Abstract
The advent of genome editing techniques based on the clustered regularly interspersed short palindromic repeats (CRISPR)–Cas9 system has revolutionized research in the biological sciences. CRISPR is quickly becoming an indispensible experimental tool for researchers using genetic model organisms, including the nematode Caenorhabditis elegans. Here, we provide an overview of CRISPR-based strategies for genome editing in C. elegans. We focus on practical considerations for successful genome editing, including a discussion of which strategies are best suited to producing different kinds of targeted genome modifications.
Keywords: Caenorhabditis elegans, CRISPR/Cas9, genome editing, WormBook
A fundamental goal of biological research is to understand the functions of genes. One common strategy for studying gene function is to observe the phenotypes of mutants to deduce the biological processes in which a gene participates and, sometimes, details of its mechanism of action. This basic idea is the foundation of classical genetics and also underlies reverse genetic approaches including RNAi. A second strategy is to observe the localization and dynamics of a gene's protein product within a cell or animal, either by antibody staining or by expressing a fluorescent protein (FP) fusion. Together, these two basic strategies form the backbone of much research in Caenorhabditis elegans and other model systems.
The use of the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system for genome engineering (Hsu et al. 2014) has greatly facilitated the study of gene function in Caenorhabditis elegans and other organisms. By making precisely targeted mutations in endogenous genes, an investigator can examine the relationship between gene function and phenotype. By inserting coding sequence for a fluorescent protein, the expression and localization of endogenous proteins can be monitored. In both cases, one avoids the caveats of overexpression and silencing that are associated with conventional transgenes. Moreover, for fluorescent protein (FP) fusions, insertion of the FP into the endogenous locus allows one to use phenotypic assays to quickly determine whether the resulting fusion protein is functional. Together, these advantages permit more carefully controlled experiments to be done and thus allow greater confidence in the results. As an added benefit, current CRISPR-based approaches (Arribere et al. 2014; Dickinson et al. 2015; Paix et al. 2015; Ward 2015) are faster and require less labor than either conventional transgenesis (Mello et al. 1991) or microparticle bombardment (Praitis et al. 2001), and they eliminate the need for specialized strain backgrounds that are required for these methods and those based on the Mos1 transposon (Robert and Bessereau 2007; Frøkjaer-Jensen et al. 2008, 2010, 2012).
Many different CRISPR approaches have been developed for C. elegans and are being widely adopted by the research community. In general, all of these methods work well, with different strategies being best suited to different experimental goals. By choosing the appropriate strategy, one can now make essentially any desired change to the C. elegans genome in a matter of days to weeks, with <1 day of hands-on labor (Dickinson et al. 2013, 2015; Arribere et al. 2014; Zhao et al. 2014; Paix et al. 2015; Ward 2015). The goal of this article is to aid users in choosing the best strategy for a given application. We provide an overview of CRISPR-based methods for C. elegans, including a discussion of which strategies are most appropriate for generating different kinds of modifications.
Overview of the CRISPR-Cas9 system
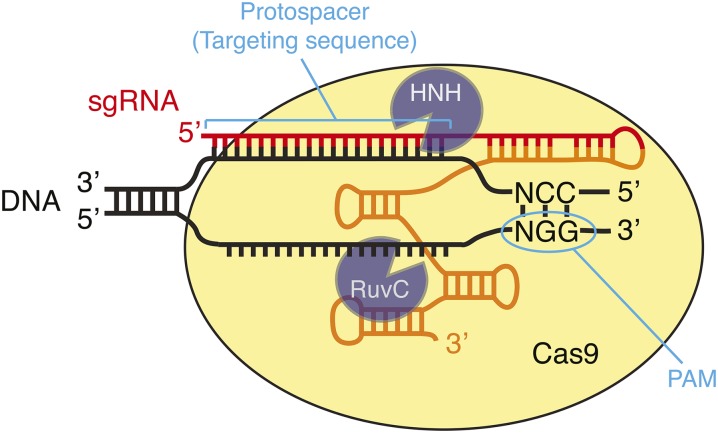
Cas9 is an endonuclease found in Archaea and some bacteria, where it is involved in adaptive immunity against phages and plasmids (Hsu et al. 2014). Unlike restriction endonucleases, whose protein structures recognize particular DNA sequences (e.g., EcoRI recognizes GAATTC), the specificity of Cas9 is determined by the sequence of an associated small RNA molecule (Figure 1) (Jinek et al. 2012). In its native context, bacterial Cas9 binds two small RNAs: a CRISPR RNA (crRNA) that determines target specificity and a trans-activating CRISPR RNA (tracrRNA) that base pairs with the crRNA and activates the Cas9 enzyme. The two RNA molecules can be fused to generate a chimeric single guide RNA (sgRNA) that supports Cas9 cleavage of DNA substrates (Jinek et al. 2012). The 20-bp guide sequence at the 5′ end of the sgRNA directly determines the sequence cleaved by Cas9, by forming Watson–Crick base pairs with the DNA target (Figure 1). In addition to this base-pairing interaction, Cas9 must interact with a protospacer-adjacent motif (PAM) on the target DNA molecule. The PAM sequence NGG is recognized by Streptococcus pyogenes (Sp) Cas9, which is the Cas9 most frequently used in the laboratory. Thus, SpCas9 can be programmed to cleave any desired nucleotide sequence that contains a GG dinucleotide, by simply changing the sequence at the 5′ end of the sgRNA. It is this ease of programming that makes Cas9 such a powerful and flexible tool for genome engineering.
Figure 1.
DNA recognition by the Cas9–sgRNA complex. Cas9 identifies its substrates by first binding to the PAM (NGG motif) and subsequently by base pairing of the sgRNA cofactor to the substrate DNA. HNH and RuvC are the two Cas9 nuclease domains that cleave the sgRNA-complementary and noncomplementary strand of the target DNA, respectively. Red and orange in the sgRNA indicate the portions derived from the bacterial crRNA and tracrRNA, respectively.
More recently, engineered derivatives of SpCas9 have been described that recognize alternate PAMs including NGA, NGAG, and NGCG (Kleinstiver et al. 2015), and some of these have been tested and shown to be effective in C. elegans (Bell et al. 2015). Cas9 homologs from bacterial species other than S. pyogenes have also been found to recognize alternate PAMs (Ran et al. 2015). Also, the unrelated CRISPR nuclease Cpf1 recognizes its targets differently from Cas9 and has been successfully used for genome editing in mammalian cells (Zetsche et al. 2015). Although Cpf1 and non-Sp Cas9 homologs have not yet been tested in C. elegans to our knowledge, it seems likely that a growing collection of RNA-guided nucleases recognizing a wider variety of PAMs than the conventional NGG will become available in the next few years.
It is important for a user of Cas9 to have some understanding of the different roles that the guide sequence and PAM play in determining Cas9 specificity. When searching for a substrate, Cas9 first binds to the PAM and only then interrogates the adjacent DNA to look for a match to the guide sequence (Sternberg et al. 2014). Thus, even DNA sequences that perfectly match the guide sequence are not recognized or cleaved if they do not contain a PAM. The requirement for an NGG PAM sequence appears fairly stringent (Jinek et al. 2012; Kuscu et al. 2014; Sternberg et al. 2014; Wu et al. 2014), although an NAG sequence may be able to support low-efficiency cleavage in some instances (Hsu et al. 2013; Jiang et al. 2013). In contrast to its strict requirement for the PAM sequence, Cas9 is somewhat tolerant of mismatches between the guide sequence and the target, especially when they occur near the 5′ end of the guide sequence (i.e., distal to the PAM) (Jinek et al. 2012; Fu et al. 2013; Hsu et al. 2013; Pattanayak et al. 2013; Kuscu et al. 2014; Ren et al. 2014; Wu et al. 2014). The practical consequences of this mismatch tolerance are discussed in Addressing Cas9 Specificity, below.
Genome engineering via double-strand break repair
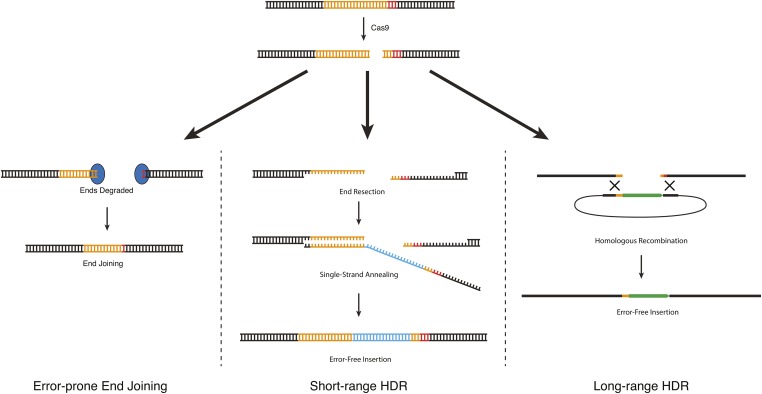
As described above in Overview of the CRISPR-Cas9 system, Cas9 can be used to generate a DNA double-strand break at a defined location in the genome. These double-strand breaks are useful because they allow an investigator to make use of endogenous cellular DNA repair machinery to generate custom modifications in the genome. Three different types of DNA repair strategies have been used to produce custom modifications in C. elegans (Figure 2):
Figure 2.
DNA repair approaches for CRISPR-based genome engineering. DNA double-strand breaks introduced by Cas9 can be repaired via three different mechanisms. End joining produces random insertion/deletion mutations. HDR produces error-free edits using an exogenous DNA molecule as a repair template. Although the mechanisms of HDR in C. elegans are not known, efficiency data suggest the existence of two different HDR pathways (see text). Short-range HDR is hypothesized to occur via a synthesis-dependent strand-annealing mechanism and can accommodate insertions of up to 1–2 kb, with the highest efficiency within 10 bp of the cut site. Long-range HDR is hypothesized to occur via a double-crossover mechanism and can accommodate insertions of at least 12 kb, at distances up to at least 1 kb from the cut site.
Error-prone repair via end joining: When Cas9 cleaves genomic DNA, some of the resulting DNA double-strand breaks are repaired by an error-prone pathway that produces small insertions or deletions (indels) at the site of the break. Mechanistically, these indels arise not via canonical nonhomologous end joining (NHEJ) as had been widely assumed, but from an alternative end-joining pathway that requires DNA polymerase Θ (van Schendel et al. 2015). When generated in protein-coding sequence, indels can shift the reading frame, resulting in a premature stop codon. Thus, error-prone repair can be used to produce loss-of-function alleles (C. Chen et al. 2013; Chiu et al. 2013; Cho et al. 2013; Friedland et al. 2013; Katic and Großhans 2013; Lo et al. 2013; Waaijers et al. 2013).
-
Homology-directed repair: In homology-directed repair (HDR), an exogenous DNA molecule is introduced along with Cas9 and serves as a template for DNA repair. Modifications present in the repair template are copied into the genome in an error-free manner. Different kinds of repair templates have been reported to yield different HDR efficiencies in C. elegans (Arribere et al. 2014; Paix et al. 2014, 2015; Dickinson et al. 2015; Ward 2015). For insertions up to ∼1 kb, repair was most efficient when the repair template contained 30–40 bp of homology to the genome, and longer homology arms led to reduced efficiency (Paix et al. 2014). On the other hand, insertions of ∼6 kb were readily obtained using 500- to 700-bp homology arms, but occurred rarely or not at all when using 30- to 40-bp homology arms (Dickinson et al. 2015). Based on these observations, there appear to be two distinct HDR pathways in C. elegans, which we call short-range HDR and long-range HDR. For convenience, we discuss these two repair pathways as if they occur via different mechanisms (as proposed in Figure 2), although the actual molecular mechanisms are not yet known.
2a. Short-range HDR is a highly local repair mechanism that occurs most efficiently within10 bp of the Cas9 cut site (Arribere et al. 2014; Paix et al. 2015; Ward 2015) and when the repair template carries 30- to 40-bp homology arms flanking the desired modification (Arribere et al. 2014; Paix et al. 2014; Ward 2015). Short-range HDR can be very efficient in C. elegans: in the best cases, >50% of F1 progeny that received active Cas9 and the repair template can carry short-range HDR events. Short-range HDR can be used to introduce point mutations, precise deletions, and small epitope tags by using a single-stranded DNA oligonucleotide as the repair template (Paix et al. 2014; Zhao et al. 2014). Larger insertions such as GFP insertion can also be made via short-range HDR, using a PCR product as the repair template (Paix et al. 2014, 2015). The main advantages of short-range HDR are its high efficiency and the fact that only 30–40 bp of homology to the genome are required for efficient repair. Short-range HDR has two main limitations. First, it occurs most efficiently within 10 bp of a Cas9 cleavage site, and efficiency declines sharply at larger distances (Arribere et al. 2014; Paix et al. 2015; Ward 2015). This can make it challenging to isolate edits that are not located near an efficient sgRNA target site (see Choosing a Cas9 target site, below, for a discussion of factors governing sgRNA efficiency). Second, for reasons that remain unclear, short-range HDR cannot accommodate insertions much larger than 1–2 kb (Dickinson et al. 2015; Paix et al. 2015); thus, short-range HDR is suitable for GFP insertion but not for larger-scale modifications.
2b. Long-range HDR allows insertion of much larger sequences [at least 12 kb (Das et al. 2015)] and at a greater distance from the cut site [at least 1 kb (Dickinson et al. 2013; Das et al. 2015; Sullivan-Brown et al. 2016)]. Plasmids carrying 500–1500 bp of genomic homology flanking the desired modifications are robust substrates for this repair mechanism (Dickinson et al. 2013, 2015). On a per-F1 basis, long-range HDR is much less efficient than short-range HDR; however, because it can accommodate larger inserts, long-range HDR allows use of selectable markers, which offset the lower efficiency by facilitating quick and easy identification of repair events. Long-range HDR is relatively insensitive to variations in sgRNA efficiency (Dickinson et al. 2015), presumably because the repair process itself, rather than Cas9 cleavage, is the limiting factor.
The different properties of short-range vs. long-range HDR influence both the experimental design and the types of modifications that each strategy is best suited to generate, as discussed in the following sections.
Four basic steps for genome engineering with Cas9
To generate custom genome modifications with CRISPR/Cas9 in any experimental system, one must accomplish four basic tasks: (1) introduce Cas9 and an appropriately targeted sgRNA; (2) if using HDR, supply the appropriate repair template; (3) identify the animals that carry the desired genome modification; and (4) address specificity, since Cas9 can generate off-target mutations under some conditions.
The next four sections discuss how each of these steps can be accomplished in C. elegans. Then, in Recommended Strategies for Different Types of Modifications, we recommend approaches to generate different kinds of custom alleles with minimal time and labor.
Using Cas9 to Generate DNA Double-Strand Breaks
Expression of Cas9 and sgRNA in C. elegans
Cas9 can be easily expressed in the C. elegans germline by injecting an expression plasmid (Dickinson et al. 2013; Friedland et al. 2013) or messenger RNA (mRNA) (Chiu et al. 2013; Katic and Großhans 2013; Lo et al. 2013). Alternatively, purified Cas9 protein may be reconstituted with its RNA cofactors and directly injected into the gonad of the worm (Cho et al. 2013; Paix et al. 2015). For plasmid-based germline expression of Cas9, the eft-3 promoter (Frøkjaer-Jensen et al. 2012) has been widely used. By substituting heat-shock or tissue-specific promoters for Peft-3, it is possible to generate indels in somatic tissue, producing tissue specific loss-of-function phenotypes (Liu et al. 2014; Shen et al. 2014). Generally speaking, it appears that transgenic expression of Cas9 can be easily achieved using the same basic approaches that are well established for other transgenes.
Similarly, sgRNA can be either expressed from a plasmid or synthesized in vitro and injected. A third option is to feed the worms bacteria expressing sgRNA, which has low efficiency compared to other methods but may be useful for high-throughput screening (Liu et al. 2014). Plasmid-based sgRNA expression constructs use a U6 promoter, which directs transcription by RNA polymerase III (C. Chen et al. 2013; Dickinson et al. 2013; Friedland et al. 2013). U6 snRNA is an essential component of the mRNA splicing machinery and thus would be expected to be ubiquitously expressed. Consistent with this prediction, PU6::sgRNA constructs have been successfully used to produce mutations both in the germline and in somatic tissues (C. Chen et al. 2013; Dickinson et al. 2013; Friedland et al. 2013; Shen et al. 2014). Note that two independently identified U6 promoters have been used in published work (Dickinson et al. 2013; Friedland et al. 2013). Although CRISPR mutations have been successfully isolated using sgRNAs expressed from both promoters, two studies have reported conflicting observations of higher efficiency with one promoter or the other (Farboud and Meyer 2015; Katic et al. 2015), suggesting that the choice of promoter might influence editing efficiency in some cases. For direct RNA injection, the RNA may be synthesized or purchased commercially. Note that if RNA is chemically synthesized commercially, it is more cost-effective to purchase separate crRNA and tracrRNA rather than the longer chimeric sgRNA, because only the crRNA is specific to a given experiment, while the tracrRNA sequence is constant.
The choice of whether to use plasmid-based Cas9 and sgRNA expression or direct Cas9 and RNA injection will depend on the needs of each individual user. Plasmid injection is simple, reliable, and familiar to most C. elegans researchers. However, this approach requires cloning each new guide sequence into an expression construct, and a relatively large number of animals (∼50–60 in our experience) need to be injected to consistently obtain the desired modification. Direct injection of Cas9 ribonucleoprotein complexes yields a higher frequency of successful injections compared to plasmid-based expression, thus reducing the number of animals that need to be injected to as few as 10 (Paix et al. 2015). The trade-off is that the user must either purchase Cas9 protein and the required small RNAs or purify them in house. Purchasing Cas9 protein, tracrRNA, and crRNA is currently quite costly (∼$200 per target, with most of the cost going to the synthetic RNAs), but the cost may drop as more commercial sources become available, and the ability to inject fewer worms may justify the cost for some users.
Cas9 and sgRNA expression plasmids from several different laboratories are available from Addgene (http://www.addgene.org/CRISPR/worm/). Escherichia coli expression vectors for producing Cas9 protein are also available (http://www.addgene.org/crispr/bacteria/).
Choosing a Cas9 target site
The first step in any CRISPR strategy is to choose the Cas9 target site. First, one needs to identify the general region to be targeted. To generate loss-of-function indel mutations, one should target a region close to the 5′ end of the coding region of the gene of interest, to maximize the chances that an indel will abolish the function of the gene. For HDR-based strategies, it is best to choose a site as close as possible to where the desired modification will be made.
Once the general region to be targeted has been identified, the next step is to identify the actual guide sequence within the target region. Three considerations govern the choice of a guide sequence: activity, specificity, and proximity to the desired modification. The relative importance of these considerations depends on the repair mechanism and screening strategy being used (see Strategies for Identifying CRISPR Modifications, below, for discussion of screening strategies). For long-range HDR with a selectable marker, specificity is the primary concern; for short-range HDR, activity and proximity to the desired modification are more important.
Specificity:
Ideally, one should select a guide sequence that is unique in the genome, to minimize the chances of generating off-target mutations. We identify specific guide sequences, using a CRISPR design tool developed by Feng Zhang’s laboratory (Hsu et al. 2013) and available at http://crispr.mit.edu. This tool lists all possible guide sequences within a 100- to 200-bp target region and identifies potential off-target cleavage sites for each guide. Each guide is assigned a specificity score from 0 to 100 (with a score 100 indicating perfect specificity). In our experience, for most 100- to 200-bp target regions in the C. elegans genome there are at least two to three possible guides with a score >95, indicating very good specificity. If more than one highly specific guide is available, we choose from among these based on predicted activity and proximity to our desired modification.
Activity:
Different guide sequences support different Cas9 cleavage efficiencies (Doench et al. 2014; Wang et al. 2014; Farboud and Meyer 2015; Xu et al. 2015). Whether cleavage efficiency is an important experimental consideration depends on the screening strategy being used (see Strategies for Identifying CRISPR Modifications, below). When using long-range HDR with a selectable marker, variations in cleavage efficiency are of no practical consequence because the repair process, rather than Cas9 cleavage efficiency, is the limiting factor. On the other hand, for short-range HDR, cleavage activity is a critical determinant of efficiency, and so it may be worthwhile to choose a slightly less specific guide to achieve higher cleavage efficiency.
Guide sequences ending in GG (not to be confused with the NGG PAM motif) have been shown to have consistently high cleavage efficiency in C. elegans (Farboud and Meyer 2015). However, these “3′GG guides” occur only once every 128 bp in random sequence (and even more infrequently in the AT-rich C. elegans genome), so using a 3′GG guide is usually not feasible. As an alternative, several prediction algorithms have been developed that may be useful for identifying the most active guide sequences (Doench et al. 2014; Liu et al. 2015; Xu et al. 2015). As of this writing, our preferred prediction tool is SSC, which is available at http://crispr.dfci.harvard.edu/SSC/. In general, guide sequences that are rich in G residues and lack pyrimidines in the last four bases before the PAM tend to be most active. Guides containing four or more consecutive T/U bases should be avoided, as these stretches can prematurely terminate PolIII transcription. Cleavage efficiency can also be improved by using an engineered sgRNA, termed sgRNA(F+E) (B. Chen et al. 2013; Ward 2015).
Proximity to the desired modification:
For short-range HDR using an oligonucleotide or PCR product repair template (see PCR screening and Co-CRISPR), the Cas9 target site should ideally be within 10 bp of the desired modification (Arribere et al. 2014; Paix et al. 2014, 2015). It is sometimes necessary to choose a less specific and/or less active guide to achieve this degree of proximity. For long-range HDR with a selectable marker (see Positive selectable markers), proximity is much less important, since efficient editing can be achieved at least 1 kb from the Cas9 target site (Dickinson et al. 2013; Das et al. 2015; Sullivan-Brown et al. 2016).
Once a guide sequence has been selected, it must be either cloned into an appropriate sgRNA vector (a U6 promoter vector for plasmid-based expression in C. elegans or a T7 promoter vector for in vitro transcription) or synthesized commercially for direct injection. The U6 promoter requires a G residue as the first base of the sgRNA sequence to initiate transcription, while for the T7 promoter, the sgRNA should typically begin with GG. If these guanine residues are not present in the chosen guide sequence, they can either be substituted for the most 5′ residues in the guide, since mismatches at these positions are well tolerated (Jinek et al. 2012; Fu et al. 2013; Hsu et al. 2013; Pattanayak et al. 2013; Kuscu et al. 2014; Ren et al. 2014; Wu et al. 2014), or simply appended to the 5′ end of the guide, since extensions of the sgRNA beyond 20 bp do not affect cleavage activity (Ran et al. 2013; Farboud and Meyer 2015). Both of these approaches have succeeded in our laboratory.
Strategies for Identifying CRISPR Modifications
Choosing an appropriate selection or screening approach is perhaps the most critical step in planning a new CRISPR genome modification. Different strategies have been devised that vary greatly in their applicability, efficiency, and difficulty. Each approach has strengths that are appropriate for different applications. We summarize each strategy here; Recommended Strategies for Different Types of Modifications provides recommendations for which strategy to use for different applications.
Screening based on mutant phenotype
The first demonstrations of Cas9-induced mutations in C. elegans involved simple visual screening for obvious mutant phenotypes such as Dpy or Unc, benomyl resistance conferred by ben-1 mutations, or loss of fluorescence from a bright GFP transgene (C. Chen et al. 2013; Chiu et al. 2013; Cho et al. 2013; Friedland et al. 2013; Katic and Großhans 2013; Lo et al. 2013; Waaijers et al. 2013). While these were useful proof-of-principle experiments, many genome modifications that are of biological interest do not confer a visible plate-level phenotype. Nevertheless, phenotype-based screening for edits at one locus can be used to enrich for edits at a second locus in “co-CRISPR” approaches (described in Co-CRISPR section).
There have also been reports of isolation of GFP knock-in strains by visually screening for fluorescence of the introduced GFP (Kim et al. 2014; Paix et al. 2014, 2015). Although fluorescence-based screening can clearly be effective in these reported cases, it requires that the gene being tagged be expressed at a high enough level that the GFP fusion protein is easily visible on a dissecting microscope at reasonably low magnification. Fluorescence-based screening is also greatly facilitated when the pattern of protein expression is known in advance. In our experience, the majority of C. elegans genes do not meet these criteria, and so screening for knock-ins based on visual examination of fluorescence is not an advisable strategy in general. It is possible in principle that dimmer knock-ins could be isolated using a flow-sorting system (Pulak 2006) or another automated system, but we are unaware of any reports of such an approach.
PCR screening
For mutations that do not produce an obvious plate phenotype, directly screening the F1 progeny of injected animals by single-worm PCR is the simplest, but also by far the most labor-intensive strategy. Several hundred F1 animals are singled to new plates, allowed to lay eggs, and then processed for PCR to identify animals heterozygous for the desired genome modification. F2 progeny of positive F1’s are then singled and the process is repeated to identify homozygotes. Direct PCR screening has now been essentially replaced by co-CRISPR (see Co-CRISPR section), which greatly reduces the number of animals that need to be screened.
Primer design for PCR screening depends on the nature of the genome modification (Paix et al. 2014). HDR insertions large enough to accommodate a PCR primer can be detected using a primer inside the insertion and a second primer outside the homology arm. Large deletions can be detected with flanking primers. For small indels or point mutations, it is best if the mutation introduces (or deletes) a unique restriction site, which enables screening by restriction fragment length polymorphism (RFLP). When performing HDR, a restriction site can often be introduced into the repair template by making silent substitutions in addition to the mutation of interest. If RFLP is not possible, the final choice is to screen by looking for a mobility shift of PCR products on polyacrylamide gels (Kim et al. 2014) or by using a nuclease that detects mismatches when wild-type and mutant PCR products are annealed (Cong et al. 2013; Ward 2015).
Co-CRISPR
Co-CRISPR (Arribere et al. 2014; Kim et al. 2014; Ward 2015) is a screening strategy that uses a visible phenotype at one locus to help identify edits at a second locus. Two loci are edited simultaneously: the locus of interest and an unlinked marker locus that produces a visible phenotype (Figure 3). The marker mutation is used to identify F1 animals derived from oocytes that received active Cas9. Among all F1 progeny of injected animals, those that received active Cas9 are most likely to carry the desired modification (Arribere et al. 2014; Kim et al. 2014; Ward 2015). By restricting PCR screening to these animals, co-CRISPR can substantially reduce the number of animals that need to be screened [to only a few dozen in the best cases (Farboud and Meyer 2015; Paix et al. 2015)]. Co-CRISPR is the screening strategy of choice for modifications generated using short-range HDR.
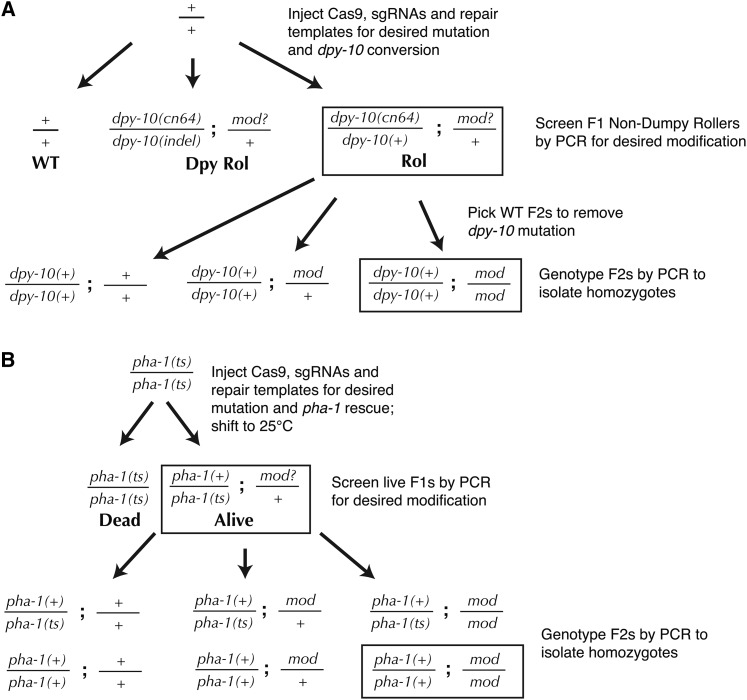
Figure 3.
Genetic schemes for co-CRISPR screening. (A) dpy-10 co-CRISPR (Arribere et al. 2014) makes use of the dominant Roller phenotype conferred by the cn64 mutation to identify a desired modification at an unlinked locus. Because dpy-10(cn64)/dpy-10(o) animals have a different phenotype than dpy-10(cn64)/+, a wild-type copy of dpy-10 can be carried through the screening, eliminating the need for outcrossing to remove the marker mutation. The sqt-1(e1350) marker mutation can be used in place of dpy-10(cn64). (B) pha-1 co-CRISPR (Ward 2015) uses repair of the temperature-sensitive lethal mutation pha-1(e2123) for live/dead screening of F1’s. No outcrossing is required because the converted pha-1 allele is wild type; however, F2’s need to be PCR genotyped for both the desired modification and pha-1(+).
For co-CRISPR to work well, the desired modification needs to occur with high efficiency relative to the marker mutation; if the marker mutation is efficient but the desired modification is inefficient, most marked F1’s will lack the mutation of interest. Thus, co-CRISPR is best suited to generating modifications that are (1) produced by short-range HDR, which is efficient on a per-F1 basis; (2) induced by a highly active sgRNA; and (3) introduced as close as possible to the cut site.
Several different marker mutations have been tested for co-CRISPR applications (Arribere et al. 2014; Kim et al. 2014; Ward 2015). The most effective of these are the gain-of-function dpy-10(cn64) and sqt-1(e1350) mutations (Figure 3A) (Arribere et al. 2014) or rescue of the temperature-sensitive lethal pha-1(e2123) mutation (Figure 3B) (Ward 2015). Since these marker mutations produce dominant phenotypes, they can be recognized in the F1 progeny of the injected animals, which are then screened by PCR (see PCR screening section) to identify animals carrying the desired modification. Then, F2 progeny of successfully edited animals are genotyped to identify homozygotes. During this F2 screening step, the dpy-10(cn64) or sqt-1(e1350) marker mutations can be eliminated by picking wild-type animals (Figure 3A), provided the desired edit and marker mutation are unlinked. When using pha-1 for co-CRISPR, the marker “mutation” is the wild-type allele of pha-1, which must be genotyped along with the desired mutation to identify homozygotes (Figure 3B).
A unique advantage of co-CRISPR compared to other screening strategies reported to date is the ability to multiplex: that is, to simultaneously generate edits at two different loci (Paix et al. 2015; Ward 2015) or two different edits at a single locus (Paix et al. 2014) from one batch of injections. Although one can also obtain doubly edited worms by editing two loci sequentially (for example, Arribere et al. 2014) or by generating two alleles separately and then crossing them together, multiplexing may save time in some cases.
Positive selectable markers
To identify genome modifications produced by long-range HDR, a selectable marker is typically introduced into the genome along with the desired modifications. Selection allows one to interrogate all progeny from a batch of injections (on the order of 10,000 in a typical experiment) without PCR screening, and thus it is the least labor-intensive strategy for identifying relatively rare long-range HDR events. Selection has a very high success rate in our experience (>95% of projects have succeeded in producing the desired edit, with ∼80% of these succeeding on the first batch of injections, for >50 different loci targeted in our laboratory). The high success rate is probably due to at least two factors. First, selection-based strategies use the long-range HDR mechanism, which is insensitive to variations in sgRNA efficiency (Dickinson et al. 2015) and to distance from the cut site up to at least 1 kb (Dickinson et al. 2013; Das et al. 2015; Sullivan-Brown et al. 2016; and our unpublished results). Second, selection allows recovery even of rare edits.
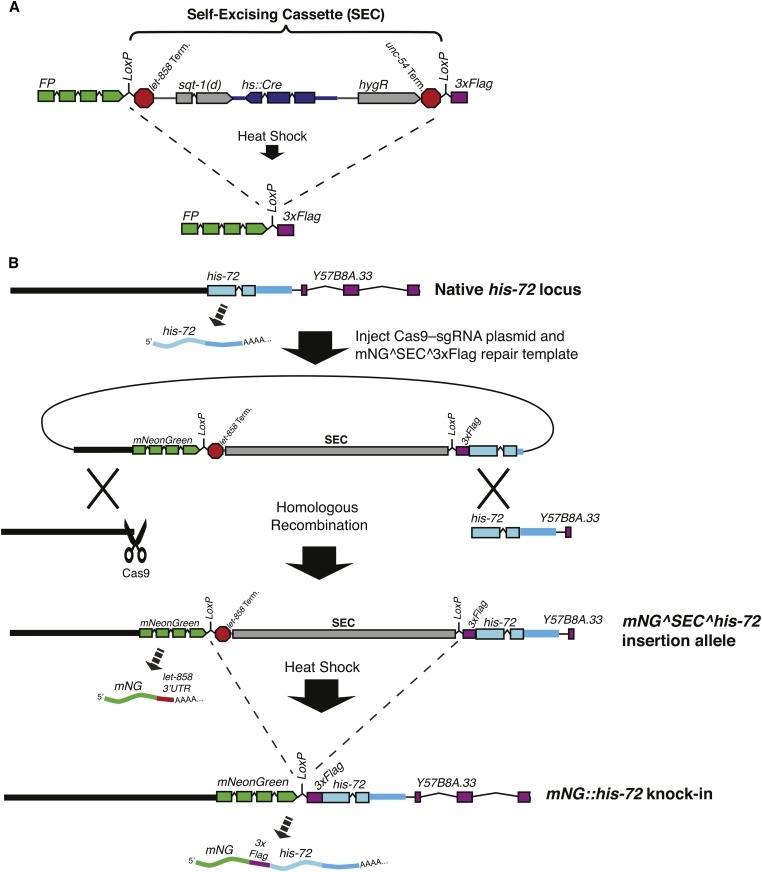
Several different selectable markers have been used in genome editing experiments, including unc-119(+) (Dickinson et al. 2013; Kim et al. 2014), blasticidin resistance (Kim et al. 2014), hygromycin resistance (C. Chen et al. 2013; Dickinson et al. 2015), and neomycin (G418) resistance (Norris et al. 2015). Using the hygromycin resistance gene (Greiss and Chin 2011) as a starting point, we recently developed a selectable marker that we refer to as a self-excising cassette (SEC) (Dickinson et al. 2015) (Figure 4). SEC consists of three parts: (1) a drug resistance gene, which allows genome modifications to be made directly in a wild-type background (or any genetic background desired), using selection; (2) a dominant phenotypic marker [we used sqt-1(e1350)] that allows one to identify homozygous insertions and marker excision events easily based on plate phenotype alone; and (3) an inducible Cre recombinase. Upon induction of Cre expression by heat shock, the entire selection cassette is removed from the genome (hence the term “self-excising”). This eliminates the need for a second injection step to deliver Cre.
Figure 4.
Gene tagging with a self-excising selection cassette. (A) Design of a self-excising cassette for drug selection. SEC consists of a drug resistance gene (hygR), a visible marker [sqt-1(d)], and an inducible Cre recombinase (hs::Cre). SEC is flanked by LoxP sites and placed within a synthetic intron in an FP::3xFlag tag, so that the LoxP site that remains after marker excision is within an intron. (B) Illustration of the organization of the his-72 locus and the predicted transcripts from this gene before editing (top), after homologous recombination (middle), and after SEC removal (bottom).
We and others have generated publicly available vectors in which SEC is placed within a synthetic intron of a fluorescent protein tag. This creates an FP–SEC module that can be inserted at any desired location in the genome, and after SEC removal, the remaining LoxP site is left in a synthetic intron within the fluorescent protein tag. Thus, no residual sequence is left in the genome outside of the fluorescent protein. These vectors also include ccdB negative selection markers for efficient insertion of homology arms (see Producing dsDNA repair templates from preexisting vectors, below). Taking the design principles of SEC as a starting point, it should be straightforward to substitute other markers for the hygromycin resistance gene and sqt-1(d) marker used in our vectors.
Because SEC contains transcriptional terminators, insertion of a fluorescent protein–SEC module at the 5′ end produces a loss-of-function mutation that is also a transcriptional reporter. The resulting allele converts to an N-terminal fluorescent protein tag after SEC removal. Thus, this approach can be used to generate a loss-of-function mutation, a promoter fusion, and a protein fusion in a single injection step.
Construction of Repair Templates for HDR
HDR is used to produce precise genome edits, in contrast to the random indels that are generated by error-prone repair. HDR can be performed using either single-stranded DNA oligonucleotides or double-stranded DNA molecules as homologous repair templates (Figure 2). Single-stranded DNA (ssDNA) repair templates are used to produce small, precise edits (e.g., point mutations), while double-stranded DNA (dsDNA) repair templates can be used to produce larger modifications. Linear repair templates with 30- to 40-bp homology arms are substrates for short-range HDR, while plasmid repair templates with 500- to 1500-bp homology arms are used for long-range HDR. Design considerations for each type of repair template are discussed separately.
Designing ssDNA oligo repair templates
ssDNA repair templates consist of the genome modification(s) of interest flanked by 30-80 nt of unmodified DNA sequence (Paix et al. 2014; Zhao et al. 2014; Ward 2015). The longest commercially available ssDNA oligonucleotides available as of this writing are Ultramer oligos from Integrated DNA Technologies, which can be up to 200 nt in length. Thus, ssDNA repair templates can in principle be used to produce insertions or substitutions up to ∼140 nt in size (200 nt minus 30 nt for each homology arm) or precise deletions of at least several kilobases (Paix et al. 2014).
A published protocol (Paix et al. 2014) includes a detailed set of instructions for designing ssDNA repair templates. In brief, an ssDNA repair template has four parts:
The homology arms are designed similarly regardless of the modification being made and comprise 30–80 nt of unmodified sequence at each end of the ssDNA oligo.
One needs to ensure that Cas9 cannot cut the modified locus; otherwise, after HDR occurs, repeated rounds of cleavage and repair will ultimately lead to the formation of an indel or random mutation rather than the precise genome modification desired. In some cases, the desired mutation already disrupts the Cas9 target site (for example, an insertion or deletion can disrupt or eliminate the target sequence). If the desired mutation leaves the Cas9 target site intact, then additional mutations must be introduced to block Cas9 cleavage. In these cases, it is best to select a Cas9 target site that resides within a protein-coding sequence, since silent (synonymous) substitutions can be introduced to block the Cas9 cleavage without otherwise affecting the activity of the gene of interest. Where possible, the simplest approach is to mutate the PAM, since a single substitution in the PAM is sufficient to completely block cleavage. If the PAM cannot be mutated without introducing an amino acid substitution, then the next best choice is to make multiple synonymous substitutions in the guide sequence. We generally make as many mutations as possible, and we consult a codon usage table (Carbone et al. 2003) to ensure that the mutations we make minimally perturb the codon optimality of the target sequence.
If one intends to screen by RFLP (see PCR screening, above), then a unique restriction site must be included.
Finally, the repair template must include the desired genome modification.
PAGE purification of repair oligos is not essential but has been reported to increase efficiency (Ward 2015).
Producing dsDNA repair templates from preexisting vectors
To produce insertions or substitutions > ∼140 bp in length, a double-stranded homologous repair template is required. PCR products carrying 30- to 60-bp homology arms are efficient substrates for short-range HDR (Paix et al. 2014, 2015), while long-range HDR requires homology arms 500–1500 bp that are typically cloned into a plasmid (C. Chen et al. 2013; Dickinson et al. 2013, 2015; Arribere et al. 2014; Kim et al. 2014). In either case, the repair template must include mutations to prevent Cas9 cleavage (see Designing ssDNA oligo repair templates).
Fluorescent protein insertion (the most common application that requires dsDNA repair templates) can be accomplished via either short-range or long-range HDR (Dickinson et al. 2013, 2015; Paix et al. 2014, 2015). For short-range HDR, homology arms can be incorporated into PCR primers that amplify the DNA to be inserted, and the resulting PCR product can be purified and used as the repair template (Paix et al. 2014, 2015).
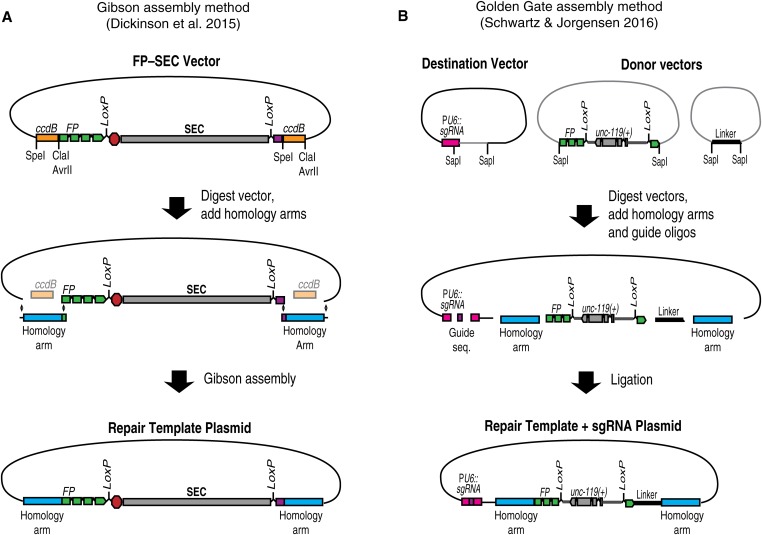
For long-range HDR, homology arms must be cloned into a vector to produce a plasmid repair template (Dickinson et al. 2015). To simplify the process of cloning homology arms, we developed a cloning procedure based on ccdB negative selection (Figure 5) (Dickinson et al. 2015). Vectors carrying different fluorescent protein–SEC modules flanked by ccdB markers are available via Addgene. To insert homology arms into one of these constructs, the vector is first digested with restriction enzymes to release the ccdB markers. Then, homology arms are inserted in place of the ccdB markers, using Gibson assembly (Gibson et al. 2009). Because ccdB is toxic to standard cloning strains of E. coli, only clones in which the ccdB markers have been replaced by the homology arms will grow. These clones can be identified by direct sequencing, without screening for inserts.
Figure 5.
(A) Gibson assembly-based strategy (Dickinson et al. 2015). An FP–SEC vector is digested to release ccdB markers, and homology arms are inserted by Gibson assembly to generate the repair template plasmid. Since the ccdB-containing parent vector does not transform, correct clones make up a majority of transformants and can be identified directly by sequencing. (B) SapTrap assembly strategy (Schwartz and Jorgensen, 2016). Homology arms and the sgRNA sequence are assembled, along with pre-existing tag and selectable marker building blocks, into a destination vector. The Type II restriction enzyme SapI generates unique overhangs that ensure ligation of the various fragments in the correct order.
Since any sequence can be cloned in place of the ccdB markers (Figure 5), this same basic cloning strategy can be used to build a repair template for any genome engineering project that will utilize SEC selection. To include additional modifications beyond the built-in FP tag, one simply needs to insert a larger DNA fragment comprising both the homology arm and any additional modifications in place of the ccdB marker. In general, any sequence located between the Cas9 cleavage site and the selectable marker is guaranteed to be copied into the genome. Therefore, when designing complex genome modifications, choose the Cas9 target site in such a way that the desired modifications lie between the cut site and the selectable marker.
An alternative, high-throughput method for assembling repair template plasmids was recently described by Schwartz and Jorgensen (2016; Figure 5B). Their approach, referred to as “SapTrap,” is based on the Golden Gate assembly method (Engler et al. 2008), which allows multiple DNA fragments to be joined together in a single reaction tube. SapTrap adds homology arms to pre-existing building blocks that can include various FP and epitope tags, selectable markers, and modules for sophisticated applications such as conditional tagging. The SapTrap destination vector also contains a second acceptor site for the guide sequence, eliminating the need to clone a separate sgRNA expression plasmid. A significant advantage of the modular SapTrap approach is that it allows a large variety of different repair constructs to be built by simply selecting the appropriate building blocks for a given application. The original SapTrap publication did not incorporate SEC selection (Schwartz and Jorgensen 2016), but a SapTrap-compatible SEC module is under construction as of this writing.
Building more complex repair templates using Gibson assembly
Although modular SEC constructs simplify the construction of repair templates for many genome engineering projects, some very complex custom modifications might still require generation of a new homologous repair template from scratch. When designing novel repair strategies, again the cardinal rule is that any sequence located between the Cas9 cleavage site and the selectable marker is guaranteed to be copied into the genome. Our preferred method for building new homologous repair templates is Gibson assembly (Gibson et al. 2009). In this cloning method, linear DNA fragments (most commonly PCR products) that overlap by 20–30 bp at their ends are covalently joined together. The requisite 20- to 30-bp overlaps are easily incorporated into the PCR primers that are used to generate the individual fragments. We prefer Gibson assembly over other cloning methods for two reasons: first, Gibson assembly does not require the addition of any extra sequences such as restriction sites or recombination targets; and second, up to six fragments can be assembled in a single step.
For researchers who are new to Gibson assembly, the following tips may be helpful:
We obtain the highest rates of successful assembly using fragments that overlap by 30 bp.
We often use PCR to amplify the vector backbone and include it as one of the fragments in the assembly. The most common cause of failure with this approach is large amounts of parent vector that carry through to the transformation. To avoid this, treat the vector PCR product with DpnI and gel purify it. If vector background still persists, reduce the amount of plasmid template used in the PCR reaction that generates the vector backbone.
When gel purifying DNA fragments for use in a Gibson assembly reaction, avoid using ethidium bromide or similar stains to visualize bands, since both ethidium bromide and UV radiation cause DNA damage that can significantly reduce cloning efficiencies. Instead, add 8 µg/ml crystal violet to the agarose gel, which allows DNA bands to be visualized under ambient light without UV exposure.
Since Gibson assembly joins DNA fragments covalently, 1 µl of a completed Gibson assembly reaction can be used as template for PCR to amplify the assembled product. We sometimes get better results by amplifying an assembled product and then ligating it into a vector, rather than including the vector directly in the assembly reaction.
If a multifragment Gibson assembly fails, try a sequential assembly strategy: assemble pairs of fragments, amplify and gel purify the resulting products, and then use those products as fragments in another assembly reaction.
Addressing Cas9 Specificity
The ability of Cas9 to recognize a specific target in the context of a complex genome is remarkable. Nevertheless, this specificity is not absolute; in mammalian systems and in vitro, Cas9 has been observed to cleave substrates that do not perfectly match the guide sequence (Jinek et al. 2012; Fu et al. 2013; Hsu et al. 2013; Pattanayak et al. 2013). These results call for an appropriate degree of caution when using Cas9 as an experimental tool.
Two studies have examined Cas9 specificity in C. elegans via whole-genome sequencing of mutant animals (Chiu et al. 2013; Paix et al. 2014), and two additional studies looked for evidence of off-target activity of Cas9 by sequencing candidate loci that closely matched the guide sequence (Dickinson et al. 2013; Friedland et al. 2013). None of these experiments detected any evidence of bona fide off-target mutations induced by Cas9, suggesting that in C. elegans, off-target mutations generated by Cas9 are uncommon. However, the two whole-genome sequencing studies both found evidence of other “passenger” mutations in CRISPR strains, at sites with no sequence similarity to the Cas9 target site. These second-site lesions are most likely spontaneous mutations that arose before or during strain construction. To avoid confounding effects of these passenger mutations on subsequent experiments, it should be standard practice to outcross mutant alleles and to isolate and characterize at least two independent alleles of every experimental genome modification. The ease and efficiency of Cas9-based approaches are such that isolating multiple alleles of each modification does not represent a significant burden. As variant Cas9 proteins recognizing different PAMs become available (Bell et al. 2015; Kleinstiver et al. 2015; Ran et al. 2015), the specificity of these enzymes will need to be carefully characterized.
When performing HDR, a second potential confounding issue is the incomplete or incorrect copying of the repair template into the genome. Rearrangements have been reported during homologous recombination from dsDNA templates (Berezikov et al. 2004; Frøkjaer-Jensen et al. 2008; Dickinson et al. 2013, 2015). Using plasmid repair templates, the frequency of rearrangements is ∼5–10% that of recombinant alleles in our experience. With short-range HDR, the repair template may be incompletely copied into the genome. Partial copying appears to occur stochastically (Arribere et al. 2014; Ward 2015) but is more frequent at larger distances from the cut site. Point mutations can occur in the repair template, due to mistakes in oligo synthesis (for the ssDNA oligo repair template), PCR errors (for dsDNA templates generated by PCR), or the DNA repair machinery responsible for HDR. Again, a straightforward solution to all of these issues is to isolate and characterize multiple independent alleles for each genome modification.
Recommended Strategies for Different Types of Modifications
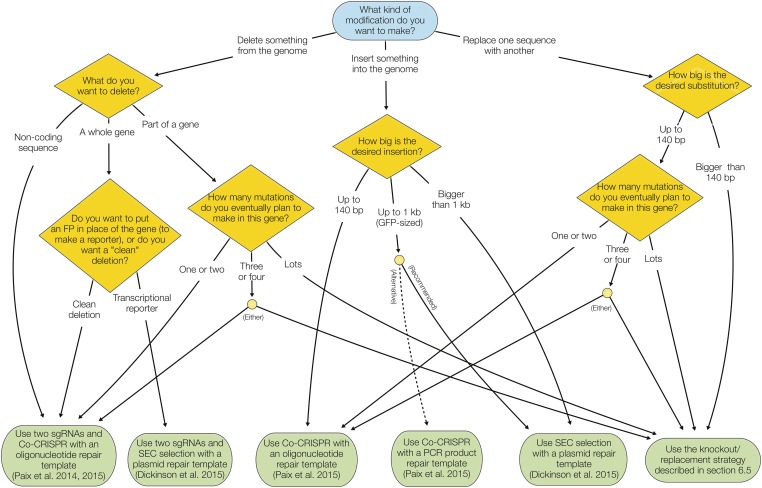
In this section, we provide our recommendations for generating common types of genome edits, taking into account all of the information from the preceding sections. These recommendations are based on published information, but also reflect our personal preferences to some extent. There are now multiple valid strategies to generate most kinds of edits, and these recommendations should be taken only as our suggestions for “what to try first.” Figure 6 shows a flow chart summarizing the recommendations.
Figure 6.
Flow chart summarizing recommended CRISPR techniques for different applications. See Recommended Strategies for Different Types of Modifications for details and rationale behind these recommendations.
Null mutations
A null mutation is a useful starting point for the analysis of almost any gene. By ascertaining the null phenotype of a gene, one establishes a basis for comparison when making targeted mutations later on. In addition, it is valuable to know the null phenotype of a gene when evaluating fluorescent protein knock-in strains: if a knock-in strain exhibits a phenotype similar to the null, this indicates that fusion to the fluorescent protein compromises the gene’s function. At least four different strategies can produce null (or strong loss-of-function) mutations:
Error-prone end-joining repair can be used to produce indels near the 5′ end of a gene, resulting in frameshift and early termination. This approach is useful for generating tissue-specific phenotypes (Shen et al. 2014) and might be adaptable to high-throughput screening based on feeding (Liu et al. 2014). However, an end-joining event leaves the majority of the gene’s coding sequence intact, and thus it is difficult to guarantee a priori that an indel mutation will be a bona fide null allele. Random indel mutations are also more difficult to screen for by PCR than HDR mutations (which can incorporate a restriction site to facilitate RFLP). Therefore, as a general rule, we prefer to use HDR to produce null mutations in which the entire coding sequence of a gene is deleted.
Paix et al. (2014, 2015) showed that gene-sized deletions could be generated by end joining, by using two sgRNAs that cut at opposite ends of the region to be deleted. More precise deletions could be generated by adding an ssDNA oligo with homology to the two ends of the deletion. In either case, the entire coding sequence of the gene is eliminated, which formally eliminates the possibility that any gene products will be produced. The same approach can also be used to delete portions of genes.
When a fluorescent protein–SEC module is inserted at the 5′ end of a gene of interest, the SEC separates the promoter from the protein-coding sequence of the gene, resulting in a loss-of-function allele (Dickinson et al. 2015). This loss-of-function allele is a useful intermediate in the construction of an N-terminal protein tag. However, N-terminal fluorescent protein–SEC insertions are not true genetic null mutations, in part because spontaneous SEC excision (resulting in expression of the gene of interest) occurs in certain tissues (Dickinson et al. 2015).
The SEC-based strategy can also be used to generate a bona fide null mutation by replacing the entire coding sequence of a gene with the fluorescent protein–SEC module. We have generated deletions of up to 9 kb using a single sgRNA and selection, but using two sgRNAs (one at each end of the region to be deleted) is expected to increase efficiency. The visible phenotype conferred by SEC makes it trivial to maintain the null allele as a heterozygote, which facilitates isolation and subsequent balancing of null mutations in essential genes. SEC can be used to facilitate mutant isolation and balancing and then eliminated once a stable strain is in hand, yielding an allele in which the coding sequence of the gene of interest is replaced by a fluorescent protein. The resulting allele functions both as a null mutation and as a transcriptional reporter (promoter fusion).
Recommendation:
Use two sgRNAs and an oligonucleotide repair template, with co-CRISPR screening, when a clean deletion of a gene (or part of a gene) without insertion of any exogenous sequence is desired (Paix et al. 2015). Use the SEC-based strategy when insertion of a fluorescent protein in place of the gene’s coding sequence is desired (Dickinson et al. 2015).
Point mutations
By “point mutations” we mean substitutions, insertions, or deletions of one or a few amino acids that can be easily templated by an ssDNA oligo. Although a selection-based strategy with long-range HDR can produce point mutants (Dickinson et al. 2013), this approach is overkill since short-range HDR with co-CRISPR can efficiently produce point mutations with minimal need for PCR screening (Kim et al. 2014; Arribere et al. 2014). Also, the co-CRISPR strategy allows one to make substitutions in the middle of genes, where integration of a selectable marker could be problematic. Finally, a co-CRISPR approach could allow multiple point mutations to be produced simultaneously (Paix et al. 2015; Ward 2015).
Recommendation:
For point mutations, use ssDNA-based HDR, with dpy-10, sqt-1, or pha-1 co-CRISPR and RFLP for screening (Arribere et al. 2014; Paix et al. 2015; Ward 2015).
Fluorescent protein fusions
Fluorescent protein fusions can be produced either by using the SEC-based strategy (Dickinson et al. 2015) or by short-range HDR with a PCR product repair template and co-CRISPR (Arribere et al. 2014; Kim et al. 2014; Paix et al. 2015). In our experience, these two approaches are very similar in terms of both the total time and hands-on labor required. The SEC strategy requires more work up front because of the need to clone homology arms in to the SEC vector, but the actual isolation of knock-in animals is easier. The co-CRISPR approach is quicker initially because the repair template is a PCR product and no cloning is required, but isolating knock-ins takes more work because PCR screening of 50–100 animals is needed. Thus, which strategy one chooses is largely a matter of personal preference. We prefer the SEC strategy in most cases, for two reasons: first, when used to generate N-terminal tags, the SEC-based strategy produces both a fluorescent protein fusion and a loss-of-function mutation from a single injection step. The loss-of-function intermediate is useful because one can quickly determine whether the tagged protein is functional by comparing the loss-of-function phenotype to the phenotype of the fluorescent protein fusion. Second, because it employs long-range HDR, the SEC strategy is insensitive to sgRNA efficiency and can produce insertions at a greater distance from the cut site, allowing more flexibility in experimental design.
Recommendation:
Use the SEC-based strategy for fluorescent protein fusions (Dickinson et al. 2015). We suggest making an N-terminal fusion unless there is a specific reason to choose a C-terminal fusion instead, because the process of generating an N-terminal fusion also yields a useful loss-of-function intermediate.
Other changes
Although the kinds of modifications above are the most common, they only scratch the surface of what is possible using Cas9-triggered homologous recombination. For example, simultaneous cutting on two chromosomes can produce custom translocations that function as balancer chromosomes (Chen et al. 2015). We can also imagine, for example, inserting LoxP or FLP recombinase target (FRT) sites to generate conditional alleles or replacement of whole genes by their homologs from other species to probe evolutionary questions. The methods one chooses to use for these or other kinds of experiments will depend on the details of the experiment, but as a general rule, we suggest that short-range HDR with co-CRISPR screening be used for all modifications that can be templated by an ssDNA oligo, while long-range HDR with SEC selection is best suited for making larger changes. SEC or other selectable markers can be incorporated into custom repair templates, using Gibson assembly.
A general strategy for dissecting gene function
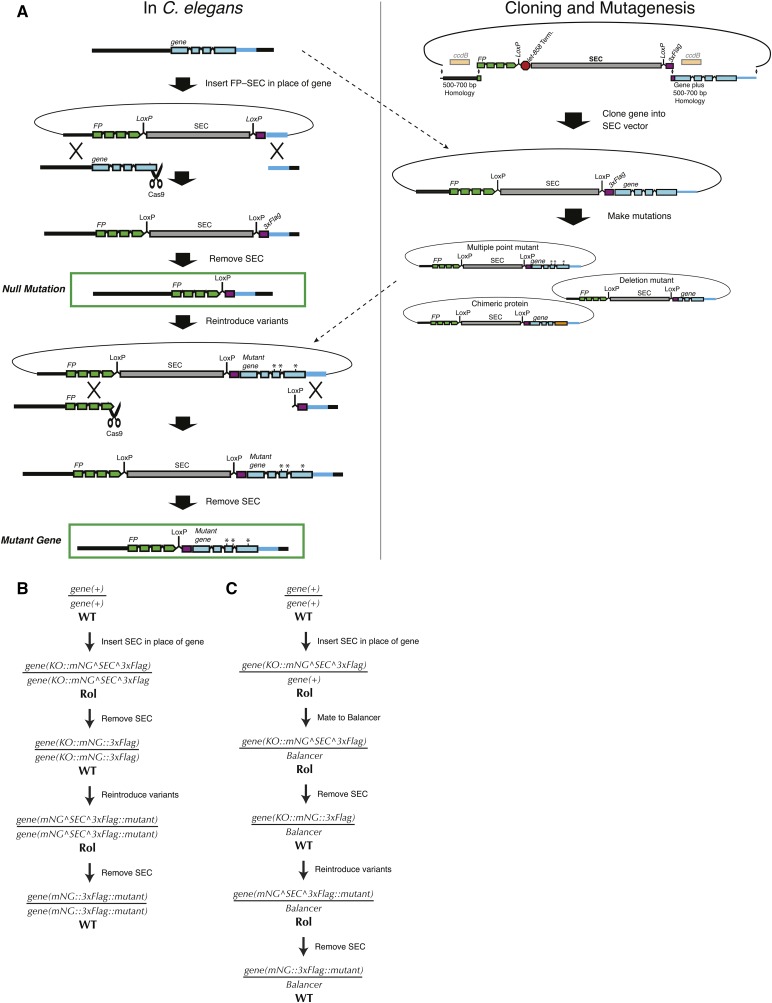
A common task for any protein of interest is to determine how different domains, binding sites, sequence motifs, or other features contribute to the function of the protein as a whole. Often this involves making many different mutants in a gene of interest and assaying their function. C. elegans is especially well suited to such “structure–function” analysis because of its short generation time, rich cell biology, defined lineage, and now with CRISPR, the ease of generating many mutants in the endogenous locus, without the need for overexpression. We have devised a simple, general strategy for performing structure–function analysis of C. elegans genes, which we demonstrated in Das et al. (2015) and describe here.
Briefly, we begin by making a null mutation and then reinsert either a wild-type or a mutant version of the gene of interest at the endogenous locus (Figure 7). The advantage of this strategy is that one can generate variants of the gene of interest in vitro, using standard cloning procedures, rather than designing a new CRISPR approach to produce each variant. Once the initial null allele is in hand, the same sgRNA, homology arms, and screening strategy can be used to reinsert each variant back into the endogenous locus. The phenotype of each variant can then be examined and compared directly to the null.
Figure 7.
A general strategy for structure–function analysis. (A) Illustration of the strategy. First, a null mutation is generated by inserting FP–SEC in place of the gene of interest; then, variants are reintroduced into the mutant background. Once the null mutant is made, multiple different variants (as many as desired) can be introduced using the same homology arms and sgRNA for the second HDR step. (B) Genetic scheme for executing this strategy for a nonessential gene. (C) Genetic scheme for executing this strategy for an essential gene. The only additional step is the introduction of a balancer chromosome during isolation of the initial null mutant.
We generate the null mutation by inserting a fluorescent protein–SEC module in place of the coding sequence (see Null mutations section, strategy 4) and then removing SEC. In principle, the null allele could also be generated by co-CRISPR with two sgRNAs and an oligonucleotide repair template. In parallel, we clone the genomic sequence of the gene of interest into an SEC-containing vector to generate a rescue construct. Mutations can be made to this rescue construct, using standard cloning techniques such as site-directed mutagenesis. If the gene of interest is nonessential, the (possibly mutated) rescue construct can be introduced directly into the homozygous null mutant in a second homologous recombination step. To generate multiple variant versions of a gene, one needs only to repeat the second recombination step for each variant. Figure 7, A and B, shows a detailed schematic of this procedure.
For essential genes, the workflow requires only one simple modification (Figure 7C). After generating the null allele, we mate it to a balancer and then remove the SEC. Variant versions of the gene are then introduced directly into the balanced null mutant background. By using an sgRNA targeting the fluorescent protein present in the null allele, we ensure that recombination occurs only on the null chromosome and not on the balancer. The resulting variant, like the parent null allele, is immediately balanced; any phenotypic assays are done using the fraction of progeny that have lost the balancer and are homozygous mutant.
Where to Go for More Information and Detailed Protocols
Detailed protocols are provided by several of the primary articles that established these methods (Dickinson et al. 2013, 2015; Arribere et al. 2014; Paix et al. 2014, 2015). In addition, we maintain a website (http://wormcas9hr.weebly.com) with up-to-date protocols that have been tested in our laboratory.
Acknowledgments
We thank the members of the Goldstein laboratory, especially Ariel Pani, Jennifer Heppert, and Christopher Higgins, for helpful discussions. We are also grateful to Geraldine Seydoux for sharing results prior to publication and to Jordan Ward for helpful discussions. We thank Amy Maddox, Geraldine Seydoux, and Jordan Ward for comments on the manuscript. Our work on CRISPR methods development was made possible by National Institutes of Health (NIH) grant T32 CA009156 and a Howard Hughes postdoctoral fellowship from the Helen Hay Whitney Foundation (to D.J.D.) and by NIH grant R01 GM083071 and National Science Foundation grant IOS 0917726 (to B.G.).
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Arribere J. A., Bell R. T., Fu B. X. H., Artiles K. L., Hartman P. S., et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, R. T., B. X. H. Fu, and A. Z. Fire, 2015 Cas9 variants expand the target repertoire in Caenorhabditis elegans Genetics 202: 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Bargmann C. I., Plasterk R. H. A., 2004. Homologous gene targeting in Caenorhabditis elegans by biolistic transformation. Nucleic Acids Res. 32: e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone A., Zinovyev A., Képès F., 2003. Codon adaptation index as a measure of dominating codon bias. Bioinformatics 19: 2005–2015. [DOI] [PubMed] [Google Scholar]
- Chen B., Gilbert L. A., Cimini B. A., Schnitzbauer J., Zhang W., et al. , 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155: 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Fenk L. A., de Bono M., 2013. Efficient genome editing in Caenorhabditis elegans by CRISPR-targeted homologous recombination. Nucleic Acids Res. 41: e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Li M., Feng X., Guang S., 2015. Targeted chromosomal translocations and essential gene knockout using CRISPR/Cas9 technology in Caenorhabditis elegans. Genetics 201: 1295–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H., Schwartz H. T., Antoshechkin I., Sternberg P. W., 2013. Transgene-free genome editing in Caenorhabditis elegans using CRISPR-Cas. Genetics 195: 1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Lee J., Carroll D., Kim J.-S., Lee J., 2013. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 195: 1177–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Dickinson D. J., Wood C. C., Goldstein B., Slep K. C., 2015. Crescerin uses a TOG domain array to regulate microtubules in the primary cilium. Mol. Biol. Cell 26: 4248–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Pani A. M., Heppert J. K., Higgins C. D., Goldstein B., 2015. Streamlined genome engineering with a self-excising drug selection cassette. Genetics 200: 1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J. G., Hartenian E., Graham D. B., Tothova Z., Hegde M., et al. , 2014. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol. 32: 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C., Kandzia R., Marillonnet S., 2008. A one-pot, one-step, precision cloning method with high throughput capability. PLoS One 3: e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B., Meyer B. J., 2015. Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics 199: 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiácovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Hollopeter G., Taylor J., Harris T. W., et al. , 2010. Targeted gene deletions in C. elegans using transposon excision. Nat. Methods 7: 451–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Ailion M., Jorgensen E. M., 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9: 117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., et al. , 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31: 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A., et al. , 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Greiss S., Chin J. W., 2011. Expanding the genetic code of an animal. J. Am. Chem. Soc. 133: 14196–14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., et al. , 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Lander E. S., Zhang F., 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Bikard D., Cox D., Zhang F., Marraffini L. A., 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic I., Großhans H., 2013. Targeted heritable mutation and gene conversion by Cas9-CRISPR in Caenorhabditis elegans. Genetics 195: 1173–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic I., Xu L., Ciosk R., 2015. CRISPR/Cas9 genome editing in Caenorhabditis elegans: evaluation of templates for homology-mediated repair and knock-ins by homology-independent DNA repair. G3 5: 1649–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ishidate T., Ghanta K. S., Seth M., Conte D., et al. , 2014. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197: 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B. P., Prew M. S., Tsai S. Q., Topkar V. V., Nguyen N. T., et al. , 2015. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523: 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C., Arslan S., Singh R., Thorpe J., Adli M., 2014. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 32: 677–683. [DOI] [PubMed] [Google Scholar]
- Liu H., Wei Z., Dominguez A., Li Y., Wang X., et al. , 2015. CRISPR-ERA: a comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics 31: 3676–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Long L., Xiong K., Yu B., Chang N., et al. , 2014. Heritable/conditional genome editing in C. elegans using a CRISPR-Cas9 feeding system. Cell Res. 24: 886–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo T.-W., Pickle C. S., Lin S., Ralston E. J., Gurling M., et al. , 2013. Heritable genome editing using TALENs and CRISPR/Cas9 to engineer precise insertions and deletions in evolutionarily diverse nematode species. Genetics 195: 331–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A. D., Kim H.-M., Colaiácovo M. P., Calarco J. A., 2015. Efficient genome editing in Caenorhabditis elegans with a toolkit of dual-marker selection cassettes. Genetics 201: 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Wang Y., Smith H. E., Lee C.-Y. S., Calidas D., et al. , 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 sites in Caenorhabditis elegans. Genetics 198: 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Folkmann A., Rasoloson D., Seydoux G., 2015. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V., Lin S., Guilinger J. P., Ma E., Doudna J. A., et al. , 2013. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31: 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J., 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R., 2006. Techniques for analysis, sorting, and dispensing of C. elegans on the COPAS flow-sorting system. Methods Mol. Biol. 351: 275–286. [DOI] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Lin C.-Y., Gootenberg J. S., Konermann S., et al. , 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Cong L., Yan W. X., Scott D. A., Gootenberg J. S., et al. , 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Yang Z., Xu J., Sun J., Mao D., et al. , 2014. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 9: 1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V., Bessereau J.-L., 2007. Targeted engineering of the Caenorhabditis elegans genome following Mos1-triggered chromosomal breaks. EMBO J. 26: 170–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. L., Jorgensen E. M., 2016. SapTrap, a Toolkit for High-Throughput CRISPR/Cas9 Gene Modification in Caenorhabditis elegans. Genetics DOI: genetics.115.184275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Zhang X., Chai Y., Zhu Z., Yi P., et al. , 2014. Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of Coronin in C. elegans neural development. Dev. Cell 30: 625–636. [DOI] [PubMed] [Google Scholar]
- Sternberg S. H., Redding S., Jinek M., Greene E. C., Doudna J. A., 2014. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan-Brown J. L., Tandon P., Bird K. E., Dickinson D. J., Tintori S. C., et al. , 2016. Identifying regulators of morphogenesis common to vertebrate neural tube closure and Caenorhabditis elegans gastrulation. Genetics 202: 123–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schendel R., Roerink S. F., Portegijs V., van den Heuvel S., Tijsterman M., 2015. Polymerase Θ is a key driver of genome evolution and of CRISPR/Cas9-mediated mutagenesis. Nat. Commun. 6: 7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waaijers S., Portegijs V., Kerver J., Lemmens B. B. L. G., Tijsterman M., et al. , 2013. CRISPR/Cas9-targeted mutagenesis in Caenorhabditis elegans. Genetics 195: 1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Wei J. J., Sabatini D. M., Lander E. S., 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. D., 2015. Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics 199: 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Scott D. A., Kriz A. J., Chiu A. C., Hsu P. D., et al. , 2014. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 32: 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Xiao T., Chen C.-H., Li W., Meyer C. A., et al. , 2015. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 25: 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B., Gootenberg J. S., Abudayyeh O. O., Slaymaker I. M., Makarova K. S., et al. , 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Zhang Z., Ke H., Yue Y., Xue D., 2014. Oligonucleotide-based targeted gene editing in C. elegans via the CRISPR/Cas9 system. Cell Res. 24: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]