Abstract
Single-strand annealing (SSA) is an important homologous recombination mechanism that repairs DNA double strand breaks (DSBs) occurring between closely spaced repeat sequences. During SSA, the DSB is acted upon by exonucleases to reveal complementary sequences that anneal and are then repaired through tail clipping, DNA synthesis, and ligation steps. In baker’s yeast, the Msh DNA mismatch recognition complex and the Sgs1 helicase act to suppress SSA between divergent sequences by binding to mismatches present in heteroduplex DNA intermediates and triggering a DNA unwinding mechanism known as heteroduplex rejection. Using baker’s yeast as a model, we have identified new factors and regulatory steps in heteroduplex rejection during SSA. First we showed that Top3-Rmi1, a topoisomerase complex that interacts with Sgs1, is required for heteroduplex rejection. Second, we found that the replication processivity clamp proliferating cell nuclear antigen (PCNA) is dispensable for heteroduplex rejection, but is important for repairing mismatches formed during SSA. Third, we showed that modest overexpression of Msh6 results in a significant increase in heteroduplex rejection; this increase is due to a compromise in Msh2-Msh3 function required for the clipping of 3′ tails. Thus 3′ tail clipping during SSA is a critical regulatory step in the repair vs. rejection decision; rejection is favored before the 3′ tails are clipped. Unexpectedly, Msh6 overexpression, through interactions with PCNA, disrupted heteroduplex rejection between divergent sequences in another recombination substrate. These observations illustrate the delicate balance that exists between repair and replication factors to optimize genome stability.
Keywords: heteroduplex rejection, DNA mismatch repair, Sgs1, Rmi1, Top3, PCNA, Msh6 overexpression
HOMOLOGOUS recombination between nonallelic, divergent sequences in the genome can lead to chromosomal rearrangements. Multiple cellular mechanisms contribute to suppressing such deleterious events. For example, the organization of the interphase nucleus helps to limit physical interactions between distant regions of the genome, and cell cycle regulation of homologous recombination factors suppresses recombination events at times when distant genomic regions and nonallelic sequences tend to be unprotected or closer to each other (reviewed in George and Alani 2012). Despite these lines of defense, physical interactions between nonallelic sequences can still frequently occur, and when damage or replication stalling occurs in the vicinity of these interactions, nonallelic homologous recombination (NAHR) can be initiated (reviewed in Liu et al. 2012). Several mechanisms have been proposed to understand how recombination events between divergent DNA sequences, known as homeologous recombination, are prevented. One such mechanism, heteroduplex rejection, requires helicase-mediated unwinding of recombination intermediates (Sugawara et al. 2004; Surtees et al. 2004).
The DNA mismatch repair (MMR) system acts to repair polymerase errors incurred during DNA replication. DNA mismatch repair factors also play critical roles in maintaining the fidelity of homologous recombination in both prokaryotes and eukaryotes by inhibiting recombination between divergent sequences, and this function is directly related to levels of sequence divergence (Rayssiguier et al. 1989; Shen and Huang 1989; Petit et al. 1991; de Wind et al. 1995; Selva et al. 1995; Chambers et al. 1996; Datta et al. 1996; Hunter et al. 1996; Porter et al. 1996; Elliott and Jasin 2001; Nicholson et al. 2000). In the yeast Saccharomyces cerevisiae, MMR is initiated by either the Msh2-Msh6 (MutSα) or Msh2-Msh3 (MutSβ) heterodimer binding to DNA containing mismatches; Msh2-Msh6 shows high specificity for single base–base mismatches and single nucleotide insertions/deletions, whereas Msh2-Msh3 shows high specificity for insertion/deletion loops up to 16 nucleotides in size (reviewed in Kunkel and Erie 2005). Msh2-Msh6 has been shown in baker’s yeast to colocalize with the DNA replication machinery (Hombauer et al. 2011a). Mlh heterodimers (primarily Mlh1-Pms1) then interact with Msh-mismatch complexes to recruit downstream factors that complete MMR through excision, resynthesis, and ligation steps. These downstream factors include the Exo1 exonuclease, the PCNA processivity clamp, replication factor C (RFC), DNA polymerases δ and ε, and RPA single-strand binding protein (reviewed in Kunkel and Erie 2005).
Antirecombination has been hypothesized to occur by regulation of branch migration of recombination intermediates to limit heteroduplex extension, rejection of recombination intermediates through nucleolytic degradation, or by unwinding heteroduplex DNA intermediates (Sugawara et al. 2004; Surtees et al. 2004; Goldfarb and Alani 2005; Waldman 2008). The single-strand annealing (SSA) pathway provides a relatively simple system to study antirecombination mechanisms (Figure 1A). This Rad52-dependent pathway is a specialized type of homologous recombination that is initiated by a double-strand break (DSB) between closely spaced repeat sequences. Resection of single-strand DNA (ssDNA) at the break, followed by annealing of homologous sequences, tail clipping, DNA synthesis, and ligation, results in repair of the DSB involving a deletion between the repeat sequences (Lin and Sternberg 1984; Fishman-Lobell et al. 1992: Sugawara and Haber 1992). A critical step required to complete SSA is the removal of 3′ nonhomologous tails, which occurs in steps requiring Msh2-Msh3 and the Rad1-Rad10 endonuclease (Fishman-Lobell et al. 1992; Sugawara et al. 1997).
Figure 1.
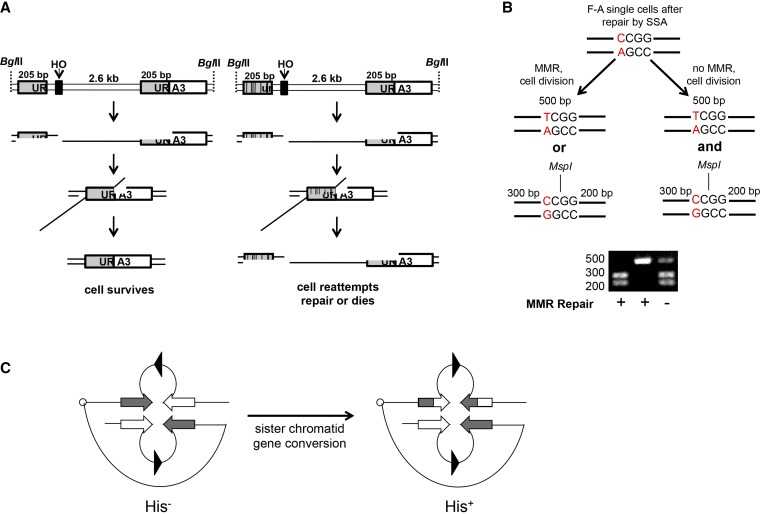
Schematics of SSA and inverted repeat recombination assays. (A) Cell survival assay to measure heteroduplex rejection efficiency. Two strains, A-A (identical) and F-A (3% divergent), possess a partial duplication of URA3, an HO cut site, and 2.6 kb of λ DNA upstream of the endogenous URA3 locus. Galactose induction of HO endonuclease results in a unique DSB between URA3 sequence repeats that is repaired efficiently by SSA in the A-A strain. Repair in F-A strains is inefficient due to rejection of the heteroduplex intermediate created by annealing of the 3% divergent URA3 repeats. Successful SSA will lead to high cell survival, but cells that try to repair by SSA (perhaps multiple times) but ultimately fail due to heteroduplex rejection will suffer the lethality caused by a persistent DSB. (B) MspI digestion assay to assess MMR. Colonies obtained from single unbudded F-A cells induced for HO expression were analyzed for the presence of an MspI site present in the URA3 “F” allele but not the URA3 “A” allele (Materials and Methods). MMR has occurred if PCR products amplifying this restriction site contain MspI digested or undigested bands, but not both. MMR has failed to occur if the PCR products contain a mixture of digested and undigested bands. (C) Intron-based intramolecular recombination assay involving a HIS3 reporter (adapted from Nicholson et al. 2000). In this assay, His+ recombinants are thought to result from gene conversion events involving inverted repeat sequences (open and gray boxes) present on sister chromatids. Intron substrates predicted to form base–base (cβ2/cβ2-ns) and 4-nt loop (cβ2/cβ2-4L) mismatches in heteroduplex DNA were analyzed in this study.
SSA is thought to be the predominant form of DSB repair within highly repetitive regions of the genome, such as the ribosomal DNA (Kobayashi 2006; Li et al. 2008; George and Alani 2012), and limits unavoidable loss of genetic information to local deletions rather than large-scale rearrangements. It could also offset repeat expansions in these regions. Studies using an SSA cassette in which two 205-bp repeats show 3% divergence have identified an antirecombination mechanism that is dependent on the Msh2-Msh6 complex and the RecQ family helicase Sgs1 (Figure 2) (Sugawara et al. 2004; Goldfarb and Alani 2005). In this system mismatch binding and ATP binding and hydrolysis activities of Msh proteins were found to be critical for antirecombination, suggesting roles for Msh proteins in both mismatch recognition and signaling (Goldfarb and Alani 2005). In contrast Mlh proteins play a less critical role in antirecombination during SSA, and other MMR/DNA repair factors such as Exo1 and Srs2, appear to be dispensable (Selva et al. 1995; Chambers et al. 1996; Datta et al. 1996; Hunter et al. 1996; Chen and Jinks-Robertson 1999; Nicholson et al. 2000; Sugawara et al. 2004). PCNA, the processivity clamp for DNA replication, has been shown to interact with Msh6 and enhance mismatch recognition by Msh2-Msh6 in vitro (Flores-Rozas et al. 2000). It has also been shown to be required at an early step as well as during the later DNA resynthesis step in MMR (Umar et al. 1996; Gu et al. 1998). However it is unclear if PCNA plays a stimulatory role during heteroduplex rejection in SSA.
Figure 2.
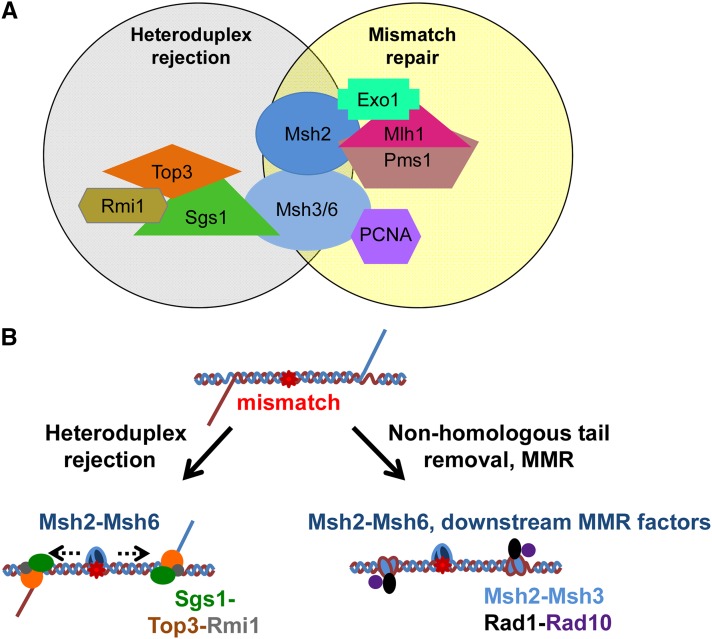
Factors that act in heteroduplex rejection and MMR. (A) Heteroduplex rejection and MMR are both initiated by Msh heterodimers binding to DNA mismatches, but subsequent steps involve interactions with different sets of factors. (B) Model for heteroduplex rejection during SSA involving Msh proteins and the Sgs1-Top3-Rmi1 complex. Following strand annealing, Msh proteins bind to mismatches (red star) in heteroduplex DNA and undergo a conformational change that, in the left panel, licenses recruitment of Sgs1-Top3-Rmi1, loaded at the junction between the heteroduplex and the 3′ nonhomologous tail, to promote heteroduplex rejection. In the right panel, recruitment of nonhomologous tail removal factors instead of Sgs1-Top3-Rmi1, results in the removal of the 3′ nonhomologous tail, followed by MMR. Adapted from George and Alani (2012).
Studies in several labs have implicated the helicase activity of Sgs1 in antirecombination (Myung et al. 2001; Spell and Jinks-Robertson 2004; Sugawara et al. 2004; Goldfarb and Alani 2005). Consistent with these findings, a study by Sugawara et al. (2004), using a three-repeat SSA competition assay, showed that heteroduplex rejection is likely to occur through an unwinding mechanism rather than by nucleolytic degradation, suggesting that heteroduplex rejection during SSA involves mismatch recognition by Msh proteins followed by recruitment of Sgs1 to stimulate heteroduplex DNA unwinding. Sgs1 is known to interact with the topoisomerase complex Top3-Rmi1 in a variety of processes including 5′ to 3′ strand resection during homologous recombination and dissolution of double Holliday junctions (Bennett et al. 2000; Fricke et al. 2001; Chang et al. 2005; Mullen et al. 2005; Ui et al. 2005; Chen and Brill 2007; Cejka et al. 2010; Cejka and Kowalczykowski 2010; Niu et al. 2010). Recently Sgs1 and Top3-Rmi1 were shown to have interdependent and independent functions in meiosis (Kaur et al. 2015; Tang et al. 2015). However an antirecombination role for the Top3-Rmi1 complex has not been clearly established, though previous studies showed that top3Δ mutants increased recombination between divergent DNA sequences (Wallis et al. 1989; Bailis et al. 1992).
Very little is known about how cells decide between heteroduplex rejection and MMR, which are both initiated by Msh proteins recognizing mismatches. This decision is critical because implementing MMR instead of rejection during homeologous recombination could lead to gene conversion, loss of heterozygosity, and genomic rearrangements, whereas triggering rejection instead of MMR when polymerase errors are encountered by Msh proteins during DNA replication, would likely disrupt the replication fork. Thus there must be mechanisms in place that ensure appropriate recruitment of downstream rejection or MMR proteins. At present, very little is known about what factors might regulate this decision.
In this study, we tested several interacting partners of Msh6 and Sgs1 for their role in heteroduplex rejection. We found that Top3 and Rmi1 are important for rejection during SSA. Additionally, the replicative clamp proliferating cell nuclear antigen (PCNA) appeared to be dispensable for rejection, but is important for repairing mismatches generated during SSA. Importantly, we provide evidence that 3′ tail clipping during SSA acts as a temporal commitment step, after which the heteroduplex substrate can no longer be rejected and is subject to MMR. Finally, we demonstrate that the levels of Msh proteins are tightly regulated and, although altering their relative levels can be beneficial in certain scenarios, they can be deleterious in others. These observations illustrate the delicate balance between repair and replication factors required to optimize genome stability.
Materials and Methods
Yeast strains
Yeast strains used in this study are shown in Table 1 and were constructed and grown using standard techniques (Rose et al. 1990; Geitz and Schiestl 1991). Strains bearing the SSA cassette are derived from EAY1141 and EAY1143 (Sugawara et al. 2004). The A or F abbreviations represent the upstream URA3 repeat sequence derived from S288c in EAY1141 (A-A), or from strain FL100 in EAY1143 (F-A).
Table 1. Strains and plasmids used in this study.
| Strains | Genotype |
| BJ5464 | MATα ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL |
| FY23 (S288c background) | MATa ura3-52 leu2Δ1 trp1Δ63 |
| EAY2402 | FY23, top3Δ::KANMX |
| EAY3623 | FY23, rmi1Δ::KANMX |
| EAY1269 | FY23, lys2::insE-A14 |
| EAY1375 | FY23, msh6Δ::hisG, lys2::insE-A14 |
| EAY1373 | FY23, msh3Δ::hisG, lys2::insE-A14 |
| J1022 (W303 background) | MATa top3ts (E447K,S583L), ade2-1, can1-100, his3-11,15, leu2-3,112, trp1-1, ura3-1 |
| EAY1141 | ho HMLα mat∆::leu2::hisG hmr-3∆, mal2, leu2, trp1, thr4::[THR4 ura3-A(205bp) HOcs URA3-A], ade3::GAL-HO::NAT |
| EAY2881 | EAY1141, top3ts::HPHMX |
| EAY3206 | EAY1141, rmi1ts-2 (N103K W168R, L192S, F215Y)::KANMX |
| EAY3203 | EAY1141, rm1ts-3 (L50I E60G N103K S137G, R211G, K236R)::KANMX |
| EAY3280 | EAY1141, msh6-KQFF > AAAA::KANMX |
| EAY3652 | EAY1141, POL30::KANMX |
| EAY3657 | EAY1141, pol30-201::KANMX |
| EAY3662 | EAY1141, pol30-204::KANMX |
| EAY3669 | EAY1141, pol30-216::KANMX |
| EAY 1387 | EAY1141, msh6Δ::KANMX |
| EAY1143 | ho HMLα, mat∆::leu2::hisG, hmr-3∆, mal2, leu2 trp1 thr4::[THR4 ura3-F(205bp) HOcs URA3-A], ade3::GAL-HO::NAT |
| EAY2916 | EAY1143, top3ts::HPHMX |
| EAY3210 | EAY1143, rmi1ts-2::KANMX |
| EAY3214 | EAY1143, rmi1ts-3::KANMX |
| EAY3281 | EAY1143, msh6-KQFF > AAAA::KANMX |
| EAY3655 | EAY1143, POL30::KANMX |
| EAY3660 | EAY1143, pol30-201::KANMX |
| EAY3666 | EAY1143, pol30-204::KANMX |
| EAY3671 | EAY1143, pol30-216::KANMX |
| SJR328 | MATα, ade2-101, his3Δ200, ura3-Nhe, lys2ΔRV::hisG, leu2-R |
| SJR769 | SJR328, cβ2/cβ2:LEU2, homologous 350 bp substrates |
| GCY615 | SJR328, cβ2/cβ2-ns:LEU2, 1.3% sequence divergent, predicted to form four, base-base mismatches in the 350 bp cβ2 substrates |
| GCY559 | SJR328, cβ2/cβ2-4L:LEU2, predicted to form four 4-nt loops in the 350 bp cβ2 substrates |
| 2μ Plasmids | |
| pRS424 | 2μ, TRP1 |
| pRS425 | 2μ, LEU2 |
| pRS426 | 2μ, URA3 |
| pEAM249 | MSH2, 2μ, TRP1 |
| pEAM42 | MSH2, 2μ, URA3 |
| pEAM251 | MSH3, 2μ, TRP1 |
| pEAM56 | MSH3, 2μ, LEU2 |
| pEAM252 | MSH3, 2μ, URA3 |
| pEAM99 | MSH6, 2μ, TRP1 |
| pEAM101 | MSH6, 2μ, LEU2 |
| pEAM250 | MSH6, 2μ, URA3 |
| pEAM261 | msh6-F337A, 2μ, TRP1 |
| pEAM257 | msh6-KQFF > AAAA, 2μ, TRP1 |
| pEAM258 | msh6-KQFF > AAAA, 2μ, URA3 |
| pEAM58 | MLH1, PMS1, 2μ, TRP1 |
| pEAM254 | POL30, 2μ, TRP1 |
| pEAM253 | SGS1, 2μ, TRP1 |
| pEAM259 | TOP3, RMI1, 2μ, TRP1 |
| pEAM63 | EXO1, 2μ, TRP1 |
| pEAM256 | RAD1, RAD10, 2μ, TRP1 |
| pEAM255 | SAW1, 2μ, TRP1 |
FY23 was obtained from Winston et al. (1995); J1022 from Wagner et al. (2006); EAY1141 and EAY1143 from Sugawara et al. (2004); and SJR328, SJR769, GCY615, GCY559 from Nicholson et al. (2000). The indicated genes were all cloned into pRS424-426 2 micrometer plasmids (Christianson et al. 1992).
The top3Δ::KANMX was obtained by PCR amplification of chromosomal sequences derived from the yeast knockout collection (Brachmann et al. 1998). These DNA fragments and the single integrating vectors described below (details provided upon request) were introduced into yeast strains using standard transformation procedures (Gietz and Schiestl 1991), and the presence of all mutant alleles in the genome was confirmed by PCR analysis followed by DNA sequencing.
The top3ts (E447K S583L) allele was a gift from Rodney Rothstein and was amplified by PCR from strain J1022 and then linked to an HPHMX cassette in the single step integration vector pEAI299 (top3ts allele was confirmed by Sanger sequencing). pEAI299 was restriction digested to integrate the top3ts::HPHMX allele into the SSA strains.
The rmi1 temperature-sensitive alleles were identified by transforming an ARS-CEN-LEU2 library containing mutagenized RMI1 into the rmi1Δ strain EAY3623. Transformants were screened for their failure at 37°, but not 22°, to complement the slow growth and methylmethane sulfonate (MMS) sensitivity phenotypes exhibited by rmi1Δ strains. The library was created by mutagenic PCR amplification of RMI1 using pJM7161 as a template (RMI1, ARS CEN LEU2, generously provided by Steven Brill). PCR was performed for 35 cycles, with each cycle consisting of a 30-sec denaturation step at 95°, a 30-sec annealing step at 56°, and a 3-min extension step at 72°. The Taq polymerase PCR reactions were performed in polymerase buffer recommended by the manufacturer (Promega), but containing 5 mM MgCl2, 20 μM dATP, 200 μM dGTP, 200 μM dCTP, 200 μM dTTP). Mutagenized rmi1 fragments were then subcloned into a pJM7161 backbone to create libraries that were transformed into EAY3623. Two rmi1 clones were identified from ∼1500 transformants that showed clear temperature-sensitive phenotypes, rmi1ts-2 (N103K W168R, L192S, F215Y), and rmi1ts-3 (L50I E60G N103K S137G, R211G, K236R). Both alleles were linked to a KANMX cassette to create the single-step integration vectors pEAI382 for rmi1ts-2, and pEAI381 for rmi1ts-3.
The pol30 alleles and msh6-KQFF > AAAA were kindly provided by Richard Kolodner (Lau et al. 2002) and Tom Kunkel (Clark et al. 2000). The pol30 alleles were amplified by PCR from RDKY3857 (pol30-201), RDKY3860 (pol30-204), and RDKY3872 (pol30-216) and then inserted into single-step integrating vectors that resulted in linkage to the KANMX marker (pEAA580 for pol30-201, pEAA581 for pol30-204, and pEAA583 for pol30-216). The msh6-KQFF > AAAA allele was obtained by PCR from YIplac211 (Clark et al. 2000) and then inserted into a single-step integrating vector that resulted in the msh6-KQFF > AAAA allele marked with KANMX (pEAI387).
Construction of 2μ plasmids
Genes listed in Table 1 were inserted into pRS 2μ vectors (Christianson et al. 1992). Briefly, these genes were derived from existing plasmids or were PCR amplified using chromosomal DNA derived from EAY1141 as a template. In all cases, at least 300 bp of DNA sequence upstream of the start codon was included. The sequence of PCR amplified DNA was confirmed by Sanger sequencing.
Purification and coimmunoprecipitation of Msh2-Msh6 and Sgs1400–1268
Msh2-Msh6 was purified using previously described methods from BJ5464 containing the 2μ plasmids pEAE9 (GAL10-MSH2) and pEAE218 (GAL10-MSH6) (Kijas et al. 2003).
HA-Sgs1400–1268 was purified from yeast using previously described methods with some minor modifications. Briefly, the yeast strain BJ5464 was transformed with pRB222 (Bennett et al. 1998; kindly provided by Robert Lahue), and induced with galactose to overproduce 6-His-Sgs1400–1268. Cells from a 6-liter induced culture were lysed by the coffee grinder method, resuspended into 10 ml lysis buffer [20 mM Tris pH 7.5, 200 mM KCl, 2% (v/v) glycerol, 1 mM PMSF] and after centrifugation, imidazole was added to the supernatant to a final concentration of 20 mM and the resulting lysate was incubated for 1 hr at 4° on an orbital rocker with 1 ml of Ni-NTA beads (Qiagen). The lysate/bead suspension was added to a disposable 0.8 × 4 cm Poly-Prep column and then washed with 10 ml of 20 mM Tris pH 7.5, 200 mM NaCl, 40 mM imidazole. Sgs1 was eluted with 20 mM Tris pH 7.5, 200 mM NaCl, 80 mM imidazole. Fractions containing high amounts of Sgs1 were pooled, concentrated using Centricon 50 kDa-cutoff spin concentrators (Amicon), and dialyzed into 20 mM Tris pH 8.0, 50 mM NaCl, 25% (v/v) glycerol, 1 mM β-mercaptoethanol. Protein aliquots were snap frozen in liquid nitrogen and stored at −80° until use. Protein concentrations were determined using the BioRad Protein Assay reagent.
Equimolar amounts of purified Msh2-Msh6 and Sgs1400–1268 proteins were incubated with 20 units of DNase I in 20 mM Tris pH 7.5, 100 mM NaCl, 3 mM MgCl2 for 30 min at room temperature. DNase I activity was confirmed by digestion of 1 μg of control DNA and agarose gel analysis. A total of 1 μl of 12CA5 mouse monoclonal anti-HA antibody (Roche) or 0.5 μl of anti-Msh2 polyclonal antibody (Studamire et al. 1998) were added per reaction and incubated for 1 hr at 4°. Protein A agarose beads were suspended 1:1 (v/v) in 50 mM Tris pH 7.5, 100 mM NaCl, 1 mM EDTA, and 20 μl of the suspension was incubated with each sample for 1 hr. Beads were washed three times with 200 μl of 50 mM Tris pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.1% NP-40 and twice with 200 μl of 50 mM Tris pH 7.5, 50 mM NaCl, 1 mM EDTA, 0.1% NP-40. Beads were boiled in SDS/PAGE loading dye for 10 min, and samples were run on 8% SDS/PAGE gels followed by staining with Coomassie blue.
Phenotypic analysis of rmi1 and top3 mutants in hydroxyurea and MMS sensitivity assays
Single rmi1ts and top3ts colonies were inoculated into 5 ml YPD and incubated for ∼36–48 hr at room temperature (22°) to bring the cultures to saturation. Saturated cultures were diluted in sterile dH2O to OD600 of 0.5 and 100 µl of each was transferred to a 96-well plate. They were subsequently serially diluted five times in 1:10 increments, and 5 µl of each were spotted onto YPD plates containing 7.5 mM erythrosine B and 0.02% methylmethane sulfonate, 20 mM hydroxyurea (HU), or no drug. Plates were incubated at either room temperature for 4 days or 37° for 3 days.
Single-strand annealing cell survival assays
SSA assays were performed as described (Sugawara et al. 2004; Goldfarb and Alani 2005; George et al. 2011). Briefly, cultures derived from single colonies were grown to mid-log phase in yeast peptone (YP), 2% lactate, diluted 1:2500, and plated (100 µl/plate) on both YP, 2% glucose and YP, 2% galactose. For the strains containing 2μ plasmids, single colonies grown on minimal dropout plates (Rose et al. 1990) were inoculated into minimal drop-out liquid media containing 2% glucose and grown overnight. Cultures were diluted into YP, 2% lactate media and then grown to mid-log phase. Appropriate dilutions of this culture were plated onto minimal dropout media containing 2% glucose and minimal dropout media containing 2% galactose. SSA efficiency was determined by taking the average of the number of colony forming units on galactose vs. glucose for each strain, and heteroduplex rejection efficiency was calculated as the ratio of SSA efficiency of the A-A strain to F-A strain. It is important to note that this ratio increased by approximately twofold when wild-type strains (EAY1141, EAY1143; Table 1) were grown on minimal compared to YP media. Because of this difference, all experiments presented in a specific table were performed under identical growth and plating conditions. A t-test (differences were considered significant when P <0.05) was performed to compare the A-A and F-A SSA efficiencies of all strains with the corresponding wild-type SSA efficiency. The URA3 product formed by SSA was analyzed by PCR (500-bp product) using primers AO3194 (5′AACCTCTGACACATGCAGCTC) and AO3195 (5′TGGTGGTACGAACATCCAATG), and the organization of the SSA substrate prior to double-strand break induction was confirmed by using the same primers (3400-bp product). PCR was performed for 35 cycles, with each cycle consisting of a 30-sec denaturation step at 95°, a 30-sec annealing step at 52°, and a 4-min extension step at 72°.
Efficiency of MMR during SSA
To assay the repair of DNA mismatches in heteroduplex DNA formed during SSA (Sugawara et al. 2004), EAY1143 and derivative cultures were grown to mid-log phase in YP, 2% lactate. Single unbudded cells obtained from these cultures were placed onto YP, 2% galactose plates. The plates were then incubated at 30° for 3 days. Chromosomal DNA was isolated from the resulting colonies and PCR was performed with the chromosomal DNA and primers AO3194 and AO3195 using conditions described above. PCR products were digested with MspI and then analyzed on a 1% agarose gel. Fisher’s exact test (P < 0.05 considered significant) was performed to assess whether MMR efficiency of mutants was significantly different from wild type.
Homeologous recombination using an inverted repeat reporter assay
Strains used to measure homeologous recombination are listed in Table 1. Strains containing 2µ plasmids were struck onto minimal dropout media plates. A total of 13–25 single colonies per strain were then inoculated into 5 ml of minimal dropout medium containing 4% galactose and 2% glycerol and grown for 2 days at 30°. Appropriate dilutions of cells were plated onto minimal media (2% galactose, 2% glycerol) plates lacking histidine and the amino acid required to maintain the 2µ plasmid (selective) and onto minimal media (2% glucose) plates lacking the amino acid required to maintain the 2µ plasmid (permissive). Plates were incubated for 4 days at 30° and then scored for frequency of His+ colonies. The rate of homeologous recombination was calculated as described (Nicholson et al. 2000).
Pairwise Mann–Whitney tests were performed between mutant and corresponding wild type of each strain, since the recombination rates are calculated from the median of all the colonies tested. Differences were considered significant when P <0.05.
Measuring mutation rates using the lys2A14 reversion assay
The lys2A14 reversion assay was performed as described (Heck et al. 2006).
Western blot analysis
Cell pellets from mid-log phase cell cultures (OD600 of 0.5–0.6) were resuspended in 0.5 ml lysis buffer (150 mM NaCl, 25 mM Tris pH 8.0, 1 mM EDTA, 10 mM β-mercaptoethanol, 1 mM PMSF) and lysed by vortexing with glass beads. Lysates containing 20 µg protein, measured using the Bradford (1976) assay, were run on an 8% SDS/PAGE gel. Contents of the gel were transferred onto a BioRad nitrocellulose membrane using a Mini Trans-Blot cell (BioRad). The membrane was then blocked overnight at 4° and probed with 1:4000 diluted rabbit anti-Msh6 (Studamire et al. 1998; Kumar et al. 2011) for 1 hr and 1:15,000 diluted horseradish peroxidase-conjugated goat antirabbit antibody for 1 hr. HRP signal was detected using the BioRad Clarity Western ECL substrate kit and exposed to CL-XPosure film (Thermo Scientific).
Results
Rationale for the experiments presented in this study
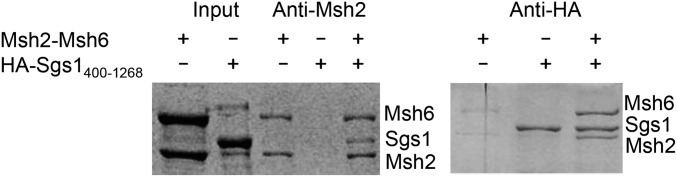
Studies in yeast and human cells indicate that Msh6 and RecQ family helicases work together to disrupt recombination intermediates (Figure 2B) (Wang et al. 2000; Myung et al. 2001; Pedrazzi et al. 2003; Spell and Jinks-Robertson 2004; Sugawara et al. 2004; Yang et al. 2004; Saydam et al. 2007). To provide further evidence for this idea in yeast, we purified Msh2-Msh6 protein and a fragment of Sgs1, Sgs1400–1268, which contains the helicase domain of Sgs1, but lacks other known or predicted domains (Bennett et al. 1998, 1999; Mullen et al. 2000), and showed by coimmunoprecipitation that Msh2-Msh6 and Sgs1 form a complex (Figure 3). These results also suggest that an Msh6 interaction site on Sgs1 lies within amino acids 400 and 1268. Though this coimmunoprecipitation does not distinguish whether Sgs1 interacts with Msh2 or Msh6, work analyzing mammalian homologs (Pedrazzi et al. 2003) suggests that Sgs1 most likely interacts directly with Msh6. Since Sgs1 does not appear to function in postreplicative MMR (Sugawara et al. 2004), and Msh6 is not known to participate in any other pathways that require Sgs1, we hypothesize that this interaction is likely to be important for heteroduplex rejection.
Figure 3.
Msh6 and Sgs1 interact by coimmunoprecipitation (Co-IP). Co-IP of Sgs1400–1268-3HA with Msh2-Msh6 in vitro using Msh2 antibody (Left), and co-IP of Msh2-Msh6 with Sgs1400–1268-3HA in vitro using HA antibody (Right). See Materials and Methods for details.
The detection of physical interactions between Sgs1 and Msh2-Msh6 encouraged us to think more critically about how the decision is made to undergo heteroduplex rejection vs. MMR, a decision step that has not been carefully explored. We find this of particular interest because mismatch recognition by Msh proteins initiates both processes. We focused on answering the following questions: (1) Does Sgs1 helicase act alone in heteroduplex rejection or does it function with Top3 and Rmi1? (2) Can overexpression of MMR or heteroduplex rejection factors alter the choice to undergo MMR vs. rejection? (3) Does the Msh6-PCNA interaction affect pathway choice?
Top3 and Rmi1 act in heteroduplex rejection
To measure heteroduplex rejection, we utilized an SSA cell survival assay (Figure 1A; Sugawara et al. 2004) in which a double-strand break is induced between two 205-bp direct repeats that are either identical (A-A strain) or 3% divergent (F-A strain, forms six base–base and a single nucleotide insertion/deletion mismatch in heteroduplex DNA). Repair of this substrate requires removal of the 3′ tail in a process dependent on Msh2-Msh3 and the Rad1-Rad10 endonuclease, in addition to other factors (Fishman-Lobell and Haber 1992; Sugawara et al. 1997; Li et al. 2008). The efficiency of SSA repair, as measured by cell viability in the two strains, was then compared. In wild-type strains, SSA efficiently repairs the break (77–93%) only when the repeats are identical (A-A; Table 2). Survival of wild-type strains bearing the F-A repeats is significantly lower (22–28%). Survival of F-A strains that lack Msh6 or Sgs1 approaches that seen in A-A strains, indicating that Msh6 and Sgs1 prevent recombination between divergent DNA sequences [Sugawara et al. 2004; data from Goldfarb and Alani (2005) reprinted in Table 2].
Table 2. Heteroduplex rejection efficiency of top3ts, rmi1ts, pol30, and msh6 mutants as determined in SSA survival assays.
| Relevant genotype | Temp. | A-A | F-A | A-A/F-A |
|---|---|---|---|---|
| Wild type | 22° | 0.93 ± 0.09 | 0.26 ± 0.05 | 3.6 |
| 25° | 0.77 ± 0.13 | 0.28 ± 0.06 | 2.8 | |
| 30° | 0.79 ± 0.11 | 0.24 ± 0.06 | 3.3 | |
| 32° | 0.84 ± 0.07 | 0.22 ± 0.05 | 3.9 | |
| 37° | 0.79 ± 0.10 | 0.25 ± 0.07 | 3.2 | |
| top3ts | 22° | 0.70 ± 0.09* | 0.38 ± 0.01** | 1.9 |
| 25° | 0.74 ± 0.18 | 0.38 ± 0.08* | 2.0 | |
| 30° | 0.75 ± 0.05 | 0.48 ± 0.05** | 1.6 | |
| 32° | 0.78 ± 0.04 | 0.42 ± 0.04** | 1.9 | |
| 37° | 0.67 ± 0.16 | 0.50 ± 0.14** | 1.4 | |
| rmi1ts-2 | 22° | 0.98 ± 0.16 | 0.58 ± 0.16* | 1.7 |
| 25° | 0.95 ± 0.13* | 0.60 ± 0.18** | 1.6 | |
| 37° | 0.91 ± 0.15 | 0.64 ± 0.10** | 1.4 | |
| rmi1ts-3 | 22° | 0.77 ± 0.05* | 0.41 ± 0.15 | 1.9 |
| 25° | 0.83 ± 0.11 | 0.41 ± 0.10* | 2.0 | |
| 37° | 0.96 ± 0.08* | 0.55 ± 0.08** | 1.8 | |
| msh6-KQFF > AAAA | 30° | 0.77 ± 0.08 | 0.21 ± 0.03 | 3.6 |
| pol30-204 | 30° | 0.70 ± 0.08* | 0.21 ± 0.03 | 3.4 |
| pol30-201 | 30° | 0.56 ± 0.10** | 0.17 ± 0.03* | 3.2 |
| pol30-216 | 30° | 0.48 ± 0.08** | 0.13 ± 0.02** | 3.7 |
| Goldfarb and Alani 2005 | ||||
| Wild type | 30° | 0.61 ± 0.12 | 0.19 ± 0.03 | 3.4 |
| msh6Δ | 30° | 0.87 ± 0.02 | 0.61 ± 0.06 | 1.4 |
| sgs1Δ | 30° | 0.79 ± 0.16 | 0.75 ± 0.18 | 1.1 |
Survival of indicated strains expressed as mean of colony forming units on galactose/glucose ± SD for 3–21 experiments. Heteroduplex rejection efficiency is shown as the ratio of A-A to F-A survival. Significant differences were calculated for data obtained in this study and are indicated as follows: *Significantly different from wild type using t test (P < 0.05). **Significantly different from wild type using t test (P < 0.01). Published data of msh6Δ and sgs1Δ along with wild-type strains from Goldfarb and Alani (2005) are shown for comparison.
Top3 is a type I topoisomerase that forms a stable complex with Sgs1 and depends on association with the small protein Rmi1 for its activity (Gangloff et al. 1994; Bennett et al. 2000; Fricke et al. 2001; Chang et al. 2005; Mullen et al. 2005; Chen and Brill 2007). The Sgs1-Top3-Rmi1 (STR) complex serves in several roles in the maintenance of genome stability, including DSB end resection, rescue of stalled replication forks, D-loop disruption, and dissolution of double Holliday junctions (Cejka et al. 2010; Niu et al. 2010; Hickson and Mankouri 2011; Mimitou and Symington 2011; Fasching et al. 2015). However, Sgs1 also displays functions that do not appear to require Top3 and Rmi1. For example, Top3 and Rmi1 are not required for DNA end resection, although they serve a stimulatory role (Cejka et al. 2010), and Sgs1 functions independently of Top3 in stimulating the Rad53 checkpoint kinase in response to HU-dependent replication fork stalling, though this checkpoint function does not require the helicase activity of Sgs1 (Bjergbaek et al. 2005).
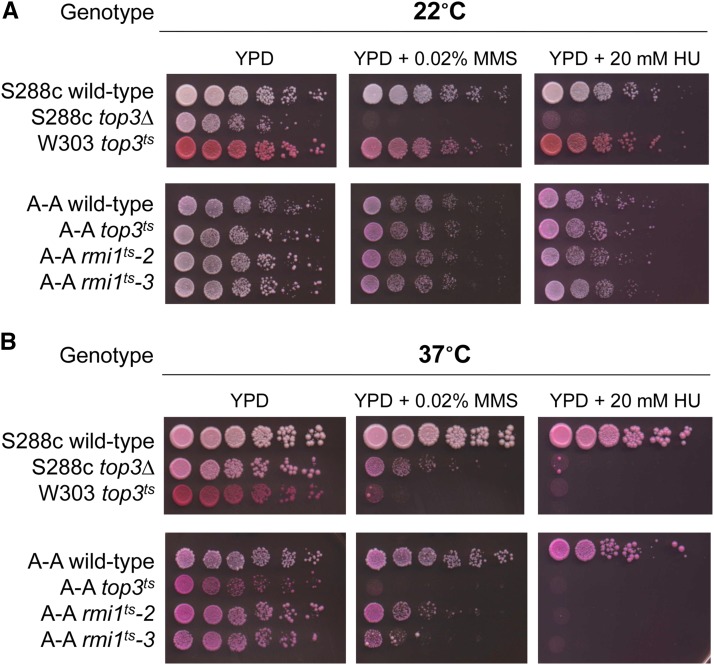
To test whether Top3 and Rmi1 play a role in heteroduplex rejection during SSA, we performed an analysis of temperature-sensitive alleles of TOP3 and RMI1 that display top3-null- and rmi1-null-like phenotypes at 37°, but not 22°. As shown in Figure 4, the top3ts allele confers slow growth and sensitivity to the DNA damaging agents MMS and HU at the restrictive temperature, similar to top3Δ (Shor et al. 2002; Wagner et al. 2006). rmi1ts-2 and rmi1ts-3 were obtained from a screen (Materials and Methods) for rmi1 mutants that failed to complement slow growth and MMS sensitivity of an rmi1Δ strain and are similar in phenotype to the rmi1-1 mutant described by Ashton et al. (2011). Both alleles confer phenotypes similar to the top3ts allele, though with less severity (Figure 4).
Figure 4.
Temperature-sensitive alleles of RMI1 display rmi1-null-like phenotypes at 37° but not 22°, similar to a previously characterized top3ts mutant. Ten-fold serial dilutions of the indicated strains spotted onto YPD plates containing 7.5 µm erythrosine B to stain dead cells pink and with or without the indicated DNA damaging agents. Both the top3ts mutant [as described by Wagner et al. (2006)] and two rmi1ts alleles isolated in this study display slow growth, increased cell death, and sensitivity to DNA damaging agents at 37° (B) but not 22° (A). Note that the W303 top3ts strain appears pink due to an ade2 mutation; thus an assessment of cell death by erythrosine B staining cannot be determined in this strain.
Heteroduplex rejection efficiency was not affected by temperature in wild-type strains (22° to 37°; Table 2). However, in strains bearing the top3ts allele or either one of the rmi1ts alleles, heteroduplex rejection was reduced at all temperatures. These data suggest that Top3 and Rmi1 rejection functions are more sensitive to mutation than the DNA repair functions. Although heteroduplex rejection was reduced in all top3 and rmi1 mutants, the A-A/F-A ratio (1.4–2.0) in these strains never reached the levels seen in sgs1Δ (1.1). The A-A/F-A ratio in these strains was similar to that seen for msh6Δ, indicating a critical, but perhaps not exclusive role for Top3-Rmi1 in heteroduplex rejection. It is important to note that heteroduplex rejection efficiency of top3 and rmi1 alleles cannot be compared to msh2Δ in this assay because the Msh2-Msh3 complex is required for SSA, where it interacts with Rad1-Rad10 to remove nonhomologous 3′ single-stranded DNA tails that form during repair (reviewed in Lyndaker and Alani 2009).
PCNA-Msh6 interactions appear dispensable for rejection during SSA
PCNA, encoded by the POL30 gene in baker’s yeast, is a homotrimeric clamp required for processivity of DNA replication and is associated with replication forks (reviewed in Kelman 1997). PCNA forms a stable interaction with the N terminus of Msh6, and this interaction involves a KQFF motif in Msh6 (Clark et al. 2000; Shell et al. 2007). Umar et al. (1996) presented data suggesting roles for PCNA in mismatch repair prior to DNA synthesis steps, and Flores-Rozas et al. (2000) and Lau and Kolodner (2003) demonstrated that interaction with PCNA enhances mismatch recognition by Msh2-Msh6. Also, Stone et al. (2008) showed that Msh-PCNA interactions play a minor role in preventing recombination between divergent sequences using inverted repeat substrates. Finally Hombauer et al. (2011b) presented data suggesting that heteroduplex rejection involving inverted repeat substrates does not require coupling to DNA replication.
The above observations encouraged us to test whether a PCNA-Msh6 interaction is important for heteroduplex rejection during SSA. We used both an msh6 allele, which disrupts Msh2-Msh6 binding to PCNA (msh6-KQFF > AAAA), as well as three pol30 alleles: a mild mutator allele pol30-204, which disrupts PCNA-Msh6 interactions; pol30-201, which maintains PCNA-Msh6 interactions but otherwise shares a similar phenotypic profile to pol30-204; and pol30-216, which displays a range of moderate-to-severe phenotypes in the presence of DNA damaging agents (Lau et al. 2002). In each of the three pol30 mutants and the msh6-KQFF > AAAA mutant, the rejection efficiency was similar to that of wild type, indicating that heteroduplex rejection was intact and that Msh6 performed its role in rejection, independent of PCNA (Table 2). Interestingly, all pol30 strains, especially pol30-216 and pol30-201, showed significantly reduced SSA efficiency between completely homologous DNA sequences (A-A). At present, we favor the idea that the SSA defects seen in pol30 mutants are due to the mutant PCNA being defective in DNA replication processivity steps during SSA (e.g., gap filling, see Discussion).
We then tested the msh6 and pol30 mutant alleles for their effect on repairing mismatches formed when F-A substrates escape rejection and undergo repair (Figure 1B). Consistent with a previous study that examined meiotic MMR (Stone et al. 2008), the pol30-201 allele caused a significant MMR defect (Table 3). We also tested msh6Δ as a control and found that it displays a significant MMR defect as published previously (Table 3) (Lau et al. 2002; Sugawara et al. 2004). These data suggest that PCNA does not influence rejection through mismatch recognition or other mismatch-specific mechanisms, but plays an important role in the repair of mismatches generated during homeologous recombination. It is important to note that the pol30-204 and msh6KQFF > AAAA mutations did not confer MMR defects, consistent with previous work showing that these mutations confer subtle defects in MMR (Clark et al. 2000; Lau et al. 2002; Shell et al. 2007). In addition, sgs1, top3, and rmi1 mutations did not confer MMR defects (Table 3), supporting the idea that heteroduplex rejection and mismatch repair are distinct pathways involving common initiation steps (mismatch binding by Msh heterodimers) that are then carried out by distinct factors (Figure 2A; Sugawara et al. 2004).
Table 3. Mismatch correction of heteroduplexes arising during SSA between F and A flanking sequences.
| Number of coloniesa | ||||
|---|---|---|---|---|
| Genotype | MspI+ (F) | MspI− (A) | Mixed (F-A) | Percent mixed |
| Wild-type | 28 | 8 | 2 | 5 |
| msh6Δ | 1 | 4 | 18 | 78** |
| pol30-201 | 17 | 3 | 15 | 43** |
| pol30-204 | 18 | 10 | 5 | 15 |
| msh6-KQFF > AAAA | 30 | 8 | 3 | 7 |
| top3ts-25° | 10 | 6 | 3 | 16 |
| top3ts-37° | 14 | 5 | 4 | 17 |
| rmi1ts-25° | 20 | 0 | 3 | 13 |
| rmi1ts-37° | 39 | 9 | 12 | 20 |
Significantly different from wild type using Fisher’s exact test (P < 0.01).
Colonies derived from single unbudded cells grown on media containing galactose were analyzed for the presence of the MspI site (Materials and Methods) present in the ura3-FL100 (F) allele. Mixed colonies contain cells with and without the MspI site.
Overexpression of Msh6 and Sgs1 improves heteroduplex rejection during SSA
The finding that Msh6 and Sgs1 physically interact and that Top3 and Rmi1 participate in heteroduplex rejection encouraged us to test whether overexpression of these factors and a set of MMR proteins could alter the heteroduplex rejection vs. MMR decision. As shown in Table 4, overexpression of Msh6, Msh3, and Sgs1, but none of the other components tested conferred significant changes in heteroduplex rejection efficiency. Overexpression of Msh6, which resulted in a roughly eightfold increase in cellular Msh6 levels (Figure 5), led to a decrease in F-A survival (P < 0.01, t-test) and consequently a sevenfold higher efficiency in heteroduplex rejection (Table 4).
Table 4. Heteroduplex rejection efficiency of overexpression strains as determined in survival assays.
| 2μ vector | A-A | F-A | A-A/F-A |
|---|---|---|---|
| pRS424 | 0.66 ± 0.04 | 0.09 ± 0.01 | 7.6 |
| MSH6 | 0.39 ± 0.04** | 0.008 ± 0.002** | 49 |
| msh6-KQFF > AAAA | 0.37 ± 0.05** | 0.012 ± 0.003** | 30 |
| MSH3 | 0.55 ± 0.02* | 0.24 ± 0.02** | 2.3 |
| MSH2 | 0.60 ± 0.07 | 0.14 ± 0.02* | 4.2 |
| MLH1, PMS1 | 0.64 ± 0.05 | 0.08 ± 0.02 | 8.3 |
| EXO1 | 0.48 ± 0.02* | 0.08 ± 0.01 | 5.7 |
| POL30 | 0.35 ± 0.01** | 0.05 ± 0.01 | 6.6 |
| SGS1 | 0.57 ± 0.03 | 0.050 ± 0.004* | 11 |
| TOP3, RMI1 | 0.62 ± 0.06 | 0.099 ± 0.004 | 6.2 |
| RAD1, RAD10 | 0.52 ± 0.03* | 0.10 ± 0.01 | 5.3 |
| SAW1 | 0.58 ± 0.04 | 0.07 ± 0.01 | 7.7 |
| pRS424, pRS425 | 0.50 ± 0.01 | 0.10 ± 0.01 | 5.0 |
| MSH2, pRS425 | 0.52 ± 0.08 | 0.12 ± 0.04 | 4.2 |
| MSH6, pRS424 | 0.51 ± 0.03 | 0.009 ± 0.005** | 54 |
| MSH3, pRS424 | 0.52 ± 0.10 | 0.20 ± 0.04* | 2.6 |
| MSH2, MSH6 | 0.56 ± 0.06 | 0.06 ± 0.01 | 9.0 |
| MSH2, MSH3 | 0.56 ± 0.05 | 0.12 ± 0.03 | 4.5 |
| MSH3, MSH6 | 0.40 ± 0.05 | 0.04 ± 0.01** | 10 |
Survival of indicated strains expressed as colony forming units on galactose/glucose ± SE of mean for 4 to 18 experiments. Heteroduplex rejection efficiencies are shown as the ratio of A-A to F-A survival. *Significantly different from wild type using t test (P < 0.05). **Significantly different from wild type using t test (P < 0.01).
Figure 5.
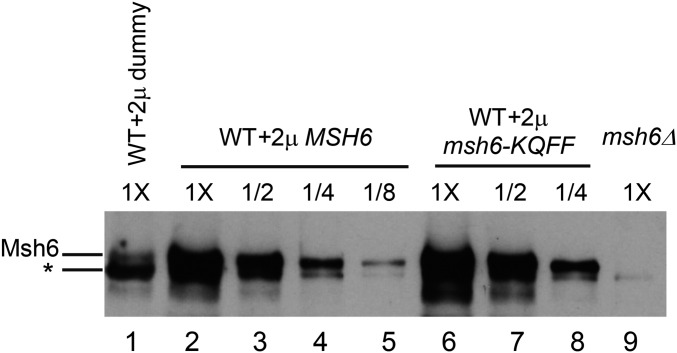
Msh6 and msh6-KQFF > AAAA are overexpressed to similar levels when present on 2μ vectors. Western blot analysis (Materials and Methods) using Msh6 antibody was performed on cell extracts derived from wild-type and msh6Δ strains. (Lane 1) Wild type containing a 2μ dummy vector. (Lanes 2–8) Wild type containing MSH6-2μ (2–5) and msh6-KQFF > AAAA-2μ (6–8) vectors. (Lane 9) msh6Δ strain lacking a 2μ vector. A twofold dilution series of extracts derived from wild-type strains containing MSH6-2μ and msh6-KQFF > AAAA-2μ are shown as indicated. With the exception of the dilution series, 20 μg of each protein extract was loaded. * indicates a nonspecific band that is seen in strains containing 2μ vectors.
Overexpression of the msh6-KQFF > AAAA protein, which was stable and expressed at a level similar to the overexpression of wild-type Msh6 (Figure 5), resulted in a phenotype similar to Msh6 overexpression, again suggesting that a PCNA-Msh6 interaction is not required for rejection during SSA (Table 4). When Msh2 and Msh6 were cooverexpressed, heteroduplex rejection returned close to wild-type levels, suggesting that the increased rejection seen in Msh6 overexpression strains was due to a reduction in Msh2-Msh3 levels as a result of Msh6 sequestering Msh2. In this model, Msh6 overexpression would lead to less efficient Msh2-Msh3-dependent 3′ tail clipping, and thus provide a longer time window for Msh2-Msh6 to promote heteroduplex rejection. In support of this, we found that Msh6 overexpression resulted in reduced repair of the A-A substrate (68 to 39%, P < 0.01, t-test; Table 4), presumably due to lack of a sufficient amount of Msh2-Msh3 that is required to clip the 3′ tails to complete SSA. Also, when Msh6 and Msh3 were cooverexpressed, the rejection efficiency was similar to wild type, presumably because the relative levels of Msh2-Msh6 and Msh2-Msh3 would be almost unchanged. It is important to note that we did not see a similar decrease in SSA efficiency in the A-A substrate when Msh6 was overexpressed along with a 2μ empty vector (Table 4). We believe that this is due to a reduction in copy number of the 2μ-MSH6 plasmid when an additional 2μ plasmid is also selected for. In this scenario, we hypothesize that SSA repair efficiency is less sensitive to Msh6 overexpression levels relative to heteroduplex rejection. We also overexpressed other proteins involved in 3′ tail clipping such as Rad1-Rad10 and Saw1, with the goal of learning whether they would affect the tail clipping efficiency and thereby heteroduplex rejection (Table 4). However, strains overexpressing these proteins displayed rejection efficiencies similar to wild type; one explanation for this is that Msh2-Msh3 binding to 3′ nonhomologous tails is a rate-limiting step.
Msh3 overexpression resulted in an increase in F-A survival (P < 0.01, t-test) and a decrease in heteroduplex rejection efficiency, which was restored to wild-type levels when Msh2 and Msh3 were cooverexpressed (Table 4). This observation is consistent with Msh3 overexpression sequestering Msh2, resulting in lower levels of Msh2-Msh6 that can participate in heteroduplex rejection. As indicated above, overexpression of Sgs1 also led to a subtle decrease in F-A survival compared to wild type (P < 0.05) and thus an increase in heteroduplex rejection. It is important to note that overexpression of all three components, Sgs1, Top3, and Rmi1, could not be done because the resulting transformants grew poorly. Our data support the idea that during recombination, yeast cells are in a rejection mode prior to 3′ tail removal, where the 3′ tails serve as a platform for STR to act on heteroduplex intermediates. Such a model also predicts that heteroduplex rejection would be further activated by increasing the expression of STR components, as was seen when Sgs1 was overexpressed. Together these data suggest that altering the levels of specific Msh complexes has a significant effect on heteroduplex rejection during SSA and that the tail clipping functions exhibited by Msh2-Msh3 play a critical regulatory role in the heteroduplex rejection vs. MMR decision.
Msh6 overexpression disrupts heteroduplex rejection in an assay likely coupled to replication
We examined the effect of Msh overexpression on homeologous recombination using an inverted repeat assay in which recombination is thought to be initiated via DNA lesions arising during or shortly after replication of the recombination substrate (Chen and Jinks-Robertson 1998; Nicholson et al. 2000). In this assay recombination between identical or divergent DNA sequences reorients intron and HIS3 sequences to generate a functional HIS3 gene (Figure 1C). Such His+ recombinants have been hypothesized to result from gene conversion events involving repeat sequences present on sister chromatids. While both Msh2-Msh6 and Msh2-Msh3 were required for suppressing recombination between substrates predicted to form base–base mismatches in heteroduplex DNA (cβ2/cβ2-ns substrate), only Msh2-Msh3 was shown to play such a role for a substrate predicted to form 4-nt insertion/deletion loops (cβ2/cβ2-4L; Chen and Jinks-Robertson 1998; Nicholson et al. 2000; Lee et al. 2007).
We introduced an MSH6-2μ plasmid into strains bearing homologous, base–base, and 4-nt loop inverted repeat substrates. Msh6 overexpression conferred a modest increase in recombination between identical repeat sequences. Surprisingly, such overexpression promoted homeologous recombination involving substrates predicted to form both base–base and 4-nt loop mismatches. In wild-type strains, the ratio of homologous to homeologous recombination rates was 16 and 6.4 for base–base and 4-nt loop mismatches, respectively. When Msh6 was overexpressed, this ratio decreased to 1.9 and 0.96 for base–base and 4-nt loop mismatches, respectively, approaching that seen in msh2Δ strains where antirecombination is severely compromised (Table 5; Lee et al. 2007). In contrast, Msh3 overexpression improved antirecombination in the 4-nt loop strains by three-fold, consistent with Msh2-Msh3 being important for its antirecombination functions. Msh2 overexpression had no effect, consistent with it acting as a common partner with both Msh6 and Msh3, and not specifying mismatch recognition.
Table 5. Recombination rates in strains overexpressing MMR proteins as measured in the inverted repeat reporter assay.
| Strain | 2μ vector | Rate of His+ recombination (×10−6) | Homologous rate/homeologous ratea |
|---|---|---|---|
| Cβ2-Cβ2 | pRS426 | 0.64 (0.47–0.90)b | |
| MSH6 | 1.5 (0.68–2.44)* | ||
| msh6 KQFF > AAAA | 2.1 (1.38–2.96)** | ||
| MSH3 | 1.1 (0.66–1.65)** | ||
| MSH2 | 0.54 (0.26–0.72) | ||
| Cβ2/Cβ2-ns | pRS426 | 0.04 (0.02–0.08) | 16 |
| MSH6 | 0.80 (0.60–1.31)** | 1.9 | |
| msh6 KQFF > AAAA | 0.07 (0.03–0.12) | 30 | |
| MSH3 | 0.08 (0.03–0.16) | 14 | |
| MSH2 | 0.04 (0.01–0.06) | 14 | |
| Cβ2/Cβ2-4L | pRS426 | 0.10 (0.05–0.56) | 6.4 |
| MSH6 | 1.57 (1.12–1.81)** | 0.96 | |
| msh6 KQFF > AAAA | 0.29 (0.11–0.36) | 7.2 | |
| MSH3 | 0.06 (0.05–0.09) | 18 | |
| MSH2 | 0.09 (0.04–0.13) | 6.0 | |
| Lee et al. 2007 | |||
| Cβ2-Cβ2 | Wild type | 2.7 (2.5–3.8) | |
| msh2Δ | 5.5 (4.7–9.2) | ||
| msh6Δ | 2.0 (1.3–2.4) | ||
| msh3Δ | 6.6 (6.2–9.6) | ||
| Cβ2/Cβ2-ns | Wild type | 0.16 (0.15–0.22) | 16.9 |
| msh2Δ | 8.7 (5.2–10) | 0.6 | |
| msh6Δ | 2.2 (1.9–2.4) | 0.9 | |
| msh3Δ | 1.8 (1.2–2.4) | 3.7 | |
| Cβ2/Cβ2-4L | Wild type | 0.25 (0.15–0.30) | 10.8 |
| msh2Δ | 6.7 (6.5–8.4) | 0.8 | |
| msh6Δ | 0.27 (0.21–0.34) | 7.4 | |
| msh3Δ | 7.6 (6.6–9.1) | 0.9 |
Homeologous recombination rates and 95% confidence intervals were calculated as described in Materials and Methods from 13–25 individual cultures. The genotypes of the strains are shown in Table 1. Cβ2/Cβ2, homologous substrate; Cβ2/Cβ2-ns, base–base mismatch substrate; Cβ2/Cβ2-4L, 4-nt loop mismatch substrate.
Homologous rate (Cβ2-Cβ2)/homeologous rate for strains with the same overexpression plasmid. bNumbers in parentheses indicate 95% confidence intervals. *Significantly different from wild type of the same strain (P < 0.05, Mann–Whitney test). **Significantly different from wild type of the same strain (P < 0.01, Mann–Whitney test). Published data of msh2Δ, msh6Δ, and msh3Δ along with wild-type strains from Lee et al. 2007 are provided for Cβ2-Cβ2, Cβ2/Cβ2-ns, and Cβ2/Cβ2-4L strain backgrounds for reference purposes.
We also tested whether overexpression of msh6-KQFF > AAAA protein, which is disrupted for its interactions with PCNA, would, like Msh6 overexpression, increase homeologous recombination. This was of interest because this mutant allele conferred phenotypes similar to wild-type MSH6 in the SSA assay (Table 2 and Table 4). However, as shown in Table 5, msh6-KQFF > AAAA overexpression did not increase homeologous recombination; overexpression improved antirecombination by twofold in the base–base mismatch strains but did not affect antirecombination in the 4-nt loop strains. These results indicate that Msh6 overexpression disrupts antirecombination in a system where recombination occurs during replication, and this reduction depends on its interaction with PCNA. Curiously, both Msh6 and msh6-KQFF > AAAA overexpression conferred a mutator phenotype in a wild-type strain (Table 6). These rates were higher than seen in msh3Δ strains, but lower than in msh6Δ strains, indicating that Msh6 overexpression confers a mutator phenotype independent of Msh6-PCNA interactions. Additionally, overexpression of msh6-F337A, an allele that disrupts Msh2-Msh6 mismatch recognition, conferred a severe mutator phenotype, indicating that it is a dominant negative allele (Table 6). For this reason, we did not include it in our overexpression experiments to test rejection efficiency.
Table 6. Yeast overexpressing Msh6 display a mutator phenotype in the lys2-A14 reversion assay.
| Strain | Reversion rate (×10−7), (95% C.I.) | Normalized |
|---|---|---|
| Wild type | 9.9 (5.8–67) | 1 |
| Wild type +MSH6-2μ | 110 (31–647)** | 11.1 |
| Wild type + msh6KQFF > AAAA-2μ | 92.5 (67.6–149)** | 9.3 |
| Wild type + msh6F337A-2μ | 4610 (3270–5650)** | 464 |
| msh6Δ | 585 (207–2030)** | 59.1 |
| msh3Δ | 37.7 (22.3–49.4)* | 3.8 |
FY23-derived strains were analyzed for lys2-A14 reversion as described in Materials and Methods and Table 1. Rates are presented per cell division. Numbers in parentheses indicate 95% confidence intervals. Rates and 95% confidence intervals were calculated from 10 independent cultures. *Significantly different from wild type using Mann–Whitney test (P < 0.05). **Significantly different from wild type using Mann–Whitney test (P < 0.01).
Discussion
Roles for Top3, Rmi1, and PCNA in SSA
In this study, we showed that both Top3 and Rmi1 are required for heteroduplex rejection during SSA. Based on previous studies showing that Sgs1, Top3, and Rmi1 form a complex, and that Sgs1 mediated unwinding of 3ʹ tailed substrates is strongly stimulated by Top3-Rmi1 (Bennett et al. 1998, 1999; Cejka et al. 2010; Cejka and Kowalczykowski 2010), we hypothesize two possible roles for Top3-Rmi1 in heteroduplex rejection: (1) Top3-Rmi1 plays a catalytic role in rejection during SSA by relieving torsional strain caused by Sgs1 unwinding from both ends of a heteroduplex substrate. (2) Top3-Rmi1 physically stabilizes Sgs1 so that it can unwind heteroduplex DNA. The latter idea is supported by previous data showing that Sgs1 levels decrease in the absence of Top3 (Chang et al. 2005).
We also tested whether PCNA plays a role in heteroduplex rejection by promoting mismatch recognition by Msh proteins. pol30 and msh6 mutants defective in PCNA-Msh6 interactions (pol30-204, msh6-KQFF > AAAA) did not show any effect on rejection efficiency, indicating that such interactions are dispensable for heteroduplex rejection during SSA. Our results are consistent with those of Stone et al. (2008), who showed in an inverted-repeat recombination system that PCNA-Msh6 interactions played a minor role in regulating the fidelity of recombination. Additionally, we observed a decrease in the viability of A-A cells in the pol30 mutant backgrounds. This indicates a role for PCNA in the SSA pathway, most likely in the DNA synthesis step following 3′ tail removal. Consistent with this idea, PCNA was shown to stabilize binding of various polymerases to the 3′-OH of a DNA template during replication and to stimulate DNA synthesis (Maga and Hübscher 1995; Einolf and Guengerich 2000; Maga et al. 2002), and human PCNA has been shown to stimulate DNA synthesis during microhomology-mediated end joining, a process similar to SSA that is initiated on terminal microsatellite sequences on ssDNA (Crespan et al. 2012).
Finally, we tested whether PCNA plays a role in repairing mismatches generated during SSA between divergent sequences that escape rejection. We found that the pol30-201 mutant, which has a moderate mutator phenotype but is proficient in PCNA-Msh6 interactions, showed a defect in the repair of mismatches generated between divergent substrates during SSA. A similar observation was made by Stone et al. (2008), who showed that the pol30-201 mutation decreased the efficiency of MMR in heteroduplex DNA generated during meiotic recombination. It is important to note that the pol30-201 mutation alters an amino acid located on the inner surface of the PCNA ring that slides on DNA; thus the observed MMR defect could result from a faulty interaction between a pcna-201-Msh2-Msh6 complex and mismatch DNA, as suggested by Lau et al. (2002). The pol30-204 mutation, however, alters an amino acid located in the monomer–monomer interface region of PCNA and disrupts the Msh6-PCNA interaction (Lau et al. 2002) as does msh6-KQFF > AAAA. Since these alleles did not confer MMR defects, the PCNA-Msh6 interaction is not likely required for the repair of mismatches generated during SSA. These data also suggest that the MMR defect conferred by the pol30-201 mutation is at an early stage in MMR. If the defect occurred during resynthesis steps, the DSB would not be fully repaired and the unbudded pol30-201 F-A cells would fail to form colonies on galactose plates. Together our data indicate that during recombination, PCNA does not act in antirecombination but promotes MMR in heteroduplex DNA.
How is the commitment to reject heteroduplex intermediates made?
In the SSA system used here, we observed an ∼80% heteroduplex rejection efficiency in wild-type strains, indicating a clear preference toward rejecting heteroduplex substrates. The rejection vs. repair decision is critical because repair involving divergent sequences would propagate specific alleles as gene conversion events, and if the substrates are on different chromosomes, SSA between them will generate chromosomal rearrangements.
These issues encouraged us to obtain a better understanding of the factors that play a role in the rejection/repair decision, especially since the same set of Msh proteins act in both processes. We used a protein overexpression approach and learned the following: First, the repair/rejection decision does not appear to involve a simple competition between MMR and rejection factors for recombination intermediates containing DNA mismatches. We found that overexpressing MMR factors such as Mlh1-Pms1, Exo1, and PCNA did not affect the efficiency of heteroduplex rejection, whereas overexpressing Sgs1 only slightly improved the rejection efficiency and Top3-Rmi1 overexpression had no effect. Second, our data are consistent with tailed DNA intermediates playing an important role in the commitment to reject heteroduplex intermediates. Our data in the SSA assay argue for Msh6 overexpression sequestering Msh2, resulting in less efficient 3′ tail clipping due to reduced levels of Msh2-Msh3. We hypothesize that lowering levels of Msh2-Msh3 creates an increased opportunity for rejection factors to be loaded onto 3′ tails and unwind heteroduplex substrates. In support of this idea, we observed a decrease in A-A survival when Msh6 was overexpressed, and cooverexpression of Msh2 and Msh6 or Msh3 and Msh6, eliminated the increased rejection phenotype seen when Msh6 was overexpressed by itself, presumably because Msh2-Msh3 levels would not be specifically compromised.
The above data indicate a temporal commitment step for heteroduplex rejection, where prior to the clipping of the 3′ tail, rejection is favored until the 3′ tail is clipped (Figure 2B). This model is also supported by biochemical studies showing that Sgs1 and its homologs in other species preferentially bind to, and unwind, “Y” shaped forked DNA substrates (Brosh et al. 2002; Saydam et al. 2007; Cejka et al. 2010). Thus the tail may be important for the loading of the STR complex onto the heteroduplex substrate. Our findings using the SSA substrates could extend to other homologous recombination pathways. For example, during the strand invasion step, partial homology could give rise to 3′ nonhomologous tails that could then act as loading sites for the STR complex. These tails would have to be removed before proceeding to DNA synthesis steps and could potentially act as a commitment step for rejection vs. repair. However, it is likely that in addition to the structure of the DNA substrate, there are additional mechanisms that play a role in driving the rejection vs. repair decision. For example, the STR complex participates in 5′ to 3′ strand resection following DSB formation (Gravel et al. 2008; Mimitou and Symington 2008; Zhu et al. 2008; Cejka et al. 2010; Niu et al. 2010). An early recruitment and localization to sites of homologous recombination may give the STR complex an advantage (cis-effect) over downstream MMR proteins in terms of a more rapid recruitment to heteroduplex recombination intermediates. Additionally, recombination factors such as Rad52, which function in SSA (Fishman-Lobell and Haber 1992; Sugawara and Haber 1992), are likely to aid, perhaps through protein–protein interactions, in the recruitment of Sgs1-Top3-Rmi1 to 3′ tails during strand annealing steps. Interestingly, Honda et al. (2014) showed that hMsh2-hMsh6 is capable of recognizing mismatch DNA in D-loop recombination intermediates, and suggest that the release of the ATP-bound hMsh2-hMsh6 sliding clamp at the D-loop branch point “is the result of branch point-induced ATP hydrolysis.” Furthermore, they hypothesize that upon encountering the branch point, hMsh2-hMsh6 undergoes conformational changes that are important for recruiting/loading downstream rejection factors such as antirecombinogenic helicases. Consistent with their work, our data emphasize the importance of the structure of the DNA substrate; the 3′ tail creates a duplex-single strand DNA junction, which is likely to play an important role, following mismatch recognition by Msh proteins, in regulating the decision to recruit rejection vs. MMR factors.
Msh3/Msh6 mismatch repair protein ratios are critical to maintain genome integrity
Several studies have demonstrated the consequences of changing the cellular ratio of Msh6 to Msh3: (1) We showed here that Msh6 overexpression prevented recombination between divergent DNA sequences at the expense of Msh2-Msh3 function during SSA. (2) Previously, the Modrich and Jiricny groups showed that amplification of the MSH3 gene in methotrexate-resistant leukemia cells caused a deficiency in MMR of base–base mismatches due to sequestration of Msh2 and consequently a reduction in Msh2-Msh6 levels (Drummond et al. 1997; Marra et al. 1998). Together these data demonstrate that a balance in Msh3/Msh6 protein levels is required to maintain various aspects of genome integrity. In support of this, Msh2-Msh6 and Msh2-Msh3 levels in the mouse vary in a tissue (and cell proliferation)-specific manner (Tomé et al. 2013), suggesting that this balance is regulated on a tissue-specific basis.
Why does Msh6 overexpression impact the two antirecombination systems differently?
In this study, we also overexpressed Msh proteins in strains bearing an inverted repeat substrate (Nicholson et al. 2000). This was done to determine if the effect of Msh overexpression on homeologous recombination during SSA could be generalized to other recombination pathways. When Msh6 was overexpressed during inverted repeat recombination, an increase was seen in recombination between divergent sequences forming base–base or 4-nt loop mismatches. This effect was not seen when a mutant msh6 allele defective in PCNA interactions, msh6-KQFF > AAAA, was overexpressed. This result caught our attention because although Msh2-Msh6 did not act in antirecombination for substrates predicted to form 4-nt loops (Nicholson et al. 2000), overexpression of Msh6 increased homeologous recombination in both base mismatch and 4-nt loop mismatch substrates. On the other hand, overexpression of Msh3, which interacts with Msh2 and plays a role in antirecombination of 4-nt loop substrates, did not disrupt antirecombination in the inverted repeat assay, but in fact improved it for the 4-nt loop substrate (Table 5).
The above observations can be explained by recombination in the inverted repeat system occurring within the context of DNA replication, when sister chromatids are in close proximity (Nicholson et al. 2000). In this scenario, a high concentration of Msh6 subunit tethered to the replication fork via its interaction with PCNA, could prevent localized deployment to heteroduplex intermediates of Msh2-Msh6 and subsequently other antirecombination factors such as Sgs1-Top3-Rmi1 or Mlh1-Pms1. Excess Msh6 at the replication fork could also sequester Sgs1 or Mlh1-Pms1, thus reducing its availability for heteroduplex rejection or more efficiently recruit downstream MMR proteins that may trigger the MMR pathway. Consistent with this, overexpressing msh6-KQFF > AAAA, which does not interact with PCNA, was functional for antirecombination. If this model is correct, why would Msh3 overexpression not confer a similar phenotype? One possibility is that Msh2-Msh6 and Msh2-Msh3 interact with PCNA differently and that Msh2-Msh3 does not have the same type of access to the replication fork. Supporting this idea is the finding that Msh2-Msh6 has distinct binding sites for PCNA and Mlh factors, whereas Msh2-Msh3 has a common binding site for both (Lau et al. 2002; Iyer et al. 2010). It is important to note that such a model is not likely to be relevant for the SSA system because in this set up, DSBs are artificially induced and not likely to be associated with the replication fork (Fishman-Lobell et al. 1992: Sugawara and Haber 1992; Sugawara et al. 1997, 2004).
Analyzing antirecombination in SSA and inverted repeat assays
Most antirecombination components identified in yeast, such as Msh proteins and Sgs1, are required in both the SSA and inverted repeat assays (Nicholson et al. 2000; Spell and Jinks-Robertson 2004; Sugawara et al. 2004). However, there appear to be differences in the way the two systems repair DNA and reject divergent DNA substrates. For example, mutations in PCNA that disrupt MMR and/or disrupt interactions with Msh6 have either no (SSA system) or minor (inverted repeats system; Stone et al. 2008) effects on heteroduplex rejection. Additionally, Mlh1-Pms1 and Exo1 appear to be partially required for rejection in the inverted repeat system (Nicholson et al. 2000), but play minor if any roles in regulating SSA between divergent sequences (Sugawara et al. 2004). Furthermore, the mechanism(s) of repairing damaged DNA in the two systems is different. The SSA assay involves repair of an induced DSB that does not require Rad51 but requires Rad59 (Sugawara et al. 2000). In contrast, the inverted repeat assay likely measures repair of spontaneous lesions that form during DNA replication through a Rad51- or Rad59-dependent sister chromatid gene conversion mechanism (Spell and Jinks-Robertson 2003). Due to differences in the type of lesions initiating recombination, it is possible that unlike in SSA, 3′ tailed substrates are not formed during recombination between inverted repeats. While we cannot completely rule out the possibility that different strain backgrounds used in the two assays caused the different phenotypes seen when Msh6 was overexpressed, a more likely explanation is that the two assays involve different repair mechanisms regulated by both common and distinct antirecombination factors.
Relevance to human disease
Repetitive and nonallelic sequences of common ancestral origin pose risks to eukaryotic cells because they have the potential to recombine and cause genome rearrangements that can lead to diseases including CMT1A, HNPP, Potocki-Lupski syndrome, segmental neurofibromatosis, and many cancers (Gu and Lupski 2008). For example, segmental duplications >1 kb in size (88 to 99% identity, making up 5–10% of primate genomes) can serve as substrates for chromosomal rearrangements via NAHR (George and Alani 2012; Liu et al. 2012). Heteroduplex rejection is likely to be a critical mechanism by which these NAHR events can be prevented. Identifying new factors and steps that regulate this process is likely to be critical to understand and predict how and when the above human disorders arise.
Acknowledgments
We are grateful to the Alani lab and Jennifer Surtees, Michael Lichten, Jim Haber, Neal Sugawara, and Gray Crouse for fruitful discussions; Madhura Raghavan for conducting some of the SSA assays; and Steven Brill, Sue Jinks-Robertson, Ian Hickson, Richard Kolodner, Rodney Rothstein, and Tom Kunkel for reagents. U.C., C.M.G., A.M.L., and E.A. were supported by NIH GM53085. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of General Medical Sciences or the National Institutes of Health.
Footnotes
Communicating editor: J. Nickoloff
Literature Cited
- Ashton T. M., Mankouri H. W., Heidenblut A., McHugh P. J., Hickson I. D., 2011. Pathways for Holliday junction processing during homologous recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 31: 1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis A. M., Arthur L., Rothstein R., 1992. Genome rearrangement in top3 mutants of Saccharomyces cerevisiae requires a functional RAD1 excision repair gene. Mol. Cell. Biol. 112: 4988–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R. J., Sharp J. A., Wang J. C., 1998. Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J. Biol. Chem. 273: 9644–9650. [DOI] [PubMed] [Google Scholar]
- Bennett R. J., Keck J. L., Wang J. C., 1999. Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J. Mol. Biol. 289: 235–248. [DOI] [PubMed] [Google Scholar]
- Bennett R. J., Noirot-Gros M. F., Wang J. C., 2000. Interaction between yeast Sgs1 helicase and DNA topoisomerase III. J. Biol. Chem. 275: 26898–26905. [DOI] [PubMed] [Google Scholar]
- Bjergbaek L., Cobb J. A., Tsai-Pflugfelder M., Gasser S. M., 2005. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 24: 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., et al. , 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Bradford M. M., 1976. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Brosh R. M., Jr, Waheed J., Sommers J. A., 2002. Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J. Biol. Chem. 277: 23236–23245. [DOI] [PubMed] [Google Scholar]
- Cejka P., Kowalczykowski S. C., 2010. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds Holliday junctions. J. Biol. Chem. 285: 8290–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., et al. , 2010. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467: 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S. R., Hunter N., Louis E. J., Borts R. H., 1996. The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Mol. Cell. Biol. 16: 6110–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., Bellaoui M., Zhang C., Desai R., Morozov P., et al. , 2005. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 24: 2024–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. F., Brill S. J., 2007. Binding and activation of DNA topoisomerase III by the Rmi1 subunit. J. Biol. Chem. 282: 28971–28979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Jinks-Robertson S., 1998. Mismatch repair proteins regulate heteroduplex formation during mitotic recombination in yeast. Mol. Cell. Biol. 18: 6525–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Jinks-Robertson S., 1999. The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics 151: 1299–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P., 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. [DOI] [PubMed] [Google Scholar]
- Clark A. B., Valle F., Drotschmann K., Gary R. K., Kunkel T. A., 2000. Functional interaction of proliferating cell nuclear antigen with MSH2–MSH6 and MSH2–MSH3 complexes. J. Biol. Chem. 275: 36498–36501. [DOI] [PubMed] [Google Scholar]
- Crespan E., Czabany T., Maga G., Hübscher U., 2012. Microhomology-mediated DNA strand annealing and elongation by human DNA polymerases λ and β on normal and repetitive DNA sequences. Nucleic Acids Res. 40: 5577–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A., Adjiri A., New L., Crouse G. F., Jinks-Robertson S., 1996. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wind N., Dekker M., Berns A., Radman M., te Riele H., 1995. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell 82: 321–330. [DOI] [PubMed] [Google Scholar]
- Drummond J. T., Genschel J., Wolf E., Modrich P., 1997. DHFR/MSH3 amplification in methotrexate-resistant cells alters the hMutSalpha/hMutSbeta ratio and reduces the efficiency of base-base mismatch repair. Proc. Natl. Acad. Sci. USA 94: 10144–10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott B., Jasin M., 2001. Repair of double-strand breaks by homologous recombination in mismatch repair-defective mammalian cells. Mol. Cell. Biol. 21: 2671–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einolf H. J., Guengerich F. P., 2000. Kinetic analysis of nucleotide incorporation by mammalian DNA polymerase delta. J. Biol. Chem. 275: 16316–16322. [DOI] [PubMed] [Google Scholar]
- Fasching C. L., Cejka P., Kowalczykowski S. C., Heyer W. D., 2015. Top3-Rmi1 dissolve Rad51-mediated D loops by a topoisomerase-based mechanism. Mol. Cell 57: 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman-Lobell J., Haber J. E., 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258: 480–484. [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell J., Rudin N., Haber J. E., 1992. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol. 12: 1292–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Rozas H., Clark D., Kolodner R. D., 2000. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat. Genet. 26: 375–378. [DOI] [PubMed] [Google Scholar]
- Fricke W. M., Kaliraman V., Brill S. J., 2001. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J. Biol. Chem. 276: 8848–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., McDonald J. P., Bendixen C., Arthur L., Rothstein R., 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14: 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geitz R. D., Schiestl R. H., 1991. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast 7: 253–263. [DOI] [PubMed] [Google Scholar]
- George C. M., Alani E., 2012. Multiple cellular mechanisms prevent chromosomal rearrangements involving repetitive DNA. Crit. Rev. Biochem. Mol. Biol. 47: 297–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George C. M., Lyndaker A. M., Alani E., 2011. The DNA damage checkpoint allows recombination between divergent DNA sequences in budding yeast. DNA Repair (Amst.) 10: 1086–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T., Alani E., 2005. Distinct roles for the Saccharomyces cerevisiae mismatch repair proteins in heteroduplex rejection, mismatch repair and nonhomologous tail removal. Genetics 169: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Chapman J. R., Magill C., Jackson S. P., 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22: 2767–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L., Hong Y., McCulloch S., Watanabe H., Li G. M., 1998. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res. 26: 1173–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Lupski J. R., 2008. CNV and nervous system diseases: What’s new? Cytogenet. Genome Res. 123: 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck J. A., Argueso J. L., Gemici Z., Reeves R. G., Bernard A., et al. , 2006. Negative epistasis between natural variants of the Saccharomyces cerevisiae MLH1 and PMS1 genes results in a defect in mismatch repair. Proc. Natl. Acad. Sci. USA 103: 3256–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson I. D., Mankouri H. W., 2011. Processing of homologous recombination repair intermediates by the Sgs1-Top3-Rmi1 and Mus81-Mms4 complexes. Cell Cycle 10: 3078–3085. [DOI] [PubMed] [Google Scholar]
- Hombauer H., Campbell C. S., Smith C. E., Desai A., Kolodner R. D., 2011a Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell 147: 1040–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombauer H., Srivatsan A., Putnam C. D., Kolodner R. D., 2011b Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science 334: 1713–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M., Okuno Y., Hengel S. R., Martín-López J. V., Cook C. P., et al. , 2014. Mismatch repair protein hMSH2-hMSH6 recognizes mismatches and forms sliding clamps within a D-loop recombination intermediate. Proc. Natl. Acad. Sci. USA 111: E316–E325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., Chambers S. R., Louis E. J., Borts R. H., 1996. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 15: 1726–1733. [PMC free article] [PubMed] [Google Scholar]
- Iyer R. R., Pluciennik A., Genschel J., Tsai M. S., Beese L. S., et al. , 2010. MutLalpha and proliferating cell nuclear antigen share binding sites on MutSbeta. J. Biol. Chem. 285: 11730–11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H., De Muyt A., Lichten M., 2015. Top3-Rmi1 DNA single-strand decatenase is integral to the formation and resolution of meiotic recombination intermediates. Mol. Cell 57: 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman Z., 1997. PCNA: structure, functions and interactions. Oncogene 14: 629–640. [DOI] [PubMed] [Google Scholar]
- Kijas A. W., Studamire B., Alani E., 2003. msh2 Separation of function mutations confer defects in the initiation steps of mismatch repair. J. Mol. Biol. 331: 123–138. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., 2006. Strategies to maintain the stability of the ribosomal RNA gene repeats-collaboration of recombination, cohesion, and condensation. Genes Genet. Syst. 81: 155–161. [DOI] [PubMed] [Google Scholar]
- Kumar C., Piacente S. C., Sibert J., Bukata A. R., O’Connor J., et al. , 2011. Multiple factors insulate Msh2-Msh6 mismatch repair activity from defects in Msh2 domain I. J. Mol. Biol. 411: 765–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Erie D. A., 2005. DNA mismatch repair. Annu. Rev. Biochem. 74: 681–710. [DOI] [PubMed] [Google Scholar]
- Lau P. J., Kolodner R. D., 2003. Transfer of the MSH2.MSH6 complex from proliferating cell nuclear antigen to mispaired bases in DNA. J. Biol. Chem. 278: 14–17. [DOI] [PubMed] [Google Scholar]
- Lau P. J., Flores-Rozas H., Kolodner R. D., 2002. Isolation and characterization of new proliferating cell nuclear antigen, POL30 mutator mutants that are defective in DNA mismatch repair. Mol. Cell. Biol. 22: 6669–6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. D., Surtees J. A., Alani E., 2007. Saccharomyces cerevisiae MSH2–MSH3 and MSH2–MSH6 complexes display distinct requirements for DNA binding domain I in mismatch recognition. J. Mol. Biol. 366: 53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Dong J., Pan X., Oum J. H., Boeke J. D., et al. , 2008. Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Mol. Cell 30: 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sternberg N., 1984. Homologous recombination between overlapping thymidine kinase gene fragments stably inserted into a mouse cell genome. Mol. Cell. Biol. 4: 852–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Carvalho C. M., Hastings P. J., Lupski J. R., 2012. Mechanisms for recurrent and complex human genomic rearrangements. Curr. Opin. Genet. Dev. 22: 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyndaker A. M., Alani E., 2009. A tale of tails: insights into the coordination of 3′ end processing during homologous recombination. BioEssays 31: 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G., Hübscher U., 1995. DNA polymerase epsilon interacts with proliferating cell nuclear antigen in primer recognition and elongation. Biochemistry 34: 891–901. [DOI] [PubMed] [Google Scholar]
- Maga G., Villani G., Ramadan K., Shevelev I., Tanguy Le Gac N., et al. , 2002. Human DNA polymerase lambda functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis. J. Biol. Chem. 277: 48434–48440. [DOI] [PubMed] [Google Scholar]
- Marra G., Iaccarino I., Lettieri T., Roscilli G., Delmastro P., et al. , 1998. Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc. Natl. Acad. Sci. USA 95: 8568–8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E. P., Symington L. S., 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E. P., Symington L. S., 2011. DNA end resection: unraveling the tail. DNA Repair (Amst.) 10: 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J. R., Kaliraman V., Brill S. J., 2000. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics 154: 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J. R., Nallaseth F. S., Lan Y. Q., Slagle C. E., Brill S. J., 2005. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol. Cell. Biol. 25: 4476–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Datta A., Chen C., Kolodner R. D., 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27: 113–116. [DOI] [PubMed] [Google Scholar]
- Nicholson A., Hendrix M., Jinks-Robertson S., Crouse G. F., 2000. Regulation of mitotic homeologous recombination in yeast: functions of mismatch repair and nucleotide excision repair genes. Genetics 154: 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Chung W. H., Zhu Z., Kwon Y., Zhao W., et al. , 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467: 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzi G., Bachrati C. Z., Selak N., Studer I., Petkovic M., et al. , 2003. The Bloom’s syndrome helicase interacts directly with the human DNA mismatch repair protein hMSH6. Biol. Chem. 384: 1155–1164. [DOI] [PubMed] [Google Scholar]
- Petit M. A., Dimpfl J., Radman M., Echols H., 1991. Control of large chromosomal duplications in Escherichia coli by the mismatch repair system. Genetics 129: 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter G., Westmoreland J., Priebe S., Resnick M. A., 1996. Homologous and homeologous intermolecular gene conversion are not differentially affected by mutations in the DNA damage or the mismatch repair genes RAD1, RAD50, RAD51, RAD52, RAD54, PMS1 and MSH2. Genetics 143: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayssiguier C., Thaler D. S., Radman M., 1989. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342: 396–401. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P., 1990. Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Saydam N., Kanagaraj R., Dietschy T., Garcia P. L., Pena-Diaz J., et al. , 2007. Physical and functional interactions between Werner syndrome helicase and mismatch-repair initiation factors. Nucleic Acids Res. 35: 5706–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selva E. M., New L., Crouse G. F., Lahue R. S., 1995. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics 139: 1175–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P., Huang H., 1989. Effect of base pair mismatches on recombination via the RecBCD pathway. Mol. Gen. Genet. 18: 358–360. [DOI] [PubMed] [Google Scholar]
- Shell S. S., Putnam C. D., Kolodner R. D., 2007. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Mol. Cell 26: 565–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E., Gangloff S., Wagner M., Weinstein J., Price G., et al. , 2002. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics 162: 647–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spell R. M., Jinks-Robertson S., 2003. Role of mismatch repair in the fidelity of Rad51- and Rad59-dependent recombination in Saccharomyces cerevisiae. Genetics 165: 1733–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spell R. M., Jinks-Robertson S., 2004. Examination of the roles of Sgs1 and Srs2 helicases in the enforcement of recombination fidelity in Saccharomyces cerevisiae. Genetics 168: 1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J. E., Ozbirn R. G., Petes T. D., Jinks-Robertson S., 2008. Role of proliferating cell nuclear antigen interactions in the mismatch repair-dependent processing of mitotic and meiotic recombination intermediates in yeast. Genetics 178: 1221–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studamire B., Quach T., Alani E., 1998. Saccharomyces cerevisiae Msh2p and Msh6p ATPase activities are both required during mismatch recognition. Mol. Cell. Biol. 18: 7590–7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Haber J. E., 1992. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 12: 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Pâques F., Colaiácovo M., Haber J. E., 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. USA 94: 9214–9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Ira G., Haber J. E., 2000. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20: 5300–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Goldfarb T., Studamire B., Alani E., Haber J. E., 2004. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl. Acad. Sci. USA 101: 9315–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surtees J. A., Argueso J. L., Alani E., 2004. Mismatch repair proteins: key regulators of genetic recombination. Cytogenet. Genome Res. 107: 146–159. [DOI] [PubMed] [Google Scholar]
- Tang S., Wu M. K., Zhang R., Hunter N., 2015. Pervasive and essential roles of the Top3-Rmi1 decatenase orchestrate recombination and facilitate chromosome segregation in meiosis. Mol. Cell 57: 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomé S., Simard J. P., Slean M. M., Holt I., Morris G. E., et al. , 2013. Tissue-specific mismatch repair protein expression: MSH3 is higher than MSH6 in multiple mouse tissues. DNA Repair (Amst.) 12: 46–52. [DOI] [PubMed] [Google Scholar]
- Ui A., Seki M., Ogiwara H., Onodera R., Fukushige S., et al. , 2005. The ability of Sgs1 to interact with DNA topoisomerase III is essential for damage-induced recombination. DNA Repair (Amst.) 4: 191–201. [DOI] [PubMed] [Google Scholar]
- Umar A., Buermeyer A. B., Simon J. A., Thomas D. C., Clark A. B., et al. , 1996. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87: 65–73. [DOI] [PubMed] [Google Scholar]
- Wagner M., Price G., Rothstein R., 2006. The absence of Top3 reveals an interaction between the Sgs1 and Pif1 DNA helicases in Saccharomyces cerevisiae. Genetics 174: 555–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman A. S., 2008. Ensuring the fidelity of recombination in mammalian chromosomes. BioEssays 30: 1163–1171. [DOI] [PubMed] [Google Scholar]
- Wallis J. W., Chrebet G., Brodsky G., Rolfe M., Rothstein R., 1989. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell 58: 409–419. [DOI] [PubMed] [Google Scholar]
- Wang Y., Cortez D., Yazdi P., Neff N., Elledge S. J., et al. , 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14: 927–939. [PMC free article] [PubMed] [Google Scholar]
- Winston F., Dollard C., Ricupero-Hovasse S. L., 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55. [DOI] [PubMed] [Google Scholar]
- Yang Q., Zhang R., Wang X. W., Linke S. P., Sengupta S., et al. , 2004. The mismatch DNA repair heterodimer, hMSH2/6, regulates BLM helicase. Oncogene 23: 3749–3756. [DOI] [PubMed] [Google Scholar]
- Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G., 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]