Abstract
The inference of demographic history from genome data is hindered by a lack of efficient computational approaches. In particular, it has proved difficult to exploit the information contained in the distribution of genealogies across the genome. We have previously shown that the generating function (GF) of genealogies can be used to analytically compute likelihoods of demographic models from configurations of mutations in short sequence blocks (Lohse et al. 2011). Although the GF has a simple, recursive form, the size of such likelihood calculations explodes quickly with the number of individuals and applications of this framework have so far been mainly limited to small samples (pairs and triplets) for which the GF can be written by hand. Here we investigate several strategies for exploiting the inherent symmetries of the coalescent. In particular, we show that the GF of genealogies can be decomposed into a set of equivalence classes that allows likelihood calculations from nontrivial samples. Using this strategy, we automated blockwise likelihood calculations for a general set of demographic scenarios in Mathematica. These histories may involve population size changes, continuous migration, discrete divergence, and admixture between multiple populations. To give a concrete example, we calculate the likelihood for a model of isolation with migration (IM), assuming two diploid samples without phase and outgroup information. We demonstrate the new inference scheme with an analysis of two individual butterfly genomes from the sister species Heliconius melpomene rosina and H. cydno.
Keywords: maximum likelihood, population divergence, gene flow, structured coalescent, generating function
GENOMES contain a wealth of information about the demographic and selective history of populations. However, efficiently extracting this information to fit explicit models of population history remains a considerable computational challenge. It is currently not feasible to base demographic inference on a complete description of the ancestral process of coalescence and recombination, and so inference methods generally rely on making simplifying assumptions about recombination. In the most extreme case of methods based on the site frequency spectrum (SFS), information contained in the physical linkage of sites is ignored altogether (Gutenkunst et al. 2009; Excoffier et al. 2013). Because the SFS is a function only of the expected length of genealogical branches (Griffiths and Tavaré 1998; Chen 2012), this greatly simplifies likelihood computations. However, it also sacrifices much of the information about past demography (Terhorst and Song 2015). Other methods approximate recombination along the genome as a Markov process (Li and Durbin 2011; Harris and Nielsen 2013; Rasmussen et al. 2014). However, this approach is computationally intensive, limited to simple models (Schiffels and Durbin 2014) and/or pairwise samples (Li and Durbin 2011; Mailund et al. 2012), and requires phase information and well-assembled genomes that are still only available for a handful of species.
A different class of methods assumes that recombination can be ignored within sufficiently short blocks of sequence (Yang 2002; Hey and Nielsen 2004). The benefit of this “multilocus assumption” is that it gives a tractable framework for analyzing linked sites and so captures the information contained in the distribution of genealogical branches. Multilocus methods are also attractive in practice because they naturally apply to RAD data or partially assembled genomes that can now be generated for any species (e.g., Davey and Blaxter 2011; Hearn et al. 2014).
For small samples, the probability of seeing a particular configuration of mutations at a locus can be obtained analytically. For example, Wilkinson-Herbots (2008) and Wang and Hey (2010) derived the distribution of pairwise differences under a model of isolation with migration (IM) and Wilkinson-Herbots (2012) extended this to a history where migration is limited to an initial period. Yang (2002) derived the probability of mutational configurations under a divergence model for three populations and a single sample from each and Zhu and Yang (2012) included migration between the most recently diverged pair of populations in this model. However, all of these particular cases can be calculated using a general procedure based on the generating function for the genealogy (Lohse et al. 2011). Here we explain how the GF and—from it—model likelihoods can be efficiently computed for larger samples than have hitherto been possible.
Assuming an infinite-sites mutation model and an outgroup to polarize mutations, the information in a nonrecombining block of sequence can be summarized as a vector of counts of mutations on all possible genealogical branches Both and are labeled by the individuals that descend from them. We have previously shown that the probability of seeing a particular configuration of mutations can be calculated directly from the Laplace transform or generating function (GF) of genealogical branches (Lohse et al. 2011). Given a large sample of unlinked blocks, this gives a framework for computing likelihoods under any demographic model and sampling scheme. Full details are given in Lohse et al. (2011). Briefly, the GF is defined as where is a vector of dummy variables corresponding to Setting the to zero necessarily gives one, the total probability; differentiating with respect to and setting the to zero gives (minus) the expected coalescence time. If we assume an infinite-sites mutation model, the probability of seeing mutations on a particular branch S is (Lohse et al. 2011, equation 1)
| (1) |
This calculation extends to the joint probability of mutations Using the GF rather than the distribution of branches itself to compute is convenient because we avoid the Felsenstein (1988) integral and because the GF has a very simple form: going backward in time, the GF is a recursion over successive events in the history of the sample (Lohse et al. 2011, equation 4),
| (2) |
where, going backward in time, Ω denotes the sampling configuration (i.e., the location and state of lineages) before some event i and the sampling configuration afterward. Events during this interval occur with a total rate The numerator is a sum over all the possible events i, each weighted by its rate Equation 2 applies to any history that consists of independently occurring events. As outlined by Lohse et al. (2011), the GF for models involving discrete events (population splits, bottlenecks) can be found by inverting the GF of the analogous continuous model. In other words, if we know the GF for a model that assumes an exponential rate of events at rate then taking the inverse Laplace transform with respect to Λ gives the GF for any fixed time of the event.
In principle, the GF recursion applies to any sample size and model and can be automated using symbolic software (such as Mathematica). In practice, however, likelihood calculations based on the GF have so far been limited to pairs and triplets: Lohse et al. (2011) computed likelihoods for an IM model with unidirectional migration for three sampled genomes and Lohse et al. (2012) and Hearn et al. (2014) derived likelihoods for a range of divergence histories for a single genome from each of three populations with instantaneous admixture, including the model used by Green et al. (2010) to infer Neanderthal admixture into modern humans (Lohse and Frantz 2014).
There are several serious challenges in applying the GF framework to larger samples of individuals. First, the number of sample configurations (and hence GF equations) grows superexponentially with sample size. Thus, the task of solving the GF and differentiating it to tabulate probabilities for all possible mutational configurations quickly becomes computationally prohibitive. Second, models involving reversible-state transitions, such as two-way migration or recombination between loci, include a potentially infinite number of events. Solving the GF for such cases involves matrix inversions (Hobolth et al. 2011; Lohse et al. 2011). Third, while assuming infinite-sites mutations may be convenient mathematically and realistic for closely related sequences, this assumption becomes problematic for more distantly related outgroups that are used to polarize mutations in practice. Finally, being able to uniquely map mutations onto genealogical branches assumes phased data that are rarely available for diploid organisms, given the limitations of current sequencing technologies.
In the first part of this article, we discuss each of these problems in turn and introduce several strategies to remedy the explosion of terms and computation time. These arguments apply generally, irrespective of the peculiarities of particular demographic models and sampling schemes, and suggest a computational “pipeline” for likelihood calculations for nontrivial samples of individuals (up to ). The accompanying Mathematica notebook (File S1) implements this scheme for a wide range of demographic histories that may involve arbitrary divergence, admixture, and migration between multiple populations, as well as population size changes. As a concrete example, we describe likelihood calculations for the two population IM model for unphased and unpolarized data from two diploid individuals. We compare the power of this scheme to that of minimal samples of a single haploid sequence per population. Finally, to illustrate the new method, we estimate divergence and migration between the butterfly species Heliconius melpomene and H. cydno (Martin et al. 2013).
Models and Methods
Partitioning the GF into equivalence classes
Since the GF is defined in terms of genealogical branches and each topology is specified by a unique set of branches, an intuitive strategy for computing likelihoods is to partition the GF into contributions from different topologies. To condition on a certain topology, we simply set GF terms that are incompatible with it to 0 (Lohse et al. 2011). Importantly however, such incompatible events still contribute to the total rate of events in the denominator of Equation 2. Then, setting all ω in the topology-conditioned GF to zero gives the probability of that particular topology. Although conditioning on a particular topology gives a GF with a manageable number of terms, it is clearly not practical to do this for all possible topologies, given their sheer number even for moderate n (Table 1).
Table 1. Fundamental quantities of genealogies for small samples (n).
| n | Branches | Ranked topologies | Unranked topologies | ECa, 1 population | EC, 2 populations | No. configb |
|---|---|---|---|---|---|---|
| Felsenstein (2003) | ||||||
| 3 | 6 | 3 | 3 | 1 | 2 | 625 |
| 4 | 14 | 18 | 15 | 2 | 6 | 15,625 |
| 6 | 62 | 2,700 | 945 | 6 | 49 | 9,765,625 |
| 8 | 254 | 1,587,600 | 135,135 | 23 | 560 | 6,103,515,625 |
| 10 | 1,022 | 2,571,912,000 | 34,469,425 | 98 | 7,139 | 3,814,697,265,625 |
aThe number of equivalence classes.
bThe number of mutational configurations for a sample from two populations with up to mutations per branch.
In the following, we distinguish between ranked and unranked topologies. The GF is a sum over all possible sequences of events in the history of a sample; Edwards (1970) called them “labeled histories.” Considering only coalescence events, each labeled history corresponds to a ranked topology, i.e., a genealogy with unique leaf labels and a known order of nodes. A fundamental property of the standard coalescent, which follows directly from the exchangeability of genes sampled from the same population, is that all ranked topologies are equally likely (Kingman 1982; Hudson 1983). In other words, if we could somehow assign each mutation to a particular coalescence (i.e., internode) interval, we could use a much simpler GF, defined in terms of the coalescence intervals rather than the branches for inference. This logic underlies demographic methods that use the branch length information contained in well-resolved genealogies (e.g., Nee et al. 1995; Pybus et al. 2002) and coalescent-based derivations of the site frequency spectrum (Griffiths and Tavaré 1998; Chen 2012).
Unfortunately, however, when analyzing sequence data from sexual organisms, we are generally limited by the number of mutations on any one genealogical branch and so often cannot resolve nodes or their order. Although unranked topologies are not equiprobable, even under the standard coalescent, their leaf labels are still exchangeable. Therefore, each unranked, unlabeled topology, or “tree shape” sensu Felsenstein (1978, 2003), is an equivalence class that defines a set of identically distributed genealogies (Figure 1). This means that we need to work out the GF only for one representative (random labeling) per equivalence class. The full GF can then be written as a weighted sum of the GFs for such class representatives,
| (3) |
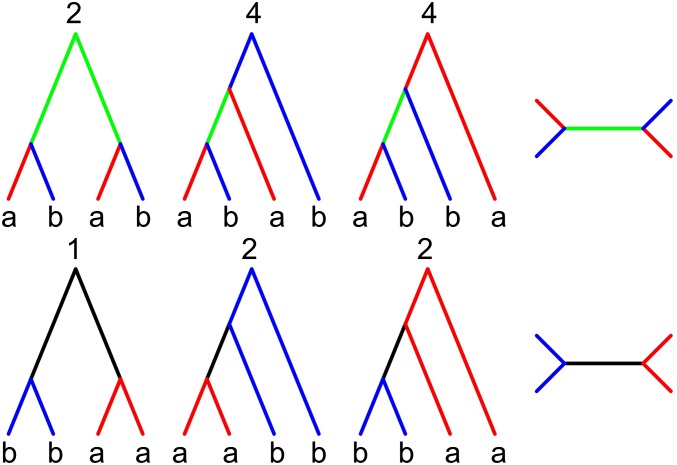
where denotes the size of equivalence class h and is the set of dummy variables that corresponds to the branches of a single class representative in h. There are necessarily many fewer equivalence classes than labeled topologies (Table 1). For example, given a sample of size from a single population, there are 945 unranked topologies, but only six equivalence classes (Figure 1).
Figure 1.
Unranked, unlabeled topologies define equivalence classes of genealogies. For a sample of from a single population there are six equivalence classes. Their size, i.e., the number of labeled genealogies in each class (), is shown above.
Crucially, the idea of tree shapes as equivalence classes extends to any demographic model and sampling scheme. For samples from multiple populations, the equivalence classes are just the permutations of population labels on (unlabeled) tree shapes. It is straightforward to generate and enumerate the equivalence classes for any sample (Felsenstein 2003). For example, for a sample of from each of two populations (three per population), there are 49 equivalence classes (partially labeled shapes), which can be found by permuting the two population labels on the unlabeled tree shapes in Figure 1.
In general, the size of each equivalence class is a function of the number of permutations of individuals on population labels. For individuals from population i, there are permutations. Since the orientation of nodes is irrelevant, each symmetric node in the equivalence class halves the number of unique permutations. Symmetric nodes are connected to identical subclades; that is, there exists an isomorphism ensuring that they have the same topology and the same population labels at the leaves (see Figure 1),
| (4) |
where is the number of symmetric nodes. Note also that the total number of unranked topologies.
Any tree shape contains at least one further symmetry: there is at least one node that connects to two leaves. Because the branches descending from that node have the same length by definition, we can combine mutations (and hence ω terms) falling on them: e.g., for a triplet genealogy with topology we can combine mutations on branches b and c without loss of information. The joint probability of seeing a configuration with and mutations can be retrieved from
| (5) |
We previously made use of this in implementing likelihood calculations for triple samples (Lohse et al. 2011). Although in principle this combinatorial argument extends to arbitrary genealogies, we can show that, for larger samples, computing from mutational configurations defined in terms of internode intervals is computationally wasteful compared to the direct calculation (see Supporting Information, File S1).
Approximating models with reversible events
Migration and recombination events are fundamentally different from coalescence and population divergence. Going backward in time, they do not lead to simpler sample configurations. Thus, the GF for models involving migration and/or recombination is a system of coupled equations, the solution of which involves matrix inversion and higher-order polynomials and quickly becomes infeasible for large n (Hobolth et al. 2011). As an example, we consider two populations connected by symmetric migration at rate Since we are often interested in histories with low or moderate migration in practice, it seems reasonable to consider an approximate model in which the number of migration events is limited. Using a Taylor series expansion, the full GF can be decomposed into histories with migration events (Lohse et al. 2011). The same argument applies to recombination between discrete loci and can be used to derive the GF for the sequential Markov coalescent (McVean and Cardin 2005). It is crucial to distinguish between M terms in the numerator and denominator. In other words, even if we stop including sampling configurations involving multiple migration events, M still contributes to the total rate in the denominator. To see how this works, consider the simplest case of a pair of genes a and b sampled from two populations connected by symmetric migration. Following Lohse et al. (2011), denotes the sampling configuration where both genes are in different populations and that where they are in the same population. We modify the GF (Lohse et al. 2011, equation 9) to include an indicator variable γ that counts the number of migration events:
| (6) |
Expanding in γ, the coefficients of correspond to histories with migration events. This is analogous to conditioning on a particular topology: the truncated GF does not sum to one (if we set the ω to zero), but rather gives the total probability of seeing no more than events. This is convenient because it immediately gives an estimate of the accuracy of the approximation. Expanding the solution of Equation 6 around gives
| (7) |
The GF conditional on there being at most one migration event is
| (8) |
The error of this approximation is
| (9) |
which is just the chance that a migration event occurs before coalescence (see Figure 2). An analogous expansion for the pairwise GF for the IM model (Lohse et al. 2011, equation 13) gives
Figure 2.
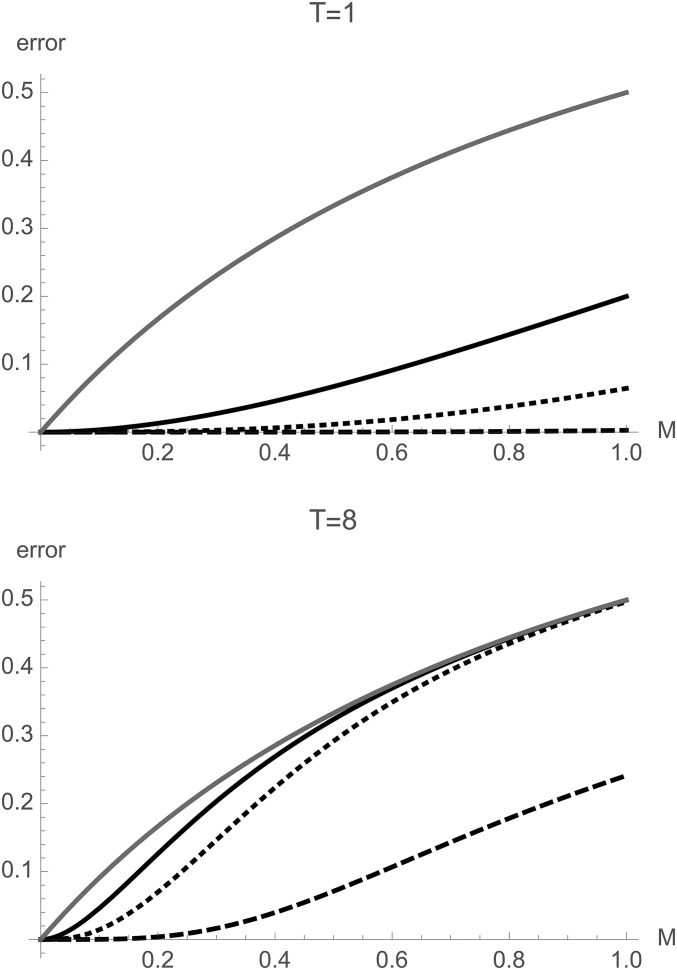
The error (Equation 9) in limiting the number of migration events to = 1 (solid line), 2 (dashed line), and 4 (dotted line) for a pairwise sample in the IM model plotted against M for different divergence times T. The results for a model of equilibrium migration without divergence are shown for comparison (shaded line).
| (10) |
Expressions for the GF conditional on a maximum of migration events and for larger samples can be found by automating the GF recursion. While these do not appear to have a simple form, plotting the error against M and T (Figure 2) shows that for recent divergence () and moderate gene flow (), histories involving more than two migration events are extremely unlikely () and can be ignored to a good approximation. Considering that for large n, coalescence [at rate ] becomes much more likely than migration (at rate ), this approximation should be relatively robust to sample size.
Unknown phase and root
There are at least two further complications for blockwise likelihood computations in practice: first, we have so far assumed that mutations can be polarized without error, i.e., that the infinite-sites mutation model holds between in- and outgroup, which is often unrealistic in practice. Second, given the current limitations of short read sequencing technology, genomic data are often unphased and one would ideally want to incorporate phase ambiguity explicitly rather than ignore it (e.g., Lohse and Frantz 2014) or rely on computational phasing.
Both unknown phase and root can be incorporated via a simple relabeling of branches. In generating the GF, we have labeled branches and corresponding ω variables by the tips (leaf nodes) they are connected to. Crucially, the full GF expressed as a sum over equivalence class representatives (Equation 3) still has unique labels for all individuals. That is, we distinguish genes sampled from the same population. To incorporate unknown phase, we simply label leaf nodes by the population they were sampled from (Figure 3). Because the genealogical branches are labeled by the set of leaf nodes they are connected to, this relabeling of leaf nodes defines branch types that correspond to categories of the (joint) SFS. In other words, in the absence of phase information branch types are defined by the number of descendants in each population. To see how this works, consider, for example, two genes from each of two populations. There are six equivalence classes of rooted genealogies (Figure 3). Combining branches with the same population labels gives seven ω variables that correspond to site types and In the absence of root information, we further combine the two branches on either side of the root. Denoting ω variables for unrooted branches by * and the two sets of individuals they are connected to, we have and The rooted branches contributing to each unrooted branch are indicated in color in Figure 3. The terms correspond to the four types of variable sites defined by the folded SFS for two populations: (heterozygous sites unique to a), (heterozygous sites unique to b), (heterozygous sites shared by both), and (fixed differences between a and b). Note also that without the root, the six equivalence classes collapse to two unrooted equivalence classes (defined by branches and ) (Figure 3).
Figure 3.
For a sample of two sequences (a diploid genome) from each of two populations (a and b), there are six classes of equivalent, rooted genealogies (left); their sizes are shown above. Without root information, these collapse to two unrooted genealogies (right). Without phase information, there are four mutation types that map to specific branches in the rooted genealogy: heterozygous sites unique to one sample ( and red and blue, respectively), shared heterozygous sites ( green), and fixed, homozygous differences ( black).
The combinatorial arguments outlined above extend to arbitrary sample sizes and numbers of populations. The GFs for unphased data are given by combining ω variables with the same number of descendants in each population. We modify Equation 3 to write the GF of an unrooted genealogy as a sum over unrooted equivalence classes (denoted ), each of which is in turn a sum over rooted equivalence classes:
| (11) |
We can use this simplified GF and Equation 1 to compute the probability of blockwise counts of mutation types defined by the joint SFS. We refer to this extension of the joint SFS to blockwise data as the blockwise site frequency spectrum (bSFS), following Bunnefeld et al. (2015) who have used the bSFS to fit bottleneck histories in a single population.
Limiting the total number of mutational configurations
In principle, we can compute the probability of seeing arbitrarily many mutations on a particular branch from Equation 1. In practice, however, the extra information gained by explicitly distinguishing configurations with large numbers of mutations (which are very unlikely for short blocks) is limited, while the computational cost increases. An obvious strategy is to tabulate exact probabilities only up to a certain maximum number of mutations per branch and combine residual probabilities for configurations involving mutations on one or multiple branches. As described by Lohse et al. (2011, 2012), the residual probability of seeing mutations on a particular branch s is given by
i.e., we subtract the sum of exact probabilities for configurations involving up to mutations from the marginal probability of seeing branch S.
Assuming that we want to distinguish between all branches in a given equivalence class and use a global for all branches, there are possible mutation counts per branch (including those with no mutations or mutations on a branch), which gives mutational configurations in total. For example, for and there are 9,765,625 mutational configurations per equivalence class (Table 1). Although this may seem daunting, most of these configurations are extremely unlikely, so a substantial computational saving can be made by choosing branch-specific We have implemented functions in Mathematica to tabulate for an arbitrary vector of (File S1).
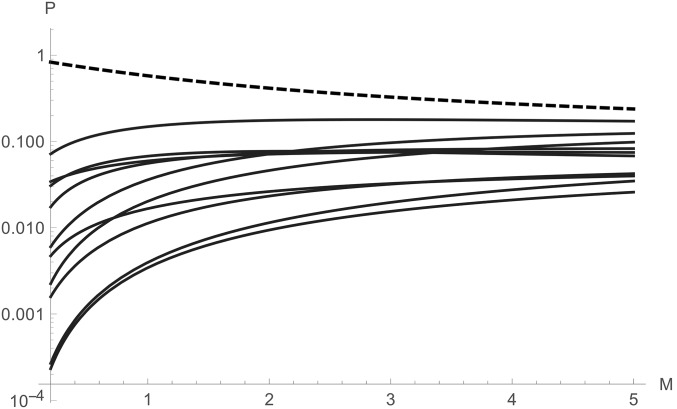
The bSFS with constitutes an interesting special case: it defines mutational configurations by the joint presence and/or absence of SFS types in a block, irrespective of their number. In the limit of very long blocks, i.e., if we assume an unlimited supply of mutations, this converges to the topological probabilities of equivalence classes that can be obtained directly from the partitioned GF by setting all We can think of this set of probabilities as the “topology spectrum.” For a sample of three genes from each of two populations this consists of 49 equivalence classes, which reduce to 11 unrooted topologies (Figure 4). Under the IM model with unidirectional migration, the GF of each class is solvable using Mathematica (File S2). The most likely topology is reciprocal monophyly, i.e., . As expected, its probability decreases with M and increases with T (Figure 4).
Figure 4.
The topology spectrum for a sample of from a two-population IM model with asymmetric migration and (in 2Ne generations). The probabilities of all 11 unrooted topologies are plotted against M. The probability of the most likely topology of reciprocal monophyly is shown as a dashed line.
Data availability
File S1 is a Mathematica notebook that contains the code to generate the GF and tabulate likelihoods under arbitrary demographic models. File S2 contains the code used for the analyses of the IM model, including the analyses of the Heliconius data and the power test. File S3 contains the processed input data for Heliconius and the python script used; raw sequence data are published by Martin et al. (2013) and available from www.datadryad.com DOI: 10.5061/dryad.dk712.
Results
The various combinatorial strategies for simplifying likelihood calculations based on the GF outlined above suggest a general “pipeline,” each component of which can be automated:
Generate all equivalence classes h and enumerate their sizes for a given sampling scheme.
Generate and solve the GF conditional on one representative of each h.
Take the inverse Laplace transform with respect to the time parameters of discrete events (e.g., divergence, admixture, bottlenecks). These processes are initially modeled as occurring with a continuous rate.
Relabel variables to combine branches and equivalence classes that are indistinguishable in the absence of root and/or phase information.
Find a sensible for each mutation type from the data.
Tabulate probabilities for all mutational configurations in each equivalence class.
In the accompanying Mathematica notebook we have implemented this pipeline as a set of general functions. These can be used to automatically generate, solve, and simplify the GF (steps 1–3), and—from this—tabulate the likelihood of a large range of demographic models (involving population divergence, admixture, and bottlenecks) (step 6). In principle, this automation works for arbitrary sample sizes. In practice, however, the inversion step (step 3) and the tabulation of probabilities (step 6) become infeasible for
To give a concrete example, we derive the GF for a model of isolation at time T (scaled in generations) with migration at rate migrants per generation (IM) between two populations (labeled a and b). We further assume that migration is unidirectional, i.e., from a to b forward in time, and that both populations and their common ancestral population are of the same effective size (we later relax this assumption when analyzing data). As above, we consider the special case of a single diploid sample per population without root and phase information. We first derive some basic properties of unrooted genealogies under this model. We then investigate the power of likelihood calculations based on the bSFS. Finally, we apply this likelihood calculation to genome data from two species of Heliconius butterflies.
The distribution of unrooted branches under the IM model
We can find the expected length of any branch (or combination of branches) S from the GF as (Lohse et al. 2011). The expressions for the expected lengths of rooted branches are cumbersome (File S2). Surprisingly, however, the expected lengths of the four unrooted branches and each of which is a sum over the underlying rooted branches (Figure 3), have a relatively simple form (Figure 5):
| (12) |
Similarly, the probability of the two unrooted topologies reduces to
Figure 5.
The expected length of unrooted genealogical branches (Equation 12) for a sample of under the IM model of two populations (a and b) with asymmetric migration and population divergence time ( generations). Colors correspond to those in Figure 3.
| (13) |
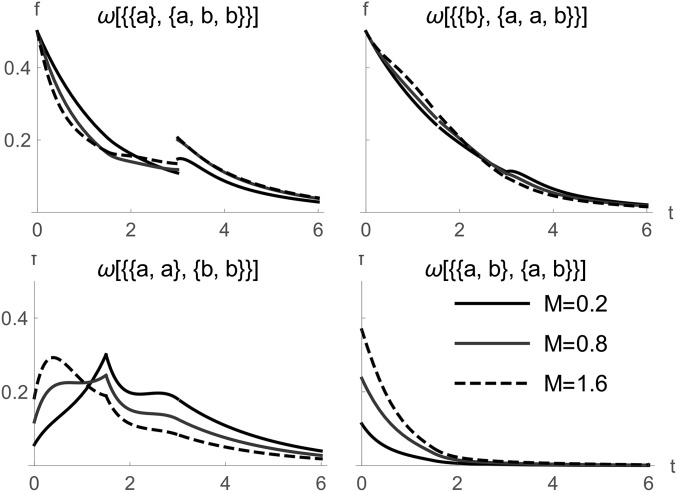
We can recover the full distribution of rooted branches from the GF by taking the inverse Laplace transform (using Mathematica) with respect to the corresponding While this does not yield simple expressions (File S2), examining Figure 6 illustrates that much of the information about population history is contained in the shape of the branch length distribution rather than its expectation (Figure 5). For example, the length of branches carrying fixed differences has a multimodal distribution with discontinuities at T and the relative size of the first mode strongly dependent on M.
Figure 6.
The length distribution of unrooted genealogical branches for a sample of under the IM model of two populations (a and b) with asymmetric migration and population divergence at (in generations).
Power analysis
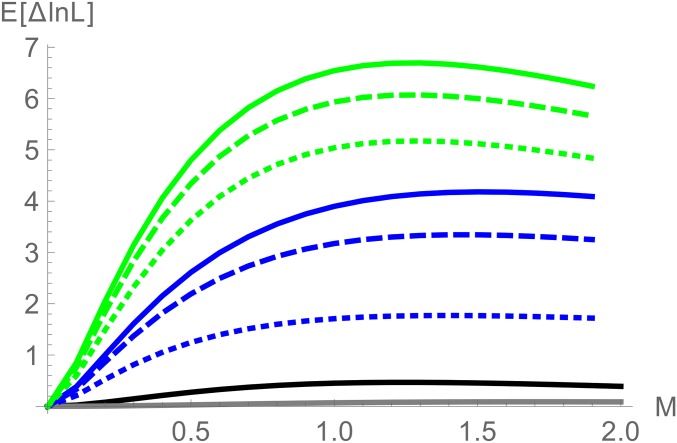
We compared the power to detect postdivergence gene flow between two different blockwise likelihood calculations: the bSFS for a diploid genome per population () and a minimal sample of a single haploid sequence () per population. As a proxy for power, we computed the expected difference in support () between the IM model and a null model of strict divergence without gene flow and arbitrarily assumed data sets of 100 blocks. However, since we are assuming that blocks are unlinked, i.e., statistically independent, scales linearly with the number of blocks.
Figure 7 shows the power to detect gene flow for a relatively old split () and sampling blocks with an average of heterozygous sites within each species. Without gene flow, this corresponds to a total number of 5.2 mutations per block on average (using Equation 12 and ). Unsurprisingly, sampling a diploid sequence from each population gives greater power to detect gene flow than pairwise samples (compare black and blue lines in Figure 7). However, contrasting this with the power of a simpler likelihood calculation for that is based only on the total number of mutations in each block (gray line in Figure 7) illustrates that the additional information does not stem from the increase in sample size per se, but rather from the addition of topology information. Perhaps counterintuitively, we find that there is less information in the distribution of for larger samples than in pairwise samples. This clearly shows that most information about postdivergence gene flow is contained in the topology, i.e., being able to assign mutations to specific branches. Similarly, adding root information almost doubles power (green lines in Figure 7).
Figure 7.
The power () to distinguish between an IM model and a null model of strict divergence () from 100 unlinked blocks of length for different sample sizes and data summaries: the total number of mutations in a sample of (black) and (gray) and the bSFS for unphased data for two diploids () with root (green) and without root (blue). Dotted, dashed, and solid lines correspond to different maximum numbers of mutations per branch type, 1, and 3, respectively.
In comparison, the threshold has relatively little effect on power. In other words, for realistically short blocks, most of the information is contained in the joint presence and absence of mutation types (regardless of their number).
Heliconius analysis
To illustrate likelihood calculation based on the bSFS, we estimated divergence and gene flow between two species of Heliconius butterflies. The sister species H. cydno and H. melpomene rosina occur in sympatry in parts of Central and South America, are known to hybridize in the wild at a low rate (Mallet et al. 2007), and have previously been shown to have experienced postdivergence gene flow (Martin et al. 2013). We sampled 225-bp blocks of intergenic, autosomal sequence for one individual genome of each species from the area of sympatry in Panama (chi565 and ro2071). These data are part of a larger resequencing study involving high-coverage genomes for four individuals of each H. cydno and H. m. rosina species as well as an allopatric population of H. melpomene from French Guiana (Martin et al. 2013). We excluded CpG islands and sites with low-quality (GQ < 30 and MQ < 30), excessively low (<10) or high (>200) coverage and considered only sites that passed these filtering criteria in all individuals.
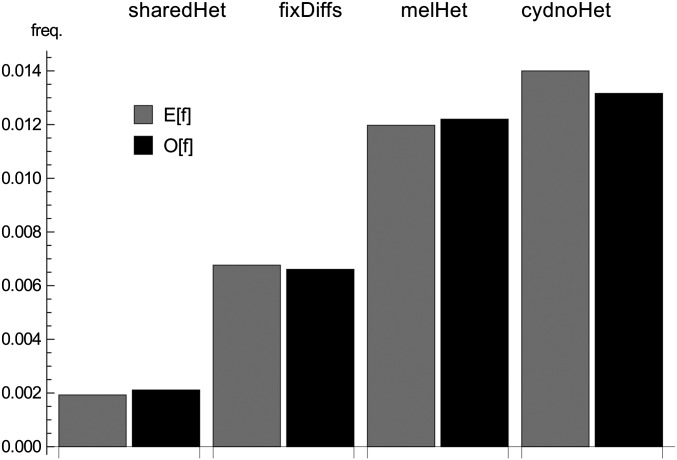
We partitioned the intergenic sequence into blocks of 225 bp length. To allow some sites to violate our filtering criteria in each block while keeping the block length postfiltering fixed, we sampled the first 150 bases passing filtering in each block (blocks with fewer remaining sites were excluded from the analysis). A total of 6.3% of blocks violated the four-gamete criterion (i.e., contained both fixed differences and shared heterozygous sites) and were removed. This sampling and filtering strategy yielded 161,726 blocks with an average per site heterozygosity of 0.017 and 0.015 in H. m. rosina and H. cydno, respectively (Figure 8). Summarizing the data by counting the four mutation types in each block gave a total of 2337 unique mutational configurations, 1743 of which occurred more than once.
Figure 8.
The folded SFS has four site types: (i and ii) heterozygous sites unique to either H. m. melpomene (i) or H. cydno (ii), (iii) heterozygous sites shared by both species, and (iv) fixed differences. Solid bars show the observed genome-wide SFS, and shaded bars show the expectation under the IM history estimated from the bSFS (Table 2) (Equation 9).
We initially used all blocks (regardless of linkage) to obtain point estimates of parameters under three models: (i) strict isolation without migration, (ii) isolation with migration from H. cydno into H. m. rosina (), and (iii) isolation with migration from H. m. rosina into H. cydno (). In all cases, we assumed that the common ancestral population shared its with one descendant species while the other descendant was assumed to have a different To keep computation times manageable, we did not consider more complex histories involving bidirectional gene flow or three parameters.
We maximize under each model. using Nelder–Mead simplex optimization implemented in the Mathematica function NMaximize. Confidence intervals (C.I.) for parameter estimates were obtained from 100 parametric bootstrap replicates. We used ms (Hudson 2002) to simulate 0.3 Mb of contiguous sequence for each of the 20 Heliconius autosomes, assuming a per site recombination rate of (Jiggins et al. 2005) and the best-fitting IM history. We partitioned each simulated data set into 150-bp blocks and estimated 95% (C.I.) as two standard deviations of estimates across bootstrap replicates (see Discussion).
We find strong support for a model of isolation with migration from H. cydno into H. m. rosina () (Table 2) with a larger in H. cydno. This model fits significantly better than simpler nested models of strict divergence or an model with a single (Table 2). Our results agree with earlier genomic analyses of these species that showed support for postdivergence gene flow based on D statistics (Martin et al. 2013), IMa analyses based on smaller numbers of loci (Kronforst et al. 2013), and genome-wide SNP frequencies analyzed using approximate Bayesian computation. Asymmetrical migration from H. cydno into H. m. rosina has also been reported previously and could be explained by the fact that F1 hybrids resemble H. m. rosina more closely due to dominance relationships among wing-patterning alleles, possibly making F1’s more attractive to H. m. rosina (Kronforst et al. 2006; Martin et al. 2015).
Table 2. Maximum-likelihood estimates of divergence and migration between H. m. rosina and H. cydno.
| θ | T (τ) | M | ||
|---|---|---|---|---|
| IM estimatesa | 1.25 | 3.24 | 1.90 | 1.50 |
| Scaled IM estimatesb | 1.10 (1.02–1.18) | 2.85 (2.55–3.23) | 1.04 (0.91–1.18) MY | (1.32–1.68) |
| Expected estimatesc | 1.22 | 3.53 | 1.97 | 1.40 |
aUnder the best model is the scaled mutation rate in H. cydno.
b in individuals, τ in MY, 95% C.I. in parentheses.
cMean across parametric bootstrap replicates.
We applied a recent direct, genome-wide estimate of the mutation rate for H. melpomene of per site and generation (Keightley et al. 2015) to convert parameter estimates into absolute values. Assuming that synonymous coding sites are evolving neutrally, we used the ratio of divergence between H. m. rosina and the more distantly related “silvaniform” clade of Heliconius at synonymous coding sites and the intergenic sites our analysis is based on to estimate the selective constraint on the latter. This gives an “effective mutation” rate of (Martin et al. 2015). Applying this corrected rate to our estimate of θ and assuming four generations per year, we obtain an estimate of for H. m. rosina and the common ancestral population and for H. cydno. We estimate species divergence to have occurred ∼0.91–1.18 MYA. Note that this is more recent than previous estimates of 1.5 MY that were obtained using approximate Bayesian computation and a different calibration based on mitochondrial genealogies (Kronforst et al. 2013; Martin et al. 2015).
Discussion
Irrespective of any particular demographic history, the possible genealogies of a sample can be partitioned into a set of equivalence classes, which are given by permuting population labels on tree shapes. We show how this fundamental symmetry of the coalescent can be exploited when computing likelihoods from blockwise mutational configurations. We have implemented this combinatorial partitioning in Mathematica to automatically generate and solve the GF of the genealogy and, from this, compute likelihoods for a wide range of demographic models (File S2). Given a particular sample of genomes, we first generate a set of equivalence classes of genealogies and condition the recursion for the GF (Lohse et al. 2011) on a single representative from each class. This combinatorial strategy brings a huge computational saving. Importantly however, it does not sacrifice any information. In contrast, approximating the GF for models that include reversible events in particular migration involves a trade-off between computational efficiency and accuracy. For example, given our high estimates for unidirectional M for Heliconius (Table 2), it would have been unrealistic to fit a history of bidirectional migration to these data without allowing for multiple migration events in each genealogy (Figure 2).
Although these approaches make it possible to solve the GF for surprisingly large samples and biologically interesting models, the number of mutational configurations (which explodes with the number of sampled genomes) remains a fundamental limitation of such likelihood calculations in practice. Given outgroup and phase, the full information is contained in a vast table of mutational configurations that are defined in terms of the branches of each equivalence class. For samples from two populations, the number of mutational configurations we need to calculate is the product of the last two columns of Table 1. For example, given a sample of three haploid genomes per population and allowing for up to mutations per branch, there are possible mutational configurations, an unrealistic number of probabilities to calculate.
The blockwise site frequency spectrum
Our initial motivation for studying the bSFS was to deal with unphased data in practice. The GF of the bSFS can be obtained from the full GF simply by combining branches with equivalent leaf labels. As well as being a lossless summary of blockwise data (in the absence of phase information), the bSFS is a promising summary in general for several reasons. First, it is extremely compact compared to the full set of (phased) mutational configurations. Unlike the latter, the size of the bSFS does not depend on the number of equivalence classes (which explodes with n, Table 1), but only on n. Given a sample of individuals from population i and assuming a global maximum number of mutations for all mutation types, the (unfolded) bSFS comprises a maximum of mutational configurations. For a sample of three haploid genomes from each of two populations and the bSFS has entries. Second, because equivalence classes of genealogies are defined by the presence and absence of SFS types, much of the topology information contained in the full data should still be captured in the bSFS. Finally, and perhaps surprisingly, at least for the IM model the expressions for the total length of branches contributing to unphased and unpolarized mutation types (Equations 12 and 13) are much simpler than those for the underlying rooted branches, which suggests that it may be possible to find general results.
Despite the strategies developed here, it is clear that full-likelihood calculations will rarely be feasible for samples given the rapid increase in the number of equivalence classes. However, a separation of timescales exists for many models of geographic and genetic structure (Wakeley 1998, 2009), and so full-likelihood solutions for moderate () samples may be sufficient for computing likelihoods for much larger samples if these contribute mainly very short branches with no mutations in the initial scattering phase during which lineages from the same population either coalesce or trace back to unsampled demes.
Dealing with linkage
A key assumption of the blockwise likelihood calculations is that there is no recombination within sequence blocks and that different blocks are independent of each other. This latter assumption is especially problematic when we analyze whole-genome data. If we divide the genome into blocks that are small enough for recombination within them to be negligible, our method gives an unbiased estimate of the likelihood of a demographic model. However, the accuracy of the model fit will be grossly overestimated if we simply multiply likelihoods across blocks, because adjacent blocks are strongly correlated. Ignoring this correlation amounts to a composite-likelihood calculation.
A common practice (e.g., Wang and Hey 2010; Excoffier et al. 2013; Lohse and Frantz 2014) is to assume a “safe distance” at which blocks (or SNPs) are statistically independent. This is equivalent to a rescaling of the suppose that we multiply likelihoods across every kth block, k being chosen large enough that blocks are uncorrelated. This procedure is valid starting at any block and so can be repeated k times, such that the whole genome is included in the analysis. Taking the average across all k analyses is equivalent to simply multiplying the likelihoods across all blocks and then dividing the total by k. However, because it is not clear how to choose k, this procedure is quite arbitrary. On the one hand, successive blocks or SNPs are not completely correlated, suggesting that this considerably underestimates the accuracy of estimates. On the other hand, however, there may be weak, long-range correlations, due to a small fraction of long regions that coalesced recently, and these may increase the variance of parameter estimates.
The safest way to account for LD is via a parametric bootstrap. Although computationally intensive, this has the added benefit that it also checks whether parameter estimates are biased (due to the assumption of no recombination within blocks). It is reassuring that in the case of the Heliconius data, we find that the biases in parameter estimates are very small indeed (last row in Table 2). We note that the confidence intervals for the Heliconius estimates we derived from the bootstrap are conservative, given the current limitations of coalescent simulators. Given the limited length of continuous recombining sequence that can be simulated, the simulated data sets were over four times smaller than the real data set. An interesting alternative to full parametric bootstrap, which we hope to implement in the future, is to use the variance of the gradient of across bootstrap replicates to adjust the Fisher information matrix (Godambe 1960; Coffman et al. 2015).
An advantage of direct-likelihood calculations is that one can easily check the absolute fit of the data to a model by asking how well the observed frequency of mutational configurations or some summary such as the SFS is predicted by the model. For example, the IM history we estimated for the two Heliconius species fits the observed genome-wide SFS reasonably well (Figure 8). The fact that we slightly underestimate the heterozygosity in H. cydno may suggest that some process (e.g., demographic change after divergence or admixture from an unsampled ghost population/species) is not captured by our model.
In general, the GF framework makes it possible to derive the distribution of any summary statistic that can be defined as a combination of genealogical branches and understand its properties under simple demographic models and small n. Although explicit calculations based on such summaries are not feasible for large n, summary statistics such as the bSFS may still have wide applicability for fitting complex models and analyzing larger samples of individuals, for example using approximate-likelihood methods, or simply as a way to visualize how genealogies vary along the genome.
Acknowledgments
We thank Lynsey Bunnefeld for discussions throughout the project and Joshua Schraiber and one anonymous reviewer for constructive comments on an earlier version of this manuscript. This work was supported by funding from the United Kingdom Natural Environment Research Council (to K.L.) (NE/I020288/1) and a grant from the European Research Council (250152) (to N.H.B.).
Footnotes
Communicating editor: Y. S. Song
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.183814/-/DC1.
Literature Cited
- Bunnefeld L., Frantz L. A. F., Lohse K., 2015. Inferring bottlenecks from genome-wide samples of short sequence blocks. Genetics 201: 1157–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., 2012. The joint allele frequency spectrum of multiple populations: a coalescent theory approach. Theor. Popul. Biol. 81: 179–195. [DOI] [PubMed] [Google Scholar]
- Coffman A. J., Hsieh P., Gravel S., Gutenkunst R. N., 2015. Computationally efficient composite likelihood statistics for demographic inference. Mol. Biol. Evol. DOI: 10.1093/molbev/msv255. Available at: http://mbe.oxfordjournals.org/content/early/2015/12/08/molbev.msv255.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey J. W., Blaxter M. L., 2011. RADseq: next-generation population genetics. Brief. Funct. Genomics 9: 416–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A. W. F., 1970. Estimation of the branch points of a branching diffusion process (with discussion). J. R. Stat. Soc. B 32: 155–174. [Google Scholar]
- Excoffier L., Dupanloup I., Huerta-Sánchez E., Sousa V. C., Foll M., 2013. Robust demographic inference from genomic and SNP data. PLoS Genet. 9: e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J., 1978. The number of evolutionary trees. Mol. Phylogenet. Evol. 27: 27–33. [Google Scholar]
- Felsenstein J., 1988. Phylogenies from molecular sequences: inference and reliability. Annu. Rev. Genet. 22: 521–565. [DOI] [PubMed] [Google Scholar]
- Felsenstein J., 2003. Inferring Phylogenies. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Godambe, V. P., 1960 An optimum property of regular maximum likelihood estimation. Ann. Math. Stat. 31: 1208–1211.
- Green R. E., Krause J., Briggs A. W., Maricic T., Stenzel U., et al. , 2010. A draft sequence of the Neanderthal genome. Science 328: 710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R., Tavaré S., 1998. The age of a mutation in a general coalescent tree. Commun. Stat. Stoch. Models 14: 273–295. [Google Scholar]
- Gutenkunst R. N., Hernandez R. D., Williamson S. H., and C. D. Bustamente, 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5: e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K., Nielsen R., 2013. Inferring demographic history from a spectrum of shared haplotype lengths. PLoS Genet. 9: e1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearn J., Stone G. N., Bunnefeld L., Nicholls J. A., Barton N. H., et al. , 2014. Likelihood-based inference of population history from low-coverage de novo genome assemblies. Mol. Ecol. 23: 198–211. [DOI] [PubMed] [Google Scholar]
- Hey J., Nielsen R., 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167: 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobolth A., Andersen L. N., Mailund T., 2011. On computing the coalescent time density in an isolation-with-migration model with few samples. Genetics 187: 1241–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 1983. Testing the constant-rate neutral allele model with protein sequence data. Evolution 37: 203–217. [DOI] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- Jiggins C. D., Mavarez J., Beltrán M., McMillan W. O., Johnston J. S., et al. , 2005. A genetic linkage map of the mimetic butterfly Heliconius melpomene. Genetics 171: 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Pinharanda A., Ness R. W., Simpson F., Dasmahapatra K. K., et al. , 2015. Estimation of the spontaneous mutation rate in Heliconius melpomene. Mol. Biol. Evol. 32: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman J. F. C., 1982. The coalescent. Stoch. Proc. Appl. 13: 235–248. [Google Scholar]
- Kronforst M., Hansen M., Crawford N., Gallant J., Zhang W., et al. , 2013. Hybridization reveals the evolving genomic architecture of speciation. Cell Rep. 5: 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronforst M. R., Young L. G., Blume L. M., Gilbert L. E., 2006. Multilocus analyses of admixture and introgression among hybridizing Heliconius butterflies. Evolution 60: 1254–1268. [PubMed] [Google Scholar]
- Li H., Durbin R., 2011. Inference of human population history from individual whole-genome sequences. Nature 475: 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse K., Frantz L. A. F., 2014. Neanderthal admixture in Eurasia confirmed by maximum-likelihood analysis of three genomes. Genetics 196: 1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse K., Harrison R. J., Barton N. H., 2011. A general method for calculating likelihoods under the coalescent process. Genetics 58: 977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse K., Barton N. H., Melika N., Stone G. N., 2012. A likelihood-based comparison of population histories in a parasitoid guild. Mol. Ecol. 49: 832–842. [DOI] [PubMed] [Google Scholar]
- Mailund T., Halager A. E., Westergaard M., Dutheil J. Y., Munch K., et al. , 2012. A new isolation with migration model along complete genomes infers very different divergence processes among closely related great ape species. PLoS Genet. 8: e1003125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J., Beltran M., Neukirchen W., Linares M., 2007. Natural hybridization in heliconiine butterflies: the species boundary as a continuum. BMC Evol. Biol. 7: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. H., Dasmahapatra K. K., Nadeau N. J., Salazar C., Walters J. R., et al. , 2013. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 23: 1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. H., A. Eriksson, K. M. Kozak, A. Manica, and C. D. Jiggins, 2015 Speciation in Heliconius butterflies: minimal contact followed by millions of generations of hybridisation. bioRxiv: DOI: 10.1101/015800.
- McVean G. A., Cardin N. J., 2005. Approximating the coalescent with recombination. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360: 1387–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nee S., Holmes E. C., Rambaut A., Harvey P. H., 1995. Inferring population history from molecular phylogenies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 349: 25–31. [DOI] [PubMed] [Google Scholar]
- Pybus O. G., Rambaut A., Holmes E. C., Harvey P. H., 2002. New inferences from tree shape: numbers of missing taxa and population growth rates. Syst. Biol. 51: 881–888. [DOI] [PubMed] [Google Scholar]
- Rasmussen M. D., Hubisz M. J., Gronau I., Siepel A., 2014. Genome-wide inference of ancestral recombination graphs. PLoS Genet. 10: e1004342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S., Durbin R., 2014. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 46: 919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhorst J., Song Y. S., 2015. Fundamental limits on the accuracy of demographic inference based on the sample frequency spectrum. Proc. Natl. Acad. Sci. USA 112: 7677–7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., 1998. Segregating sites in Wright’s island model. Theor. Popul. Biol. 53: 166–174. [DOI] [PubMed] [Google Scholar]
- Wakeley J., 2009. Coalescent Theory. Roberts & Company, Greenwood Village, CO. [Google Scholar]
- Wang Y., Hey J., 2010. Estimating divergence parameters with small samples from a large number of loci. Genetics 184: 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson-Herbots H., 2012. The distribution of the coalescence time and the number of pairwise nucleotide differences in a model of population divergence or speciation with an initial period of gene flow. Theor. Popul. Biol. 82: 92–108. [DOI] [PubMed] [Google Scholar]
- Wilkinson-Herbots H. M., 2008. The distribution of the coalescence time and the number of pairwise nucleotide differences in the “isolation with migration” model. Theor. Popul. Biol. 73: 277–288. [DOI] [PubMed] [Google Scholar]
- Yang Z., 2002. Likelihood and Bayes estimation of ancestral population sizes in hominoids using data from multiple loci. Genetics 162: 1811–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T., Yang Z., 2012. Maximum likelihood implementation of an isolation-with-migration model with three species for testing speciation with gene flow. Mol. Biol. Evol. 49: 832–842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
File S1 is a Mathematica notebook that contains the code to generate the GF and tabulate likelihoods under arbitrary demographic models. File S2 contains the code used for the analyses of the IM model, including the analyses of the Heliconius data and the power test. File S3 contains the processed input data for Heliconius and the python script used; raw sequence data are published by Martin et al. (2013) and available from www.datadryad.com DOI: 10.5061/dryad.dk712.