Abstract
Sleep is evolutionarily conserved and required for organism homeostasis and survival. Despite this importance, the molecular and cellular mechanisms underlying sleep are not well understood. Caenorhabditis elegans exhibits sleep-like behavioral quiescence and thus provides a valuable, simple model system for the study of cellular and molecular regulators of this process. In C. elegans, epidermal growth factor receptor (EGFR) signaling is required in the neurosecretory neuron ALA to promote sleep-like behavioral quiescence after cellular stress. We describe a novel role for VAV-1, a conserved guanine nucleotide exchange factor (GEF) for Rho-family GTPases, in regulation of sleep-like behavioral quiescence. VAV-1, in a GEF-dependent manner, acts in ALA to suppress locomotion and feeding during sleep-like behavioral quiescence in response to cellular stress. Additionally, VAV-1 activity is required for EGF-induced sleep-like quiescence and normal levels of EGFR and secretory dense core vesicles in ALA. Importantly, the role of VAV-1 in promoting cellular stress–induced behavioral quiescence is vital for organism health because VAV-1 is required for normal survival after cellular stress.
Keywords: behavioral quiescence, Caenorhabditis elegans, sleep, Vav
DESPITE being a subject of formal study for over 150 years, sleep is not clearly understood. Moreover, various explanations for the functional role of sleep have been proposed, such as allowing “recharging” of cells following high metabolic activity (Benington and Heller 1995; Tu and McKnight 2006; Scharf et al. 2008) and remodeling of synapses built during wakefulness (Tononi and Cirelli 2006). Yet sleep problems have a significant impact on human health and are a leading reason for seeking medical attention (Mahowald and Schenck 2005). Therefore, there is a great need for understanding the cellular and molecular mechanisms that regulate sleep–wake cycles.
Simple model organisms such as Caenorhabditis elegans have the potential to provide valuable information regarding sleep regulation. C. elegans is known for its easily manipulated genetics, small nervous system with mapped neuronal connectivity, stereotypical behaviors, and the ability to be studied efficiently in large numbers. Numerous studies have shown that C. elegans lethargus, a restful period that occurs before each molt of the cuticle during larval development, is neuronally regulated and likely orthologous to sleep in mammals (Van Buskirk and Sternberg 2007; Raizen et al. 2008; Van Buskirk and Sternberg 2010; Choi et al. 2013; Iwanir et al. 2013; Nelson et al. 2013; Turek et al. 2013; Cho and Sternberg 2014; Singh et al. 2014). Lethargus quiescence in C. elegans shares several characteristics with mammalian sleep: inactivity (decreased locomotion and cessation of pharyngeal pumping, or feeding), a specific posture, reduced response to aversive stimuli, and rapid reversibility (Cassada and Russell 1975; Raizen et al. 2008; Schwarz et al. 2012; Iwanir et al. 2013; Cho and Sternberg 2014). In addition, lethargus quiescence is under homeostatic regulation (e.g., depriving animals in lethargus or rest by stimulation increases sleep drive), and prolonged deprivation of rest is lethal (Raizen et al. 2008; Driver et al. 2013), as it is in rats (Rechtschaffen and Bergmann 2002). Recently, C. elegans has further been shown to display sleep-like quiescence following exposure to noxious stimuli that result in cellular stress (Hill et al. 2014; Nelson et al. 2014). Fascinatingly, molecules that regulate behavioral quiescence in C. elegans and Drosophila are conserved and influence sleep in mammals, indicating evolutionarily conserved origins of sleep and the potential utility of studying sleep in such simple animals (Zimmerman et al. 2008; Sehgal and Mignot 2011; Nelson and Raizen 2013). These molecules include the cAMP protein kinase A and cAMP response element-binding protein (CREB) signaling axis (Graves et al. 2003; Nelson et al. 2013), cGMP-dependent protein kinase (Raizen et al. 2008; Langmesser et al. 2009), neuropeptides (van den Pol 2012; Choi et al. 2013; Nelson et al. 2013, 2014), dopamine (Singh et al. 2014), transcription factor AP2 (Mani et al. 2005; Turek et al. 2013), and epidermal growth factor receptor (EGFR) (Snodgrass-Belt et al. 2005; Van Buskirk and Sternberg 2007). Strikingly, EGFR has been shown to be required in only one neuron of C. elegans (the ALA interneuron) for proper induction of sleep-like quiescence behavior during lethargus and cellular stress–induced sleep-like quiescence (Van Buskirk and Sternberg 2007; Hill et al. 2014).
Previously, we demonstrated that VAV-1, an evolutionarily conserved guanine nucleotide exchange factor (GEF) for Rho-family GTPases, is an important factor in a neural circuit that regulates C. elegans locomotion (Fry et al. 2014). We found that VAV-1 is required in the ALA interneuron to reduce locomotory speed in active adult animals. Here we show that VAV-1 in the ALA interneuron also serves a critical function to suppress locomotion and feeding behavior during sleep-like quiescence. VAV-1 is required for LIN-3/EGF-induced quiescence and cellular stress–induced quiescence in adulthood. We also show that VAV-1 regulates levels of LET-23/EGFR and IDA-1, which is a conserved dense core vesicle (DCV) transmembrane protein implicated in peptide release (Cai et al. 2004, 2009) in the ALA interneuron. Importantly, the role of VAV-1 in quiescence induction after cellular stress is biologically significant because loss of vav-1 results in animals with impaired quiescence that correlates with impaired survival after stress. We also find that overexpression of constitutively active VAV-1 is sufficient to induce behavioral quiescence in adult animals. Together our results suggest that VAV-1 is a component of ALA neuronal signaling that mediates behavioral quiescence, which is critical for survival of the animal after cellular stress.
Material and Methods
Strains and maintenance
C. elegans strains were maintained and handled according to standard procedures (Stiernagle 2006). Animals were grown on nematode growth medium (NGM) plates seeded with OP50 Escherichia coli at 20°, and experiments were performed at ∼24° unless otherwise noted. Day 1 adult hermaphrodites were selected for analysis in all experiments except for lethargus assays, for which mid-L4 larval-stage animals were selected. N2 (Bristol) was the wild-type strain. vav-1 mutants are vav-1(ak41) homozygotes with selective expression of wild type vav-1 in the pharynx, which restores normal pharyngeal activity, unless otherwise noted (Norman et al. 2005). The following mutant and transgenic strains were used in this study: N2: Bristol wild-type strain; AML10: otIs355 (Prab-3::NLS::mCherry); otIs45 (Punc-119::GFP); BL5752: inIs181; inIs182 (Pida-1::GFP); CG21: egl-30(tg26); him-5(e1490); CL2070: dvIs70 (Phsp-16::GFP); DA521: egl-4(ad450sd); KM246: Pida-1::ida-1::GFP; KRN118: vav-1(ak41) takIs5 (Ppha-4::vav-1::GFP); KRN149: takEx6 (Pvav-1::2xNLS::mCherry); KRN314: takEx81 [Pvav-1::vav-1(GEF Dead)::GFP]; KRN320: takEx84 [Pvav-1(minimal promoter)::vav-1ΔSH3B::GFP]; KRN350: vav-1(ak41); takEx84 [Pvav-1(minimal promoter)::vav-1ΔSH3B::GFP]; KRN396: vav-1(ak41); takEx101 [Pvav-1(minimal promoter)::vav-1ΔSH3A::GFP]; KRN407: vav-1(ak41); takEx106 [Pvav-1(minimal promoter)::vav-1::GFP]; KRN423: vav-1(ak41) takIs5; takEx81 [Pvav-1::vav-1(GEF dead)::GFP]; KRN459: vav-1(ak41) takIs5; takEx107 [Pvav-1(minimal promoter)::vav-1ΔSH2::GFP]; KRN595: vav-1(ak41) takIs5; takEx67 (Pver-3::vav-1::GFP); KRN663: rgEx235 (Pplc-3::YFP); KRN738: vav-1(ak41); takEx215 (Pvav-1::vav-1); KRN875: takEx295 [Phsp-16::vav-1(gf)::GFP]; NY2066: ynIs66 (Pflp-7::GFP); him-5(e1490); OH10690: otIs356 (Prab-3::NLS::mCherry); PS4886: plc-3(tm1340)/mIn1; PS5970: syIs197(hs::lin-3C) him-5(e1490); TJ375: gpIs1(hsp-16::GFP); and VM4240: vav-1(ak41); akEx87 (Pvav-1::vav-1::GFP).
Molecular biology and transgenesis
To generate SH-domain deletion constructs of VAV-1, we first generated a vav-1 construct that consisted of 3 kb of the 5′ cis-regulatory region of vav-1 gene that drives a vav-1 complementary DNA (cDNA) containing a C-terminal GFP tag [the vav-1 cDNA and GFP tag were isolated from the Ppha-4::vav-1::GFP construct (pRF84) previously described (Norman et al. 2005)]. Next, we used site-directed mutagenesis using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent) to generate SH-domain deletion constructs. Primers used for deletion generation are listed in Supporting Information, Table S1. Plasmids were sequenced to rule out PCR-induced errors. Correct expression of these constructs was confirmed by GFP expression in the pharynx, ALA neuron, and somatic gonad.
To generate an hsp-16.41::vav-1(gf) construct, we swapped the myo-3 5′ cis-regulatory region out of pKN19 [Pmyo-3::vav-1(gf)::GFP] (Norman et al. 2005; Spooner et al. 2012) for the hsp-16.41 found in pPD49.83 (kind gift from A. Fire) using SphI and NheI sites.
Transgenic strains were generated by microinjection using standard protocols (Jin 1999). The host strain was wild type (N2, Bristol).
Fluorescence microscopy
Animals were immobilized using 300 μM sodium azide on 2% agarose pads. Fluorescence was determined and differential interference contrast (DIC) imaging was conducted with an inverted Zeiss AxioObserver microscope equipped with an Andor Clara charged-couple device (CCD) camera and a 40× or 60× objective lens (NA1.4). Images of the ALA neuron cell body were taken specifically in animals with ALA oriented toward the objective (cell body between the objective and midwidth of the animal but not deeper) for fluorescence of the highest intensity. Images of the ALA axon were taken in the posterior region of the animal (between the vulva and the bend of the gonad), and 20 µm was selected in which the ida-1::ida-1::GFP puncta were most in focus (because the strain carries a dominant roller mutation that complicates selection of regions of interest). In each imaging session, mutant strains were analyzed along with wild-type animals. For analysis of ALA translational markers (GFP fusion proteins), in comparisons between wild-type, vav-1, and vav-1; vav-1 rescue strains (Figure 2), images were analyzed using ImageJ (Schneider et al. 2012) and a Wacom Bamboo tablet and stylus. Freehand regions of interest (ROIs) were drawn around ALA neuron cell bodies or axon puncta, and the mean fluorescence (for axons, area of puncta) was measured. For each image, mean background fluorescence of a nearby ROI was subtracted from the ALA cell body or punctal mean fluorescence. Mutant data were normalized to wild type to correct for differences in light-source strength because imaging was conducted over months. Between 21 and 49 animals were analyzed per genotype (displayed in figure legends).
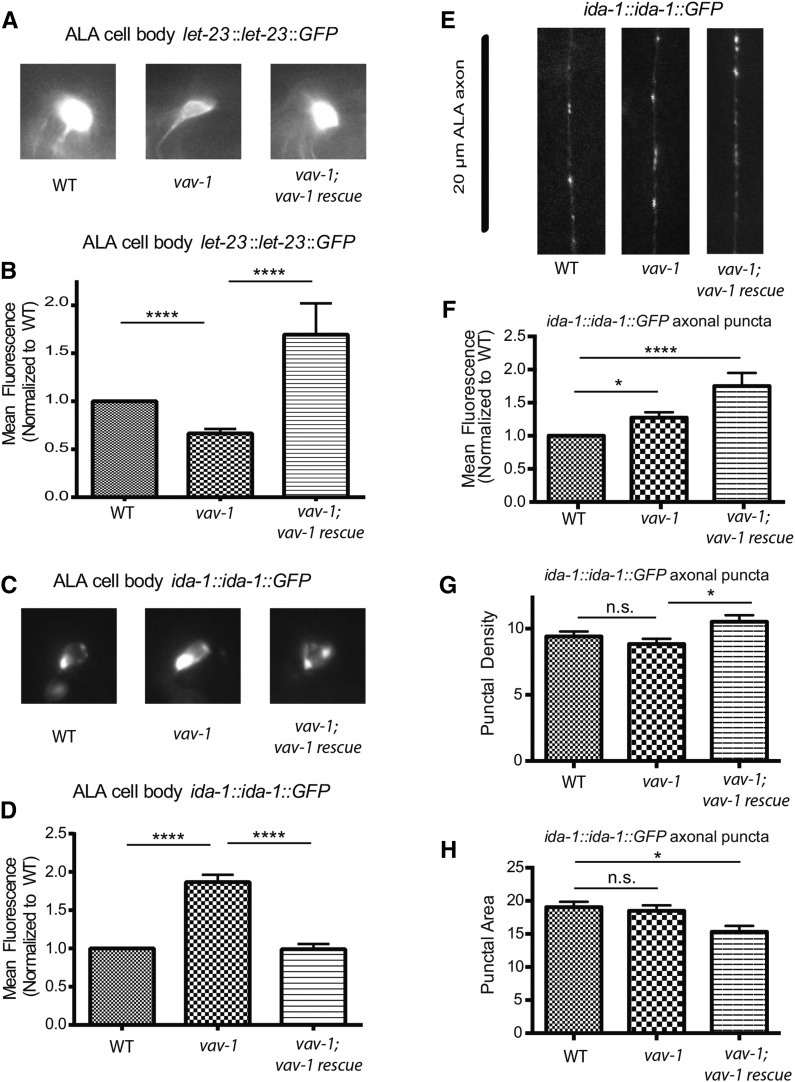
Figure 2.
VAV-1 regulates LET-23/EGFR and IDA-1/phogrin in the cell body of ALA. (A) Representative images of LET-23::GFP in the ALA neuron cell body. (B) Quantification of the mean fluorescence of LET-23::GFP in the ALA cell body of wild-type animals, vav-1 mutants, and vav-1 mutants expressing a rescuing vav-1 construct (vav-1; vav-1 rescue = vav-1; takEx215). n = 43 (wild type), 49 (vav-1), and 26 (vav-1; vav-1 rescue). (C) Representative images of IDA-1::GFP in the cell body of ALA. (D) Quantification of the mean fluorescence of IDA-1::GFP in the ALA cell body of wild-type animals, vav-1 mutants, and vav-1 mutants expressing a rescuing vav-1 construct (vav-1; vav-1 rescue = vav-1; takEx215). n = 25 (wild type), 27 (vav-1), and 18 (vav-1; vav-1 rescue). (E) Representative images of IDA-1::GFP along 20 µm of one ALA axon in the posterior of the animal. (F–H) Quantification of the mean fluorescence, density, and area of ALA axon puncta in wild-type animals, vav-1 mutants, and vav-1; vav-1 (takEx215) rescue animals. n = 44 (wild type), 37(vav-1), and 21 (vav-1; vav-1 rescue). Data are represented as mean ± SEM. Data in B, D, and F were normalized to wild type and analyzed by Kruskal-Wallis one-way ANOVA followed by Dunn’s multiple comparisons (****P < 0.0001). Data in G and H were analyzed by one-way ANOVA followed by Tukey’s multiple comparisons (*P < 0.05; ****P < 0.0001; n.s., not significant).
Quiescence assays
Lethargus quiescence assays were performed essentially as described previously with microfluidic chambers (Singh et al. 2011). Briefly, overnight OP50 E. coli culture was concentrated (sevenfold), treated with kanamycin, and resuspended in liquid NGM. This OP50 medium was applied to the microfluidic chambers (6- or 10-well format), and mid-L4 stage animals were loaded into chambers with a worm pick (only one animal per chamber and at least one blank chamber). Animals were staged based on L4 vulval morphology, active locomotion, and active feeding (pharyngeal pumping). A coverslip was placed over the chambers and sealed with molten 2% agarose. Time-lapse video recording (1 frame/10 sec) of all chambers was conducted using an Olympus dissecting microscope, Sony CCD camera, and Wormlab software. Quiescence data were generated using a Matlab image-analysis program that uses pixel differences to denote movement between frames (value of 0) or no movement (value of 1) and plots a rolling average (over 60 frames) called fractional quiescence (Singh et al. 2011). Recordings were 6- to 12-hr long, and for each animal, 6 hr of data was centered on clear peaks of fractional quiescence surrounded on either side by low background fractional quiescence values (below 0.2). Total quiescence was the total time an animal was quiescent during lethargus (each frame with a quiescence value of 1 represents 10 sec), while the duration of quiescence was the number of minutes within each 6-hr plot with a fractional quiescence value of 0.2 or more (in a consolidated bout of quiescence).
Heat shock–induced overexpression of LIN-3/EGF was carried out essentially as described previously (Van Buskirk and Sternberg 2007). Briefly, parafilmed agar plates bearing test animals were fully submerged in a 34.5° water bath for 30 min (topped with weights). Young-adult animals were then immediately transferred to fresh NGM plates seeded with OP50 to recover at 20° for 2 hr, and all quiescence measurements were conducted within the following 3 hr. For locomotory quiescence assays, each plate of heat-shocked and recovered animals was placed on an Olympus SZ61 stereomicroscope equipped with a Sony XCD-V60 CCD camera, and animals were allowed to equilibrate for 2 min. A 1-min movie was taken (at 7.5 frames/sec) of each plate. An animal was considered quiescent if its behavior fulfilled two criteria: the animal did not move three body bends or more within 1 min (both forward and backward movements were considered), and the animal spent at least one period of 10 sec or more absolutely still (not crawling and not foraging, i.e., moving just the head). For Figure 3, 50–120 total animals were analyzed per genotype in at least three independent experiments (except for non-heat-shocked hs:LIN-3 and hs:LIN-3; vav-1 controls, of which 10–20 animals were analyzed in one or two independent experiments). For Figure S2, 50–60 animals were analyzed per genotype.
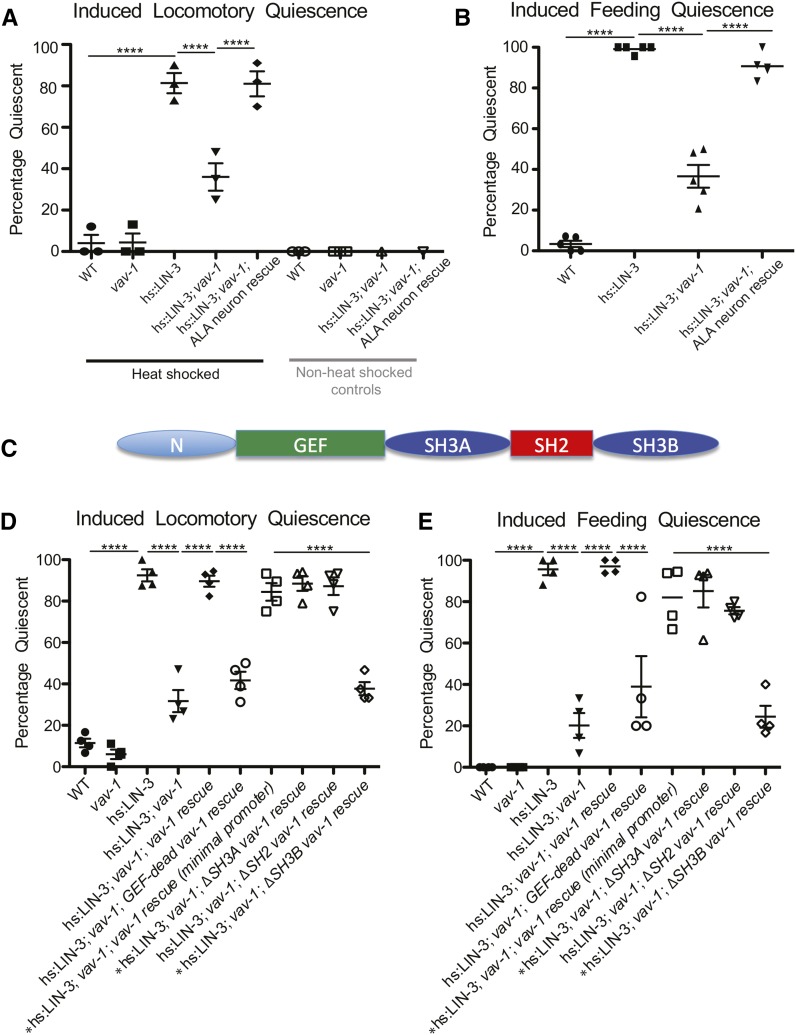
Figure 3.
VAV-1 is required in the ALA interneuron for EGF-induced sleep-like quiescence. LIN-3/EGF was conditionally expressed by a heat shock–induced construct, and behavioral quiescence (locomotory and feeding quiescence) was measured. (A) Percentage of animals exhibiting locomotory quiescence and (B) percentage of animals displaying feeding quiescence (lack of pharyngeal pumping) are drastically increased on expression of LIN-3/EGF. The LIN-3/EGF-induced quiescence is disrupted by vav-1 mutation and restored in vav-1 mutants by VAV-1 expression in the ALA neuron (driven by the ver-3 5′ cis-regulatory element). Non-heat-shocked animals do not display sleep-like quiescence. (C) The multidomain structure of the VAV-1 protein is shown. (D) Percentages of animals exhibiting locomotory quiescence and (E) feeding quiescence show that the defective EGF-induced quiescence of vav-1 mutants can be restored on expression of vav-1 by the full ∼8-kb vav-1 5′ cis-regulatory element (vav-1 rescue) or a minimal ∼3kb vav-1 5′ cis-regulatory element [vav-1 rescue (minimal promoter)]. Rescue of quiescence behavior in vav-1 mutants requires GEF activity and the SH3B domain of VAV-1. An asterisk (*) before the genotype label indicates vav-1 mutants that do not contain takIs5 (Ppha-4::vav-1::GFP). Three to four separate trials of 20–30 animals per genotype were analyzed for locomotory quiescence (except for non-heat-shocked hs:LIN-3 and hs:LIN-3; vav-1 controls, for which one to three separate trials with 10–20 animals were analyzed), and four to five separate trials of > 30 animals were analyzed per genotype for feeding quiescence. Data are represented as mean ± SEM and were analyzed by one-way ANOVA with Tukey posttests (****P < 0.0001).
Feeding quiescence was determined by cessation of pharyngeal pumping, observed by eye using a dissecting microscope (Van Buskirk and Sternberg 2007). Each animal was observed for 5 sec, and if any full contraction of the pharyngeal terminal bulb occurred, the animal was scored as “pumping.” In total, 120–150 animals per strain were scored for pharyngeal pumping in four to six independent experiments. For Figure S1 and Figure S2, 50–60 animals were analyzed per genotype.
Feeding quiescence following heat shock–induced overexpression of constitutively active VAV-1 [hsp-16.41::vav-1(gf)] was performed as described earlier (Van Buskirk and Sternberg 2007). Young-adult animals were exposed to 33° for 30 min and allowed to recover for 2 hr before scoring for feeding quiescence. Between 40 and 60 animals were analyzed per genotype.
Heat stress–induced quiescence was induced essentially as described previously (Hill et al. 2014). Parafilm-sealed plates bearing test animals were fully submerged in a 35.5° water bath for 30 min. Heat shock of each strain in an assay was staggered by 2.5–3 min to allow precise 30-min exposure and assessment at recovery time points. After heat exposure, animals were quickly removed from plates to fresh ∼24° plates seeded with OP50 E. coli and assessed for feeding quiescence (lack of pharyngeal pumping observed for ∼5 sec per animal) every 10 min (at earlier time points), 30 min, or 60 min (at later time points) during recovery. Between 80 and 210 animals per strain were tested in total in four to seven independent experiments.
Heat stress survival
Parafilmed plates bearing test animals were fully submerged in a 40° water bath for 20 min (Hill et al. 2014). Survival of each strain was assessed daily after heat exposure. Animals were tested in three independent experiments. Thirty animals for each genotype were assayed per experiment.
Acute heat response
Unseeded NGM agar plates were warmed to 37°. Each test animal was transferred to the warmed plate, and body bends were immediately counted for 1 min. Control animals were transferred to room temperature (∼24°) unseeded NGM plates, and body bends were counted for 1 min. Ten animals were analyzed for each group (genotype + temperature).
Heat shock reporter analysis was carried out by placing animals containing hsp-16.2::GFP reporter constructs in a 33° water bath for 30 min and allowing them to recover for 2 hr. Subsequently, animals were mounted on agar pads, and whole-body fluorescence intensity was examined on a Zeiss AxioObserver microscope equipped with an Andor Clara CCD camera using a 10× objective lens.
Statistics and sharing
Statistical comparisons between two groups were conducted by unpaired two-tailed t-tests. Comparisons of three groups or more were done by one-way ANOVA followed by Tukey posttests. However, Figure 2, B, D, and F, and Figure 4, D–G display data analyzed by Kruskal-Wallis one-way ANOVA followed by Dunn’s multiple comparisons because population variances differed significantly (in Figure 2, B, D, and F, due to normalization to wild type). X-Y curves of quiescence or survival (Figure 5 and Figure 6) were analyzed by repeated-measures two-way ANOVA and Tukey posttests.
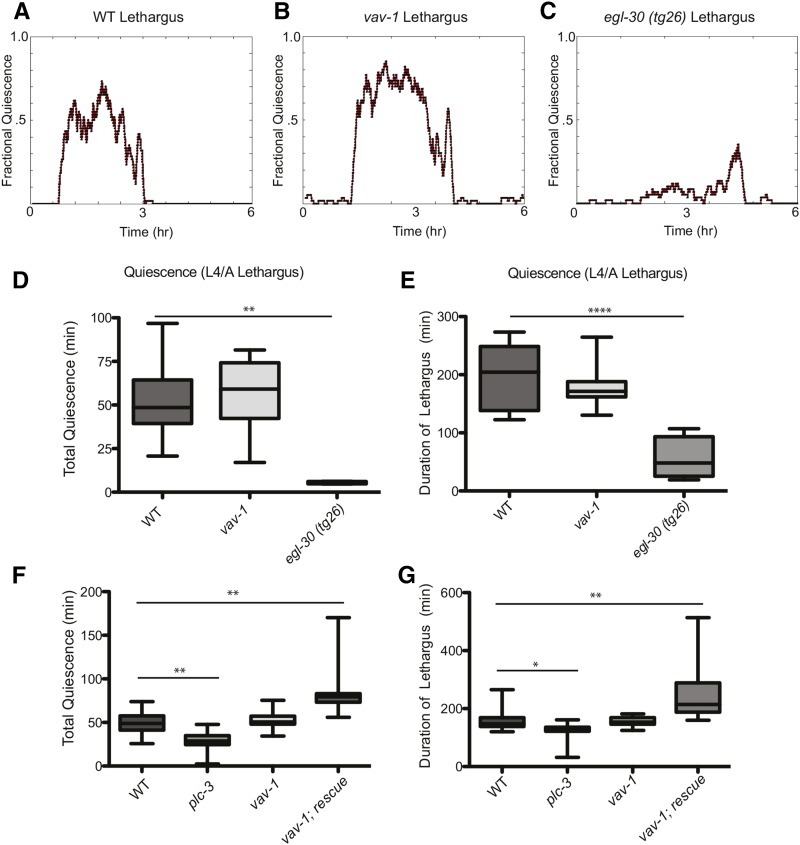
Figure 4.
VAV-1 is dispensable for normal lethargus quiescence. (A–C) Representative plots of fractional quiescence during L4/adult lethargus of wild-type animals, vav-1 mutants, and egl-30 (tg26, gf) animals in microfluidic chambers. Six hours of movement data are shown, centered on a consolidated bout of locomotory quiescence before transition to active adulthood. (D) Quantification of the total lethargus quiescence over 6 hr, and (E) quantification of the duration of lethargus quiescence of wild-type animals, vav-1 mutants, and egl-30(tg26) animals show that vav-1 mutants do not have impaired locomotory quiescence during lethargus, unlike lethargus quiescence defective egl-30 mutants. n = 12 (wild type), 10 (vav-1), and 5 (egl-30). (F) Quantification of the total lethargus quiescence and (G) duration of lethargus in wild-type animals and plc-3, vav-1, and vav-1; rescue (vav-1; akEx87–vav-1 overexpression) mutants. n = 22 (wild type), 15 (plc-3), 14 (vav-1), and 11 (vav-1; rescue). Data are represented as mean ± SEM and were analyzed by Kruskal-Wallis one-way ANOVA followed by Dunn’s multiple comparisons (*P < 0.05; **P < 0.01; ****P < 0.0001).
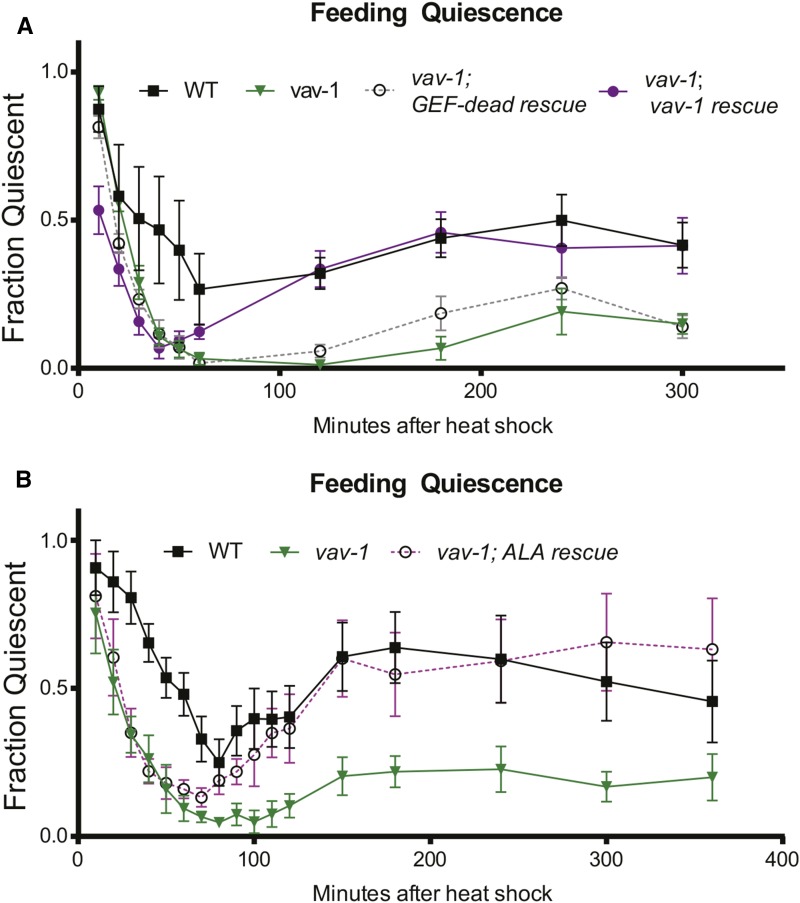
Figure 5.
VAV-1 is required in the ALA interneuron for heat stress–induced quiescence. Feeding quiescence after a 30-min exposure to 35.5° heat stress is shown; the curves display behavioral quiescence immediately on transfer to room temperature and slow recovery (10 min to ∼80 min), followed by a second, longer bout of behavioral quiescence (∼120–300 min). (A) vav-1 mutants are defective in behavioral quiescence after heat stress. The extended second bout of behavioral quiescence can be rescued by expression of wild-type vav-1, but this rescue requires VAV-1 GEF activity. (B) Similarly, expression of vav-1 in the ALA interneuron rescues the vav-1 mutant defect in the extended second bout of behavioral quiescence. Data are represented as mean ± SEM, and statistical analyses were conducted using repeated-measures two-way ANOVA followed by Tukey posttests. P-values are detailed in File S1.
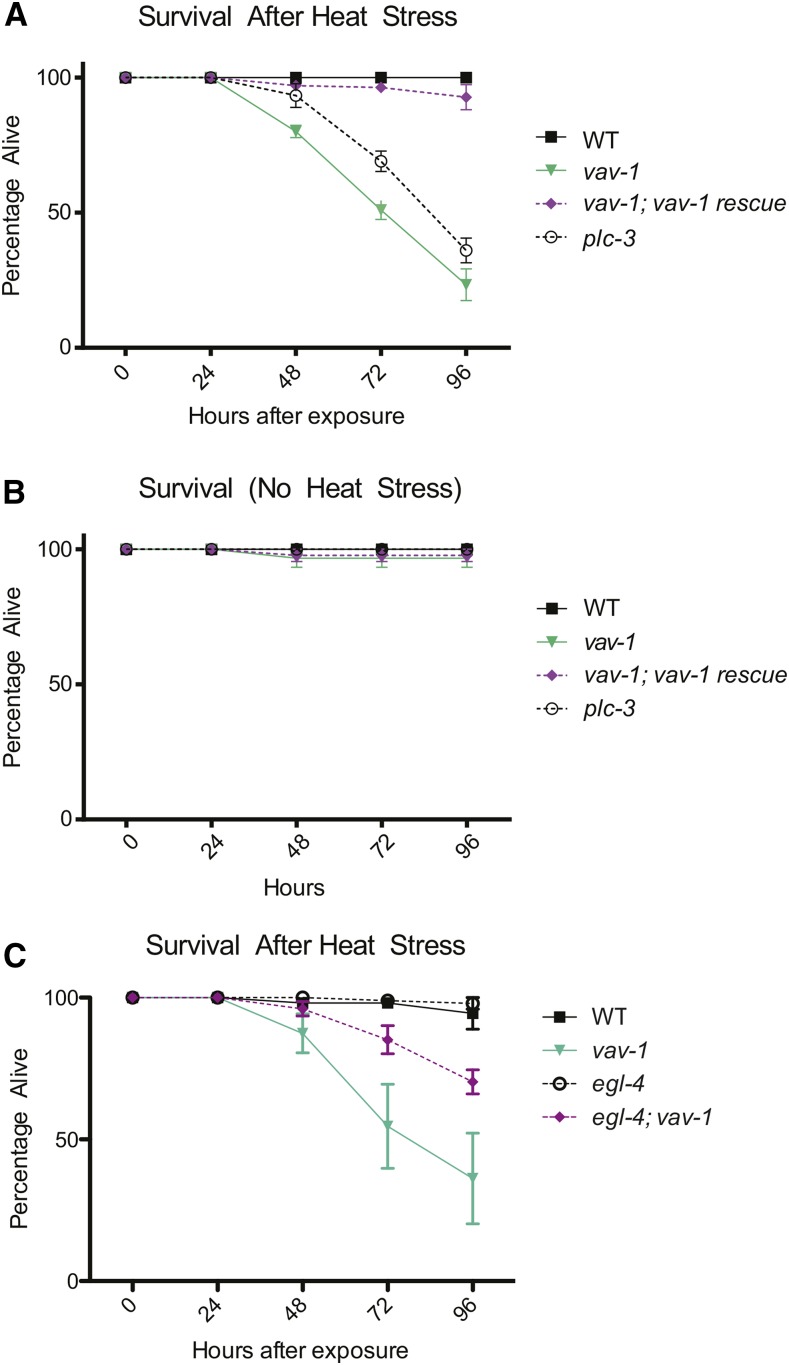
Figure 6.
VAV-1 is required for survival after noxious heat stress. (A) Animals were exposed to 40° heat stress for 20 min, and survival was assessed over the following 4 days. Like plc-3(tm1340) mutants, which are defective for heat stress–induced quiescence (Hill et al. 2014), vav-1 mutants show a loss of survival after exposure to noxious heat. This survival defect is rescued by expression of wild-type vav-1. (B) None of the tested strains showed differences in survival without heat stress exposure. (C) Survival of wild-type animals and vav-1, egl-4(ad450sd), and egl-4(ad450sd); vav-1 animals after 20 min of exposure to 40° heat stress. egl-4(ad450sd) significantly suppresses the survival defect observed in vav-1 mutants. Data are represented as mean ± SEM, and statistical analyses were conducted using repeated-measures two-way ANOVA followed by Tukey posttests. P-values are detailed in File S1.
Relevant statistical comparisons showing significant differences and P-values are listed in File S1. Asterisks indicating statistical differences are not displayed in Figure 5 and Figure 6 for simplicity of the graphs.
Data availability
All strains and computer resources are available on request.
Results
VAV-1 is critical for normal accumulation of LET-23/EGFR and DCVs in the ALA interneuron
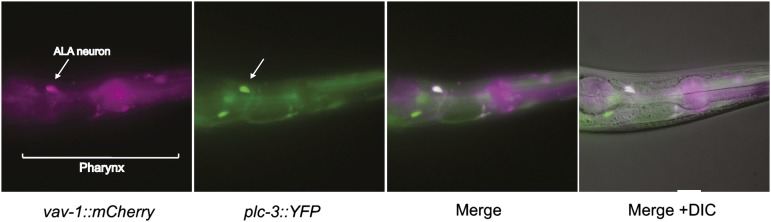
The C. elegans genome encodes a single Vav family member, VAV-1. VAV-1 is essential for pharyngeal function, and complete loss of vav-1 results in lethality in the first larval stage (L1). Pharynx-specific expression of vav-1 rescues L1 lethality but reveals defects in other tissues (Norman et al. 2005). For example, VAV-1 was recently shown to regulate the rate of locomotion by functioning in the ALA interneuron to reduce the speed of animals crawling on an agar surface (Fry et al. 2014). Here, using several independent vav-1 fluorescent reporter transgenic lines, we confirm the expression of vav-1 in the ALA interneuron, which is readily identifiable by its shape and position in the dorsal ganglion and its colocalization with a transcriptional reporter of a known ALA signaling molecule, PLC-3/PLCγ (Figure 1).
Figure 1.
VAV-1 is expressed in the ALA interneuron. Fluorescence and differential interference contrast (DIC) images of an adult animal expressing a vav-1 reporter (Pvav-1::2xNLS-mCherry) and a plc-3 reporter (Pplc-3::YFP), which is known to be expressed in the ALA interneuron (Van Buskirk and Sternberg 2010). Bracket marks the pharynx, and arrow points to the ALA neuron. Anterior is to the right. Scale bar, 10 µm.
ALA is a single interneuron that extends a small process into the dorsal nerve cord and sends two lateral processes down the entire length (of the animal) (White et al. 1986). Previously, ALA cell position and axon processes were shown to develop and differentiate normally in vav-1 mutants (Fry et al. 2014). To investigate ALA differentiation in greater detail, we examined several transcriptional reporters in vav-1 mutants. Unless otherwise stated, from this point onward, vav-1 mutants described in this paper are animals that are homozygous for a vav-1 deletion and carry an integrated transgene that expresses the vav-1 cDNA in the pharynx [vav-1(ak41) takIs5 (Ppha-4::vav-1::GFP)], which rescues L1 larval lethality (Norman et al. 2005; Fry et al. 2014). First, to test whether loss of vav-1 affects neuronal differentiation of ALA in vav-1 mutants, we examined the expression of two pan-neuronal markers, rab-3 (Nonet et al. 1997) and unc-119 (Maduro and Pilgrim 1995), in the ALA neuron of vav-1 mutants. In agreement with VAV-1 not having a role in ALA neuronal development, we observed consistent ALA expression of mCherry driven by the 5′ cis-regulatory element of the rab-3 gene and GFP driven by the 5′ cis-regulatory element of the unc-119 gene in vav-1 animals (Table 1). Next, using their 5′ cis-regulatory elements, we examined the expression of several ALA-expressed genes, flp-7, ida-1, and plc-3, that have more restricted neuronal expression patterns (Zahn et al. 2001; Cai et al. 2004; Kim and Li 2004; Van Buskirk and Sternberg 2007). Examination of Pflp-7:GFP, Pida-1:GFP, and Pplc-3:YFP in vav-1 mutants revealed consistent ALA expression (Table 1), suggesting that VAV-1 does not have a role in ALA development or differentiation. To further explore the role of VAV-1 in ALA, we examined the cellular distribution of two proteins shown to function in ALA, LET-23 and IDA-1 (Van Buskirk and Sternberg 2010). let-23 encodes an EGFR and has been implicated in the function of ALA in promoting behavioral quiescence during lethargus and on cellular stress (Van Buskirk and Sternberg 2007; Hill et al. 2014). ida-1 encodes a phogrin/islet antigen-2b, which is a transmembrane DCV protein (Zahn et al. 2001; Cai et al. 2004) that is implicated DCV release (Zhou et al. 2007) and may have a role in EGF-induced quiescence (Van Buskirk and Sternberg 2010). To investigate the role VAV-1 may have in EGFR activity in ALA, we used a LET-23::GFP translational reporter driven by the 5′ cis-regulatory element of the let-23 gene (Chang and Sternberg 1999; Van Buskirk and Sternberg 2010). From our examination of ALA, we found that while LET-23::GFP was expressed in all vav-1 mutants analyzed, its fluorescence intensity was significantly reduced (Figure 2A). Importantly, this reduced fluorescence could be rescued by expression of wild-type vav-1 by the 5′ cis-regulatory element of the vav-1 gene (Figure 2, A and B). These data suggest that VAV-1 regulates the accumulation of LET-23/EGFR in ALA. Next, because the ALA neuron has been shown to be peptidergic (Nelson et al. 2014) and IDA-1 has been shown to function in ALA, we used an IDA-1::GFP translational fusion protein expressed by the 5′ cis-regulatory element of the ida-1 gene, which has been used to examine DCV function in ALA (Zhou et al. 2007), to investigate the localization and expression of DCVs in vav-1 mutants. In addition to examining the localization and fluorescence intensity in the ALA cell body, we investigated IDA-1::GFP punctate distribution and fluorescence intensity in the ALA neuron lateral processes, which could uncover defects in DCV trafficking (Zahn et al. 2004). We found that IDA-1::GFP fluorescence in the cell body of ALA is increased in vav-1 mutants and can be rescued by expression of wild-type vav-1 by the 5′ cis-regulatory element of the vav-1 gene (Figure 2, C and D). We hypothesized, therefore, that IDA-1::GFP might accumulate in the ALA cell body as a result of a perturbation in IDA-1::GFP trafficking to or within the axons. Surprisingly, we found that the fluorescence intensity of IDA-1::GFP axonal puncta is modestly increased in vav-1 mutants, while the number of puncta (punctal density) and the size of the puncta along the ALA axon are not significantly different between wild-type and vav-1 mutant animals (Figure 2, E–H). Unlike IDA-1::GFP fluorescence in the cell body of ALA (Figure 2, C and D), the change in fluorescence intensity of IDA-1::GFP axonal puncta is not rescued by wild-type vav-1 expression (Figure 2F). Additionally, expression of the wild-type vav-1 rescue construct leads to slight changes in punctal density and area (Figure 2, G and H). These data suggest that the changes in ALA axon puncta may not be related to the function of VAV-1 and that the changes in ALA cell body LET-23 and IDA-1 (which are more striking and rescued by wild-type vav-1 expression) are more likely to be biologically relevant. Nevertheless, together our data indicate that VAV-1 regulates molecules within the ALA interneuron cell body that are critical for signal transduction within ALA and DCV signaling from ALA to other tissues (i.e., released peptides).
Table 1. Percentage of animals showing reporter expression in ALA.
| ALA reporter | Wild type | vav-1 mutants |
|---|---|---|
| unc-119::GFP | 100% (20/20) | 100% (40/40) |
| rab-3::mCherry | 100% (40/40) | 100% (80/80) |
| ida-1::GFP | 100% (20/20) | 100% (30/30) |
| flp-7::GFP | 100% (25/25) | 100% (45/45) |
| plc-3::YFP | 96% (25/26) | 97% (32/33) |
VAV-1 is required for LIN-3/EGF-induced behavioral quiescence
The ALA interneuron and LET-23/EGFR were previously found to be required for lethargus and cellular stress– and EGF-induced sleep-like quiescence (Van Buskirk and Sternberg 2007; Hill et al. 2014). Because we have found consistent expression of vav-1 in ALA (Figure 1) and altered accumulation of LET-23/EGFR in ALA of vav-1 mutants (Figure 2, A and B), we hypothesized that VAV-1 is involved in behavioral sleep-like quiescence. We investigated this possibility by conditional expression of the EGF-like ligand LIN-3 using heat shock induction of LIN-3/EGF expression (Van Buskirk and Sternberg 2010). This method has been shown to induce sleep-like quiescence in adult animals. As expected, induced expression of LIN-3/EGF in young-adult animals resulted in both locomotory (Figure 3A) and feeding quiescence (Figure 3B), as measured by the proportions of animals exhibiting cessation of movement and lack of pharyngeal pumping behavior, respectively. Strikingly, the behavioral quiescence induced by LIN-3/EGF expression is strongly suppressed in vav-1 mutants. On induction of LIN-3/EGF expression, vav-1 mutants show significant locomotory activity and pharyngeal pumping (Figure 3, A and B). To confirm that the loss of quiescence behaviors is due to a loss of VAV-1 in ALA, we expressed VAV-1 specifically in the ALA neuron (and two muscle cells, the saucer cell of the pharynx and the anal sphincter, using the ver-3 5′ cis-regulatory element) of vav-1 mutants. We found that restoring VAV-1 expression in ALA rescues sensitivity to LIN-3/EGF-induced quiescence because these animals exhibit similar quiescence to wild-type animals (Figure 3, A and B). These data indicate that VAV-1 acts in the ALA interneuron to mediate LIN-3/EGF-induced quiescence in adults.
Because ALA requires the secretion of FLP-13 peptides to mediate LIN-3/EGF-induced quiescence (Nelson et al. 2014) and we have observed abnormal accumulation of IDA-1::GFP, a marker that labels DCVs, in the ALA neuron of vav-1 mutants (Figure 2, C and D), we investigated whether overexpression of IDA-1 can rescue the failure of vav-1 mutants to respond to LIN-3/EGF-induced quiescence. Thus, we induced LIN-3/EGF in vav-1 mutants overexpressing IDA-1 and examined quiescence. In these experiments, we did not observe a suppression of the quiescence behavior defect in vav-1 mutants (Figure S1). These data indicate that simple overexpression of IDA-1 in vav-1 mutants is not sufficient to rescue the vav-1 defect in LIN-3/EGF-induced quiescence.
VAV-1 requires GEF activity and the C-terminal SH3 domain to mediate LIN-3/EGF-induced behavioral quiescence
VAV-1 is a multidomain signaling molecule that contains an evolutionarily conserved array of protein-protein interaction domains in addition to the classical Dbl homology–Pleckstrin (DH-PH) homology domains found in GEFs (Norman et al. 2005). In the C-terminal region of VAV-1, an SH2 domain is flanked by two SH3 domains (SH3A and SH3B, respectively) (Figure 3C). Previously, GEF activity of VAV-1 was shown to be required for L1 larval survival (Norman et al. 2005) and in ALA for regulating locomotory behavior in active animals (Fry et al. 2014). Using this vav-1 GEF-dead construct (Fry et al. 2014), we explored whether GEF activity was required for LIN-3/EGF-induced quiescence. We introduced the VAV-1 GEF-dead construct into the vav-1 mutant background and analyzed LIN-3/EGF-induced quiescence. As shown in Figure 3 (D and E), although the wild-type vav-1 rescuing construct restores locomotory and feeding quiescence to vav-1 mutants expressing induced LIN-3/EGF, the GEF-dead vav-1 construct does not. These data indicate that GEF activity of VAV-1 is required for mediating behavioral quiescence.
Next, we generated different vav-1 domain deletion constructs to determine which, if any, protein-protein interaction domains are required for quiescence induction by LIN-3/EGF expression. First, we found that vav-1 constructs with deletion of SH3A or SH3B were able to rescue the vav-1 mutant L1 lethality, unlike the SH2 deletion construct. These data suggest that the SH2 domain is critical for the function of VAV-1 in the pharynx. While the SH2 deletion construct could not rescue L1 lethality, we found that it completely rescued the LIN-3/EGF-induced quiescence in vav-1 mutants (Figure 3, D and E). Because the SH3A and SH3B deletion constructs rescued the vav-1 mutant L1 lethality, analysis of LIN-3/EGF-induced quiescence was conducted in the vav-1(ak41) homozygous background lacking the takIs5 (Ppha-4::vav-1::GFP) integrated transgene. From these analyses, we found that while the SH3A deletion could fully rescue the LIN-3/EGF-induced quiescence in vav-1 mutants, the SH3B deletion could not (Figure 3, D and E). These data indicate that although the SH2 domain is important for VAV-1 pharyngeal activity, it is dispensable for LIN-3/EGF-induced quiescence. Conversely, the SH3B domain is required for LIN-3/EGF-induced quiescence but is dispensable for VAV-1 pharyngeal function.
GEF-dead and SH3B deletion vav-1 constructs did not rescue LIN-3/EGF-induced quiescence in vav-1 mutants, suggesting that these domains are required for VAV-1 function. However, it is possible that these constructs may function in a dominant-negative manner in ALA and disrupt normal quiescence induction by LIN-3/EGF. To investigate this possibility, we tested whether animals expressing either vav-1 mutant construct in an otherwise wild-type background could respond to LIN-3/EGF-induced quiescence. Wild-type animals expressing the vav-1 (GEF-dead) or the vav-1 SH3B deletion construct responded normally to LIN-3/EGF-induced quiescence (Figure S2). These data indicate that these constructs likely do not have a dominant-negative impact on LIN-3/EGF-induced quiescence.
VAV-1 function is dispensable for sleep-like behavior during lethargus
The ALA interneuron has been shown to influence C. elegans behavioral quiescence in several different scenarios. For instance, ablation of ALA disrupts quiescence during lethargus (Van Buskirk and Sternberg 2007), the larval period characterized by cessation of movement and feeding (Raizen et al. 2008). Additionally, LET-23/EGFR function in ALA is required for normal quiescence during lethargus (Van Buskirk and Sternberg 2007). Therefore, because VAV-1 acts in ALA to regulate locomotion in adult animals (Fry et al. 2014), regulates the accumulation of LET-23/EGFR (Figure 2), and is critical for LIN-3/EGF-induced quiescence (Figure 3), we hypothesized that VAV-1 acts in ALA to suppress locomotion during lethargus. To analyze animals in lethargus, we selected animals in the middle of the L4 larval stage that were actively moving and feeding (pharyngeal pumping was observed). We then measured the locomotory activity of these animals across L4/adult lethargus, the transition period between the L4 larval and adult stages. We used specialized microfluidic chambers designed to keep animals confined to an area within a camera field of view for recording the entirety of the lethargus period while still allowing animals to move freely within the chamber (Singh et al. 2011). Surprisingly, when we observed locomotory quiescence during lethargus of wild-type animals (Figure 4A) and vav-1 mutants (Figure 4B), we found no difference in the total quiescence (Figure 4D) or the duration of quiescence (Figure 4E), unlike lethargus quiescence defective egl-30 mutants (Figure 4, C–E) (Schwarz and Bringmann 2013; Nagy et al. 2014). Because ALA has been reported to play a minor role in lethargus quiescence (Van Buskirk and Sternberg 2007; Trojanowski et al. 2015), we tested a mutant, plc-3, that has been shown previously to have a role in ALA to regulate quiescence during lethargus to determine whether our assay is sensitive enough to detect a minor lethargus quiescence defect. Indeed, plc-3 mutants show a significant reduction in quiescence behavior during L4 to adult lethargus (Figure 4, F and G). Interestingly, when we examined vav-1 mutants overexpressing wild-type vav-1, we found an extended lethargus quiescence period (Figure 4, F and G), indicating that excessive VAV-1 signaling can increase the lethargus quiescence period.
VAV-1 is required for sleep-like quiescence and survival after cellular stress
Recent publications have revealed an additional type of sleep-like quiescence behavior mediated by ALA: on exposure to cellular stress (e.g., noxious heat, 35–40°), C. elegans displays sleep-like quiescence (Hill et al. 2014; Nelson et al. 2014). Indeed, when wild-type animals are exposed to noxious heat, sleep-like behavioral quiescence is readily observed, and recovery occurs over 60–80 min after heat shock (Figure 5, A and B). Importantly, a second, longer bout of ∼2–5 hr (depending on heat stress temperature) of sleep-like behavioral quiescence occurs and has been proposed to be required for maintaining cellular proteostasis and survival after cellular stress (Figure 5, A and B)(Hill et al. 2014). Like lethargus quiescence and LIN-3/EGF-induced quiescence, proteins expressed in ALA are required to mediate this type of quiescence, including CEH-17, a transcription factor needed for differentiation of ALA, as well as LET-23/EGFR and PLC-3/PLCγ (Hill et al. 2014). Therefore, we investigated whether VAV-1 is required for this type of cellular stress–induced quiescence. We found that vav-1 mutants are indeed impaired in both the early and later, long phase of quiescence following noxious heat stress (Figure 5, A and B). The longer phase of impaired quiescence observed in vav-1 mutants can be rescued by wild-type vav-1 gene expression but not the GEF-dead vav-1 gene under the control of its 5′ cis-regulatory element (Figure 5A). Additionally, the second, longer phase defect observed in vav-1 mutants also can be rescued by driving expression of VAV-1 in the ALA neuron using the ver-3 5′ cis-regulatory element (Figure 5B), indicating a cell-autonomous role of VAV-1 in ALA in regulating cellular stress–induced sleep-like behavior. The failure of rescue in the initial phase may be due to transgene expression efficiency. While vav-1 mutants show defective quiescence induced by noxious heat, they sense noxious heat and respond normally by increasing the rate of locomotion on transfer to prewarmed 37° plates (Figure S3A). Furthermore, vav-1 mutants undergo a normal heat shock response, as visualized by GFP, under the control of the heat shock response element (5′ cis-regulatory element of the hsp-16.2 gene) (Figure S3, B–D).
Interestingly, it was shown previously that impairment in the longer phase of quiescence is correlated with decreased organism survival after cellular stress from noxious heat (Hill et al. 2014). Thus, we tested the survival of vav-1 mutants after noxious heat exposure. We found that vav-1 mutants have significantly impaired survival after heat stress (like positive-control plc-3/PLCγ mutants) (Hill et al. 2014), which we rescued by expression of wild-type vav-1 (Figure 6A). None of these strains had decreased survival without heat stress (Figure 6B). Together our results suggest that VAV-1 is an important factor mediating quiescence from the ALA neuron and that the inability of vav-1 mutants to rest after cellular stress, as observed in wild-type animals, is detrimental to their long-term survival. In agreement with this conclusion, we found that introducing a mutation that increases the frequency of sleep-like behavior can increase the survival of vav-1 mutants after noxious heat stress. This mutation, a gain-of-function allele of the cyclic GMP–dependent protein kinase, egl-4, causes spontaneous bouts of sleep-like quiescence (Raizen et al. 2008). Moreover, egl-4 is proposed to act downstream of ALA to promote LIN-3/EGF- and cellular stress–induced quiescence (Van Buskirk and Sternberg 2007; Hill et al. 2014). Analysis of egl-4(gf); vav-1 double mutants showed a significant increased survival over vav-1 mutants after exposure to noxious heat (Figure 6C).
Induction of activated VAV-1 is sufficient to induce quiescence behavior
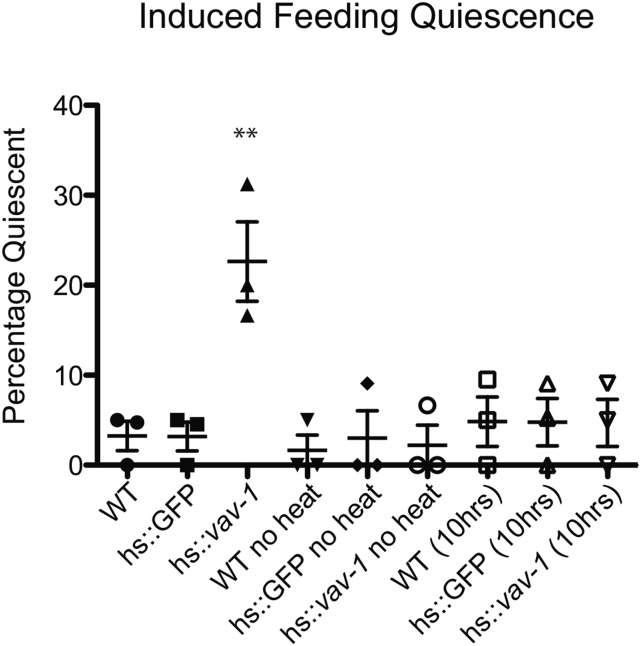
Our data thus far indicate that VAV-1 is required to promote ALA-mediated quiescence. However, it could be that VAV-1 has a more general role in ALA function and is not directly involved in quiescence signaling. To test the hypothesis that activated VAV-1 has a direct role in inducing sleep-like quiescence behavior, we generated an inducible transgene using the hsp-16.41 5′ cis control element (Jones et al. 1989) to induce the expression of a constitutively active VAV-1 mutant. The constitutively active VAV-1 protein contains three tyrosine-to-phenylalanine mutations in the regulatory region, which have a conserved role in regulating GEF activity in Vav family members (Aghazadeh et al. 2000; Norman et al. 2005). Heat shock activation of this construct in otherwise wild-type young-adult active animals caused a significant increase in quiescence behavior, whereas non-heat-shocked animals showed normal active behavior (Figure 7). Moreover, 10 hr after heat shock induction, the heat-shocked animals resumed normal active behavior (Figure 7). These results indicate that VAV-1 has a direct role in promoting sleep-like quiescence.
Figure 7.
Induction of constitutively active VAV-1 is sufficient to induce behavioral quiescence. A constitutively active VAV-1 mutant was placed under the control of the 5′ cis-regulatory element of the inducible heat shock promoter gene hsp-16.41. Young-adult animals were exposed to 33° for 30 min and allowed to recover for 2 hr. While wild-type and hs::GFP animals displayed normal active behavior, induction of constitutively active vav-1 (hs::vav-1) produces a significant increase in the percentage of animals displaying quiescence behavior. At 10 hr after heat shock, hs::vav-1 animals showed a complete recovery. Non-heat-shocked animals (no heat) do not display quiescence behavior. Three separate trials with 20–30 animals for each genotype were analyzed. Data are presented as mean ± SEM and were analyzed by one-way ANOVA with Tukey posttest analysis (**P < 0.01).
Discussion
VAV-1 was shown previously to have a crucial role in modulation of rhythmic behaviors in C. elegans (e.g., the feeding behavior of pharyngeal pumping, ovulation, and the defecation cycle) (Norman et al. 2005). Recently, VAV-1 also was shown to regulate the rhythmic behavior of locomotion, providing an inhibitory input from the ALA interneuron to the neural circuit regulating locomotion and thus reducing locomotory speed in wakeful, active adults (Fry et al. 2014). We show here that VAV-1, in a GEF-dependent manner, is necessary for sleep-like quiescence behaviors; VAV-1 is needed for suppression of rhythmic locomotion and feeding during EGF-induced and cellular stress–induced behavioral quiescence. Further, we demonstrate that activation of VAV-1 is sufficient to induce quiescence behavior in active adult animals. Moreover, we discovered that VAV-1 requires GEF activity and acts in the neurosecretory ALA interneuron, a neuron previously shown to be critical for sleep-like behavior, to mediate behavioral quiescence (Van Buskirk and Sternberg 2007; Hill et al. 2014). We also found that VAV-1 regulates the protein levels of two critical ALA signaling molecules, LET-23/EGFR and IDA-1/phogrin, required for ALA function (Van Buskirk and Sternberg 2007; Zhou et al. 2007; Van Buskirk and Sternberg 2010). Significantly, we demonstrate that loss of VAV-1–dependent behavioral quiescence results in reduced organism survival after cellular stress.
Previous work has shown that ablation of ALA results in a reduction of behavioral quiescence during lethargus (Van Buskirk and Sternberg 2007). However, because removal of ALA produces only a minor defect in lethargus quiescence, this suggests that ALA is not the only cell required for inducing behavioral quiescence associated with lethargus (Van Buskirk and Sternberg 2007). Indeed, RIA and RIS neurons, for instance, are also involved in lethargus quiescence (Nelson et al. 2013; Turek et al. 2013). Moreover, subsequent publications have shed light on perhaps a more vital role of ALA: induction of cellular stress–induced quiescence that is correlated with survival rates after cellular stress (e.g., noxious heat) (Hill et al. 2014; Nelson et al. 2014). Mutation of ceh-17, a transcription factor required for LET-23/EGFR and PLC-3/PLCγ expression in ALA, and mutation of flp-13, a gene encoding FMRFamide-like neuropeptides expressed in ALA, both disrupt EGF- and cellular stress–induced quiescence. Interestingly, FLP-13, like VAV-1, is not required for endogenous lethargus quiescence. These observations suggest that the signaling mechanisms within ALA mediating sleep-like quiescence behaviors diverge at some point, with requirements of separate signaling molecules for the regulation of lethargus sleep-like quiescence and cellular stress–induced sleep-like quiescence and that FLP-13 and VAV-1 are crucial in the cellular stress–induced quiescence pathway. Interestingly, we found that overexpression of vav-1 results in an extended quiescent bout in animals undergoing lethargus, suggesting that VAV-1, if overly abundant, can affect endogenous lethargus quiescence and prolong this quiescent state. Furthermore, our data demonstrate that IDA-1::GFP levels in the ALA cell body are regulated by VAV-1, suggesting that VAV-1 may be involved in DCV function in this neuron. Perhaps VAV-1 influences localization or release of FLP-13 (and/or other neuropeptides); further studies are needed to investigate these possibilities.
Cellular stress induced by noxious heat causes perturbation of cellular homeostasis, which drives sleep-like behavioral quiescence (Hill et al. 2014; Nelson et al. 2014). The sleep-like behavioral quiescence induced by cellular stress results in an initial short-lived quiescent state (∼1 hr) followed by brief recovery and a second, prolonged quiescent state (several hours) (Hill et al. 2014). Hill et al. (2014) proposed that the prolonged bout of quiescence is critical for regulating cellular homeostasis. From our analyses, we have found that while vav-1 mutants are defective in both cellular stress–induced sleep-like behavioral quiescence periods, we observed complete rescue of the second bout only on wild-type vav-1 expression from either its 5′ cis-regulatory control element or the ALA-specific 5′ cis-regulatory element. It is unclear why we did not observe rescue in the initial phase of quiescence. This could be due to transgene expression levels and/or genetic background differences that lead to defects in the initial period of quiescence. Nevertheless, our studies confirm the notion that the second, prolonged bout of sleep-like behavioral quiescence is critical for cellular homeostasis because the rescued animals, unlike vav-1 mutants, showed wild-type survival after exposure to noxious heat stress.
Of the three mammalian Vav genes, Vav2 and Vav3 are expressed in the nervous system (Cowan et al. 2005; Sauzeau et al. 2010a). Although the functions of Vav proteins in the nervous system are far from clear, several studies have found critical roles for Vav family members in nervous system function. For example, Vav proteins have been shown to be important for axonal development (Cowan et al. 2005; Malartre et al. 2010; Sauzeau et al. 2010a; Fernandez-Espartero et al. 2013), synaptic plasticity (Hale et al. 2011), and negative regulation of sympathetic nervous system activity (Sauzeau et al. 2006, 2007, 2010b, 2011; Zhu et al. 2015). Moreover, Vav3 has been identified as a candidate schizophrenia gene (Ikeda et al. 2011; Aleksic et al. 2013). Here we have found that VAV-1 is required in the ALA interneuron for promoting both EGF- and cellular stress–induced sleep-like behavioral quiescence. We provide evidence implicating VAV-1 in neuronal EGF signaling: (1) vav-1 mutants fail to respond to an EGF quiescence signal and (2) VAV-1 regulates levels of the EGFR/LET-23 in the ALA neuron. Previous studies have demonstrated that EGFR/LET-23 and its downstream effector phospholipase C gamma, PLC-3/PLCγ, act in the ALA neuron to promote both EGF- and stress-induced sleep-like behavioral quiescence (Van Buskirk and Sternberg 2007; Hill et al. 2014). Our data are consistent with VAV-1 acting in this LET-23–PLC-3 signaling pathway. Notably, Vav proteins have been identified as positive effectors of EGFR signaling pathways in other tissues of C. elegans, as well as in Drosophila and mammalian cells (Margolis et al. 1992; Dekel et al. 2000; Pandey et al. 2000; Hornstein et al. 2003; Norman et al. 2005). However, our data are the first to suggest Vav interaction with EGFR or PLCγ in neurons. While our analyses demonstrate that vav-1 mutants are defective in responding to EGF-induced quiescence and show reduced EGFR levels in the ALA cell body, the role of VAV-1 in mediating EGFR signaling is unclear. Notably, Vav proteins have been implicated in promoting EGFR and other receptor tyrosine kinase (RTK) signaling by regulating the endocytosis of the RTKs (Cowan et al. 2005; Thalappilly et al. 2010). Indeed, Vav2, in a GEF-dependent manner, has been shown to have a critical role in EGFR signaling by delaying EGFR receptor internalization and degradation via an interaction with endosomal-associated proteins (Thalappilly et al. 2010). Whether VAV-1 has a role in mediating endocytosis and degradation of the LET-23/EGFR in the ALA interneuron will require further studies.
Although this is the first study to uncover a role for a Vav protein in regulation of a sleep-like or quiescence behavior, a Rho-family GTPase guanine nucleotide–activating protein encoded by the crossveinless-c gene was recently implicated in sleep homeostasis in Drosophila (Donlea et al. 2014), suggesting a conserved role for Rho-family GTPases in sleep behavior. Furthermore, Vav2 and Vav3 transcripts are expressed in regions of the human and mouse brain associated with sleep (Lein et al. 2007; Hawrylycz et al. 2012), such as the thalamus (Fuentealba and Steriade 2005). Because EGF has a conserved role in sleep (Kushikata et al. 1998; Snodgrass-Belt et al. 2005; Foltenyi et al. 2007; Van Buskirk and Sternberg 2007) and we demonstrate that VAV-1 acts in an EGFR signaling pathway and has a role in sleep-like behaviors in C. elegans, it would be interesting to explore whether Vav2 and/or Vav3 has an evolutionarily conserved role in human sleep.
Acknowledgments
We thank the people in the Norman laboratory for help with cloning and transgenesis and valuable discussions. Many strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40-OD010440). This work was supported by NIH grants GM-088213 (K.N.) and NS-055813 (A.H.), as well as a Brown Institute for Brain Science and the Norman Prince Neurosciences Institute Postdoctoral Fellowship in Translational Neuroscience (H.H.).
Footnotes
Communicating editor: P. Sengupta
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.183038/-/DC1.
Literature Cited
- Aghazadeh B., Lowry W. E., Huang X. Y., Rosen M. K., 2000. Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell 102: 625–633. [DOI] [PubMed] [Google Scholar]
- Aleksic B., Kushima I., Hashimoto R., Ohi K., Ikeda M., et al. , 2013. Analysis of the VAV3 as candidate gene for schizophrenia: evidences from voxel-based morphometry and mutation screening. Schizophr. Bull. 39: 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benington J. H., Heller H. C., 1995. Restoration of brain energy metabolism as the function of sleep. Prog. Neurobiol. 45: 347–360. [DOI] [PubMed] [Google Scholar]
- Cai T., Fukushige T., Notkins A. L., Krause M., 2004. Insulinoma-associated protein IA-2, a vesicle transmembrane protein, genetically interacts with UNC-31/CAPS and affects neurosecretion in Caenorhabditis elegans. J. Neurosci. 24: 3115–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T., Hirai H., Fukushige T., Yu P., Zhang G., et al. , 2009. Loss of the transcriptional repressor PAG-3/Gfi-1 results in enhanced neurosecretion that is dependent on the dense-core vesicle membrane protein IDA-1/IA-2. PLoS Genet. 5: e1000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassada R. C., Russell R. L., 1975. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 46: 326–342. [DOI] [PubMed] [Google Scholar]
- Chang C., Sternberg P. W., 1999. C. elegans vulval development as a model system to study the cancer biology of EGFR signaling. Cancer Metastasis Rev. 18: 203–213. [DOI] [PubMed] [Google Scholar]
- Cho J. Y., Sternberg P. W., 2014. Multilevel modulation of a sensory motor circuit during C. elegans sleep and arousal. Cell 156: 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Chatzigeorgiou M., Taylor K. P., Schafer W. R., Kaplan J. M., 2013. Analysis of NPR-1 reveals a circuit mechanism for behavioral quiescence in C. elegans. Neuron 78: 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan C. W., Shao Y. R., Sahin M., Shamah S. M., Lin M. Z., et al. , 2005. Vav family GEFs link activated Ephs to endocytosis and axon guidance. Neuron 46: 205–217. [DOI] [PubMed] [Google Scholar]
- Dekel I., Russek N., Jones T., Mortin M. A., Katzav S., 2000. Identification of the Drosophila melanogaster homologue of the mammalian signal transducer protein, Vav. FEBS Lett. 472: 99–104. [DOI] [PubMed] [Google Scholar]
- Donlea J. M., Pimentel D., Miesenbock G., 2014. Neuronal machinery of sleep homeostasis in Drosophila. Neuron 81: 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver R. J., Lamb A. L., Wyner A. J., Raizen D. M., 2013. DAF-16/FOXO regulates homeostasis of essential sleep-like behavior during larval transitions in C. elegans. Curr. Biol. 23: 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Espartero C. H., Ramel D., Farago M., Malartre M., Luque C. M., et al. , 2013. GTP exchange factor Vav regulates guided cell migration by coupling guidance receptor signalling to local Rac activation. J. Cell Sci. 126: 2285–2293. [DOI] [PubMed] [Google Scholar]
- Foltenyi K., Greenspan R. J., Newport J. W., 2007. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat. Neurosci. 10: 1160–1167. [DOI] [PubMed] [Google Scholar]
- Fry A. L., Laboy J. T., Norman K. R., 2014. VAV-1 acts in a single interneuron to inhibit motor circuit activity in Caenorhabditis elegans. Nat. Commun. 5: 5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba P., Steriade M., 2005. The reticular nucleus revisited: intrinsic and network properties of a thalamic pacemaker. Prog. Neurobiol. 75: 125–141. [DOI] [PubMed] [Google Scholar]
- Graves L. A., Hellman K., Veasey S., Blendy J. A., Pack A. I., et al. , 2003. Genetic evidence for a role of CREB in sustained cortical arousal. J. Neurophysiol. 90: 1152–1159. [DOI] [PubMed] [Google Scholar]
- Hale C. F., Dietz K. C., Varela J. A., Wood C. B., Zirlin B. C., et al. , 2011. Essential role for vav Guanine nucleotide exchange factors in brain-derived neurotrophic factor-induced dendritic spine growth and synapse plasticity. J. Neurosci. 31: 12426–12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M. J., Lein E. S., Guillozet-Bongaarts A. L., Shen E. H., Ng L., et al. , 2012. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489: 391–399. Available at: Allen Institute for Brain Science. Allen Human Brain Atlas, 2015, http://human.brain-map.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A. J., Mansfield R., Lopez J. M., Raizen D. M., Van Buskirk C., 2014. Cellular stress induces a protective sleep-like state in C. elegans. Curr. Biol. 24: 2399–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein I., Mortin M. A., Katzav S., 2003. DroVav, the Drosophila melanogaster homologue of the mammalian Vav proteins, serves as a signal transducer protein in the Rac and DER pathways. Oncogene 22: 6774–6784. [DOI] [PubMed] [Google Scholar]
- Ikeda M., Aleksic B., Kinoshita Y., Okochi T., Kawashima K., et al. , 2011. Genome-wide association study of schizophrenia in a Japanese population. Biol. Psychiatry 69: 472–478. [DOI] [PubMed] [Google Scholar]
- Iwanir S., Tramm N., Nagy S., Wright C., Ish D., et al. , 2013. The microarchitecture of C. elegans behavior during lethargus: homeostatic bout dynamics, a typical body posture, and regulation by a central neuron. Sleep 36: 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., 1999. Transformation, pp. 66–96 in Caenorhabditis elegans: A Practical Approach, edited by Hope I. A. Oxford University Press, New York. [Google Scholar]
- Jones D., Dixon D. K., Graham R. W., Candido E. P., 1989. Differential regulation of closely related members of the hsp16 gene family in Caenorhabditis elegans. DNA 8: 481–490. [DOI] [PubMed] [Google Scholar]
- Kim K., Li C., 2004. Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J. Comp. Neurol. 475: 540–550. [DOI] [PubMed] [Google Scholar]
- Kushikata T., Fang J., Chen Z., Wang Y., Krueger J. M., 1998. Epidermal growth factor enhances spontaneous sleep in rabbits. Am. J. Physiol. 275: R509–R514. [DOI] [PubMed] [Google Scholar]
- Langmesser S., Franken P., Feil S., Emmenegger Y., Albrecht U., et al. , 2009. cGMP-dependent protein kinase type I is implicated in the regulation of the timing and quality of sleep and wakefulness. PLoS One 4: e4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E. S., Hawrylycz M. J., Ao N., Ayres M., Bensinger A., et al. , 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445: 168–176. Available at: Allen Institute for Brain Science. Allen Mouse Brain Atlas, 2015, http://mouse.brain-map.org. [DOI] [PubMed] [Google Scholar]
- Maduro M., Pilgrim D., 1995. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141: 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald M. W., Schenck C. H., 2005. Insights from studying human sleep disorders. Nature 437: 1279–1285. [DOI] [PubMed] [Google Scholar]
- Malartre M., Ayaz D., Amador F. F., Martin-Bermudo M. D., 2010. The guanine exchange factor vav controls axon growth and guidance during Drosophila development. J. Neurosci. 30: 2257–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani A., Radhakrishnan J., Farhi A., Carew K. S., Warnes C. A., et al. , 2005. Syndromic patent ductus arteriosus: evidence for haploinsufficient TFAP2B mutations and identification of a linked sleep disorder. Proc. Natl. Acad. Sci. USA 102: 2975–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis B., Hu P., Katzav S., Li W., Oliver J. M., et al. , 1992. Tyrosine phosphorylation of vav proto-oncogene product containing SH2 domain and transcription factor motifs. Nature 356: 71–74. [DOI] [PubMed] [Google Scholar]
- Nagy S., Raizen D. M., Biron D., 2014. Measurements of behavioral quiescence in Caenorhabditis elegans. Methods 68: 500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. D., Raizen D. M., 2013. A sleep state during C. elegans development. Curr. Opin. Neurobiol. 23: 824–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. D., Trojanowski N. F., George-Raizen J. B., Smith C. J., Yu C. C., et al. , 2013. The neuropeptide NLP-22 regulates a sleep-like state in Caenorhabditis elegans. Nat. Commun. 4: 2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. D., Lee K. H., Churgin M. A., Hill A. J., Van Buskirk C., et al. , 2014. FMRFamide-like FLP-13 neuropeptides promote quiescence following heat stress in Caenorhabditis elegans. Curr. Biol. 24: 2406–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M. L., Staunton J. E., Kilgard M. P., Fergestad T., Hartwieg E., et al. , 1997. Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J. Neurosci. 17: 8061–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman K. R., Fazzio R. T., Mellem J. E., Espelt M. V., Strange K., et al. , 2005. The Rho/Rac-family guanine nucleotide exchange factor VAV-1 regulates rhythmic behaviors in C. elegans. Cell 123: 119–132. [DOI] [PubMed] [Google Scholar]
- Pandey A., Podtelejnikov A. V., Blagoev B., Bustelo X. R., Mann M., et al. , 2000. Analysis of receptor signaling pathways by mass spectrometry: identification of vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc. Natl. Acad. Sci. USA 97: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen D. M., Zimmerman J. E., Maycock M. H., Ta U. D., You Y. J., et al. , 2008. Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451: 569–572. [DOI] [PubMed] [Google Scholar]
- Rechtschaffen A., Bergmann B. M., 2002. Sleep deprivation in the rat: an update of the 1989 paper. Sleep 25: 18–24. [DOI] [PubMed] [Google Scholar]
- Sauzeau V., Sevilla M. A., Rivas-Elena J. V., de Alava E., Montero M. J., et al. , 2006. Vav3 proto-oncogene deficiency leads to sympathetic hyperactivity and cardiovascular dysfunction. Nat. Med. 12: 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauzeau V., Jerkic M., Lopez-Novoa J. M., Bustelo X. R., 2007. Loss of Vav2 proto-oncogene causes tachycardia and cardiovascular disease in mice. Mol. Biol. Cell 18: 943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauzeau V., Horta-Junior J. A., Riolobos A. S., Fernandez G., Sevilla M. A., et al. , 2010a Vav3 is involved in GABAergic axon guidance events important for the proper function of brainstem neurons controlling cardiovascular, respiratory, and renal parameters. Mol. Biol. Cell 21: 4251–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauzeau V., Sevilla M. A., Montero M. J., Bustelo X. R., 2010b The Rho/Rac exchange factor Vav2 controls nitric oxide-dependent responses in mouse vascular smooth muscle cells. J. Clin. Invest. 120: 315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauzeau V., Carvajal-Gonzalez J. M., Riolobos A. S., Sevilla M. A., Menacho-Marquez M., et al. , 2011. Transcriptional factor aryl hydrocarbon receptor (Ahr) controls cardiovascular and respiratory functions by regulating the expression of the Vav3 proto-oncogene. J. Biol. Chem. 286: 2896–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf M. T., Naidoo N., Zimmerman J. E., Pack A. I., 2008. The energy hypothesis of sleep revisited. Prog. Neurobiol. 86: 264–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz J., Bringmann H., 2013. Reduced sleep-like quiescence in both hyperactive and hypoactive mutants of the Galphaq Gene egl-30 during lethargus in Caenorhabditis elegans. PLoS One 8: e75853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz J., Spies J. P., Bringmann H., 2012. Reduced muscle contraction and a relaxed posture during sleep-like lethargus. Worm 1: 12–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A., Mignot E., 2011. Genetics of sleep and sleep disorders. Cell 146: 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K., Chao M. Y., Somers G. A., Komatsu H., Corkins M. E., et al. , 2011. C. elegans Notch signaling regulates adult chemosensory response and larval molting quiescence. Curr. Biol. 21: 825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K., Ju J. Y., Walsh M. B., DiIorio M. A., Hart A. C., 2014. Deep conservation of genes required for both Drosophila melanogaster and Caenorhabditis elegans sleep includes a role for dopaminergic signaling. Sleep 37: 1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snodgrass-Belt P., Gilbert J. L., Davis F. C., 2005. Central administration of transforming growth factor-alpha and neuregulin-1 suppress active behaviors and cause weight loss in hamsters. Brain Res. 1038: 171–182. [DOI] [PubMed] [Google Scholar]
- Spooner P. M., Bonner J., Maricq A. V., Benian G. M., Norman K. R., 2012. Large isoforms of UNC-89 (obscurin) are required for muscle cell architecture and optimal calcium release in Caenorhabditis elegans. PLoS One 7: e40182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle, T., 2006 Maintenance of C. elegans (February 11, 2006), pp. 1–11 in WormBook, ed. The C. elegans Research Community, /10.1895/wormbook.1.7.1, http//www.wormbook.org.
- Thalappilly S., Soubeyran P., Iovanna J. L., Dusetti N. J., 2010. VAV2 regulates epidermal growth factor receptor endocytosis and degradation. Oncogene 29: 2528–2539. [DOI] [PubMed] [Google Scholar]
- Tononi G., Cirelli C., 2006. Sleep function and synaptic homeostasis. Sleep Med. Rev. 10: 49–62. [DOI] [PubMed] [Google Scholar]
- Trojanowski N. F., Nelson M. D., Flavell S. W., Fang-Yen C., Raizen D. M., 2015. Distinct mechanisms underlie quiescence during two Caenorhabditis elegans sleep-like states. J. Neurosci. 35: 14571–14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu B. P., McKnight S. L., 2006. Metabolic cycles as an underlying basis of biological oscillations. Nat. Rev. Mol. Cell Biol. 7: 696–701. [DOI] [PubMed] [Google Scholar]
- Turek M., Lewandrowski I., Bringmann H., 2013. An AP2 transcription factor is required for a sleep-active neuron to induce sleep-like quiescence in C. elegans. Curr. Biol. 23: 2215–2223. [DOI] [PubMed] [Google Scholar]
- Van Buskirk C., Sternberg P. W., 2007. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat. Neurosci. 10: 1300–1307. [DOI] [PubMed] [Google Scholar]
- Van Buskirk C., Sternberg P. W., 2010. Paired and LIM class homeodomain proteins coordinate differentiation of the C. elegans ALA neuron. Development 137: 2065–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol A. N., 2012. Neuropeptide transmission in brain circuits. Neuron 76: 98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. G., Southgate E., Thomson J. N., Brenner S., 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314: 1–340. [DOI] [PubMed] [Google Scholar]
- Zahn T. R., Macmorris M. A., Dong W., Day R., Hutton J. C., 2001. IDA-1, a Caenorhabditis elegans homolog of the diabetic autoantigens IA-2 and phogrin, is expressed in peptidergic neurons in the worm. J. Comp. Neurol. 429: 127–143. [DOI] [PubMed] [Google Scholar]
- Zahn T. R., Angleson J. K., MacMorris M. A., Domke E., Hutton J. F., et al. , 2004. Dense core vesicle dynamics in Caenorhabditis elegans neurons and the role of kinesin UNC-104. Traffic 5: 544–559. [DOI] [PubMed] [Google Scholar]
- Zhou K. M., Dong Y. M., Ge Q., Zhu D., Zhou W., et al. , 2007. PKA activation bypasses the requirement for UNC-31 in the docking of dense core vesicles from C. elegans neurons. Neuron 56: 657–669. [DOI] [PubMed] [Google Scholar]
- Zhu S., Zhao C., Wu Y., Yang Q., Shao A., et al. , 2015. Identification of a Vav2-dependent mechanism for GDNF/Ret control of mesolimbic DAT trafficking. Nat. Neurosci. 18: 1084–1093. [DOI] [PubMed] [Google Scholar]
- Zimmerman J. E., Naidoo N., Raizen D. M., Pack A. I., 2008. Conservation of sleep: insights from non-mammalian model systems. Trends Neurosci. 31: 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains and computer resources are available on request.