Abstract
Receptor-like kinases (RLKs) mediate cell-signaling pathways in Arabidopsis thaliana, including those controlling growth and development, immune response, and cell death. The RLK coreceptor BRI1-ASSOCIATED KINASE-1 (BAK1) partners with multiple ligand-binding RLKs and contributes to their signaling in diverse pathways. An additional RLK, BAK1-INTERACTING RECEPTOR-1 (BIR1), physically interacts with BAK1, and loss-of-function mutations in BIR1 display constitutive activation of cell death and immune response pathways and dwarfism and a reduction in lateral root number. Here we show that bir1 plants display defects in primary root growth, characterize bir1 lateral root defects, and analyze expression of BIR1 and BAK1 promoters within the root. Using an allelic series of bak1 mutations, we show that loss of BAK1 function in immune response pathways can partially suppress bir1 cell death, immune response, and lateral root phenotypes and that null bak1 alleles enhance bir1 primary root phenotypes. Based on our data, we propose a model in which BIR1 functions to regulate BAK1 participation in multiple pathways.
Keywords: receptor-like kinases, genetic suppression, cell death, lateral roots
RECEPTOR-like kinases (RLKs) are a major component of signaling pathways in plants. Composed of an extracellular domain, single-pass transmembrane region, and a cytoplasmic kinase domain, RLKs share structural similarity with the animal receptor tyrosine kinases (RTKs) yet share sequence similarity with the Toll-like receptors (TLRs) and their associated kinases (Shiu and Bleecker 2001a, Asai et al. 2002). The RLK family is greatly expanded in plants compared to RTK and TLR families, with over 600 RLKs in Arabidopsis as opposed to 58 RTKs and 13 TLRs in humans (Nurnberger et al. 2004). This may reflect an increased need for receptor-mediated signaling in plants owing to their sessile nature and lack of cell motility, as well as their sole reliance on an innate immune system for pathogen recognition [reviewed in Nurnberger et al. (2004)].
RLKs in Arabidopsis are essential components of signaling pathways in diverse processes from growth and development to the recognition of conserved pathogen-associated molecular patterns (PAMPs) in pathogen-triggered immunity (PTI) [reviewed in Schwessinger and Ronald (2012) and Shiu and Bleecker (2001b)]. Perhaps the most well-characterized RLK-mediated pathway is the pathway containing BRASSINOSTEROID INSENSITIVE 1 (BRI1), a leucine-rich repeat domain containing RLK (LRR-RLK), which serves as the major ligand-binding receptor for the plant steroid hormone brassinosteroid (BR). BR signaling functions to promote both cell division and cell expansion and influences many processes in plants, including germination, vascular differentiation, vegetative and reproductive growth, root growth and gravitropism, photomorphogenesis, flowering time, biotic and abiotic stress response, and male fertility [reviewed in Zhu et al. (2013)].
Within PTI signaling, LRR-RLK-mediated pathways trigger transcriptional changes in the plant to produce defense compounds such as defensins, lytic enzymes, and secondary metabolites, as well as factors to strengthen cell walls and increase concentrations of reactive oxygen species (ROS) [reviewed in Nurnberger et al. (2004)]. The LRR-RLK FLAGELLIN SENSING 2 (FLS2) recognizes a conserved PAMP from bacterial flagellin, and the LRR-RLK EF-Tu RECEPTOR (EFR) recognizes the bacterial elongation factor EF-Tu to activate defense responses against biotrophic plant pathogens (Kunze et al. 2004; Zipfel et al. 2004).
In addition to roles as primary ligand-binding receptors, RLKs function as coreceptors in signaling pathways. Foremost of these is BRI1-ASSOCIATED KINASE 1 (BAK1), originally identified as a physical and genetic interactor of BRI1 in BR signaling pathways and later identified as an interactor of FLS2 and EFR in PTI pathways (Li et al. 2002; Nam and Li 2002; Chinchilla et al. 2007). In both BR and PTI signaling, BAK1 association is ligand dependent and is characterized by auto- and trans-phosphorylation events between BAK1 and the primary receptor, followed by activation of downstream components (Chinchilla et al. 2007; Heese et al. 2007; Wang et al. 2008; Oh et al. 2010). Reflecting its secondary role in these pathways, bak1 mutant phenotypes are less severe than those in the primary ligand-binding receptor (Li et al. 2002; Nam and Li 2002; Kemmerling et al. 2007).
Studies of BAK1 [also named SOMATIC EMBRYOGENESIS RECEPTOR KINASE 3 (SERK3); for simplicity, BAK1 will be used in this paper) and the other members of the SERK family have shown that they share overlapping function [reviewed in Li (2010)]. SERK1 and BAK1-LIKE 1 (BKK1, also named SERK4; for simplicity, BKK1 will be used in this paper) can suppress bri1 hypomorphs when overexpressed and function with BRI1 in the BR signaling pathway (Karlova et al. 2006; He et al. 2007). In addition, BAK1 and BKK1 have been identified as negative regulators of cell death, with bak1 single mutants showing defects in containment of necrotic lesions caused by pathogen infection and bak1; bkk1 double mutants displaying spontaneous cell death phenotypes accompanied by constitutive defense activation, increased ROS accumulation, and seedling lethality (He et al. 2007; Kemmerling et al. 2007). bak1 single-mutant cell death phenotypes are visible only in response to necrotizing pathogens and are believed to be independent of BAK1 function in BR signaling (Kemmerling et al. 2007). bak1; bkk1 cell death phenotypes are also thought to be BR independent because microarray analysis showed no significant overlap between upregulated defense-related transcripts present in bak1; bkk1 double mutants and bri1 null mutants, and pathogen resistance phenotypes were not rescued with exogenous brassinolide (BL) application (Kemmerling et al. 2007). However, this does not completely rule out a role for BR signaling in the bak1; bkk1 cell death phenotype. Prior studies have hypothesized that BAK1/BKK1 may function redundantly as coreceptors for an as yet unidentified RLK or RLKs controlling cell death, similar to the role of BAK1 in PTI and BR signaling pathways (He et al. 2007). The PRK5 LRR-RLK has been found recently to bind the GRIM REAPER cell death–inducing peptide (Wrzaczek et al. 2015) and is one candidate RLK that may interact with BAK1/BKK1, but this interaction has not been tested yet.
A role for SERK receptors in root development has been described recently (Du et al. 2012; Gou et al. 2012). Formation of the cell layers and distinct cell types of the primary root occurs through oriented cell divisions in the root apical meristem (RAM) at the tip of the root. Further “transit-amplifying” cell divisions occur in the meristematic zone of the root, followed by cell elongation within the elongation zone, and finally, cells take on their mature functions within the differentiation zone. The RAM contains a group of three to four infrequently dividing cells, the quiescent center (QC), surrounded by initial cells that give rise to the various root cell types. The distal RAM consists of the columella root cap initial cells (CSCs) directly below the QC that divide to form the columella cells (CCs), which, when mature, contain starch granules that function in root gravitropism. The proximal RAM contains specific initial cells that give rise to lateral root cap cells; the files of cells that make up the epidermal, cortical, endodermal, and pericycle radial layers; and additional initials that form the core vasculature of the root [reviewed in Wierzba and Tax (2013)].
Lateral roots (LRs) form from differentiated cells of the primary root and contain their own meristematic tissues when mature (Malamy and Benfey 1997). Pericycle cells within the meristematic zone are primed prior to differentiation as lateral root founder cells and then reenter the cell cycle in the elongation zone to form lateral root primordia (LRP) (Benková and Bielach 2010). Initiated LRP then can mature via a series of seven distinguishable stages (I–VII) before emerging via the endodermal, cortical, and epidermal layers of the primary root and acquiring meristematic capability as mature LRs (Malamy and Benfey 1997). Not all initiated LRP mature into LRs, and this maturation is controlled in part by hormone and nutrient concentration (Malamy 2005). Major plant hormones, including BR, affect LR development, and recently, it was shown that treatment with the PAMP flg22, perceived by FLS2, results in a reduction in LR number (Fukaki and Tasaka 2009; Beck et al. 2014).
serk1; bak1; bkk1 triple mutants are insensitive to BR treatment and cannot be rescued by BRI1 overexpression, suggesting that BR signaling is completely impaired in their absence (Gou et al. 2012). Both bri1 mutants and serk1; bak1; bkk1 triple mutants exhibit a reduction in the size and cell number of the root meristematic zone; however, serk1; bak1; bkk1 defects are more severe than those seen in bri1 or BR biosynthetic mutants, suggesting that these receptors function via an additional BR-independent pathway in the root. In addition, serk1; bak1; bkk1 triple mutants do not form any starch granules in CCs of the root tip (Du et al. 2012).
An additional LRR-RLK, BAK1-INTERACTING RECEPTOR-LIKE KINASE 1 (BIR1), was identified as a negative regulator of cell death pathways (Gao et al. 2009). Like bak1; bkk1 double mutants, bir1-1 mutants show constitutive expression of defense-related transcripts and increased ROS accumulation; however, unlike bak1; bkk1 double mutants, bir1-1 plants are not seedling lethal but are extremely dwarfed, with reduced rosette leaf size and premature senescence. Unlike bak1; bkk1 double mutants, bir1-1 phenotypes are partially suppressed by growth at high temperature (28°) and by mutations affecting salicylic acid (SA) biosynthesis and R-protein-mediated resistance (Gao et al. 2009). Mass spectrometric analysis of co-immunoprecipitating proteins of BIR1-FLAG identified peptides shared by SERK1, SERK2, and BAK1, and BAK1-BIR1 interaction was confirmed by bimolecular fluorescence complementation (Gao et al. 2009).
BIR1, as well as BAK1, was identified in a yeast two-hybrid screen for interactors of BONZAI1 (BON1), a copine protein with roles in plant growth and a negative regulator of defense response through R proteins (Wang et al. 2011). Double mutants of bon1 and related homologs of BON1, bon2 and bon3, show similar phenotypes to bir1-1, and these phenotypes are also suppressed under high-temperature conditions. bir1-1; bon1 double mutants showed additive effects: overexpression of BIR1 partially suppressed bon1 growth phenotypes and, reciprocally, overexpression of BON1 partially suppressed bir1-1 growth defects (Wang et al. 2011). Biochemical analysis demonstrated that BON1 and BIR1 are both in vitro kinase substrates of BAK1 (Wang et al. 2011). The resulting model includes three major components: BON1 and BIR1 act in parallel to guard R-protein defense pathways that are also regulated by high temperature, BON1 and BIR1 positively regulate each other at a downstream transcriptional level, and BAK1 may activate defense response by negatively regulating BON1 and BIR1 via phosphorylation (Wang et al. 2011).
The previous research, summarized earlier, proposes both positive and negative interactions between BIR1 and BAK1. In this study, we use a genetic approach to clarify the relationship between BIR1 and BAK1 in different cell types and under different environmental conditions. Here we show through genetic interactions that bir1 immune response phenotypes can be suppressed by bak1 mutations, while bir1 root phenotypes, which have not been previously characterized, are enhanced by bak1 mutations. Previous models of BIR1-BAK1 interaction are insufficient to explain these complex relationships, and we propose a new model for their interactions.
Materials and Methods
Plant growth
For soil-grown plants, approximately five seeds were planted in 2-square-inch pots with soil [four parts Sunshine Mix 3 (Sungro Horticulture) to one part vermiculite (CAS #318-00-9, Therm-O-Rock West)] presoaked in water. Trays containing the pots were covered in plastic wrap and cold treated at 4° for 3–4 days before transfer to a Conviron MTR30 at 22° or an E7 Plant Growth Chamber at 28° set to 16 hr of light and 8 hr of dark with 75% relative humidity and a light intensity of approximately 120 µE/m2 from cool-white fluorescent tube lamps (Philips F96/CW/VHO or F48/CW/VHO) supplemented by 40-W incandescent bulbs (Philips). Plastic wrap was removed after the seedlings were established (3–5 days), and one representative seedling was allowed to grow per pot. The pots were subirrigated with Hoagland nutrient solution as necessary.
Plate assays were performed using surface-sterilized seeds, which were soaked in 70% ethanol and 0.1% Triton X-100 for 10 min and then rinsed three times with 95% ethanol. Seedlings were grown vertically on 1% agar plates with 0.5× Murashige and Skoog (MS) medium (Sigma-Aldrich #M5524) and 0.05% 2-(N-morpholino)ethane sulfonic acid (MES) (Sigma-Aldrich #M2933), pH 5.7. Plates were sealed with Micropore surgical tape (3M), stratified in the dark at 4° for 72 hr, and grown vertically at 22° in a controlled-temperature room with 16 hr of light and 8 hr of dark under cool-white fluorescent tubes (Phillips Alto F40CW/RS/EW) with an average output of 90 µE/m2.
For seedling growth-inhibition assays, seeds were sterilized and grown vertically as earlier, except for the addition of 1% sucrose to the medium. At 5 days after germination, seedlings were transferred, two per well, to 24-well dishes containing 500 µl 0.5× MS + MES + 1% sucrose medium with either 100 nM flg22, 1 µM elf18, or an equal volume of 100 mM NaCl, 0.1% BSA as mock treatment and grown for an additional 8 days. flg22 (QRLSTGSRINSAKDDAAGLQIA) peptide (Mimotopes, Clayton, Victoria, Australia) was dissolved in water to 1 mM and diluted to 500 µM in 100 mM NaCl, 0.1% BSA before addition to medium cooled to 45° at a concentration of 100 nM. For seedling growth inhibition, seedlings were blotted with filter paper to remove medium and weighed in pools of four seedlings on an Ohaus Adventurer AR3130 balance.
bir1-1, bak1-1, bak1-3, bak1-4, bak1-5, bri1-301, and pBAK1:GUS lines were described previously (Li et al. 2002; Kemmerling et al. 2007; Gao et al. 2009; Kang et al. 2010; Roux et al. 2011; Schwessinger et al. 2011). Consistent with published data, bir1-1 (Gao et al. 2009) contains a transfer DNA (T-DNA) insertion within the coding DNA sequence (CDS) 1453 bp downstream of the ATG. bir1-3 is in the Ws ecotype and was isolated from the FLAG_193B06 line obtained from the Versailles Arabidopsis Stock Center (IJPB-INRA, Versailles, France) (Samson et al. 2002). bir1-3 contains a T-DNA insertion within the putative 5′ UTR 343 bp upstream of the ATG, as confirmed by PCR and sequence analysis (Supporting Information, Figure S1A).
Cloning and transformation
For pBIR1:BIR1:GFP, the region from 1516 bp upstream of the ATG up to, but not including, the stop codon of BIR1 was amplified from BAC clone MJE7 (Arabidopsis Biological Resource Center, Ohio State University) using TaKaRa Ex Taq polymerase (TaKaRa Bio, Inc.) and cloned into the pCR8/GW-TOPO vector (Life Technologies), followed by Gateway LR recombination (Life Technologies) into the pBIBKAN-GWR-GFP vector (Gou et al. 2010). For pBIR1:GUS, the region 1516 bp upstream of and including the start codon of BIR1 was amplified using PrimeSTAR polymerase (TaKaRa Bio, Inc.) and cloned into the pENTR/D-TOPO vector (Life Technologies), followed by Gateway LR recombination (Life Technologies) into the pBIB-GUS-BASTA vector (Gou et al. 2010). Constructs were transformed into the Col accession or indicated mutant backgrounds using Agrobacterium GV3103 and the floral dip method (Clough and Bent 1998).
Histology and histochemical analysis
Seedlings containing pBIR1:GUS and pBAK1:GUS were stained with GUS solution containing X-Gluc [2 mM X-Gluc (Gold Bio Technology) dissolved in DMSO, 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, 0.2% Triton X-100, and 50 mM sodium phosphate, pH 7.2] at 37° for 1–16 hr. For DIC imaging, samples then were fixed in 90% ethanol:10% acetic acid for 12 hr, cleared, and mounted on slides in chloral hydrate solution [8:2:1 chloral hydrate:double-distilled H2O(ddH2O):glycerol]. For transverse sectioning of roots, GUS-stained seedlings were embedded in 1% agarose; fixed in 4% gluteraldehyde, 3% paraformaldehyde, and 50 mM sodium phosphate buffer, pH 7.2, for 3 hr; rinsed twice in 50 mM sodium phosphate, pH 7.2; dehydrated in an increasing series of 10, 20, 50, 70, 90, and 100% ethanol; infiltrated with increasing concentrations of Technovit 7100 (Heraeus, Germany); and cured in BEEM capsules (SI Supplies) according to the manufacturer’s instructions. Then 12- to 16-µm sections were cut using a Sorval MT2B ultramicrotome equipped with glass knives.
For analysis of root apical meristem morphology, roots were fixed and stained with a modified-Schiff propidium iodide method (mS-PI) (Truernit et al. 2008), cleared, and mounted in chloral hydrate solution.
For analysis of LR morphology, roots were fixed and cleared using an acid/base method according to Malamy and Benfey (1997) or a modified acid/base method incorporating mS-PI staining by incubating rehydrated samples in 1% periodic acid for 40 min; rinsing with ddH2O; incubating in 100 mM sodium metabisulfite, 0.15 N HCl, and 100 µg/ml propidium iodide for 1 hr; and finally, clearing and mounting in chloral hydrate solution. For bir1-1 mutants, only seedlings from the L or M class were analyzed for LR phenotypes because the S-class primary roots were too short.
Trypan blue staining was performed by boiling leaf samples for 1 min in trypan blue solution [trypan blue (EMD Chemicals #368) dissolved in lactophenol solution (EMD Chemical #R03266) to a concentration of 2.5 mg/ml and then diluted with two volumes of 100% ethanol], incubating overnight in trypan blue solution at room temperature, clearing with chloral hydrate solution at 37° for 3 days, and mounting in chloral hydrate solution.
3,3′-Diaminobenzidine (DAB) staining was performed by vacuum infiltrating samples with DAB solution [1 mg/ml 3,3′-diaminobenzidine (Sigma-Aldrich #D5637) in 50 mM Tris, pH 5.0, and 0.05% Tween 20 (Sigma-Aldrich #P9416)], followed by incubation for 24 hr at room temperature in the dark. Samples then were cleared in boiling ethanol for 10 min and further cleared and mounted in chloral hydrate solution.
Microscopy, image acquisition, and measurements
DIC imaging of LPR, root tips, and transverse sections was performed using 10× or 20× magnification as indicated on a Zeiss Axioplan compound microscope equipped with a Q-Imaging Micro Publisher 3.3 RTV camera and Q-CapturePro 5.0 software.
Confocal microscopy was performed using a Zeiss LSM 510 microscope and AxioVision software. For mS-PI-stained tissues, samples were excited at 488 nm and emission collected through an LP 505 filter.
Mature plant and rosette images were taken individually with a Canon Power Shot SX110 digital camera; images of seedlings for root measurements were obtained by scanning plates at 720 dpi on an Epson Perfection 2400 Photo Scanner. Images of individual plants were scaled to each other, the backgrounds were removed, brightness and contrast were adjusted, and the images were merged into a composite image in Adobe Photoshop.
To measure plant height and leaf length and width, the first four rosette leaves, axillary leaves, and primary bolt of 5-week-old plants were affixed to paper and scanned at 720 dpi. Leaf length was measured along the midvein and width at the widest point. The primary bolt was measured from its base to the youngest inflorescence. Roots were measured from the root-hypocotyl junction to the tip.
All measurements were made using ImageJ 1.42q software (NIH). Data were analyzed and charts produced in Microsoft Excel. Student’s t-test was used for calculating statistical differences between sample populations with P-values as indicated in the text.
Data availability
Strains are available on request.
Results
bir1 phenotypes
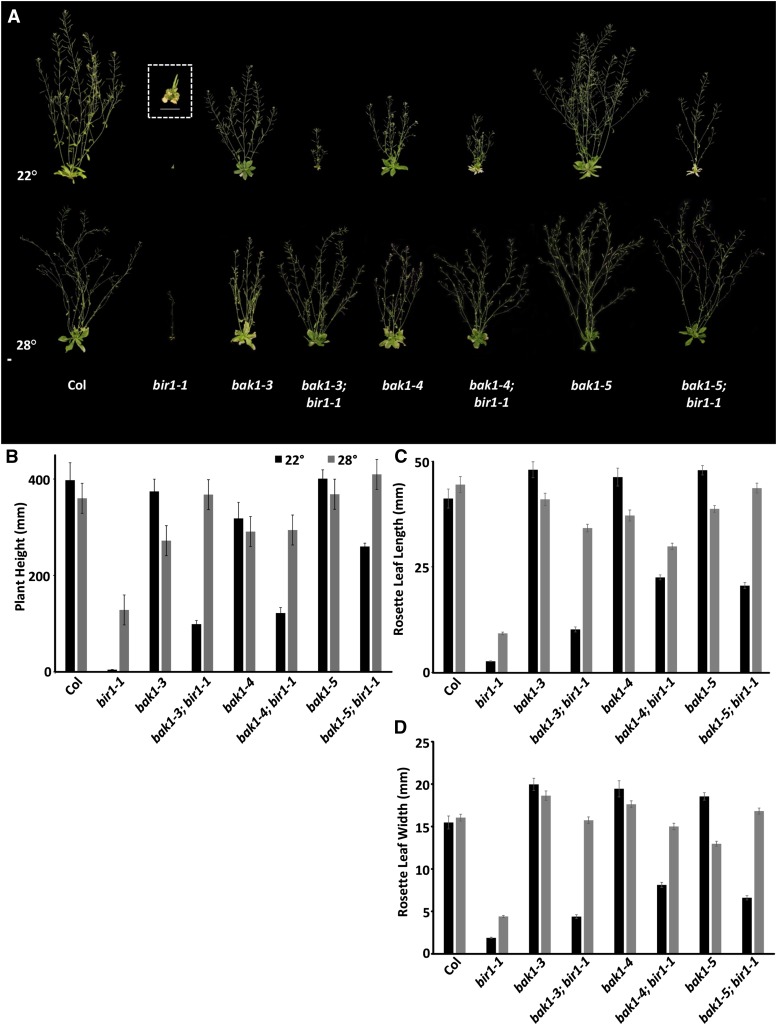
Analysis of the preceding ground phenotypes of bir1-1 mutants is consistent with a previous characterization (Gao et al. 2009); homozygous plants are extremely dwarfed at about 1% of the height of wild-type Col, with epinastic cotyledons and compact rosettes, containing leaves that show premature senescence and are 7% of the length and 12% of the width of Col leaves (Figure 1, A–D). When grown under standard conditions at 22°, bir1-1 plants rarely survive to bolting, and if they do, they produce a single, very short bolt bearing only one or two siliques. These siliques do produce viable homozygous seed. Homozygous bir1-3 plants, which are in the WS ecotype, present similar phenotypes to those of bir1-1 (Figure S1B). As previously reported for bir1-1 and other mutants exhibiting constitutive cell death phenotypes, growth at 28° partially suppresses growth phenotypes in bir1-1 and bir1-3 plants, restoring plant height to 36% of that of Col for bir1-1 and to 40% of that of Ws for bir1-3 (Figure 1, A and B, and Figure S1, A and B). Rosette leaf length and width are increased to 21 and 27% of wild type, respectively, for bir1-1 (Figure 1, C and D). Transgenic pBIR1:BIR1-GFP plants containing the genomic sequence corresponding to the putative promoter and CDS, translationally fused to the CDS of GFP, were able to complement bir1-1 defects, including plant height (Figure S2, A and B), primary root growth (Figure 2A), and the density of the LRP (Figure 2, A and B, and Figure S3).
Figure 1.
Suppression of bir1-1 by an allelic series of bak1 mutants and high temperature. (A) Five-week-old plants of indicated genotypes. Inset is a magnified view of bir1-1 homozygous plants. Scale bar, 1 cm. Average plant height (B), rosette leaf length (C), and width (D) of 5-week-old plants of indicated genotypes grown at 22 and 28°. Error bars, average ± SE.
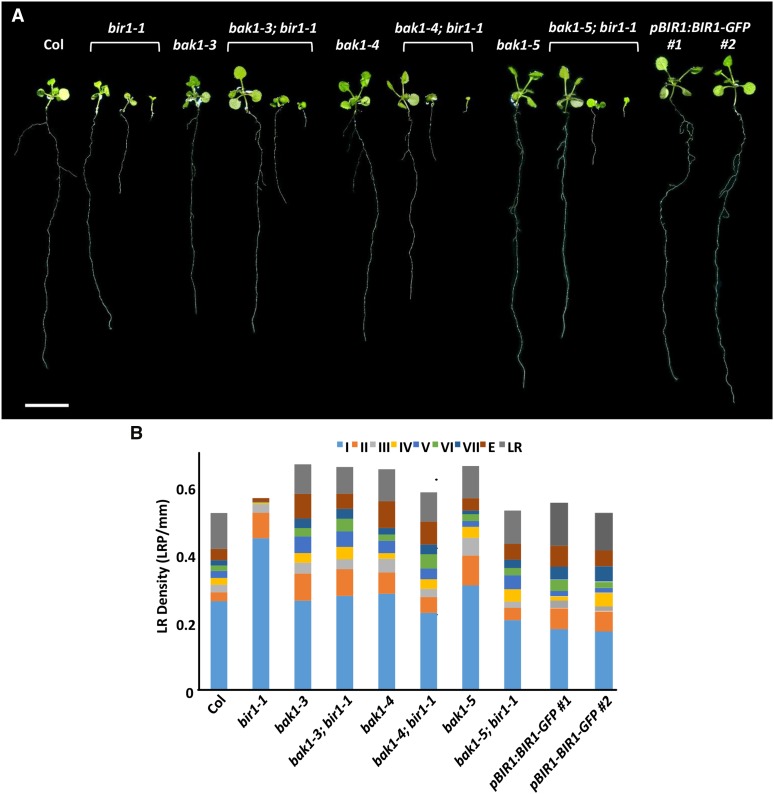
Figure 2.
bak1 interaction with bir1-1 primary and LR growth phenotypes . (A) Representative 14-dag seedlings grown on 0.5× MS medium. Scale bar, 1 cm. (B) Total LRP density of 14-dag seedling for the indicated genotypes.
Previous studies of BIR1 have focused primarily on its physical interaction with BAK1 and bir1-1 phenotypes in cell death and immune response pathways (Gao et al. 2009; Wang et al. 2011). In addition to these roles, we have identified novel root phenotypes for bir1-1 mutants. When grown on MS medium, slightly more than half of bir1-1 primary roots (52%) are near to the Col primary root length at about 80% of wild type but are statistically different (P < 0.001). To more precisely characterize bir1-1 phenotypes, we have divided root defects into three classes: this group of seedlings is classified as long. Furthermore, 12% of bir1-1 seedlings [designated bir1-1 (medium)] display primary roots that are between 50 and 10% of the length of Col, and 36% of bir1-1 seedlings [bir1-1 (short)] are less than 10% of the length of Col (averaging 6.8%) (Table 1). Only minor increases in bir1-1 (short) root growth are seen between 3 and 14 days, suggesting that most of these roots have ceased growing (data not shown).
Table 1. Distribution of root length classes for bir1 and bak1 single and double mutants.
| Class | Col | bir1-1 | bak1-3 | bak1-3; bir1-1 | bak1-4 | bak1-4; bir1-1 | bak1-5 | bak1-5; bir1-1 | pBIR1:BIR1-GFP 1 | pBIR1:BIR1-GFP 2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Long | 89% (25) | 52% (22) | 84% (26) | 56% (30) | 77% (23) | 63% (38) | 78% (21) | 57% (20) | 75% (15) | 79% (15) |
| Medium | 0% (0) | 12% (5) | 6% (2) | 22% (12) | 17% (5) | 17% (10) | 11% (3) | 11% (4) | 10% (2) | 16% (3) |
| Short | 11% (3) | 36% (15) | 10% (3) | 22% (12) | 6% (2) | 20% (12) | 11% (3) | 31% (11) | 15% (3) | 5% (1) |
Percentage of germinated 14-dag roots that are long, medium, and short for each genotype (≤50%, 50 > 10%, and <10% average Col length, respectively) (n).
It was reported previously that bir1-1 mutants display reduced numbers of LRs, but no quantification has been published (Wang et al. 2011). In addition, it has been shown recently that treatment of seedlings with the pathogen elicitor peptide flg22 leads to a reduction in overall LR number (Beck et al. 2014). To determine the basis of the apparent reduction of mature LRs in bir1-1, 14-dag roots were fixed, cleared, and analyzed microscopically to assess the density of LRP at stages I–VII of development, as well as to characterize the quantity of LRs at the emergence and mature stages.
Examination of LRP density in bir1-1 mutants shows significant increases in stage I and II primordia compared with Col (Table 2). Stage III LRP density did not differ significantly from Col, and bir1-1 roots contain significant decreases in all stages beyond stage III compared with Col, with no stage VI or VII or mature LRs observed for bir1-1 in this experiment (Figure 2B and Figure S3). In addition to alteration of the proportion of LRP in individual stages, bir1-1 roots contain a slight increase in the total density of LRP compared with Col (Figure 2C, Figure S3, and Table 2). Similar results were seen in repeated experiments; results from a single assay are presented for clarity.
Table 2. Comparison of Col and bir1-1 LRP density.
| Stage | Col (LRP/mm) | bir1-1 (LRP/mm) | P-value |
|---|---|---|---|
| I | 0.26 | 0.45 | <0.001 |
| II | 0.026 | 0.075 | <0.01 |
| III | 0.022 | 0.025 | >0.5 |
| IV | 0.020 | 0.0052 | <0.05 |
| V | 0.021 | 0.0024 | <0.05 |
| VI | 0.016 | 0 | <0.01 |
| VII | 0.015 | 0 | <0.01 |
| Emergence | 0.034 | 0.01 | <0.05 |
| LR | 0.11 | 0 | <0.001 |
| Overall | 0.52 | 0.57 |
Here we have described novel root phenotypes for bir1-1 mutants; a significant number of bir1-1 seedlings exhibit defects in primary root growth, and in addition to a reduction in mature LRs, bir1-1 roots contain lowered densities of stage IV–VII and emerging LRP, as well as an increase in stage I and II primordia.
Genetic interactions of BIR1 and BAK1 mutants
Given the physical interaction between BIR1 and BAK1 (Gao et al. 2009; Halter et al. 2014), as well as shared aspects of bir1-1 and bak1-4; bkk1-2 cell death phenotypes (He et al. 2007), we hypothesized that BIR1, BAK1, and BKK1 act in a common pathway to control cell death and predicted that bir1 mutations would either additively enhance bak1-4; bkk1 phenotypes or be epistatic and have no increased effect. Therefore, we crossed bir1 mutants to bak1 mutants to test for a genetic interaction. Surprisingly, rather than bak1; bir1 double mutants enhancing or having no effect on bir1 dwarf phenotypes, bak1; bir1 double mutants showed suppression of bir1 phenotypes. To further examine their genetic interactions, bak1; bir1 double mutants were created using an allelic series of bak1 mutations: bak1-4, a null allele that displays defects in BL signaling, immune response activation, pathogen-induced cell death, and, as a double with bkk1, general loss of cell death regulation; bak1-3, a hypomorphic intronic T-DNA insertion that greatly reduces BAK1 transcript level but is not fully null and shares all bak1-4 phenotypes, including an enhanced cell death phenotype that is not as severe as bak1-4; bkk1 double mutants; bak1-5, a single-amino-acid-substitution mutant of BAK1 that results in hypoactive kinase activity and has been reported to show defects in positive regulation of immune response pathways but appears not to affect BL signaling or cell death regulation pathways; and bak1-1, an intronic T-DNA insertion allele in the WS background (Li et al. 2002; He et al. 2007; Albrecht et al. 2008; Roux et al. 2011; Schwessinger et al. 2011; Du et al. 2012; Gou et al. 2012).
bak1 suppression of bir1 aerial growth defects:
When grown at 22°, bak1-3; bir1-1, bak1-4; bir1-1, and bak1-5; bir1-1 double mutant combinations all show partial suppression of bir1-1 reduced height and rosette leaf length and width defects (Figure 1, A–D). The heights of bak1-3; bir1-1 and bak1-4; bir1-1 plants are 25 and 31% of that of wild-type Col compared with 1% for bir1-1 single mutants; this does not differ statistically from high-temperature suppression of bir1-1 single mutants, which restores bir1-1 height to 36% of Col. bak1-5; bir1-1 double mutants, whose height is 65% of Col, showed the most pronounced suppression of bir1-1 growth defects (Figure 1, A and B). When grown at 28°, bak1-5; bir1-1 plants are statistically the same as wild-type plants in height, and bak1-4; bir1-1 plants are indistinguishable from bak1-4 single mutants in height. At 22°, rosette leaf length and width are restored in bak1-3; bir1-1 plants to a similar level as bir1-1 plants grown under high temperature (Figure 1, C and D). bak1-4; bir1-1 and bak1-5; bir1-1 plants show a greater suppression of bir1-1 rosette leaf length and width at 22° than bak1-3; bir1-1 plants, at 55 and 50% of Col, respectively, though still intermediate of the wild type. At 28°, plants of all three double-mutant combinations are fully restored to wild-type rosette leaf width, but only bak1-5; bir1-1 plants are similar to wild type in rosette leaf length (Figure 1, C and D). Because bak1-3 and bak1-4 plants are known to have altered rosette leaf width and length owing to impairment of BL signaling, this is not surprising (Albrecht et al. 2008). Similar to the Col allele, plants homozygous for the WS bir1-3 allele, when combined with the null bak1-1 allele, resemble bak1-1 single-mutant plants in height when grown at 28° (Figure S1, A and B).
bak1 suppression of bir1 cell death phenotypes:
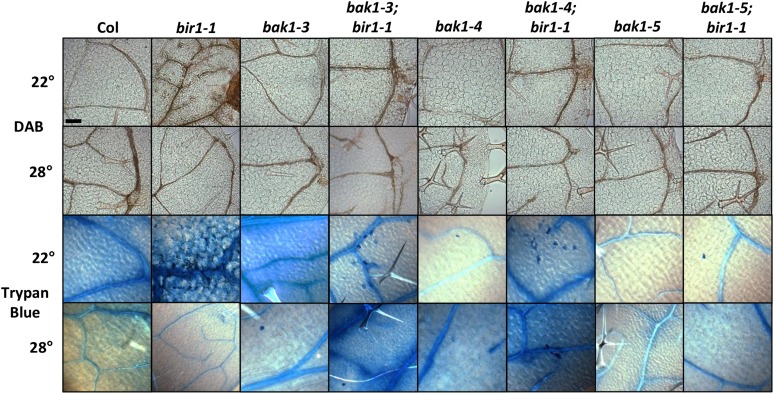
It was reported previously that bir1-1 phenotypes include accumulation of ROS and increased cell death (Gao et al. 2009; Wang et al. 2011). To test suppression of these phenotypes by bak1, double-mutant plants grown at 22 and 28° were stained with DAB, an indicator of hydrogen peroxide within cells, and the vital stain trypan blue, which cannot be readily absorbed by live cells. When stained with DAB, 22° grown 2-week-old bir1-1 leaves show a pronounced increase in precipitate formation, especially surrounding the vasculature, compared with Col (Figure 3). bak1-3; bir1-1 and bak1-4; bir1-1 leaves also show increased DAB staining compared with Col, but with less severity than bir1-1. bak1-5; bir1-1 leaves are indistinguishable from wild type, as are bak1-3, bak1-4, and bak1-5 single-mutant leaves. Prior studies have shown increased ROS production for bak1-3 and bak1-4, but not bak1-5, when inoculated with bacterial and fungal pathogens or elicitors and not in the absence of infection unless in the bak1-4; bkk1 double mutant (He et al. 2007; Kemmerling et al. 2007; Schwessinger et al. 2011). At 28°, bir1-1 plants do not exhibit an increase in DAB staining compared with Col, nor do any of the double-mutant combinations or bak1 single mutants (Figure 3).
Figure 3.
bak1 suppression of bir1-1 cell death phenotypes . Leaves from 2-week-old seedlings grown at 22 and 28° and stained with DAB and trypan blue.
Trypan blue staining of bir1-1 shows extensive staining of dead cells within 2-week-old leaves grown at 22° compared to wild type, as indicated by solid-blue-stained cells in Figure 3. Two-week-old leaves from 22° grown bak1-3; bir1-1, bak1-4; bir1-1, and bak1-5; bir1-1 seedlings show greatly reduced staining compared with bir1-1 but still contain occasional dead cells; however, dead cells are observed least frequently in bak1-5; bir1-1 leaves. Of note, occasional dead cells are seen in bak1-3 and bak1-4 single-mutant leaves but not as often as in their respective double-mutant combinations with bir1-1, and they are not seen in bak1-5. For all genotypes tested, cells of leaves from plants grown at 28° do not show substantial trypan blue staining.
From these results, we conclude that bir1-1 growth and cell death phenotypes are at least partially dependent on BAK1-mediated pathways and that bir1-1 plants grown under high-temperature conditions are fully reverted to a wild-type phenotype when combined with bak1-5.
bak1 interaction with bir1-1 root growth phenotypes:
To assess the effect of bak1 mutants on bir1-1 root phenotypes, seedlings were grown on MS medium to measure primary root length. bak1-3 and bak1-4 single mutants, like bir1-1 single mutants, show a modest but significant decrease in primary root growth compared with Col (bak1-3: 75% of Col, P < 0.001; bak1-4: 77% of Col, P < 0.001) and do not differ significantly from bir1-1 (long) roots (Table 1). bak1-5 single mutants did not differ significantly from Col. Medium and short root classes are present for all three bak1 alleles, with 6, 17, and 11% medium and 10, 6, and 11% short roots for bak1-3, bak1-4, and bak1-5 genotypes, respectively (Table 1). Most bak1-3; bir1-1, bak1-4; bir1-1, and bak1-5; bir1-1 double mutants are similar to bir1-1 (long) single-mutant roots at 72, 75, and 89% of the length of Col (P < 0.01 for bak1-3; bir1-1, bak1-4; bir1-1, and bak1-5; bir1-1) (Table 1). As in bir1-1 single mutants, all double mutants containing bir1-1 have medium and short root classes, with 22, 15, and 11% medium roots and 22, 22, and 31% short roots for bak1-3; bir1-1, bak1-4; bir1-1, and bak1-5; bir1-1 genotypes, respectively (Table 1).
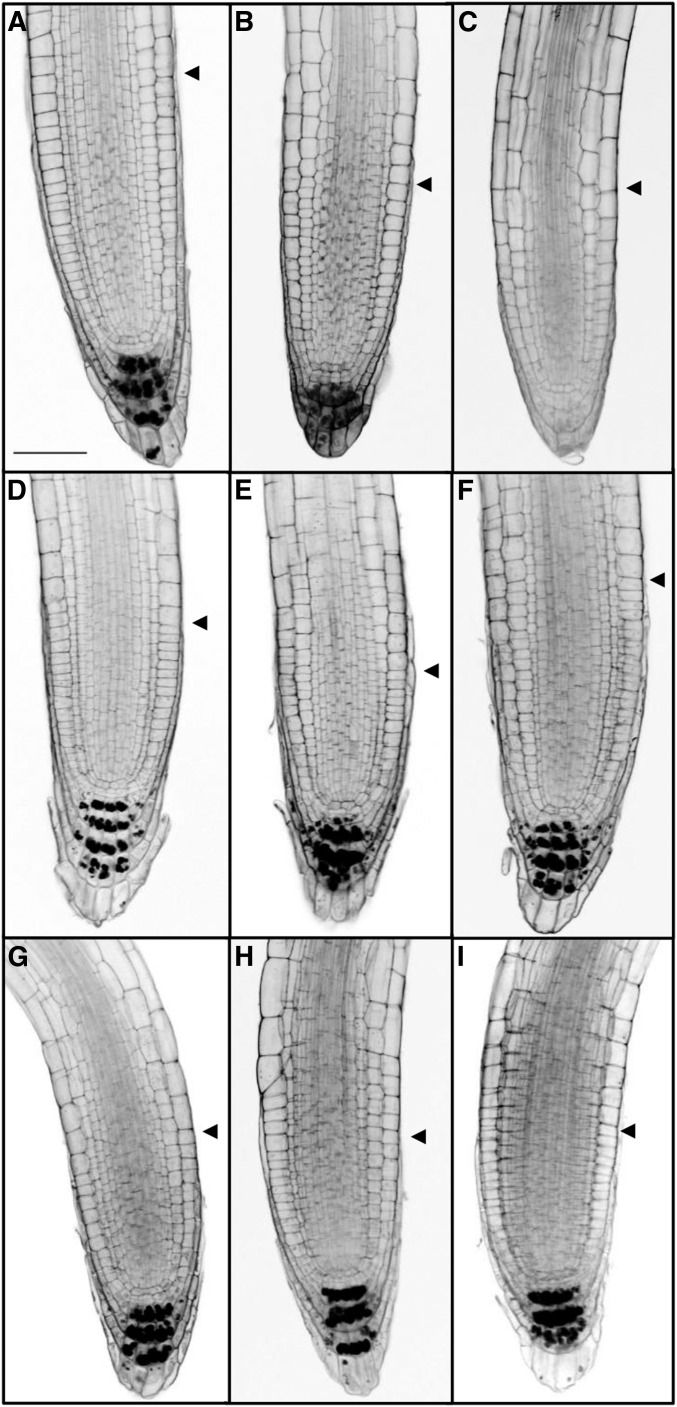
To more closely examine the basis of bir1-1 root defects, bir1-1 (long), bir1-1 (medium) and (short), bak1, and bak1; bir1-1 roots were stained with a mS-PI method for high-resolution imaging and staining of starch granules within mature columella (Truernit et al. 2008). Wild-type roots typically contain three to four tiers of starch granule–containing mature columella cells. bak1-3, bak1-4, and bak1-5 roots display a similar pattern to wild-type Col (Figure 4, A, D–F). bir1-1 (long) roots predominantly have only three layers of starch granule–containing columella tiers, and bir1-1 (medium) and (short) roots contain fewer or none at all (Figure 4, B and C). bak1-3; bir1-1, bak1-4; bir1-1, and bak1-5; bir1-1 roots all resemble wild-type Col (Figure 4L). From this we conclude that bir1-1 (medium) and (short) root defects are likely accompanied by morphologic changes within the columella.
Figure 4.
Analysis of bir1-1 root apical meristem defects. Representative samples of mS-PI-stained 7-dag root apical meristems of (A) Col, (B) bir1-1 (long), (C) bir1-1 (medium) and (short), (D) bak1-3, (E) bak1-4, (F) bak1-5, (G) bak1-3; bir1-1, (H) bak1-4; bir1-1, and (I) bak1-5; bir1-1. Scale bar, 50 µm. Arrowhead indicates beginning of elongation zone.
bir1-1 lateral root development phenotypes and suppression by bak1:
Visually, bak1; bir1-1 plate-grown roots appear to suppress the reduction in LR formation seen in bir1-1 roots (Figure 2A). To quantify LRP staging and density, single-mutant bak1 and bak1; bir1-1 double-mutant roots were fixed at 14 dag and stained.
All bak1 single mutants appear similar to Col in stage I LRP density but contain significant increases in stage II LRP to a level that does not significantly differ from bir1-1 single mutants (Figure 2B). bak1-3 and bak1-4 single mutants are similar to Col in stage III LRP density, while bak1-5 single mutants show an increase to 0.052 LRP/mm compared to 0.022 LRP/mm in Col. Stage IV LRP density does not differ from Col for any bak1 single-mutant allele. bak1-3 is increased in stage V LRP density (Col: 0.021 LRP/mm; bak1-3: 0.049 LRP/mm; P < 0.01), while bak1-4 and bak1-5 are similar to Col. Stages VI and VII do not differ from Col for any bak1 single-mutant allele, and only bak1-3 shows an increase in emergence-stage LRP density to 0.073 LRP/mm compared to 0.034 LRP/mm in Col. No statistically significant difference is seen in mature LR density among bak1 single-mutant alleles and Col (Figure 2B and Figure S3).
All bak1; bir1-1 double mutants tested show complete suppression of bir1-1 stage I LRP increases and do not differ significantly from Col but are significantly different from bir1-1 (P < 0.001). However, bak1-3; bir1-1 double mutants still differ significantly from Col for stage II LRP density (P < 0.001), whereas bak1-4; bir1-1 and bak1-5; bir1-1 double mutants show suppression of stage II increases seen in bir1-1, including suppression of the stage II increase seen in bak1-5 single mutants (Figure 2B and Figure S3). No difference was seen in stage III LRP density between bak1; bir1-1 double mutants and Col or bir1-1. Stage IV LRP density decreases seen in bir1-1 single mutants are completely suppressed in all bak1; bir1-1 double mutants. For stage V LRP density, bak1; bir1-1 double mutants show significant increases compared with Col and bir1-1 and resemble their respective bak1 single mutants with the exception of bak1-5; bir1-1, which is similar to the other bak1; bir1-1 double mutants, bak1-3, and bak1-4 but not bak1-5. Stage VI, VII, and emergence LRP and mature LR density in all bak1; bir1-1 double-mutant combinations show suppression of bir1-1 single-mutant decreases. Emergence-stage density increases seen in bak1-3 and bak1-4 single mutants are also present in their respective bak1; bir1-1 double mutants (Figure 2B and Figure S3). Staging of LRP density within bir1-1 (medium) and bak1; bir1-1 (medium) roots shows a similar trend to that seen in the long classes of these genotypes, with the exception of mature LR density, which is decreased compared with Col (Figure S3).
Assessing overall LRP and LR density, bir1-1 is similar to Col, but all three bak1 alleles show an increase in total LRP and LR density, suggesting that more primordia are initiated in these mutants (bak1-3: 0.67 LRP/mm; bak1-4: 0.65 LRP/mm; bak1-5: 0.66 LRP/mm) (Figure 2B). bak1-3; bir1-1 and bak1-4; bir1-1 double mutants, at 0.66 and 0.58 LRP/mm, respectively, are increased in total LRP density relative to Col, but bak1-5; bir1-1 double mutants are only slightly increased (0.53 LRP/mm) (Figure 2B).
From this we conclude that bir1-1 LR development defects appear to affect early stages of LRP development after initiation, between stages II and IV, which prevents the formation of most mature LRs. We have shown that all three bak1 alleles are sufficient to alleviate late-stage LRP developmental defects in bir1-1. Furthermore, a previously undescribed phenotype in LRP development, increases in stage II LRP density and overall density, has been identified for bak1, suggesting a novel function of BAK1 in LRP development.
BIR1 and BAK1 expression within roots:
To put the observed root phenotypes of bir1-1, bak1, and bak1; bir1-1 seedlings into a functional context, we generated constructs containing the promoter region of BIR1 fused to GUS and analyzed transgenic lines containing this construct in parallel with previously published pBAK1:GUS transgenic lines.
In the aerial portions of the plant, pBIR1:GUS is expressed strongly within developing stipules (Figure 5B), as well as in the vasculature of cotyledons and leaves, stomata, and the hydathodes (Figure 5A). Strong expression is noticeable after mechanical wounding of leaf tissue (Figure 5C).
Figure 5.
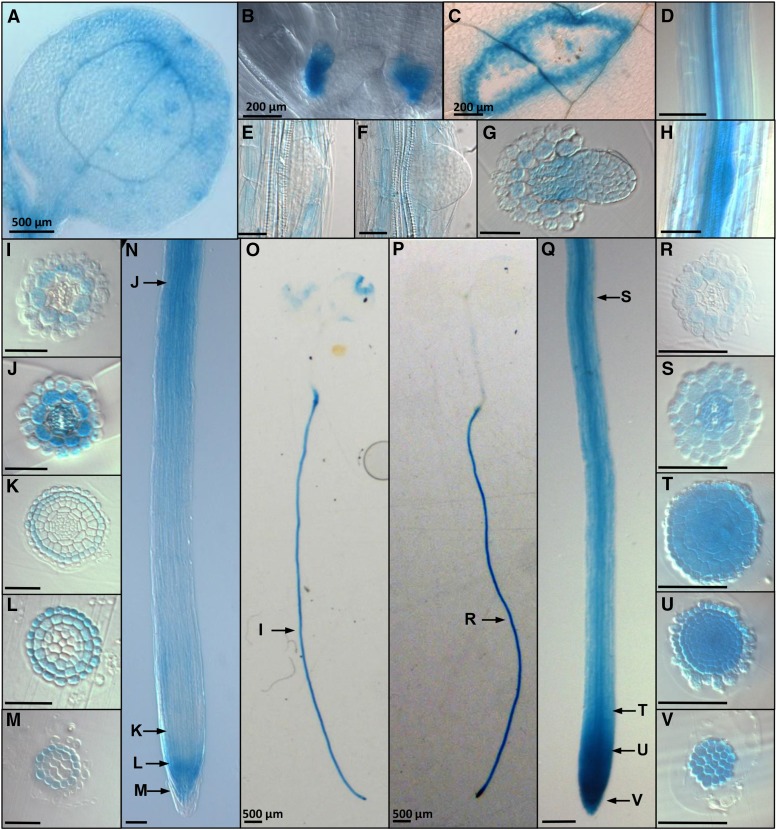
BIR1 and BAK1 expression . X-gluc-stained pBIR1:GUS plants: (A) 3-dag cotyledon, (B) stipules (scale bar, 200 µm), (C) wounded leaf, (E) stage VII LRP, and (F) emergence-stage LRP. Transverse sections of plastic embedded roots at (G) emerging LR, (I) mature region, (J) differentiation region, and (K–M) root apical meristem. (N) Meristematic, elongation, and differentiation zones. (O) A 7-dag seedling. X-gluc-stained pBAK1:GUS plants: (D) mature region of root, (H) stage IV LRP, (P) 7-dag seedling, and (Q) meristematic, elongation, and differentiation zones, Transverse sections of plastic embedded roots at (R) mature region, (S) differentiation region, and (T–V) root apical meristem. Arrows indicate the representative regions that transverse sections correspond to. Scale bar, 50 µm (unless otherwise noted).
Within the root, pBIR1:GUS shows strong expression throughout mature regions of the root at 3, 7, 10, and 14 dag (7 dag: Figure 5, N and O). Transverse sectioning of plastic embedded root tissue reveals that within mature root tissue, pBIR1:GUS is expressed primarily within the cortical cell layer (Figure 5I). Within the differentiation zone, pBIR1:GUS expression remains strongest in the cortex but is also seen within epidermal cells and strongly within pericycle, procambium, and protoxylem cells (Figure 5J). pBIR1:GUS expression appears reduced within the elongation zone, and within the RAM, it is seen primarily within LRC cells (Figure 5, K–M). pBIR1:GUS is not seen preferentially within developing LRP (Figure 5, E and F), although transverse sections of emerging LRs show pBIR1:GUS expression throughout the primary root vasculature adjoining the emerging LRP (Figure 5G). The auxin influx carrier LIKE AUXIN-3 (LAX3) is important for proper LR development and is expressed not in developing LRP but in cortical cells (Swarup et al. 2008); likewise, BIR1 may function in flanking cell layers.
pBAK1:GUS-expressing plants show extensive staining throughout the root, including strong expression within the meristematic zone of the root tip, consistent with previous studies (Figure 5, P and Q) (Du et al. 2012). Expression is also noticeable within developing LRP (Figure 5H). In mature root tissue, pBAK1:GUS plants exhibited an expression pattern similar to pBIR1:GUS plants, and primarily the cortical cell layer stained (Figure 5R).
Within the differentiation zone, pBAK1:GUS expression extended throughout the radial layers and the vasculature except for the metaxylem (Figure 5S). RAM tissue showed strong staining throughout all cell layers, including the columella (Figure 5, T–V).
We conclude that BIR1 and BAK1 have overlapping expression domains within the root, specifically expression within the vasculature of the differentiation zone and within the cortical cell layer of more mature tissue, as well as overlapping expression within the LRC of the RAM.
Effects of bak1; bir1 mutants on BR and defense signaling pathways
Because BAK1 is a critical component in positive regulation of immune response pathways and contributes significantly to steroid signaling pathways, bir1-1 single mutants and bak1; bir1-1 double mutants were tested for sensitivity to altered steroid levels and induction of immune response by pathogen elicitor. To test sensitivity to a reduction in endogenous BR levels, plants were grown on medium containing the BR biosynthesis inhibitor brassinazole (BRZ) or DMSO control, followed by growth vertically to assess effects on primary root growth.
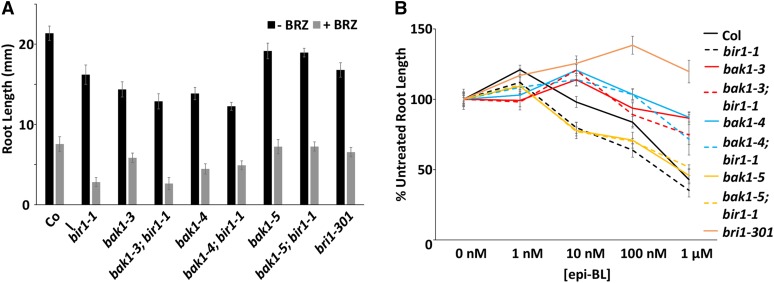
BRZ significantly reduces primary root growth of wild-type plants. Growth on BRZ-containing medium has a greater effect on bir1-1 than on Col (Figure 6A). This is similar to what is seen for bak1-4, as well as bak1-3; bir1-1 and bak1-4; bir1-1; however, bak1-5; bir1-1 roots are similar to wild type in the presence of BRZ (Figure 6A).
Figure 6.
Effect of BL and BRZ on bir1-1 and bak1 hypocotyl and root growth. (A) Average root length of 7-dag seedlings grown on medium containing 1 µM BRZ or control. (B) Average root length of 7-dag seedlings grown on medium containing 1, 10, and 100 nM and 1 µM epi-brassinolide or control normalized to control length. Error bars, SE.
Treatment of seedlings with the steroid epi-brassinolide (epi-BL) causes stimulatory effects at low concentrations and inhibitory effects at high concentration in primary root growth assays. Stimulatory effects of epi-BL on bak1-3 and bak1-4 primary root growth is shifted to a higher concentration, in this case from 1 to 10 nM epi-BL, and shows reduced inhibition at 100 nM and 100 µM (Figure 6B). bri1-301 exhibits only stimulation of root growth, as expected. bir1-1 and bak1-5 are similar in trend to Col, but the stimulatory effect at 1 nM is somewhat reduced, as is the case for bak1-5; bir1-1 (Figure 6B). bak1-3; bir1-1 responses resemble those of bak1-3 plants (Figure 6B). Unlike bir1-1, bak1-4; bir1-1 plants display a dampened response with a less distinct stimulatory peak (Figure 6B). These results suggest that BIR1 may play a minor role in BL stimulation of root growth at low concentrations. Based on these results, we suggest that either BIR1 has BAK1-dependent functions in hypocotyl elongation or that BIR1 may affect the interaction of BAK1 with another receptor or receptors that also function in hypocotyl elongation.
To ascertain the effect of bir1-1 and bak1; bir1 on PTI pathways, seedlings were grown in the presence or absence of the pathogen elicitor flg22 peptide. In wild-type plants, flg22 treatment activates the immune response through its ligand-binding receptor FLS2. Activation of this pathway results in seedling growth inhibition (SGI), which can be quantified by measuring the fresh weight of treated seedlings.
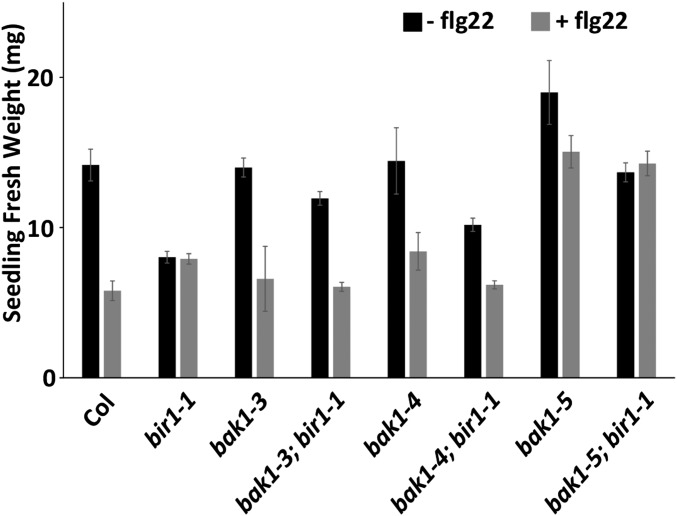
Col seedlings average a reduction in fresh weight to 41% of untreated seedlings in the presence of flg22 (Figure 7). For bir1-1 single-mutant plants, treated and untreated fresh weights do not differ significantly from each other and are 57 and 56% of that of untreated Col, respectively. Both treated and untreated bir1-1 fresh weights are significantly greater than that of treated Col (P < 0.01). flg22-treated bak1-3 and bak1-4 single-mutant plants show a reduction to 47 and 58% of their untreated fresh weights and do not differ significantly from treated Col. As reported previously, bak1-5 single mutants are partially insensitive to flg22 treatment, with a treated bak1-5 fresh weight that is 79% of the untreated fresh weight. Furthermore, bak1-5 untreated fresh weight tended to be greater than untreated Col fresh weight at 134%. For all bak1; bir1-1 double-mutant plants, untreated fresh weight is restored to a level similar to that of Col and significantly different from the bir1-1 untreated fresh weight (bak1-3; bir1-1 and bak1-4;bir1-1: P < 0.05; bak1-5; bir1-1: P < 0.001) (Figure 7). In addition, treated bak1-3; bir1-1 and bak1-4; bir1-1 fresh weights reverted to levels similar to that of treated Col and are significantly less than treated bir1-1 average fresh weight (bak1-3; bir1-1: P < 0.05; bak1-4; bir1-1: P < 0.01). bak1-5; bir1-1 treated fresh weight resembled treated bak1-5 fresh weight (Figure 7).
Figure 7.
Flg22-induced seedling growth inhibition . Seedling growth inhibition triggered by flg22 or mock treatment for the indicated genotypes. Results are average ± SE (n ≤ 4 pools of four seedlings).
These data show that bir1-1 single mutants are insensitive to flg22-induced SGI and, furthermore, that bir1-1 fresh weight is more similar to the fresh weight of wild-type plants treated with flg22 than wild-type plants in the absence of peptide. In all bak1; bir1-1 double-mutant combinations, loss of BAK1 function is sufficient to restore untreated fresh weight to a level similar to untreated Col fresh weight. In addition, bak1; bir1-1 treated seedlings resemble their respective treated bak1 single mutants in fresh weight, suggesting that the pathways stimulated by flg22 may already be active in bir1-1 seedlings.
Discussion
In this study, we have characterized novel root phenotypes of bir1 mutants. First, a significant number of bir1-1 seedlings are defective in primary root elongation, which is accompanied by changes in the columella cells (Figure 2A and Figure 4). In addition, analysis of bir1-1 LR development shows that bir1-1 seedlings accumulate early-stage LRP and, in addition to a lack of apparent mature LRs, show a reduction in later-stage LRP (Figure 2B and Figure S3).
Given the physical interaction of BIR1 and BAK1, and in order to put these, as well as previously published bir1-1 phenotypes, into a functional context, we tested the genetic relationship between bir1 and bak1. Examining bak1; bir1 double mutants generated using an allelic series of BAK1 mutations, we found that bak1 alleles acted either to suppress or enhance bir1-1 phenotypes in bak1; bir1 double-mutant plants. Specifically, bak1-3, bak1-4, and bak1-5 mutations suppressed bir1-1 plant height, leaf length and width, cell death, and LR phenotypes. For PTI pathways, bak1 mutations suppressed the insensitivity of bir1-1 to seedling growth inhibition by the pathogen elicitor flg22 in bak1; bir1-1 double mutants. In the steroid signaling pathway, bak1-3 and bak1-4 mutations had an enhancing effect on bir1-1, with bak1-3; bir1-1 and bak1-4; bir1-1 genotypes showing hypersensitivity to BRZ-mediated steroid reduction affecting primary root growth and a loss of BL stimulatory effects on primary root growth at low concentrations.
A previously published model of BIR function as a negative regulator of R-proteins is not sufficient to explain the suppression of bir1-1 by bak1 seen in this study (Gao et al. 2009). In the previous model, BIR1 acted to negatively regulate an R-protein, and in bir1-1, this R-protein activated cell death EVR (Gao et al. 2009; Wang et al. 2011). Suppression of bir1-1 phenotypes by bak1 mutations would require that control of this R-protein is regained owing to loss of BAK1-mediated PTI signaling or that BAK1 acts downstream of this R-protein. Because BAK1 is targeted by AvrPto, the possibility exists that BAK1 and BKK1 may function as R-protein guardees, and in bak1-4; bkk1 double mutants, when both BAK1 and BKK1 are lost, defense response is activated, but this does not explain the suppression of bir1-1 by bak1 and, in particular, the ability of bak1-5 to suppress bir1-1.
In the previously proposed model for BAK1/BKK1 control of cell death, BAK1 and BKK1 are redundantly required to regulate an unknown RLK (He et al. 2007). If this unknown RLK regulates cell death, in bak1-4; bkk1 double mutants this unknown RLK could be either a negative regulator of cell death that cannot function or a positive regulator of cell death that BAK1 and BKK1 inhibit. In this scenario, BIR1 might be required for interaction between BAK1/BKK1 and this unknown RLK. bak1 suppression of bir1-1 excludes the unknown RLK as a negative regulator because it would be nonfunctional in both bir1-1 and bak1; bir1-1. Thus far, no negative regulatory role of BAK1 has been identified in BAK1-RLK interactions, making BAK1/BKK1 inhibition of a positive regulator unlikely. Furthermore, suppression of cell death in bak1-5; bir1-1 would require BAK1-5 to be defective in this interaction, but bak1-5; bkk1 double mutants do not show cell death phenotypes. That bak1-5; bkk1 double mutants do not show cell death phenotypes and that bak1-5; bir1-1 double mutants show suppression of bir1-1 cell death phenotypes argue that bir1-1 phenotypes are not due to an unknown LRR-RLK that regulates cell death in a BAK1-dependent manner.
If BIR1, BAK1, and BKK1 acted together in a complex to negatively regulate cell death, it would be expected that bir1-1 phenotypes would be (1) as severe as bak1-4; bkk1 double mutants, (2) would not be suppressed by high temperature, or (3) bak1-4; bkk1 double mutants would be suppressed by high temperature, but none of these are the case.
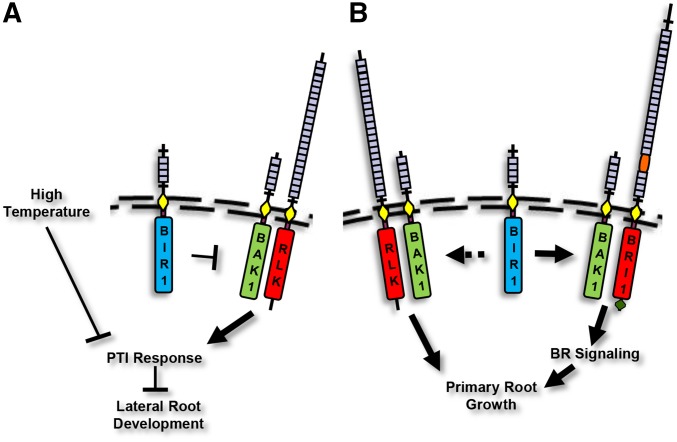
Based on our data, we propose that BIR1 negatively regulates a cell death pathway that, in part, depends on BAK1-mediated PTI signaling and in part on a separate pathway that is negatively regulated by high-temperature stress (Figure 8A). These pathways are independent of each other because neither high-temperature stress nor loss of BAK1 PTI function in bak1-5 is sufficient to suppress bir1-1; however, when combined, bak1-5; bir1-1 plants are fully suppressed. The ability of bak1-5 to suppress bir1-1 indicates that BAK1 is downstream of BIR1. Given that BAK1 is ubiquitously expressed, functions as a coreceptor positively regulating PTI through several pathways (Postel et al. 2010), and is likely acting in putative RLK-mediated PTI pathways for other pathogens, BAK1 participation in these pathways must be tightly regulated. BIR1 may function to negatively regulate BAK1 interaction in PTI signaling pathways to prevent precocious or inappropriate activation.
Figure 8.
Graphical model of BIR1 and BAK1 function . (A) BIR1 acts as a negative regulator of BAK1 function as a coreceptor for ligand-binding RLKs, such as FLS2, that control pathogen-triggered immune response pathways that include EDS1 and PAD4. High temperature also negatively regulates this response, and LR development is negatively affected by PTI response. (B) BIR1 positively regulates BAK1 function as a coreceptor for BRI1, positively regulating primary root growth, and BIR1 also may positively affect BR-independent root growth via BAK1/SERK interaction with an unknown RLK.
It has been proposed that BIR2, a closely related receptor to BIR1, functions to sequester BAK1 from interaction with PTI RLKs in the absence of ligand. bir2 knockdown plants exhibit increased PTI signaling, increased FLS2-BAK1 association, and enhanced cell death on pathogen infection compared to wild type, and overexpression of BIR2 leads to decreased FLS2-BAK1 association (Halter et al. 2014). BIR1 may be acting similarly in negatively regulating BAK1 association with primary receptors for PTI. Given the recent data that flg22 treatment leads to a reduction in LR density (Beck et al. 2014), the observed defect in bir1-1 LR development, and the suppression of this defect in bak1-5; bir1-1 plants, it is likely that this defect may be caused by inappropriate BAK1-dependent PTI signaling. Further analysis will be necessary to uncover this link.
Unlike the bir1-1 LR defect, bir1-1 primary root defects are enhanced rather than suppressed by bak1-3 and bak1-4 mutations. In addition, bir1-1 dampens responsiveness to BL root growth stimulation. From these data, we cannot yet discern whether BIR1 functions in mediating BAK1 participation in BL-dependent and -independent root growth but propose that BIR1 acts to positively regulate BAK1 in these pathways (Figure 8B). As our knowledge of RLK function in plants increases, BIR1 and, from recent studies, BIR2 appear to represent a novel role for RLKs as regulators of the interaction between coreceptors and primary ligand-binding receptors.
Acknowledgments
This research was supported by NSF IOS-1257316. Thanks to Jaimie Van Norman and Birgit Kemmerling for their comments on the manuscript, and to Cyril Zipfel for providing bak1-5 seeds.
Footnotes
Communicating editor: S. Poethig
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.180380/-/DC1
Literature Cited
- Albrecht C., Russinova E., Kemmerling B., Kwaaitaal M., de Vries S. C., 2008. Arabidopsis somatic embryogenesis receptor kinase proteins serve brassinosteroid-dependent and -independent signaling pathways. Plant Physiol. 148: 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M. R., Chiu W. L., et al. , 2002. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. [DOI] [PubMed] [Google Scholar]
- Beck M., Wyrsch I., Strutt J., Wimalasekera R., Webb A., et al. , 2014. Expression patterns of flagellin sensing 2 map to bacterial entry sites in plant shoots and roots. J. Exp. Bot. 65: 6487–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Bielach A., 2010. Lateral root organogenesis—from cell to organ. Curr. Opin. Plant Biol. 13: 677. [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nurnberger T., et al. , 2007. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F., 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Du J., Yin H., Zhang S., Wei Z., Zhao B., et al. , 2012. Somatic embryogenesis receptor kinases control root development mainly via brassinosteroid-independent actions in Arabidopsis thaliana. J. Integr. Plant Biol. 54: 388. [DOI] [PubMed] [Google Scholar]
- Fukaki H., Tasaka M., 2009. Hormone interactions during lateral root formation. Plant Mol. Biol. 69: 437. [DOI] [PubMed] [Google Scholar]
- Gao M., Wang X., Wang D., Xu F., Ding X., et al. , 2009. Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6: 34–44. [DOI] [PubMed] [Google Scholar]
- Gou X., He K., Yang H., Yuan T., Lin H., et al. , 2010. Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genomics 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X., Yin H., He K., Du J., Yi J., et al. , 2012. Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 8: e1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter T., Imkampe J., Mazzotta S., Wierzba M., Postel S., et al. , 2014. The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24: 134–143. [DOI] [PubMed] [Google Scholar]
- He K., Gou X., Yuan T., Lin H., Asami T., et al. , 2007. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 17: 1109–1115. [DOI] [PubMed] [Google Scholar]
- Heese A., Hann D. R., Gimenez-Ibanez S., Jones A. M., He K., et al. , 2007. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B., Wang H., Nam K. H., Li J., Li J., 2010. Activation-tagged suppressors of a weak brassinosteroid receptor mutant. Mol. Plant 3: 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R., Boeren S., Russinova E., Aker J., Vervoort J., et al. , 2006. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18: 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerling B., Schwedt A., Rodriguez P., Mazzotta S., Frank M., et al. , 2007. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr. Biol. 17: 1116–1122. [DOI] [PubMed] [Google Scholar]
- Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., et al. , 2004. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., 2010. Multi-tasking of somatic embryogenesis receptor-like protein kinases. Curr. Opin. Plant Biol. 13: 509–514. [DOI] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K. A., Doke J. T., Tax F. E., et al. , 2002. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222. [DOI] [PubMed] [Google Scholar]
- Malamy J. E., 2005. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 28: 67. [DOI] [PubMed] [Google Scholar]
- Malamy J. E., Benfey P. N., 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33. [DOI] [PubMed] [Google Scholar]
- Nam K. H., Li J., 2002. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212. [DOI] [PubMed] [Google Scholar]
- Nurnberger T., Brunner F., Kemmerling B., Piater L., 2004. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198: 249–266. [DOI] [PubMed] [Google Scholar]
- Oh M. H., Wang X., Wu X., Zhao Y., Clouse S. D., et al. , 2010. Autophosphorylation of Tyr-610 in the receptor kinase BAK1 plays a role in brassinosteroid signaling and basal defense gene expression. Proc. Natl. Acad. Sci. USA 107: 17827–17832. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Postel S., Kufner I., Beuter C., Mazzotta S., Schwedt A., et al. , 2010. The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur. J. Cell Biol. 89: 169–174. [DOI] [PubMed] [Google Scholar]
- Roux M., Schwessinger B., Albrecht C., Chinchilla D., Jones A., et al. , 2011. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson F., Brunaud V., Balzergue S., Dubreucq B., Lepiniec L., et al. , 2002. FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30: 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B., Ronald P. C., 2012. Plant innate immunity: perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63: 451–482. [DOI] [PubMed] [Google Scholar]
- Schwessinger B., Roux M., Kadota Y., Ntoukakis V., Sklenar J., et al. , 2011. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S. H., Bleecker A. B., 2001a Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98: 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.-H., Bleecker A. B., 2001b Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE 2001: re22. [DOI] [PubMed] [Google Scholar]
- Swarup K., Benkova E., Swarup R., Casimiro I., Peret B., et al. , 2008. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10: 946–954. [DOI] [PubMed] [Google Scholar]
- Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., et al. , 2008. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kota U., He K., Blackburn K., Li J., et al. , 2008. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15: 220–235. [DOI] [PubMed] [Google Scholar]
- Wang Z., Meng P., Zhang X., Ren D., Yang S., 2011. BON1 interacts with the protein kinases BIR1 and BAK1 in modulation of temperature-dependent plant growth and cell death in Arabidopsis. Plant J. 67: 1081–1093. [DOI] [PubMed] [Google Scholar]
- Wierzba M. P., Tax F. E., 2013. Notes from the underground: receptor-like kinases in Arabidopsis root development. J. Integr. Plant Biol. 55: 1224–1237. [DOI] [PubMed] [Google Scholar]
- Wrzaczek M., Vainonen J. P., Stael S., Tsiatsiani L., Help-Rinta-Rahko H., et al. , 2015. GRIM REAPER peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. EMBO J. 34: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. Y., Sae-Seaw J., Wang Z. Y., 2013. Brassinosteroid signalling. Development 140: 1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E. J., Jones J. D., et al. , 2004. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available on request.