Abstract
Most standard QTL mapping procedures apply to populations derived from the cross of two parents. QTL detected from such biparental populations are rarely relevant to breeding programs because of the narrow genetic basis: only two alleles are involved per locus. To improve the generality and applicability of mapping results, QTL should be detected using populations initiated from multiple parents, such as the multiparent advanced generation intercross (MAGIC) populations. The greatest challenges of QTL mapping in MAGIC populations come from multiple founder alleles and control of the genetic background information. We developed a random-model methodology by treating the founder effects of each locus as random effects following a normal distribution with a locus-specific variance. We also fit a polygenic effect to the model to control the genetic background. To improve the statistical power for a scanned marker, we release the marker effect absorbed by the polygene back to the model. In contrast to the fixed-model approach, we estimate and test the variance of each locus and scan the entire genome one locus at a time using likelihood-ratio test statistics. Simulation studies showed that this method can increase statistical power and reduce type I error compared with composite interval mapping (CIM) and multiparent whole-genome average interval mapping (MPWGAIM). We demonstrated the method using a public Arabidopsis thaliana MAGIC population and a mouse MAGIC population.
Keywords: best linear unbiased prediction, empirical Bayes, mixed model, polygene, restricted maximum likelihood, multiparental populations, Multiparent Advanced Generation Inter-Cross (MAGIC), MPP
THERE is an urgent need to develop and study multiparent advanced generation intercross (MAGIC) populations (Rakshit et al. 2012). Along with nested association mapping populations (Yu et al. 2008), the MAGIC population is called a second-generation mapping resource (Rakshit et al. 2012). Using MAGIC populations to perform QTL mapping was first proposed for mice by Threadgill et al. (2002). Such a population is called the Collaborative Cross (CC) population (Churchill et al. 2004; Collaborative Cross Consortium 2012). Simulation studies showed that an eight-parent CC population with 1000 progenies is capable of increasing mapping resolution to the sub-centimorgan range (Valdar et al. 2006). MAGIC populations in Drosophila melanogaster are called Drosophila Synthetic Population Resources (DSPR) (MacDonald and Long 2007; King et al. 2012a, et al.b). A review of MAGIC populations in crops can be found in Huang et al. (2015). The first plant MAGIC population was developed in Arabidopsis thaliana by Kover et al. (2009). The population will be described later. Subsequently, MAGIC populations have been developed in wheat (Huang et al. 2012; Mackay et al. 2014), rice (Bandillo et al. 2013), and other crop species (Gaur et al. 2012; Pascual et al. 2015; Sannemann et al. 2015). One key difference between MAGIC populations and other multiparent populations is that all MAGIC lines have experienced multiple generations of inbreeding and thus all are inbred lines. As a result, they are also considered genetic reference populations whose particular genome arrangement can be replicated indefinitely. MAGIC populations in plants undoubtedly will become more popular in the future of plant genetics and breeding (Varshney and Dubey 2009; Rakshit et al. 2012; Huang et al. 2015), which calls attention to the need for improvements in statistical methods to analyze and interpret data derived from these populations. A recent call for papers on QTL mapping in MAGIC populations by GENETICS and G3 (http://www.genetics.org/) further indicates the urgent need for new technologies in MAGIC population QTL mapping.
Current methods of QTL mapping for MAGIC populations are adopted primarily from methods used in biparental populations. For example, composite interval mapping (CIM) (Zeng 1994), originally developed for biparental populations, has been used in QTL mapping for MAGIC populations to control genomic background. Other methods and programs of QTL mapping in MAGIC populations include MCQTL (Jourjon et al. 2005), R/qtl (Broman et al. 2003), R happy (Mott et al. 2000), and R/mpMap (Huang and George 2011), most of which have an option to perform CIM. However, there is an intrinsic limitation in cofactor selection, which is more problematic in MAGIC populations than in biparental populations. In an eight-parent-initiated MAGIC population, each marker has 8 − 1 = 7 founder effects to estimate. The total number of effects will soon saturate the linear model as the number of cofactors increases. For example, a MAGIC population of size 500 will allow only fewer than 500/7 ≈ 71 cofactors to be included in the model. When the number of cofactors is small, the CIM procedure is sensitive to the selection of cofactors. Ideally, a model should include all markers in a single model. However, when the marker density is high, genome scanning (a single-QTL model) provides a better alternative method for QTL mapping, but the cofactors should be replaced by a polygenic effect, as done in genome-wide association studies (GWAS) (Yu et al. 2006). We recently developed a QTL mapping procedure by fitting a polygene using a marker-inferred relationship matrix (replacing cofactors) and demonstrated the robustness of the method (Xu 2013b).
Recently, Gatti et al. (2014) developed a mixed model for QTL mapping in Diversity Outbred (DO) mice by treating the effects of scanned markers as fixed and a polygenic effect as random. The polygenic effect essentially replaced cofactors to control the genetic background. The method tends to have a low power because part of the effect of the marker currently scanned is absorbed by the polygene. Our simulation studies showed that dramatic improvement can be achieved in terms of resolution and statistical power of mapped QTL if the effect of the current QTL captured by the polygene is taken into account. Verbyla et al. (2014) developed a multiple-QTL model for QTL mapping in MAGIC populations. The method is called multiparent whole-genome QTL analysis (MPWGAIM), and several steps are involved in selecting markers for inclusion in the model. First, a polygenic base model is implemented to detect the whole-genome effect on the traits of interest. If the polygenic variance is significantly larger than zero, then markers are subject to selection under a random-model approach; i.e., the founder allelic effects of a marker are treated as random effects, and the variance of those founder effects is estimated and the marker is then selected if the variance is sufficiently large. The final model will include all markers selected (forward selection). This is a variable-selection approach and may be costly if the number of markers and the number of QTL found are large. We will treat this model as the “gold standard” for simulation and comparison. Another recent study of QTL mapping in MAGIC populations is the Bayesian modeling of haplotype effects (Zhang et al. 2014), where the founder haplotype effects are estimated via Markov chain Monte Carlo (MCMC) sampling or importance sampling (IS). One important feature of the Bayesian method is the ability to handle uncertainty of the founder allelic inheritance. The only concern with the Bayesian method is the high computational cost when the sample size and the number of markers are very large because Monte Carlo sampling is involved. It is recommended to use the Bayesian method to fine-tune the model after markers are selected using some simple methods such as interval mapping (IM) and CIM.
In this study, we extended the mixed-model methodology of QTL mapping in MAGIC populations by fitting a polygenic effect as random and a scanned marker effect either as fixed or random. Furthermore, we released the polygenic counterpart of a scanned marker effect back to the model to avoid competition between the marker effect and its polygenic counterpart. This improved mixed-model methodology has significantly improved the statistical power of QTL detection. We used a CC mouse population (Collaborative Cross Consortium 2012) to perform simulations to examine the properties of the new methods (there are no phenotypic values available for the CC mouse population). The Arabidopsis MAGIC population of Kover et al. (2009) and the pre-CC mouse population of Rutledge et al. (2014) were reanalyzed using the new methods to demonstrate the differences between the new and existing methods.
Materials and Methods
MAGIC populations
Three MAGIC populations were used in this study to demonstrate the new methods of QTL mapping, two populations in mice and one in A. thaliana. The first MAGIC population in mice does not have phenotypes available on the website (http://www.csbio.unc.edu/CCstatus) and was used only for simulation studies. The second MAGIC population of mice has both genotype and phenotype information and was used as a real application example. The MAGIC population in A. thaliana also has both genotype and phenotype information and was reanalyzed to compare the results of the different methods.
First MAGIC population of mice:
This MAGIC population is called the CC population (Churchill et al. 2004). The genotype data were published by the Collaborative Cross Consortium (2012). No phenotype information is available in the 458 CC mice, and thus the data were used only for simulation study. The CC population is an eight-parent MAGIC population derived from a funnel mating design. We downloaded the recombination breakpoint data of 19 autosomes from 458 CC mice posted on the University of North Carolina (UNC) System Genetics website (http://www.csbio.unc.edu/CCstatus). Using the breakpoint information, we inferred 6683 bins (intact chromosome segments). A bin is defined as a segment that contains no breakpoints across all lines within the segment. Within a bin, all markers segregate in exactly the same pattern across lines (perfect LD). Therefore, a single marker can represent the whole bin. For detailed information on bin data analysis, see Xu (2013a). The bin data are available in Supporting Information, File S1.
Second MAGIC population of mice:
The second MAGIC population was derived from the same eight parents as the first CC population, but the CC mice were not fully inbred, and therefore, the population is called the pre-CC population. The data were obtained from Rutledge et al. (2014) and consist of 151 individuals. This data set includes 27,039 SNPs evenly distributed among the 20 chromosomes (including the X chromosome). Probabilities of the parental origins of the SNPs were calculated using the HAPPY program based on the hidden Markov model (Mott et al. 2000). In the original study of this population, the authors focused on two traits associated with severe asthma and decrements in lung function, including airway polymorphonuclear neutrophil (PMN) recruitment and the concentration of CXCL1 in lung lavage fluid. Here we reanalyzed the first trait, PMN.
MAGIC population of Arabidopsis:
The MAGIC population of A. thaliana (Kover et al. 2009) consists of 527 lines descended from a heterogeneous stock of 19 intermated parents. These lines and the 19 founders were genotyped with 1260 SNP markers [minor allele frequency (MAF) > 5%] and phenotyped for two development-related traits, the number of days between bolting and flowering (DBF) and growth rate (GR), where GR was measured as the residual of regression by fitting the number of leaves to the number of days to germination. The 527 lines were derived from the 19 founder accessions of A. thaliana, intermating for four generations, and then inbreeding for six additional generations, forming nearly homozygous lines. The authors further updated the database after the initial publication. We downloaded the updated genotypes and phenotypes from http://mus.well.ox.ac.uk/magic/. There were only 426 lines having both the genotype and phenotype information. In this analysis, we included the 426 lines and 1254 markers distributed among five chromosomes (total length of the genome is 118 Mb). The founder strain probabilities for all loci were calculated using the HAPPY program. We analyzed both DBF and GR.
Statistical methods
Polygenic model:
The polygenic model is the null model used to scan the entire genome for QTL identification. We now use an eight-parent MAGIC population as an example to demonstrate the model. The method holds for any p-parent MAGIC populations. Let y be an n × 1 vector of phenotypic values for n individuals. Define Zk as an n × 8 matrix of founder allele inheritance indicators for locus k. The jth row of matrix Zk is defined as a 1 × 8 vector. If this individual is a heterozygote carrying the first and second founder alleles, then we define
If the individual is a homozygote inheriting both alleles from the fifth founder, then Zjk is defined as
The general rule for defining Zjk is that there are at most two nonzero elements, and the sum of all eight elements equals 2. We then define the following polygenic model, which is the null model used to test significance of an individual marker:
| (1) |
where X is an n × r design matrix for r fixed effects; β is an r × 1 vector for the r fixed effects; ξ is an n × 1 vector of polygenic effects with an assumed multivariate normal distribution , where K is a marker-derived kinship matrix and φ2 is a polygenic variance; and is a vector of residual errors with an unknown error variance σ2. The marker inferred kinship matrix is defined as
| (2) |
where is a normalization factor. The expectation of y is , and the variance-covariance matrix is
| (3) |
where is the variance ratio, and . After absorbing β and σ2, the restricted maximum likelihood is only a function of λ, which is
| (4) |
where
| (5) |
The restricted maximum likelihood solution of λ was obtained by maximizing the preceding likelihood function using the Newton iteration algorithm. The eigen-decomposition algorithm proposed by Kang et al. (2008) was used to evaluate the likelihood function for fast computation. The estimated variance ratio is denoted by and will be used as a known constant in the genomic scanning model that follows. File S2 describes the method of estimating λ along with the effect of the marker scanned, the so-called exact method (Zhou and Stephens 2012).
Fixed model:
To test the significance of the kth marker, we first used the fixed-model approach proposed by Gatti et al. (2014). The model is
| (6) |
where Zk is the allelic inheritance matrix for marker k, as defined earlier, and
| (7) |
is an 8 × 1 vector for the eight founder allelic effects. Under this model, the γk vector is assumed to be fixed effects. The model is in fact a mixed model because it contains both fixed and random effects. We call it the fixed model because later on we will treat γk as random effects. Under the fixed model, we can only estimate and test seven (8 – 1 = 7) effects by deleting the last founder allele from the model. The maximum-likelihood method was used to estimate γk, and the result turned out to be identical to the weighted-least-squares estimate after premultiplying all variables (y, X, and Zk) by the eigenvectors of the K matrix and the weight for the jth individual being , where δj is the jth eigenvalue of the K matrix (Xu 2013b). Note that is the estimated variance ratio under the polygenic model, as described earlier. This method is called the approximate method (Zhou and Stephens 2012). The likelihood-ratio test was used as the test statistic and is defined as
| (8) |
where L0 is the log likelihood function under the null model (γk = 0), and L1 is the log likelihood function under the alternative model. Note that the estimated β and σ2 under the two models are different. The P-value was calculated from the chi-square distribution with seven degrees of freedom. This method is called FIXED-A when compared with other methods.
Recall that Zk contributes to the calculation of the kinship matrix K, as shown in equation 2. Although not explicitly estimated in the polygene, the effect of marker k has a polygenic counterpart that may compete with the estimated γk when marker k is scanned. Let be the estimated polygenic effect contributed by marker k, and is the estimated effect for this marker under the polygenic model. We can release this effect from the polygene back to the model to avoid this competition. The revised model is
| (9) |
Rearranging this equation leads to
| (10) |
Further defining , we now have a new model
| (11) |
which is the same as equation 6 except that the y vector changes every time a marker is scanned. Note that , the polygenic component from marker k, is calculated only once under the null model. Therefore, this revised method does not present much additional computational burden. The method to obtain is called the best linear unbiased prediction (BLUP) and is described in File S2. This revised method is called FIXED-B when compared with other methods.
Random model:
The fixed-model approach may not be stable when the number of founders is large (Gatti et al. 2014), and the design matrix Zk may have variable ranks across different markers. Under the null model, the likelihood-ratio test statistic follows a chi-square distribution with degrees of freedom depending on the number of founders. We propose to treat the eight founder effects as random variables following a normal distribution with mean zero and a common variance. Although it is still a mixed model, we call it a random model to distinguish it from the fixed model described earlier. The linear model remains the same as equation 6, but is assumed, where is a locus-specific variance. The expectation of y remains , and the variance-covariance matrix is
| (12) |
where λ in H is replaced by the estimated value under the polygenic model. A restricted-maximum-likelihood (REML) estimate of is obtained by maximizing the restricted likelihood function. Woodbury matrix identities (Golub and Van Loan 1996) are applied along with the eigen-decomposition to ease the computational burden (File S2). The null hypothesis for marker k is , which is tested using the likelihood-ratio test
| (13) |
Under the null model, this test statistic follows approximately a mixture of and distributions with an equal weight (Chernoff 1954; Visscher 2006). This method is called RANDOM-A when compared with other methods.
We also developed a revised version of the random model by avoiding competition between the current marker scanned and its polygenic counterpart using model 11 as we did for the fixed model. This revised random model is called RANDOM-B to distinguish it from other methods.
Multiparent whole-genome average interval mapping (MPWGAIM):
Here we also performed the analysis using the MPWGAIM approach proposed by Verbyla et al. (2014) for comparison using their R package mpwgaim. In the mpwgaim package, only detected markers are reported without test statistics attached. For comparison with our methods, we calculated the Wald test statistics of detected markers based on their estimated effects and variances and then obtained the P-value from the chi-square distribution with 8 − 1 = 7 degrees of freedom. For the simulated data analysis, we also applied the MPWGAIM method. The empirical critical value for hypothesis test was inferred from multiple (1000) simulations under the null model. The 95th percentile of the highest Wald test from each of the multiple simulations was chosen as the empirical critical value. The P-value was transformed by −log10 and used to determine whether or not a marker exceeds the empirical critical value.
IM and CIM:
IM (Lander and Botstein 1989) and CIM (Zeng 1994) also were used to analyze the data to compare the results with the new methods. These two methods are called IM and CIM-x, respectively, where x indicates the number of cofactors included in the model for background control. The statistical model for IM differs from model 6 by ignoring the polygenic effect. The model for CIM differs from model 6 by replacing the polygenic effect with selected cofactors. The IM method was implemented in the HAPPY program (Mott et al. 2000). The CIM method was implemented using our own R program. For the CIM-x method, the number of cofactors x was set at the following levels for the first MAGIC population of mice: 65, 50, and 30. For a sample size of 458, the maximum number of cofactors cannot be higher than 458/7 ≈ 65; otherwise, there will not be any degrees of freedom left to estimate the residual error variance. For the second MAGIC population of mice (the pre-CC population), the number of cofactors was set at 20, 10, and 5. The population size is 151, and thus the number of cofactors cannot be higher than 151/7 ≈ 20. For the Arabidopsis population, the number of cofactors was set at 20, 15, and 10. The maximum number of possible cofactors cannot be greater than 428/18 ≈ 23. The likelihood-ratio test statistic also was used for the IM and CIM methods.
P-value and permutation:
We now have a total of seven methods to compare: FIXED-A, MPWGAIM, IM, and CIM are existing methods, and FIXED-B, RANDOM-A, and RANDOM-B are new methods proposed in this study. The P-value of a marker was calculated from the central chi-square distribution with 8 − 1 = 7 degrees of freedom for the two mouse populations and 19 − 1 = 18 degrees of freedom for the Arabidopsis population under the FIXED-A, FIXED-B, MPWGAIM, IM, and CIM methods. For the RANDOM-A and RANDOM-B methods, the P-value for each marker was calculated from a mixture of two chi-square distributions, denoted by , where is just a fixed number of 0 (Chernoff 1954; Visscher 2006). Let Pk be the P-value for marker k, it was calculated using
| (14) |
where is the likelihood-ratio test statistic calculated using equation 13, and is a chi-square variable with one degree of freedom. In the real data analysis, we permuted the data 1000 times to generate a null distribution of the test statistics []. From this null distribution, we determined the 95% quantile and used it as an empirical critical value of a test statistic. A marker with the test statistic [] greater than this critical value is claimed to be significant at the 0.05 genome-wide type I error rate. For the IM and CIM methods, a permuted sample was generated by randomly shuffling the phenotypes and keeping the genotypes intact. For the four methods with polygenic background control, the labels of the kinship matrix go with the reshuffled phenotypes so that the polygenic covariance structure remains the same as that in the original data set. This kind of permutation will not destroy the polygenic variance (Cheng and Palmer 2013). Note that permutation was used only in real data analysis to generate empirical critical values for significance tests. In power calculation of the simulated data analysis, empirical critical values were generated from multiple simulations under the null model.
Simulation experiment
The simulation experiment was conducted based on the genotypic data of the first MAGIC population of mice (the CC population). As a result, the sample size was fixed at 458. We used genotypes of the first five chromosomes as the true genotypes to conduct the simulation experiment. The five chromosomes contain 490, 503, 428,423, and 406 bins, respectively, leading to a total of 2250 bins. The design of the simulated QTL mimicked closely that of Verbyla et al. (2014). We simulated a total of seven QTL distributed on the five chromosomes. Information about the seven simulated QTL is shown in Table 1. The simulated allelic effects of the eight founders are given in Table 2. The polygenic and residual error variances were set at φ2 = 0.5 and σ2 = 0.5, respectively. The seven QTL collectively have a total variance of 1.1752, which is partitioned into the sum of variances for all seven QTL (1.80) plus twice the sum of all covariances −0.6248 (1.80 − 0.6248 = 1.1752). The total phenotypic variance is 1.1752 + 0.5 + 0.5 = 2.1752. Therefore, the proportion contributed to the phenotypic variance by all seven QTL is 1.1752/2.1752 = 0.5403. The proportion of the polygenic variance contributed to the phenotypic variance is 0.5/2.1752 = 0.2298. The total genetic contribution (QTL + polygene) is 0.5403 + 0.2298 = 0.7701. QTL-1 and -7 are small in terms of the proportions contributed to the trait phenotypic variance. The remaining four QTL are relatively large.
Table 1. Information for the seven simulated QTL using genotypes of the first MAGIC population of mice.
| QTL | Chromosome | Position (cM) | Bin | Variancea | Proportionb |
|---|---|---|---|---|---|
| QTL-1 | 1 | 41.35 | 209 | 0.10 | 0.046 |
| QTL-2 | 2 | 21.16 | 602 | 0.20 | 0.092 |
| QTL-3 | 3 | 58.79 | 1313 | 0.30 | 0.138 |
| QTL-4 | 3 | 65.18 | 1348 | 0.30 | 0.138 |
| QTL-5 | 4 | 27.42 | 1564 | 0.40 | 0.185 |
| QTL-6 | 4 | 41.19 | 1641 | 0.40 | 0.185 |
| QTL-7 | 5 | 28.65 | 1994 | 0.10 | 0.046 |
Variance of a QTL, which is defined as , and the variance is taken across all individuals in the MAGIC population.
Proportion of the total phenotypic variance explained by the QTL.
Table 2. Founder effects for the seven simulated QTL using genotypes of the first MAGIC population of mice.
| Founder namea | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| QTL | Chr. | Position | A/J | C57BL | 129S1 | NOD | NZO | CAST | PWK | WSB |
| QTL-1 | 1 | 41.35 | −0.174 | −0.015 | 0.145 | −0.409 | 0.046 | −0.281 | −0.058 | −0.073 |
| QTL-2 | 2 | 21.16 | −0.473 | −0.095 | −0.063 | 0.352 | 0.052 | 0.161 | −0.074 | 0.303 |
| QTL-3 | 3 | 58.79 | 0.21 | −0.181 | −0.398 | −0.414 | 0.391 | −0.422 | −0.089 | −0.174 |
| QTL-4 | 3 | 65.18 | −0.294 | 0.116 | 0.58 | 0.067 | −0.111 | 0.172 | −0.443 | 0.267 |
| QTL-5 | 4 | 27.42 | 0.549 | 0.113 | 0.595 | 0.266 | −0.13 | 0.161 | −0.43 | −0.265 |
| QTL-6 | 4 | 41.19 | −0.287 | 0.225 | −0.027 | 0.104 | −0.123 | −0.227 | 0.809 | 0.061 |
| QTL-7 | 5 | 28.65 | −0.252 | −0.042 | 0.042 | −0.083 | 0.028 | 0.346 | −0.202 | 0.132 |
Strain names of the 8 founder strains initiating the CC population.
Under the preceding parameter setups, we generated 1000 independent data sets to evaluate the empirical powers under a 0.05 type I error. We also generated 1000 additional data sets under the null model (no QTL were simulated but the polygene). Results of the data analysis from the null model were used to generate the empirical distribution of the test statistics [] and draw the empirical thresholds of the test statistics for hypothesis tests. The statistical powers from the 1000 replicated simulation experiments were reported by comparing the results with the empirically drawn thresholds of the test statistics. For each simulated QTL, a ±5-cM window around the true position was reserved for power calculation, as done by Verbyla et al. (2014). If any bin within this window was detected, the QTL covered by this window was claimed to be detected. Any detected bins beyond this window were counted as false positives. All seven methods mentioned earlier were used to analyze the simulated data. The empirical powers were compared for the seven methods.
Data availability
The new methods of QTL mapping for MAGIC populations were implemented in an R package called MagicQTL, which is provided in the Supporting Information and downloadable from the journal article website (see File S3 for the R package and File S4 for the user instruction of the R package). R codes for data simulation, data preparation, and data analysis are downloadable from https://github.com/JulongWei/MagicQTL. This website also provides the R code for calling the MPWGAIM package.
Results
Simulation studies
Statistical powers and false discovery rate (FDR):
The empirical statistical powers drawn from 1000 replicated simulations are given in Table 3. In general, the RANDOM-A and RANDOM-B methods have slightly higher powers than the FIXED-A and FIXED-B methods for the five large simulated QTL. The FIXED-B and RANDOM-B methods have substantially higher powers than the FIXED-A and RANDOM-A methods. The MPWGAIM method has lower power for the first four large QTL (QTL-2 to QTL-5) than that of the FIXED-A, FIXED-B, RANDOM-A, and RANDOM-B methods. The MPWGAIM method has an advantage over the other methods for detecting the following three QTL: QTL-1, QTL-6, and QTL-7. Except for the MPWGAIM method, no methods have sufficient power to detect the two small QTL (QTL-1 and QTL-7). Overall, the new methods (i.e., FIXED-B, RANDOM-A, and RANDOM-B) are more powerful than the existing methods (i.e., FIXED-A, MPWGAIM, IM, and CIM) for large QTL.
Table 3. Statistical powers for the seven simulated QTL and FDR drawn from 1000 replicated simulation experiments.
| Method | QTL-1 | QTL-2 | QTL-3 | QTL-4 | QTL-5 | QTL-6 | QTL-7 | FDR |
|---|---|---|---|---|---|---|---|---|
| FIXED-A | 0.001 | 0.699 | 0.827 | 0.911 | 0.985 | 0.628 | 0.003 | 0.0015 |
| FIXED-B | 0.003 | 0.868 | 0.919 | 0.957 | 0.993 | 0.750 | 0.014 | 0.0034 |
| RANDOM-A | 0.002 | 0.704 | 0.849 | 0.916 | 0.985 | 0.636 | 0.004 | 0.0014 |
| RANDOM-B | 0.003 | 0.868 | 0.923 | 0.959 | 0.993 | 0.754 | 0.014 | 0.0036 |
| MPWGAIM | 0.150 | 0.500 | 0.690 | 0.700 | 0.670 | 0.770 | 0.340 | 0.0179 |
| IM | 0.003 | 0.352 | 0.326 | 0.355 | 0.494 | 0.389 | 0.001 | 0.0608 |
| CIM-30 | 0.002 | 0.476 | 0.620 | 0.761 | 0.986 | 0.754 | 0.015 | 0.0113 |
| CIM-50 | 0.001 | 0.136 | 0.043 | 0.015 | 0.512 | 0.009 | 0.003 | 0.0317 |
| CIM-65 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.002 | 0.000 | 0.940 |
We also compared the FDR for the seven methods (see the last column of Table 3). Here we define the FDR as the proportion of detected QTL that are not true (±5.00 cM away from a simulated QTL). Clearly, the FIXED-A, FIXED-B, RANDOM-A, and RANDOM-B methods achieve better control of the FDR than the MPWGAIM method, which, in general, is better than the IM and CIM methods.
Behaviors of the methods:
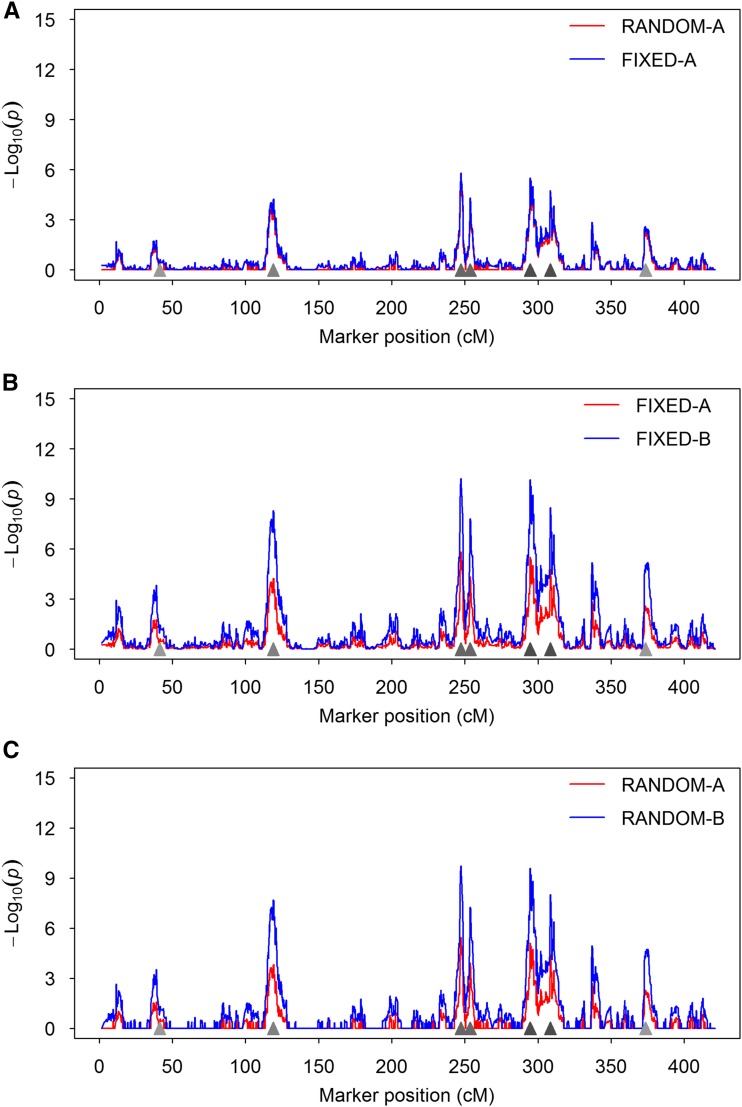
We first demonstrate the difference between the random model and the fixed model in terms of the test statistic expressed as of scanned markers using a single simulated data set (Figure 1). Figure 1A shows the difference between the RANDOM-A and FIXED-A methods. Clearly, the test statistic of the FIXED-A method is slightly higher than that of the RANDOM-A method. We also noticed that the statistic for the RANDOM-A method is very close to zero in regions where no QTL was simulated. This demonstrates the shrinkage property of the random method. In either case, the test-statistic profiles show clear peaks at positions where simulated QTL reside, and the heights of the peaks are proportional to the sizes of the simulated QTL. Figure 1B compares the FIXED-A and FIXED-B methods, where the profile of the FIXED-B method shows higher peaks than the FIXED-A method. This implies that the FIXED-B method may have a higher power than the FIXED-A method. Figure 1C compares the RANDOM-A and RANDOM-B methods. This also implies that releasing the polygenic counterpart of a marker back to the model may help to increase the power of detecting this marker. These types of behaviors are expected to be observed in data analyses of real experiments.
Figure 1.
Test-statistic profiles of different methods from a simulated data set. The test statistics are presented as . Locations of the simulated QTL are represented by the filled triangles on the x-axis. This figure demonstrates the common behaviors of the different methods that are expected in a real data analysis. (A) Comparison between RANDOM-A and FIXED-A. (B) Comparison between FIXED-A and FIXED-B. (C) Comparison between RANDOM-A and RANDOM-B.
Average test-statistic profiles:
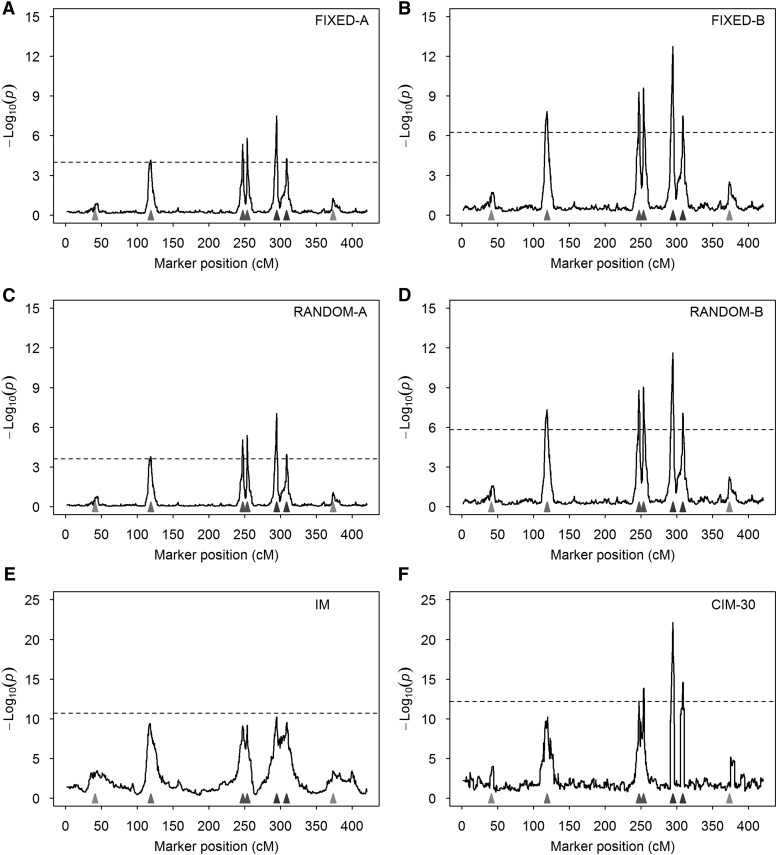
We replicated the simulation experiment 1000 times under both the null model (without QTL effects) and the alternative model (with simulated QTL). The average test-statistic profiles [] over the 1000 replicates and the 95% threshold values are illustrated in Figure 2. Comparing the fixed models (Figure 2, A and B) with the random models (Figure 2, C and D), we found that the test statistics are slightly higher for the fixed models than for the random models, but the former are also associated with higher threshold values in the test statistics. Comparing -A models (Figure 2, A and C) with -B models (Figure 2, B and D), the latter have higher peaks at positions where simulated QTL reside. For the four models, peaks corresponding to the five large QTL are higher than the threshold values, but peaks corresponding to the two small QTL are below the thresholds. The peaks for the second QTL barely touch the thresholds for -A models (Figure 2, A and C), indicating that the modified models (releasing the polygenic effect back to the model) help to boost the power. None of the peaks in IM reaches the threshold value (Figure 2E). CIM with 30 cofactors only detected four of the five large QTL (Figure 2F). When we increased the number of cofactors to 50 and 65, the CIM method behaved very badly (Figure S1). For the MPWGAIM method, owing to the lack of the test statistics in the package, we only reported the power and FDR.
Figure 2.
Average test-statistic profiles [] of six methods from 1000 replicated simulation experiments. The horizontal dashed lines represent the 95% thresholds drawn from 1000 simulated samples under the null model. The true locations of the seven simulated QTL are represented by the filled triangles on the x-axis. (A) Result of FIXED-A from the simulated data. (B) Result of FIXED-B from the simulated data. (C) Result of RANDOM-A from the simulated data. (D) Result of RANDOM-B from the simulated data. (E) Result of IM from the simulated data. (F) Result of CIM-30 from the simulated data.
Average estimated founder effects:
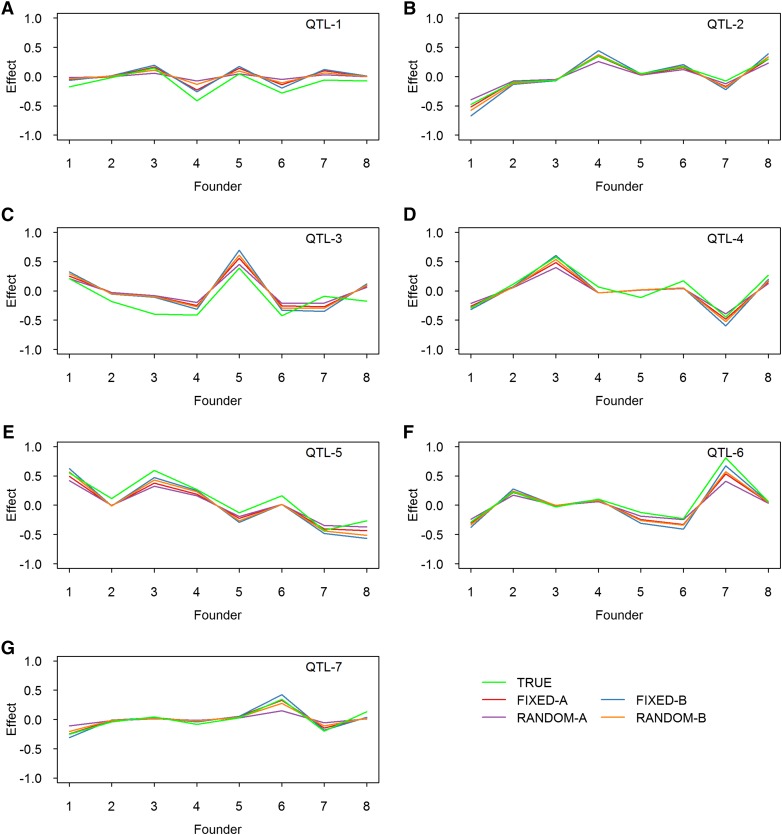
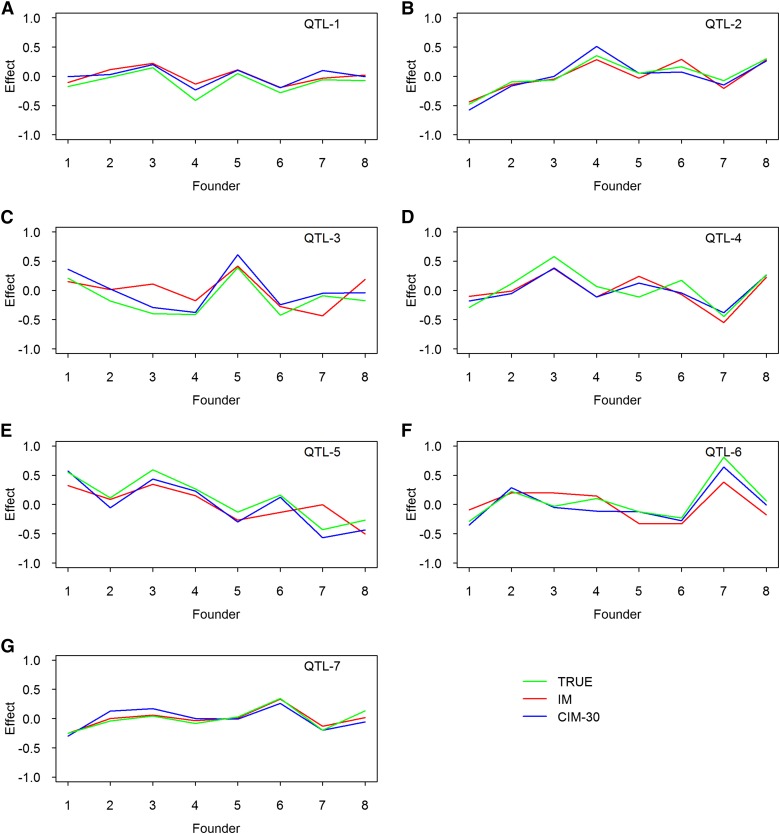
We also estimated the founder effects for the seven simulated QTL based on all simulations, and they are illustrated in Figure 3 for the fixed and random models and in Figure 4 for the IM and CIM procedures. The true effects also were plotted along with the estimated effects. All methods provided good estimates of the founder effects. The random models tend to shrink the estimated effects toward zero when the simulated QTL sizes are small (Figure 3, A and G). Although the IM and CIM methods are not as good as the other methods in terms of statistical power, both gave very good estimated founder effects. Figure S2 shows the average estimated effects of the founders when 50 and 65 markers were used as cofactors for the CIM method.
Figure 3.
True and estimated allelic effects of eight founders for seven simulated QTL in the simulation experiment. The estimated effects are the average effects of 1000 replicated experiments. Results from four methods are presented: FIXED-A, FIXED-B, RANDOM-A, and RANDOM-B. (A) Effect of QTL-1. (B) Effect of QTL-2. (C) Effect of QTL-3. (D) Effect of QTL-4. (E) Effect of QTL-5. (F) Effect of QTL-6. (G) Effect of QTL-7.
Figure 4.
True and estimated allelic effects of eight founders for seven simulated QTL in the simulation experiment. The estimated effects are the average effects of 1000 replicated experiments. Results from two methods are presented: IM and CIM-30, where -30 means that 30 markers are used as cofactors. (A) Effect of QTL-1. (B) Effect of QTL-2. (C) Effect of QTL-3. (D) Effect of QTL-4. (E) Effect of QTL-5. (F) Effect of QTL-6. (G) Effect of QTL-7.
Results of experimental data analyses:
MAGIC population in A. thaliana:
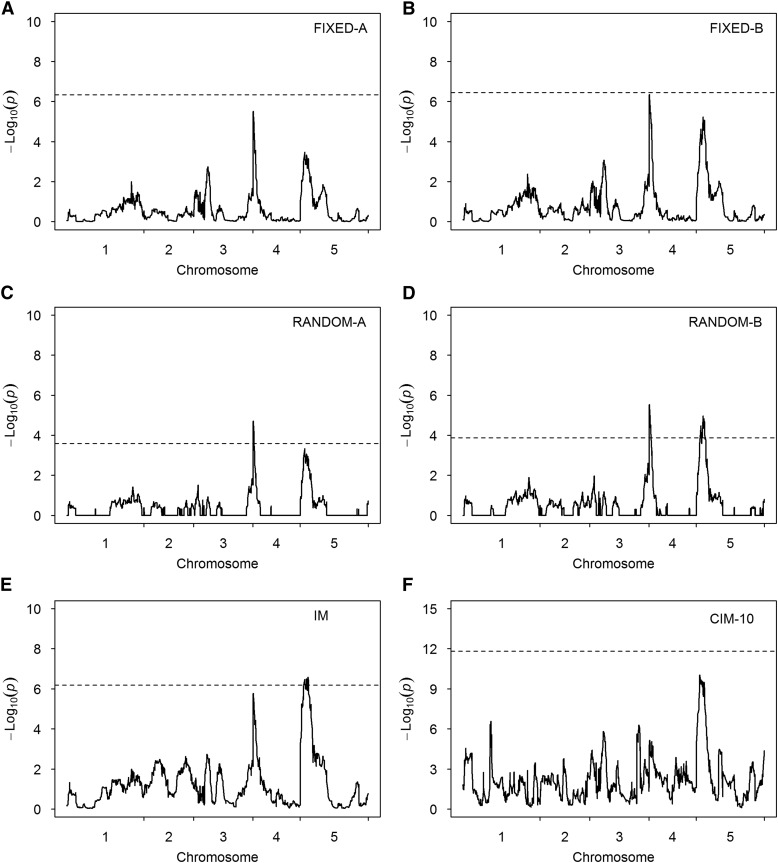
Under the polygenic model, we estimated the variance and heritability for each of the two traits, the days between bolting and flowering (DBF), and the growth rate (GR). The results are shown in Table 4. The heritability of the two traits is 0.342 and 0.473, respectively. The variance ratios for DBF and GR are and , respectively, which were used as known values and incorporated into the covariance structures for genomic scanning of all markers. Figure 5 illustrates the test-statistic profiles [] along with the 95% thresholds generated from 1000 permuted samples for DBF. Markers with test-statistic values greater than the thresholds were claimed to be statistically significant. There are two peaks standing out on chromosomes 4 and 5, respectively, for all methods except CIM-10. These two regions also showed up in the original analysis of Kover et al. (2009). However, the only method that detected both peaks is RANDOM-B, implying that this method may be the most powerful method. The detected QTL on chromosome 4 is located near a known gene called FRIGIDA. No related genes were found near the detected QTL on chromosome 5. The MPWGAIM method also detected the two QTL in the same regions (Table 5). In addition, the MPWGAIM method detected three more QTL, one on chromosome 1 (PERL0236029) and two on chromosome 3 (MASC00175 and MN3_22843506). Figure S3 (A and B) shows the results of this data analysis using the CIM method when 15 and 20 markers were used as cofactors.
Table 4. Estimated variance components and heritability of two traits in the A. thaliana population and one trait in the mouse population.
| Population | Trait | Polygenic variance | Residual variance | Heritability |
|---|---|---|---|---|
| A. thaliana | Bolt to flower | 2.187 | 4.203 | 0.342 |
| Growth rate | 1.989 | 2.215 | 0.473 | |
| Mouse | PMN | 1.373 | 1.127 | 0.549 |
Figure 5.
Test-statistic profiles [] for DBF of the A. thaliana MAGIC population obtained from six methods. The horizontal dotted lines represent the 95% thresholds generated from 1000 permuted samples. (A) Result of the FIXED-A method. (B) Result of the FIXED-B method. (C) Result of the RANDOM-A method. (D) Result of the RANDOM-B method. (E) Result of the IM method. (F) Result of the CIM-10 method.
Table 5. Significant SNPs associated with two traits in the A. thaliana population and one trait in the mouse population.
| Population | Trait | Method | SNP | Chr. | Position (kb) | P-valuea | Varianceb | Candidate gene (kb) |
|---|---|---|---|---|---|---|---|---|
| A. thaliana | Bolt to flower | RANDOM-A | MN4_142943 | 4 | 143 | 0.011 | 0.437 | FRIGIDA |
| RANDOM-B | MN4_142943 | 4 | 143 | 0.005 | 0.517 | Chr. 4: 269–272 | ||
| MN5_2707605 | 5 | 2,708 | 0.023 | 0.598 | ||||
| IM | MASC02783 | 5 | 2,522 | 0.041 | 0.874 | |||
| MPWGAIM | MN4_142943 | 4 | 143 | —c | 0.527 | |||
| MN5_1931248 | 5 | 1,931 | —c | 0.629 | ||||
| Growth rate | FIXED-B | GA1_3232 | 4 | 1,243 | 0.013 | 0.626 | FRIGIDA | |
| RANDOM-B | GA1_8429 | 4 | 1,238 | 0.010 | 0.299 | Chr. 4: 269–272 | ||
| MPWGAIM | GA1_8429 | 4 | 1,238 | —c | 0.253 | AT4G02990 (RUG2) | ||
| IM | FRI_1888 | 4 | 270 | 0.005 | 0.493 | Chr. 4: 1322–1324 | ||
| CIM-10 | GA1_7762 | 4 | 1,239 | 0.011 | 0.853 | AT4G02780 (GA1) | ||
| Chr. 4: 1238–1245 | ||||||||
| Mouse | PMN | FIXED-B | M2.887 | 2 | 87,583 | 0.040 | 0.471 | |
| RANDOM-B | M2.887 | 2 | 87,583 | 0.030 | 0.292 | |||
| MPWGAIM | M2.887 | 2 | 87,583 | —c | 0.263 | |||
| IM | M2.887 | 2 | 87,583 | 0.013 | 0.450 | |||
| CIM-5 | M2.824 | 2 | 80,663 | 0.028 | 0.496 |
P-value was obtained from a permutation test.
Denotes the variance of the effect of the detected marker combining the founder allele inheritance indicators.
Owing to the high computational cost, no permutation analysis was conducted.
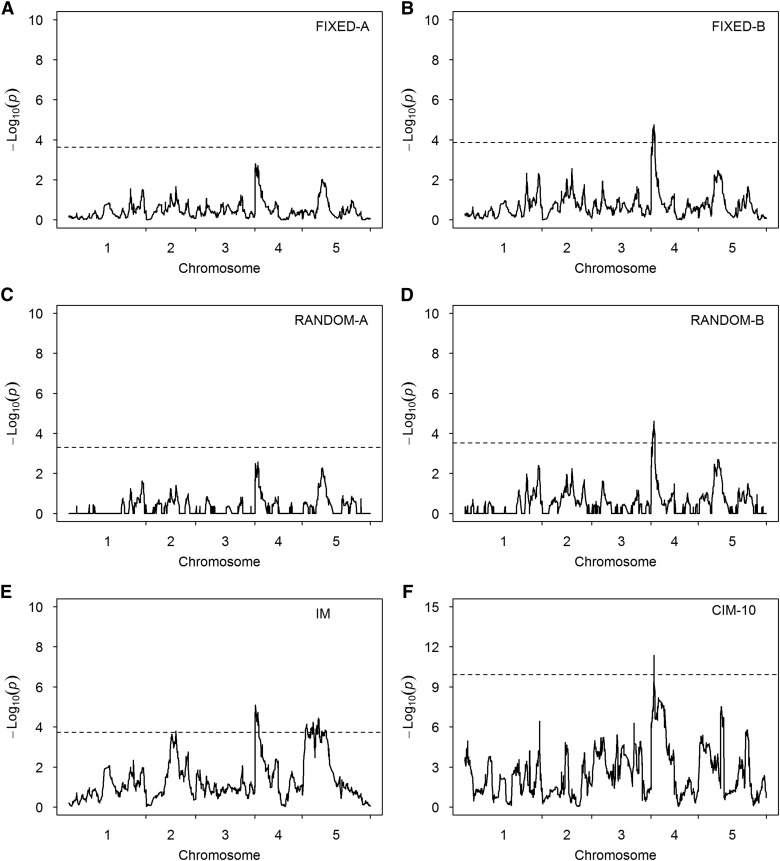
The test-statistic profiles along with permutation-generated thresholds are illustrated in Figure 6 for GR. There are many bumps in the test-statistic profiles below the thresholds, indicating that this trait is mostly polygenic. One peak in the beginning of chromosome 4 appears to be common to all six methods. Except the FIXED-A and RANDOM-A methods, all other methods have detected the peak as statistically significant. We list the SNPs exceeding the threshold values in Table 5. Three candidate genes are found in this area (about ±200 kb around the detected marker), FRIGIDA, AT4G02990 (RUG2), and AT4G02780 (GA1). The first candidate gene (FRIGIDA) is known to affect flowering time. This gene is also related to growth rate in the original study (Kover et al. 2009), where it was pointed out that this gene not only plays an important role in plan reproduction but also is a major determinant of the plant developmental process. The second candidate gene (RUG2) is important for leaf development in A. thaliana, and its loss of function leads to a pleiotropic phenotype, including leaf variegation, reduced growth, and perturbed mitochondrial and chloroplastic gene expression and development (Quesada et al. 2011). The third candidate gene (GA1) codes for the enzyme ent-kaurene synthase A. In GA1 mutants, the gibberellin biosynthesis pathway is inactivated. As a result, these mutants are deficient in bioactive Gas (Sun and Kamiya 1994). Some additional markers are detected by the IM and MPWGAIM methods, and they are listed in Table S1. The markers on chromosomes 2 and 5 detected by the IM method overlap with the additional markers detected by the MPWGAIM method. The other methods also show some bumps in regions near the additional markers detected by the IM and MPWGAIM methods. These regions (about ±200 kb around the peaked markers) harbored several candidate genes, which are not related to GR in terms of gene function. Figure S3 (C and D) shows the results when 15 and 20 markers were used as cofactors for the CIM method.
Figure 6.
Test-statistic profiles [] for GR of the A. thaliana MAGIC population obtained from six methods. The horizontal dotted lines represent the 95% thresholds generated from 1000 permuted samples. (A) Result of the FIXED-A method. (B) Result of the FIXED-B method. (C) Result of the RANDOM-A method. (D) Result of the RANDOM-B method. (E) Result of the IM method. (F) Result of the CIM-10 method.
Pre-CC population of mice:
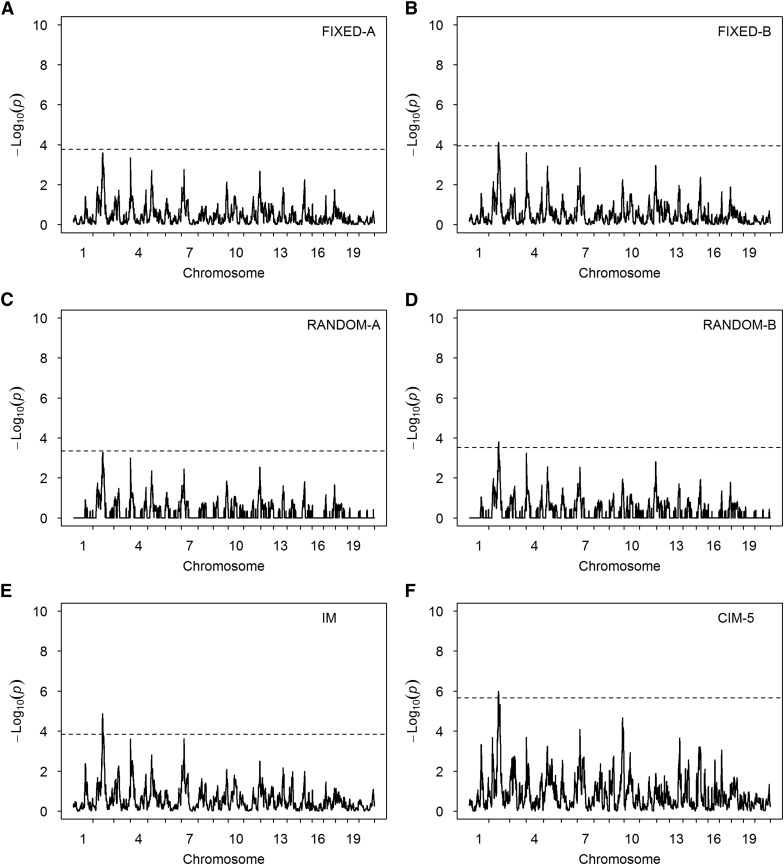
We analyzed a trait named PMN from this population. The phenotypic values were log transformed prior to the analysis, as done in the original study. We estimated the genetic variance and heritability of the trait, which are presented in Table 3. The trait is highly heritable, with a heritability of 0.55. The variance ratio is , which was used along with the kinship matrix to control the polygenic effect in QTL mapping. We scanned the entire genome using all seven methods. The test-statistic profiles are illustrated in Figure 7. Except for the FIXED-A and RANDOM-A methods, all other methods detected a marker on chromosome 2 (Table 5). This marker also was detected by Rutledge et al. (2014) in the original study. They found a candidate gene (Dpn1) near this marker. No other candidate genes were found in the neighborhood of this marker. Figure S3 (E and F) shows the results when 10 and 20 markers were used as cofactors for the CIM method.
Figure 7.
Test-statistic profiles [] for PMN of the pre-CC mouse population obtained from six methods. The horizontal dotted lines represent the 95% thresholds generated from 1000 permuted samples. (A) Result of the FIXED-A method. (B) Result of the FIXED-B method. (C) Result of the RANDOM-A method. (D) Result of the RANDOM-B method. (E) Result of the IM method. (F) Result of the CIM-5 method.
Discussion
A key difference between QTL mapping in MAGIC and biparental populations is the difference in the number of effects to be estimated and tested per locus. Under the fixed-model framework, for an eight-parent MAGIC population, the number of effects per locus is 8 − 1 = 7, while it is always 2 − 1 = 1 for a biparental population. Under the null model, the likelihood-ratio text follows a chi-square distribution with 7 degrees of freedom. In a p-parent MAGIC population, p − 1 is the degrees of freedom. When p is large, this test is not convenient and sometimes can be unstable (Gatti et al. 2014). For example, if some founder alleles fail to appear in the progeny for some loci, the Z matrices for these loci will not have the same rank as those loci with full representation of all founders. This variable-rank situation will cause some difficulty in programming. More important, the degree of freedom will vary across loci, so the likelihood-ratio text statistic will not be comparable across loci. We developed a random-model approach to estimate and test the variance among all founder effects per locus. As a result, we only need to estimate and test a single parameter (the variance) regardless how large the number of founders is in a MAGIC population. Simulation studies showed that the random-model approach is slightly more powerful than the fixed-model approach.
Some investigators also considered founder allelic effects as random in MAGIC population QTL mapping (Verbyla et al. 2014; Zhang et al. 2014). The MPWGAIM procedure of Verbyla et al. (2014) assumes that all founder allelic effects of the same locus share a common variance and that this variance varies across loci. A forward variable selection approach was adopted by adding one locus at a time to the model until no further improvement was achieved. For consistency of comparison, we adopted the critical value generated from the null model, similar to the other methods, to evaluate the power of QTL detection by the MPWGAIM method using the same test criterion. We demonstrated lower powers (for large QTL) and higher FDR for the MPWGAIM method. The MPWGAIM method can be time consuming if the numbers of markers and QTL included in the model are large. Table 6 compares the computational times of our methods with that of the MPWGAIM method under several different scenarios. Clearly, the new genome-scanning approaches proposed in this study are substantially faster than the MPWGAIM method. The Bayesian method of Zhang et al. (2014) also treats founder allelic effects as random, and it is a multiple-QTL model. Because the method is implemented via an MCMC sampling scheme, it is also computationally expensive. The authors suggested that the method is better used to fine-tune the results after an initial genome scan of all markers.
Table 6. Computational performances of different methods with different sample sizes and different numbers of markers.
| Method | Mouse-458-2250a | Mouse-458-6683b | Mouse-151-27309 | Arabidopsis-426-1254 |
|---|---|---|---|---|
| FIXED-A | 22 sec | 53 sec | 1 min 43 sec | 23 sec |
| FIXED-B | 41 sec | 1 min 44 sec | 4 min 25 sec | 34 sec |
| RANDOM-A | 36 sec | 1 min 42 sec | 5 min 21 sec | 27 sec |
| RANDOM-B | 55 sec | 2 min 31 sec | 8 min 10 sec | 39 sec |
| MPWGAIM | 32 min 29 sec | 2 h 33 min | 1h 33 min | 11 min 51 sec |
The first number after the species name is the sample size, and the second number is the number of markers.
The number of bins is 6683, which is the total number of bins of the entire 19 chromosomes of the mouse genome.
When cofactors are replaced by the polygene for background control, there is a potential competition between a currently scanned marker and its counterpart in the polygene, which is detrimental to the power. The competition can be very serious when the number of markers used to calculate the kinship matrix is small, although it may be negligible when a very large number of markers are used to calculate the kinship matrix. To prevent such a competition, we proposed releasing the polygenic component corresponding to the scanned marker back to the model. This has dramatically increased the statistical power of QTL detection. The BLUP estimate of a marker effect in the polygene is calculated only once prior to the marker scanning step, and thus little additional computational cost is present. We could have removed the currently scanned marker from the kinship matrix to avoid the competition. However, this would substantially increase the computational burden because a new kinship matrix would have to be provided for each marker scanned. Special algorithms, such as the spectrally transformed linear mixed model (FaST-LMM) proposed by Lippert et al. (2011), may be used to ease the computational intensity. However, the fast speed is not achieved without a cost. One has to use markers with a number substantially smaller than the sample size to gain the fast speed. When the number of markers used to construct the kinship matrix is too small, optimal control of the polygene may not be guaranteed (Zhou and Stephens 2012).
The genotype coding system of QTL mapping in MAGIC populations is different from that in biparental populations. We used the Zk variable (an n × 8 matrix) to indicate the founder allelic inheritances for the kth marker. This variable also was used to calculate the marker-inferred kinship matrix K. The kinship matrix was eventually rescaled by a normalization factor, which is the average of the diagonal elements of the original unnormalized kinship matrix. After normalization, the diagonal elements of the kinship matrix are all around unity. Such normalization will bring the estimated polygenic variance into the same scale as the residual error variance. Our normalization factor is different from that proposed by VanRaden (2008), which is the sum of heterozygosity across all loci. The normalization factor only changes the scale of the estimated polygenic variance; it affects neither the hypothesis tests nor the results of QTL mapping. In GWAS, where the Zk variable is simply a vector, Kang et al. (2008) placed a weight variable for each marker in calculating the kinship matrix to take into account variable information contents (allele frequencies) across different marker loci. It is not obvious how to evaluate information contents when the genotype indicator variable Zk for each marker is a matrix. In CC and pre-CC mice, all founders contributed equally to the mapping population, and thus, the weight variable can be safely ignored (e.g., taking the default value of 1 from all markers). In the 19-parent MAGIC population of Arabidopsis, where the parental contribution varies across founders, a weighted kinship matrix may be more appropriate. Further study is needed to develop an appropriate weight matrix. Alternatively, the method of Gatti et al. (2014) for calculating the kinship matrix may be adopted here. The relationship between each pair of individuals is a kind of average “scaled similarity” over all loci. In our notation, the relationship between individuals i and j (the ith row and the jth column of the kinship matrix) is expressed as
| (15) |
We did not use this kinship matrix because the polygenic counterpart of marker k (used in the FIXED-B and RANDOM-B methods) would be difficult to interpret when this K matrix is used. Furthermore, whether or not such a kinship matrix can adjust unbalanced contributions from different founders is still questionable.
The random-model approach is a kind of Bayesian analysis if the founder effects are considered as parameters and the variance of the founder effects is considered as a prior variance. Because the prior variance is estimated from the data, it is called empirical Bayes (Xu 2007).The random model developed for QTL mapping in MAGIC populations can be used in a number of other situations. The method can be extended to QTL mapping in DO populations, such as the DO population of mice developed from the same eight parents as the CC mice (Gatti et al. 2014).
The random-model approach is computationally more intensive than the fixed-model approach, where the QTL effects are treated as fixed effects because it requires estimation of a variance component for each marker scanned. We adopted the eigen-decomposition algorithms for the polygenic (null) model and combined them with the Woodbury matrix identity for estimation of QTL variance. It would not be realistic to perform such a random-model QTL mapping without resort to these special algorithms. There may be room for further improvement in the computational speed. However, we emphasize the concept and the novelty of the method, which are far more important than technical improvement in computational speed. Finally, all analyses were performed using an R program written by the authors. We developed an R package named MagicQTL, which is provided on the journal website.
Acknowledgments
We thank Arunas P. Verbyla for sharing the mpwgaim program and for the tremendous help in running this program. We also thank Samir N. P. Kelada for providing the pre-CC mouse data. This project was supported by National Science Foundation grant 005400 to S.X. and a China Scholarship Council Award to J.W.
Footnotes
Communicating editor: I. Hoeschele
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.179945/-/DC1
Literature Cited
- Bandillo N., Raghavan C., Muyco P. A., Sevilla M. A. L., Lobina I. T., et al. , 2013. Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice 6: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen Ś., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Cheng R., Palmer A. A., 2013. A simulation study of permutation, bootstrap, and gene dropping for assessing statistical significance in the case of unequal relatedness. Genetics 193: 1015–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff H., 1954. On the distribution of the likelihood ratio. Ann. Math. Statist. 25: 573–578. [Google Scholar]
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D., et al. , 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137. [DOI] [PubMed] [Google Scholar]
- Collaborative Cross Consortium , 2012. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 190: 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti D. M., Svenson K. L., Shabalin A., Wu L.-Y., Valdar W., et al. , 2014 Quantitative trait locus mapping methods for Diversity Outbred mice. G3 4: 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur P. M., Jukanti A. K., Varshney R. K., 2012. Impact of genomic technologies on chickpea breeding strategies. Agronomy 2: 199–221. [Google Scholar]
- Golub G. H., Van Loan C. F., 1996. Matrix Computations, Ed. 3 Johns Hopkins University Press, Baltimore. [Google Scholar]
- Huang B. E., George A. W., 2011. R/mpMap: a computational platform for the genetic analysis of multiparent recombinant inbred lines. Bioinformatics 27: 727–729. [DOI] [PubMed] [Google Scholar]
- Huang B. E., George A. W., Forrest K. L., Kilian A., Hayden M. J., et al. , 2012. A multiparent advanced generation inter‐cross population for genetic analysis in wheat. Plant Biotechnol. J. 10: 826–839. [DOI] [PubMed] [Google Scholar]
- Huang B. E., Verbyla K. L., Verbyla A. P., Raghavan C., Singh V. K., et al. , 2015. MAGIC populations in crops: current status and future prospects. Theor. Appl. Genet. 128: 999–1017. [DOI] [PubMed] [Google Scholar]
- Jourjon M.-F., Jasson S., Marcel J., Ngom B., Mangin B., 2005. MCQTL: multi-allelic QTL mapping in multi-cross design. Bioinformatics 21: 128–130. [DOI] [PubMed] [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., MacDonald S. J., Long A. D., 2012a Properties and power of the Drosophila Synthetic Population Resource for the routine dissection of complex traits. Genetics 191: 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Merkes C. M., McNeil C. L., Hoofer S. R., Sen S., et al. , 2012b Genetic dissection of a model complex trait using the Drosophila Synthetic Population Resource. Genome Res. 22: 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., et al. , 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Botstein D., 1989. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C., Listgarten J., Liu Y., Kadie C. M., Davidson R. I., et al. , 2011. FaST linear mixed models for genome-wide association studies. Nat. Methods 8: 833–835. [DOI] [PubMed] [Google Scholar]
- MacDonald S. J., Long A. D., 2007. Joint estimates of quantitative trait locus effect and frequency using synthetic recombinant populations of Drosophila melanogaster. Genetics 176: 1261–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay I. J., Bansept-Basler P., Barber T., Bentley A. R., Cockram J., et al. , 2014. An eight-parent multiparent advanced generation inter-cross population for winter-sown wheat: creation, properties, and validation. G3 4: 1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R., Talbot C. J., Turri M. G., Collins A. C., Flint J., 2000. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. USA 97: 12649–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual L., Desplat N., Huang B. E., Desgroux A., Bruguier L., et al. , 2015. Potential of a tomato MAGIC population to decipher the genetic control of quantitative traits and detect causal variants in the resequencing era. Plant Biotechnol. J. 13: 565–577. [DOI] [PubMed] [Google Scholar]
- Quesada V., Sarmiento‐Mañús R., González‐Bayón R., Hricová A., Pérez‐Marcos R., et al. , 2011. Arabidopsis RUGOSA2 encodes an mTERF family member required for mitochondrion, chloroplast and leaf development. Plant J. 68: 738–753. [DOI] [PubMed] [Google Scholar]
- Rakshit S., Rakshit A., Patil J., 2012. Multiparent intercross populations in analysis of quantitative traits. J. Genet. 91: 111–117. [DOI] [PubMed] [Google Scholar]
- Rutledge H., Aylor D. L., Carpenter D. E., Peck B. C., Chines P., et al. , 2014. Genetic regulation of Zfp30, CXCL1, and neutrophilic inflammation in murine lung. Genetics 198: 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannemann W., Huang B. E., Mathew B., Léon J., 2015. Multi-parent advanced generation inter-cross in barley: high-resolution quantitative trait locus mapping for flowering time as a proof of concept. Mol. Breed. 35: 1–16. [Google Scholar]
- Sun T. P., Kamiya Y., 1994. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill D. W., Hunter K. W., Williams R. W., 2002. Genetic dissection of complex and quantitative traits: from fantasy to reality via a community effort. Mamm. Genome 13: 175–178. [DOI] [PubMed] [Google Scholar]
- Valdar W., Flint J., Mott R., 2006. Simulating the collaborative cross: power of quantitative trait loci detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics 172: 1783–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423. [DOI] [PubMed] [Google Scholar]
- Varshney R. K., Dubey A., 2009. Novel genomic tools and modern genetic and breeding approaches for crop improvement. J. Plant Biochem. Biotechnol. 18: 127–138. [Google Scholar]
- Verbyla A. P., George A. W., Cavanagh C. R., Verbyla K. L., 2014. Whole-genome QTL analysis for MAGIC. Theor. Appl. Genet. 127: 1753–1770. [DOI] [PubMed] [Google Scholar]
- Visscher P. M., 2006. A note on the asymptotic distribution of likelihood ratio tests to test variance components. Twin Res. Hum. Genet. 9: 490–495. [DOI] [PubMed] [Google Scholar]
- Xu S., 2007. An empirical Bayes method for estimating epistatic effects of quantitative trait loci. Biometrics 63: 513–521. [DOI] [PubMed] [Google Scholar]
- Xu S., 2013a Genetic mapping and genomic selection using recombination breakpoint data. Genetics 195: 1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., 2013b Mapping quantitative trait loci by controlling polygenic background effects. Genetics 195: 1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W. H., Vroh Bi I., Yamasaki M., et al. , 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208. [DOI] [PubMed] [Google Scholar]
- Yu J., Holland J. B., McMullen M. D., Buckler E. S., 2008. Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z.-B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wang W., Valdar W., 2014. Bayesian modeling of haplotype effects in multiparent populations. Genetics 198: 139–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M., 2012. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44: 821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The new methods of QTL mapping for MAGIC populations were implemented in an R package called MagicQTL, which is provided in the Supporting Information and downloadable from the journal article website (see File S3 for the R package and File S4 for the user instruction of the R package). R codes for data simulation, data preparation, and data analysis are downloadable from https://github.com/JulongWei/MagicQTL. This website also provides the R code for calling the MPWGAIM package.