Abstract
Dosage-balance selection preserves functionally redundant duplicates (paralogs) at the optimum for their combined expression. Here we present a model of the dynamics of duplicate genes coevolving under dosage-balance selection. We call this the compensatory drift model. Results show that even when strong dosage-balance selection constrains total expression to the optimum, expression of each duplicate can diverge by drift from its original level. The rate of divergence slows as the strength of stabilizing selection, the size of the mutation effect, and/or the size of the population increases. We show that dosage-balance selection impedes neofunctionalization early after duplication but can later facilitate it. We fit this model to data from sodium channel duplicates in 10 families of teleost fish; these include two convergent lineages of electric fish in which one of the duplicates neofunctionalized. Using the model, we estimated the strength of dosage-balance selection for these genes. The results indicate that functionally redundant paralogs still may undergo radical functional changes after a prolonged period of compensatory drift.
Keywords: duplication, expression evolution, dosage balance, neofunctionalization, whole-genome duplication
THE fate of duplicate genes is characterized by two extremes: degeneration and the origin of biological novelty. Early models for the evolutionary dynamics of duplicates suggested that typically one member of a duplicate pair would quickly degenerate into a nonfunctional pseudogene (Haldane 1933; Ohno 1970). More rarely, a duplicate instead may evolve a novel function in a process called neofunctionalization (Muller 1936; Ohno 1970; Ohta 1987). The time scale for either pseudogenization or neofunctionalization is expected to be on the order of a few million years (Lynch and Conery 2000).
Recent research indicates, however, that the evolutionary dynamics for many duplicates are not so simple (Walsh 1995, 2003; Force et al. 1999; Papp et al. 2003; He and Zhang 2005; Rastogi and Liberles 2005; Scannell and Wolfe 2008; Qian et al. 2010; Kondrashov 2012). Some genes are dosage sensitive, meaning that a change in their copy number alters expression and disrupts the stoichiometric balance of their gene products with those of other genes. Duplicates of dosage-sensitive genes typically will fix in a population only if they originate in a whole-genome duplication (WGD), where all interacting partners duplicate together. Selection to maintain the stoichiometric relations between the products of duplicate genes, termed dosage-balance selection, can preserve duplicates as functionally redundant copies for prolonged periods of time (Birchler et al. 2001, 2005; Veitia 2002; Papp et al. 2003; Aury et al. 2006; Blomme et al. 2006; Freeling and Thomas 2006; Stranger et al. 2007; Qian and Zhang 2008; Edger and Pires 2009; Makino and McLysaght 2010; Konrad et al. 2011; Birchler and Veitia 2012; McGrath et al. 2014a).
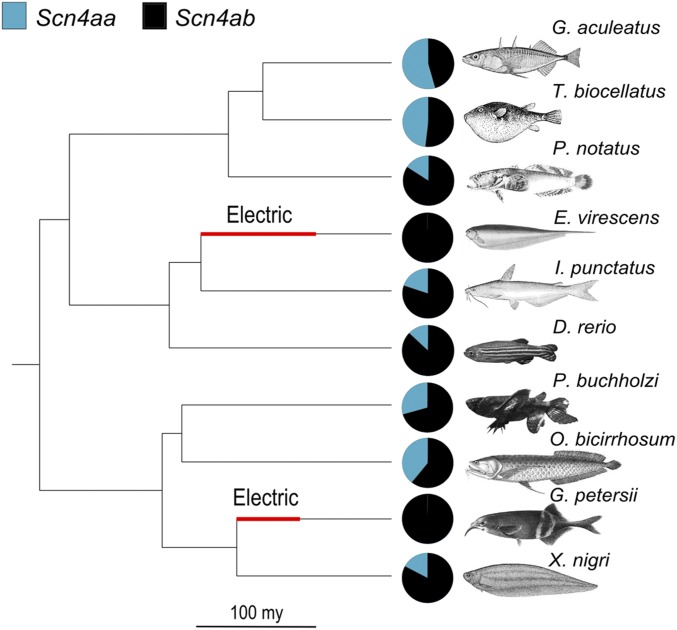
Recent data on a pair of sodium channel duplicates in teleost fish are consistent with the expectations of the dosage-balance hypothesis (Thompson et al. 2014). The two duplicates, also called paralogs, have been conserved in muscle cells for over 300 million years since the teleost-specific WGD. In two independent lineages of electric fish, however, only one of the sodium channels is expressed in muscle cells. The other duplicate neofunctionalized and now plays a key role in the electric organ (Novak et al. 2006; Zakon et al. 2006; Arnegard et al. 2010). These convergent neofunctionalization events happened on a very slow time scale, more than 100 million years after duplication (Arnegard et al. 2010; Lavoué et al. 2012; Betancur-R et al. 2013). The phylogenetic context for the evolution of the duplicates is shown in Figure 1.
Figure 1.
Sodium channel expression and phylogenetic relationships of 10 teleost fish species. The families represented here span almost the entire teleost clade. The relative expression in skeletal muscle of the voltage-gated sodium channel genes Scn4aa and Scn4ab is represented with pie charts for each of the 10 species. Thick red segments on the branches leading to the two electric fish Eigenmannia veriscens and Gnathonemus petersii indicate the likely times when electric organs evolved and Scn4aa neofunctionalized (Arnegard et al. 2010; Betancur-R et al. 2013).
Thompson et al. (2014) proposed that in the teleost ancestor the duplicates were preserved after WGD by dosage-balance selection. They hypothesized that under this selective constraint, one paralog gradually drifted to lower expression levels, while the other compensated by evolving higher expression. Eventually, one paralog contributed so little to its original function that it could be neofunctionalized in the electric organ without major compromise to muscles. This mode of evolution also may explain comparative expression patterns observed in some ciliates (Gout and Lynch 2015) as well as some mammals (Lan and Pritchard 2015). This hypothesis raises theoretical and quantitative issues not previously explored. Can dosage-balance selection in fact maintain duplicates for hundreds of millions of years? Will this mode of evolution produce comparative patterns in a phylogeny that are distinct from other models? And how does this evolutionary process affect the likelihood of neofunctionalization?
Here we develop a model for the evolution of paralog expression under dosage-balance selection. It envisions a process, which we call compensatory drift, in which paralogs diverge by weakly selected mutations that fix largely by drift. The model shows how key genetic parameters determine the time scale over which duplicates are maintained before one is lost or neofunctionalizes. The evolutionary dynamics are determined by just two compound parameters. The first is a speed parameter that relates mutation, selection, and random genetic drift to the rate at which the duplicates’ expression diverges. The second is a threshold parameter that determines the point at which expression of one duplicate is sufficiently low that it is largely relieved from dosage-balance constraints and free to evolve a novel function. The model predicts two phases of evolution. In the initial phase, the difference in expression between functionally identical paralogs drifts randomly while their combined expression remains nearly constant. In the second phase, the expression threshold is reached, and one of the duplicates quickly accrues function-altering substitutions.
We fit the compensatory drift model to data from Thompson et al. (2014) on the expression of sodium channel duplicates in 10 families of teleost fish. Our estimate for the speed parameter is consistent with what is known about the biological parameters that feed into it, suggesting that compensatory drift is a plausible model for sodium channel evolution. Our estimate for the threshold parameter is, to our knowledge, the first available. Finally, we demonstrate that dosage-balance selection can greatly enhance the probability of neofunctionalization compared to the classic neutral scenario. These results suggest that WGD, as well as contexts in which dosage-balance selection acts, may be a particularly rich source of genetic novelty for geologically long periods of time.
Materials and Methods
The model
After duplication, stabilizing selection favors an optimal total expression of two paralogs. A mutation that affects expression of either one will either increase or decrease fitness depending on whether it brings total expression closer to or further from the optimum. Mutations also experience random genetic drift, so there is a nonzero probability that both mildly deleterious and beneficial mutations will be established.
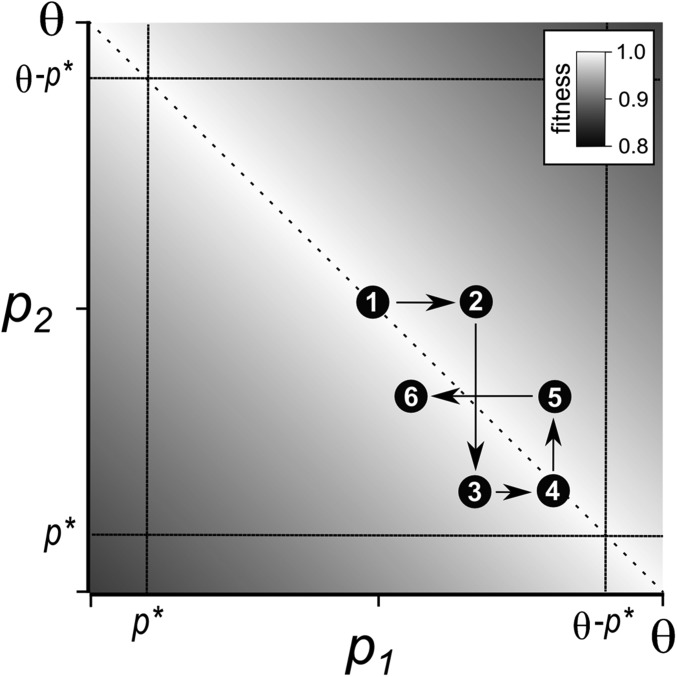
We visualize compensatory drift as a series of fixation events that change the expression of the duplicates. A schematic of the process is shown in Figure 2. The two paralogs evolve in an anticorrelated pattern. Mutations in one duplicate can move the total expression away from its optimum. Compensatory mutations in the other duplicate tend to move total expression closer to the optimum. The result is that total expression remains close to the optimum, while the difference in their expression fluctuates randomly. The state of the population at any time is described by the total expression of the two duplicates and the difference in expression between them. If expression evolves to a point at which one of the duplicates produces the bulk of the gene product, selection is no longer strong enough to prevent function-altering substitutions from accruing in the paralog with lower expression. This threshold can be interpreted as either the point where pseudogenization quickly occurs or where the benefit of neofunctionalization outweighs the fitness tradeoff from loss of the ancestral function.
Figure 2.
Schematic of the coevolution of paralog expression under compensatory drift. Axes show the expression of the duplicate genes, and fitness is represented with a gray scale. The dashed diagonal line shows the maximal fitness where p1 + p2 = θ. The dotted vertical and horizontal lines show the expression thresholds where a duplicate loses its original function. A sequence of several consecutive expression changes is shown with numbered circles.
Assumptions
The expression levels of two duplicates are denoted as p1 and p2. We assume that stabilizing selection acts on the sum of expression . The fitness function acting on A is proportional to a normal distribution with mean equal to the optimum for total expression θ and with variance ω2 (which are assumed to be constant in time). The variance parameter determines the strength of selection, where larger values of ω2 indicate weaker stabilizing selection. No selection acts on the difference in expression .
Mutations occur in the regulatory regions of each of the four gene copies at a rate µ per generation. They evolve according to a Fisher-Wright model of drift and selection. Mutations enter the population at a rate of 4Nμ, where N is the population size. Their effects on expression are additive. The effect of a given mutation on p1 or p2, which we denote as δ, is normally distributed with mean 0 and variance . We therefore assume that the distribution of mutational effects is constant in time and independent of a gene’s current level of expression. Biologically, this means that the regulation of expression is free of complicated forms of epistasis.
New mutations are either lost or fixed under the combined forces of selection and drift. We assume that mutation is weak (), so there is a negligible chance that more than one mutation will be segregating. (We will return to this point in the Results section, which suggests that the model also may be a good approximation when that assumption is violated.) Evolution thus proceeds by a series of fixation events at the two loci. This is a Poisson process, and the waiting time between mutations is exponentially distributed with mean of generations.
We calculate the fixation probability for each mutation using the diffusion approximation of Kimura (1962)
| (1) |
where s is the selection coefficient of the new mutation
| (2) |
Equation 2 is an approximation that neglects the deviation of the population from the optimum θ. The approximation is valid when the SD of mutational effects is large relative to the typical deviation from the optimum. We verified the accuracy of the approximation using parameter values consistent with the data on teleost sodium channels from Thompson et al. (2014) (Supporting Information, File S1).
We assume that when the duplication occurs, the two paralogs have equal expression, and their total expression is optimal (D = 0 and A = θ). As evolution proceeds, expression of the duplicates eventually will fall to a threshold level, denoted p*, while its paralog rises to θ − p*. At this point, the paralog with lower expression rapidly either becomes a pseudogene or neofunctionalizes. This threshold is represented in our model by a critical difference in the expression of the duplicates D* = θ − 2p*. If D reaches either D* or −D*, then one or the other duplicate loses its original function.
Evolutionary dynamics
Our goal is to determine the probability distribution for expression levels at times following the duplication event. Simulations reveal that under plausible parameter values, evolutionary trajectories are confined to values of A that are very close to θ (File S1). This suggests that the dynamics can be approximated by a one-dimensional diffusion in the expression difference D = p1 − p2. We write the probability density of D at time t following the duplication as . The evolution of the density function is described by
| (3) |
This is the heat equation (Cox and Miller 1965), where is the diffusion parameter. This parameter determines the speed at which D evolves, and it equals the rate of increase in variance of the probability distribution D per generation. File S2 shows that
| (4) |
where k is a constant that is independent of the model’s parameters. It is difficult to calculate analytically, so we determined its value (k ≈ 1.543) numerically (File S2).
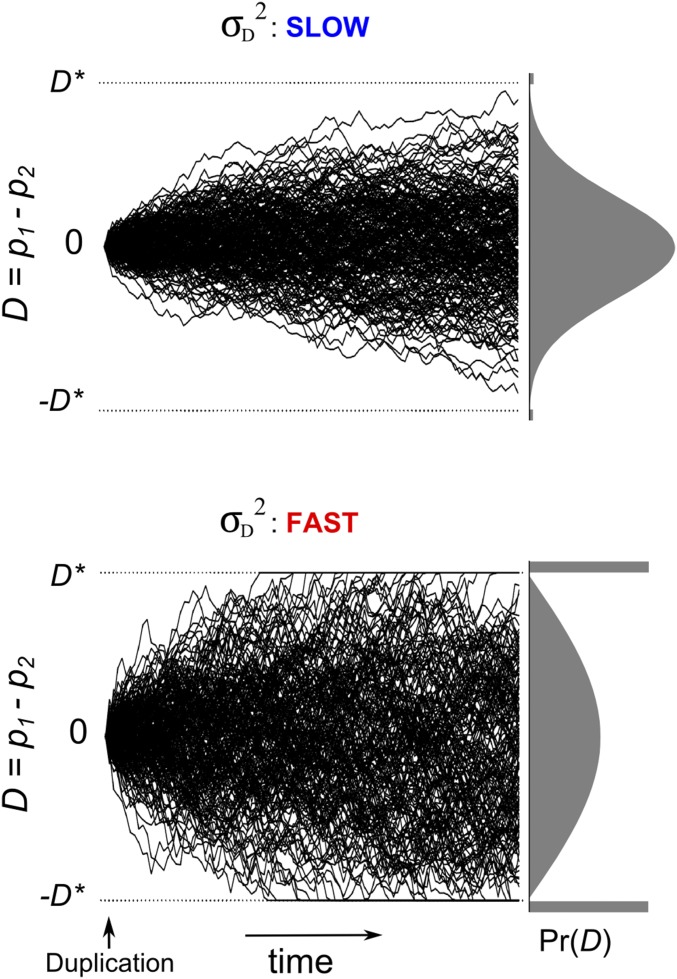
From Equation 4, we gain insight into the impact of biological parameters on the speed at which D evolves. Imagine that we follow a set of evolutionary lineages that began to diverge independently after the duplication event. The variance in the distribution of D initially increases at a constant rate and is equal to at t generations after duplication. Thus, the diffusion rate sets the speed of divergence, as illustrated in Figure 3. Equation 4 shows how the biological parameters affect this speed. The speed is reduced by larger population sizes. Larger values of N cause a greater number of mutations to enter the population in each generation but also increase the efficiency of purifying selection; the net result is that a smaller number of mutations fix (see Equation 1). Equation 4 also shows that the speed of divergence increases with higher mutation rates (larger μ) and decreased strengths of selection (larger ω2). A somewhat counterintuitive result is that the speed of divergence declines as the average effect size of mutations σm increases. This is so because larger mutations are more likely to be strongly deleterious and therefore very unlikely to fix.
Figure 3.
The speed of divergence in the expression of duplicate genes. Examples of the evolution of expression difference D (= p1 − p2) when the diffusion parameter is small (top) and large (bottom). Each panel shows sample trajectories and the final probability distribution for D (at right). In the top panel, the final distribution of D is approximately normal; neofunctionalization and pseudogenization are rare. In the bottom panel, neofunctionalization is frequent (large rectangles outside the thresholds at ±D*).
To summarize the model, the probability density of D evolves according to Equation 3, with initial condition D = 0 at t = 0 and with absorbing barriers at ±D*. Before doing any further analysis, Equation 3 tells us a simple but important fact: although the model is based on six underlying biological parameters (μ, ω, σm, N, θ, and p*), the evolutionary dynamics are governed by only two: the speed parameter and the threshold D*.
The solution for the density function of D is
| (5) |
for −D* < D < D* (Cox and Miller 1965). The probability that one of the duplicates has either been lost as a pseudogene or has neofunctionalized after t generations is
| (6) |
where Φ denotes the standard normal cumulative distribution function (Cox and Miller 1965).
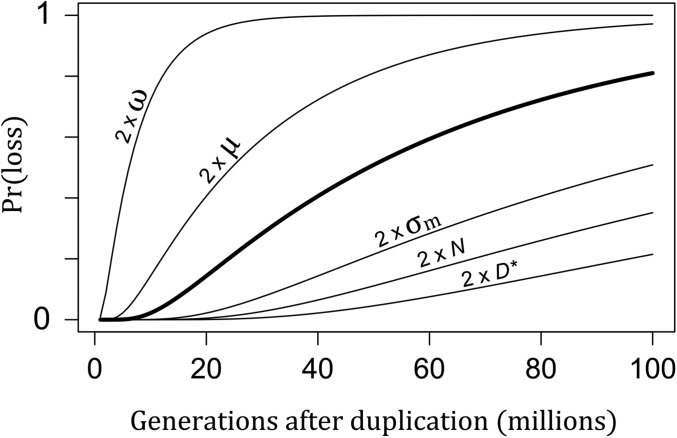
With Equation 6, we can infer how varying the biological parameters in Equation 4 affect the expected life span of duplicate genes. Figure 4 shows the result of varying each biological parameter of the model on the time scale of duplicate loss. These results imply that the time between duplication and loss can be very long, especially for large populations, genes under strong dosage constraints (small ω), and genes with high expression (large D*). To get some idea of the time scale, we can calculate the number of generations needed to reach a probability of 1/2 that one of the duplicates is lost using parameter values that are plausible for the electric fish clades: a population size N = 104 and a mutation rate μ = 10−5. The strength of stabilizing selection () is such that 90% of mutations are strongly deleterious () and so have negligible chance of fixation. The threshold is D* = 5σm, which means that following duplication, the threshold p* could in principle be reached with the fixation of just five mutations of typical size. (As we will see, however, this does not happen because most mutations are eliminated by dosage-balance selection.) Under these assumptions, we find from Equation 6 that after 1.7 billion generations, there is still a 50% probability that neither gene will have been lost. Thus, dosage-balance selection can maintain functional paralogs for very long evolutionary periods. If we decrease the strength of dosage-balance selection such that half the mutations are nearly neutral (), the amount of time decreases dramatically to just 11 million generations.
Figure 4.
The probability of gene loss after duplication. The thick curve shows the probability of loss, either through pseudogenization or neofunctionalization, over time with standard parameter values: the selection strength is ω2 = 104, the SD of the mutation effect size is σm = 1, the population size is N = 104, the mutation rate is μ = 10−5, and the expression threshold is D* = 100. Other curves show results when individual parameter values are doubled.
Data availability
Simulation code is available upon request.
Results
Fitting the model to data
To assess the plausibility of this model and to estimate parameters of biological interest, we fit this model to data on the expression of sodium channel duplicates from Thompson et al. (2014). The data are the relative expression levels of the two teleost-specific paralogs in 10 families of fish sampled broadly across the entire teleost clade and the phylogenetic relations between those families (Figure 1). The parameters being fit are the diffusion rate and the threshold for gene loss D*.
We used approximate Bayesian computation (ABC) because it allows inferences about models that are too complicated for statistical frameworks such as likelihood (Tavare et al. 1997; Beaumont et al. 2002; Beaumont 2010). The basic approach is to compare summary statistics measured from simulated data to the same statistics measured from real data. Estimates for the parameters are given by the values that produce simulated data sets that are most similar to the real data. In practice, this is accomplished by choosing values for the model parameters from prior distributions, simulating data using the model with those values, and comparing the summary statistics that result with those from the real data. The parameter values used in the simulation are rejected from the posterior distribution if the summary statistics from the real and simulated data sets are not sufficiently similar.
We simulated the evolution of expression on the phylogeny under the model described earlier. The output of the simulation gave the identities of the lineages (if any) that lost one of the paralogs to muscle function and the relative expression of the two paralogs for those lineages that have not. These results were compared to the actual data using two types of summary statistics. The first, which is binary, is determined by whether neofunctionalization occurred in the same locations on the tree as observed in the data. We rejected all simulations in which this pattern was not observed. The second kind of summary statistic was the independent contrasts (Felsenstein 1985) at the nodes of the phylogeny for the relative expression of the duplicates in the nonelectric fish. We rejected simulations if the Euclidean distance of the independent contrasts between the real and simulated data exceeded a threshold. Further details are given in File S3.
Including the electric fish data in the analysis upwardly biases our estimate of the probability of neofunctionalization. (The families of fish in the data set are not randomly chosen: it intentionally includes the only two families in which neofunctionalization is known.) To address this issue, we performed ABC analysis both with the electric fish and without them. Excluding the electric fish biases the estimate in the opposite direction, and therefore, the two analyses give boundaries for our estimates of model parameters.
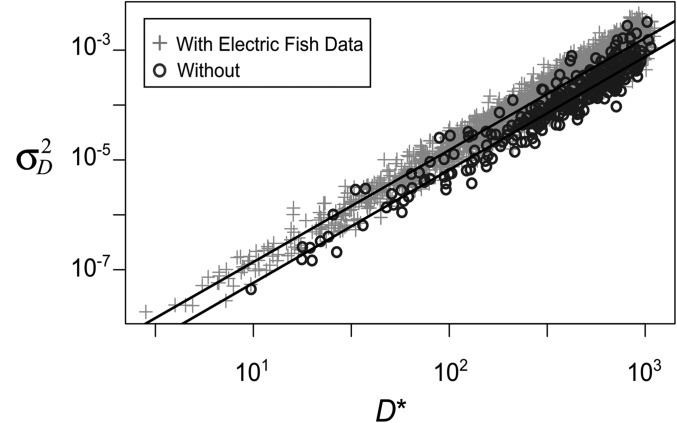
The joint posterior distributions for the diffusion rate and the threshold for gene loss D* from the two analyses are shown in Figure 5. The distributions are quite similar. On a log-log plot, the values of and log D* are strongly correlated. The data are consistent with either small values of the speed parameter and the threshold D* or with large values of both parameters.
Figure 5.
Joint posterior distribution of the diffusion rate parameter and the expression threshold D* estimated from sodium channel expression in teleost fish. The joint distributions are from two ABC analyses using the expression data from Thompson et al. (2014). In one analysis, the two lineages of electric fish are included, and in the other, they are not. The linear relationship between the parameters from the two analyses is very similar. The regression lines are log = 2.04 log D* −20.5 with the electric fish and log = 2.06 log D* − 21.3 without them.
We can use published information about absolute gene expression levels to refine the likely range of values for these parameters. Promoter and enhancer mutation studies suggest that gene expression levels may be on the order of 10σm to 100σm (Melnikov et al. 2012; Patwardhan et al. 2012; Metzger et al. 2015). The data from Thompson et al. (2014), in conjunction with estimates of the distribution of transcript levels in eukaryotic cells (Mortazavi et al. 2008; Islam et al. 2010; Schwanhäusser et al. 2011; Marguerat et al. 2012), suggest that a conservative lower limit for D* is 3σm (see File S3 for details). Letting D* vary between 3σm and 102σm, we used the linear regressions shown in Figure 5 to determine a range of plausible values for . We estimate that if only three substitutions of typical size are needed to reduce a paralog’s expression to the threshold (D* = 3σm), then the expected value of is 5.4 × 10−9σm per year. If expression is much larger, such that 100 substitutions of typical size are required to reach threshold, then the expected value of is 1.5 × 10−5σm per year.
We explored what these results imply about the biological parameters on which the model is based. We began by estimating the strength of dosage-balance selection on the sodium channels. We assumed the range of values for just described, that μ lies between 10−6 and 10−4 per allele per generation, that N lies between 104 and 106, and that there is one generation per year. Equation 4 and these parameter values then imply that the variance of the fitness function ω2 is between and . We can use plausible expression levels derived from the studies cited earlier to estimate how efficient dosage-balance selection is at removing expression mutations in terms of transcripts per cell. If the sodium channels are expressed at 50 transcripts per cell and σm is 5% of that expression level, for example, then the estimated values of ω2 imply that mutations that change sodium channel expression by more than 5.3 transcripts per cell are efficiently eliminated by dosage-balance selection.
Next, we asked about the properties of mutations that fix. We simulated the compensatory drift process using the parameter values cited in the preceding paragraph (see File S1 for details). These results show that for small values of D* (= 3σm) and strong dosage-balance selection (ω2 = ), 97% of mutations are removed by selection that otherwise would fix. On average, mutations that fix change expression by only 0.02σm, and some 9000 substitutions occur before one of the duplicates becomes a pseudogene or neofunctionalizes. For a larger value of D* (100σm) and very weak selection (ω2 = ), only 17% of mutations are prevented from fixing by dosage-balance selection. The effect of the average mutation that fixes is 0.7σm, and 8000 substitutions occur before the threshold is reached. We emphasize that these estimates are very rough, but they are, to our knowledge, the first for these important evolutionary parameters.
We find that if dosage-balance selection is strong (ω not very much bigger than σm), then the parameter estimates for the sodium channels are consistent with the assumptions of one-dimensional diffusion approximation. With weak selection, however, the approximation breaks down. This is so because total expression can deviate substantially from the optimum so that the dynamics are not well approximated by a one-dimensional diffusion. Our model therefore describes the evolutionary dynamics of these sodium channel duplicates if D* and ω are not very much larger than σm but would be more accurately modeled by a two-dimensional diffusion model if they are not. It may be difficult to develop analytic results for this model, but it could be studied numerically.
Stochastic simulations suggest that our results are surprisingly robust to the assumption that no more than one mutation segregates at any given time (i.e., ). Simulations of a Wright-Fisher model show that mutations that fix do so largely as a neutral process. The distribution of fitness effects for fixed mutations is shown in File S1. For the parameter values simulated, the mean value of is between 0.15 and 0.24, and it is very rare for mutations to fix with . We ran simulations in which the mutation rate varied over more than four orders of magnitude. When Nµ = 1, the most common allele is typically at a frequency of only about 50% (File S2). Nevertheless, the substitution rate is very close to what our model predicts (File S3). This behavior is also consistent with a model in which mutations that segregate at appreciable frequencies are entirely neutral. The results of the simulations begin to significantly depart from the expectations of our model only when Nµ > 1. In sum, our analytic results may apply when mutation rates are higher than the approximations assume.
Neofunctionalization and compensatory drift
Because dosage-balance selection can maintain duplicates for long evolutionary periods, it may be more likely that neofunctionalization will occur than it does when dosage balance is weak or absent (Force et al. 1999; Papp et al. 2003; Aury et al. 2006; Hughes et al. 2007; Scannell and Wolfe 2008; Thompson et al. 2014; Gout and Lynch 2015). To explore this idea further, we extended our model by adding two new kinds of mutations. The first is a loss-of-function mutation that renders one of the duplicates a pseudogene. The probability that it fixes is again given by the fitness function used in the main model. The second kind of mutation neofunctionalizes one of the duplicates. It suffers the same fitness cost as a loss-of-function mutation but also benefits from a 0.1% fitness gain from its new function.
We compared the frequency of neofunctionalization in three simulated populations (File S1) evolving under dosage-balance selection that ranged from strong to very weak: ω2 = , , and . For all three simulations, mutations that alter expression were 10 times more frequent than pseudogenizing mutations, and pseudogenizing mutations were 103 times more frequent than neofunctionalizing mutations. The population size was N = 104, the mutation rate was μ = 10−5 mutations per allele per generation, and the optimal expression was θ = 5σm.
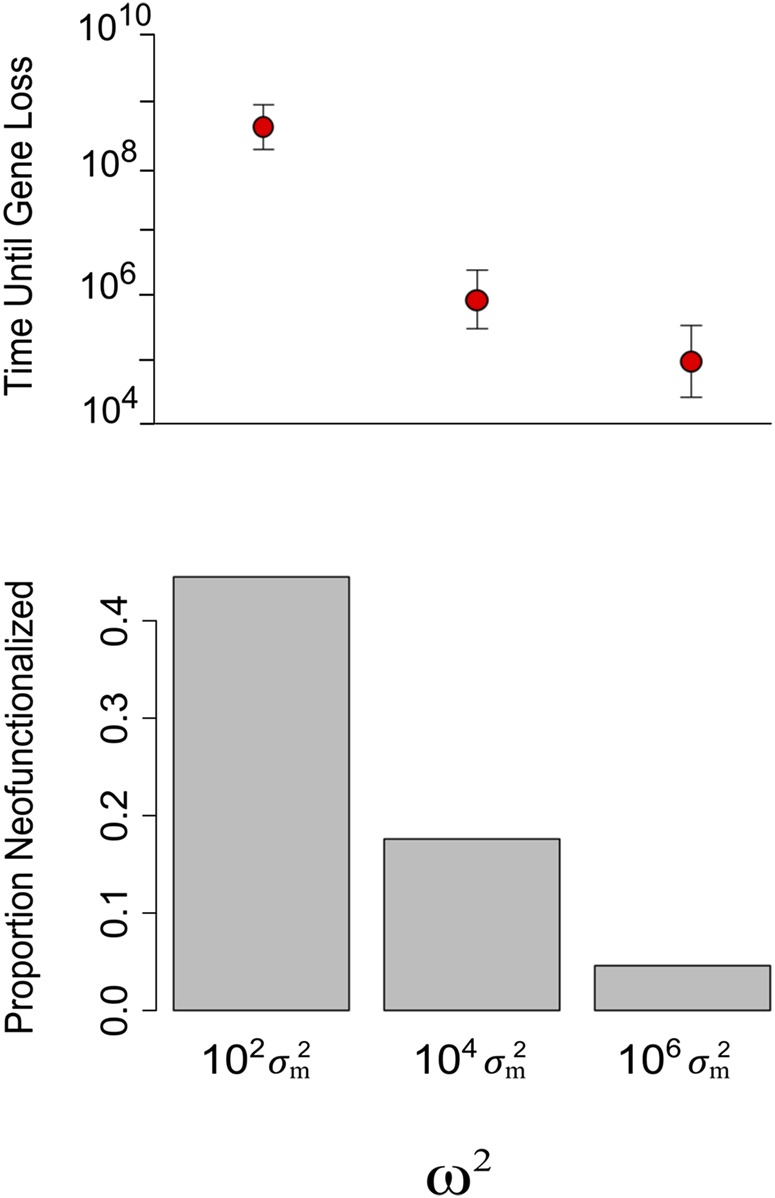
We found that neofunctionalization is greatly facilitated by dosage-balance selection. Figure 6 shows that when dosage-balance selection is stronger, duplicate genes are preserved for longer, and more mutations occur before a duplicate is lost. In consequence, neofunctionalization happens nearly 10 times more often than when dosage-balance selection is very weak. Neofunctionalization is most likely when expression falls inside a window of values in which the cost of losing the original function is smaller than the benefit of gaining the new function. In this window, a mutation that causes pseudogenization is still too deleterious to fix. Equation 4 shows that stronger dosage-balance selection slows the rate of compensatory drift and thus increases the amount of time the population spends in this evolutionary window. These results suggest that dosage-balance selection greatly diminishes the evolutionary potential of paralogs early after duplication but, after a long period of compensatory drift, greatly facilitates the acquisition of a new adaptive function.
Figure 6.
Dosage-balance selection and the probability of neofunctionalization. The top panel shows the number of mutations (a proxy for time) that occur before one of the duplicates neofunctionalizes or pseudogenizes. Dots show the mean, and the whiskers show 1 SD. The bottom panel compares the frequency of neofunctionalization for three strengths of dosage-balance selection, from strong to very weak. Results are based on 103 simulations.
Discussion
We formalized a model of the evolutionary process that we call compensatory drift. This model shows how dosage-balance selection on duplicate genes (paralogs) can lead to neofunctionalization some tens to hundreds of millions of years after duplication. Dosage-balance selection constrains the combined expression of both paralogs to an optimum, but not the expression of the individual genes. This allows the relative expression of the paralogs to drift apart by the fixation of mutations with small effects. The speed at which this divergence occurs is determined by the diffusion rate , which, in turn, is a function of several biological parameters. Our results show that stronger dosage-balance selection and larger mutational effects on expression slow divergence because a greater fraction of mutations is strongly deleterious and so has virtually no chance of fixation. Larger populations also decrease divergence because they enhance the efficiency of selection and so eliminate a larger fraction of mutations.
Simulations of compensatory drift reveal that dosage-balance selection can improve the odds that neofunctionalization occurs rather than pseudogenization. If a novel function yields a slight advantage while having a large tradeoff with the ancestral function, dosage-balance selection can still improve the chances of neofunctionalization, but only after a long period of compensatory drift. As expression of one paralog declines, the strength of selection to maintain its original function diminishes. It reaches a level at which mutations that pseudogenize the gene are still strongly deleterious, but mutations that neofunctionalize are beneficial. Our results show that the probability of neofunctionalization is increased when the added expression of duplicates is high, dosage-balance constraints are strong, and population sizes are large.
We fit the model to data on sodium channel duplicates in teleost fish. We estimate that the diffusion rate lies between and per year, where is the variance of mutation effect sizes. The square root of is roughly equal to the amount of divergence that accumulates in a lineage per year. This implies that duplicates diverge between and per year, where is the average effect that a mutation has on the amount of gene product produced by a duplicate. About 8000–9000 substitutions occur before the threshold is reached. This number seems large, but it is not inconceivable. Summing up all the genetic elements that can affect expression (e.g., promoters, enhancers, microRNAs, post-translational regulators, etc.), there are many mutational targets for expression changes. Indeed, high rates of enhancer gain and loss (enhancer turnover) have been seen in several taxa (Schmidt et al. 2010; Domene et al. 2013; Paris et al. 2013; Arnold et al. 2014). Dosage-balanced duplicates may undergo more rapid enhancer turnover than singleton genes because compensation is possible at two different loci. A last consideration is that the time span involved is long, on the order of 108 generations. In any event, our inferences about numbers of substitutions are very imprecise, and the actual number may be much smaller. In the future, we expect that larger data sets of comparative paralog expression will emerge and will allow greater precision in parameter estimates using methods of analysis such as ABC.
This work builds on earlier hypotheses about the evolution of dosage-sensitive duplicates. Aury et al. (2006) proposed that expression of duplicates evolves by compensatory changes, which can greatly delay the pseudogenization or neofunctionalization of one of the pair. Later work suggested that this process leads to a “random walk” along a line of equal combined expression, a process that could explain comparative gene expression patterns observed in disparate lineages of organisms (Thompson et al. 2014; Gout and Lynch 2015; Lan and Pritchard 2015). Other researchers suggested that gene loss in a duplicated network would cause imbalances and thus put positive selective pressure for loss of other duplicates in the same network, leading to concerted duplicate inactivation (Papp et al. 2003; Hughes et al. 2007; Konrad et al. 2011). Under compensatory drift, the eventual loss of a duplicate may not have much impact on other genes in its network because its paralog will already be producing (almost) all the gene product needed.
Several lines of evidence are consistent with dosage-balance selection after WGD. In contrast to classical models in which redundant duplicates evolve neutrally (Ohno 1970; Walsh 1995; Force et al. 1999; Lynch and Conery 2000), dosage-balance selection will cause both genes to be essential immediately after duplication. WGD does not disrupt dosage balance, and therefore, many preserved duplicates originating in a WGD may evolve under dosage-balance selection. Paramecium tetraurelia has undergone three WGDs in its evolutionary history. In a large proportion of the duplicates from the most recent WGD, both members of the pair are evolving under strong purifying selection, and this proportion declines over time (Aury et al. 2006). This pattern indicates that many genes are dosage sensitive and evolve under dosage-balance selection but that eventually selection to conserve function is lost for one of the duplicates. Other examples come from vertebrates. Some 100 million years after a WGD in the ancestor of salmonid fish, about half the duplicates are retained, and one-quarter of those are still similar in expression and sequence (Berthelot et al. 2014). In a WGD that happened in the ancestor of teleost fish about 300 million years ago, many duplicate pairs persisted for over 200 million years before a member of the pair was lost (Blomme et al. 2006; Brunet et al. 2006; Sato et al. 2009). Delayed loss of duplicates long after a WGD is also seen in Paramecium species (McGrath et al. 2014b). Together these patterns indicate that many duplicates after WGDs are dosage sensitive and evolve in two phases: an initial prolonged phase where both duplicates evolve under selection that conserves function and a later phase in which a duplicate is lost. This later phase could be due to a paralog drifting to low expression and may be the stage at which a redundant gene is most likely to evolve a new function.
Additional predictions flow from the compensatory drift model. Duplicate pairs should persist longer if their total expression is high because more mutations must fix to reach the expression threshold p* (i.e., D* is larger). (Figure 4 shows the impact of increasing D* on the time until duplicate loss.) Both yeast and paramecia show just this pattern: there is a positive correlation between expression levels and the longevity of duplicated genes following WGD (Seoighe and Wolfe 1999; Aury et al. 2006; Gout et al. 2010; McGrath et al. 2014b). To explain this pattern, Gout et al. (2010) argued that stabilizing selection on total expression is stronger on dosage-sensitive duplicates that have high levels of expression. This idea is consistent with our model: the speed at which expression of paralogs diverges becomes slower as the strength of selection increases. The model also makes predictions about patterns of subfunctionalization of dosage-balanced duplicates. When duplicates are expressed in different cell types under different regulation, compensatory drift can occur in parallel in the two cell types, occasionally leading to subfunctionalized expression. Finally, our model makes predictions about phylogenetic patterns. We expect the member of a duplicate pair that has neofunctionalized in a lineage to have lower expression than its paralog in closely related lineages where neofunctionalization has not occurred (Anderson and Evans 2009; Thompson et al. 2014). Recent data support this prediction (Gout and Lynch 2015).
Compensatory drift may play an important role in two other evolutionary contexts. Dosage-balance selection can act on gene duplicates that do not arise by WGD. Selection for increased expression can fix a duplicated gene in a population (Kondrashov 2012). Subsequently, there is stabilizing selection favoring the new, higher-expression optimum. Once this level is reached, the expression can diverge by compensatory drift, as described by our model. Second, compensatory drift can act on the transcription and translation rates for a gene evolving under stabilizing selection for expression. An important difference with duplicate genes is that transcription and translation rates cannot completely compensate for each other. Qualitatively, however, we expect to see similar evolutionary dynamics.
In our model, neofunctionalization happens after a long period of compensatory drift. Alternatively, a novel gene function could predate the duplication event as a minor pleiotropic effect that is not optimized because of tradeoffs. Under the escape-from-adaptive-conflict model, duplicates are freed from these tradeoffs, allowing one of them to become rapidly optimized for the alternative function (Conant and Wolfe 2008; Des Marais and Rausher 2008). However, if one of the gene’s functions requires both duplicates to contribute expression, then compensatory drift would have to occur before one duplicate can escape from the adaptive conflict.
Compensatory drift is related to but distinct from quantitative subfunctionalization (QS). This process describes how, following duplication, degenerative mutations accumulate by drift in each paralog until their total expression declines to a minimum total level necessary for viability (Force et al. 1999, Stoltzfus 1999, Lynch and Force 2000, Hahn 2009; Qian et al. 2010). Compensatory drift, in contrast, is the divergence of expression in paralogs that have already reached optimal expression under dosage-balance selection. A second difference between the processes is that under compensatory drift, half the mutations that fix increase expression, while under QS, none of them do. Despite these differences, there are also important similarities. Both processes can dramatically increase the probability that a gene neofunctionalizes. The two processes could operate in succession. Following the tandem duplication of a gene, expression of each duplicate can decline until both paralogs are necessary to produce the minimal expression needed. The duplicates then can diverge through compensatory drift.
Dosage-balance selection may provide opportunities for adaptation long after WGD occurs. When one of a duplicate pair of genes drifts to a low level of expression, a period of incubation occurs during which it can evolve a new function. As illustrated by duplicates of sodium channel genes in teleost fish, downregulation of dosage-sensitive duplicates may be a common preadaptation in many diversifying gene families. Compensatory drift thus still may be facilitating adaptation very long after the two WGDs that occurred near the root of the vertebrate tree.
Acknowledgments
We thank Laura Crothers for comments on the manuscript. We also acknowledge the Texas Advanced Computing Center (TACC) at the University of Texas at Austin for providing HPC resources that have contributed to the research results reported in this paper. This research was funded by National Science Foundation grants DEB-1311521 to A.T. and DEB-0819901 to M.K.
Footnotes
Communicating editor: J. Wakeley
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.178137/-/DC1
Literature Cited
- Arnold C. D., Gerlach D., Spies D., Matts J. A., Sytnikova Y. A., et al. , 2014. Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nat. Genet. 46: 685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. W., Evans B. J., 2009. Regulatory evolution of a duplicated heterodimer across species and tissues of allopolyploid clawed frogs (Xenopus). J. Mol. Evol. 68: 236–247. [DOI] [PubMed] [Google Scholar]
- Arnegard M. E., Zwickl D. J., Lu Y., Zakon H. H., 2010. Old gene duplication facilitates origin and diversification of an innovative communication system—twice. Proc. Natl. Acad. Sci. USA 107: 22172–22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury J., Jaillon O., Duret L., Noel B., Jubin C., et al. , 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444: 171–178. [DOI] [PubMed] [Google Scholar]
- Beaumont M. A., 2010. Approximate Bayesian computation in evolution and ecology. Annu. Rev. Ecol. Evol. Syst. 41: 379–406. [Google Scholar]
- Beaumont M. A., Zhang W., Balding D. J., 2002. Approximate Bayesian computation in population genetics. Genetics 162: 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot C., Brunet F., Chalopin D., Juanchich A., Bernard M., et al. , 2014. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 5: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur-R R., Broughton R. E., Wiley E. O., Carpenter K., López J. A., et al. , 2013 The tree of life and a new classification of bony fishes. PLoS Curr. 5: pii: ecurrents.tol.53ba26640df0ccaee75bb165c8c26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J. A., Veitia R. A., 2012. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc. Natl. Acad. Sci. USA 109: 14746–14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J. A., Bhadra U., Bhadra M. P., Auger D. L., 2001. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploidy syndromes and quantitative traits. Dev. Biol. 234: 275–288. [DOI] [PubMed] [Google Scholar]
- Birchler J. A., Riddle N. C., Auger D. L., Veitia R. A., 2005. Dosage balance in gene regulation: biological implications. Trends Genet. 21: 219–226. [DOI] [PubMed] [Google Scholar]
- Blomme T., Vandepoele K., De Bodt S., Simillion C., Maere S., et al. , 2006. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 7: R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet F. G., Crollius H. R., Paris M., Aury J., Gilbert P., et al. , 2006. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol. Biol. Evol. 23: 1808–1816. [DOI] [PubMed] [Google Scholar]
- Conant G. C., Wolfe K. H., 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9: 938–950. [DOI] [PubMed] [Google Scholar]
- Cox D. R., Miller H. D., 1965. Markov processes in continuous time with continuous state space, pp. 203–251 in The Theory of Stochastic Processes. Wiley, New York. [Google Scholar]
- Des Marais D. L., Rausher M. D., 2008. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454: 762–765. [DOI] [PubMed] [Google Scholar]
- Domene S., Bumaschy V. F., de Souza F. S. J., Franchini L. F., Nasif S., et al. , 2013. Enhancer turnover and conserved regulatory function in vertebrate evolution. Phil. Trans. R. Soc. Lond. B Biol. Sci. 368: 20130027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger P. P., Pires J. C., 2009. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 17: 699–717. [DOI] [PubMed] [Google Scholar]
- Felsenstein J., 1985. Phylogenies and the comparative method. Am. Nat. 125: 1–15. [Google Scholar]
- Force A., Lynch M., Pickett F. B., Amores A., Yan Y., et al. , 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., Thomas B. C., 2006. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 16: 805–814. [DOI] [PubMed] [Google Scholar]
- Gout J.-F., Lynch M., 2015. Maintenance and loss of duplicated genes by dosage subfunctionalization. Mol. Biol. Evol. 32: 2141–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout J., Kahn D., Duret L., Paramecium Post-Genomics Consortium , 2010. The relationship among gene expression, the evolution of gene dosage, and the rate of protein evolution. PLoS Genet. 6: e1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. W., 2009. Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 100: 605–617. [DOI] [PubMed] [Google Scholar]
- Haldane J. B. S., 1933. The part played by recurrent mutation in evolution. Am. Nat. 67: 5–19. [Google Scholar]
- He X., Zhang J., 2005. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169: 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T., Ekman D., Ardawatia H., Elofsson A., Liberles D. A., 2007. Evaluating dosage compensation as a cause of duplicate gene retention in Paramecium tetraurelia. Genome Biol. 8: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S., Kjällquist U., Moliner A., Zajac P., Fan J.-B., et al. , 2011. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 21: 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1962. On the probability of fixation of mutant genes in a population. Genetics 47: 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov F. A., 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. Biol. Sci. 279: 5048–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad A., Teufel A. I., Grahnen J. A., Liberles D. A., 2011. Toward a general model for the evolutionary dynamics of duplicate genes. Genome Biol. Evol. 3: 1197–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, X., and J. K. Pritchard, 2015 Long-term survival of duplicate genes despite absence of subfunctionalized expression. bioRxiv DOI: 10.1101/019166
- Lavoué S., Miya M., Arnegard M. E., Sullivan J. P., Hopkins C. D., et al. , 2012. Comparable ages for the independent origins of electrogenesis in African and South American weakly electric fishes. PLoS One 7: e36287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J. S., 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch M., Force A., 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino T., McLysaght A., 2010. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc. Natl. Acad. Sci. USA 107: 9270–9274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S., Schmidt A., Codlin S., Chen W., Aebersold R., et al. , 2012. Quantitative Analysis of Fission Yeast Transcriptomes and Proteomes in Proliferating and Quiescent Cells. Cell 151: 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath C. L., Gout J. F., Doak T. G., Yanagi A., Lynch M., 2014a Insights into three whole-genome duplications gleaned from the Paramecium caudatum genome sequence. Genetics 197: 1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath C. L., Gout J.-F., Johri P., Doak T. G., Lynch M., 2014b Differential retention and divergent resolution of duplicate genes following whole-genome duplication. Genome Res. 24: 1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A., Murugan A., Zhang X., Tesileanu T., Wang L., et al. , 2012. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 30: 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger B. P. H., Yuan D. C., Gruber J. D., Duveau F., Wittkopp P. J., 2015. Selection on noise constrains variation in a eukaryotic promoter. Nature 521: 344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B., 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1936. Bar duplication. Science 83: 528–530. [DOI] [PubMed] [Google Scholar]
- Novak A., Jost M., Lu Y., Taylor A., Zakon H. H., et al. , 2006. Gene duplications and evolution of vertebrate voltage-gated sodium channels. J. Mol. Evol. 63: 208–221. [DOI] [PubMed] [Google Scholar]
- Ohno S., 1970. Evolution by Gene Duplication. Springer-Verlag, New York. [Google Scholar]
- Ohta T., 1987. Simulating evolution by gene duplication. Genetics 115: 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B., Pal C., Hurst L. D., 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197. [DOI] [PubMed] [Google Scholar]
- Paris M., Kaplan T., Li X. Y., Villalta J. E., Lott S. E., et al. , 2013. Extensive divergence of transcription factor binding in Drosophila embryos with highly conserved gene expression. PLoS Genet. 9: e1003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan R. P., Hiatt J. B., Witten D. M., Kim M. J., Smith R. P., et al. , 2012. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 30: 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W., Zhang J., 2008. Gene dosage and gene duplicability. Genetics 179: 2319–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W., Liao B., Chang A. Y., Zhang J., 2010. Maintenance of duplicate genes and their functional redundancy by reduced expression. Cell 26: 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi S., Liberles D. A., 2005. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol. Biol. 5: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Hashiguchi Y., Nishida M., 2009. Temporal pattern of loss/persistence of duplicate genes involved in signal transduction and metabolic pathways after teleost-specific genome duplication. BMC Evol. Biol. 9: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell D. R., Wolfe K. H., 2008. A burst of protein sequence evolution and a prolonged period of asymmetric evolution follow gene duplication in yeast. Genome Res. 18: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D., Wilson M. D., Ballester B., Schwalie P. C., Brown G. D., et al. , 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., et al. , 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342. [DOI] [PubMed] [Google Scholar]
- Seoighe C., Wolfe K. H., 1999. Yeast genome evolution in the post-genome era. Curr. Opin. Microbiol. 2: 548–554. [DOI] [PubMed] [Google Scholar]
- Stoltzfus A., 1999. On the possibility of constructive neutral evolution. J. Mol. Evol. 49: 169–181. [DOI] [PubMed] [Google Scholar]
- Stranger B. E., Forrest M. S., Dunning M., Ingle C. E., Beazley C., et al. , 2007. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315: 848–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavare S., Balding D. J., Griffiths R. C., Donnelly P., 1997. Inferring coalescence times from DNA sequence data. Genetics 145: 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A., Vo D., Comfort C., Zakon H. H., 2014. Expression evolution facilitated the convergent neofunctionalization of a sodium channel gene. Mol. Biol. Evol. 31: 1941–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia R. A., 2002. Exploring the etiology of haploinsufficiency. BioEssays 24: 175–184. [DOI] [PubMed] [Google Scholar]
- Walsh B., 2003. Population-genetic models of the fates of duplicate genes. Genetica 118: 279–294. [PubMed] [Google Scholar]
- Walsh J. B., 1995. How often do duplicated genes evolve new functions? Genetics 139: 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakon H. H., Lu Y., Zwickl D. J., Hillis D. M., 2006. Sodium channel genes and the evolution of diversity in communication signals of electric fishes: convergent molecular evolution. Proc. Natl. Acad. Sci. USA 103: 3675–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Simulation code is available upon request.