Abstract
Cell signaling pathways that control proliferation and determine cell fates are tightly regulated to prevent developmental anomalies and cancer. Transcription factors and coregulators are important effectors of signaling pathway output, as they regulate downstream gene programs. In Caenorhabditis elegans, several subunits of the Mediator transcriptional coregulator complex promote or inhibit vulva development, but pertinent mechanisms are poorly defined. Here, we show that Mediator’s dissociable cyclin dependent kinase 8 (CDK8) module (CKM), consisting of cdk-8, cic-1/Cyclin C, mdt-12/dpy-22, and mdt-13/let-19, is required to inhibit ectopic vulval cell fates downstream of the epidermal growth factor receptor (EGFR)-Ras-extracellular signal-regulated kinase (ERK) pathway. cdk-8 inhibits ectopic vulva formation by acting downstream of mpk-1/ERK, cell autonomously in vulval cells, and in a kinase-dependent manner. We also provide evidence that the CKM acts as a corepressor for the Ets-family transcription factor LIN-1, as cdk-8 promotes transcriptional repression by LIN-1. In addition, we find that CKM mutation alters Mediator subunit requirements in vulva development: the mdt-23/sur-2 subunit, which is required for vulva development in wild-type worms, is dispensable for ectopic vulva formation in CKM mutants, which instead display hallmarks of unrestrained Mediator tail module activity. We propose a model whereby the CKM controls EGFR-Ras-ERK transcriptional output by corepressing LIN-1 and by fine tuning Mediator specificity, thus balancing transcriptional repression vs. activation in a critical developmental signaling pathway. Collectively, these data offer an explanation for CKM repression of EGFR signaling output and ectopic vulva formation and provide the first evidence of Mediator CKM-tail module subunit crosstalk in animals.
Keywords: Mediator complex, CDK8, MED23, MED15, EGFR, Notch
PRECISE regulation of transcription is required to execute developmental programs such as proliferation and cell fate determination. The Mediator complex (“Mediator”) is a conserved eukaryotic transcriptional coregulator of RNA polymerase II (Pol II) transcription (Malik and Roeder 2010; Poss et al. 2013). Mediator consists of ∼30 subunits that assemble into four modules. “Core” Mediator consists of three of the four modules: the head and middle modules, which contact Pol II, and the tail module, which serves as a docking site for transcription factors. The fourth module, the dissociable cyclin dependent kinase 8 (CDK8) kinase module (CKM), interacts with transcription factors, core Mediator, chromatin, and the Pol II machinery to either repress or activate transcription (Malik and Roeder 2010; Nemet et al. 2014). Whereas many head or middle Mediator subunits are broadly required for Pol II transcription, tail and CKM subunits regulate specific transcriptional programs in animal development or physiology (Malik and Roeder 2010; Nemet et al. 2014).
The CKM consists of enzymatic subunits CDK8 and cyclin C, and structural subunits MED12 and MED13 that tether the CKM to core Mediator (Tsai et al. 2013). CKM subunits regulate many transcriptional programs important for development and/or tumorigenesis, often by directly binding to and influencing the activity of key transcription factors (e.g., β-catenin, Notch, etc.) (Fryer et al. 2004; Donner et al. 2007; Firestein et al. 2008; Zhou et al. 2012). Furthermore, in Saccharomyces cerevisiae, the CKM regulates the activity of the Mediator tail module subunits MED2, MED3, and MED15 (van de Peppel et al. 2005; Gonzalez et al. 2014). However, whether such intra-Mediator signaling effects occur in metazoans and affect e.g., animal development has not yet been tested.
Several Mediator subunits including at least one CKM subunit regulate vulva development in Caenorhabditis elegans (Tuck and Greenwald 1995; Singh and Han 1995; Kwon and Lee 2001; Moghal and Sternberg 2003a). The study of cell fate specification in the C. elegans vulva has proven a powerful way to identify the components and regulatory interactions of several evolutionarily conserved signaling pathways (Félix and Barkoulas 2012; Schmid and Hajnal 2015). Thus, this organogenesis event provides an ideal paradigm to study Mediator subunit specificity and cooperation in a metazoan.
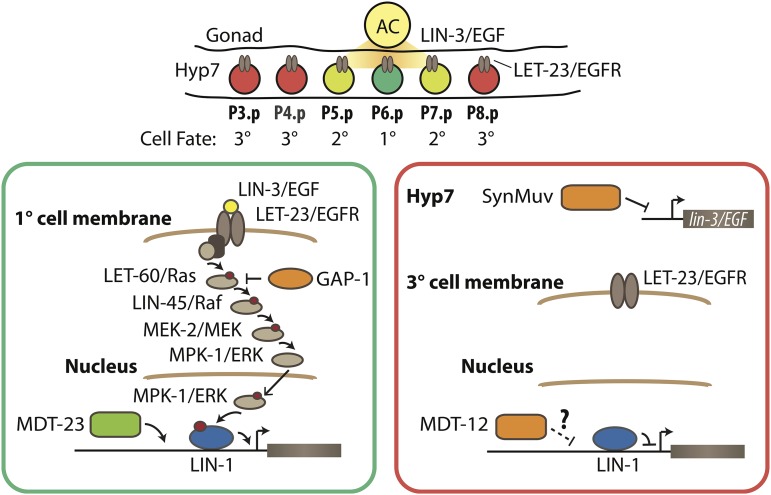
C. elegans vulval organogenesis is induced by epidermal growth factor receptor (EGFR) signaling (Moghal and Sternberg 2003b), a prominent pathway in animal development that is frequently activated in human cancers (Normanno et al. 2006; Baselga and Swain 2009). The C. elegans vulva develops from six ventral vulva precursor cells (VPCs), named P3.p through P8.p from anterior to posterior (Figure 1). The VPCs form an equivalence group, meaning that all six cells are able to adopt the primary (1°) vulval cell fate (producing eight descendants), the secondary (2°) vulval cell fate (producing seven descendants), or the tertiary (3°) nonvulval fate (producing two descendants that fuse with the surrounding hypodermis). A signaling cell in the somatic gonad, called the anchor cell, emits a LIN-3/EGF-like ligand in close proximity to P6.p (Hill and Sternberg 1992); therefore, LET-23/EGFR and the downstream LET-60/Ras, MPK-1/extracellular signal-regulated kinase (ERK) cascade is strongly activated in P6.p (Aroian et al. 1990). MPK-1/ERK activation in P6.p modulates the activity of effector transcription factors such as the ELK1/Ets-family transcription factor LIN-1 and the FoxB transcription factor LIN-31, thereby specifying the 1° vulval fate in P6.p (Miller et al. 1993; Tan et al. 1998; Jacobs et al. 1998). The neighboring P5.p and P7.p cells are thought to receive a weaker LIN-3/EGF signal from the anchor cell (Katz et al. 1995) as well as a lateral Notch signal emitted from the 1° cell P6.p, inducing them to adopt a 2° vulval fate (Chen and Greenwald 2004). Located furthest from the anchor cell, P3.p, P4.p, and P8.p do not receive sufficient EGF signal, and adopt the 3° nonvulval cell fate (Sternberg and Horvitz 1986). Mutations that enhance or reduce EGFR or Notch signaling induce ectopic vulval cell fates (multivulva phenotype, Muv) or loss of vulval cell fates (vulvaless phenotype, Vul), respectively (Sternberg and Horvitz 1989). These phenotypes are thus powerful indicators of EGFR and Notch signaling pathway activity.
Figure 1.
Transcriptional regulators in C. elegans vulval induction. The C. elegans vulva is derived from an equivalence group consisting of six vulva precursor cells (VPCs), named P3.p through P8.p from anterior to posterior. A localized LIN-3/EGF signal from the anchor cell (AC) in the somatic gonad activates a LET-23/EGFR-LET-60/Ras-MPK-1/ERK signaling cascade strongly in P6.p. ERK activation in P6.p modulates transcription factor activity in the nucleus (only LIN-1 is shown here for simplicity), leading to induction of the 1° cell fate. In P5.p and P7.p, a weak LIN-3/EGF signal combined with lateral Notch signaling from P6.p (not depicted) instead produces the 2° cell fate. In P3.p, P4.p, and P8.p, the EGFR signaling cascade is not activated by LIN-3/EGF, and cells adopt the nonvulval 3° cell fate. Transcriptional regulators of the EGFR signaling pathway are critical for correct vulval cell fate specification: e.g., the MDT-23/SUR-2 Mediator subunit is a coactivator of EGFR signaling-induced transcription, the SynMuv corepressor complexes are required to inhibit ectopic lin-3/EGF transcription in the hypodermis (Hyp7) surrounding the VPCs, and the MDT-12/DPY-22 Mediator subunit is required to inhibit vulva development by mechanisms that remain unclear (dashed arrow). The GTPase activating protein GAP-1 that negatively regulates LET-60/Ras activity post-translationally is also shown.
Transcriptional regulation is important in maintaining appropriate EGFR signaling pathway output (Figure 1). For example, transcription factors such as LIN-1/Ets and LIN-31/Forkhead are required to repress 1° cell fate specification in VPCs other than P6.p (Miller et al. 1993; Beitel et al. 1995). In addition, multiple chromatin-modifying complexes, encoded by the synthetic multivulva (synMuv) genes, redundantly repress ectopic lin-3/EGF transcription in the hypodermis and other tissues to inhibit 1° cell fate specification in VPCs other than P6.p (Myers and Greenwald 2005; Cui et al. 2006; Saffer et al. 2011). Furthermore, the Mediator subunits mdt-23/sur-2, mdt-24/lin-25, and mdt-6 promote vulva development, whereas the CKM subunit mdt-12/dpy-22 inhibits vulva development in an anchor cell-independent manner (reviewed in Grants et al. 2015); for standardized Mediator subunit nomenclature, please see Bourbon et al. (2004). The mechanism by which mdt-23/sur-2 promotes vulva development has been partially elucidated, as it is a critical coactivator of a target gene downstream of the EGFR signaling pathway, the lag-2 Notch ligand gene (Zhang and Greenwald 2011). The lin-1/Ets effector transcription factor is similarly required to repress the lag-2 gene (Zhang and Greenwald 2011), raising the question of whether and how Mediator and LIN-1 interact to control common target genes. The other three Mediator subunits implicated in vulva development (mdt-6, mdt-12/dpy-22, and mdt-24/lin-25) interact genetically with components of the EGFR signaling pathway, but their mode of action within this pathway remain poorly understood.
Here, we used the vulva organogenesis paradigm to study the requirements of all four CKM subunits in this process and to interrogate functional interactions with other transcriptional regulators, including the synMuv genes, the key transcription factors lin-1/Ets and lin-31/FoxB, and the Mediator subunit mdt-23/sur-2, an essential effector of EGFR signaling output. We show that all four CKM subunits inhibit ectopic vulval cell fates in C. elegans. We demonstrate that the CKM catalytic subunit cdk-8 acts downstream of let-23/EGFR and mpk-1/ERK in VPCs, in a kinase-dependent manner. Our data implicate cdk-8 as a corepressor for the LIN-1/Ets repressive transcription factor to inhibit EGFR signaling-induced transcription. Furthermore, our data indicate that vulval induction in CKM mutants is independent of the mdt-23/sur-2 coactivator, and instead requires the Mediator tail module subunits mdt-15, mdt-27, and mdt-29 for induction of ectopic vulval cell fates.
Materials and Methods
Microarrays and data analysis
Microarray gene expression profiling was performed at the University of California San Francisco SABRE Functional Genomics Facility. We used Agilent C. elegans (V2) 4x44K Gene Expression Microarrays (G2519F-020186) and single color labeling. Total RNA was extracted from developmentally synchronized mid-L4 stage worms as assessed by vulval morphology [wild-type N2 worms and cdk-8(tm1238) mutants], as described (Taubert et al. 2008). RNA quality was assessed on an Agilent 2100 Bioanalyzer using a Pico Chip (Agilent). RNA was amplified and labeled with Cy3-CTP using the Agilent low RNA input fluorescent linear amplification kit. Labeled cRNA was assessed using the Nanodrop ND-100, and equal amounts of Cy3-labeled target were hybridized to the microarrays for 14 hr, according to the manufacturer’s protocol. Arrays were scanned using the Agilent microarray scanner and raw signal intensities were extracted with Feature Extraction v9.1 software. The dataset was normalized using quantile normalization (Bolstad et al. 2003). No background subtraction was performed, and median feature pixel intensity was used as raw signal before normalization. All arrays were of good quality and had similar foreground and background signal distributions for both messenger RNA (mRNA) and control probes. This suggests that quantile normalization is appropriate. To identify differentially expressed genes, a linear model was fit to the comparison to estimate the mean M-values and calculate moderated t-statistic, B-statistic, false discovery rate, and P-value for each gene. Adjusted P-values (AdjP) were produced as described (Holm 1979). All procedures were carried out using functions in the R package limma in Bioconductor (Gentleman et al. 2004; Smyth 2004). Using this approach, we identified a total of 1860 spots with an AdjP of <0.05 and a fold change of ≥2 (representing 461 downregulated and 829 upregulated genes) (Supporting Information, Table S1). Microarray data have been deposited in Gene Expression Omnibus (GSE68520).
Differentially expressed genes were compared to published gene expression datasets using EASE (Hosack et al. 2003). For best comparison to our data, we reanalyzed published lin-35 data (Kirienko and Fay 2007) to define a set of genes deregulated twofold or more in L4 larvae, yielding 132 downregulated and 367 upregulated genes. We compared this set to our cdk-8 targets and calculated the significance of the overlap using Fisher’s exact test.
C. elegans strains, culture, and genetic methods
C. elegans strains were cultured as described (Brenner 1974) at 20° or 23°, as indicated. We used nematode growth medium (NGM)-lite (0.2% NaCl, 0.4% tryptone, 0.3% KH2PO4, 0.05% K2HPO4) agar plates seeded with Escherichia coli strain OP50 unless otherwise indicated.
Strains are listed in Table S4. Wild type was Bristol N2. cdk-8(tm1238) and cic-1(tm3740) are likely null alleles that abolish cdk-8 expression (Figure S1B) and cic-1 function, respectively (see also Steimel et al. 2013). For allele details, see www.wormbase.org. mdt-12/dpy-22 mutants were identified as Dpy, GFP-negative progeny of rescued dpy-22(os38); osEx89[dpy-22(+)] mothers, and homozygous mdt-13/let-19 mutants were identified as Dpy, GFP-negative progeny of balanced let-19(mn19)/mIn1 mothers.
VPC induction
VPC induction was scored as described (Han et al. 1990), in synchronous mid-L4 animals under DIC optics at ×1000 magnification. In wild-type animals, P5.p–P7.p are induced to give a VPC induction score of 3.0. In Vul animals, these VPCs are not fully induced (VPC induction <3.0); in Muv animals, P3.p, P4.p, or P.8p are induced (VPC induction >3.0).
Multivulva and vulvaless phenotype penetrance
Muv and Vul morphologies have been described (Horvitz and Sulston 1980; Sulston and Horvitz 1981). To facilitate scoring a large number of worms to accurately assess low-penetrance phenotypes, Muv phenotype penetrance was scored in synchronous day 1 adult animals in a dissection microscope at ×200 magnification (mdt-13/let-19 mutants) or ×56 magnification (all other strains). To corroborate Muv penetrances scored in adult animals, we also conducted VPC induction analysis in L4 animals (see above). To assess Vul phenotypes, both Vul and Muv penetrances were extrapolated from VPC induction scores: animals were scored as Vul if VPC induction was <3.0 in P5.p–P7.p and were scored as Muv if VPC induction occurred in P3.p, P4.p or P8.p; using these criteria, animals were occasionally scored as simultaneously Vul and Muv.
RNA isolation and quantitative real-time PCR
Total RNA was extracted from developmentally synchronized mid-L4 stage worms as assessed by vulval morphology. RNA isolation and qPCR were performed as described (Goh et al. 2014). We used t-tests (two-tailed, equal variance) to calculate statistical significance of gene expression changes between mutants (Gaussian distribution). qPCR primers were designed with Primer3web (bioinfo.ut.ee/primer3/) and tested on serial cDNA dilutions to analyze PCR efficiency (primer sequences in Table S5), except lin-3 (analyzed by TaqMan assay, Invitrogen 4448892, assay ID Ce02418781_m1).
Fluorescent reporter analysis
Synchronous worms were imaged using DIC optics and fluorescence microscopy on a Zeiss Axioplan 2 microscope. Analysis of fluorescence intensity was conducted using ImageJ software, normalizing for cell size and background fluorescence.
Generation of transgenic rescue strains
cdk-8 rescue transgenes (steEx43, 45–47) were generated by gonad microinjection of a mixture of 50 ng/μl rescue plasmid [cdk-8(+), SPD732; lin-31P::cdk-8, SPD793; dpy-7P::cdk-8, SPD772; cdk-8(KD), SPD789; 5 ng/μl pCFJ90[myo-2p::mCherry], and 95 ng/μl pPD95.77 empty vector into N2 worms, then selecting transgenic mCherry-positive progeny. These were then crossed to cdk-8 and/or cdk-8; lin-15A mutants (Table S4). Cloning primer sequences are provided (Table S5).
Feeding RNAi knockdown
Feeding RNAi was performed as described (Goh et al. 2014), with the following modifications: synchronous mid-L4 hermaphrodites were allowed to lay eggs at 20° overnight on RNAi plates (Ahringer Library 96-well format; mdt-15: plate 74, well C09; lin-1: 94, G02; Vidal Library 96-well format: mdt-27: GHR-11064@H02; mdt-29: GHR-11007@D05; all clones were sequenced to confirm identity; negative control was empty vector L4440), after which embryos were isolated by bleach treatment and transferred to fresh RNAi plates. F1 progeny were grown on RNAi plates (20° or 23°) until they reached the desired developmental stage.
Western blot
Immunoblot using standard lysis, SDS/PAGE and Western blot techniques was performed, with α-MDT-15 (Taubert et al. 2006) and α-GAPDH (Calbiochem, CB1001) antibodies, as described (Goh et al. 2014).
Data availability
Primer sequences are listed in Table S5. Strains are listed in Table S4 and are available upon request. Gene expression data are available at GEO with the accession number GSE68520.
Results
cdk-8-dependent transcripts overlap with targets of a synMuv gene
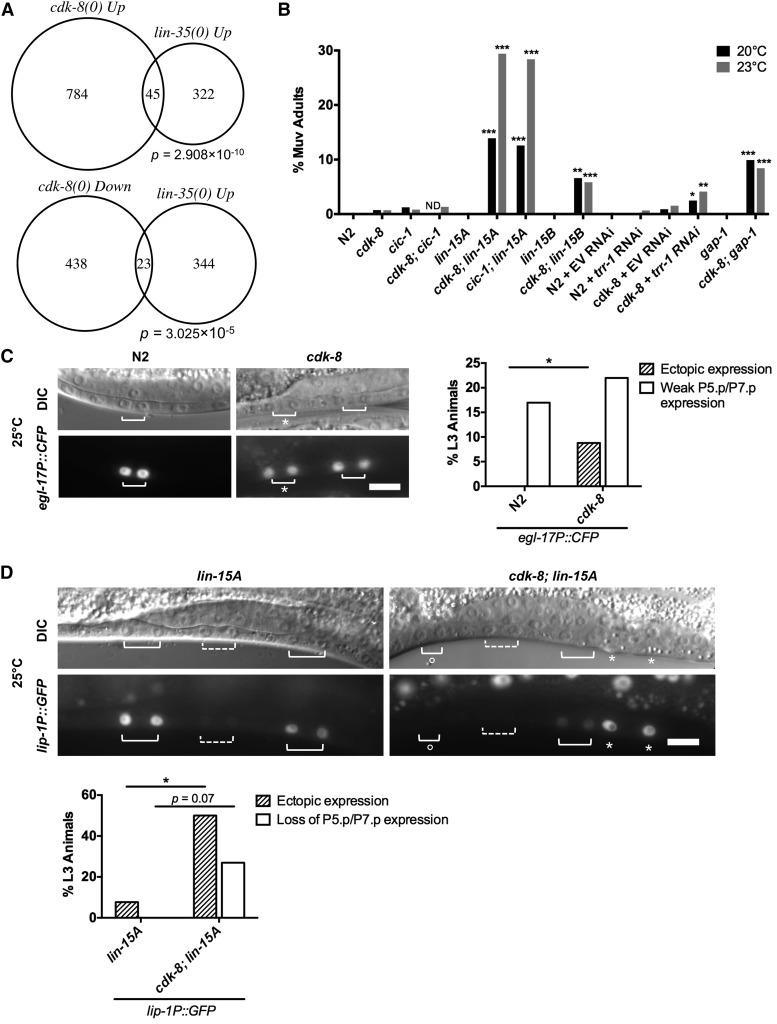
To define the role of the CKM in metazoan development, we compared transcriptional profiles of developmentally synchronized L4 larval stage cdk-8(tm1238) null mutants to wild-type N2 worms using microarrays (see Materials and Methods and Figure S1A for cdk-8 mutant information). We found that 829 genes were upregulated and 461 genes were downregulated more than twofold in cdk-8 null mutants, representing ∼6.7% of all C. elegans genes (Table S1). To identify cdk-8-dependent gene programs, we compared our lists of cdk-8 regulated genes to other gene lists using EASE (Hosack et al. 2003; Engelmann et al. 2011). The top hit among genes upregulated in cdk-8 mutants was a set of genes upregulated in lin-35/Retinoblastoma (RB) synMuv gene mutants (Kirienko and Fay 2007), and lin-35-repressed genes also overlapped significantly with genes downregulated in cdk-8 mutants (Figure 2A). Importantly, the mRNA levels of lin-35 and the efl-1/dpl-1 transcription factor heterodimer that is repressed by LIN-35 were not altered in cdk-8 mutants (Figure S1B), indicating that cdk-8 does not affect lin-35 target gene expression by altering the abundance of lin-35 or its partners. Together, these data suggest that cdk-8 could act in parallel to lin-35 as they regulate similar gene sets.
Figure 2.
cdk-8 represses vulval cell fates redundantly with synMuv genes. (A) Overlap of cdk-8 regulated genes with lin-35/RB repressed genes. P values were determined by Fisher’s exact test. (B) Adult Muv penetrance in cdk-8 and mutants with synMuv genes (n ≥ 76). *P < 0.05, **P < 0.01, ***P < 0.001 vs. cdk-8 mutant or vs. cdk-8 + empty vector (EV), Fisher’s exact test. ND, not determined. See Table S2 for raw data. (C) Micrographs show wild-type or cdk-8 mutant worms expressing the arIs92[egl-17P::CFP] 1° fate marker. Bracket: Pn.px cells expressing reporter. Asterisk: ectopic reporter expression. Bar: 10 μm. The graph displays the percentage of animals (n > 50) with ectopic egl-17P::CFP expression (defined as expression in P3.p/P4.p/P8.p, or expression in P5.p/P7.p of equal intensity to P6.p) or weak P5.p/P7.p expression (defined as expression in P5.p/P7.p that is weaker intensity than P6.p). *P < 0.05, Fisher’s exact test. (D) Micrographs show lin-15A and cdk-8; lin-15A mutants expressing the Notch-inducible reporter zhIs4[lip-1P::GFP]. Dashed bracket: P6.px. Solid bracket: P5.px and P7.px. Open circle: loss of expression. Asterisk: ectopic expression. Bar: 10 μm. The graph shows the percentage of animals (n ≥ 13) with ectopic expression (defined as strong expression in P3.p, P4.p, or P8.p) or loss of expression (defined as weak or no expression in P5.p or P7.p) of the lip-1P::GFP reporter. *P < 0.05, Fisher’s exact test.
cdk-8 represses EGFR signaling-dependent primary vulval cell fate specification
We next investigated whether cdk-8, like mdt-12/dpy-22 (Moghal and Sternberg 2003a), also represses vulval induction. Indeed, cdk-8 and cic-1(tm3740) null mutants displayed a low penetrance Muv phenotype, as measured both by VPC induction analysis in L4 animals (Table 1) and by scoring the occurrence of ectopic vulval protrusions in adult worms (Figure 2B; see statistical comparisons between L4 and adult Muv scores, Table S2). cdk-8 and cic-1 appeared to function redundantly in vulva formation, as cdk-8; cic-1 double mutants showed no significant increase in Muv penetrance compared to either single mutant (Figure 2B). We then tested if the cdk-8 mutant Muv phenotype was associated with ectopic EGFR signaling-induced 1° vulval cell fates using an egl-17P::CFP reporter (arIs92) (Yoo et al. 2004). In wild-type worms, we observed strong egl-17P::CFP expression in the 1° cell descendants, P6.px, and occasional weak expression in the 2° cell descendants, P5.px or P7.px (Figure 2C), as reported (Yoo et al. 2004). cdk-8 mutants expressed egl-17P::CFP strongly in P6.ps and weakly in P5.px and P7.px, like wild type (Figure 2C). However, some cdk-8 mutants exhibited egl-17P::CFP expression in P5.px or P7.px that was equal in intensity to the expression level in the 1° cell descendants P6.px, or exhibited ectopic induction of egl-17P::CFP in VPCs that normally adopt the nonvulval fate (Figure 2C). The ectopic expression of egl-17::CFP suggested that the 1° cell fate had been derepressed in presumptive 2° or nonvulval cells. This reflects that the low penetrance Muv phenotype of cdk-8 mutants is caused in part through derepressed ERK signaling output.
Table 1. VPC induction scores and Muv/Vul penetrance in L4 animals (20°).
| Genotype | VPC induction ± SEM | % Muv L4a | % Vul L4b | % WT L4 vulva | n | |
|---|---|---|---|---|---|---|
| N2 | 3.00 ± 0.00 | 0 | 0 | 100 | 15 | |
| CKM | cdk-8(tm1238) | 3.02 ± 0.02 | 2 | 0 | 98 | 50 |
| mdt-13(mn19) | 2.96 ± 0.07 | 15 | 27 | 65 | 26 | |
| SynMuv | lin-15A(n767) | 3.00 ± 0.00 | 0 | 0 | 100 | 18 |
| cdk-8(tm1238); lin-15A(n767) | 3.20 ± 0.08 | 21 | 0 | 79 | 28 | |
| lin-15B(n744) | 3.00 ± 0.00 | 0 | 0 | 100 | 12 | |
| cdk-8(tm1238); lin-15B(n744) | 3.07 ± 0.05 | 14 | 0 | 86 | 14 | |
| N2 + EV RNAi | 3.00 ± 0.00 | 0 | 0 | 100 | 48 | |
| N2 + trr-1 RNAi | 3.00 ± 0.00 | 0 | 0 | 100 | 15 | |
| cdk-8(tm1238) + EV RNAi | 3.00 ± 0.00 | 0 | 0 | 100 | 37 | |
| cdk-8(tm1238) + trr-1 RNAi | 3.02 ± 0.02 | 4 | 0 | 96 | 26 | |
| gap-1(ga133) | 3.00 ± 0.00 | 0 | 0 | 100 | 15 | |
| cdk-8(tm1238); gap-1(ga133) | 3.03 ± 0.02 | 7 | 0 | 93 | 29 | |
| let-23 | let-23(sy97) | 0.00 ± 0.00 | 0 | 96 | 4 | 9 |
| cdk-8(tm1238); let-23(sy97) | 2.44 ± 0.27 | 0 | 37 | 63 | 8 | |
| mpk-1 | mpk-1(oz14) | 2.03 ± 0.17 | 0 | 80 | 20 | 15 |
| cdk-8(tm1238); mpk-1(oz140) | 2.95 ± 0.03 | 0 | 10 | 90 | 15 | |
| mdt-13(mn19); mpk-1(oz140) | 2.85 ± 0.27 | 23 | 31 | 46 | 13 | |
| lin-1 | lin-1(n1790) | 2.98 ± 0.08 | 20 | 25 | 65 | 20 |
| cdk-8(tm1238); lin-1(n1790) | 3.58 ± 0.16 | 67 | 0 | 34 | 12 | |
| mdt-13(mn19); lin-1(n1790) | 5.33 ± 0.15 | 100 | 0 | 0 | 12 | |
| N2 + lin-1 RNAi | 3.10 ± 0.06 | 7 | 0 | 93 | 40 | |
| cdk-8(tm1238) + lin-1 RNAi | 3.25 ± 0.06 | 31 | 0 | 69 | 61 | |
| lin-31 | lin-31(n301) | 3.65 ± 0.14 | 73 | 20 | 13 | 30 |
| cdk-8(tm1238); lin-31(n301) | 2.46 ± 0.23 | 33 | 53 | 30 | 30 | |
| mdt-23 | mdt-23 (ku9) | 1.13 ± 0.16 | 0 | 100 | 0 | 23 |
| mdt-13(mn19); mdt-23(ku9) | 3.57 ± 0.21 | 64 | 18 | 23 | 22 | |
| mdt-15 | N2 + mdt-15 RNAi | 3.00 ± 0.00 | 0 | 0 | 100 | 33 |
| lin-1(n1790) + EV RNAi | 3.00 ± 0.12 | 17 | 19 | 70 | 46 | |
| lin-1(n1790) + mdt-15 RNAi | 3.01 ± 0.14 | 18 | 22 | 67 | 45 |
Percentage of L4 animals with ectopic vulval invagination at P3.p, P4.p, or P8.p.
Percentage of L4 animals with vulval induction <3.0 at P5.p-P7.p. Note: it is possible for an animal to be scored as both Vul and Muv.
cdk-8 interacts genetically with the synMuv repressors of lin-3/EGF transcription
The CKM subunit mdt-12/dpy-22 was previously shown to act downstream of let-23/EGFR to modulate vulva development (Moghal and Sternberg 2003a). However, the developmental roles of all four CKM subunits are not equivalent (Loncle et al. 2007), and our gene expression profiling suggested that cdk-8 might interact genetically with the synMuv genes to alter vulval cell fate decisions. As synMuv genes encode three redundant chromatin-modifying complexes, a Muv phenotype results when genes in any two complexes are simultaneously mutated (Myers and Greenwald 2005; Cui et al. 2006; Saffer et al. 2011). We therefore studied the simultaneous inactivation of cdk-8 or cic-1 and representative synMuv class A (lin-15A), B (lin-15B), or C (trr-1) genes. Mutation or RNA interference (RNAi) depletion of all representative synMuv genes enhanced the Muv phenotype of cdk-8 mutants (Figure 2B, Table 1). In addition, loss of the Ras GTPase-activating protein gene gap-1, which causes weak LET-60/Ras derepression, also enhanced the cdk-8 mutant Muv penetrance (Figure 2B, Table 1). Our microarray analysis did not reveal significant downregulation of any known synMuv genes in cdk-8 mutants (Table S3), indicating that cdk-8 does not simply regulate synMuv mRNA levels. Taken together, these results suggest that cdk-8 and cic-1 act redundantly with synMuv genes to repress vulval cell fates.
We observed adjacent VPCs expressing a 1° cell fate marker in cdk-8 mutants (Figure 2C), which is uncharacteristic of synMuv gene mutants. This phenotype instead suggests defects in Notch signaling, which inhibits adjacent 1° fates by inducing EGFR signaling inhibitor genes (Sternberg 1988; Berset et al. 2001; Yoo et al. 2004; Chen and Greenwald 2004). Therefore, we examined the expression of the Notch-inducible EGFR signaling inhibitor, lip-1/ERK phosphatase, using a lip-1P::GFP reporter (zhIs4) (Berset et al. 2001). We used the sensitized lin-15A mutant background to increase the frequency of ectopic VPC induction events. lin-15A single mutants expressed lip-1P::GFP strongly in P5.px and P7.px, but expression was weak or absent in other Pn.px cells (Figure 2D), as reported for wild-type worms (Berset et al. 2001). In contrast, some cdk-8; lin-15A mutants lost strong lip-1P::GFP expression in P5.px and P7.px, consistent with loss of the 2° fate (Figure 2D). Furthermore, some cdk-8; lin-15A mutants ectopically expressed lip-1P::GFP strongly in nonvulval P3.px, P4.px, or P8.px, suggesting ectopic 2° fates (Figure 2D). Thus, cdk-8 mutants display hallmarks of both down- and upregulated Notch signaling, suggesting that CDK-8 action on the Notch pathway may occur indirectly via the EGFR signaling pathway upstream.
cdk-8 regulates lin-3/EGF transcription in the anchor cell
We next tested if cdk-8 acts redundantly with the synMuv genes to repress lin-3/EGF transcription (Cui et al. 2006; Saffer et al. 2011). As the synMuv genes act primarily in the hypodermis to repress lin-3 transcription (Myers and Greenwald 2005; Saffer et al. 2011), derepression of lin-3 in synMuv double mutants is detectable by quantitative PCR in whole-animal preparations (Cui et al. 2006). We used Taqman quantitative PCR analysis to quantify whole-animal lin-3 mRNA levels in cdk-8, lin-15A, and lin-15B single mutants, and in cdk-8; lin-15A and cdk-8; lin-15B double mutants; the lin-15AB(n309) mutant served as a positive control, as it is known to upregulate lin-3 expression (Figure 3A) (Cui et al. 2006). Compared to wild-type worms, cdk-8 single mutants, cdk-8; lin-15A double mutants, and cdk-8; lin-15B double mutants showed no statistically significant change in lin-3 mRNA levels (Figure 3A). Thus, the enhanced Muv penetrance of cdk-8; lin-15A and cdk-8; lin-15B mutants compared to cdk-8 single mutants (Figure 2B) likely does not arise from hypodermal lin-3 derepression.
Figure 3.
cdk-8 acts upstream and downstream of let-23/EGFR. (A) qPCR analysis of lin-3 mRNA levels in cdk-8 single mutants and mutants with synMuv genes, relative to wild-type worms. lin-15AB mutant is shown as a positive control (hatched bar). Error bars represent standard error of the mean (SEM, n = 3 independent trials). No statistically significance differences, Wilcoxon signed rank test by the Pratt method. (B) hdEx508[cdk-8P::GFP] expression in the anchor cell (arrow) during early vulval induction. Top: VPCs divided once (Pn.px). Middle: VPCs divided twice (Pn.pxx). Bottom: Invagination of Pn.pxx epithelium. The fluorescent signal visible near VPCs localizes to neuron cell bodies. Bar: 10 μm. (C) Average fluorescence intensity of syIs107[lin-3ACEL::GFP] in anchor cell of wild-type worms and cdk-8 mutants. *P < 0.05 vs. N2, t-test. A.U., arbitrary units. (D) L4 Vul penetrance in cdk-8; let-23/EGFR and cdk-8; mpk-1/ERK mutants (n ≥ 8). ****P < 0.0001, Fisher’s exact test. See Table 1 for raw data. (E) Tissue specificity of Muv phenotype in cdk-8; lin-15A mutant adults expressing wild-type cdk-8 driven by its own promoter cdk-8(+), the Pn.p promoter lin-31P, or the hypodermal promoter dpy-7P, compared to nontransgenic siblings (Sib) in each strain (n ≥ 76). ****P < 0.0001 vs. nontransgenic sibling, Fisher’s exact test. See Table S2 for raw data.
Next, we investigated whether cdk-8 is required to regulate lin-3 transcription in the signal-emitting anchor cell, which would not be detectable in whole-animal quantitative PCR analysis. In line with this hypothesis, we observed expression of a transcriptional cdk-8P::GFP reporter (hdEx508) in the anchor cell (Figure 3B). To assess lin-3 transcription in the anchor cell alone, we used a lin-3 anchor cell-specific enhancer element (ACEL) GFP reporter (syIs107) (Hwang and Sternberg 2004). We detected a small but significant upregulation of lin-3 ACEL reporter expression in cdk-8 mutants compared to wild-type worms at the L3 larval stage (Figure 3C), suggesting that cdk-8 is required to repress lin-3/EGF transcription in the anchor cell.
cdk-8 acts downstream of mpk-1/ERK to regulate VPC induction cell autonomously
As the effect size of cdk-8 loss on lin-3/EGF anchor cell expression was small, and as mdt-12/dpy-22 has been found to act downstream of let-23/EGFR (Moghal and Sternberg 2003a), we next investigated cdk-8’s role in the EGFR signaling pathway downstream of lin-3. We conducted genetic epistasis analyses with strong loss-of-function alleles of EGFR, let-23(sy97), and ERK, mpk-1(oz140); both caused highly penetrant Vul phenotypes due to blockade of the EGFR-Ras-ERK pathway (Figure 3D, Table 1). cdk-8 inactivation significantly rescued the Vul phenotype of let-23 or mpk-1 single mutants (Figure 3D, Table 1). These data suggest that cdk-8 primarily acts downstream or parallel to mpk-1/ERK to repress vulval cell fate specification by the EGFR signaling pathway.
cdk-8’s position downstream of mpk-1/ERK suggested a cell-autonomous role in VPCs (Figure 1). Nuclear expression of the MDT-12/DPY-22 protein in VPCs and in the anchor cell had previously been observed, and gonad-independent vulval induction in mdt-12/dpy-22 mutants suggested an anchor cell-independent role for MDT-12/DPY-22 (Moghal and Sternberg 2003a). However, the tissue-specific requirements for MDT-12/DPY-22 in VPCs vs. the hypodermis, two important drivers of VPC cell fate (Fay and Yochem 2007; Schmid and Hajnal 2015), had not been tested. We used the lin-15A sensitized background to analyze tissue-specific requirements for cdk-8 in VPCs vs. the hypodermis. First, we demonstrated that a transgene expressing cdk-8 from its own promoter [cdk-8(+)] rescued the cdk-8; lin-15A mutant Muv phenotype compared to nontransgenic siblings (Figure 3E). This transgene appeared to be broadly expressed and functional, as it rescued two additional phenotypes observed in cdk-8 mutants: decreased body length (Dumpy phenotype, Dpy) and the low brood size of the cdk-8 mutant (Figure S2, A and B). Expression of cdk-8 from the lin-31 promoter (lin-31P::cdk-8), which drives transgene expression in Pn.ps and some neurons (Tan et al. 1998; Kishore and Sundaram 2002), also significantly rescued the cdk-8; lin-15A Muv phenotype (Figure 3E). In contrast, expression of cdk-8 from the hypodermis-specific dpy-7 minimal promoter (dpy-7P::cdk-8) (Gilleard et al. 1997) did not significantly rescue the Muv penetrance of cdk-8; lin-15A mutant worms (Figure 3E), although it was able to rescue the Dpy phenotype (Figure S2A). Unexpectedly, the lin-31P::cdk-8 transgene partially rescued the cdk-8; lin-15A Dpy phenotype compared to nontransgenic worms, albeit to a lesser extent than cdk-8(+) or dpy-7P::cdk-8 (Figure S2A). In sum, these experiments provide evidence that cdk-8 is required cell autonomously in VPCs but not in the hypodermis to suppress ectopic vulval induction.
cdk-8 activity is kinase dependent
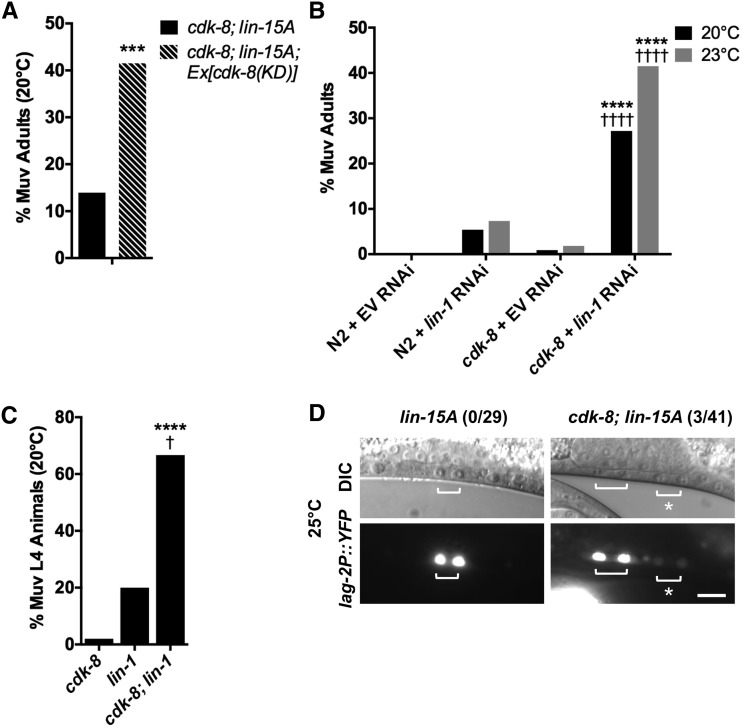
We next addressed how cdk-8 functions downstream of mpk-1/ERK. First, we studied CDK-8’s kinase requirement using a kinase-dead CDK-8(D182A) transgene [CDK-8(KD)]. The D182A mutation is homologous to the previously reported D173A mutation in human CDK8 and the D290A mutation in budding yeast CDK8, both of which result in loss of enzymatic activity (Liao et al. 1995; Gold and Rice 1998); however, we note that the kinase activity of C. elegans CDK-8(D182A) has not been tested directly. CDK-8(KD) did not rescue the cdk-8; lin-15A mutant Muv phenotype, and actually enhanced Muv penetrance (Figure 4A), suggesting that kinase activity is required for transgenic rescue of the Muv phenotype of cdk-8 null mutants.
Figure 4.
CDK-8 is a LIN-1/Ets corepressor. (A) Adult Muv penetrance in cdk-8; lin-15A mutants expressing kinase dead (KD) cdk-8, compared to nontransgenic cdk-8; lin-15A (n ≥ 41). ***P < 0.001 vs. nontransgenic, Fisher’s exact test. See Table 1 for raw data. (B) Adult Muv penetrance in cdk-8 mutants on empty vector (EV) or lin-1/Ets RNAi (n ≥ 410) ****P < 0.0001 vs. cdk-8 + empty vector (EV), ††††P < 0.0001 vs. N2 + lin-1 RNAi, Fisher’s exact test. See Table S2 for raw data. (C) L4 Muv penetrance in cdk-8; lin-1(n1790) mutants (n ≥ 12). ****P < 0.0001 vs. cdk-8, †P < 0.05 vs. lin-1(n1790), Fisher’s exact test. See Table 1 for raw data. (D) Wild-type expression pattern of arEx1098[lag-2P(min)::GFP] in lin-15A mutants, and ectopic expression in cdk-8; lin-15A mutants. Bracket: Pn.px. Asterisk: ectopic expression. Bar: 10 μm. Number of ectopic expression events over total sample size noted next to genotype.
cdk-8 promotes lin-1/Ets repressor activity
Next, we investigated transcription factors that repress VPC induction, i.e., the ELK1/Ets-family transcription factor LIN-1 and the FoxB/Forkhead-family transcription factor LIN-31 (Miller et al. 1993; Beitel et al. 1995). We hypothesized that the CKM may coregulate LIN-1 and/or LIN-31 and thus tested their genetic interactions with cdk-8. Loss of cdk-8 did not enhance the lin-31 mutant Muv phenotype (Table 1); therefore, we did not investigate lin-31 further. In contrast, cdk-8 mutation strongly enhanced the low Muv penetrance caused by lin-1 RNAi depletion (Figure 4B, Table 1). To corroborate the RNAi experiment, we also studied the lin-1(n1790) mutant, which displays both reduction-of-function (ectopic vulval induction of P3.p, P4.p, and P8.p due to reduced lin-1 mRNA levels) and gain-of-function (reduced vulval induction in P5.p–P7.p due to impaired LIN-1–ERK binding) phenotypes (Jacobs et al. 1998). As seen in lin-1 RNAi worms, loss of cdk-8 significantly enhanced the Muv penetrance of the lin-1(n1790) mutant (Figure 4C, Table 1). Together, these results suggest that residual LIN-1 requires cdk-8 for efficient repression of vulval induction.
To test whether cdk-8 promotes transcriptional repression by LIN-1, we investigated whether the direct LIN-1 target gene lag-2 (Zhang and Greenwald 2011) was derepressed in cdk-8 mutants. In wild-type animals, a lag-2P(min)::YFP minimal promoter reporter (arEx1098) is induced by EGFR signaling in P6.p, whereas in lin-1 null mutants, it is ectopically induced in additional VPCs (Zhang and Greenwald 2011). We again used the lin-15A sensitized background to study cdk-8 requirements for lag-2P(min) repression. In all lin-15A single mutant animals examined, lag-2P(min)::YFP exhibited a wild-type expression pattern, as expected (n = 29; Figure 4D). In contrast, in cdk-8; lin-15A mutants, lag-2P(min)::YFP was occasionally ectopically expressed in VPCs other than P6.p (in 3/41 animals; Figure 4D). Thus, cdk-8 is partially required to repress a direct LIN-1 target gene. Taken together with the requirement for cdk-8 for lin-1 repressor function in vulval induction, these data suggest that CDK-8 may function as a LIN-1 corepressor, perhaps acting redundantly with other corepressors.
The CKM subunit mdt-13/let-19 represses primary vulval fate specification
Having shown that the enzymatic CKM subunit represses EGFR signaling-induced transcription in the C. elegans vulva, we next investigated whether the structural CKM subunits mdt-12/dpy-22 and mdt-13/let-19 (Tsai et al. 2013) have similar molecular functions. C. elegans mdt-12/dpy-22 represses ectopic vulva formation downstream of let-23/EGFR (Moghal and Sternberg 2003a), but whether and how it affects vulval cell fate specification is not understood. Thus, we investigated the requirements and mechanisms of structural CKM subunits in the EGFR signaling pathway, focusing primarily on mdt-13/let-19.
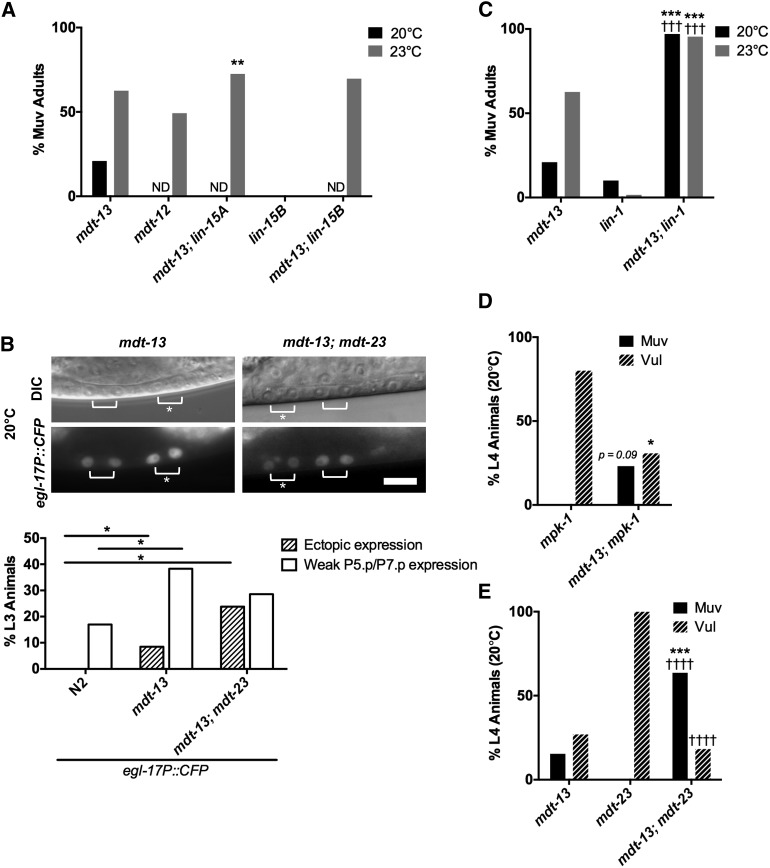
We first investigated the vulval phenotypes of mdt-12/dpy-22 and mdt-13/let-19 mutants. mdt-13/let-19 mutants exhibited a temperature-sensitive Muv phenotype (Figure 5A) of much higher penetrance than cdk-8 mutants (Figure 2B). We observed a similarly high penetrance Muv phenotype in mdt-12/dpy-22 reduction-of-function mutants (Figure 5A). We note that the os38 mutant used in this study shows higher Muv penetrance than the 2–3% Muv penetrance reported for dpy-22(sy622) and dpy-22(sy665) mutants (Moghal and Sternberg 2003a). os38 may cause stronger loss of function than these alleles due to truncation closer to the N terminus and/or the presence of an additional missense mutation (Yoda et al. 2005). Overall, these results demonstrate that mdt-12/dpy-22 and mdt-13/let-19 are more strongly required than cdk-8 and cic-1 to repress vulval induction.
Figure 5.
mdt-13/let-19 represses vulval cell fates downstream of mpk-1/ERK. (A) Adult Muv penetrance in mdt-13/let-19 mutants and mutants with representative synMuv genes (n ≥ 99). **P < 0.01 vs. mdt-13/let-19 single mutant, Fisher’s exact test. ND, not determined. See Table S2 for raw data. (B) Micrographs show mdt-13/let-19 and mdt-13/let-19; mdt-23/sur-2 mutants expressing the arIs92[egl-17P::CFP] 1° fate marker. Bracket: Pn.px cells expressing reporter. Asterisk: ectopic reporter expression. Bar: 10 μm. Graphs show the percentage of animals (n > 20) with ectopic egl-17P::CFP expression (defined as expression in P3.p/P4.p/P8.p, or expression in P5.p/P7.p of equal intensity to P6.p) or weak P5.p/P7.p expression (defined as expression in P5.p/P7.p that is weaker intensity than P6.p). *P < 0.05, Fisher’s exact test. (C) Adult Muv penetrance in mdt-13/let-19; lin-1(n1790) mutants (n ≥ 44). mdt-13/let-19 single mutant Muv penetrance from Figure 5A is included for reference. ***P < 0.001 vs. mdt-13/let-19 single mutant, †††P < 0.001 vs. lin-1(n1790) single mutant, Fisher’s exact test. See Table S2 for raw data. (D) L4 Muv and Vul penetrance in mdt-13/let-19; mpk-1/ERK mutants (n ≥ 13). P = 0.09 (Muv) and *P < 0.05 (Vul) vs. mpk-1 single mutant, Fisher’s exact test. See Table 1 for raw data. (E) L4 Muv and Vul penetrance in mdt-13/let-19; mdt-23/sur-2 mutants (n ≥ 22). ***P < 0.001 vs. mdt-13/let-19 single mutant, ††††P < 0.0001 vs. mdt-23/sur-2 single mutant, Fisher’s exact test. See Table 1 for raw data.
We next tested whether mdt-13/let-19 adopts roles similar to cdk-8 in vulva development. mdt-13/let-19 mutants induced egl-17P::CFP expression in 46/47 P6.px cells examined (Figure 5B). In addition, mdt-13/let-19 mutants displayed increased proportions of P.5px and P7.px weakly expressing egl-17P::CFP compared to wild-type worms (Figure 5B). Furthermore, mdt-13/let-19 mutants displayed strong egl-17P::CFP expression in some P5.px and P7.px cells, or ectopic expression of egl-17P::CFP outside of P5.px–P7.px, suggesting derepression of the 1° fate in these cells (Figure 5B). Overall, these data indicated that the Muv phenotype of mdt-13/let-19 mutants was due in part to derepressed ERK signaling output.
In support of a role similar to cdk-8 in regulation of vulval cell fate specification, mdt-13/let-19 interacted genetically with the synMuv genes and with lin-1/Ets. Specifically, mdt-13/let-19 mutants interacted genetically with the class A synMuv transcriptional repressor lin-15A, and the class B synMuv gene lin-15B(n374) showed a similar trend (Figure 5A). In addition, loss of mdt-13/let-19 significantly enhanced the Muv penetrance of lin-1(n1790) mutant worms (Figure 5C, Table 1), in line with a role for MDT-13/LET-19 as a LIN-1 corepressor. Thus, like cdk-8, mdt-13/let-19 interacts genetically with transcriptional regulators in the EGFR signaling pathway.
Finally, we investigated the genetic position of mdt-13/let-19 in the EGFR-Ras-ERK pathway. Similarly to cdk-8, loss of mdt-13/let-19 suppressed the mpk-1 loss-of-function Vul phenotype (Figure 5D, Table 1). Furthermore, the mdt-13/let-19 mutant Muv phenotype appeared epistatic to the mpk-1 mutant Vul phenotype, although this trend was not statistically significant (Figure 5D, Table 1). Together, these data support a model in which the CKM acts downstream of mpk-1/ERK to inhibit vulval cell fate specification.
The CKM restricts the specificity of core Mediator subunits
As several Mediator subunits positively regulate vulva development (reviewed in Grants et al. 2015), we hypothesized that the CKM might function in part by inhibiting these subunits. In wild-type worms, the mdt-23/sur-2 tail module subunit is critical for vulva development downstream of Ras (Singh and Han 1995), and it is required for activation of the EGFR signaling target gene lag-2 (Zhang and Greenwald 2011); therefore, we tested how a mutation in a CKM subunit affected vulval induction in mdt-23/sur-2 mutants. All mdt-23/sur-2 single mutants examined exhibited a Vul phenotype (Figure 5E, Table 1). VPC induction analysis of mdt-13/let-19 single mutants demonstrated that, in addition to the Muv phenotype, some animals also displayed decreased proliferation of P5.p or P7.p (mild Vul; Figure 5E, Table 1). Unexpectedly, mdt-13/let-19; mdt-23/sur-2 mutants exhibited a significantly stronger Muv penetrance than mdt-13/let-19 single mutants, and loss of mdt-13/let-19 function significantly suppressed the Vul phenotype of mdt-23/sur-2 single mutants (Figure 5E, Table 1), suggesting that loss of mdt-13/let-19 derepresses vulval induction independently of mdt-23/sur-2. As seen in mdt-13/let-19 single mutants, mdt-13/let-19; mdt-23/sur-2 double mutants showed strong egl-17P::CFP expression in some P5.px and P7.px cells, or ectopic expression of egl-17P::CFP outside of P5.px–P7.px, suggesting derepression of the 1° fate (Figure 5B). Overall, these data indicated that the Muv phenotype of mdt-13/let-19; mdt-23/sur-2 double mutants was due in part to derepressed ERK signaling output. Together, these findings indicate that loss of the CKM allows activation of EGFR signaling-driven cell fate specification independently of mdt-23/sur-2 activity.
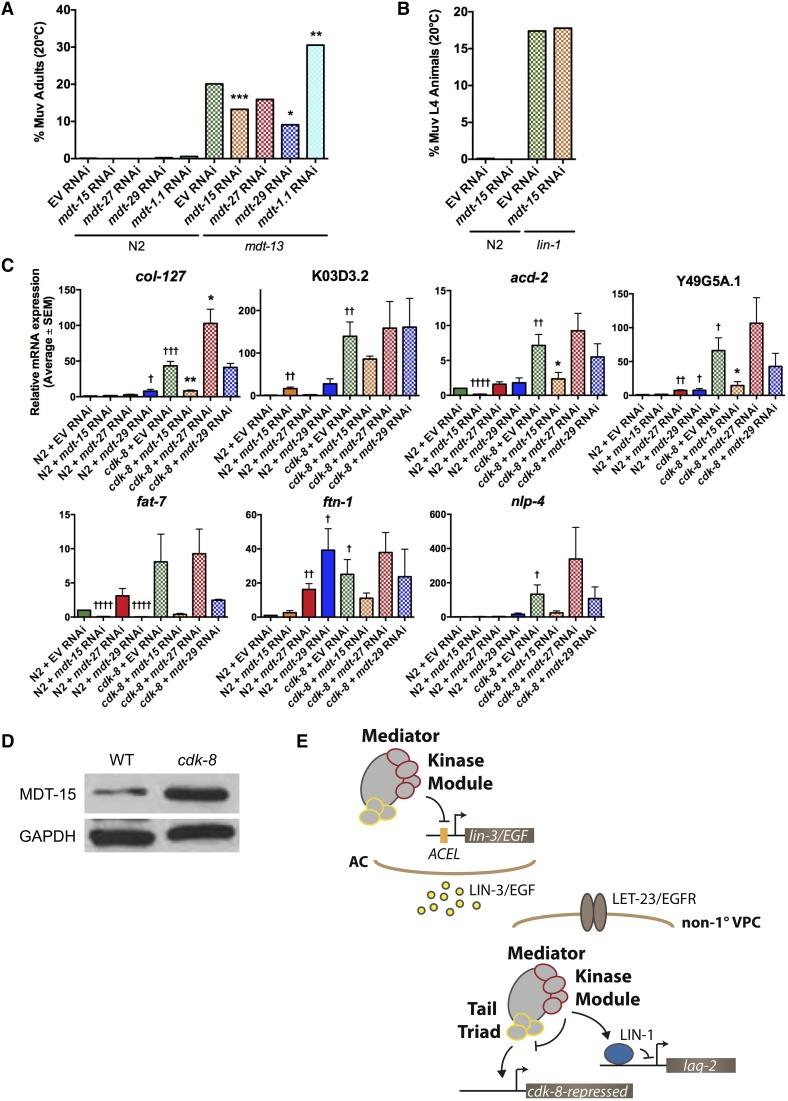
Our results suggested that the CKM might influence Mediator subunit(s) other than mdt-23/sur-2. As S. cerevisiae CDK8 inhibits the Mediator tail module triad composed of MED2, MED3, and MED15 (van de Peppel et al. 2005; Gonzalez et al. 2014), we hypothesized that their putative C. elegans orthologs MDT-29, MDT-27, and MDT-15 (Bourbon 2008), might be targets for CKM inhibition. mdt-15 and mdt-29 knockdown had no effect on vulva formation in wild-type animals (i.e., causing neither Muv nor Vul phenotypes and displaying normal VPC induction; Table 1), but significantly reduced the Muv penetrance of mdt-13/let-19 mutants (Figure 6A); mdt-27 RNAi caused a similar trend (Figure 6A). This effect was specific to the tail module triad, as knockdown of the mdt-1.1 tail module subunit in fact increased the Muv penetrance of mdt-13/let-19 mutants (Figure 6A). Furthermore, requirement for the tail module triad in ectopic vulva formation appeared to be specific to CKM mutants, as mdt-15 RNAi had no effect on ectopic vulva formation in lin-1(n1790) mutants (Figure 6B, Table 1). Thus, tail module triad activity appears to be derepressed in a CKM mutant, causing aberrant activation of vulval fate specification.
Figure 6.
The CKM inhibits activation of vulval induction by the mdt-15, mdt-27, mdt-29 triad. Adult Muv penetrance in mdt-13/let-19 mutants on mdt-15, mdt-27, or mdt-29 triad subunit RNAi or mdt-1.1 nontriad subunit RNAi (n ≥ 198). *P < 0.05, **P < 0.01, *** P < 0.001 vs. mdt-13/let-19 + empty vector (EV), Fisher’s exact test. See Table S2 for raw data. (B) L4 Muv penetrance in lin-1(n1790) mutants on mdt-15 RNAi (n ≥ 45). No statistically significant difference, Fisher’s exact test. See Table 1 for raw data. (C) qPCR analysis of mRNA levels of cdk-8-repressed genes upon mdt-15, mdt-27, or mdt-29 RNAi. Error bars represent SEM, n = 3 independent trials. *P < 0.05, **P < 0.01 vs. cdk-8 + empty vector (EV); †P < 0.05, ††P < 0.01, †††P < 0.001, ††††P < 0.0001 vs. N2 + empty vector (EV), Fisher’s exact test. (D) α-MDT-15 Western blot in wild-type vs. cdk-8 mutants, with α-GAPDH loading control. Representative immunoblot from one of three trials. (E) Model of CKM inhibition of EGFR-Ras-MAPK signaling-dependent cell fate specification by repressing lin-3/EGF in the anchor cell (AC), promoting LIN-1 repressor activity (e.g., at lag-2), and inhibiting tail module triad activity (e.g., at cdk-8-repressed genes) in non-1° VPCs (i.e., VPCs other than P6.p).
Next, we investigated whether the CKM modifies the target gene specificity of the triad. We used qPCR to quantify the expression of cdk-8-repressed genes identified by our microarray analysis (Table S1) in wild-type worms and cdk-8 mutants treated with empty vector (EV), mdt-15, mdt-27, or mdt-29 RNAi. On EV RNAi, seven of nine genes tested were upregulated in cdk-8 mutants compared to wild-type worms, as expected (Figure 6C). Upregulation of these cdk-8-repressed genes was strongly attenuated by mdt-15 depletion, whereas mdt-29 knockdown only affected fat-7, and mdt-27 depletion caused no significant changes (Figure 6C). Thus, for the genes investigated, induction in cdk-8 mutants appears to specifically require mdt-15, but not the other predicted tail module triad subunits. However, we cannot rule out unequal RNAi efficiency accounting for these differing requirements, although RNAi knockdown of all three genes appeared successful, as we observed partial sterility (not shown) consistent with the essential nature of these core Mediator subunits (Fernandez et al. 2005; Sönnichsen et al. 2005; Taubert et al. 2006). Notably, only two cdk-8-repressed genes, acd-2 and fat-7, displayed mdt-15 and/or mdt-29-dependent activation in wild-type worms (Figure 6C). Thus, as seen in the genetic analysis of vulva induction, loss of cdk-8 appears to cause unrestrained tail module activity, i.e., mdt-15 activates novel target genes when cdk-8 is deleted.
Finally, we investigated the molecular cause of unrestrained mdt-15 activity in CKM mutants. Loss of cdk-8 did not alter mRNA levels of any triad subunits (Figure S3). Western blot analysis showed elevated MDT-15 protein levels in cdk-8 mutants compared to wild type (Figure 6D). Taken together, these results demonstrate that cdk-8 is required for post-transcriptional regulation of MDT-15.
Discussion
EGFR signaling is critical for cell proliferation and cell fate determination in animal development. Several Mediator subunits positively or negatively regulate EGFR signaling-driven developmental processes (reviewed in Grants et al. 2015), but pertinent mechanisms remain incompletely understood. Here, we used the well-characterized vulva development paradigm in C. elegans to delineate the role of the Mediator CKM module. Our results suggest a model whereby the CKM acts within the vulval precursor cells, in a kinase-dependent manner, to fine tune EGFR transcriptional output by modulating two transcriptional regulators: the key downstream transcription factor LIN-1/Ets and core Mediator (Figure 6E). This model is based on four key observations: First, we demonstrate that the primary site of action for the CKM is in the VPCs, as cdk-8 and mdt-13/let-19 repress vulva formation downstream of mpk-1/ERK, a key component of the EGFR signaling cascade inside VPCs, and cdk-8 expression in VPCs is sufficient for this repression. Second, cdk-8 repression of ectopic vulva formation is kinase dependent. Third, the CKM appears to act as a corepressor of the Ets-family transcription factor LIN-1, as loss of cdk-8 or mdt-13/let-19 enhances the ectopic vulval induction caused by lin-1 reduction of function, and cdk-8 is required for full repression of a direct LIN-1 target promoter. Fourth, ectopic vulva formation in mdt-13/let-19 is independent of the Mediator subunit mdt-23/sur-2, which is critical for EGFR signaling-driven transcription and vulval development in wild-type worms (Singh and Han 1995; Zhang and Greenwald 2011); instead, mdt-13/let-19 modulates the specificity of the tail module triad subunits mdt-15, mdt-29, and mdt-27, preventing aberrant activation of downstream transcription. By implicating all CKM subunits and by connecting the CKM to lin-1 and to core Mediator, our data substantially expand on the prior finding that loss of CKM subunit mdt-12 caused ectopic vulva formation by unknown molecular mechanisms (Moghal and Sternberg 2003a). Additionally, our genetic and molecular analysis provide first evidence that CKM-tail module crosstalk, akin to that seen in yeast Mediator (van de Peppel et al. 2005; Gonzalez et al. 2014), occurs in metazoan Mediator, an important experimental finding as tail module subunit sequence conservation between species is extremely poor (Bourbon 2008).
The CKM inhibits vulva development in a kinase-dependent manner
We performed unbiased gene expression profiling to define gene programs that depend on cdk-8 in vivo, which revealed that only 6.7% of C. elegans genes are regulated by cdk-8 (Figure 2A, Table S1). This number F agrees with studies in yeast, wherein CDK8 regulates only 3% of genes (Holstege et al. 1998). Thus, CDK8 appears to be a gene program-specific transcriptional coregulator across species.
Among cdk-8-dependent genes, we identified a significant overlap with genes regulated by lin-35/RB, a synMuv transcriptional repressor (Figure 2A) (Kirienko and Fay 2007). We note that as synMuv genes act redundantly, lin-35 single mutants do not exhibit any defects in EGFR signaling or vulval induction (Myers and Greenwald 2005; Cui et al. 2006; Kirienko and Fay 2007). Thus, the overlap between cdk-8- and lin-35-dependent genes suggested that the CKM and lin-35 cooperate in multiple aspects of C. elegans development. Similarly, in Drosophila, CDK8 and RB act in parallel in the Wnt signaling pathway (Morris et al. 2008). Therefore, we explored whether the CKM and synMuv genes cooperate in the EGFR signaling pathway to regulate C. elegans vulva development. Both cdk-8 and mdt-13/let-19 were required to repress C. elegans vulva formation, in a partially redundant manner with the synMuv genes (Figure 2B and Figure 5A). However, cdk-8 did not act redundantly with the synMuv genes to repress lin-3/EGF transcription (Figure 3A), suggesting that the CKM and the synMuv genes regulate EGFR signaling at different junctions, as discussed below.
Comparing the vulval phenotypes of CKM mutants, we found evidence that cdk-8 and cic-1 act redundantly to repress vulval induction, as a cdk-8; cic-1 double mutant displayed the same Muv penetrance as cdk-8 or cic-1 single mutants (Figure 2B). In addition, we found that mdt-12/dpy-22 and mdt-13/let-19 were more strongly required to repress vulva development than cdk-8 or cic-1. In S. cerevisiae Mediator, MED12 and MED13 enable CDK8 and cyclin C docking to Mediator (Tsai et al. 2013). Loss of MDT-12/DPY-22 or -13 in C. elegans may similarly disrupt CDK-8 and CIC-1 function, as well as considerably reducing the size of the CKM. Although CDK-8’s kinase activity is required to inhibit vulva development (Figure 4A), this does not rule out the possibility that the CKM also employs kinase-independent steric mechanisms, as observed in other systems (Knuesel et al. 2009). Thus, additional kinase-independent mechanisms could account for the stronger requirement for mdt-12/dpy-22 and -13 in vulva development.
The CKM inhibits the primary vulval cell fate
Vulva formation in C. elegans requires both EGFR and Notch signaling (Félix and Barkoulas 2012), and human CDK8 represses Notch signaling-driven transcription by promoting turnover of the Notch intracellular domain (Fryer et al. 2004). Therefore, we examined whether the vulval phenotypes in CKM mutants occur due to defects in EGFR signaling, Notch signaling, or both. Using an EGFR-Ras-ERK signaling-induced 1° cell fate reporter, we demonstrated that cdk-8 and mdt-13/let-19 are required to repress ectopic vulva formation in part by repressing the 1° cell fate (Figure 2C and Figure 5B). Using a Notch signaling-induced 2° cell fate reporter, we showed that cdk-8 is required to represses ectopic 2° fates in nonvulval VPCs, as well as to promote the 2° fate in P5.p and P7.p (Figure 2D). However, cdk-8 action in the Notch pathway might occur indirectly in this context. EGFR signaling in P6.p induces expression of Notch ligands, e.g., lag-2, which promote the 2° fate in the neighboring cells, P5.p and P7.p (Chen and Greenwald 2004). We observed evidence of possible cell fate transformations from 2° to 1° in P5.p or P7.p in cdk-8 and mdt-13 mutants, as these cells occasionally exhibited strong expression of the 1° cell fate marker egl-17P::CFP (Figure 2C and Figure 5B), expression of the EGFR signaling target gene lag-2 (Figure 4D) or loss of the strong lip-1P::GFP expression characteristic of 2° cells (Figure 2D). It is possible that VPCs transformed to the 1° fate could then induce 2° fates in neighboring VPCs, accounting for our observation of ectopic 2° cells.
CKM subunits have been implicated as regulators of canonical Wnt signaling (Zhang and Emmons 2000; Firestein et al. 2008; Morris et al. 2008) and cell cycle quiescence (Clayton et al. 2008), processes which also contribute to vulva development. Activation of Wnt signaling can bypass requirements for let-23/EGFR in vulva development (Gleason et al. 2002). However, the Muv phenotype of mdt-12/dpy-22 mutants is independent of bar-1/β-catenin (Moghal and Sternberg 2003b), suggesting that the CKM does not repress vulva development through the canonical Wnt signaling pathway. Deregulation of cell cycle quiescence can expand the VPC equivalence group, which are competent to form ectopic vulvae if presented with the appropriate signals [e.g., lin-12/Notch gain of function employed by Clayton et al. (2008)]. Although CKM subunits are required for VPC cell cycle quiescence (Clayton et al. 2008), this alone is unlikely to account for the ectopic vulvae observed in these animals. First, the ectopic vulval invaginations observed in cdk-8 and mdt-13/let-19 animals while scoring VPC induction (Table 1) were positioned in the correct location for P3.p, P4.p, and P8.p descendants. Second, ectopic expression of 1° and 2° cell fate markers in cdk-8 and mdt-13/let-19 mutants (Figure 2, C and D, and Figure 5B) suggests that EGFR and/or Notch signaling indeed drives ectopic vulva formation in these mutants.
The CKM promotes LIN-1/Ets repressor activity
We observed derepression of the lin-3/EGF ACEL in cdk-8 mutants (Figure 3C), implicating cdk-8 as a novel repressor of lin-3/EGF transcription in the anchor cell. Albeit interesting, genetic epistasis analysis with let-23/EGFR and mpk-1/ERK loss-of-function alleles clearly demonstrated that cdk-8 is primarily required downstream of mpk-1/ERK to repress vulval induction (Figure 3D). The let-23(sy97) mutant protein is ligand insensitive (Aroian and Sternberg 1991; Aroian et al. 1994); therefore, weak lin-3/EGF activation in the anchor cell due to loss of cdk-8 cannot account for the vulval phenotypes observed in cdk-8; let-23(sy97) mutants (Figure 3D). Furthermore, epistasis analysis with mpk-1/ERK confirmed that cdk-8 acts downstream of the core EGFR-Ras-ERK pathway to regulate vulval induction (Figure 3D). In line with a position downstream of mpk-1/ERK, we showed that cdk-8 is required in VPCs to suppress ectopic vulval induction (Figure 3E). A previous report demonstrated that repression of vulval induction by the CKM subunit mdt-12/dpy-22 is gonad independent, and thus anchor cell independent, and that an MDT-12/DPY-22::GFP transgene is expressed in VPCs (Moghal and Sternberg 2003a), supporting a role for the CKM in VPCs.
Downstream of mpk-1/ERK, we found evidence that the CKM promotes LIN-1/Ets-mediated repression of vulval induction (Figure 4, B and C, and Figure 5C), and that cdk-8 promotes transcriptional repression of a direct LIN-1 target, the lag-2/Notch ligand minimal promoter (Figure 4D). The lag-2 minimal promoter contains activator and repressor elements, VPCact and VPCrep, that cooperatively restrict expression to P6.p (Zhang and Greenwald 2011). On its own, VPCact is sufficient to drive transcription in all VPCs (P3.p–P8.p) in a LIN-3/EGF ligand-independent manner. VPCrep represses this basal VPCact-driven transcription in VPCs other than P6.p, thereby restricting expression of the lag-2 minimal promoter to the 1°-fated VPC. VPCrep contains an Elk1 consensus site, which is bound by LIN-1 in vitro (Miley et al. 2004), and requires lin-1/Ets for repression of transcription in VPCs other than P6.p (Zhang and Greenwald 2011). Our results indicate that cdk-8 is partially required for transcriptional repression of the lag-2 minimal promoter (Figure 4D), suggesting that the CKM promotes LIN-1-mediated repression at VPCrep. An alternative explanation for the ectopic expression of the lag-2 minimal promoter observed in cdk-8 mutants is that the CKM might inhibit a factor that activates transcription through VPCact. The transcription factor(s) that acts at VPCact remains poorly defined; however, the mdt-23/sur-2 Mediator subunit is required for VPCact-driven transcription in P3.p–P8.p (Zhang and Greenwald 2011). As we demonstrated that vulval induction in mdt-13/let-19 mutants does not require mdt-23/sur-2 (Figure 5, B and E), this implies that the CKM likely does not inhibit MDT-23/SUR-2 activity at VPCact. Overall, our findings suggest that the CKM may act as a corepressor for LIN-1.
In murine embryonic stem cells, Mediator recruitment is important for transcriptional activation by Ets factors, e.g., Elk1 (Stevens et al. 2002; Balamotis et al. 2009). In this context, activation of Elk1 by ERK phosphorylation promotes binding to Mediator in a MED23/Sur2-dependent manner (Stevens et al. 2002). Similarly, in a colon cancer cell line, CDK8 promotes transcriptional elongation of serum response immediate early genes, which are targeted by multiple transcription factors including Elk1 (Donner et al. 2010). However, the role of Mediator in transcriptional repression by an Ets factor has not previously been explored. In the absence of ERK phosphorylation, Ets factors, e.g., LIN-1, can promote transcriptional repression of target genes (Jacobs et al. 1998; Zhang and Greenwald 2011). Although the Sin3A-HDAC-1 corepressor complex has been implicated in an epigenetic mechanism that attenuates transcriptional activation by ERK-phosphorylated Elk1(Yang et al. 2001), to our knowledge, corepressors of Ets factor-mediated transcriptional repression have not previously been identified. This report provides evidence that the Mediator CKM is required for repression by Ets factors, representing an advance in our understanding of Ets factor repressive mechanisms.
Our findings are also of potential clinical interest, as the human CKM is implicated in tumorigenesis (Firestein et al. 2008; Donner et al. 2010; Mäkinen et al. 2011; Lim et al. 2014). Loss of MED12 causes cellular resistance to chemotherapeutic agents that inhibit activated BRAF, the human ERK kinase kinase (Shalem et al. 2014); this suggests that MED12 represses EGFR signaling downstream of BRAF, in line with our findings for the C. elegans CKM. Furthermore, recurrent MED12 mutations are implicated in uterine leiomyomas and breast fibroadenomas (Mäkinen et al. 2011; Lim et al. 2014; Mittal et al. 2015), but the pathogenic mechanisms of these mutations have not been fully elucidated. Investigation of these mutations using the C. elegans vulva development paradigm may provide insight into their mode of action.
The CKM restrains the core Mediator tail module triad
Epistatic relationships between Mediator subunits have been identified in S. cerevisiae (van de Peppel et al. 2005; Gonzalez et al. 2014), but intra-Mediator regulation has not been demonstrated in metazoans. Previous studies (reviewed in Grants et al. 2015) and our data show that several core Mediator subunits promote C. elegans vulva development, whereas CKM subunits inhibit this process. This suggested that intra-Mediator regulation might coordinate gene expression downstream of the EGFR-Ras-ERK signaling pathway that drives vulva development. Initially, we hypothesized that the CKM may oppose mdt-23/sur-2-mediated activation of EGFR signaling, as mdt-23/sur-2 is required for vulval induction and activation of EGFR signaling-induced transcription, e.g., lag-2 (Singh and Han 1995; Zhang and Greenwald 2011). Unexpectedly, loss of mdt-13/let-19 circumvented the requirement for mdt-23/sur-2 in vulval induction (Figure 5E). We therefore explored regulatory interactions between the CKM and the metazoan orthologs of S. cerevisiae MED2, MED3, and MED15, which are subject to inhibitory post-translational regulation by CDK8 (van de Peppel et al. 2005; Gonzalez et al. 2014). Sequence conservation is weak between yeast MED2 and MED3 and their putative metazoan homologs, MED29 and MED27, respectively (Bourbon 2008). Whether MED29 and MED27 function as part of the tail module remains unclear, as structural and biochemical studies locate these subunits between the head and tail modules (Sato et al. 2003; Tsai et al. 2014). Thus, we were intrigued to find that vulva formation in a C. elegans CKM mutant required mdt-15, mdt-27, and mdt-29 (Figure 6A). This requirement appeared specific to the tail module triad, as neither mdt-1.1/MED1 nor mdt-23/sur-2 was required for vulval induction in CKM mutants (Figure 5E and Figure 6A). Furthermore, the triad did not appear to be generally required for ectopic vulval induction in animals with a wild-type CKM, as mdt-15 knockdown had no effect on ectopic vulval induction in lin-1(n1790) mutants (Figure 6B). Together, these findings suggest that the CKM restrains triad activity, preventing it from aberrantly activating vulval induction.
Gene expression analysis in cdk-8 mutants identified a requirement for mdt-15, but little or no requirement for mdt-27 or mdt-29, in transcriptional activation of cdk-8-repressed genes (Figure 6C). In the S. cerevisiae tail module triad, both MED3 and MED2 are required for overexpression of CDK8-repressed genes in CDK8 mutants, but the requirement for MED15 has not been tested directly (van de Peppel et al. 2005; Gonzalez et al. 2014). These requirements might be explained by the fact that both MED2 and MED3 are necessary to anchor the triad to the tail module (Myers et al. 1999; van de Peppel et al. 2005; Gonzalez et al. 2014). Similar requirements may not exist in metazoan Mediator, as human Mediator displays more extensive structural contacts between the head and tail modules (Tsai et al. 2014), which may result in redundancy for some tail module subunits.
Investigating the regulatory relationship between CDK-8 and MDT-15 further, we found that cdk-8 is required for post-transcriptional negative regulation of MDT-15, as MDT-15 protein but not mRNA levels increase in cdk-8 mutants (Figure 6D). This regulatory relationship resembles that seen in yeast where the three triad subunits MED2, MED3, and MED15 are negatively regulated post-translationally by CDK8-driven phosphorylation of MED3, promoting ubiquitin-proteasome-dependent turnover of all three triad subunits (Gonzalez et al. 2014). It will be interesting to delineate whether the metazoan CKM regulates MDT-15 protein levels directly, e.g., by phosphorylation leading to ubiquitin-mediated degradation, or indirectly through action upon other Mediator subunits.
In summary, our findings suggest that the Mediator CKM represses EGFR-Ras-ERK signaling-driven cell fate specification in C. elegans by regulating repressor activity of an Ets-family transcription factor and by promoting specificity of Mediator tail module subunits.
Acknowledgments
We thank all Taubert lab members for critical discussions; Shirley Chen for help with experiments; R. Barbeau, C. Eisley, A. Barczak, and D. Erle from the Sandler Asthma Basic Research Center Functional Genomics Core Facility (University of California San Francisco) for help with microarray gene expression profiling; J. Ewbank for EASE analysis of gene expression arrays; J. Escobar for advice on scoring VPC induction; I. Greenwald for GS5096 arEx1098[lag-2p(min)::YFP]; S. Mitani for cdk-8(tm1238) and cic-1(tm3740) mutants; and D. Moerman and J. Ward for comments on the manuscript. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported by grants from Canadian Institute of Health Research (MOP-93713), Natural Sciences and Engineering Research Council of Canada (RGPIN 386398-13), Canada Foundation for Innovation (all to S.T.), and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to H.S.). J.M.G. was supported by Vanier Canada Graduate Scholarship, Natural Sciences and Engineering Research Council of Canada Canada Graduate Scholarship - Masters, Child and Family Research Institute, and University of British Columbia scholarships, and S.T. by a Canada Research Chair. The authors declare no competing financial interests.
J.M.G., H.S., and S.T. conceived and designed experiments; J.M.G., L.T.L.Y., A.Y., C.C.Y., H.S., and S.T. performed experiments; J.M.G., A.Y., H.O., H.S., and S.T. analyzed data; J.M.G., A.Y., H.O., and H.S. contributed reagents/materials/analysis tools; and J.M.G., H.S., and S.T. wrote the paper.
Footnotes
Communicating editor: D. Greenstein
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.180265/-/DC1.
Literature Cited
- Aroian R. V., Sternberg P. W., 1991. Multiple functions of let-23, a Caenorhabditis elegans receptor tyrosine kinase gene required for vulval induction. Genetics 128: 251–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroian R. V., Koga M., Mendel J. E., Ohshima Y., Sternberg P. W., 1990. The let-23 gene necessary for Caenorhabditis elegans vulval induction encodes a tyrosine kinase of the EGF receptor subfamily. Nature 348: 693–699. [DOI] [PubMed] [Google Scholar]
- Aroian R. V., Lesa G. M., Sternberg P. W., 1994. Mutations in the Caenorhabditis elegans let-23 EGFR-like gene define elements important for cell-type specificity and function. EMBO J. 13: 360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamotis M. A., Pennella M. A., Stevens J. L., Wasylyk B., Belmont A. S., et al. , 2009. Complexity in transcription control at the activation domain-mediator interface. Sci. Signal. 2: ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga J., Swain S. M., 2009. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat. Rev. Cancer 9: 463–475. [DOI] [PubMed] [Google Scholar]
- Beitel G. J., Tuck S., Greenwald I., Horvitz H. R., 1995. The Caenorhabditis elegans gene lin-1 encodes an ETS-domain protein and defines a branch of the vulval induction pathway. Genes Dev. 9: 3149–3162. [DOI] [PubMed] [Google Scholar]
- Berset T., Hoier E. F., Battu G., Canevascini S., Hajnal A., 2001. Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science 291: 1055–1058. [DOI] [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P., 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193. [DOI] [PubMed] [Google Scholar]
- Bourbon H.-M., 2008. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36: 3993–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon H.-M., Aguilera A., Ansari A. Z., Asturias F. J., Berk A. J., et al. , 2004. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14: 553–557. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Greenwald I., 2004. The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev. Cell 6: 183–192. [DOI] [PubMed] [Google Scholar]
- Clayton J. E., van den Heuvel S. J. L., Saito R. M., 2008. Transcriptional control of cell-cycle quiescence during C. elegans development. Dev. Biol. 313: 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Chen J., Myers T. R., Hwang B. J., Sternberg P. W., et al. , 2006. SynMuv genes redundantly inhibit lin-3/EGF expression to prevent inappropriate vulval induction in C. elegans. Dev. Cell 10: 667–672. [DOI] [PubMed] [Google Scholar]
- Donner A. J., Szostek S., Hoover J. M., Espinosa J. M., 2007. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol. Cell 27: 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner A. J., Ebmeier C. C., Taatjes D. J., Espinosa J. M., 2010. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 17: 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann I., Griffon A., Tichit L., Montañana-Sanchis F., Wang G., et al. , 2011. A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans. PLoS One 6: e19055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay D. S., Yochem J., 2007. The SynMuv genes of Caenorhabditis elegans in vulval development and beyond. Dev. Biol. 306: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M.-A., Barkoulas M., 2012. Robustness and flexibility in nematode vulva development. Trends Genet. TIG 28: 185–195. [DOI] [PubMed] [Google Scholar]
- Fernandez A. G., Gunsalus K. C., Huang J., Chuang L.-S., Ying N., et al. , 2005. New genes with roles in the C. elegans embryo revealed using RNAi of ovary-enriched ORFeome clones. Genome Res. 15: 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein R., Bass A. J., Kim S. Y., Dunn I. F., Silver S. J., et al. , 2008. CDK8 is a colorectal cancer oncogene that regulates [bgr]-catenin activity. Nature 455: 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer C. J., White J. B., Jones K. A., 2004. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 16: 509–520. [DOI] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., et al. , 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard J. S., Barry J. D., Johnstone I. L., 1997. cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Mol. Cell. Biol. 17: 2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason J. E., Korswagen H. C., Eisenmann D. M., 2002. Activation of Wnt signaling bypasses the requirement for RTK/Ras signaling during C. elegans vulval induction. Genes Dev. 16: 1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh G. Y. S., Martelli K. L., Parhar K. S., Kwong A. W. L., Wong M. A., et al. , 2014. The conserved Mediator subunit MDT-15 is required for oxidative stress responses in Caenorhabditis elegans. Aging Cell 13: 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold M. O., Rice A. P., 1998. Targeting of CDK8 to a promoter-proximal RNA element demonstrates catalysis-dependent activation of gene expression. Nucleic Acids Res. 26: 3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D., Hamidi N., Del Sol R., Benschop J. J., Nancy T., et al. , 2014. Suppression of Mediator is regulated by Cdk8-dependent Grr1 turnover of the Med3 coactivator. Proc. Natl. Acad. Sci. USA 111: 2500–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grants J. M., Goh G. Y. S., Taubert S., 2015. The Mediator complex of Caenorhabditis elegans: insights into the developmental and physiological roles of a conserved transcriptional coregulator. Nucleic Acids Res. 43: 2442–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Aroian R. V., Sternberg P. W., 1990. The let-60 locus controls the switch between vulval and nonvulval cell fates in Caenorhabditis elegans. Genetics 126: 899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill R. J., Sternberg P. W., 1992. The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature 358: 470–476. [DOI] [PubMed] [Google Scholar]
- Holm S., 1979. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6: 65–70. [Google Scholar]
- Holstege F. C., Jennings E. G., Wyrick J. J., Lee T. I., Hengartner C. J., et al. , 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728. [DOI] [PubMed] [Google Scholar]
- Horvitz H. R., Sulston J. E., 1980. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96: 435–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack D. A., Dennis G., Jr, Sherman B. T., Lane H. C., Lempicki R. A., 2003. Identifying biological themes within lists of genes with EASE. Genome Biol. 4: R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B. J., Sternberg P. W., 2004. A cell-specific enhancer that specifies lin-3 expression in the C. elegans anchor cell for vulval development. Development 131: 143–151. [DOI] [PubMed] [Google Scholar]
- Jacobs D., Beitel G. J., Clark S. G., Horvitz H. R., Kornfeld K., 1998. Gain-of-function mutations in the Caenorhabditis elegans lin-1 ETS gene identify a C-terminal regulatory domain phosphorylated by ERK MAP kinase. Genetics 149: 1809–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz W. S., Hill R. J., Clandinin T. R., Sternberg P. W., 1995. Different levels of the C. elegans growth factor LIN-3 promote distinct vulval precursor fates. Cell 82: 297–307. [DOI] [PubMed] [Google Scholar]
- Kirienko N. V., Fay D. S., 2007. Transcriptome profiling of the C. elegans Rb ortholog reveals diverse developmental roles. Dev. Biol. 305: 674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore R. S., Sundaram M. V., 2002. ced-10 Rac and mig-2 function redundantly and act with unc-73 trio to control the orientation of vulval cell divisions and migrations in Caenorhabditis elegans. Dev. Biol. 241: 339–348. [DOI] [PubMed] [Google Scholar]
- Knuesel M. T., Meyer K. D., Bernecky C., Taatjes D. J., 2009. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 23: 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J. Y., Lee J., 2001. Biological significance of a universally conserved transcription mediator in metazoan developmental signaling pathways. Development 128: 3095–3104. [DOI] [PubMed] [Google Scholar]
- Liao S. M., Zhang J., Jeffery D. A., Koleske A. J., Thompson C. M., et al. , 1995. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 374: 193–196. [DOI] [PubMed] [Google Scholar]
- Lim W. K., Ong C. K., Tan J., Thike A. A., Ng C. C. Y., et al. , 2014. Exome sequencing identifies highly recurrent MED12 somatic mutations in breast fibroadenoma. Nat. Genet. 46: 877–880. [DOI] [PubMed] [Google Scholar]
- Loncle N., Boube M., Joulia L., Boschiero C., Werner M., et al. , 2007. Distinct roles for Mediator Cdk8 module subunits in Drosophila development. EMBO J. 26: 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkinen N., Mehine M., Tolvanen J., Kaasinen E., Li Y., et al. , 2011. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 334: 252–255. [DOI] [PubMed] [Google Scholar]
- Malik S., Roeder R. G., 2010. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miley G. R., Fantz D., Glossip D., Lu X., Saito R. M., et al. , 2004. Identification of residues of the Caenorhabditis elegans LIN-1 ETS domain that are necessary for DNA binding and regulation of vulval cell fates. Genetics 167: 1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. M., Gallegos M. E., Morisseau B. A., Kim S. K., 1993. lin-31, a Caenorhabditis elegans HNF-3/fork head transcription factor homolog, specifies three alternative cell fates in vulval development. Genes Dev. 7: 933–947. [DOI] [PubMed] [Google Scholar]
- Mittal P., Shin Y.-H., Yatsenko S. A., Castro C. A., Surti U., et al. , 2015. Med12 gain-of-function mutation causes leiomyomas and genomic instability. J. Clin. Invest. 125: 3280–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghal N., Sternberg P. W., 2003a A component of the transcriptional mediator complex inhibits RAS-dependent vulval fate specification in C. elegans. Development 130: 57–69. [DOI] [PubMed] [Google Scholar]
- Moghal N., Sternberg P. W., 2003b The epidermal growth factor system in Caenorhabditis elegans. Exp. Cell Res. 284: 150–159. [DOI] [PubMed] [Google Scholar]
- Morris E. J., Ji J.-Y., Yang F., Di Stefano L., Herr A., et al. , 2008. E2F1 represses [bgr]-catenin transcription and is antagonized by both pRB and CDK8. Nature 455: 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers L. C., Gustafsson C. M., Hayashibara K. C., Brown P. O., Kornberg R. D., 1999. Mediator protein mutations that selectively abolish activated transcription. Proc. Natl. Acad. Sci. USA 96: 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers T. R., Greenwald I., 2005. lin-35 Rb acts in the major hypodermis to oppose ras-mediated vulval induction in C. elegans. Dev. Cell 8: 117–123. [DOI] [PubMed] [Google Scholar]
- Nemet J., Jelicic B., Rubelj I., Sopta M., 2014. The two faces of Cdk8, a positive/negative regulator of transcription. Biochimie 97: 22–27. [DOI] [PubMed] [Google Scholar]
- Normanno N., De Luca A., Bianco C., Strizzi L., Mancino M., et al. , 2006. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 366: 2–16. [DOI] [PubMed] [Google Scholar]
- Poss Z. C., Ebmeier C. C., Taatjes D. J., 2013. The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 48: 575–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffer A. M., Kim D. H., van Oudenaarden A., Horvitz H. R., 2011. The Caenorhabditis elegans synthetic multivulva genes prevent ras pathway activation by tightly repressing global ectopic expression of lin-3 EGF. PLoS Genet. 7: e1002418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Tomomori-Sato C., Banks C. A. S., Parmely T. J., Sorokina I., et al. , 2003. A mammalian homolog of Drosophila melanogaster transcriptional coactivator intersex is a subunit of the mammalian Mediator complex. J. Biol. Chem. 278: 49671–49674. [DOI] [PubMed] [Google Scholar]
- Schmid T., Hajnal A., 2015. Signal transduction during C. elegans vulval development: a NeverEnding story. Curr. Opin. Genet. Dev. 32: 1–9. [DOI] [PubMed] [Google Scholar]
- Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., et al. , 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Han M., 1995. sur-2, a novel gene, functions late in the let-60 ras-mediated signaling pathway during Caenorhabditis elegans vulval induction. Genes Dev. 9: 2251–2265. [DOI] [PubMed] [Google Scholar]
- Smyth G. K., 2004 Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article3. [DOI] [PubMed]
- Sönnichsen B., Koski L. B., Walsh A., Marschall P., Neumann B., et al. , 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469. [DOI] [PubMed] [Google Scholar]
- Steimel A., Suh J., Hussainkhel A., Deheshi S., Grants J. M. et al., 2013 The C. elegans CDK8 Mediator module regulates axon guidance decisions in the ventral nerve cord and during dorsal axon navigation. Dev. Biol. 377: 385–398. [DOI] [PubMed]
- Sternberg P. W., 1988. Lateral inhibition during vulval induction in Caenorhabditis elegans. Nature 335: 551–554. [DOI] [PubMed] [Google Scholar]
- Sternberg P. W., Horvitz H. R., 1986. Pattern formation during vulval development in C. elegans. Cell 44: 761–772. [DOI] [PubMed] [Google Scholar]
- Sternberg P. W., Horvitz H. R., 1989. The combined action of two intercellular signaling pathways specifies three cell fates during vulval induction in C. elegans. Cell 58: 679–693. [DOI] [PubMed] [Google Scholar]
- Stevens J. L., Cantin G. T., Wang G., Shevchenko A., Shevchenko A., et al. , 2002. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296: 755–758. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1981. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev. Biol. 82: 41–55. [DOI] [PubMed] [Google Scholar]
- Tan P. B., Lackner M. R., Kim S. K., 1998. MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during C. elegans vulval induction. Cell 93: 569–580. [DOI] [PubMed] [Google Scholar]
- Taubert S., Van Gilst M. R., Hansen M., Yamamoto K. R., 2006. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 20: 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S., Hansen M., Van Gilst M. R., Cooper S. B., Yamamoto K. R., 2008. The Mediator subunit MDT-15 confers metabolic adaptation to ingested material. PLoS Genet. 4: e1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K.-L., Sato S., Tomomori-Sato C., Conaway R. C., Conaway J. W., et al. , 2013. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat. Struct. Mol. Biol. 20: 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K.-L., Tomomori-Sato C., Sato S., Conaway R. C., Conaway J. W., et al. , 2014. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157: 1430–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck S., Greenwald I., 1995. lin-25, a gene required for vulval induction in Caenorhabditis elegans. Genes Dev. 9: 341–357. [DOI] [PubMed] [Google Scholar]
- van de Peppel J., Kettelarij N., van Bakel H., Kockelkorn T. T. J. P., van Leenen D., et al. , 2005. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell 19: 511–522. [DOI] [PubMed] [Google Scholar]
- Yang S. H., Vickers E., Brehm A., Kouzarides T., Sharrocks A. D., 2001. Temporal recruitment of the mSin3A-histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol. Cell. Biol. 21: 2802–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda A., Kouike H., Okano H., Sawa H., 2005. Components of the transcriptional Mediator complex are required for asymmetric cell division in C. elegans. Development 132: 1885–1893. [DOI] [PubMed] [Google Scholar]
- Yoo A. S., Bais C., Greenwald I., 2004. Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science 303: 663–666. [DOI] [PubMed] [Google Scholar]
- Zhang H., Emmons S. W., 2000. A C. elegans mediator protein confers regulatory selectivity on lineage-specific expression of a transcription factor gene. Genes Dev. 14: 2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Greenwald I., 2011. Spatial regulation of lag-2 transcription during vulval precursor cell fate patterning in Caenorhabditis elegans. Genetics 188: 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Spaeth J. M., Kim N. H., Xu X., Friez M. J., et al. , 2012. MED12 mutations link intellectual disability syndromes with dysregulated GLI3-dependent Sonic Hedgehog signaling. Proc. Natl. Acad. Sci. USA 109: 19763–19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Primer sequences are listed in Table S5. Strains are listed in Table S4 and are available upon request. Gene expression data are available at GEO with the accession number GSE68520.