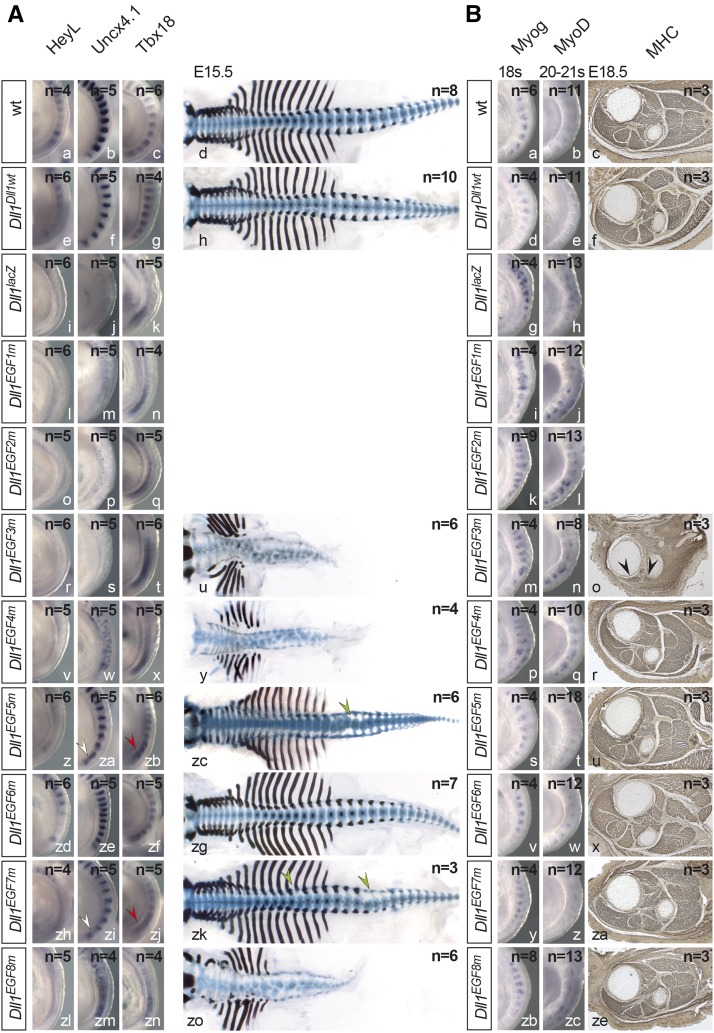
DLL1 signaling is instrumental for establishment of anterior–posterior somite polarity and subsequent axial skeleton development (Hrabe de Angelis et al. 1997; Cordes et al. 2004). To compare the impact of EGF repeat mutations on somitogenesis, we analyzed expression of the Notch target HeyL (Leimeister et al. 2000), and Uncx4.1 and Tbx18, markers for posterior and anterior somite compartments, respectively, in E9.5 embryos (Neidhardt et al. 1997; Kraus et al. 2001). The expression patterns of these genes as well as the axial skeletons of homozygous Dll1Dll1wt (Figure 3A, e–h) and Dll1EGF6m (Figure 3A, zd–zg) embryos were indistinguishable from wild type (Figure 3A, a–d). Dll1EGF1m, Dll1EGF2m, and Dll1EGF3m embryos (Figure 3A, l–t) resembled embryos lacking DLL1 (Figure 3A, i–k), and Dll1EGF3m E15.5 skeletons showed massive defects (Figure 3A, u). Dll1EGF4m and Dll1EGF8m mutants displayed somewhat milder A–P polarity defects than Dll1EGF3m, Dll1EGF4m being more severely affected than Dll1EGF8m (Figure 3A, v–x and zl–zn). Skeletal preparations revealed misshapen vertebrae and ribs similar to Dll1EGF3m mutants (Figure 3A, y and zo). Dll1EGF5m (Figure 3A, zc) and Dll1EGF7m (Figure 3A, zk) mutants displayed mild skeletal defects, which might underlie the slightly reduced breeding performance of these mutant lines (Figure S1G). Although HeyL expression was clearly reduced (Figure 3A, z and zh), Uncx4.1 and Tbx18 showed only minor irregularities (arrowheads in Figure 3A, za, zb, and zi, zj).
Figure 3.
Somite patterning and skeletal muscle development in mutants homozygous for individual Dll1 EGF alleles. (A) Whole-mount in situ hybridizations (WISH) of E9.5 and skeletal preparations of E15.5 embryos. Alleles are indicated at the left, probes at the top. White and red arrowheads point to irregularities of Uncx4.1 and Tbx18 expression patterns, respectively; green arrowheads point to malformed vertebrae. (B) Muscle differentiation in mutants homozygous for individual EGF alleles. WISH of 18 and 20–21 somite-stage embryos and anti-MHC antibody staining of hind-limb sections of E18.5 embryos. Alleles are indicated at the left, probes/antibodies at the top. Arrowheads in o point to remnants of skeletal muscles. For My32 staining, we analyzed three individual embryos with a minimum of six consecutive sections per genotype.
Myogenesis
Loss of DLL1 leads to premature differentiation of myoblasts and severely reduced skeletal muscles (Schuster-Gossler et al. 2007). To compare the mutations in this context, we analyzed expression of Myog and MyoD, two regulators of myogenesis (Arnold and Braun 2000), in age-matched somite-stage (ss) 18 and 20–21 embryos and stained skeletal muscles in cross sections of hind limbs at similar proximo-distal positions for myosin heavy chain (MHC). In wild-type 18 ss embryos, Myog was expressed in the anterior 7–8 somites, and at ss 20–21 faint MyoD expression was detected (Figure 3B, a and b). Dll1Dll1wt, Dll1EGF5m, Dll1EGF6m, and Dll1EGF7m embryos expressed Myog and MyoD virtually identical to wild-type embryos (Figure 3B, d, e, s, t, v, w, y, z) and had apparently normal skeletal muscles at E18.5 (compare Figure 3B, c, and Figure 3B, f, u, x, za). In Dll1 knockout embryos, Myog and MyoD expression was upregulated (Figure 3B, g and h), which was similarly observed in Dll1EGF1m, Dll1EGF2m, and Dll1EGF3m embryos (Figure 3B, i–n). Hind limbs of motionless Dll1EGF3m mutants lacked skeletal muscles with the exception of a few MHC-positive remnants (arrowheads in Figure 3B, o) resembling the phenotype of embryos that are heteroallelic for the Dll1 null and a hypomorphic allele (Schuster-Gossler et al. 2007). Myog and MyoD expression in Dll1EGF4m and Dll1EGF8m embryos appeared slightly upregulated (Figure 3B, p, q and zb, zc); however, in cross sections through the hind limbs skeletal muscles were indistinguishable from wild type (Figure 3B, r, ze).
Neurogenesis
DLL1-mediated Notch activation represses neuronal differentiation (de la Pompa et al. 1997; Rocha et al. 2009). To compare the EGF-repeat mutations in this context, we analyzed expression of Neurog1 and NeuroD, a neuronal determination and differentiation gene, respectively, in 23 ss embryos (Ma et al. 1996). To analyze whether the EGF mutations lead to obvious overall alterations of the architecture of the central nervous system at later stages, we stained cross sections of the cervical spinal cord of E18.5 embryos for NeuN, a neuron-specific nuclear protein (Mullen et al. 1992) that labels all neurons. Like wild type, Dll1Dll1wt and Dll1EGF6m embryos showed normal expression of Neurog1 and NeuroD in the brain and dorsal neural tube and in cranial and spinal ganglia (Figure 4A, a, b, d, e, v, w) and an indistinguishable cytoarchitecture of the cervical spinal cord (Figure 4A, c, f, x). In Dll1EGF1m, Dll1EGF2m, and Dll1EGF3m embryos, Neurog1 and NeuroD expression was upregulated in the spinal cord, and premature expression domains were detected in the nasal placodes (white and red arrowheads, respectively, in Figure 4A, i–n). Neurog1 was additionally upregulated in the mid- and forebrain (yellow and black arrowheads, respectively, in Figure 4A, i, k, m), resembling Dll1 knockout embryos (Figure 4A, g and h). Unexpectedly, despite the clearly enhanced neuronal differentiation in early embryos, the spinal cords of homozygous Dll1EGF3m embryos appeared enlarged rather than reduced in size, and cellularity was increased (Figure 4A, o). This contrasts with the reduction of neural tube size at E12.5 that was observed in embryos with cell type-specific deletion of Dll1 in neural tube progenitors (Rocha et al. 2009). The development of this phenotype is unclear at present and will require a detailed analysis in the future. A total of 23 ss Dll1EGF4m and Dll1EGF8m embryos displayed weak premature expression of Neurog and NeuroD in the nasal placodes (red arrow heads in Figure 4A, p, q, zb, zc). However, their spinal cords appeared indistinguishable from wild type (Figure 4A, r, zd).
Figure 4.
Neuronal differentiation and left–right asymmetry in mutants homozygous for individual EGF alleles. (A) WISH of E9.5 and antibody staining of spinal cord sections of E18.5 embryos. Alleles are indicated at the left, analyzed markers at the top. Arrowheads point to regions of upregulated gene expression in the nasal placode (red arrowhead), midbrain (yellow arrowhead), forebrain (black arrowhead), and spinal cord (white arrowhead). For NeuN, we analyzed three individual embryos with a minimum of three and a maximum of nine consecutive sections per genotype. (B) WISH of six ss (Nodal, dorsal views) E8.5 (Pitx2, dorsal views) and E10.5 (Tbx5, ventral views) embryos. Alleles are indicated at the left, probes at the top. For genotyping of Dll1EGF4m (s) and Dll1EGF8m (ze) 6 ss embryos, the posterior halves of embryos were removed prior to hybridization to Nodal. Black arrowheads in (b, e, t, w, z, zc, zf) point to Pitx2 expression in the left LPM. “lv” indicates the Tbx5-positive ventricle normally positioned on the left side. The numbers of analyzed embryos are summarized in Table S6 and Table S7.
Left–right asymmetry
DLL1-mediated Notch activation is essential for establishment of left–right asymmetry (Krebs et al. 2003). We analyzed expression of nodal in E8.0, Pitx2 in E8.5 embryos, and heart looping at E10.5 in embryos hybridized to a Tbx5 probe that shows strong expression in the left ventricle (Bruneau et al. 1999). In wild-type embryos, nodal is first expressed asymmetrically around the node followed by expression in the left lateral plate mesoderm (LPM) (Figure 4B, a) and (Lowe et al. 1996). Subsequently, Pitx2 expression is activated in the LPM (Figure 4B b), and (Yoshioka et al. 1998), and at E10.5 rightward looping of the heart indicates correct left–right determination (Figure 4B, c). Normal expression of nodal and Pitx2 and consecutive normal heart looping was observed in all Dll1Dll1ki, Dll1EGF4m, Dll1EGF5m, Dll1EGF6m, Dll1EGF7m, and Dll1EGF8m embryos (Figure 4B, d, e, f, s–zg; Table S6; Table S7). As described in embryos lacking Dll1 (Krebs et al. 2003), nodal expression was not initiated, Pitx2 expression was missing in the LPM or randomized, and heart looping was abnormal (Figure 4B, g, h, i). About 25% of Dll1EGF1m, Dll1EGF2m, and Dll1EGF3m embryos obtained from matings between heterozygous mutants showed no nodal expression even after prolonged staining, absent or randomized Pitx2 expression in the LPM, and randomization of heart looping (Figure 4B, j–r; Table S6; Table S7).
Discussion
To obtain insights into the significance of each EGF repeat in the ectodomain of the Notch ligand DLL1 for its function, we disrupted two disulfide bridges in each repeat individually by substituting cysteine with glycine residues. These mutations should disrupt and destabilize the domain structure of the respective repeat as has been observed for cysteine substitutions in EGF repeats of other proteins (Suk et al. 2004; Mor-Cohen et al. 2012) and should increase the intrinsic flexibility of the mutated repeats. Analyses of single cysteine substitutions (disruption of a single disulfide bridge) in fibrillin showed only localized structural effects but did not exclude the possibility of different effects if other disulfide bridges were disrupted (Suk et al. 2004). In addition, although we disrupted the same disulfide bridges in each EGF repeat, the severity of the disruption of the individual domain structure might vary between individual mutated EGF repeats due to their different amino acid compositions, which could contribute to different reductions in DLL1 function. Nonetheless, our mutations reveal the consequences of disrupting the structural integrity of individual EGF repeats in the context of the full ligand protein.
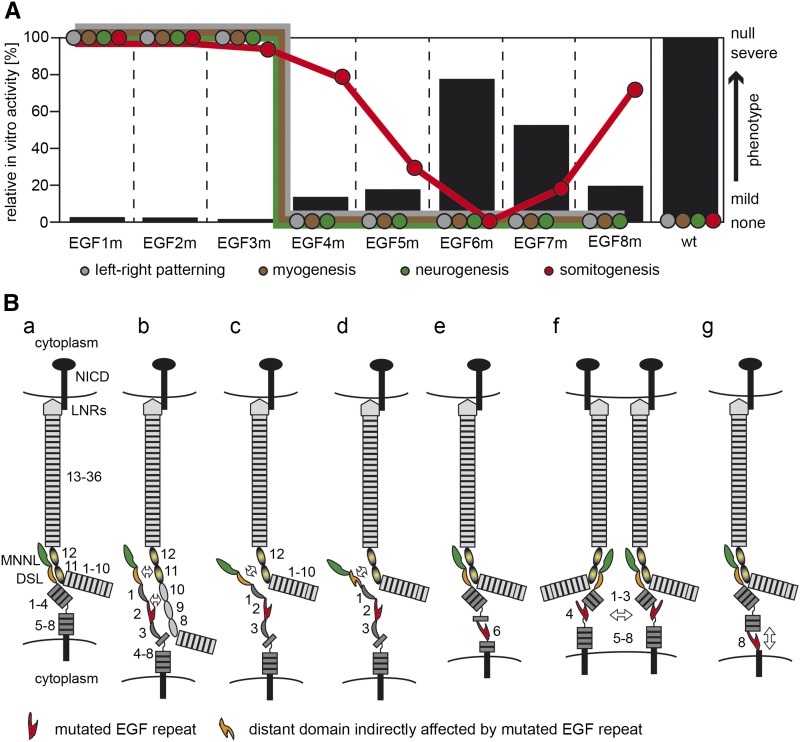
To analyze how our EGF mutations affect DLL1 ligand function, we first tested the ability of mutant proteins to activate the Notch1 and Notch2 receptors in cell-based transactivation assays and observed very similar activity patterns ranging from complete absence of ligand activity to levels up to 80% of the wild-type level (Figure 5A). This suggests that DLL1 interacts with both Notch receptors very similarly, at least under the conditions of our transactivation assays. Generating the same individual EGF mutations in endogenous DLL1, we established an allelic series in mice in vivo and observed similar effects of our EGF repeat mutations on DLL1 ligand activity: mutations in EGF repeats close to the MNNL-DSL binding interface resulted in null or strong hypomorphic mutations, mutations more distant affected ligand activity successively less up to EGF6, and mutations close to the membrane interfered with DLL1 function again more strongly (summarized in Figure 5A).
Figure 5.
Schematic summary of results and potential effects of EGF repeat mutations on DLL1. (A) Schematic representation of DLL1 activity in transactivation assays in vitro (black bars; scale on left y-axis) and severity of mutant phenotypes (colored circles; arbitrary scale on right y-axis). The position of colored circles indicates the severity of the mutant phenotype with respect to left–right patterning, myogenesis, neurogenesis, and somitogenesis. Above a certain threshold corresponding to <20% DLL1 activity measured in in vitro, left–right patterning, myogenesis, and neurogenesis proceed apparently normal. In contrast, somitogenesis shows a graded response to reduced DL1 activity. (B) Model depicting potential effects of EGF repeat mutations on DLL1 function. (a) Interaction between MNNL (green) and DSL (orange) domains of DLL1 (dark gray) with EGF 11 and 12 (graded yellow/black ovals) of Notch (light gray) based on Luca et al. (2015). The potential kink between EGF4 and 5 of DLL1 (Kershaw et al. 2015) is indicated by a curved line. (b) Mutation of EGF2 in DLL1 might disrupt the direct interaction of EGF2 with Notch outside the Notch MNNL/DSL interface and destabilize DLL1–Notch interaction. (c) Alternatively, mutation of EGF2 in DLL1 might disrupt the linear arrangement of the DSL and adjacent EGF domains and thereby interfere with efficient binding. (d) Propagation of the disruption of an EGF repeat (example shown for EGF2) might affect the structure of neighboring domains and interaction with Notch. (e) Mutation of EGF6 might be tolerated due to its proximity to a naturally occurring bent between EGF4 and 5. (f) Mutation in EGF repeats (example shown for EGF4) might affect clustering of DLL1 and thereby reduce effective Notch activation. (g) Mutated EGF repeats (example shown for EGF8) with intrinsically increased flexibility might weaken the pulling force that is assumed to be required for Notch activation. Numbers refer to EGF repeats.
As the consequences of our mutations may extend beyond the mutated EGF repeat, their impact on DLL1 activity may be explained by interference local and/or distant DLL1–Notch interactions. Mutations in EGF2 and EGF3 completely abolished activation of Notch1 and Notch2 in vitro. In mice, EGF2m behaved as a null allele indistinguishable from Dll1lacZ or Dll1EGF1m whereas EGF3m behaved like a severe hypomorph (Figure 5A). These findings are consistent with results of previous in vitro studies using purified ligand protein fragments showing that the presence of EGF2 and EGF3 in these fragments is essential for effective binding to, and activation of, Notch (Shimizu et al. 1999; Andrawes et al. 2013). Since the MNNL and the DSL domain of DLL4 (and most likely also of DLL1) are in direct contact with the ligand binding interface of the receptor (EGF 11 and 12) (Luca et al. 2015), EGF2 and EGF3 could contribute to the direct interaction of ligand and receptor by binding to Notch adjacent to EGF 11 (Figure 5B, b), which is consistent with the observation that additional EGF repeats outside the major ligand binding site of Notch receptors are essential for full Notch activation and function (Shimizu et al. 1999; Hambleton et al. 2004; Xu et al. 2005; Cordle et al. 2008b; Andrawes et al. 2013). Such an interaction could not be observed by Luca et al. (2015) since EGF repeats of Notch1 potentially involved in this interaction were not included in their study. Additionally, the linear arrangement of the DSL, EGF1, and EGF2 domains, which was detected in the crystal structure of Notch1 EGF 11–13 with the DLL4 N-terminal domains (Luca et al. 2015), or in the uncomplexed structures of DLL1 and JAG1 (Cordle et al. 2008a; Kershaw et al. 2015), might be destabilized by EGF2m and EGF3m, resulting in ineffective interaction of the MNNL/DSL domains with EGF 11 and 12 of Notch (Figure 5B, c). Furthermore, it is possible that mutations in EGF repeats close to the MNNL and DSL domains impinge on the structure of these domains and thereby affect receptor ligand interaction (Figure 5B, d). Likewise, the mutation of EGF4 (or EGF5) might spread beyond the repeat boundary and thereby perturb the likely interaction of EGF3 (and EGF2) with Notch. If the effects of our EGF mutations spread to adjacent domains, their impact on the DLL1–Notch binding interface and DLL1 activity can be expected to decrease with increasing distance from the binding interface. This possibility might be reflected by the apparent correlation between decreasing phenotypic strength and distance of our mutations from the binding interface up to EGF 6.
In our transactivation assays, NOTCH1 and NOTCH2 were activated by EGF6m at ∼80% of wild-type levels, and in vivo this allele was indistinguishable from wild type in all analyzed processes (Figure 5A). Eighty percent of DLL1 activity should be sufficient for normal development, since mice that are heterozygous for the Dll1 null allele, and presumably have 50% of DLL1 activity, are normal. EGF6 is found to be located next to a bent potentially present in DLL1 (Kershaw et al. 2015), which might explain why DLL1 tolerates the disruption of two disulfide bridges in EGF6 and the presumed increased flexibility fairly well (Figure 5B, e). Alternatively, the integrity of EGF6 is not important for DLL1 function and the mutation does not propagate into EGF repeats important for ligand receptor interaction, or the mutation affects the intradomain stability of EGF6 only mildly, possibilities that we cannot distinguish at present.
Disruption of EGF repeats close to the cell membrane are less likely to directly affect the interaction of ligand with Notch. Recycling of DLL1 molecules is thought to lead to their clustering in microdomains at the cell surface, which in certain contexts is required for Notch activation (Musse et al. 2012). It is conceivable that mutations in EGF repeats impinge on and weaken homophilic DLL1 interactions and thus affect ligand clustering and density on the cell surface leading to reduced DLL1 activity (Figure 5B, f). Upon ligand binding, the extracellular domain of Notch is endocytosed into the signal-sending cell, generating a pulling force that is necessary for a conformational change in the Notch extracellular domain and subsequent S2 cleavage (Meloty-Kapella et al. 2012; Musse et al. 2012; Gordon et al. 2015). An increased flexibility of EGF repeats might compromise DLL1’s ability to exert the necessary pulling force on the Notch extracellular domain and thus weaken Notch activation (Figure 5B, g). In addition, interactions with as-yet-unknown proteins on the cell surface might be affected.
With respect to somitogenesis, myogenesis, neurogenesis, and left–right determination—early developmental processes known to require DLL1 activity—EGF3m is virtually indistinguishable from the Dll1lacZ null allele. However, DLL1EGF3m mutants did not show the severe hemorrhages observed in the Dll1lacZ, Dll1EGF1m, and Dll1EGF2m null alleles and survived until E18.5, suggesting some residual DLL1 activity that we did not detect in our in vitro transactivation assays. Alternatively, the rescue of the early lethality might reflect a DLL1 function independent from Notch. No such activity is known for mammalian DLL1, but such a function has been suggested for Drosophila Delta (Mok et al. 2005). In the Dll1EGF4m, Dll1EGF5m, Dll1EGF7m, and Dll1EGF8m embryos, DLL1-dependent myogenesis, neurogenesis, and left–right determination proceeded apparently normally (Figure 5A), although we cannot exclude minor abnormalities, suggesting that these processes require fairly low DLL1 activity. In contrast, all these mutants displayed anterior–posterior polarity defects in somites and, consequently, skeletal malformations of varying degrees (Figure 5A).
The severity of somite defects did not strictly correlate with transactivation capacity of mutant DLL1 proteins in vitro (Figure 1, I–L, and Figure 5A). Most likely this reflects limitations of the cell-based assays in vitro compared to physiological conditions in different developmental contexts, where other cell-surface components may influence ligand activity. In addition, normalization to wild-type ligand expression levels will underestimate ligand activity if the amount of ligand on the sending cells is in the plateau of the dose–response curve; i.e., even lower amounts would elicit the same response. This possibility might be particularly evident in the case of EGF5m and might explain its low normalized activity in vitro, which is similar to EGF4m (Figure 1, J and L, and Figure 5A), but much higher in vivo, allowing for survival of homozygous mutants (Figure 5A).
Only somitogenesis appears to be affected by alterations of the structural integrity of EGF repeats distant from the ligand–receptor interface and responds in a graded manner (Figure 5A) consistent with our previous observation that somitogenesis is fairly sensitive to reduced Notch activity (Schuster-Gossler et al. 2009). Reduction of protein O-fucosyltransferase 1 (POFUT1), which links fucose to specific Ser or Thr residues in EGF repeats and is essential for Notch signaling (Shi and Stanley 2003), also affects only somite patterning (Schuster-Gossler et al. 2009). O-linked fucose can be further modified by Fringe glycosyltransferases, which modulate NOTCH activation by ligands (reviewed in Stanley 2007). Lunatic fringe (LFNG) is expressed in the presomitic mesoderm, required for normal somitogenesis (Zhang and Gridley 1998), and appears to act there as a negative regulator of Notch signaling (Morimoto et al. 2005). LFNG modification of NOTCH might affect the interaction of EGF-mutant DLL1 with NOTCH and might contribute to the specific impact of the EGF repeat mutations on somitogenesis, which requires future analyses. However, the tissue-specific differences in phenotypic outcomes clearly indicate context-dependent effects of disrupted EGF repeats in DLL1. DLL1 is also a substrate for POFUT1 (Panin et al. 2002; Müller et al. 2014). The mutations that we generated do not affect O-fucosylation sites, although the structure of mutated EGF repeats might affect O-fucosylation (Wang et al. 1996). However, in vitro POFUT1 was dispensable for DLL1 function (Müller et al. 2014), and in Drosophila O-linked fucose was dispensable in signal-sending cells (Okajima and Irvine 2002), suggesting that altered sugar modification of DLL1 does not contribute to its reduced activity.
Mutations in the Notch pathway components DLL3, LFNG, HES7, and MESP2 underlie the severe vertebral abnormalities in the autosomal recessive human condition spondylocostal dysostosis (SCD; reviewed in Penton et al. 2012). However, 70% of the patients do not have mutations in these genes (Chapman et al. 2011). Our alleles define somite patterning as particularly sensitive to disruption of the intrinsic secondary structure of EGF repeats of DLL1, raising the possibility that such mutations in DLL1 in humans can underlie cases of SCD. Collectively, our in vitro and in vivo analyses discriminate context-dependent requirements of Delta–Notch interaction and show that disruption of the intrinsic domain structure of each EGF repeat impinges on DLL1-mediated Notch activation, and hence the structural integrity of each of these EGF repeats is required for full ligand activity.
Acknowledgments
We thank Alain Israel, Shigeru Chiba, Raphael Kopan, and Andreas Kispert for providing plasmids; Thomas Klein, Dietmar Manstein, and Matthias Gaestel for critical comments and discussion; and Anatoli Heiser for technical assistance. This work was supported by grant no. GO 449/10-1 from the German Research Council (to A.G.) and by funding from the Cluster of Excellence “From Regenerative Biology to Reconstructive Therapy” (to A.G.).
Footnotes
Communicating editor: T. R. Magnuson
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.184515/-/DC1.
Literature Cited
- Alten L., Schuster-Gossler K., Eichenlaub M. P., Wittbrodt B., Wittbrodt J., et al. , 2012. A novel mammal-specific three partite enhancer element regulates node and notochord-specific Noto expression. PLoS ONE 7: e47785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson E. R., Sandberg R., Lendahl U., 2011. Notch signaling: simplicity in design, versatility in function. Development 138: 3593–3612. [DOI] [PubMed] [Google Scholar]
- Andrawes M. B., Xu X., Liu H., Ficarro S. B., Marto J. A., et al. , 2013. Intrinsic selectivity of Notch 1 for Delta-like 4 over Delta-like 1. J. Biol. Chem. 288: 25477–25489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold H. H., Braun T., 2000. Genetics of muscle determination and development. Curr. Top. Dev. Biol. 48: 129–164. [DOI] [PubMed] [Google Scholar]
- Bauer R. C., Laney A. O., Smith R., Gerfen J., Morrissette J. J. D., et al. , 2010. Jagged1 (JAG1) mutations in patients with tetralogy of Fallot or pulmonic stenosis. Hum. Mutat. 31: 594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettenhausen B., Hrabe de Angelis M., Simon D., Guénet J. L., Gossler A., 1995. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development 121: 2407–2418. [DOI] [PubMed] [Google Scholar]
- Braune E.-B., Schuster-Gossler K., Lyszkiewicz M., Serth K., Preusse K., et al. , 2014. S/T Phosphorylation of DLL1 is required for full ligand activity in vitro but dispensable for DLL1 function in vivo during embryonic patterning and marginal zone B cell development. Mol. Cell. Biol. 34: 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson S. K., Plaehn E. G., Kluckman K. D., Hagaman J. R., Maeda N., et al. , 1996. Single-copy transgenic mice with chosen-site integration. Proc. Natl. Acad. Sci. USA 93: 9067–9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau B. G., Logan M., Davis N., Levi T., Tabin C. J., et al. , 1999. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev. Biol. 211: 100–108. [DOI] [PubMed] [Google Scholar]
- Chapman G., Sparrow D. B., Kremmer E., Dunwoodie S. L., 2011. Notch inhibition by the ligand DELTA-LIKE 3 defines the mechanism of abnormal vertebral segmentation in spondylocostal dysostosis. Hum. Mol. Genet. 20: 905–916. [DOI] [PubMed] [Google Scholar]
- Chillakuri C. R., Sheppard D., Lea S. M., Handford P. A., 2012. Notch receptor-ligand binding and activation: insights from molecular studies. Semin. Cell Dev. Biol. 23: 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes R., Schuster-Gossler K., Serth K., Gossler A., 2004. Specification of vertebral identity is coupled to Notch signalling and the segmentation clock. Development 131: 1221–1233. [DOI] [PubMed] [Google Scholar]
- Cordle J., Johnson S., Tay J. Z. Y., Roversi P., Wilkin M. B., et al. , 2008a A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat. Struct. Mol. Biol. 15: 849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordle J., Redfieldz C., Stacey M., van der Merwe P. A., Willis A. C., et al. , 2008b Localization of the delta-like-1-binding site in human Notch-1 and its modulation by calcium affinity. J. Biol. Chem. 283: 11785–11793. [DOI] [PubMed] [Google Scholar]
- de la Pompa J. L., Wakeham A., Correia K. M., Samper E., Brown S., et al. , 1997. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development 124: 1139–1148. [DOI] [PubMed] [Google Scholar]
- de Vries W. N., Binns L. T., Fancher K. S., Dean J., Moore R., et al. , 2000. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis 26: 110–112. [PubMed] [Google Scholar]
- Downing A. K., Knott V., Werner J. M., Cardy C. M., Campbell I. D., et al. , 1996. Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders. Cell 85: 597–605. [DOI] [PubMed] [Google Scholar]
- Geffers I., Serth K., Chapman G., Jaekel R., Schuster-Gossler K., et al. , 2007. Divergent functions and distinct localization of the Notch ligands DLL1 and DLL3 in vivo. J. Cell Biol. 178: 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon W. R., Zimmerman B., He L., Miles L. J., Huang J., et al. , 2015. Mechanical allostery: evidence for a force requirement in the proteolytic activation of Notch. Dev. Cell 33: 729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambleton S., Valeyev N. V., Muranyi A., Knott V., Werner J. M., et al. , 2004. Structural and functional properties of the human Notch-1 ligand binding region. Structure 12: 2173–2183. [DOI] [PubMed] [Google Scholar]
- Hooper M., Hardy K., Handyside A., Hunter S., Monk M., 1987. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature 326: 292–295. [DOI] [PubMed] [Google Scholar]
- Hrabe de Angelis M., McIntyre J., Gossler A., 1997. Maintenance of somite borders in mice requires the Delta homologue DII1. Nature 386: 717–721. [DOI] [PubMed] [Google Scholar]
- Kershaw N. J., Church N. L., Griffin M. D. W., Luo C. S., Adams T. E., et al. , 2015. Notch ligand delta-like1: X-ray crystal structure and binding affinity. Biochem. J. 468: 159–166. [DOI] [PubMed] [Google Scholar]
- Kraus F., Haenig B., Kispert A., 2001. Cloning and expression analysis of the mouse T-box gene Tbx18. MOD 100: 83–86. [DOI] [PubMed] [Google Scholar]
- Krebs L. T., Iwai N., Nonaka S., Welsh I. C., Lan Y., et al. , 2003. Notch signaling regulates left-right asymmetry determination by inducing Nodal expression. Genes Dev. 17: 1207–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladi E., Nichols J. T., Ge W., Miyamoto A., Yao C., et al. , 2005. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J. Cell Biol. 170: 983–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimeister C., Schumacher N., Steidl C., Gessler M., 2000. Analysis of HeyL expression in wild-type and Notch pathway mutant mouse embryos. MOD 98: 175–178. [DOI] [PubMed] [Google Scholar]
- Lowe L. A., Supp D. M., Sampath K., Yokoyama T., Wright C. V., et al. , 1996. Conserved left-right asymmetry of nodal expression and alterations in murine situs inversus. Nature 381: 158–161. [DOI] [PubMed] [Google Scholar]
- Luca V. C., Jude K. M., Pierce N. W., Nachury M. V., Fischer S., et al. , 2015. Structural basis for Notch1 engagement of Delta-like 4. Science 347: 847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Kintner C., Anderson D. J., 1996. Identification of neurogenin, a vertebrate neuronal determination gene. Cell 87: 43–52. [DOI] [PubMed] [Google Scholar]
- Meloty-Kapella L., Shergill B., Kuon J., Botvinick E., Weinmaster G., 2012. Notch ligand endocytosis generates mechanical pulling force dependent on dynamin, epsins, and actin. Dev. Cell 22: 1299–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoguchi S., Taniguchi Y., Kato H., Okazaki T., Strobl L. J., et al. , 1997. RBP-L, a transcription factor related to RBP-Jkappa. Mol. Cell. Biol. 17: 2679–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok L.-P., Qin T., Bardot B., LeComte M., Homayouni A., et al. , 2005. Delta activity independent of its activity as a ligand of Notch. BMC Dev. Biol. 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor-Cohen R., Rosenberg N., Einav Y., Zelzion E., Landau M., et al. , 2012. Unique disulfide bonds in epidermal growth factor (EGF) domains of β3 affect structure and function of αIIbβ3 and αvβ3 integrins in different manner. J. Biol. Chem. 287: 8879–8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto M., Takahashi Y., Endo M., Saga Y., 2005. The Mesp2 transcription factor establishes segmental borders by suppressing Notch activity. Nature 435: 354–359. [DOI] [PubMed] [Google Scholar]
- Morrissette J. D., Colliton R. P., Spinner N. B., 2001. Defective intracellular transport and processing of JAG1 missense mutations in Alagille syndrome. Hum. Mol. Genet. 10: 405–413. [DOI] [PubMed] [Google Scholar]
- Mullen R. J., Buck C. R., Smith A. M., 1992. NeuN, a neuronal specific nuclear protein in vertebrates. Development 116: 201–211. [DOI] [PubMed] [Google Scholar]
- Müller J., Rana N. A., Serth K., Kakuda S., Haltiwanger R. S., et al. , 2014. O-fucosylation of the Notch ligand mDLL1 by POFUT1 is dispensable for ligand function. PLoS One 9: e88571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musse A. A., Meloty-Kapella L., Weinmaster G., 2012. Notch ligand endocytosis: mechanistic basis of signaling activity. Semin. Cell Dev. Biol. 23: 429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt L. M., Kispert A., Herrmann B. G., 1997. A mouse gene of the paired-related homeobox class expressed in the caudal somite compartment and in the developing vertebral column, kidney and nervous system. Dev. Genes Evol. 207: 330–339. [DOI] [PubMed] [Google Scholar]
- Okajima T., Irvine K. D., 2002. Regulation of notch signaling by O-linked fucose. Cell 111: 893–904. [DOI] [PubMed] [Google Scholar]
- Panin V. M., Shao L., Lei L., Moloney D. J., Irvine K. D., et al. , 2002. Notch ligands are substrates for protein O-fucosyltransferase-1 and Fringe. J. Biol. Chem. 277: 29945–29952. [DOI] [PubMed] [Google Scholar]
- Parks A. L., Stout J. R., Shepard S. B., Klueg K. M., Santos Dos A. A., et al. , 2006. Structure-function analysis of Delta trafficking, receptor binding and signaling in Drosophila. Genetics 174: 1947–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penton A. L., Leonard L. D., Spinner N. B., 2012. Notch signaling in human development and disease. Semin. Cell Dev. Biol. 23: 450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redeker C., Schuster-Gossler K., Kremmer E., Gossler A., 2013. Normal development in mice over-expressing the intracellular domain of DLL1 argues against reverse signaling by DLL1 in vivo. PLoS ONE 8: e79050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha S. F., Lopes S. S., Gossler A., Henrique D., 2009. Dll1 and Dll4 function sequentially in the retina and pV2 domain of the spinal cord to regulate neurogenesis and create cell diversity. Dev. Biol. 328: 54–65. [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Gossler K., Cordes R., Gossler A., 2007. Premature myogenic differentiation and depletion of progenitor cells cause severe muscle hypotrophy in Delta1 mutants. Proc. Natl. Acad. Sci. USA 104: 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Gossler K., Harris B., Johnson K. R., Serth J., Gossler A., 2009. Notch signalling in the paraxial mesoderm is most sensitive to reduced Pofut1 levels during early mouse development. BMC Dev. Biol. 9: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serth K., Schuster-Gossler K., Kremmer E., Hansen B., Marohn-Köhn B., et al. , 2015. O-fucosylation of DLL3 is required for its function during somitogenesis. PLoS ONE 10: e0123776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Stanley P., 2003. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc. Natl. Acad. Sci. USA 100: 5234–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K., Chiba S., Kumano K., Hosoya N., Takahashi T., et al. , 1999. Mouse jagged1 physically interacts with notch2 and other notch receptors. Assessment by quantitative methods. J. Biol. Chem. 274: 32961–32969. [DOI] [PubMed] [Google Scholar]
- Singh R., Hoogaars W. M., Barnett P., Grieskamp T., Rana M. S., et al. , 2012. Tbx2 and Tbx3 induce atrioventricular myocardial development and endocardial cushion formation. Cell. Mol. Life Sci. 69: 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P., 2007. Regulation of Notch signaling by glycosylation. Curr. Opin. Struct. Biol. 17: 530–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk J. Y., Jensen S., McGettrick A., Willis A. C., Whiteman P., et al. , 2004. Structural consequences of cysteine substitutions C1977Y and C1977R in calcium-binding epidermal growth factor-like domain 30 of human fibrillin-1. J. Biol. Chem. 279: 51258–51265. [DOI] [PubMed] [Google Scholar]
- Vidigal J. A., Morkel M., Wittler L., Brouwer-Lehmitz A., Grote P., et al. , 2010. An inducible RNA interference system for the functional dissection of mouse embryogenesis. Nucleic Acids Res. 38: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lee G. F., Kelley R. F., Spellman M. W., 1996. Identification of a GDP-L-fucose:polypeptide fucosyltransferase and enzymatic addition of O-linked fucose to EGF domains. Glycobiology 6: 837–842. [DOI] [PubMed] [Google Scholar]
- Xu A., Lei L., Irvine K. D., 2005. Regions of Drosophila Notch that contribute to ligand binding and the modulatory influence of Fringe. J. Biol. Chem. 280: 30158–30165. [DOI] [PubMed] [Google Scholar]
- Yoshioka H., Meno C., Koshiba K., Sugihara M., Itoh H., et al. , 1998. Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell 94: 299–305. [DOI] [PubMed] [Google Scholar]
- Zhang N., Gridley T., 1998. Defects in somite formation in lunatic fringe-deficient mice. Nature 394: 374–377. [DOI] [PubMed] [Google Scholar]