Abstract
Geographic patterns of genetic differentiation have long been used to understand population history and to learn about the biological mechanisms of adaptation. Here we present an examination of genomic patterns of differentiation between northern and southern populations of Australian and North American Drosophila simulans, with an emphasis on characterizing signals of parallel differentiation. We report on the genomic scale of differentiation and functional enrichment of outlier SNPs. While, overall, signals of shared differentiation are modest, we find the strongest support for parallel differentiation in genomic regions that are associated with regulation. Comparisons to Drosophila melanogaster yield potential candidate genes involved in local adaptation in both species, providing insight into common selective pressures and responses. In contrast to D. melanogaster, in D. simulans we observe patterns of variation that are inconsistent with a model of temperate adaptation out of a tropical ancestral range, highlighting potential differences in demographic and colonization histories of this cosmopolitan species pair.
Keywords: geographic variation, population genomics, Drosophila
THE geographic distribution of genetic or phenotypic variation can provide valuable insight into the process of adaptation. For example, consistent patterns of genetic variation across space have long been interpreted as evidence for local adaptation owing to spatially varying selection (Endler 1977; Adrion et al. 2015). This is well illustrated in populations of Drosophila melanogaster, a model system showing consistent phenotypic and molecular clines across environmental gradients (Hoffmann and Weeks 2007). Among these, the association of latitude with variation in ecologically relevant traits such as heat knockdown resistance, chill coma recovery, and diapause incidence (Hoffmann et al. 2002; Schmidt and Paaby 2008) provides strong support for local adaptation to climate.
Despite efforts to understand the potential adaptive nature of molecular variation in populations of Drosophila, there remains some disconnect between our understanding of allele-frequency clines and phenotypic clines, of which the latter are more easily and intuitively interpretable. For the vast majority of clinal molecular polymorphisms in Drosophila, the mechanisms underlying their maintenance are poorly understood. Nevertheless, because gene flow in Drosophila is thought to be high, strong differentiation often can be argued as evidence of local adaptation. Moreover, because the physical scale of linkage disequilibrium (LD) in Drosophlia is often smaller than the size of genes (Langley et al. 2012), differentiation between populations generally is associated with hypotheses regarding individual genes as targets of selection.
Observation of parallel patterns of differentiation furthers the argument for an adaptive basis to differentiation, and in general, comparisons of patterns of variation across independent replicate geographic transects may contribute to an understanding of the contribution of a variant to fitness under differing environmental conditions. This approach has been used often in Drosophila and other systems (Anderson and Oakeshott 1984; Colosimo et al. 2005; Hohenlohe et al. 2010; Paaby et al. 2010; Turner et al. 2010). Parallel patterns provide not only compelling evidence that a particular trait or genetic variant plays a role in adaptation but also insight into the repeatability of adaptation. Even the absence of parallel differentiation contributes to our understanding of the repeatability of adaptive differentiation and the different mechanisms and constraints that influence both phenotypic and molecular evolution.
While there has been a great focus on geographic variation in D. melanogaster, investigations of other Drosophila have demonstrated the presence of geographic patterns in a number of other species in the genus (e.g., Sturtevant and Dobzhansky 1936; Dobzhansky 1948, 1947; Huey 2000; Hallas et al. 2002; Arthur et al. 2008; Tyukmaeva et al. 2011; Price et al. 2014), revealing cross-species convergence in clines for traits such as wing size and cold tolerance. Among these species, Drosophila simulans presents an especially attractive system for further study of geographic genetic variation because of recent divergence (5 million years ago, Tamura et al. 2004) from the well-studied D. melanogaster. In addition to this shared evolutionary history, similarities in recent colonization histories and shared cosmopolitan distributions provide reason to expect that the two species have experienced recent parallel evolution, and indeed, the D. melanogaster/D. simulans pair has been a popular focus for comparative population genetics (e.g., Parsons 1975a; Singh et al. 1987; Capy and Gibert 2004; Zhao et al. 2015).
Although the two species share recent common ancestry and have broadly similar ecologies, there are several important differences between them. For example, D. melanogaster appears to be more tolerant of high ethanol concentrations, and the two species differ in their seasonal abundances and thermal tolerances (Parsons 1975b, 1977). Moreover, it is known that their geographic centers of diversity vary, with D. melanogaster being most diverse in South-Central Africa (Pool et al. 2012) and D. simulans in Madagascar (Dean and Ballard 2004). Such contrasts emphasize the possibility that the two species are historically adapted to different environments and have experienced vastly different colonization histories. Potential differences in population histories are further reflected in the contrasting patterns of genetic variation outside of Africa (Begun and Whitley 2000; Andolfatto 2001; Capy and Gibert 2004), with D. simulans exhibiting higher within-population diversity and D. melanogaster higher levels of between-population diversity (Singh 1989). Notably, while strong clines are abundant in D. melanogaster and have been the focus of extensive investigation, there seems to be less evidence for clinal variation in D. simulans. For example, Arthur et al. (2008) showed that there are no apparent clines for cold tolerance or heat shock in Australian populations of D. simulans despite the fact that these traits are strongly clinal in Australian D. melanogaster (Hoffmann et al. 2002), and Gibert et al. (2004) reported that even when present, clines in D. simulans are weak. One potential interpretation of this is a relative lack of local adaptation in D. simulans. More recently, Machado et al. (2015) found genomic evidence for clinal variation in D. simulans and verified that it is less pronounced than in D. melanogaster.
While differences in the strength of clinal variation in D. simulans compared to D. melanogaster may suggest that the two species are responding to their local environments in different ways, the findings of Machado et al. (2015), differentiation in patterns of gene expression (Zhao et al. 2015), and phenotypic clines in traits such as body size indicate that D. simulans is likely evolving, in at least some capacity, to spatially varying selection. This is further supported by the observation that there is significant overlap in differentiated genes between D. simulans and D. melanogaster (Machado et al. 2015; Zhao et al. 2015). This between-species parallel differentiation in both gene expression (Zhao et al. 2015) and allele frequency (Machado et al. 2015) raises the additional possibility that weaker phenotypic clines generally reported for D. simulans may not accurately reflect the influence of spatially varying selection on this species.
To further investigate patterns of geographic differentiation in D. simulans and similarities and differences with respect to D. melanogaster, we resequenced four D. simulans populations—one northern and one southern—in both North America and Australia. We then employed an FST outlier approach to identify putative targets of spatially varying selection. The advantages of this two-continent design are twofold. First, we are able to address our direct objective of assessing the degree of parallelism in local adaptation between the two continents and compare them to analogous patterns in D. melanogaster. Second, focusing on SNPs that are strongly differentiated on two continents will, to some degree, mitigate potential false discoveries that may arise as a consequence of sampling error, fine-scale local environmental adaptation, or demography. Such comparative population genomic approaches may inform our understanding of parallelism at various levels from the nucleotide level to gene annotations or pathways and also may provide useful information regarding constraints, repeatability, and diversity of mechanisms of adaptation in these two species.
Similar genome-wide studies of differentiation in comparable populations of D. melanogaster (Turner et al. 2008; Kolaczkowski et al. 2011; Fabian et al. 2012; Reinhardt et al. 2014) have detected signals of parallel differentiation and, in particular, a strong association of large inversions with allele-frequency differentiation. The prevalence of inversion frequency clines in D. melanogaster is thought to reflect some response to spatially varying selection, but their adaptive significance remains unclear. This is noteworthy because inversion polymorphisms are virtually absent in D. simulans (Ashburner and Lemeunier 1976), and it remains unknown what the implications are for adaptive differentiation in D. simulans. In addition to learning more about general patterns of variation and potential mechanisms of adaptation, an assessment here of genomic patterns of geographic variation in D. simulans presents the opportunity to gain further insight into general patterns of geographic variation in the two species, as well as common responses to challenges posed by novel environments.
Materials and Methods
Sampling and sequencing
Four populations are represented in this study: northern Australia, southern Australia, northern United States, and southern United States (Table 1). The two U.S. subpopulation libraries were generated from pools of single daughters of females sampled directly from the field in 2011. The two Australian subpopulations were generated by pooling a single female from isofemale lines established in 2004 (Arthur et al. 2008). Libraries were prepared according to the NEBNext DNA Library Prep Master Mix Set for the Illumina Protocol and were sequenced on the Illumina HiSeq2000 platform at two or three libraries per lane. Reads were trimmed using SolexaQA (Cox et al. 2010) with a quality-score threshold of 28, and any resulting reads shorter than 36 bp were discarded. Both subpopulations from a given continent were sequenced in the same lane, which eliminates concerns of lane effects on within-continent differentiation.
Table 1. Size, collection dates, and locations of samples.
| Key | Latitude | Chromosome | Year | Source |
|---|---|---|---|---|
| FL (US) | 25.47°N | 66 | September 2011 | Daughters of field-caught females |
| RI (US) | 41.84°N | 66 | September 2011 | Daughters of field-caught females |
| QLD (AU) | 42.77°S | 22 | February–March 2004 | Isofemale lines |
| TAS (AU) | 25.54°S | 16 | February–March 2004 | Isofemale lines |
Reads were aligned to the D. simulans w501 assembly from Hu et al. (2013) and Wolbachia pipientis strain wRi using the Burrows-Wheeler Aligner (BWA) (Li and Durbin 2009). Reads with mapping quality under 30 were discarded, and optical duplicates and reads mapping to multiple regions were removed. Initially, sites with coverage < 15 and >2 SDs from the mean were removed from the analysis because these sites are, respectively, prone to inflated FST from sampling error and potential duplications or paralogy. Because of substantially smaller sample sizes, estimated allele frequencies in the Australian populations have greater variance than those in the North American population; that is,
where n, m, and p are sample size, coverage, and allele frequency, respectively (Futschik and Schlötterer 2010). To reduce noise and potential biases from smaller sample sizes, minimum cutoffs in Queensland and Tasmania were increased to 20 and 29, respectively, for outlier-based analyses at the SNP level.
Repetitive regions, defined by Hu et al. (2013), were removed after alignment. To minimize the effect of sequencing error on population genetics analyses, for each continent, a variant was considered only if it was supported by two or more reads and was segregating at a frequency > 0.05. Because they are likely to correspond to regions of low recombination, regions of low heterozygosity on the proximal and distal ends of each chromosome arm were removed. These regions were determined by defining uninterrupted sequences of 100-kb windows (sliding by 50 kb) on the ends of the chromosome arms that were below half the chromosomal average for either mean or number of segregating sites.
Outlier SNPs and regions
Both and were calculated at each position in the genome using estimators described in Kolaczkowski et al. (2011). Within each continent, SNPs in the upper tail of the distribution were considered to be highly differentiated. Where indicated, further refinement of candidate loci took place by considering only SNPs that were outliers on both continents and were differentiated in the same direction with respect to latitude because these are more likely to be under parallel differential selection. Because sites with lower coverage will have greater variance in as a result of sampling error, outliers may be enriched for sites with lower coverage. To address this effect of coverage, polymorphic sites were binned based on the minimum coverage of the two populations in a continent. These sites then were ranked by within each bin, and SNPs were required to be outliers with respect to both genome-wide and coverage-based rank to be classified as strongly differentiated sites. and rank were highly correlated by Spearman’s rank-order correlation (ρ = 0.98 on both continents). Statistical significance for enrichment was calculated using Fisher’s exact test (FET).
Derived alleles
Gene alignments for D. melanogaster, D. simulans, and D. yakuba from Hu et al. (2013) were used to determine the ancestral allele at a polymorphic site. If either allele at a biallelic site matched the D. melanogaster and D. yakuba sequence, it was considered to be the ancestral state, and the other allele was considered to be derived. For a given SNP within a continent, the allele present at higher frequency in the higher-latitude population was considered to be the temperate-adapted allele. As the threshold was increased, the number of temperate-adapted alleles that were ancestral was compared to the number of alleles that were derived. The null expectation is for the proportion of temperate-adapted derived alleles to be unchanged across thresholds, and statistical tests for over/underrepresentation of temperate-adapted alleles for a given threshold were based on a binomial expectation, with rate given by the proportion of temperate-adapted alleles across the whole genome.
Annotations
All analyses of functional regions used the annotations accompanying Hu et al. (2013). These annotations were augmented using the assembled transcriptome from Rogers et al. (2014). Transcripts from Rogers et al. (2014) were matched to D. melanogaster annotations by aligning predicted translations to D. melanogaster translations in FlyBase Release 5.9 using BLAST (Altschul et al. 1990) under default parameters. The top BLAST hits were retained only if protein sequences aligned at the first residue and the final residue of the D. simulans protein aligned to within five residues of the D. melanogaster stop codon. Some analyses focus on different “annotation classes”: upstream (region 500 bp upstream of transcription start site), exon, 3′ UTR, coding sequence (CDS), intron, 5′ UTR, and intergenic (unannotated).
Circular permutation
Because the positions of outliers are autocorrelated throughout the genome, generating a null expectation for non-SNP-based analyses (e.g., the expected number of shared outlier genes based on random sampling of SNPs or genes) can be a challenge. To address this issue, for analyses involving annotations, we generated a null distribution of enrichments by iteratively shifting the relative position of each SNP along a concatenation of all chromosomes by one randomly selected number; the positions at which SNPs occur remain unchanged for all permutations. For each iteration, a new random number was selected, and a list of outlier annotations was generated. This approach provides an alternative to explicitly defining independent differentiated regions because the local autocorrelation of is conserved in each iteration and is similar to the strategy used in Nordborg et al. (2005).
Gene ontologies
To account for any bias in overrepresented Gene Ontology (GO) categories owing to gene size, we performed a circular permutation of the values at each genic site by shifting the relative position of each base by a randomly chosen number and recalculating the GO enrichment P-value under a hypergeometric model. By iterating this process, we obtained a distribution of enrichment P-values for each GO category, which then was used to obtain a quantile value for the P-value that was observed in the nonpermuted data. This preserves the autocorrelation in the distribution of . The R Bioconductor (Gentleman et al. 2004) and org.Dm.eg.db (M. Carlson, org.Dm.eg.db: Genome wide annotation for Fly. R package v2.6.4; available at: http://www.bioconductor.org/) packages were used to map genes to GOs.
Data availability
Genomic data are available as raw sequence reads from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under PRJNA307610.
Results
The four study populations were sequenced to mean coverages ranging from 43 to 70 (Table 1). Heterozygosity does not differ significantly across autosomal arms in the North American populations [Kruskal-Wallis test using 100-kb windows: P = 0.11 (FL) and P = 0.21 (RI)]. In Australia, however, there is significant heterogeneity among the autosomes in both populations [P = 3.3 × 10−6 (QLD) and P = 4.5 × 10−3 (TAS)]. Genome wide, higher-latitude populations have higher mean heterozygosity than lower-latitude populations based both on genome-wide and mean in 100-kb windows (Table S1, Figure S1), and this pattern is consistent across the genome (Wilcoxon signed rank on mean in 100-kb windows: P < 10−15 for both continents). This contrasts with observations in D. melanogaster of higher heterozygosity at lower latitudes (Kolaczkowski et al. 2011; Fabian et al. 2012; Reinhardt et al. 2014). We note that of the four populations, FL has the lowest heterozygosity genome wide, reflecting perhaps more severe drift than the other three populations. This is in contrast to the findings of Machado et al. (2015), who did not observe substantially lower levels of heterozygosity in FL populations of D. simulans compared to other populations sampled in North America.
Mean is heterogeneous across autosomal arms for both continents (Table S1) (P = 0.006 and P = 0.002, respectively), although the rank order of chromosome arms differs for the two continents. In particular, mean is highest on the X chromosome in North America [as found by Machado et al. (2015)] but highest on 3R in Australia. Furthermore, there appears to be little shared differentiation on a broad physical scale of 100-kb windows (Figure S2). Compared to populations of D. melanogaster (Fabian et al. 2012; Reinhardt et al. 2014) sampled over a similar spatial scale, D. simulans appear to exhibit less genome-wide differentiation.
Scale of differentiation
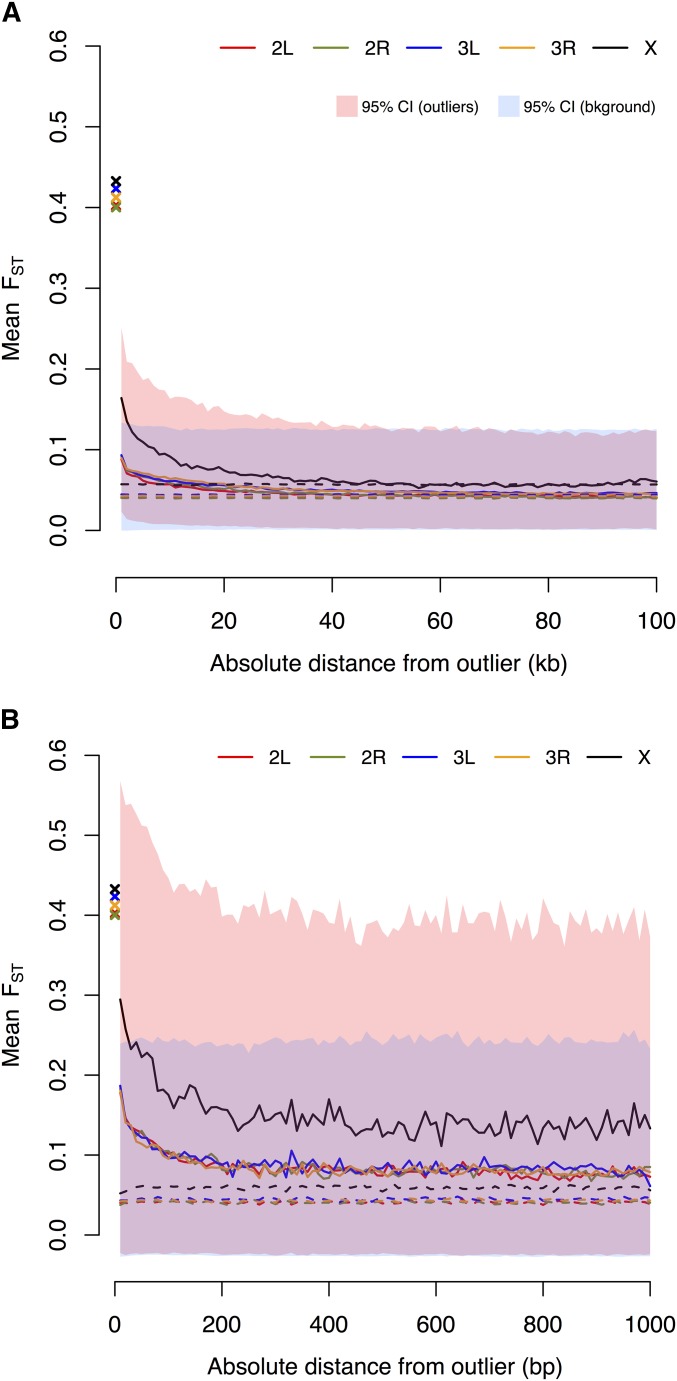
is expected to be correlated across closely linked SNPs, at least in part because of the effects of linked selection. To summarize the physical scale of differentiation that arises from this nonindependence, we measured mean as a function of distance from SNPs in the top 1% tail of . On a scale measured by 1-kb nonoverlapping windows, mean decays to background genomic levels within 60 kb in North America (Figure 1A) and on a scale of 100 kb in Australia (Figure S3A). Although lack of detailed information on recombination variation in D. simulans precludes a formal comparison of recombination rate and differentiation, the observed physical scale of genomic variation in recombination rate in D. melanogaster (Comeron et al. 2012) suggests that the relatively large scale of correlated could be influenced by genome-wide heterogeneity in recombination rate.
Figure 1.
(A) Mean in increments of nonoverlapping 1-kb windows as a function of distance from an outlier SNP in the top 1% tail. Crosses denote mean of outlier windows. Background values represent mean as a function of distance from 10,000 randomly selected nonoutlier SNPs. Confidence intervals are defined by the upper and lower 2.5% quantiles. (B) Decay measured in 10-bp nonoverlapping windows away from outliers in North America.
On a scale measured in 10-bp nonoverlapping windows, decays rapidly within 100 bp (Figure 1A). This smaller scale of decay is reminiscent of the scale of LD observed in D. melanogester (Langley et al. 2012) and is consistent with adaptation from standing variation. Although we cannot test the driving factors behind these heterogeneous scales of decay, it is clear that strongly differentiated SNPs do not occur independently throughout the genome.
Relative frequencies of derived alleles
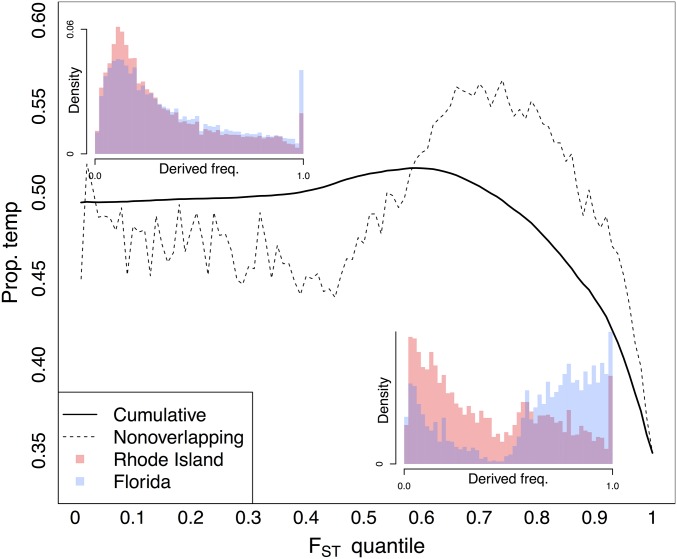
Under a model of a tropical D. simulans ancestral range, adaptation to temperate climates in recently established populations should generate a pattern, on average, of derived variants segregating at higher frequency at lower latitudes at strongly differentiated SNPs (e.g., Sezgin et al. 2004). We find that, on average, at the most differentiated SNPs, the derived allele was found to be segregating at a higher frequency in the lower-latitude population compared to the higher-latitude population. This pattern is present and significant (with P < 10−12 at the 99% cutoff; see Materials and Methods) on both continents but is more pronounced in North America (Figure 3 and Figure S9). Differences in the derived allele frequency for FL and RI populations for outlier SNPs reflect this observation, with a larger skew toward high-frequency derived alleles in FL for this subset of the genome.
Figure 3.
Proportion of derived alleles that are at higher frequency in higher-latitude populations as a function of in North America. Dotted line represents the proportion in nonoverlapping bins. Solid line represents the cumulative distribution of the dotted line. Left inset is the derived allele-frequency spectra for the two North American populations. Right inset is the genome-wide derived allele-frequency spectra for SNPs in the 0.99 tail. Only SNPs segregating at a total frequency > 0.05 in North America were considered.
To further investigate this pattern, we compared heterozygosities surrounding outlier SNPs between high- and low-latitude populations. Because selection reduces local variation within the genome (Maynard Smith and Haigh 1974; Charlesworth et al. 1993), we expected small regions that have large differences in heterozygosity between the two populations and also contain an outlier SNP to be the most likely to have experienced recent differential selection. We therefore measured the differences in heterozygosity in 100-bp nonoverlapping windows between the two populations on a given continent and identified windows that fell in the ±2.5% tail on either side of the distribution of population differences in (Figure S10). We then identified windows containing an outlier SNP (1%) and compared the number of such windows with smaller in the lower-latitude population to the number with smaller in the higher-latitude population. As shown in Figure S10, there are more windows that support recent adaptation in the lower-latitude population compared to the higher-latitude population (302 compared to 163 in North America; chi-squared test: P = 1.2 × 10−7, given an expectation scaled by the relative portion of low-heterozygosity windows).
Shared differentiation at SNPs
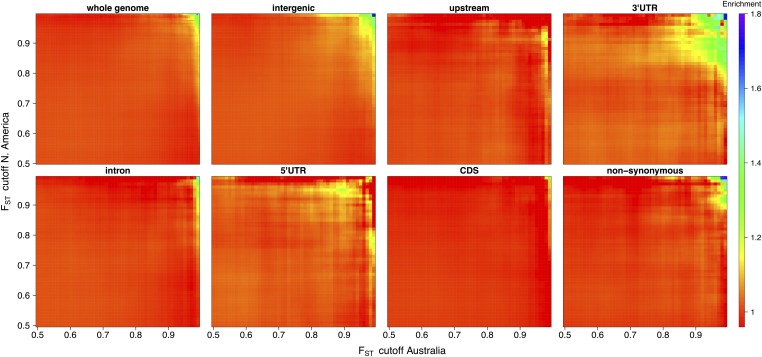
In the absence of shared differentiation between Australia and the United States, the proportion of shared outlier SNPs is expected to be roughly equal to the product of the proportions of SNPs defined as outliers on each continent (e.g., within the set of all shared SNPs, we expect 1% of SNPs to be found in the top 10% of on both continents). Because it is unknown a priori what an appropriate cutoff is, we evaluated this enrichment for a range of cutoffs (Figure 2). These results then were used to inform suitable outlier cutoffs of 5% in North America and 15% in Australia for downstream analyses. (Figure 3)
Figure 2.
Enrichment for number of shared-outlier SNPs for pairwise outlier quantiles increasing in 0.5% increments on both continents within given subsets of the genome. Values in the heat maps are cumulative (e.g., the 95th percentile is a subset of the 90th percentile).
Enrichment for shared-outlier SNPs increases as cutoffs for become more extreme, providing evidence for shared differentiation on a genome-wide scale (Figure 2). The statistical significance of this enrichment under the FET, however, is modest (Figure S5). The pattern of increased enrichment with cutoff persists at the scale of 100-bp and 1-kb windows (Figure S4), consistent with the scales of differentiation reported earlier.
In addition to an enriched sharing of outliers, if a variant is subject to latitudinally varying selection, then we expect the difference in allele frequency between low- and high-latitude populations to have the same sign on the two continents (referred to here as same-direction SNPs). In the absence of such parallel differentiation, the expectation is to observe approximately half the SNPs to be same-direction SNPs independent of . We tested for parallelism among shared outliers under a binomial model with probability 0.5 of a shared-outlier SNP being a same-direction SNP and did not observe a significant signal of same directionality and instead observed an enrichment of opposite-direction SNPs across many outlier cutoffs (Figure S6 and Figure S7).
To further investigate these patterns of enrichment and parallelism, we focused our analysis on different annotation categories (see Materials and Methods for details). Within each subset of the genome corresponding to a category, we conducted the same enrichment analyses. These indicate a potential signal of enriched parallel differentiation within the 3′ UTR regions of the genome (Figure 2) and are consistent with the observed expression-level differentiation found by Zhao et al. (2015). Consistent patterns of enriched parallel differentiation were not observed for other regions of the genome, but this could be due in part to limited power.
We now focus on same-direction SNPs that are found in both the top 5% tail in North America and the top 15% tail in Australia, referring to this subset of SNPs as same-direction shared outliers. This subset of SNPs was tested for associations with different annotation classes. As earlier, null distributions of enrichment values were generated by circular permutation to assess the significance of observed enrichments. We found a significant enrichment of same-direction shared-outlier SNPs in the 3′ UTR regions, consistent with the results of the parallel enrichment earlier (Figure S8) and similar to patterns reported by Kolaczkowski et al. (2011) and Reinhardt et al. (2014). Although significant enrichment is not observed for any other class, it is again possible that this can be attributed to insufficient power, especially considering that the number of shared-outlier SNPs in some annotation classes can be small.
Genes containing same-direction shared-outlier SNPs in the 3′ UTR, 5′ UTR, and upstream region or as a nonsynonymous variant are listed in File S1. While many of these genes have only one or two shared same-direction outliers, two genes, Dopamine transporter (DAT) and RpS14b, stand out for containing four or more differentiated SNPs within relatively short genomic regions in the UTR and upstream regions (and therefore may be associated with regulation of expression). Dopamine is a neurotransmitter that has many biological roles, one of which is the circadian rhythm (Hirsh et al. 2010), a clinally varying trait in D. melanogaster (Svetec et al. 2015). A mutation in D. melanogaster DAT is associated with decreased sleep duration and arousal threshold (Kume et al. 2005; Kume 2006), as well as metabolic rate and thermal preference and tolerance (Ueno et al. 2012). Little is known about the function of RpS14b. Another gene, lava lamp (lva), containing four same-direction nonsynonymous SNPs, is a golgin protein involved in transmembrane secretion during development (Sisson et al. 2000; Papoulas et al. 2005).
Differentiated genes
Convergence in adaptation is also possible through selection on different variants within the same gene (Rosenblum et al. 2010). Given the autocorrelation of the position of outlier SNPs, and because larger genes are by chance likely to contain an outlier SNP on both continents, we permuted (10,000 iterations) the positions among genic SNPs to assess the extent of enrichment of shared genes (see Materials and Methods). In this instance, new outlier gene sets were generated for North America, and the proportion of outlier genes shared with Australia was used as a measure of sharing. As earlier, the significance of the enrichment was evaluated by comparing the proportion of shared genes to the distribution generated by the permutations. We tested a range of cutoffs but found no significant enrichment of shared genes (when requiring P < 0.01). The strongest signal of enrichment is present at the 1% cutoffs on both continents, with P = 0.05. We compared this subset of genes to genes identified by Reinhardt et al. (2014) as differentiated on both continents in D. melanogaster and by Zhao et al. (2015) as differentially expressed between Maine and Panama populations of D. simulans (File S2). These genes, which are strongly differentiated on two continents in two species, may be among the most promising candidates for further study on potential targets of spatially varying selection in cosmopolitan Drosophlia.
Between-species parallelism in genes associated with insecticide resistance
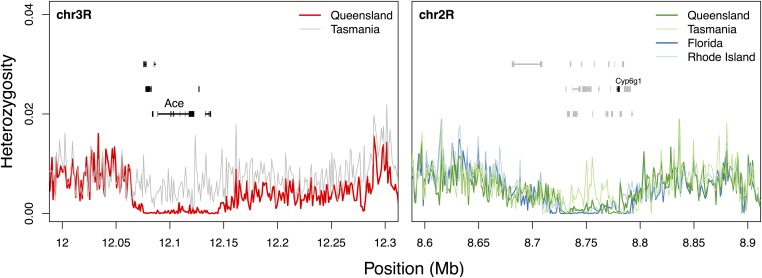
Two genes, Cyp6g1 and Acetylcholine esterase (Ace), known to be involved in resistance to insecticides in D. melanogaster, appear to have undergone a selective sweep in one or more D. simulans populations (Figure 4). The sweep in Cyp6g1 recapitulates the result of Schlenke and Begun (2004) and appears to be global, although the Tasmanian population appears to retain some diversity in this region. In contrast, the sweep surrounding Ace is apparent only in the QLD population. Kolaczkowski et al. (2011) found an overlapping region containing a putative copy-number variant segregating at higher frequency in QLD populations of D. melanogaster, pointing to the possibility that the two species are responding in a parallel manner to insecticides through different molecular mechanisms. This gene also was identified by Fabian et al. (2012) as a highly differentiated gene in North America. Both Ace and Cyp6g1 have been identified as targets of recent strong selection in D. melanogaster (Garud et al. 2015).
Figure 4.
Large regions of reduced diversity around known insecticide resistance loci, shown in nonoverlapping 1-kb windows. (Left) Region of reduced diversity surrounding Ace in Queensland. (Right) Region of reduced diversity around Cyp6g1.
In D. melanogaster, six Ace amino acid polymorphisms have been implicated in insecticide resistance (Fournier et al. 1992; Mutero et al. 1994). We looked for amino acid polymorphisms in D. simulans Ace genes that were fixed in QLD but at intermediate frequency in TAS and found three candidate residues that are identical to those found in D. melanogaster (Table S2). Moreover, these are due to the same DNA polymorphisms, presumably because there is only one substitution in the respective ancestral codons that can produce these specific amino acid polymorphisms. Such specificity in convergence has been observed in the Resistance to dieldrin (Rdl) gene, in which a replacement of Ala302 associated with cyclodiene resistance in D. melanogaster has been identified in multiple insect species (Ffrench-Constant et al. 2000). There is also evidence to suggest differentiation in expression of Ace between high- and low-latitude populations of D. simulans; the 3′ UTR region of the gene contains a same-direction shared SNP (File S1) and has been identified by Zhao et al. (2015) as a differentially expressed gene between Maine and Panama populations of D. simulans and D. melanogaster.
Reinhardt et al. (2014) reported a large continent-specific differentiated region surrounding Cyp6g1 in Australian populations of D. melanogaster. A nearby region in D. simulans shows continent-specific differentiation in Australia (Figure S2) but is located adjacent downstream of Cyp6g1. Because of this, it is unclear whether or not this is an example of parallel differentiation between species, and if it is, it raises the possibility that the common target is not Cyp6g1.
Gene ontology
For each continent, we performed independent GO enrichment analyses on the subsets of genes containing a SNP in the 1% and 4% tails in Australia and North America, respectively. To account for any bias in enrichment of GOs introduced by gene size, we permuted the values of SNPs present in annotated genes (see Materials and Methods). GOs that had a P-value (hypergeometric test) of <0.005 and a quantile value, based on circular permutation, of <0.01 are listed in File S3. One GO term—GO:0016021 (integral component of membrane)—is enriched on both continents.
Discussion
General patterns of differentiation
Here we have presented a genome-wide analysis of geographic variation in D. simulans to compare populations from high and low latitudes. Our results, consistent with previous studies of spatial variation in this species (Choudhary and Singh 1987; Singh 1989; Long and Singh 1992; Machado et al. 2015), indicate that on a genomic scale, is lower in D. simulans than in D. melanogaster, even when the effects of inversions are removed (Kolaczkowski et al. 2011; Fabian et al. 2012; Reinhardt et al. 2014). The X chromosome is an exception to this, with mean comparable to that of North American populations of D. melanogaster (Reinhardt et al. 2014). It should be noted, however, that differences in sampling, sequence quality, and criteria for retaining sites for analysis are seen between studies, casting some uncertainty on the interpretation of these comparisons.
Average was approximately twofold higher between the North American populations compared to the Australian populations. This genome-wide difference could be explained by a more recent colonization of Australia, lower rates of gene flow in North America, or demographic processes (e.g., a bottleneck) in North America. Pairwise comparisons of indicate that the FL population is the most differentiated compared to the others (Figure S11). Given that the FL population also has the lowest estimated genome-wide heterozygosity of the four populations, it is possible that some recent demographic history of the FL population may be contributing to the overall higher levels of differentiation observed in the North American samples, but technical effects related to library construction or sequencing cannot be ruled out. Our findings here are in contrast to those of Machado et al. (2015), who observed that their northernmost population sampled in Maine seemed to be an outlier relative to the other populations. Combined, these results support the role of local perturbation in shaping geographic patterns of variation in D. simulans.
Curiously, between North American populations of D. simulans, the X chromosome is most differentiated, but the converse is true in D. melanogaster, in which the X chromosome is the least differentiated arm (Kolaczkowski et al. 2011; Reinhardt et al. 2014; Machado et al. 2015). This is consistent with the results of Machado et al. (2015) and is perhaps a continent-specific effect because the X chromosome is not relatively more differentiated in Australia. While it seems likely that such a chromosome-wide effect could be due to demography, such as sex-biased dispersal, or extreme bottlenecks, these hypotheses cannot be addressed with the currently available data.
Within chromosomes, sites with high are not uniformly distributed. Mean around outliers decays to approximately 5% more than background levels within 200 bp (Figure 1), which is roughly consistent with the scale of LD in D. melanogater (and the therefore assumed scale of LD in D. simulans). However, mean decays completely to background levels on the much larger scale of 100 kb. This is consistent with large-scale heterogeneity of across the genome, perhaps associated with recombination-rate variation (Begun et al. 2007; Comeron et al. 2012). This pattern also could reflect reduced heterozygosity as a result of recent adaptation in one population resulting in elevated local . It is therefore possible that recent population-specific sweeps are contributing to larger-scale patterns of differentiation. This is distinct from differentiation due to selection against migrants, although an argument could be made for attributing most strong differentiation to differential selection if migration is sufficiently high in D. simulans. We did not observe megabase-scale regions of elevated such as those present in D. melanogaster, perhaps because of the lack of large-inversion polymorphisms in D. simulans.
Patterns of parallel differentiation
Parallelism and convergence can occur at many functional levels ranging from phenotype to nucleotide (Manceau 2010; Rosenblum et al. 2010). Here we examined potential patterns of parallelism in SNPs, genes, and GOs. The enrichment of shared SNPs at extreme cutoffs is consistent with the two continents sharing some mechanisms of local adaptation to latitudinally varying selective pressures. While it is difficult to assess how much of the excess sharing of outliers is driven by high caused by linkage to true targets of selection, which is likely to be driving some autocorrelation in outlier SNP positions, the decay we see on a relatively small scale (∼100 bp) provides support for some local adaptation from standing variation. The significant number of shared SNPs along similar outlier classes (along the diagonal of Figure S5) indicates that beyond the most differentiated sites, there is substantial correlation in the patterns of differentiation across the genome.
Among different genomic regions, the 3′ UTR regions have the strongest patterns of shared differentiation, consistent with differential selection acting on regulatory variation. This result is consistent with evidence from Zhao et al. (2015) of adaptive gene expression differentiation between Maine and Panama populations of D. simulans and with enrichment for clinal SNPs found by Machado et al. (2015). We detected relatively little evidence for between-continent parallel differentiation within other regions of the genome, but this does not necessarily indicate that structural variation or variation in other genomic regions does not play an important role in adaptation because these analyses reflect genome-wide patterns and also could be affected by a lack of power. Additionally, we note that the set of same-direction shared outliers found in 3′ UTR regions comprises a very small subset of the genome, and as such, these enrichment results should be treated with caution.
The modest enrichment of shared SNPs did not translate into a strong pattern of sharing at the gene or GO level: between the two continents, only one GO term—GO:0016021 (integral component of membrane)—is shared. While acknowledging the dangers of post hoc interpretation of GO analysis (Pavlidis et al. 2012), we note that within continents, terms such as GO:009416 (response to light) and GO:0045792 (negative regulation of cell size) are enriched because these could relate to phenotypic clines observed in traits that are influenced by circadian rhythm (Hut et al. 2013; Svetec et al. 2015) and body size (Zwaan et al. 2000) in Drosophila.
While we observed signals of parallel differentiation, we note that the enrichment across all levels seems at best weak, especially in comparison to patterns of differentiation in D. melanogaster (Fabian et al. 2012; Bergland et al. 2014; Reinhardt et al. 2014). This would suggest that while there may be some shared differentiation resulting from parallel adaptation, populations on the two continents are largely responding differently on the molecular level to their local environments. This is consistent with our current understanding of adaptation in Drosophila; given that many ecologically relevant phenotypes (e.g., body size) are likely to be polygenic, we should not expect to detect strong differentiation at loci of relatively small effect. This is especially true for this study, which investigates differentiation only between population pairs. However, in D. melanogaster, large inversions such as In(3R)P are likely to have a substantial effect on fitness and, accordingly, show strong patterns of parallel differentiation. Our results, in light of earlier findings of similar studies in D. melanogaster, reiterate the important contribution of inversion frequency clines in shaping patterns of shared differentiation and suggest that local adaptation from selection on large-effect loci may be relatively uncommon in D. simulans. Lastly, we note that incomplete annotation of the D. simulans genome, especially in comparison to D. melanogaster, may have influenced the results of all annotation-based analyses, mostly by reducing power to detect shared differentiation.
Recent adaptation in D. simulans
Although one objective of this study was to gain insight into potential mechanisms of local adaptation in D. simulans, it is possible that high between two populations is driven by reduced diversity in one population rather than selection against migrants, as described in classic cline models (Haldane 1948). Because D. simulans has a large population size, however, and because it is thought that rates of gene flow are high, we have assumed that the most differentiated sites are or are closely linked to targets of spatially varying selection. Even if we are detecting strong differentiation due to selection in a single population, these differentiated sites can provide valuable information about recent adaptation.
The population-specific sweep in a region surrounding Ace—a gene associated with insecticide resistance—is a case in which strong selection in one population (QLD) has driven high levels of differentiation. Given that there is no reduced diversity around Ace in TAS populations, the differentiation of SNPs within the region is likely to have been driven by differences in pesticide application in the Australian populations. While there is evidence that insecticide resistance can have a negative pleiotropic effect on fitness in the absence of insecticide (e.g., Lenormand et al. 1999), whether or not differentiation at this locus is maintained by selection against migrants or migration selection is in a nonequilibrium state remains unknown. Nevertheless, this speaks to the point that some portion of the differentiation we have observed may not be driven by latitudinally varying climatic factors but also may be influenced by much more localized variation in environment, such as agriculture. These local factors are unlikely to drive a signal of parallel latitudinal differentiation between continents and may perhaps account for some of the differences in differentiated loci between the two continents.
Very highly differentiated and nonsynonymous SNPs identified in Ace are associated with insecticide resistance in D. melanogaster, presenting a compelling example of convergent evolution between species at the nucleotide level. However, additional patterns of differentiation surrounding this gene indicate that there may be more to its role in adaptation: Zhao et al. (2015) reported that Ace in D. simulans and D. melanogaster is differentially expressed between Maine and Panama populations, and in our study, we identified a same-direction SNP in the 3′ UTR of the gene. Furthermore, Kolaczkowski et al. (2011) identified a putative duplication spanning Ace in D. melanogaster to be segregating at a higher frequency in QLD compared to TAS. This suggests that both structural and regulatory variation in Ace may be responding to selection in Drosophila.
Lastly, the signals of selection surrounding Ace provide evidence that patterns of variation are influenced by recent human activity. This is consistent with the findings of Wurmser et al. (2013) that some of the most pronounced differences in expression profiles of African and non-African D. simulans are potentially attributable to adaptation to insecticides outside of Africa and our observation that there is reduced variation around Cyp6g1 in all our sampled populations. It should be noted that the strong signals indicating responses to selection from insecticides reflect the effect sizes and initial frequencies of the loci contributing to resistance, and the fact that they are easily detected should not downplay the relative importance of adaptation to other environmental variables. Our understanding of patterns of genetic variation pertaining to other ecologically relevant traits will improve with a better understanding of the underlying mechanisms of ecologically important phenotypes.
Adaptation out of ancestral range
Given its ancestral range of East Africa/Madagascar (Lachaise et al. 1988; Dean and Ballard 2004; Kopp et al. 2006), we looked for evidence that D. simulans has been experiencing adaptation to temperate environments by comparing the frequencies of derived alleles in high- and low-latitude populations. We found that derived variants are at higher frequency in the lower-latitude populations (Figure 3) and that the diversity around high- SNPs is lower in these populations compared to the higher-latitude populations (Figure S10). Furthermore, the genome-wide mean heterozygosity on both continents is lower at lower latitudes. This is in contrast to observations in populations of D. melanogaster, which show reduced genomic diversity (Kolaczkowski et al. 2011; Fabian et al. 2012) and higher frequency of derived alleles in temperate populations for some strongly differentiated loci (Sezgin et al. 2004; Turner et al. 2008; Kolaczkowski et al. 2011; Reinhardt et al. 2014).
Combined, our results indicate that unlike D. melanogaster, there is little support that D. simulans is ancestrally tropical adapted with recent adaptation to temperate climates outside of Africa. Rather, the smaller population size and increased frequency of derived alleles in lower-latitude populations are consistent with these populations experiencing greater environmental stresses than their more temperate counterparts. This is supported by studies suggesting that D. simulans may be better adapted to cold temperatures (Petavy et al. 2001; Chakir et al. 2002) and less adapted to hot temperatures (Jenkins and Hoffmann 1994; Kellermann et al. 2012), although results across studies are somewhat equivocal in their conclusions (Parsons 1977; Boulétreau-Merle et al. 2003; David et al. 2004). Our own results, in contrast to the genomic results of Machado et al. (2015), would require confirmation from comparing patterns of diversity among additional populations along latitudinal clines.
While the mechanism may remain unclear, contrasting patterns between D. melanogaster and D. simulans emphasize potential differences in the biogeographic histories of the two species. While both are considered to be African in origin, the ancestral ranges may have differed substantially (Lachaise et al. 1988). Specifically, the D. simulans ancestral range is believed to be in Madacascar/East Africa (Dean and Ballard 2004; Rogers et al. 2014), while D. melanogaster is thought to have an ancestral range further to the west (Pool et al. 2012). It is conceivable that these two regions have historically experienced substantially different climates, leading to phenotypic differences between ancestral populations of the two species. It also has been proposed that there are substantial differences in adaptive strategies to similar environments between the two species (Choudhary and Singh 1987).
Continent-specific adaptation and clinal variation
The analysis presented here highlights the differences between two cosmopolitan species and suggests that within D. simulans, Australian and North American populations are adapting to their local environments via both shared and different mechanisms. These results point to several aspects of biology that are potentially important for local adaptation in this species, including regulation of expression, light response, and insecticide resistance.
With the current data set, we are likely to detect either genome-wide patterns or signatures of selection at specific loci of large effect. While this has provided us with some additional insight into recent adaptation in D. simulans, a substantially larger data set would be required to gain a deeper and more detailed understanding of the demographic and adaptive processes influencing this species. In light of this and our observation that much of the differentiation within continents is not shared between continents, it seems that dense sampling of a single clinal transect, perhaps with replicate clines within a continent, would be the best strategy for understanding the genetics of local adaptation. This also would address whether differentiation reflects a continuously varying environment or is influenced by local, discontinuous environmental heterogeneity. Lastly, we have assumed, like many others before us, that gene flow is high in D. simulans and that clines are stable (such that allele frequencies do not change substantially over time). Based on temporal sampling of a single population, Machado et al. (2015) found that while somewhat stable, allele-frequency clines in D. simulans are less stable than clines in D. melanogaster. Although evidence for temporally stable clines indicates that this assumption is appropriate for some variants, the extent to which this is true on a genome-wide scale will be addressed as data sets with denser temporal and spatial sampling become available.
Acknowledgments
We thank A. Hoffmann for providing us with D. simulans stock lines and Y. Brandvain, G. Coop, B. Cooper J. Cridland, N. Svetec, L. Zhao, T. Seher and anonymous reviewers for valuable insights and comments. This study was funded by a National Institutes of Health grant to D.J.B. (GM-084056) and a National Science Foundation Graduate Research Fellowship to A.S. (1148897).
Footnotes
Communicating editor: W. Stephan
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.185496/-/DC1.
Literature Cited
- Adrion J. R., Hahn M. W., Cooper B. S., 2015. Revisiting classic clines in Drosophila melanogaster in the age of genomics. Trends Genet. 31: 434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S., Gish W., Miller W., Myers E. W., Lipman D., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Anderson P. R., Oakeshott J. G., 1984. Parallel geographical patterns of allozyme variation in two sibling Drosophila species. Nature 308: 729–731. [Google Scholar]
- Andolfatto P., 2001. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 18: 279–290. [DOI] [PubMed] [Google Scholar]
- Arthur A. L., Weeks A. R., Sgrò C. M., 2008. Investigating latitudinal clines for life history and stress resistance traits in Drosophila simulans from eastern Australia. J. Evol. Biol. 21: 1470–1479. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Lemeunier F., 1976. Relationships within the melanogaster species subgroup of the genus Drosophila (Sophophora). I. Inversion polymorphisms in Drosophila melanogaster and Drosophila simulans. Proc. R. Soc. Lond. B Biol. Sci. 193: 137–157. [DOI] [PubMed] [Google Scholar]
- Begun D. J., Whitley P., 2000. Reduced X-linked nucleotide polymorphism in Drosophila simulans. Proc. Natl. Acad. Sci. USA 97: 5960–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun D. J., Holloway A. K., Stevens K., Hillier L. W., Poh Y.-P., et al. , 2007. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 5: e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland A. O., Behrman E. L., O’Brien K. R., Schmidt P. S., Petrov D. A., 2014. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 10: e1004775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulétreau-Merle J., Fouillet P., Varaldi J., 2003. Divergent strategies in low temperature environment for the sibling species Drosophila melanogaster and D. simulans: overwintering in extension border areas of France and comparison with African populations. Evol. Ecol. 17: 523–548. [Google Scholar]
- Capy P., Gibert P., 2004. Drosophila melanogaster, Drosophila simulans: so similar yet so different. Genetica 120: 5–16. [DOI] [PubMed] [Google Scholar]
- Chakir M., Chafik A., Moreteau B., Gibert P., David J. R., 2002. Male sterility thermal thresholds in Drosophila: D. simulans appears more cold-adapted than its sibling D. melanogaster. Genetica 114: 195–205. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Morgan M. T., Charlesworth D., 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary M., Singh R., 1987. A comprehensive study of genetic variation in natural populations of Drosophila melanogaster. III. Variations in genetic structure and their causes between Drosophila melanogaster and its sibling species Drosophila simulans. Genetics 117: 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo P. F., Hosemann K. E., Balabhadra S., Jr. G. V., Dickson M., et al. , 2005. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307: 1928–1933. [DOI] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. P., Peterson D. A., Biggs P. J., 2010. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David J. R., Allemand R., Capy P., Chakir M., Gibert P., et al. , 2004. Comparative life histories and ecophysiology of Drosophila melanogaster and D. simulans. Genetica 120: 151–163. [DOI] [PubMed] [Google Scholar]
- Dean M. D., Ballard J. W. O., 2004. Linking phylogenetics with population genetics to reconstruct the geographic origin of a species. Mol. Phylogenet. Evol. 32: 998–1009. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T., 1947. Adaptive changes induced by natural selection in wild populations of Drosophila. Evolution 1: 1–16. [Google Scholar]
- Dobzhansky T., 1948. Genetics of natural populations. XVI. Altitudinal and seasonal changes produced by natural selection in certain populations of Drosophila pseudoobscura and Drosophila persimilis. Genetics 33: 158–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler J. A., 1977. Geographic Variation, Speciation, and Clines. Princeton University Press, Princeton, NJ. [PubMed] [Google Scholar]
- Fabian D. K., Kapun M., Nolte V., Kofler R., Schmidt P. S., et al. , 2012. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol. Ecol. 21: 4748–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ffrench-Constant R., Anthony N., Aronstein K., Rocheleau T., Stilwell G., 2000. Cyclodiene insecticide reistance: from molecular to population genetics. Annu. Rev. Entomol. 48: 449–466. [DOI] [PubMed] [Google Scholar]
- Fournier D., Bride J.-M., Hoffmann F., Karch F., 1992. Two types of modifications confer resistance to insecticide. J. Biol. Chem. 267: 14270–14274. [PubMed] [Google Scholar]
- Futschik A., Schlötterer C., 2010. The next generation of molecular markers from massively parallel sequencing of pooled DNA samples. Genetics 186: 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garud N. R., Messer P. W., Buzbas E. O., Petrov D. A., 2015. Recent selective sweeps in North American Drosophila melanogaster show signature of soft sweeps. PLoS Genet. 23; 11(2): e1005004. DOI: 10.1371/journal.pgen.1005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., et al. , 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert P., Capy P., Imasheva A., 2004. Comparative analysis of morphological traits among Drosophila melanogaster and D. simulans: genetic variability, clines and phenotypic plasticity. Genetica 120: 165–179. [DOI] [PubMed] [Google Scholar]
- Haldane S., 1948. The theory of a cline. J. Genet. 48: 277–284. [DOI] [PubMed] [Google Scholar]
- Hallas R., Schiffer M., Hoffmann A. A., 2002. Clinal variation in Drosophila serrata for stress resistance and body size. Genet. Res. 79: 141–148. [DOI] [PubMed] [Google Scholar]
- Hirsh J., Riemensperger T., Coulom H., Iché M., Coupar J., et al. , 2010. Roles of dopamine in circadian rhythmicity and extreme light sensitivity of circadian entrainment. Curr. Biol. 20: 209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Weeks A. R., 2007. Climatic selection on genes and traits after a 100 year-old invasion: a critical look at the temperate-tropical clines in Drosophila melanogaster from eastern Australia. Genetica 129: 133–147. [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Anderson A., Hallas R., 2002. Opposing clines for high and low temperature resistance in Drosophila melanogaster. Ecol. Lett. 5: 614–618. [Google Scholar]
- Hohenlohe P. A., Bassham S., Etter P. D., Stiffler N., Johnson E. A., et al. , 2010. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 6: e1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T., Eisen M., Thornton K., Andolfatto P., 2013. A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 23: 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huey R. B., 2000. Rapid evolution of a geographic cline in size in an introduced fly. Science 287: 308–309. [DOI] [PubMed] [Google Scholar]
- Hut R. A., Paolucci S., Dor R., Kyriacou C. P., Daan S., 2013. Latitudinal clines: an evolutionary view on biological rhythms. Proc. R. Soc. B. 280: 20130433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins N., Hoffmann A., 1994. Genetic and maternal variation for heat resistance in Drosophila from the field. Genetics 137: 783–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermann V., Overgaard J., Hoffmann A. A., Fløjgaard C., Svenning J.-C., et al. , 2012. Upper thermal limits of Drosophila are linked to species distributions and strongly constrained phylogenetically. Proc. Nat. Acad. Sci. USA 109: 16228–16233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B., Kern A. D., Holloway A. K., Begun D. J., 2011. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187: 245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A., Frank A., Fu J., 2006. Historical biogeography of Drosophila simulans based on Y-chromosomal sequences. Mol. Phylogenet. Evol. 38: 355–362. [DOI] [PubMed] [Google Scholar]
- Kume K., 2006. A Drosophila dopamine transporter mutant, fumin (fmn), is defective in arousal regulation. Sleep Biol. Rhythms 4: 263–273. [Google Scholar]
- Kume K., Kume S., Park S. K., Hirsh J., Jackson F. R., 2005. Dopamine is a regulator of arousal in the fruit fly. J. Neurosci. 25: 7377–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D., Cariou M., David J., 1988. Historical biogeography of the Drosophila melanogaster species subgroup. Evol. Biol. 22: 159–225. [Google Scholar]
- Langley C. H., Stevens K., Cardeno C., Lee Y. C. G., Schrider D. R., et al. , 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192: 533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T., Bourguet D., Guillemaud T., Raymond M., 1999. Tracking the evolution of insecticide resistance in the mosquito Culex pipiens. Nature 400: 861–864. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A. D., Singh R. S., 1992. Geographic variation in Dorophila: from molecules to morphology and back. Trends Ecol. Evol. 7: 340–345. [DOI] [PubMed] [Google Scholar]
- Machado H., Bergland A. O., O’Brien K. R., Behrman E. L., Schmidt P., et al. , 2015. Comparative population genomics of latitudinal variation in D. simulans and D. melanogaster. Mol. Ecol. DOI: 10.1111/mec.13446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau M., 2010. Convergence in pigmentation at multiple levels: mutations, genes and function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favorable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- Mutero A., Pralavorio M., Bride J.-M., Fournier D., 1994. Resistance-associated point mutations in insecticide-insensitive acetylcholinesterase. Proc. Natl. Acad. Sci. USA 91: 5922–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., Hu T. T., Ishino Y., Jhaveri J., Toomajian C., et al. , 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby A. B., Blacket M. J., Hoffmann A. a., Schmidt P. S., 2010. Identification of a candidate adaptive polymorphism for Drosophila life history by parallel independent clines on two continents. Mol. Ecol. 19: 760–774. [DOI] [PubMed] [Google Scholar]
- Papoulas O., Hays T. S., Sisson J. C., 2005. The golgin Lava lamp mediates dynein-based Golgi movements during Drosophila cellularization. Nat. Cell Biol. 7: 612–618. [DOI] [PubMed] [Google Scholar]
- Parsons P., 1975a Phototactic responses along a gradient of light intensities for the sibling species Drosophila melanogaster and Drosophila simulans. Behav. Genet. 5: 17–25. [DOI] [PubMed] [Google Scholar]
- Parsons P., 1975b The comparative evolutionary biology of the sibling species, Drosophila melanogaster and D. simulans. Q. Rev. Biol. 50: 151–169. [DOI] [PubMed] [Google Scholar]
- Parsons P., 1977. Resistance to cold temperature stress in populations of Drosophila melanogaster and D. simulans. Aust. J. Zool. 25: 693–698. [Google Scholar]
- Pavlidis P., Jensen J. D., Stephan W., Stamatakis A., 2012. A critical assessment of storytelling: Gene Ontology categories and the importance of validating genomic scans. Mol. Biol. Evol. 29: 3237–3248. [DOI] [PubMed] [Google Scholar]
- Petavy G., David J., Gibert P., Moreteau B., 2001. Viability and rate of development at different temperatures in Drosophila: a comparison of constant and alternating thermal regimes. J. Therm. Biol. 26: 29–39. [DOI] [PubMed] [Google Scholar]
- Pool J. E., Corbett-Detig R. B., Sugino R. P., Stevens K. A., Cardeno C. M., et al. , 2012. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 8: e1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price T. R., Bretman A., Gradilla A. C., Reger J., Taylor M. L., et al. , 2014. Does polyandry control population sex ratio via regulation of a selfish gene? Proc. Biol. Sci. 281: 20133259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt J. A., Kolaczkowski B., Jones C. D., Begun D. J., Kern A. D., 2014. Parallel geographic variation in Drosophila melanogaster. Genetics 197: 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers R. L., Shao L., Sanjak J. S., Andolfatto P., Thornton K. R., 2014. Revised annotations, sex-biased expression, and lineage-specific genes in the Drosophila melanogaster group. G3 4: 2345–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum E. B., Römpler H., Schöneberg T., Hoekstra H. E., 2010. Molecular and functional basis of phenotypic convergence in white lizards at White Sands. Proc. Natl. Acad. Sci. USA 107: 2113–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke T., Begun D., 2004. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc. Natl. Acad. Sci. USA 101: 1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P. S., Paaby A. B., 2008. Reproductive diapause and life-history clines in North American populations of Drosophila melanogaster. Evolution 62: 1204–1215. [DOI] [PubMed] [Google Scholar]
- Sezgin E., Duvernell D. D., Matzkin L. M., Duan Y., Zhu C.-T., et al. , 2004. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics 168: 923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Choudhary M., David J., 1987. Contrasting patterns of geographic variation in the cosmopolitan sibling species Drosophila melanogaster and Drosophila simulans. Biochem. Genet. 25: 27–40. [DOI] [PubMed] [Google Scholar]
- Singh R. S., 1989. Population genetics and evolution of species related to Drosophila melanogaster. Annu. Rev. Genet. 23: 245–253. [DOI] [PubMed] [Google Scholar]
- Sisson J., Field C., Ventura R., 2000. Lava lamp, a novel peripheral golgi protein, is required for Drosophila melanogaster cellularization. J. Cell Biol. 151: 905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A., Dobzhansky T., 1936. Geographical distribution and cytology of “sex ratio” in Drosophila pseudoobscura and related species. Genetics 21: 473–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetec N., Zhao L., Saelao P., Chiu J. C., Begun D. J., 2015. Evidence that natural selection maintains genetic variation for sleep in Drosophila melanogaster. BMC Evol. Biol. 15: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Subramanian S., Kumar S., 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21: 36–44. [DOI] [PubMed] [Google Scholar]
- Turner T. L., Levine M. T., Eckert M. L., Begun D. J., 2008. Genomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics 179: 455–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Bourne E. C., Von Wettberg E. J., Hu T. T., Nuzhdin S. V., 2010. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat. Genet. 42: 260–263. [DOI] [PubMed] [Google Scholar]
- Tyukmaeva V. I., Salminen T. S., Kankare M., Knott K. E., Hoikkala A., 2011. Adaptation to a seasonally varying environment: a strong latitudinal cline in reproductive diapause combined with high gene flow in Drosophila montana. Ecol. Evol. 1: 160–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T., Tomita J., Kume S., Kume K., 2012. Dopamine modulates metabolic rate and temperature sensitivity in Drosophila melanogaster. PLoS One 7: e31513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser F., Mary-Huard T., Daudin J.-J., Joly D., Montchamp-Moreau C., 2013. Variation of gene expression associated with colonisation of an anthropized environment: comparison between African and European populations of Drosophila simulans. PLoS One 8: e79750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Wit J., Svetec N., Begun D. J., 2015. Parallel gene expression differences between low and high latitude populations of Drosophila melanogaster and D. simulans. PLoS Genet. 11: e1005184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaan B. J., Azevedo R. B., James C., Van ’t Land J., Partridge L., 2000. Cellular basis of wing size variation in Drosophila melanogaster: a comparison of latitudinal clines on two continents. Heredity 84: 338–347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genomic data are available as raw sequence reads from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under PRJNA307610.