Abstract
RNA editing refers to post-transcriptional processes that alter the base sequence of RNA. Recently, hundreds of new RNA editing targets have been reported. However, the mechanisms that determine the specificity and degree of editing are not well understood. We examined quantitative variation of site-specific editing in a genetically diverse multiparent population, Diversity Outbred mice, and mapped polymorphic loci that alter editing ratios globally for C-to-U editing and at specific sites for A-to-I editing. An allelic series in the C-to-U editing enzyme Apobec1 influences the editing efficiency of Apob and 58 additional C-to-U editing targets. We identified 49 A-to-I editing sites with polymorphisms in the edited transcript that alter editing efficiency. In contrast to the shared genetic control of C-to-U editing, most of the variable A-to-I editing sites were determined by local nucleotide polymorphisms in proximity to the editing site in the RNA secondary structure. Our results indicate that RNA editing is a quantitative trait subject to genetic variation and that evolutionary constraints have given rise to distinct genetic architectures in the two canonical types of RNA editing.
Keywords: genetics, RNA editing, Diversity Outbred, Apobec1, secondary structure, Multiparent Advanced Generation Inter-Cross (MAGIC), multiparental populations, MPP
RNA EDITING in mammals occurs through deamination of adenosine, which is converted to inosine (A-to-I editing), or deamination of cytosine, which is converted to uracil (C-to-U editing) (Davidson and Shelness 2000; Bass 2002). Other types of editing have been reported, but these findings remain controversial (Bass et al. 2012; Gu et al. 2012). The two canonical editing types, A-to-I and C-to-U editing, are mediated by distinct pathways. A-to-I editing is catalyzed on double-stranded (ds) RNA by proteins in the adenosine deaminase, RNA-specific (ADAR) family (ADAR1 and ADAR2) and is most common in neuronal tissues. However, the Adar gene family is ubiquitously expressed, and editing has been reported in many other tissues (Gu et al. 2012). Homozygous deletion of Adar genes is embryonic lethal in mice, and defects in A-to-I editing have been associated with neurodegenerative disorders and cancers (Gurevich et al. 2002; Paz et al. 2007). The C-to-U editing pathway is catalyzed by apolipoprotein B messenger RNA (mRNA) editing enzyme catalytic polypeptide 1 (Apobec1), which is expressed primarily in small intestine and liver, where it targets the transcript of apolipoprotein B (Apob), converting a CAA (glutamine) codon within the coding sequence to a stop codon (UAA). This editing event results in two APOB protein isoforms, APOB48 from the edited transcript and APOB100 from the unedited transcript. Editing of Apob is evolutionarily conserved and occurs in mice, humans, and other mammals. The edited isoform APOB48 functions in the synthesis, assembly, and secretion of chylomicrons in the small intestine; the unedited isoform APOB100 is expressed in the liver and gives rise to very low-density lipoprotein (VLDL), which is converted to LDL in the bloodstream. Although VLDL can contain either APOB48 or APOB100, LDL exclusively contains APOB100 (Davidson and Shelness 2000). Mice carrying a homozygous null allele of Apobec1 are viable but exhibit abnormal lipid homeostasis (Hirano et al. 1996).
Both A-to-I and C-to-U editing alters RNA nucleotides at specific positions in a tissue-specific manner (Nakamuta et al. 1995; Bass 2002; Dawson et al. 2004). Recent reports have described editing events at tens to thousands of sites in humans and mice (Li et al. 2009; Rosenberg et al. 2011). Editing is often incomplete, with only a proportion of available transcripts being edited. The mechanisms that determine the specificity and efficiency of RNA editing are not well understood. Previous studies in humans have shown only limited effects of genetic variation on RNA editing. In one study, six A-to-I editing sites were found to be edited consistently across 32 individuals (Greenberger et al. 2010). Another study found evidence of genetic variation for two (of 7389) A-to-I editing sites (Daneck et al. 2012). Still another study found an association between RNA editing rates and genetic variation of Apobec1 (Hassan et al. 2014). The broader impact of genetic variation on RNA editing in humans and mice remains unclear. Identification of allelic variants that alter the editing process could provide new insights into these mechanisms, and this provides the motivation for our genetic mapping study.
The Diversity Outbred (DO) population is a multiparent outbred mouse population derived from the same eight progenitor lines as the Collaborative Cross (CC) (Svenson et al. 2012). The DO population provides high levels of allelic diversity and high-resolution genetic mapping. Here we use natural allelic variants in the DO population to identify polymorphic loci that affect RNA editing with the aim of understanding the genetic factors that determine quantitative levels of editing. We consider the editing ratio—the proportion of edited reads at a site—as a quantitative trait for genetic analysis. Our findings reveal distinct modes of genetic regulation in the two editing pathways and provide insight into evolutionary constraints on the mechanisms that determine the specificity and efficiency of RNA editing.
Materials and Methods
DO mice
We obtained DO mice (J:DO, Stock #009376; 277 in total, 143 females and 134 males) from The Jackson Laboratory. Animals were received at 3 weeks of age and housed from wean age with free access to either standard rodent chow containing 6% fat by weight (73 females and 68 males; LabDiet 5K52, Scott Distributing, Hudson, NH) or high-fat chow containing 22% fat by weight (70 females and 66 males; TD.08811, Harlan Laboratories, Madison, WI). Animals were phenotyped for multiple metabolic and hematologic parameters as described by Svenson et al. (2012) and euthanized at 26 weeks of age. Liver samples were collected from each animal and stored in RNAlater solution (Life Technologies, Grand Island, NY) at −80°. All procedures on DO mice were approved by the Animal Care and Use Committee at The Jackson Laboratory (Protocol #06006).
DO founder strains
Breeder pairs for each of the eight DO founder strains, A/J, C57BL/6J, 129S1/SvImJ, NOD/ShiLtJ, NZO/HlLtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ, were purchased from The Jackson Laboratory and were bred to produce a total of 128 male mice, 16 per founder strain. Male progeny mice were maintained on standard breeder chow (Purina LabDiet 5013) until weaning at the University of Wisconsin–Madison. Beginning at 4 weeks of age, half the male mice from each strain were maintained on either a semipurified control diet containing 17% of kilocalories from fat (TD.08810) or a high-fat, high-sucrose (HF/HS) diet containing 45% of kilocalories from fat and 34% (by weight) sucrose (TD.08811); diets were from Harlan Laboratories (Madison, WI). Owing to reduced litter size or poor breeding, male CAST/EiJ and NZO/HlLtJ mice were purchased from The Jackson Laboratory at ∼3 weeks of age and switched to the control or HF/HS diet at 4 weeks. Animals were euthanized at 26 weeks of age (except for the NZO/HlLtJ mice). NZO/HlLtJ mice were euthanized at 20 weeks of age owing to high lethality in response to the HF/HS diet. Liver samples were collected, snap frozen, and shipped on dry ice to The Jackson Laboratory for RNA-seq analysis. All animal procedures were approved by the Animal Care and Use Committee at University of Wisconsin–Madison (Protocol #A00757-0-07-11).
RNA sequencing
Total liver RNA was isolated using the TRIzol Plus RNA Extraction Kit (Life Technologies) with on-column DNase digestion. Following the Illumina TruSeq standard protocol, indexed mRNA-seq libraries were generated from 1 μg total RNA followed by quality control and quantitation on an Agilent Bioanalyzer (Santa Clara, CA) and the KAPA Biosystems Library quantitative PCR (qPCR) quantitation method. Finally, 100-bp single-end reads were sequenced using the Illumina HiSeq 2000 (San Diego, CA). To minimize technical variation (lane and barcode effects in sequencing), the samples were randomly assigned to lanes and multiplexed at 12 or 24 samples per lane with randomly selected barcodes. Each DO sample was sequenced with two or four technical replicates to obtain ∼10 million reads per sample. Base calling was performed using CASAVA v1.8.0, and FASTQ files were filtered to remove low-quality reads using the Illumina CASAVA-1.8 FASTQ Filter (http://cancan.cshl.edu/labmembers/gordon/fastq_illumina_filter/).
RNA editing site prediction in founder strains
We constructed strain-specific transcriptomes for seven of the eight founder strains (except C57BL/6J) using ModTools (Huang et al. 2014). We incorporated strain-specific SNPs and short (<50 bp) indels from the Sanger Mouse Genomes Project (Keane et al. 2011; Yalcin et al. 2011) (REL-1303) into the Genome Reference Consortium Mouse Genome, Build 38 (GRCm38) reference sequence. We constructed MOD files and strain-specific genomes and adjusted transcript annotation using the vcf2mod, insilico, and modmap utilities from the ModTools suite. We extracted the transcriptome of each founder strain using rsem-prepare-reference (RSEM v1.2.15) (Li and Dewey 2011) and built Bowtie indices (Bowtie v0.12.8) (Langmead et al. 2009). We searched for RNA editing sites using a two-tiered approach in which we searched for sites in the founder strains and then evaluated those sites in DO mice. A detailed description of the process with the script commands is available in Supporting Information, File S1 and Figure S1.
All 16 replicates from each strain were aligned to their respective strain-specific transcriptome using Bowtie with parameters -a –best –strata –v 3. We converted transcriptome alignments to genome alignments using rsem-tbam2gbam (Li and Dewey 2011) and adjusted the strain-specific coordinates to GRCm38 coordinates using Lapels (Huang et al. 2014).
We piled up the reads within each transcript (Ensembl archive 68) for each of the 16 replicates in a single DO founder strain. For each founder strain, we retained sites with the following properties:
Exactly two alleles expressed;
At least two reads in at least 75% of the 16 replicates;
Minor allele frequency (MAF) > 5%;
Reference and edited allele coverage ≥ 160 reads;
No intersection with any known SNP or structural variant (Keane et al. 2011; Yalcin et al. 2012); and
Occurrence within a gene that contains no noncanonical editing sites.
Our reconstructions of founder strain and individual DO transcriptomes are based on the current mouse reference genome (GRCm38) and include annotated pseudogenes. However, some of the founder strain genomes are likely to harbor unannotated pseudogenes with paralogous SNP variants that, if they are transcribed, could appear to be editing. While we can account for known pseudogenes, unknown or unannotated pseudogenes are more challenging. The last filter (#6) was included to address these on the assumption that paralogous variants are likely to produce signatures of noncanonical editing. It is possible that some sites passing our filters are artifacts of expression of unannotated pseudogenes (File S2, File S3, File S4, and File S5). We have included counts of the number of canonical and noncanonical editing sites included throughout the process (Table S1).
We took the union of all editing sites from the eight founder strains and retained sites in genes with only canonical editing in all eight strains, coverage ≥ 160 at the edited bases, and less than 1% of reads in the nonedited bases. This produced a set of putative editing sites that we then queried and filtered in the DO samples (File S6).
RNA editing quantification in the DO samples
We built a combined transcriptome consisting of transcripts from all eight founder strains and indexed it using Bowtie (Langmead et al. 2009). We aligned RNA-seq reads from each DO animal to the combined transcriptome and separated them into individual alignments. We then converted the individual transcriptome alignments to genome alignments and then to GRCm38 coordinates using rsem-tbam2gbam and Lapels. Once all alignments were in the same coordinate system, we extracted only one instance of every read across all eight alignments and filtered any reads that were mapped to different coordinates in any two strains. This produced a BAM file for each DO sample representing alignments to all eight founder strains.
We piled up the reads in each DO sample at the putative editing sites and retained those with mean read depths ≥ 20 and a mean editing ratio ≥ 2%. We retained 186 sites (File S7).
Previous reports have noted that false-positive RNA editing sites may be due to a bias in the read position in which the putative editing site occurs (Pickrell et al. 2012). We looked at the position of each of the 186 editing sites in the 100-bp reads from each founder. We counted the frequency with which the editing site occurred at each position in the reads in each of the eight founders. We tested whether a greater proportion of reads occurred in the first and last 5 bp of each read than expected by chance using a binomial test (H0: proportion = 0.1). We discarded 54 editing sites that occurred predominantly at the ends of reads and retained 102 sites (File S8).
We queried the RADAR (Ramaswami and Li 2014) and DARNED (Kiran et al. 2013) databases for all reported mouse editing sites and found 8891 sites. We piled these sites up in the DO samples and retained 98 sites with mean coverage ≥ 20 and a mean editing ratio ≥ 2% (File S9). Of these, 17 sites were identical to the de novo sites, and we retained 81 additional editing sites from RADAR and DARNED (File S9). We combined these with the 102 de novo sites and proceeded with a total of 183 sites.
Genotyping and haplotype reconstruction of DO genomes
Genotyping of the DO mice was described in our previous study (Svenson et al. 2012). DNA was extracted from tail biopsies and genotyped using the Mouse Universal Genotyping Array (MUGA; GeneSeek, Lincoln, NE). A total of 277 animals were genotyped. The founder haplotypes were reconstructed using a hidden Markov model based on the normalized intensity values from the MUGA (Gatti et al. 2014).
QTL mapping of the RNA editing ratio
The haplotype reconstruction process produced a matrix of eight founder allele dosages for each sample at each marker. We fit a linear mixed model by regressing the edited counts on sex, diet, and total counts with an adjustment for kinship between animals (Gatti et al. 2014). When total counts in a sample at an editing site were <10, we found that many false associations were produced by low total counts in the denominator of the editing ratio. Therefore, we excluded samples with total counts <10 from the model because estimates of the editing ratio became unstable. The regression model used was
| (1) |
where yi = edited counts for animal i; βs = effect of sex; si = sex of animal i; βd = effect of diet; di = diet for animal i; βr = effect of total counts; ni = total counts for animal i; βj = effect of founder allele j; gij = founder allele dosage for founder j in animal i; and γi = random effect representing the polygenic influence of animal i. We report the LOD ratio as the mapping statistic. We determined significance thresholds by permuting the phenotype and covariate values 1000 times (Churchill and Doerge 1994, 2008). We selected the maximum association for each editing ratio and calculated the genome-wide p-value from the empirical distribution of null LOD scores. We applied a Benjamini and Hochberg false-discovery-rate (FDR) correction (Benjamini and Hochberg 1995) to these p-values and retained those with padj < 0.05 (File S10 and File S11).
Quantification of different isoforms of Apobec1
Two isoforms of Apobec1 were found from the reference genome annotation. The difference between the two isoforms consisted of differences in the length of exon 4, one with a longer exon 4 (NM_031159) and one with a shorter exon 4 (NM_001134391). We quantified the expression of the two known isoforms and of a new isoform discovered in this study—the extension of exon 5. We constructed six isoforms of Apobec1, two of which were the reference isoforms. The remaining four isoforms were to test which of the two reference isoforms the aberrant extended isoform is derived from and the full length of the aberrant isoform. We embedded the six isoforms into the annotation file downloaded from Ensembl (http://useast.ensembl.org) and then used RSEM (Li and Dewey 2011) to quantify the six isoforms, tolerating zero mismatches in the alignments. Transcripts per million (TPM) and fragments per kilobase of transcript per million aligned reads (FPKM) were used for quantification and comparison.
Secondary-structure prediction
Full-length pre-mRNAs containing introns were imputed from strain-specific transcriptomes in which Sanger SNPs and indels were incorporated into the reference sequence. Minimum-free-energy structures were predicted for full-length mRNAs using the Vienna RNAfold tool (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) with default parameters (Lorenz et al. 2011).
We calculated the most stable substructure around the edit site using RNALfold (Hofacker et al. 2004) from the Vienna RNA Package v2.1.7 (Lorenz et al. 2011), which calculates the most stable substructures within a large sequence. By running RNALfold –z, we obtained all substructures with a Z-score ≤ −1.0 (lower Z-scores indicate more stable structures), from which we retrieved the substructure containing the edit site with the lowest Z-score.
We calculated the probability that the editing site occurs in a favorable editing position (occurring in a stem or a bulge or internal loop of size ≤ 2) by summing the pairing and unpairing probabilities of all relevant nucleotides around the editing site. For instance, the probability of the editing site being in a stem is the sum of all base-pairing probabilities between it and any other nucleotide (i.e., the probability of the editing site being paired). For an editing site e to be in a bulge of size 1, the probability is that of the editing site being unpaired (1 − sum of all base-pairing probabilities for it) times the probability of nucleotide e − 1 being paired times the probability of nucleotide e + 1 being paired. An analogous calculation was performed for internal loops and larger sizes. File S13 provides the Python code for calculating the probability of the editing site being in a favorable position. Base-pairing probabilities were calculated by running RNAfold –p on the whole gene sequence.
Gene expression analysis
Gene expression was computed by summarizing the expression of all isoforms computed by RSEM (Li and Dewey 2011) from the founder strains. FPKM mapped reads were used. Similar expression results were obtained simply by summing the number of reads aligned to all the isoforms. The expression of APOB was computed by summarizing the reads that uniquely aligned to the Apob transcript from the eight founder strains.
Motif analysis
We used MEME (v4.10.0) (Bailey and Elkan 1994) to analyze the 30-nt downstream sequences adjacent to the 59 C-to-U editing sites that mapped to chromosome 6. We used the following settings: exactly one motif per sequence, motif length between 10 and 12 nt, and analysis of only the current strand. The described motif was the motif with the highest score. We searched for the motif in the same sequences using FIMO (v4.10.0) (Bailey and Elkan 1994), searching only on the given strand.
Genome assembly and annotation
We used genome coordinates from GRCm38 (Waterston et al. 2002). We obtained SNPs, indels, and structural variants for the eight DO founder strains from the Sanger Mouse Genomes Project v3 (Keane et al. 2011). We used the Ensembl archive 68 transcriptome (Flicek et al. 2012).
Data availability
The FASTQ files for the founder and DO RNA-seq runs are archived at the Gene Expression Omnibus under accession number GSE45684 (Munger et al. 2014). The alignment pipeline, including alignment and RNA editing site calling, is described in File S1, File S2, File S3, File S4, File S5, File S6, File S7, File S8, File S9, File S10, File S11, and File S12.
Results
Our study employed liver RNA-seq data from two experiments. We profiled liver RNA in males from the eight inbred founder strains of the DO population. We also profiled liver samples in a genetic mapping panel of 277 DO mice of both sexes (Svenson et al. 2012). We implemented a conservative screen to identify candidate editing sites (see Materials and Methods). Our goal was not the exhaustive identification of editing sites. Rather, we aimed to obtain reliable editing sites and then to evaluate quantitative variation in editing. We selected sites with robust evidence for editing in the DO founders (editing > 5% with at least 160 reads) and minimal evidence of sequencing or alignment errors, and we identified 192 sites in 131 genes. We retained 156 of those sites (51 A-to-I and 105 C-to-U sites) in 113 genes with mean coverage > 20 and mean editing ratio ≥ 2% in DO mice. We removed sites that had an excess of editing sites in the proximal or distal 5 bp of each read and retained 102 sites. We then queried the RADAR (Ramaswami and Li 2014) and DARNED (Kiran et al. 2013) RNA editing site databases and found 98 sites with mean coverage > 20 and mean editing ratio ≥ 2% for these sites in DO mice. Of these, 17 overlapped the 102 editing sites found in the de novo discovery pipeline. We retained a total of 183 sites (102 + 98 − 17) from the de novo discovery and the RADAR/DARNED databases (Table S2). There were 96 A-to-I sites and 87 C-to-U sites. Most of these sites (160) occur in the 3′ UTR of genes, two in intergenic regions, three in introns, three in noncoding RNAs or expressed pseudogenes, and 15 in coding regions. Among the 15 coding variants, 8 are synonymous, 6 are nonsynonymous, and 1, a well-known site in Apob, produces a stop codon (Chen et al. 1987). In earlier work, we had identified and confirmed 17 liver RNA editing sites (Gu et al. 2012); we detected 14 of these sites here, and the remaining 3 sites showed evidence of editing but were removed because they fell below our minimum read coverage criteria.
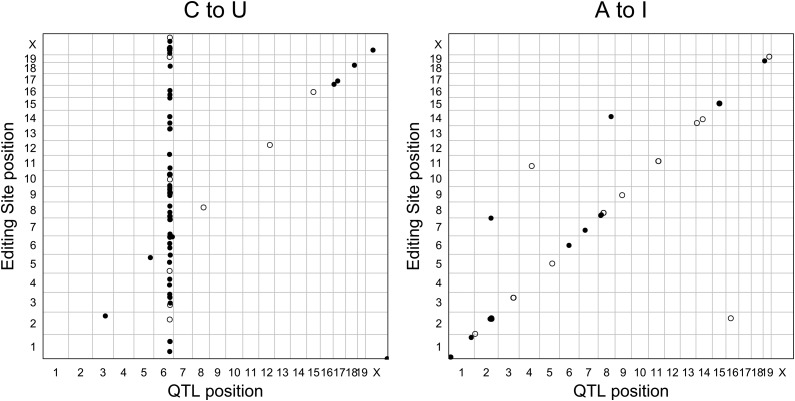
We mapped the editing ratio for each of the 183 sites and found significant associations (FDR < 5%) for 119 sites in 81 genes. Of these sites, 70 were C-to-U sites, and 59 of these mapped to a region on chromosome 6, while 11 mapped to a location near the edited site (Figure 1A and Table S1). The remaining 49 mapped associations were produced by A-to-I sites, and most of these mapped to a location near the edited site (Figure 1B).
Figure 1.
Mapping of C-to-U and A-to-I RNA editing reveals distinct patterns of genetic regulation. (A) Marker location (horizontal axis) vs. editing-site location (vertical axis) for the C-to-U editing sites shows that editing variation is due to genetic variants near the editing site (diagonal band) and at a location on chromosome 6 (vertical band). Open circles show sites from the RADAR or DARNED databases, and closed circles show sites from the de novo editing site discover. (B) Marker location vs. editing-site location for the A-to-I edit sites shows that editing variation is primarily due to genetic variants near the editing site. Note that some points represent multiple overlapping editing sites.
C-to-U editing
C-to-U editing ratios for most sites were associated with a single region on chromosome 6. We found 59 C-to-U editing sites with editing ratios that associated with a region of chromosome 6 between 119.6 and 125.6 Mb. Of these 59 sites, 50 came from the de novo pipeline, and 9 were from RADAR or DARNED. We examined the average C-to-U editing ratio in the DO population for these 59 sites and found that all but three sites in the Decorin (Dcn) gene were positively correlated with one another and shared a common pattern of allelic effects, suggesting that a single pleiotropic locus is driving variation in most C-to-U edited sites.
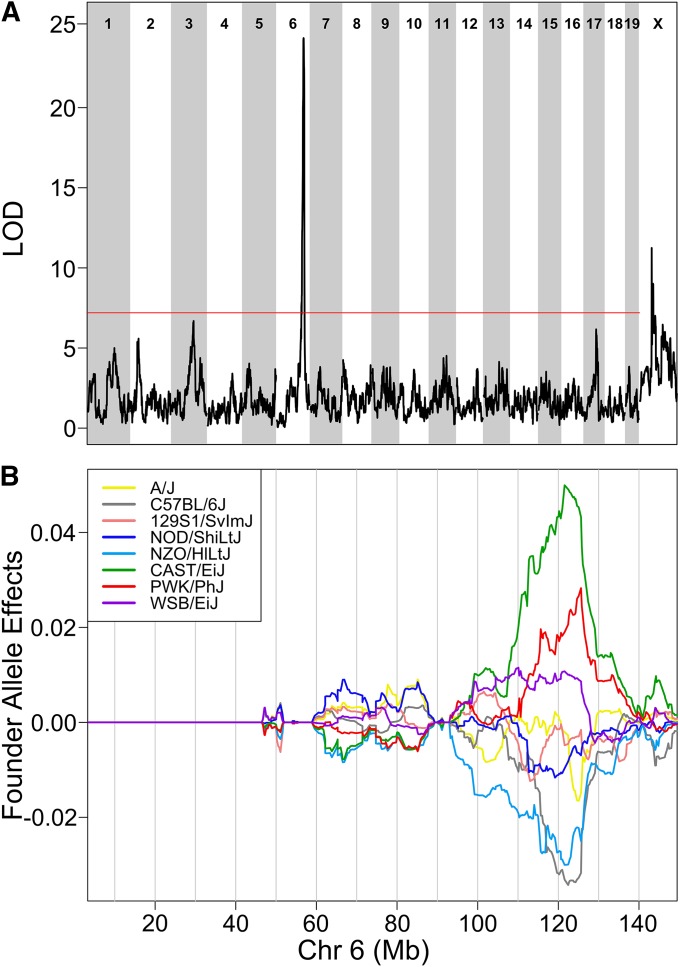
We averaged the editing ratios across all 59 positively correlated sites and mapped the average editing ratio to chromosome 6 (Figure 2A). The founder effects at the peak locus show a complex pattern (Figure 2B). DO mice carrying the CAST/EiJ, PWK/PhJ, and WSB/EiJ alleles have higher mean C-to-U editing; mice carrying the A/J, 129S1, and NOD/ShiLtJ alleles have intermediate editing; and mice carrying the C57BL/6J or NZO/HlLtJ alleles have low editing. The Bayesian credible interval for the association peak spans 2.5 Mb (121.1–123.6 Mb) and contains 39 transcripts, of which 10 are expressed in liver, Apobec1, Phc1, Slc6a12, Slc6a13, M6pr, Mug1, Mug2, Necap1, Pex26, and Gm10319. None of these genes has an obvious functional connection with RNA editing with the exception of Apobec1 (apolipoprotein B mRNA editing enzyme catalytic polypeptide 1), the cytidine deaminase that catalyzes C-to-U editing (Petersen-Mahrt and Neuberger 2003). We hypothesize that Apobec1 variants are the causal factor influencing quantitative variation in C-to-U editing.
Figure 2.
Mean C-to-U editing ratios for most editing sites map to a region on chromosome 6 at 122 Mb. (A) Genome scan of mean C-to-U editing for 70 editing sites shows a strong association on chromosome 6. Horizontal axis shows the mouse genome; vertical axis plots the LOD score. Red line is the permutation-derived α = 0.05 significance threshold. (B) Founder allele effects on chromosome 6 reveal a complex pattern of allele effects. Horizontal axis shows chromosome 6 in Mb. Vertical axis shows the founder allele effects.
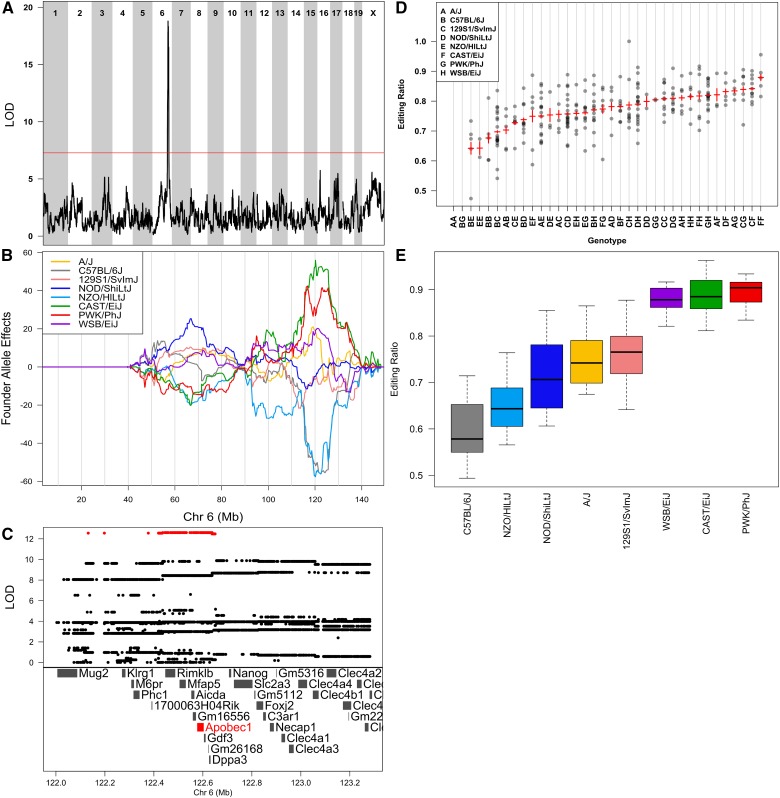
To understand the effect of segregating alleles on the C-to-U editing ratio, we examined the canonical C-to-U editing site in Apob, which has an association on chromosome 6 at 120.2 Mb (Figure 3A). Apob transcript levels do not have an association on chromosome 6 in the liver, and thus, the editing association arises as a result of editing variation, not transcriptional variation. The pattern of allele effects for Apob C-to-U editing is similar to the pattern for the 70 C-to-U sites shown earlier (Figure 3B), suggesting that the underlying genetic mechanisms are similar. We imputed the founder SNPs onto the reconstructed DO haplotypes and performed association mapping within the support interval. We found that SNPs with alleles shared by C57BL/6J and NZO/HlLtJ contribute the highest LOD scores (Figure 3C). We estimated the editing ratio for each of the 36 genotypes present in the DO samples (Figure 3D). The genotype estimates are noisier owing to the smaller number of animals in each genotype class; nevertheless, there appears to be a continuous genotype-dependent variation of editing ratios, suggesting the presence of multiple functional alleles segregating in the DO samples. We examined the Apob editing ratios in the eight founder strains and compared them with the estimated additive effect of founder haplotypes obtained in the DO population (Figure 3, B and E). In both groups of animals, the C57BL/6J and NZO/HlLtJ haplotypes are associated with the lowest editing ratios, and the CAST/EiJ and PWK/PhJ haplotypes are associated with the highest editing ratios. Editing is somewhat elevated in the presence of the WSB/EiJ haplotype, while A/J, 129S1/SvlmJ, and NOD/ShiLtJ haplotypes are intermediate.
Figure 3.
Genome scan of Apob C-to-U editing shows that editing is regulated by genetic variants on chromosome 6 that overlap Apobec1. (A) Genome scan of Apob editing shows a strong association on chromosome 6. Horizontal axis shows the mouse genome; vertical axis shows the LOD score. Red line is the permutation-derived α = 0.05 significance threshold. (B) Founder allele effect plots for Apob C-to-U editing on chromosome 6. Horizontal axis plots Mb along chromosome 6; vertical axis plots the founder allele effects from the linkage mapping model. Each colored line represents the effect of one of the eight founder alleles along the genome. (C) Association mapping within the QTL support interval shows a band of SNPs with high LOD scores (in red) that cover Rimklb, Mfap5, Aicda, Gm16556, Apobec1, Gdf3, Gm26168, and Dppa3. Apobec1 is highlighted in red because it has been associated previously with Apob C-to-U editing. (D) Genotypes of DO mice at the location of the maximum LOD score (horizontal axis) vs. the editing ratio (vertical axis). DO mice may have one of 36 unphased genotypes, and these are denoted by two letters, each referring to one founder, along the horizontal axis. (E) Apob C-to-U editing ratio in the eight DO founders shows that the editing pattern is similar to the founder allele effects near Apobec1.
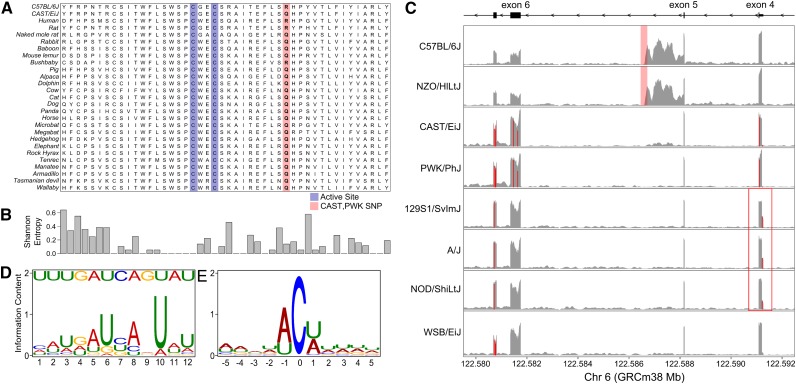
To better understand the mechanism driving the genetic variation in C-to-U editing, we examined SNPs and small insertions and deletions (indels) in Apobec1 that distinguish four haplotype groups: (1) CAST/EiJ and PWK/PhJ, (2) C57BL/6J and NZO/HlLtJ, (3) A/J, 129S1/SvlmJ, and NOD/ShiLtJ, and (4) WSB/EiJ. Fourteen SNPs or indels are shared by PWK/PhJ and CAST/EiJ, one of which creates an amino acid substitution in Apobec1 (rs50844667, 122,581,478 bp); a positively charged arginine (R), located near the catalytic domain, is replaced by a neutral glutamine (Q) (Figure 4A). The glutamine variant found in CAST/EiJ and PWK/PhJ is shared by most species of mammals (Figure 4B). Only humans, rats (reference strain BN/SsNMcw), and most laboratory mouse strains, including the reference strain C57BL/6J, share the arginine residue at this site. The high editing ratio observed in CAST/EiJ and PWK/PhJ strains suggests that the glutamine allele of APOBEC1 may have increased catalytic activity.
Figure 4.
Genomic variants in the DO founders fit the pattern of Apob editing. (A) A nonsynonymous SNP in exon 6 of Apobec1 contributed by CAST/EiJ and PWK/PhJ converts an arginine (R) residue to a glutamine (Q), highlighted in red. Residues in the active site are highlighted in blue. (B) Most mammals have a glutamine residue at this location. Shannon entropy for each base position shows that the glutamine is somewhat conserved. (C) RNA-seq pileups of Apobec1 in the eight DO founders show transcriptional variation between alleles. C57BL/6J and NZO/HlLtJ alleles carry an insertion (shaded in red) in the fifth intron that overlaps a retained intron. 129S1/SvImJ, A/J, and NOD/ShiLtJ alleles carry a SNP (red box) in the 5′ UTR of exon 4 that has increased expression in those strains. The y-axis shows the read depth normalized to library size. (D) Consensus mooring sequence motif for C-to-U edited genes. (Top) The Apobec1 mooring sequence. (Bottom) The consensus binding motif discovered using MEME. (E) Sequence information content around the edited C-to-U site shows that bases at the proximal and distal positions are either A or U. There does not appear to be sequence preference at other nearby base positions.
Three Apobec1 variants were shared by C57BL/6J and NZO/HlLtJ strains. One is a 262-nt insertion (relative to the other six strains) in the fifth intron (122,586,456–122,586,718 bp) (Figure 4C). Examination of RNA-seq reads at Apobec1 in the eight founder samples shows an aberrant splicing pattern that occurs in ∼50% of transcripts from these strains; the fifth exon is extended by ∼1245 nt. This alternatively spliced transcript introduces 15 stop codons into the reading frame; thus, the mRNA may produce both a truncated transcript and a corresponding truncated protein. Overall, there appears to be a twofold reduction of total Apobec1 expression in C57BL/6J and NZO/HlLtJ strains (Figure S2). However, when we estimated the normalized coverage of exon 6 alone, we found that the six strains that do not contain the intronic insertion have three- to fivefold higher coverage, suggesting that they may have three to five times the number of full-length Apobec1 transcripts.
There is a single SNP shared by A/J, 129S1/SvlmJ, and NOD/ShiLtJ strains in exon 4 (rs51979390, 122,591,220 bp) (Figure 4C) that occurs at the splice junction and is associated with expression of a long Apobec1 isoform (NM_031159) in these strains. The long isoform adds an extra 56 nt to the 5′ end of exon 4, which contains the 5′ UTR. The SNP is 40 nt downstream of the start of the longer exon 4 and 16 nt upstream of the shorter exon 4. Typically, the short isoform (NM_001134391) accounts for >95% of the total Apobec1 mRNA (Nakamuta et al. 1995), but we observed high levels of the long isoform in A/J, 129S1/SvlmJ, and NOD/ShiLtJ strains. While expression of the long isoform is increased in these three strains, the total expression level of functional Apobec1 is reduced compared to the WSB/EiJ strain. Expression of Apobec1 is highest in WSB/EiJ (Figure S2). These polymorphisms define an allelic series at Apobec1 with four functionally distinct groups. They combine to form 10 distinct genotypic classes in the outbred DO mice, which is consistent with the complex and continuous variation in C-to-U editing ratio (Figure 3D).
The “mooring” sequence, where the APOBEC1 complementation factor (A1CF) binds to Apob, is thought to be important in determining specificity of Apob editing (Smith et al. 2012). We searched for the mooring sequence within a 30-nt window downstream of the editing site in all 59 C-to-U editing sites with a strong association on chromosome 6 and found a consensus motif that is similar to the Apob sequence and is similar to previous reports (Figure 4D and Table S3) (Rosenberg et al. 2011). In Apob, the mooring sequence begins 3 nt downstream of the edited base. In other genes, the motif was found between 0 and 18 nt from the edited base. We did not find a correlation between the distance of the motif to the edited base and either the MEME enrichment p-value or the mean editing ratio in the DO population. Although we found SNPs in the mooring sequence of four sites and short indels (<50 nt) between the editing site and the distal end of the mooring sequence in two editing sites, we did not find any correlation between these genomic variants and the editing ratio. Thus, although A1cf may play a role in C-to-U editing, we did not find that genetic variation within the mooring sequence influences editing efficiency. Consistent with previous work, we observed enrichment of A and U bases adjacent to the edited base (Figure 4E) (Rosenberg et al. 2011). We also found that the bases adjacent to the editing site were enriched for sequences with an A in the 5′ position (χ2 test, p = 1.9 × 10−4); 44 sites (75%) contained an A, whereas 15 sites (25%) contained a U.
Our results suggest that variation in C-to-U editing ratios in the DO population is regulated by an allelic series of the editing enzyme Apobec1. These findings are consistent with a recent report of editing variation in the AXB/BXA recombinant inbred panel (Hassan et al. 2014). Of the remaining C-to-U editing sites, 11 show local genetic associations that map to the region of the edited gene. Two C-to-U editing sites map to distant loci other than chromosome 6. Six of the 11 sites have unusually high LOD scores (>30), and it is possible that some of these sites are the result of errors in the data or to unannotated paralogous variation. Of the local QTL, three genes with moderate but significant LOD scores, Aldh6a1, Gramd1c, and Lamp2, showed some evidence of regulation by the Apobec1 locus. We searched for other distant QTL that might regulate C-to-U editing but did not find a consistent signal around the 11 local C-to-U editing sites. Curiously, the three sites in Dcn are anticorrelated with the pattern of allele effects at the Apobec1 locus, an observation for which we currently have no explanation.
A-to-I editing
A very different picture emerges from genetic analysis of the 96 A-to-I edited sites detected in this study. Although approximately half (47) of these sites showed no evidence of editing ratio variation across the founder strains, a total of 49 A-to-I editing sites in 22 genes varied significantly across the founder strains and revealed significant associations in the DO population. Unlike C-to-U editing, where most of the sites displayed a similar pattern of allele effects, A-to-I editing sites were highly variable, suggesting independent genetic regulation for each site. The only exceptions were for editing sites located within the same gene where the pattern of allele dependence was similar for all sites within that gene.
These observations suggest that local genetic factors drive variation in A-to-I editing frequency. When we examined the support interval for each association, we found that the interval included the edited gene itself. It is known that the secondary structure of target mRNAs is an important factor for A-to-I editing site specificity (Dawson et al. 2004; Rieder and Reenan 2012). Thus, we hypothesized that cis-acting variants alter mRNA secondary structure near the editing site. To test this hypothesis, we examined the relationship between genetic variants, target RNA structure, and the edited sites. To do so, full-length target RNAs of each founder strain were imputed from known SNPs and indels. These RNAs were folded in silico, and their structure was examined for strain-dependent differences.
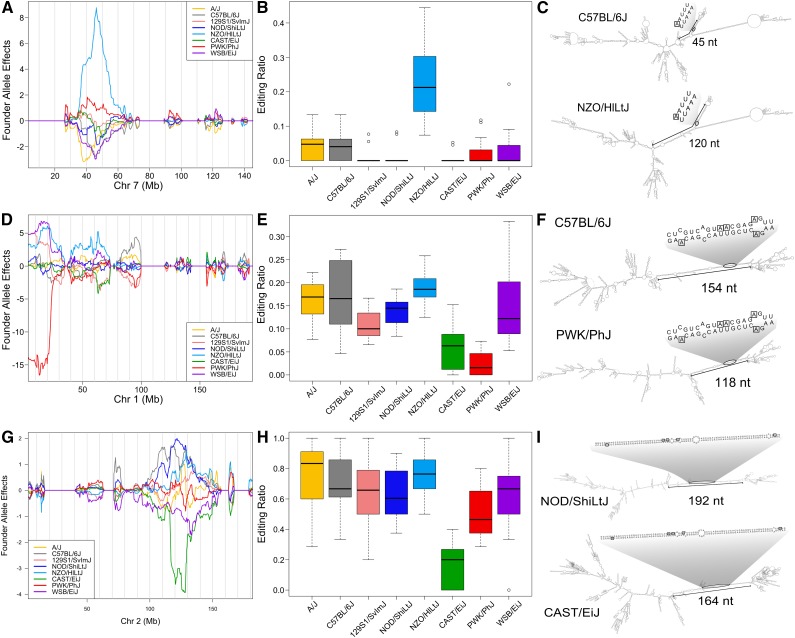
Because A-to-I editing enzymes prefer double-stranded regions of RNA (Lehmann and Bass 1999), we focused on the impact of genetic variants on double-stranded regions of the target mRNA and whether these regions were associated with edited sites. The long, noncoding (linc) RNA 0610005C13Rik contains one editing site for which the NZO/HlLtJ allele shows the highest editing (Figure 5, A and B). The NZO/HlLtJ allele carries 56 SNPs and 9 structural variants in 0610005C13Rik, and these lead to a structural change compared to the reference sequence. In allele C57BL/6J, there is a 45-nt region of predominantly dsRNA around the editing site, while the NZO/HlLtJ transcript contains a 120-nt dsRNA region. RNAfold simulations of the ensemble of structures suggest that the editing site has a high probability of occurring in the stemlike region in all strains. However, the double-stranded feature is unusually stable in the NZO/HlLtJ strain, which shows the highest editing ratio (Z = −13.77 for the most stable locally folded structure of NZO/HILtJ). Given these observations and the fact that editing efficiency is known to positively correlate with dsRNA length, we examined other A-to-I editing sites and asked whether the editing site resided within regions of dsRNA that varied across strains.
Figure 5.
Structural differences between the founder strains may influence A-to-I editing efficiency. (A) Founder allele effects for 0610005C13Rik show that DO mice carrying the NZO/HlLtJ allele have more edited transcripts. (B) Among the DO founder strains, NZO/HlLtJ has the highest editing ratio. (C) RNA secondary structure of the full mRNA for 0610005C13Rik for C57BL/6J and NZO/HlLtJ shows that the NZO/HlLtJ strain contains a longer dsRNA region around the editing sites. The location of the editing sites on the full-length RNA is circled; the nucleotides around the editing site are enlarged and outlined by rectangles. (D) Founder allele effects for Lact2b show that DO mice carrying the PWK/PhJ allele have lower editing. (E) Among the DO founder strains, PWK/PhJ has the lowest editing ratio. (F) Full-length mRNA for Lact2b in C57BL/6J and PWK/PhJ shows that PWK/PhJ carries a shorter dsRNA region near the editing sites. (G) Founder allele effects for 2010106G01Rik in DO mice show a complex pattern of editing ratios, with NOD/ShiLtJ having high editing and CAST/EiJ having low editing. (H) Editing ratios. The founders are similar to the allele effects observed in the DO population but differ in the order of editing ratios. (I) Full-length mRNA for 2010106G01Rik shows that the dsRNA region around the editing sites is slightly shorter in CAST/EiJ.
Lactamase β2 (Lactb2) contains nine edited sites, of which five have significant QTL. The editing frequency at these five sites is low compared to other strains when the PWK/PhJ allele is present in DO mice (Figure 5D), and this effect is recapitulated in the founder strains (Figure 5E). The computed structure of the full mRNA of PWK/PhJ contains a shorter length of dsRNA than the reference C57BL/6J strain (Figure 5F), and this may reduce editing efficiency. Analogous to 0610005C13Rik, the local stability of the Lact2b RNA structure trended with the editing ratios across strains. For example, the most stable local structure of PWK/PhJ is the least stable compared to the other strains, and PWK/PhJ has the lowest editing ratio. The gene Signal Peptide Peptidase Like 2A (Sppl2a) contains five A-to-I edited sites, and all have significant QTL. The founder allele effects in the DO population (Figure 5G) suggest that NOD/ShiLtJ alleles will have higher editing efficiency than CAST/EiJ alleles. Editing ratios in the founders (Figure 5H) are somewhat similar to the DO allele effects. NOD/ShiLtJ, which has higher editing than CAST/EiJ, has a longer stretch of dsRNA in the full mRNA folded structure than CAST/EiJ (Figure 5I). Recent reports have shown evidence of cis-acting intronic dsRNA elements that influence editing (Daniel et al. 2012). We searched for promiscuous editing in the three genes shown in Figure 5 but did not find evidence of editing at sites other than the target editing sites.
There were four distant associations among the A-to-I editing sites, and each association occurred in a different genomic location, suggesting that unlike C-to-U editing, there is no single gene that drives A-to-I editing in the livers of DO mice. There are some intriguing candidate genes under these associations. The association on chromosome 1 for Tmem245 contains transfer RNA (tRNA) splicing endonuclease 15 homolog (Tsen15). The association on chromosome 2 for Cd200r3 contains adenosine deaminase like (Adal). While we have no molecular evidence of distant regulation of A-to-I editing, these associations may represent interactions between edited transcripts and genes under the associations.
These observations demonstrate that variation of the A-to-I editing ratio in the DO population is driven primarily by local variants. Given the strain-dependent changes in target RNA structure, it appears that these variants play a role in modulating the local RNA structure of edited nucleotides.
Discussion
We used RNA editing ratio as a quantitative trait and found that most C-to-U editing sites in our study were controlled by a single trans-acting factor at Apboec1 that encodes an enzyme (APOBEC1) that catalyzes C-to-U editing. It is generally thought that APOBEC1 is positioned at the editing site by a mooring sequence (Smith et al. 2012) that is recognized by an APOBEC1 cofactor, A1CF. Consistent with this model, most of our C-to-U sites contain a mooring-like motif. However, we found no evidence for linkage at the A1cf locus itself, or linkage to variants within the mooring sequence, suggesting that genetic variation within the mooring sequences or A1cf does not drive variation in C-to-U editing in the DO population. The DO founder strains carry only nonsynonymous variants in A1cf, and there is little variation in transcript levels in the founder strains (Figure S2). Another RNA binding protein, RBM47, has been identified recently as a required APOBEC1 cofactor for C-to-U editing in vivo (Fossat et al. 2015). However, as with A1cf, we did not find evidence for linkage to Rbm47 for any of the C-to-U sites studied, suggesting that neither putative cofactor of APOBEC1 is a driver of editing variation in the DO population. Our analysis strongly supports a role for Apobec1 in determining quantitative variations in C-to-U editing. The C57BL/6J and NZO/HlLtJ Apobec1 alleles with an alternative mRNA isoform are associated with both lower total mRNA and lower C-to-U editing. This outcome is particularly relevant given that most studies of C-to-U editing in mice use C57BL/6J, which may not be representative of murine C-to-U editing.
Apob editing is the most well-studied C-to-U editing event. Editing produces one of two versions of APOB, APOB48 (edited) and APOB100. APOB48 is involved in the transport of dietary fat by chylomicrons. APOB100 contains the LDL receptor binding site and is essential for the efficient clearance of LDL from the circulation. High levels of APOB100 are associated with atherosclerosis (Davidson and Shelness 2000). Given the large effect of polymorphisms on editing ratios in the DO population, it will be important to characterize how human genetic variation in APOBEC1 affects C-to-U editing ratios and downstream phenotypes. There are 22 SNPs within the exon boundaries of human APOBEC1, and these may alter the APOB48-to-APOB100 ratio as well as circulating LDL/high-density lipoprotein (HDL) ratios.
In contrast to C-to-U editing, A-to-I editing in the liver seems to be regulated mostly by local factors. We found associations for 49 A-to-I editing sites, all of them in the vicinity of the edited gene. In many cases, these genetic variants appear to affect dsRNA stability and length in the region containing the edited sites of target RNAs. The major ubiquitous A-to-I editing enzyme, ADAR, shows distinct and strong preferences for long, stable dsRNA (Herbert and Rich 2001). Thus, the observed alterations in RNA structure are consistent with a model in which variant-induced dsRNA destabilization or length reduction negatively affects RNA editing efficiency. These findings are in good agreement with studies of natural genetic variation in Drosophila showing that local variants near the edited site are an important determinant of editing efficiency (Sapiro et al. 2015). While the Adar family of genes contain missense polymorphisms in the DO population, these variants do not appear to influence A-to-I editing in the liver.
ADAR-mediated editing occurs in many tissues, and the alteration of ADARs is known to be detrimental. We found little evidence for trans-acting associations despite the existence of six polymorphisms between the eight founder strains that alter amino acids in Adar1 and several large indels in introns of Adarb1. While these polymorphisms may affect other non-editing-related functions of ADARs (Ota et al. 2013), these studies show that they have little or no impact on A-to-I editing in the liver. Our findings do not preclude the possibility that editing in other tissues may be affected by ADAR polymorphisms or by other proteins. In mammals, most A-to-I editing occurs in the brain, and in Drosophila, the fragile X mental retardation protein (FMRP) has been shown to affect editing at distant sites (Bhogal et al. 2011). In future studies it will be important to use natural genetic variation in the mouse to assess long- and short-range influences on RNA editing in the brain.
We have successfully identified associations and specific polymorphic loci that influence RNA editing. Both C-to-U and A-to-I editing is variable with clear underlying genetic drivers. We mapped multiple local associations for A-to-I editing sites and a shared distant association in Apobec1 for C-to-U editing sites in the DO population. Most of the local A-to-I editing associations map to SNPs in the edited transcript, and we have identified candidate polymorphisms for some genes that are likely to affect mRNA secondary structure. The distinct genetic architectures discovered for C-to-U and A-to-I editing may reflect the different functional constraints in these two editing pathways. C-to-U editing of the transcriptome occurs at low ratios at sites other than Apob, and functional polymorphisms in the central catalytic enzyme appear to be tolerated. In contrast, A-to-I editing is widespread, and polymorphisms in central enzymes (i.e. ADARs) are more likely to have pleiotropic effects that are detrimental to the organism. Thus, when variation in A-to-I editing is present, the causal polymorphisms are local, limiting the effects to a single transcript.
Acknowledgments
This work was funded by National Institutes of Health (NIH) grants P50-GM076468 and R01-GM070683 to G.A.C. and National Cancer Institute grant CA34196 to The Jackson Laboratory in support of The Jackson Laboratory’s shared services. J.H.C. was supported by NIH grant R21-HG007554. E.S. was supported by NIH grant K99/R00-K99HD083521. M.K. and A.A. were supported by NIH grants R01-DK066369, R01-DK058037, and R24-DK091207.
Footnotes
Communicating editor: E. G. Petretto
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.179481/-/DC1
Literature Cited
- Bailey T. L., Elkan C., 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36. [PubMed] [Google Scholar]
- Bass B. L., 2002. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71: 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass B., Hundley H., Li J. B., Peng Z., Pickrell J., et al. , 2012. The difficult calls in RNA editing. Nat. Biotechnol. 30: 1207–1209. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Bhogal B., Jepson J. E., Savva Y. A., Pepper A. S., Reenan R. A., et al. , 2011. Modulation of dADAR-dependent RNA editing by the Drosophila fragile X mental retardation protein. Nat. Neurosci. 14: 1517–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. H., Habib G., Yang C. Y., Gu Z. W., Lee B. R., et al. , 1987. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science 238: 363–366. [DOI] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 2008. Naive application of permutation testing leads to inflated type I error rates. Genetics 178: 609–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneck P., Nellåker C., R. E., McIntyre J. E., Buendia-Buendia S., Bumpstead, et al. , 2012. High levels of RNA-editing site conservation amongst 15 laboratory mouse strains. Genome Biol. 13: r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel C., Veno M. T., Ekdahl Y., Kjems J., Ohman M., 2012. A distant cis-acting intronic element induces site-selective RNA editing. Nucleic Acids Res. 40: 9876–9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson N. O., Shelness G. S., 2000. APOLIPOPROTEIN B: mRNA editing, lipoprotein assembly, and presecretory degradation. Annu. Rev. Nutr. 20: 169–193. [DOI] [PubMed] [Google Scholar]
- Dawson T. R., Sansam C. L., Emeson R. B., 2004. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J. Biol. Chem. 279: 4941–4951. [DOI] [PubMed] [Google Scholar]
- Flicek P., Amode M. R., Barrell D., Beal K., Brent S., et al. , 2012. Ensembl 2012. Nucleic Acids Res. 40: D84–D90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossat N., Tourle K., Radziewic T., Liebhold D., Studdert J.B., et al. , 2014. C to U RNA editing mediated by APOBEC1 requires RNA-binding protein RBM47. EMBO Rep. 15: 903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti D. M., Svenson K. L., Shabalin A., Wu L. Y., Valdar W., et al. , 2014. Quantitative trait locus mapping methods for diversity outbred mice. G3 4: 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberger S., Levanon E. Y., Paz-Yaacov N., Barzilai A., Safran M., et al. , 2010. Consistent levels of A-to-I RNA editing across individuals in coding sequences and non-conserved Alu repeats. BMC Genomics 11: 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T., Buaas F. W., Simons A. K., Ackert-Bicknell C. L., Braun R. E., et al. , 2012. Canonical A-to-I and C-to-U RNA editing is enriched at 3′UTRs and microRNA target sites in multiple mouse tissues. PLoS One 7: e33720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich I., Tamir H., Arango V., Dwork A. J., Mann J. J., et al. , 2002. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron 34: 349–356. [DOI] [PubMed] [Google Scholar]
- Hassan M. A., Butty V., Jensen K. D., Saeij J. P., 2014. The genetic basis for individual differences in mRNA splicing and APOBEC1 editing activity in murine macrophages. Genome Res. 24: 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A., Rich A., 2001. The role of binding domains for dsRNA and Z-DNA in the in vivo editing of minimal substrates by ADAR1. Proc. Natl. Acad. Sci. USA 98: 12132–12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K., Young S. G., Farese R. V., Jr, Ng J., Sande E., et al. , 1996. Targeted disruption of the mouse apobec-1 gene abolishes apolipoprotein B mRNA editing and eliminates apolipoprotein B48. J. Biol. Chem. 271: 9887–9890. [DOI] [PubMed] [Google Scholar]
- Hofacker I. L., Priwitzer B., Stadler P. F., 2004. Prediction of locally stable RNA secondary structures for genome-wide surveys. Bioinformatics 20: 186–190. [DOI] [PubMed] [Google Scholar]
- Huang S., Holt J., Kao C., McMillan L., Wang W., 2014. A novel multi-alignment pipeline for high-throughput sequencing data. Database (Oxford), bau057: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., et al. , 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiran A. M., O’Mahony J. J., Sanjeev K., Baranov P. V., 2013. Darned in 2013: inclusion of model organisms and linking with Wikipedia. Nucleic Acids Res. 41: D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K. A., Bass B. L., 1999. The importance of internal loops within RNA substrates of ADAR1. J. Mol. Biol. 291: 1–13. [DOI] [PubMed] [Google Scholar]
- Li B., Dewey C. N., 2011. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. B., Levanon E. Y., Yoon J. K., Aach J., Xie B., et al. , 2009. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324: 1210–1213. [DOI] [PubMed] [Google Scholar]
- Lorenz R., Bernhart S. H., Honer Zu Siederdissen C., Tafer H., Flamm C., et al. , 2011. ViennaRNA Package 2.0. Algorithms Mol. Biol. 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger S. C., Raghupathy N., Choi K., Simons A. K., Gatti D. M., et al. , 2014. RNA-seq alignment to individualized genomes improves transcript abundance estimates in multiparent populations. Genetics 198: 59–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamuta M., Oka K., Krushkal J., Kobayashi K., Yamamoto M., et al. , 1995. Alternative mRNA splicing and differential promoter utilization determine tissue-specific expression of the apolipoprotein B mRNA-editing protein (Apobec1) gene in mice: structure and evolution of Apobec1 and related nucleoside/nucleotide deaminases. J. Biol. Chem. 270: 13042–13056. [DOI] [PubMed] [Google Scholar]
- Ota H., Sakurai M., Gupta R., Valente L., Wulff B. E., et al. , 2013. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 153: 575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz N., Levanon E. Y., Amariglio N., Heimberger A. B., Ram Z., et al. , 2007. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 17: 1586–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt S. K., Neuberger M. S., 2003. In vitro deamination of cytosine to uracil in single-stranded DNA by apolipoprotein B editing complex catalytic subunit 1 (APOBEC1). J. Biol. Chem. 278: 19583–19586. [DOI] [PubMed] [Google Scholar]
- Pickrell J. K., Gilad Y., Pritchard J. K., 2012. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome.” Science 335: 1302 (author reply: 1302). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G., Li J. B., 2014. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 42: D109–D113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder L. E., Reenan R. A., 2012. The intricate relationship between RNA structure, editing, and splicing. Semin. Cell Dev. Biol. 23: 281–288. [DOI] [PubMed] [Google Scholar]
- Rosenberg B. R., Hamilton C. E., Mwangi M. M., Dewell S., Papavasiliou F. N., 2011. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat. Struct. Mol. Biol. 18: 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapiro A. L., Deng P., Zhang R., Li J. B., 2015. Cis regulatory effects on A-to-I RNA editing in related Drosophila species. Cell Reports 11: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. C., Bennett R. P., Kizilyer A., McDougall W. M., Prohaska K. M., 2012. Functions and regulation of the APOBEC family of proteins. Semin. Cell Dev. Biol. 23: 258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson K. L., Gatti D. M., Valdar W., Welsh C. E., Cheng R., et al. , 2012. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics 190: 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R. H., Lindblad-Toh K., Birney E., Rogers J., Abril J. F., et al. , 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Yalcin B., Wong K., Agam A., Goodson M., Keane T. M., et al. , 2011. Sequence-based characterization of structural variation in the mouse genome. Nature 477: 326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B., Adams D. J., Flint J., Keane T. M., 2012. Next-generation sequencing of experimental mouse strains. Mamm. Genome 23: 490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The FASTQ files for the founder and DO RNA-seq runs are archived at the Gene Expression Omnibus under accession number GSE45684 (Munger et al. 2014). The alignment pipeline, including alignment and RNA editing site calling, is described in File S1, File S2, File S3, File S4, File S5, File S6, File S7, File S8, File S9, File S10, File S11, and File S12.