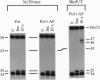
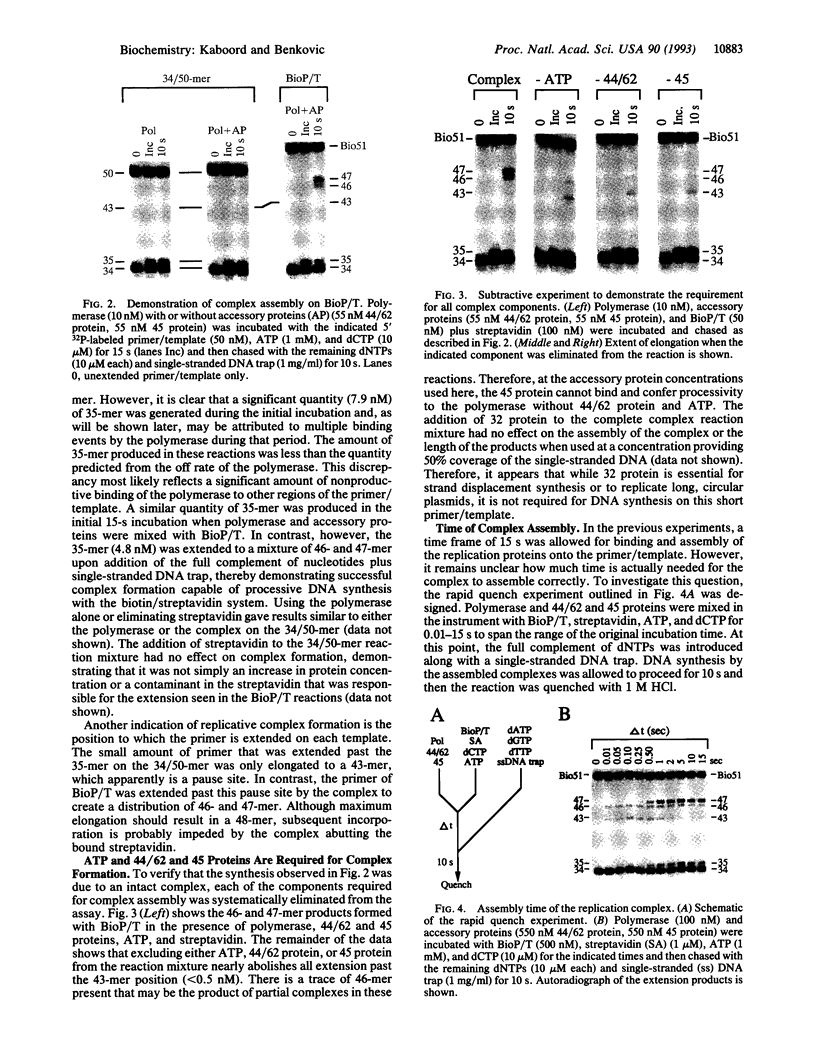
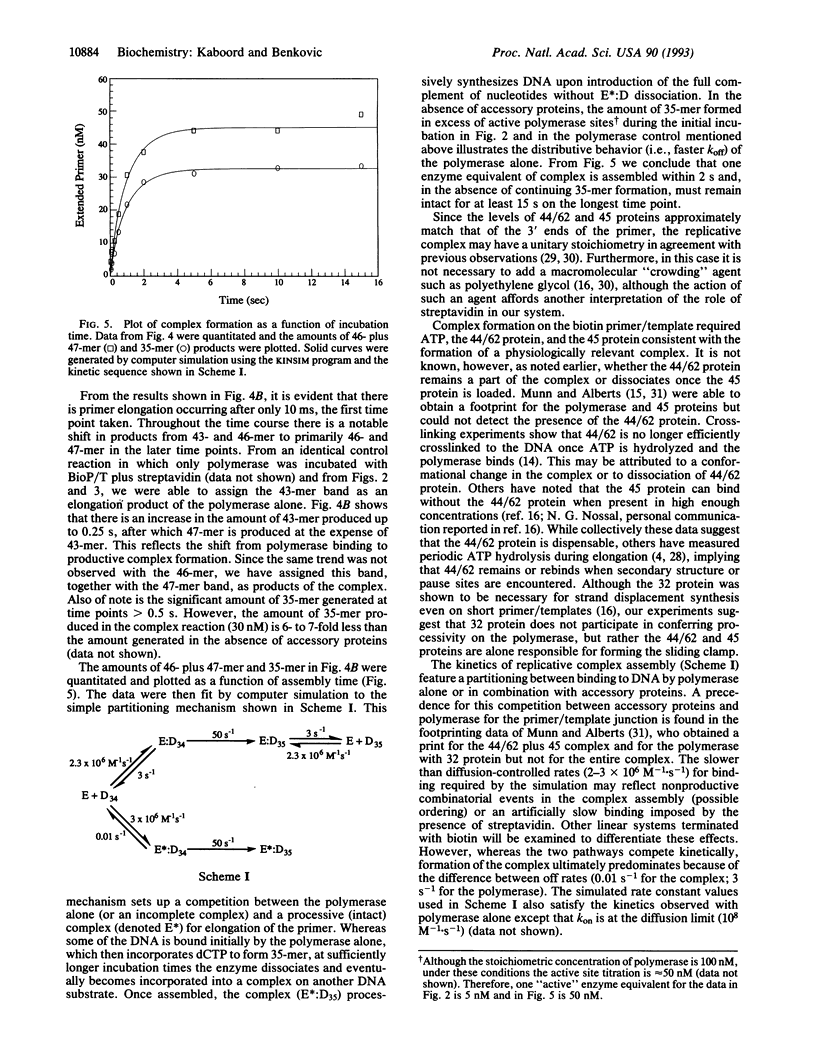
Abstract
DNA synthesis on a primed DNA substrate by bacteriophage T4 requires the assembly of a core replication complex consisting of the T4 DNA polymerase, a single-stranded binding protein (32 protein), and the accessory proteins 44/62 and 45. In this paper, we demonstrate the successful assembly of this core complex on a short linear primer/template system at levels of accessory proteins equivalent to the concentration of primer 3' ends. The key to this assembly is the presence of streptavidin molecules bound at each end of the DNA substrate via biotin moieties incorporated into the template strand. Streptavidin serves to block the ends of the primer/template, thus preventing translocation of the accessory proteins away from the site of assembly and their subsequent dissociation from the ends of the primer/template. Complex assembly on this substrate requires ATP and the presence of both the 44/62 and 45 proteins. The time required for assembly of a full enzyme equivalent of complex in our system is approximately 2 s.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M. Prokaryotic DNA replication mechanisms. Philos Trans R Soc Lond B Biol Sci. 1987 Dec 15;317(1187):395–420. doi: 10.1098/rstb.1987.0068. [DOI] [PubMed] [Google Scholar]
- Capson T. L., Benkovic S. J., Nossal N. G. Protein-DNA cross-linking demonstrates stepwise ATP-dependent assembly of T4 DNA polymerase and its accessory proteins on the primer-template. Cell. 1991 Apr 19;65(2):249–258. doi: 10.1016/0092-8674(91)90159-v. [DOI] [PubMed] [Google Scholar]
- Capson T. L., Peliska J. A., Kaboord B. F., Frey M. W., Lively C., Dahlberg M., Benkovic S. J. Kinetic characterization of the polymerase and exonuclease activities of the gene 43 protein of bacteriophage T4. Biochemistry. 1992 Nov 17;31(45):10984–10994. doi: 10.1021/bi00160a007. [DOI] [PubMed] [Google Scholar]
- Chen M., Pan Z. Q., Hurwitz J. Sequence and expression in Escherichia coli of the 40-kDa subunit of activator 1 (replication factor C) of HeLa cells. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2516–2520. doi: 10.1073/pnas.89.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M. W., Nossal N. G., Capson T. L., Benkovic S. J. Construction and characterization of a bacteriophage T4 DNA polymerase deficient in 3'-->5' exonuclease activity. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2579–2583. doi: 10.1073/pnas.90.7.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogol E. P., Young M. C., Kubasek W. L., Jarvis T. C., von Hippel P. H. Cryoelectron microscopic visualization of functional subassemblies of the bacteriophage T4 DNA replication complex. J Mol Biol. 1992 Mar 20;224(2):395–412. doi: 10.1016/0022-2836(92)91003-8. [DOI] [PubMed] [Google Scholar]
- Green N. M. Avidin. Adv Protein Chem. 1975;29:85–133. doi: 10.1016/s0065-3233(08)60411-8. [DOI] [PubMed] [Google Scholar]
- Herendeen D. R., Kassavetis G. A., Geiduschek E. P. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science. 1992 May 29;256(5061):1298–1303. doi: 10.1126/science.1598572. [DOI] [PubMed] [Google Scholar]
- Huang C. C., Hearst J. E., Alberts B. M. Two types of replication proteins increase the rate at which T4 DNA polymerase traverses the helical regions in a single-stranded DNA template. J Biol Chem. 1981 Apr 25;256(8):4087–4094. [PubMed] [Google Scholar]
- Hurwitz J., Dean F. B., Kwong A. D., Lee S. H. The in vitro replication of DNA containing the SV40 origin. J Biol Chem. 1990 Oct 25;265(30):18043–18046. [PubMed] [Google Scholar]
- Jarvis T. C., Newport J. W., von Hippel P. H. Stimulation of the processivity of the DNA polymerase of bacteriophage T4 by the polymerase accessory proteins. The role of ATP hydrolysis. J Biol Chem. 1991 Jan 25;266(3):1830–1840. [PubMed] [Google Scholar]
- Jarvis T. C., Paul L. S., Hockensmith J. W., von Hippel P. H. Structural and enzymatic studies of the T4 DNA replication system. II. ATPase properties of the polymerase accessory protein complex. J Biol Chem. 1989 Jul 25;264(21):12717–12729. [PubMed] [Google Scholar]
- Jarvis T. C., Paul L. S., von Hippel P. H. Structural and enzymatic studies of the T4 DNA replication system. I. Physical characterization of the polymerase accessory protein complex. J Biol Chem. 1989 Jul 25;264(21):12709–12716. [PubMed] [Google Scholar]
- Jarvis T. C., Ring D. M., Daube S. S., von Hippel P. H. "Macromolecular crowding": thermodynamic consequences for protein-protein interactions within the T4 DNA replication complex. J Biol Chem. 1990 Sep 5;265(25):15160–15167. [PubMed] [Google Scholar]
- Johnson K. A. Rapid kinetic analysis of mechanochemical adenosinetriphosphatases. Methods Enzymol. 1986;134:677–705. doi: 10.1016/0076-6879(86)34129-6. [DOI] [PubMed] [Google Scholar]
- Kong X. P., Onrust R., O'Donnell M., Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992 May 1;69(3):425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Kuchta R. D., Mizrahi V., Benkovic P. A., Johnson K. A., Benkovic S. J. Kinetic mechanism of DNA polymerase I (Klenow). Biochemistry. 1987 Dec 15;26(25):8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- Mace D. C., Alberts B. M. Characterization of the stimulatory effect of T4 gene 45 protein and the gene 44/62 protein complex on DNA synthesis by T4 DNA polymerase. J Mol Biol. 1984 Aug 5;177(2):313–327. doi: 10.1016/0022-2836(84)90459-5. [DOI] [PubMed] [Google Scholar]
- Mace D. C., Alberts B. M. The complex of T4 bacteriophage gene 44 and 62 replication proteins forms an ATPase that is stimulated by DNA and by T4 gene 45 protein. J Mol Biol. 1984 Aug 5;177(2):279–293. doi: 10.1016/0022-2836(84)90457-1. [DOI] [PubMed] [Google Scholar]
- McCarthy D., Minner C., Bernstein H., Bernstein C. DNA elongation rates and growing point distributions of wild-type phage T4 and a DNA-delay amber mutant. J Mol Biol. 1976 Oct 5;106(4):963–981. doi: 10.1016/0022-2836(76)90346-6. [DOI] [PubMed] [Google Scholar]
- Mesner L. D., Sutherland W. M., Hockensmith J. W. DNA-dependent adenosinetriphosphatase A is the eukaryotic analogue of the bacteriophage T4 gene 44 protein: immunological identity of DNA replication-associated ATPases. Biochemistry. 1991 Dec 10;30(49):11490–11494. doi: 10.1021/bi00113a002. [DOI] [PubMed] [Google Scholar]
- Mizrahi V., Benkovic P., Benkovic S. J. Mechanism of DNA polymerase I: exonuclease/polymerase activity switch and DNA sequence dependence of pyrophosphorolysis and misincorporation reactions. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5769–5773. doi: 10.1073/pnas.83.16.5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn M. M., Alberts B. M. DNA footprinting studies of the complex formed by the T4 DNA polymerase holoenzyme at a primer-template junction. J Biol Chem. 1991 Oct 25;266(30):20034–20044. [PubMed] [Google Scholar]
- Munn M. M., Alberts B. M. The T4 DNA polymerase accessory proteins form an ATP-dependent complex on a primer-template junction. J Biol Chem. 1991 Oct 25;266(30):20024–20033. [PubMed] [Google Scholar]
- Nossal N. G. Protein-protein interactions at a DNA replication fork: bacteriophage T4 as a model. FASEB J. 1992 Feb 1;6(3):871–878. doi: 10.1096/fasebj.6.3.1310946. [DOI] [PubMed] [Google Scholar]
- Onrust R., Stukenberg P. T., O'Donnell M. Analysis of the ATPase subassembly which initiates processive DNA synthesis by DNA polymerase III holoenzyme. J Biol Chem. 1991 Nov 15;266(32):21681–21686. [PubMed] [Google Scholar]
- Reddy M. K., Weitzel S. E., von Hippel P. H. Assembly of a functional replication complex without ATP hydrolysis: a direct interaction of bacteriophage T4 gp45 with T4 DNA polymerase. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3211–3215. doi: 10.1073/pnas.90.8.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenberg P. T., Studwell-Vaughan P. S., O'Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991 Jun 15;266(17):11328–11334. [PubMed] [Google Scholar]
- Tsurimoto T., Melendy T., Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990 Aug 9;346(6284):534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Functions of replication factor C and proliferating-cell nuclear antigen: functional similarity of DNA polymerase accessory proteins from human cells and bacteriophage T4. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1023–1027. doi: 10.1073/pnas.87.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991 Jan 25;266(3):1950–1960. [PubMed] [Google Scholar]
- Young M. C., Reddy M. K., von Hippel P. H. Structure and function of the bacteriophage T4 DNA polymerase holoenzyme. Biochemistry. 1992 Sep 22;31(37):8675–8690. doi: 10.1021/bi00152a001. [DOI] [PubMed] [Google Scholar]