Abstract
Predicting the accuracy of estimated genomic values using genome-wide marker information is an important step in designing training populations. Currently, different deterministic equations are available to predict accuracy within populations, but not for multipopulation scenarios where data from multiple breeds, lines or environments are combined. Therefore, our objective was to develop and validate a deterministic equation to predict the accuracy of genomic values when different populations are combined in one training population. The input parameters of the derived prediction equation are the number of individuals and the heritability from each of the populations in the training population; the genetic correlations between the populations, i.e., the correlation between allele substitution effects of quantitative trait loci; the effective number of chromosome segments across predicted and training populations; and the proportion of the genetic variance in the predicted population captured by the markers in each of the training populations. Validation was performed based on real genotype information of 1033 Holstein–Friesian cows that were divided into three different populations by combining half-sib families in the same population. Phenotypes were simulated for multiple scenarios, differing in heritability within populations and in genetic correlations between the populations. Results showed that the derived equation can accurately predict the accuracy of estimating genomic values for different scenarios of multipopulation genomic prediction. Therefore, the derived equation can be used to investigate the potential accuracy of different multipopulation genomic prediction scenarios and to decide on the most optimal design of training populations.
Keywords: genomic prediction, multipopulation, accuracy, prediction equation, genomic selection, GenPred, shared data resource
GENOMIC markers can be used to estimate genomic values of individuals, also known as additive genetic values or breeding values, that are used to select animals (e.g., Dekkers 2007; De Roos et al. 2011) and plants for breeding (e.g., Heffner et al. 2009; Jannink et al. 2010) and in humans to predict the genetic risk of diseases (e.g., Wray et al. 2007; De Los Campos et al. 2010). In genomic prediction, genome-wide single-nucleotide polymorphism (SNP) marker information is used to predict genomic values based on SNP effects estimated in a training population consisting of individuals with known SNP genotypes and phenotypes (Meuwissen et al. 2001). The accuracy of estimating genomic values is in general higher when the size of the training population is larger, when the level of linkage disequilibrium (LD) between the SNPs and the quantitative trait loci (QTL) underlying the trait is higher, and when the predicted individuals are more related to the individuals in the training population (e.g., Daetwyler et al. 2008; Zhong et al. 2009; De Los Campos et al. 2013; Wientjes et al. 2013).
For numerically small populations, the size of the training population is limited, which restricts the accuracy of genomic prediction. Therefore, combining different populations in one training population for estimating SNP effects is an appealing approach to increase the size of the training population and, thereby, the accuracy of predicting genomic values. The potential accuracy of combing different populations in one training population has been investigated by combining populations from different breeds (e.g., Hayes et al. 2009a; Harris and Johnson 2010), lines (e.g., Zhong et al. 2009; Calus et al. 2014; Lehermeier et al. 2014), subpopulations (e.g., De Los Campos et al. 2013), or countries (e.g., Lund et al. 2011; Haile-Mariam et al. 2015). The increase in accuracy by adding individuals from another population to the training population is in most cases much lower than the increase in accuracy obtained by adding an equal number of individuals from the same population. This is a result of differences that exist between populations, like differences in allele frequencies, LD patterns (De Roos et al. 2008; Zhong et al. 2009; De Los Campos et al. 2012), allele substitution effects of QTL (Spelman et al. 2002; Thaller et al. 2003; Wientjes et al. 2015b), environments in combination with genotype-by-environment interactions (Lund et al. 2011; Haile-Mariam et al. 2015), the presence of QTL that are segregating only in one population (Kemper et al. 2015), and the absence of close family relationships across populations.
Different deterministic equations are available to calculate the accuracy of genomic prediction when the training population is a subset from the same population as the predicted individuals (Daetwyler et al. 2008; Vanraden 2008; Goddard 2009). One type of deterministic equation is based on prediction error variance of the mixed-model equation and uses the genomic relationships within the training population and between training and predicted individuals (Vanraden 2008). This equation has been extended to enable the calculation of the accuracy when different populations are combined in one training population (Wientjes et al. 2015b). A disadvantage of this equation is, however, that individuals have to be genotyped before the accuracy can be calculated. Therefore, this equation cannot be used to decide on the most optimal design of training populations. Another type of deterministic equation is able to predict the accuracy before genotype information is available and is based on population parameters, such as the size of the training population, the heritability of the trait, and the effective number of chromosome segments (Daetwyler et al. 2008, 2010). This equation can be used to investigate the accuracy of different training population designs; however, the equation is not applicable for situations with more than one population in the training population.
The first objective of this study is to develop a deterministic equation using population parameters to predict the accuracy of genomic values when different populations are combined in one training population. The different combined populations might, for example, be populations from different lines or environments or populations measured for different traits. The second objective is to validate the derived equation. For the validation, different scenarios of multipopulation genomic prediction were considered by dividing 1033 Holstein–Friesian cows with real genotypes and simulated phenotypes into three populations, assuming different heritabilities within populations and different genetic correlations between populations. Moreover, the equation was used to investigate the potential accuracy for one specific dairy cattle scenario and one specific human scenario.
Materials and Methods
Theory
The accuracy of estimated genomic values (rEGV) is defined as the correlation between estimated and true genomic values. The overall accuracy depends on the square root of the proportion of genetic variance captured by the SNPs (rLD) and on the accuracy of estimating SNP effects (reffect) (Daetwyler 2009; Goddard 2009). The rLD depends on the strength of LD between QTL and SNPs; the stronger the LD, the higher the proportion of the genetic variance that is captured by the SNPs. The reffect depends on the characteristics of the trait, the population in which the effects are estimated, and the population in which the effects are used to predict genomic values. First, we derive reffect for a training population consisting of two distinct populations, based on the same assumptions as underlying a commonly used prediction equation for single-population genomic prediction. Thereafter, reffect is combined with rLD to account for the proportion of the genetic variance captured by the SNPs to derive the accuracy of multipopulation genomic prediction.
Using the assumptions that M independent loci are underlying the trait and that each locus is explaining an equal amount of the genetic variance, Daetwyler et al. (2008) derived the following prediction equation for reffect when considering single-population genomic prediction,
| (1) |
in which h2 is the heritability of the trait and N is the number of individuals with phenotypes and genotypes included in the training population. The original derivation of this equation is rather complex and difficult to extend to multipopulation genomic prediction. As shown by Wientjes et al. (2015b), the same equation can also be derived by partitioning the variance of the average phenotype of N individuals into a part explained by one locus and a part not explained by that locus in which is the total genetic variance and is the phenotypic variance. In general, the accuracy of predicting an effect is equal to the square root of the proportion of the total variance explained by that effect (Appendix A provides a formal proof that this result applies to estimation of gene effects). So, the accuracy of predicting the effect of one locus equals
| (2) |
Since each locus is assumed to explain only very little variance, Due to the assumption that each locus explains an equal amount of the genetic variance, the accuracy of estimating the effect of one locus is the same for each of the loci and represents the overall accuracy of estimating SNP effects (see Appendix A):
| (3) |
Thus, this approach results in the same equation to predict the accuracy as derived by Daetwyler et al. (2008). The derivation described in Equations 2 and 3 is, however, much simpler, and this derivation will be extended to derive the accuracy of multipopulation genomic prediction.
Similar to Daetwyler et al. (2008), we assume that M independent loci are underlying the trait and that each locus explains an equal amount of the genetic variance. The effects of the loci might be different in each population, which is measured by the genetic correlation between populations. Furthermore, we assume that NA individuals from population A and NB individuals from population B with phenotype and genotype information are combined into one training population to estimate SNP effects. These estimated SNP effects are then used to predict genomic values of individuals from population C that could be a sample from one of the training populations or could be from a different population. The information from populations A and B, used to estimate SNP effects, is combined in a selection index approach (Hazel 1943), using the average phenotype of NA individuals from population A (xA) and the average phenotype of NB individuals from population B (xB) as records and the genomic values of individuals from population C as breeding goal traits,
| (4) |
in which bA and bB are the regression coefficients on the average phenotype of individuals from population A (xA) and B (xB) to predict genomic values for individual i from population C ().
The regression coefficients of genomic values of individuals from population C on the average phenotype of population A and B can be calculated as
| (5) |
in which P is the (co)variance matrix of xA and xB and g is a vector with covariances between xA and xB and the true genomic value of individual i from population C (),
| (6) |
and
| (7) |
In analogy with Wientjes et al. (2015b), the variance of the average phenotype of NA individuals can be partitioned into a part explained by one locus and a part not explained by that locus in which is the total genetic variance in population A and is the total phenotypic variance in population A. So, the total variance of xA can be written as
| (8) |
Note that represents the part of the phenotypic variance not explained by that locus, i.e., the residual variance () for one locus j.
The covariance between the average phenotypes in the two populations can be partitioned into a part explained by one locus, a part not explained by that locus, and twice the covariance between the two parts. In an additive model, and the parts not explained by a locus, i.e., the residual variances, are expected to be independent across populations, indicating that only the covariance between the populations of the part explained by one locus is assumed to differ from zero. Therefore, the covariance can be written as
| (9) |
in which and are the genetic standard deviations in, respectively, populations A and B and is the genetic correlation between populations A and B. Hence,
| (10) |
in which is the total genetic variance in population B and is the total phenotypic variance in population B.
Since an additive model is assumed, the covariance between the average phenotype of population A and the true genomic value of individual i from population C is also equal to the covariance between the populations of the part explained by one locus,
| (11) |
in which is the genetic standard deviation in population C and is the genetic correlation between populations A and C. Hence,
| (12) |
in which is the genetic correlation between populations B and C. Substituting Equations 10 and 12 in Equation 5 results in
| (13) |
With some algebra (see Appendix B), it can be shown that the accuracy of this selection index, representing the accuracy of estimating SNP effects, equals
| (14) |
When only one population is included in the training population, Equation 14 reduces to
| (15) |
This equation is equivalent to the equation of Wientjes et al. (2015b) for across-population genomic prediction. When estimated SNP effects are applied in another subset of the same population as the training population, i.e., = 1, Equation 15 becomes equivalent to the equation derived by Daetwyler et al. (2008) to predict the accuracy of estimating SNP effects within a population (Equation 1).
As explained before, the accuracy of genomic prediction depends on reffect as well as on rLD, accounting for the proportion of the genetic variance captured by the SNPs. It might, for example, be that the SNP effects are accurately estimated (reffect = 1), but when LD between QTL and SNPs is not complete, not all genetic variance can be captured by the SNPs and the accuracy of genomic prediction is still not 1. Moreover, when a number of QTL are segregating in the predicted population and not in the training population, part of the genetic variance in the predicted population can never be captured by the SNPs in the training population. Altogether, this indicates that the proportion of the genetic variance in the predicted population that can be captured by the SNPs in the training population is specific for a combination of training and predicted populations. Therefore, rLD affects the covariance between the phenotypes in the training population and the aggregated genotype of the predicted individuals (Equation 12), which results in
| (16) |
in which is the square root of the proportion of the genetic variance in predicted population C captured by the SNPs in training population A, and is the square root of the proportion of the genetic variance in predicted population C captured by the SNPs in training population B. Using Equation 16 instead of Equation 12 in the remaining part of the derivation results in the following equation to predict the accuracy of genomic prediction:
| (17) |
In this study, and were assumed to be characteristics of the training and predicted populations and depending on the SNP density and the properties of the QTL underlying the trait. Therefore, an empirical approach was needed to estimate values for and The values were estimated in the scenarios when only one population (A or B) was used as training population, by calculating rLD as in which rEGV was the empirical accuracy and reffect the predicted accuracy assuming all genetic variance in the predicted population was captured by the SNPs. The empirically estimated values for and were used to predict the accuracy when populations A and B were combined in the training population to predict genomic values for individuals from population C.
Derivation of Me to replace M
An important assumption underlying the derived equation is that M independent loci are underlying the trait. In a finite population, loci do not segregate independently due to linkage disequilibrium between loci. The equation predicting the accuracy of SNP effects using a single population (Equation 1), derived by Daetwyler et al. (2008), accounts for that by replacing M by the effective number of chromosome segments, Me, in the population (Daetwyler et al. 2010). The Me within a population is a statistical concept and can be interpreted as the effective number of chromosome segments that are independently segregating in that population. In other words, it represents the effective number of effects that has to be estimated to predict genomic values for individuals from that population. In the derived equation for multipopulation genomic prediction, different populations are combined in the training population, each with different values for Me. For predicting genomic values for individuals from population C, using estimated SNP effects in population A, the effective number of estimated effects is equal to the effective number of chromosome segments shared between populations A and C (). Equivalently, when estimated SNP effects in population B are used, the effective number of estimated effects is equal to the effective number of chromosome segments shared between populations B and C (). In analogy of Me within a population, the Me across populations can be interpreted as the effective number of segments that are segregating in a combined population, when considering the differences in LD between the populations. Therefore, we propose the following adjustment to Equation 17:
| (18) |
The same equation can also be derived when a selection index is used, combining estimated genomic values for individuals from population C based on training populations of, respectively, population A or B, as shown in Appendix C.
The Me within a population can be calculated as
| (19) |
(Goddard et al. 2011), in which Gij contains the genomic relationship and E(Gij) the expected values for the genomic relationships between all individuals i and j from that population, with the variance taken over all pairwise relationships between individuals i and j. In analogy to Equation 19, the values for Me across populations can be calculated using
| (20) |
(Wientjes et al. 2015b), in which contains the genomic relationships and E() contains the expected genomic relationships between all individuals i from population 1 and individuals j from population 2, again with the variance taken over all pairwise relationships between individuals i and j. The genomic relationships can be calculated following Yang et al. (2010), by calculating the genomic relationships between individual i from population y and individual j from population z as and the genomic relationship of individual i from population y with itself as in which n is the number of SNPs; and are the genotypes at locus k coded as 0, 1, and 2; and and are the allele frequencies for the second allele (with homozygote genotype coded as 2) at locus k for, respectively, populations y and z. The genomic relationships used to calculate Me are based on population-specific allele frequencies to ensure that unrelated individuals have an expected genomic relationship of 0, which is an underlying assumption of the equation to calculate Me (Goddard et al. 2011).
In most human studies, individuals included in the data are unrelated (e.g., Yang et al. 2010; Lee et al. 2012; Maier et al. 2015). This indicates that all expected genomic relationships (E(G)) would approximately be zero and Equation 20 simplifies to In most livestock studies, individuals are related, and E(G) could be approximated by the pedigree relationship matrix A; i.e., When the G and A matrices are used to calculate Me, both matrices should be scaled to the same base population. This can be achieved by rescaling the inbreeding level in G to the inbreeding in A, for example by using the following adjustment separately for each of the within-population and across-population blocks (Powell et al. 2010),
| (21) |
in which is the average pedigree inbreeding level of individuals in population b and J is a matrix filled with ones.
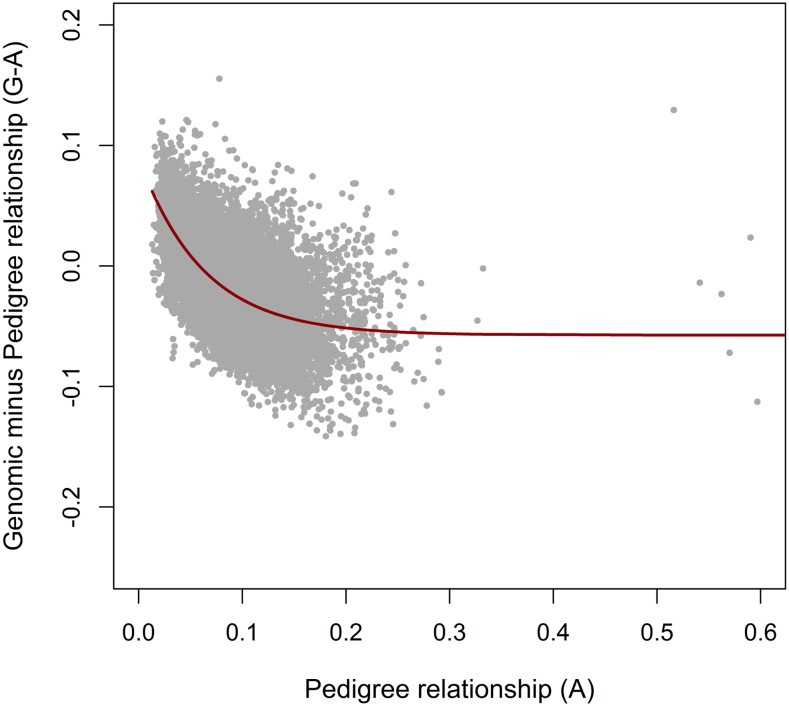
The G−E(G) values are expected to follow a normal distribution around zero for each value of E(G). The pedigree relationships between individuals in A, however, depend on the depth of the pedigree for both individuals. In general, the pedigree relationships will more closely resemble E(G) when the pedigree is deeper. When the pedigree is not deep or complete enough for all or a subset of the individuals, extra variation in G−A is introduced, resulting in an underestimation of Me when A is used to represent E(G). The impact of an insufficient pedigree depth on the calculated Me can be reduced by taking only the relationships of individuals with the most complete pedigree into account to calculate Me. To check whether selecting these individuals indeed minimized the impact of an insufficient pedigree depth, values of G−A can be plotted vs. values of A. When the values for G−A are lower for higher A values, as is shown in Figure 1, an insufficient pedigree depth is still influencing the calculation of Me. To account for this particular pattern, an exponential function was fitted through the data. For all values of A in the data, the parameters of the function were estimated in R (R Development Core Team 2011) and the fitted values of the function were subtracted from the values of G−A before calculating Me.
Figure 1.
The genomic minus pedigree relationships (G−A) vs. the pedigree relationships (A) for across-population elements between individuals of two populations. The red line is the fitted exponential function () used to correct G−A values to reduce the impact of an insufficient pedigree depth.
Validation
After deriving the equation, the aim was to validate it for a broad range of scenarios, differing in heritabilities within populations and genetic correlations between populations. These scenarios resemble the combining of populations from different environments or measured for different traits. For the validation, real genotypes and simulated phenotypes were used. A pedigree with on average 3.5 complete generations per individual was available, with a minimum of 1 complete generation and a maximum of 9 complete generations. In each of the scenarios, an empirical accuracy was calculated and compared with the predicted accuracy, using the derived equation to investigate how accurately the accuracy was predicted. The genotype and pedigree information from all individuals, as well as the simulated phenotypes, is available at http://dx.doi:10.5061/dryad.1525t.
Genotypes:
Genotypes were available for 1033 dairy cows from The Netherlands, each originating for at least 87.5% from the Holstein–Friesian breed; i.e., all animals were purebred Holstein–Friesians. Genotyping was done using the Illumina BovineSNP50 Beadchip (50k; Illumina, San Diego), after which genotypes were imputed to higher density (777k), using 3150 Holstein–Friesian animals as a reference population (Pryce et al. 2014). The accuracy of imputation across imputed loci, as reflected by the Beagle R2 value, was on average 0.96, indicating high imputation accuracy. As a quality control, SNPs with a call rate <95%, an unknown mapping position, located on the sex chromosomes, a minor allele frequency (MAF) < 0.005, for which only two genotypes were observed, and in complete linkage disequilibrium with a neighboring SNP were deleted. This quality control step reduced the number of SNPs for this study to 422,405.
A total of 50,000 candidate QTL were selected from the 422,405 SNPs, and in each replicate QTL were randomly sampled from the candidate QTL to simulate phenotypes for each individual. The candidate QTL were selected from the SNPs using two different approaches: (1) Candidate QTL were randomly selected (RANDOM) and (2) candidate QTL were selected from the SNPs with a MAF < 0.2 (LOW MAF), since the MAF of QTL underlying complex traits is expected to be lower than the MAF of SNPs (Goddard and Hayes 2009; Yang et al. 2010; Kemper and Goddard 2012) due to ascertainment bias of the SNPs on the SNP chips (Matukumalli et al. 2009). For each of the two approaches, the remaining 372,405 SNPs were used as markers. In this way, the QTL underlying a trait could be randomly sampled from the candidate QTL in each of the replicates, while the subset of SNP markers was constant across replicates for both RANDOM and LOW MAF.
Phenotypes:
The 1033 individuals were divided into three groups to represent different populations. The first two groups (populations 1 and 2) contained 450 individuals and represented the different training populations (populations A and B in the derived equation). The last group (population 3) contained 133 individuals and represented the group of predicted individuals for which genomic values were estimated (population C in the derived equation). The division over the groups was performed using pedigree information, by allocating paternal and maternal half-sib families to the same population. In this way, relationships within a population were higher than between populations, as usually would be expected for distinct populations.
For both the RANDOM and the LOW MAF approach of selecting candidate QTL, phenotypes were simulated by randomly sampling 4000 QTL from the group of 50,000 candidate QTL. The QTL underlying the trait were the same in each of the populations. For each QTL, allele substitution effects were sampled from a multivariate normal distribution, with a mean of 0 and standard deviation of 1, using different genetic correlations between the populations. Only additive effects and no dominance or epistatic interactions were assumed. True genomic values (TGVs) were calculated by multiplying the QTL genotypes, coded as 0, 1, and 2, by the simulated allele substitution effects of the population to which the individual belonged. Across populations, the TGVs were rescaled to a mean of 0 and a variance of 1. In each of the populations, the genetic variance was calculated as the variance of the TGVs for the individuals from that population. For all individuals, the environmental effect was sampled from N(0, × Var(TGVi)), in which Var(TGVi) is the variance of TGV in population i to which the individual belonged. For each individual, the simulated TGV and the environmental effect were summed to calculate the phenotype.
Scenarios:
Seven different scenarios of multipopulation genomic prediction were investigated, differing in heritabilities and genetic correlations between the populations (Table 1). The first four scenarios represent multienvironment genomic prediction, where populations in different environments were combined in one training population in which SNP effects were estimated. In these scenarios, the variances were assumed to be homogeneous; i.e., heritability was assumed to be the same in each population (0.95), but genetic correlations between populations varied from 0.4 to 1. The last three scenarios represent multitrait genomic prediction, where populations measured for different traits are combined in one training population. In these scenarios, variances were assumed to be heterogeneous; i.e., each population had a different heritability of 0.3 or 0.95, and genetic correlations between populations were 0.6 or 1. The values for the heritabilities of 0.3 and 0.95 were chosen to have a clear contrast between the populations.
Table 1. Overview of the different scenarios to simulate phenotypes.
| Heritability | Genetic correlation | ||||
|---|---|---|---|---|---|
| Scenariosa | Pop. 1 | Pop. 2 | Pop. 1 and 2 | Pop. 1 and 3 | Pop. 2 and 3 |
| Homogeneous variances | |||||
| HOM_1.0-0.6 | 0.95 | 0.95 | 0.60 | 1.00 | 0.60 |
| HOM_0.8-0.6 | 0.95 | 0.95 | 0.60 | 0.80 | 0.60 |
| HOM_0.8-0.4 | 0.95 | 0.95 | 0.60 | 0.80 | 0.40 |
| HOM_0.4-0.4 | 0.95 | 0.95 | 0.60 | 0.40 | 0.40 |
| Heterogeneous variances | |||||
| HET_1.0-1.0 | 0.95 | 0.30 | 1.00 | 1.00 | 1.00 |
| HET_1.0-0.6 | 0.95 | 0.30 | 0.60 | 1.00 | 0.60 |
| HET_0.6-1.0 | 0.95 | 0.30 | 0.60 | 0.60 | 1.00 |
Pop., population.
Scenarios are labeled as follows: The names of the scenarios assuming homogeneous variances in both training populations start with HOM, followed by the genetic correlation between populations 1 and 3 and the genetic correlation between populations 2 and 3. The names of scenarios with heterogeneous variances in the training populations start with HET, followed by the genetic correlation between populations 1 and 3 and the genetic correlation between populations 2 and 3.
In each scenario, population 1, population 2, or populations 1 and 2 were used as the training population and population 3 contained the predicted individuals. Each scenario was analyzed using both approaches of selecting QTL: RANDOM and LOW MAF. Simulations were replicated 100 times in each scenario.
Calculating Me:
Values for Me across the different populations were calculated based on the difference between the genomic and the pedigree relationship matrix. Since the subset of SNPs slightly differed between the two approaches of selecting candidate QTL, RANDOM and LOW MAF, values for Me were calculated for each of the approaches. To reduce the impact of incompleteness of the pedigree, only individuals with at least three generations of complete pedigree were taken into account, resulting in 329 individuals in population 1, 270 individuals in population 2, and 90 individuals in population 3. Thereafter, an exponential function was fitted through the data to further reduce the impact of an insufficient pedigree depth, as explained before. The G matrix was the same for all replicates, since the subset of 372,405 SNPs was constant for all replicates while QTL were resampled every replicate, resulting in the same Me for all replicates. Therefore, only one accuracy could be predicted for all replicates of the same approach of selecting candidate QTL, representing the expected average accuracy of estimating SNP effects.
Empirical accuracy of genomic prediction:
The empirical accuracies of genomic prediction were obtained both with a single-trait and with a multitrait Genomic Best Linear Unbiased Prediction (GBLUP) type of model run in ASReml (Gilmour et al. 2009), using the simulated phenotypes and including population as a fixed effect. Genomic values for the predicted individuals were estimated using a genomic relationship matrix, G, containing all training and predicted individuals and simulated phenotypes of the training individuals. The G matrix included in the models was calculated using the allele frequencies across all individuals without taking the population into account. The other steps in calculating G were the same as explained above.
In the single-trait model, variances were estimated using Residual Maximum Likelihood (REML). Therefore, the model used was termed Genomic-Relatedness-Matrix Residual Maximum Likelihood (GREML) instead of GBLUP, where variances are assumed to be known. In the single-trait model, the phenotypes of the different populations were pooled in one population, without taking the genetic correlations between the populations into account. The differences in heritability were, however, taken into account by weighting the phenotypes differently and in this way acknowledging that the phenotypes in one population were more accurately representing the genomic values of the individuals compared to the phenotypes in the other population. It was assumed that the heritability of the phenotypes from the population with the lowest heritability, i.e., a heritability of 0.3, represented the trait heritability based on one measurement. The phenotypes of individuals from this population were given a weight of 1. The heritability of the other population, i.e., a heritability of 0.95, represented the heritability based on multiple measurements of the same trait. In other words, it represented the reliability of the phenotype based on more than one record. This indicates that the genetic variance can be assumed to be the same in both populations. The weight for the phenotypes of individuals from the population with the highest reliability (r2) was equal to the ratio of the residual variances in both populations, which can be calculated as
| (22) |
Following Equation 22, a weight of 44.33 was given to the phenotypes from the population with a heritability of 0.95. One possible scenario where phenotypes could be weighted differently is in dairy cattle populations, where phenotypes of cows are generally based on one single measurement and phenotypes of bulls are based on different numbers of progeny, for which the same weights can be obtained following Garrick et al. (2009).
The multitrait model considered the phenotypes for the same trait in the different populations as different traits with a genetic correlation between the traits. Estimating all genetic correlations in the multitrait model was not possible, since phenotypes of the predicted individuals were not included in the model. Therefore, genetic correlations and variance components were assumed to be known and fixed to the simulated values, and the multitrait model was termed GBLUP.
For each of the models, the accuracy of genomic prediction was calculated as the correlation between the simulated TGVs and predicted genomic values. Note that the single-trait and multitrait GBLUP models use both SNP information and simulated phenotypes that differed across the replicates. Therefore, averages and standard errors across the replicates were calculated and compared to the predicted accuracies.
Evaluating the potential accuracies of two scenarios
The derived equation can be used to investigate the accuracy of different scenarios of multipopulation genomic prediction. To show this, we used Equation 18 to evaluate the potential accuracy for two specific scenarios, assuming that all genetic variance in the predicted population was captured by the SNPs in the training population ( = = 1). The first scenario is relevant for dairy cattle breeding, where bulls with deregressed estimated genetic values based on daughter information are in general used in the training population, with a heritability equal to the reliability of the estimated genetic values. Different studies have investigated the potential to increase the accuracy of genomic prediction by adding cows to the training population with their own phenotypes, which are in general less reliable than estimated genetic values (e.g., Calus et al. 2013; Cooper et al. 2015). In Equation 18 different numbers of cows (range 0–50,000) were added to a training population of 10,000 bulls, assuming a heritability of 0.05 for the phenotypes of cows that represents the heritability of a fertility trait in dairy cattle (e.g., Karoui et al. 2012), different reliabilities (range 0–1) for the estimated genetic values of bulls, and a genetic correlation of 1 between the estimated genetic values of bulls and the phenotypes of cows. The values for Me were set to the values derived from the cattle genotype data used in this study.
The second scenario is based on human studies, in which it was assumed that different numbers of individuals from a population of African descent (range 0–100,000) were added to a training population of 5000 individuals of European descent to increase the accuracy of predicting genetic risk for the European population. As an example, parameters for the trait schizophrenia were used, with a heritability of 0.28 in the European population, a heritability of 0.24 in the African population, and a genetic correlation of 0.66 between the populations (De Candia et al. 2013). The Me in the European population ( in Equation 18) was set to 43,000, based on the equation (Goddard 2009), an effective population size (Ne) of 10,000 (McEvoy et al. 2011), and a genome length (L) of 30 M (Venter et al. 2001). The Me across the populations ( in Equation 18) was varied (range 43,000–2,000,000).
Data availablity
The genotype and pedigree information from all individuals, as well as the simulated phenotypes, is available at http://dx.doi.org/10.5061/dryad.1525t. File Genotypes_422405SNPs contains the genotype for each individual. File Pedigree contains the pedigree for each individual. File ID_Population contains the division of the individuals over the populations. File Phenotypes_QTL_RANDOM contains the simulated phenotypes for each individual for the RANDOM scenario. File Phenotypes_QTL_LowMAF contains the simulated phenotypes for each individual for the LOW MAF scenario.
Results
In this section, the results of the prediction equation are first presented assuming that all genetic variance in the predicted population (population 3) is captured by the SNPs in the training population. These predicted accuracies were used to calculate and based on the ratio between the empirical and the predicted accuracy of genomic prediction when only one of the populations, population 1 or population 2, was used as the training population. As a next step, the calculated values for and were used to predict the accuracy of genomic prediction when populations 1 and 2 were combined in the training population.
Calculating Me
In Table 2, the different estimated Me values across populations are shown. Due to only small differences in the subset of SNPs used to calculate G, estimated Me values were very similar for the scenarios with QTL randomly sampled (RANDOM) and QTL sampled with a low MAF (LOW MAF). Using population-specific allele frequencies or allele frequencies across populations had only a very small effect on the estimated values for Me, as well as on the predicted accuracies (range −0.9%–1.3%). This indicates that, for this study, the use of population-specific allele frequencies or the allele frequency across populations did not influence the results, due to the very similar allele frequencies across the three populations. Therefore, the predicted accuracies are shown only for the Me values calculated based on a G matrix using the allele frequencies across the populations.
Table 2. Estimated Me values across populations, using population-specific allele frequencies or the allele frequency across populations to set up G.
| Scenario | Population-specific allele frequency | Allele frequency across populations |
|---|---|---|
| QTL with low MAF | ||
| Populations 1 and 3 | 1541 | 1515 |
| Populations 2 and 3 | 1616 | 1652 |
| QTL randomly sampled | ||
| Populations 1 and 3 | 1620 | 1585 |
| Populations 2 and 3 | 1694 | 1741 |
Scenarios with QTL randomly sampled (RANDOM)
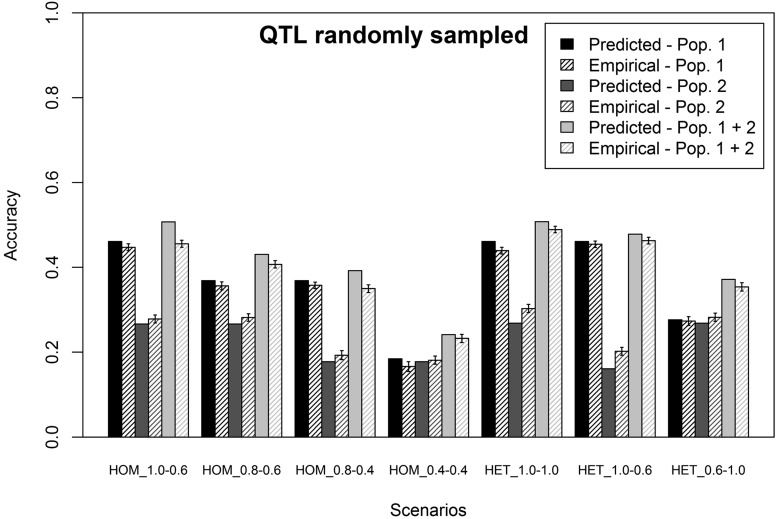
In this section, results are presented for the RANDOM scenarios of simulating phenotypes. For these scenarios, the predicted accuracies and average empirical accuracies of genomic prediction obtained with a single-trait model using either a single or a combined training population and different scenarios of simulated phenotypes are shown in Figure 2. The first four scenarios show the accuracies when different genetic correlations between the populations were simulated, with the same heritability in each of the populations. These scenarios show that when only one population was used as a training population, predicted and empirical accuracies were, as expected, higher when the genetic correlation between training and predicted individuals was higher. There was only a small difference between the accuracies obtained using population 1 or 2 as the training population when the genetic correlation with the predicted individuals was the same, because both populations were about equally related to the predicted individuals. Combining the two populations in one training population always resulted in an increase in both predicted and empirical accuracies. The magnitude of the increase in accuracy depended on the genetic correlation between the predicted individuals and the added population; the higher the genetic correlation, the higher the increase in accuracy.
Figure 2.
Predicted and empirical accuracies of genomic prediction (±SEs), using a single-trait model, one or two populations in the training population, QTL randomly sampled from the SNPs and assuming in the prediction equation that all genetic variance in the predicted population was captured by the SNPs in the training population. The different scenarios represent the different genetic correlations and heritabilities used to simulate phenotypes. The scenarios starting with HOM have homogeneous variances in both training populations, and the scenarios starting with HET have heterogeneous variances. For each scenario, HOM or HET is followed by the genetic correlation between populations 1 and 3 and the genetic correlation between populations 2 and 3.
The last three scenarios show the predicted and empirical accuracies, using different heritabilities in each of the populations and genetic correlations of 1 and 0.6 between populations. These scenarios show that when only one population was used as the training population, predicted and empirical accuracies were, as expected, higher when the heritability in the training population was higher. For this study, a heritability of 0.3 resulted in ∼60% of the accuracy obtained with a heritability of 0.95. Adding 450 individuals from the population with a low heritability to a training population of 450 individuals from the population with a high heritability, however, still resulted in an increase in accuracy. The increase in both predicted and empirical accuracies was again lower when the genetic correlation was lower, similar to the scenarios with the same heritability in each population.
For each of the scenarios, the predicted accuracy of genomic prediction shown in Figure 2 is assuming that = = 1. In general, predicted accuracies were very slightly overestimating the empirical accuracies of genomic prediction (±1%), both when the heritability was the same in each population and when the heritability was different. When population 1 was used as the training population, the overestimation was on average 4% (range 1–11%). When population 2 was used as the training population, the empirical accuracy was slightly underestimated by the predicted accuracy by on average 8% (range −20% to −2%). When both populations were combined in the training population, the overestimation was on average 6% (range 3–12%). These results indicate that when QTL were randomly sampled from the SNPs, most of the genetic variance in the predicted individuals was tagged by the SNPs in the training population, especially when population 2 was used as the training population, and the estimated value for = 0.96 and for = 1. Using these calculated values to predict the accuracy of genomic prediction for the combined training population reduced the overestimation of the empirical accuracy to 3%.
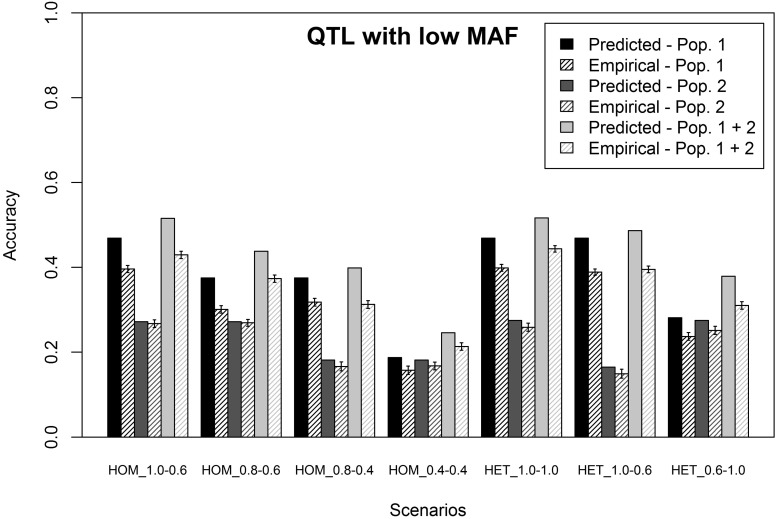
Scenarios sampling QTL with low MAF (LOW MAF)
In this section, results are presented for the LOW MAF scenarios of simulating phenotypes. For these scenarios, the predicted and average empirical accuracies of genomic prediction obtained with a single-trait model using either a single or a combined training population are shown in Figure 3, assuming = = 1. All empirical accuracies for the LOW MAF scenarios were lower than the accuracies obtained for the RANDOM scenarios. The predicted accuracies, however, were similar to the predicted accuracies for the RANDOM scenarios. So, the predicted accuracies for the LOW MAF scenarios overestimated the empirical accuracies to a greater extent. On average, the overestimation was ±15% and again higher when population 1 was used as the training population, compared to using population 2 as the training population (population 1, 20%; population 2, 7%; combined training population, 20%). These results indicate that, as expected, a smaller proportion of the genetic variance in the predicted individuals was tagged by the SNPs in the training population when QTL were sampled with a low MAF and the estimated value for = 0.84 and for = 0.94. Using these calculated values to predict the accuracy of genomic prediction for the combined training population reduced the overestimation of the empirical accuracy to 5%.
Figure 3.
Predicted and empirical accuracies of genomic prediction (±SEs), using a single-trait model, one or multiple populations in the training population, QTL sampled with a low minor allele frequency (MAF) and assuming in the prediction equation that all genetic variance in the predicted population was captured by the SNPs in the training population. The different scenarios represent the different genetic correlations and heritabilities used to simulate phenotypes. The scenarios starting with HOM have homogeneous variances in both training populations, and the scenarios starting with HET have heterogeneous variances. For each scenario, HOM or HET is followed by the genetic correlation between populations 1 and 3 and the genetic correlation between populations 2 and 3.
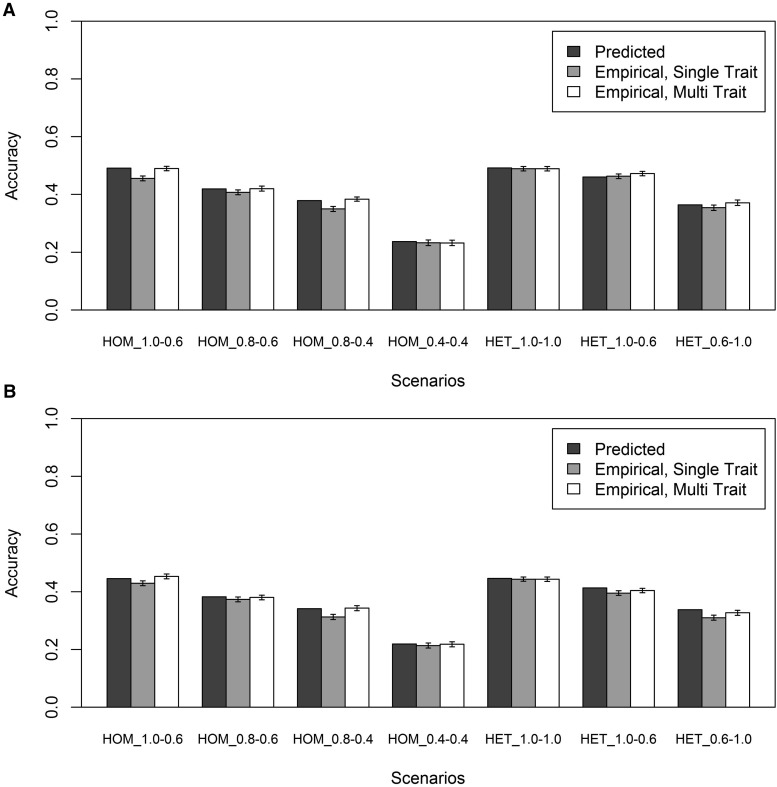
Single-trait vs. multitrait model
The analyses using a combined training population were performed using both a single-trait model and a multitrait model, where the same trait in the different populations was modeled as a different correlated trait. The accuracies from both models are shown in Figure 4, for the (Figure 4A) RANDOM and the (Figure 4B) LOW MAF scenarios. In Figure 4, the predicted accuracies for the combined training populations use the values of and estimated when only population 1 or 2 was included in the training population. In general, accuracies obtained with the multitrait model were equal to or higher than accuracies obtained with the single-trait model, depending on the genetic correlations. When the genetic correlations between both training populations and the predicted population were the same, accuracies obtained with the single-trait and the multitrait model were similar. When the genetic correlations were different, accuracies obtained with the multitrait model were higher than accuracies obtained with the single-trait model. Due to these higher empirical accuracies, the overestimation of the empirical accuracy obtained with the multitrait model by the predicted accuracy of genomic prediction using the estimated values of and reduced on average across replicates to 0% (range −2% to +2%) for the RANDOM scenarios and to 1% (range −2% to +3%) for the LOW MAF scenarios. This indicates that the equation can accurately predict the accuracy of genomic prediction when the proportion of the genetic variance in the predicted population not captured by the SNPs in the training population is known and taken into account.
Figure 4.
(A and B) Predicted and empirical accuracies of genomic prediction (±SEs), using a training population consisting of two populations and QTL (A) randomly sampled or (B) with a low minor allele frequency and accounting for the proportion of genetic variance in the predicted population captured by the SNPs in the training population in the prediction equation. Empirical accuracies were obtained with either a single-trait model or a multitrait model. The different scenarios represent the different genetic correlations and heritabilities used to simulate phenotypes. The scenarios starting with HOM have homogeneous variances in both training populations, and the scenarios starting with HET have heterogeneous variances. For each scenario, HOM or HET is followed by the genetic correlation between populations 1 and 3 and the genetic correlation between populations 2 and 3.
The potential accuracies of two scenarios
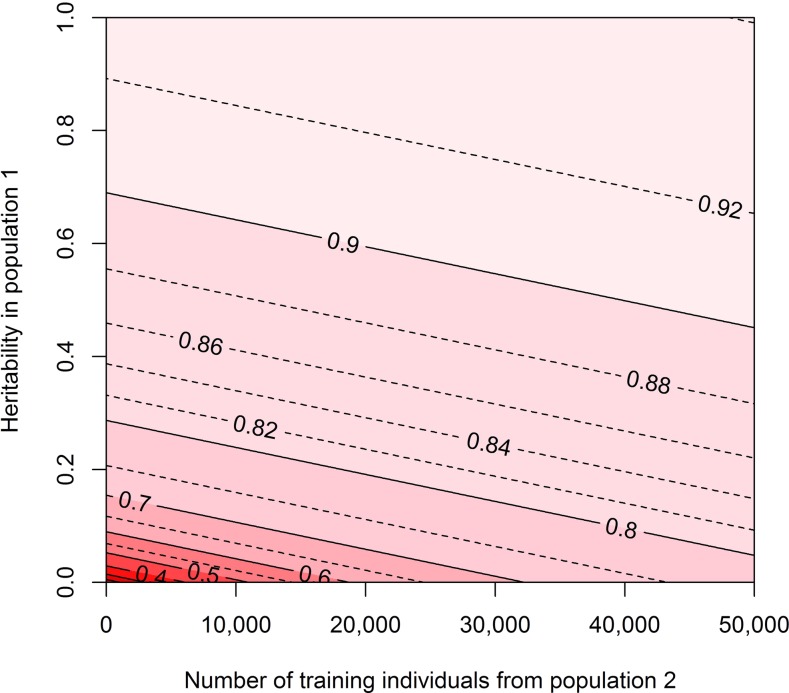
The potential accuracies when cows with their own phenotypes were added to a training population of 10,000 bulls with deregressed estimated genetic values are shown in Figure 5, for different numbers of cows added to the training population and different reliabilities for the estimated genetic values. Figure 5 shows that when the reliability of the estimated genetic values of the bulls was low, a relatively small amount of cows had to be added to the training population to see a substantial increase in accuracy. When the reliability of the estimated genetic values was high (>0.7), a high accuracy was already obtained with 10,000 bulls in the training population (accuracies were >0.9), and enlarging the training population by adding cows with their own phenotypes resulted in only a minor increase in accuracy.
Figure 5.
Predicted accuracies with different numbers of individuals from population 2 added to a training population consisting of 10,000 individuals from population 1 with different heritabilities for the trait. The input parameters represent a scenario in dairy cattle where a cow population with their own phenotypes (population 2) was added to a bull population with estimated genetic values based on daughter information (population 1). Due to different numbers of daughters used to estimate genetic values for the bulls, the heritability or reliability of the phenotype in population 1 ranged between 0 and 1. The heritability for the trait in population 2 was 0.05, and genetic correlations between the training populations and between both training populations and the predicted population were 1. The values for Me were equal to the values in the simulations ( = 1620, = 1694).
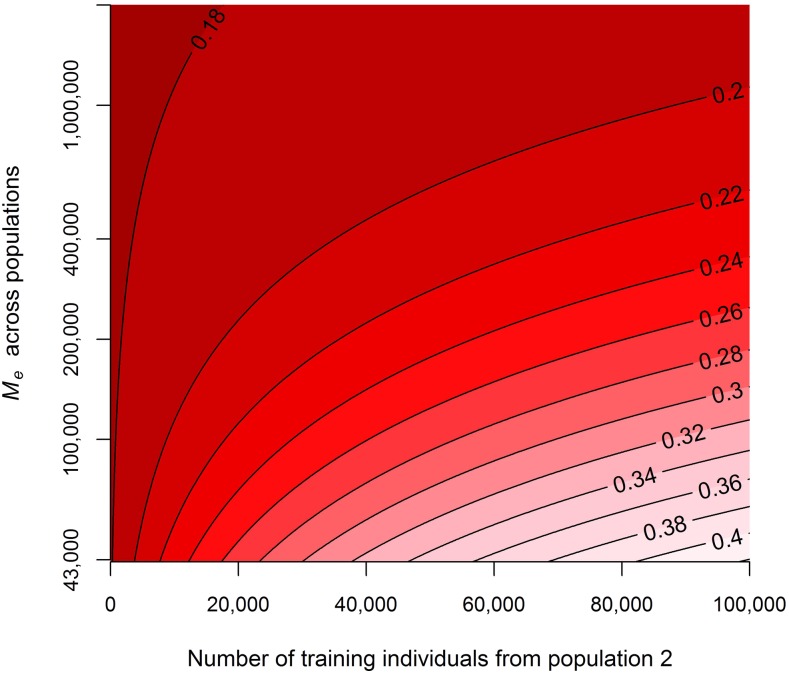
The potential accuracies for the human scenario where a population of African descent was added to a training population of European descent to predict the genetic risk of individuals from the European population are shown in Figure 6, with different numbers of individuals from the African population added to the training population and different values for Me across the populations. Figure 6 shows that when Me across the two populations was low, adding individuals from another population could substantially improve the accuracy of predicting genetic risk. When the Me across the two populations was large (>20 times the Me within the European population), adding individuals from the other population resulted in only a minor increase in accuracy. This indicates that to improve the accuracy of predicting genomic values, using training individuals from populations that are more closely related and have a more consistent LD pattern, resulting in lower values for Me across populations, is more beneficial than using training individuals from populations that are only distantly related.
Figure 6.
Predicted accuracies with different numbers of individuals from population 2 added to a training population consisting of 5000 individuals from population 1 with different values for the effective number of chromosome segments, Me, across populations 1 and 2. The input parameters represent a human scenario where a population of African descent (population 2) was added to a population of European descent (population 1) to predict the genetic risk for schizophrenia in the European population (population 3 = population 1), with heritabilities of 0.28 in population 1 and 0.24 in population 2 and a genetic correlation of 0.66 between populations 1 and 2 (De Candia et al. 2013). The Me in population 1 was set to 43,000, based on the equation (Goddard 2009) and an effective population size of 10,000 (McEvoy et al. 2011).
Discussion
In this article, a deterministic equation was derived using population parameters to predict the accuracy of genomic values when different populations are combined in the training population. The equation was able to accurately predict the accuracy of multienvironment and multitrait genomic prediction when the proportion of the genetic variance in the predicted population captured by the SNPs in the training population was known and taken into account. In addition to being able to deal with differences in heritability in each population and genetic correlations between populations different from 1, the equation can in principle handle data from more divergent populations, such as populations from different environments, breeds, or lines. The proportion of the genetic variance captured by the SNPs can, however, be expected to be lower across more divergent populations, as is discussed later. To confirm that the equation indeed gives accurate predictions for those other scenarios when the proportion of the genetic variance captured by the SNPs is known, further validation of the equation is required, using a broader range of populations, preferably with real genotype and phenotype information.
Potential of the derived equation
The equation gives insight into important parameters for multipopulation genomic prediction and can be used to compare different scenarios. The equation, for example, shows that when the Me across populations is two times higher than Me within a population, two times more individuals from the other population have to be added to obtain the same increase in accuracy when the heritabilities are the same, the genetic correlation between populations is 1, and all genetic variance can be captured. When these last criteria are not met, even more individuals from the other population have to be added to obtain the same increase in accuracy.
The equation can also be used to investigate the potential accuracy of different scenarios, as was done in Figure 5 and Figure 6. In Figure 6, the equation was applied to a scenario where human populations of European and African descent were combined in one training population to predict schizophrenia risk for the European population, a scenario that was suggested by De Candia et al. (2013). The results show that when the LD pattern is very different across populations, resulting in a high Me across populations, it is very unlikely to see an increase in prediction accuracy, even when a lot of individuals from the other population are added. Moreover, they show that the sensitivity of the accuracy for Me is much smaller at larger values of Me across populations compared to small values of Me, which is in agreement with the results found within a population (Brard and Ricard 2015). Evaluation of such scenarios requires that estimates for the input parameters, such as the Me across predicted and training populations, the heritability of the trait in each of the training populations, the genetic correlations between the populations (rG), and the part of the genetic variance in the predicted population captured by the SNPs in the training population (rLD), should, however, be known. Apart from the heritability, for which estimates are straightforward to calculate, each of the input parameters and how to estimate values for those parameters are discussed in more detail in the following paragraphs.
Effective number of chromosome segments (Me)
In the derived prediction equation, Me across populations is an important parameter. This parameter can be interpreted as a statistical concept and represents the effective number of segments that are segregating in a combined population, which is a measure for the effective number of effects that has to be estimated in one population to predict genomic values for individuals from another population. It depends on the consistency in LD between the populations; when the LD pattern is completely different between the populations, each of the segments has to be very small to segregate in both populations, resulting in a large Me across the populations.
It is of note that the derived equation assumes that Me segments are underlying the trait and that each segment explains an equal amount of the genetic variance. This indicates that the equation is basically assuming an infinitesimal model. The GBLUP model also assumes an infinitesimal model, and therefore the Me represents the number of effects that have to be estimated in a GBLUP model and the prediction equation is able to accurately predict the accuracy from a GBLUP type of model. In a Bayesian variable selection model, the number of effects that have to be estimated can be lower than Me for traits where the effective number of QTL underlying that trait is lower than Me (Daetwyler et al. 2010; Van Den Berg et al. 2015). This indicates that when the number of QTL is substantially lower than Me and a Bayesian variable selection model is used, the number of estimated effects is equal to the effective number of QTL, which is the value that should be used in the equation to predict the accuracy of genomic values.
Within a population, the value for Me can be estimated based on the effective population size (Goddard 2009; Hayes et al. 2009b; Goddard et al. 2011), as well as using the relationship matrices based on genomic information and pedigree information (Goddard et al. 2011; Wientjes et al. 2013). For the Me across populations, it is not possible to use the equations based on effective population size and a value for Me can be estimated based only on the genomic and pedigree relationship matrices. In the prediction equation, however, the Me across populations should be known for predicting the accuracy of genetic values before individuals are genotyped. For these scenarios, it is possible to estimate Me based on a small subset of individuals, for example 100 individuals from both populations, for which pedigree and genotype information is available. Another approach would be to estimate Me based on the differences between the populations, since the value for Me across populations depends on the strength of LD between loci (Goddard et al. 2011), which is at least partly different across populations (Sawyer et al. 2005; De Roos et al. 2008; Veroneze et al. 2013; Wientjes et al. 2015c). The more divergent the populations are, the higher the value for Me across populations. In this study, the estimated Me within a population was ∼1350 for all three populations and the values for Me across populations were ∼20% higher. In a study using different closely related cattle breeds, the Me values across populations were reported to be ∼10 times larger than Me within a population (Wientjes et al. 2015b). This indicates that when very closely related populations are investigated, the Me across populations can be expected to be ∼2 times the Me within a population. For closely related breeds, the Me across populations can be expected to be 10 times the Me within a population. For distantly related populations, the value for Me across populations can be even higher.
Genetic correlation between populations (rG)
Another input parameter is the genetic correlation between the populations, which is the correlation between the allele substitution effects of the QTL. In a simulation study with at least 100 individuals in each of the populations, it was shown that this parameter can accurately be estimated using a genomic multitrait model, where the same trait in different populations was treated as a different trait (Wientjes et al. 2015b). For closely related populations with an overlapping pedigree, such as populations in different countries that have some common coancestry, the genetic correlation can also be estimated using a pedigree relationship matrix (Schaeffer 1994). For more distantly related populations, such as different breeds or lines, the pedigree would probably not be deep enough to capture the relationships across populations and a relationship matrix based on genomic information is required (Karoui et al. 2012; Huang et al. 2014).
Genetic variance captured by the SNPs (rLD)
Results of this study show that the empirical accuracy of genomic prediction depended on the MAF of the QTL underlying the simulated trait; when QTL had on average a lower MAF than the SNPs, the accuracy reduced. This is in agreement with results of other studies using single-population or multipopulation genomic prediction (Daetwyler et al. 2013; Wientjes et al. 2015a). The reason for this is a decrease in the strength of LD between QTL and SNPs when the MAF of QTL is lower than the MAF of SNPs (Khatkar et al. 2008; Yan et al. 2009; Wientjes et al. 2015c), reducing the proportion of the genetic variance captured by the SNPs. As stated before, the MAF of QTL underlying complex traits is expected to be lower than the MAF of SNPs (Goddard and Hayes 2009; Yang et al. 2010; Kemper and Goddard 2012), indicating that it is highly likely that not all the genetic variance can be captured by the SNPs in real data.
The square root of the proportion of the genetic variance captured by the SNPs is represented in the prediction equation as rLD and depends on the density of the SNP chip, the characteristics of the QTL underlying the trait, and the investigated populations (Daetwyler 2009; Erbe et al. 2013). This parameter can only be estimated based on empirical data, by comparing the predicted and empirical accuracy. Using this approach, rLD was estimated to be ∼1 when QTL were randomly sampled from the SNPs and ∼0.85 when QTL had a low MAF in this study. In other studies using real data, the square of rLD, i.e., was estimated to be ∼0.8, using a 50k chip in Holstein–Friesian dairy populations for net merit (Daetwyler 2009) and production traits (Erbe et al. 2013), and was slightly lower in Brown Swiss dairy populations for production traits (Erbe et al. 2013; Román-Ponce et al. 2014). The studies estimating focused on only one population. Across populations, the value for rLD is supposed to be lower and depends on the number of generations since the separation of the populations; the higher the number of generations, the lower the consistency in LD (e.g., Andreescu et al. 2007; De Roos et al. 2008) and the higher the chance of QTL segregating in only one population (Kemper et al. 2015). Therefore, the values of = 0.89 for rLD found in the empirical studies can probably be seen as the upper limit of rLD, which can be obtained only when the predicted and training populations are subsets from the same population. The more divergent the predicted and training populations are, the lower the value of rLD and the farther away the value is from the upper limit of rLD within a population.
Single-trait vs. multitrait model
Empirical accuracies were obtained using both a single-trait model and a multitrait model. The results showed that the use of a multitrait model was beneficial when the genetic correlation between the two training populations and the predicted population was different. In an empirical study with three different chicken lines with different genetic correlations between populations, a multitrait model resulted in more or less similar accuracies compared to a single-trait model (Huang et al. 2014). In an empirical study with three dairy cattle breeds, a multitrait model using estimated genetic correlations resulted in more or less similar accuracies compared to a multitrait model with genetic correlations fixed at 0.95 (Karoui et al. 2012). Combining dairy cattle populations from three different countries, however, showed a higher accuracy for a multitrait model compared to a single-trait model (De Haas et al. 2012). So, empirical studies have shown that multitrait models yield accuracies that are similar to or slightly higher than those of single-trait models; however, genetic correlations were generally estimated with large standard errors.
The observed increase in accuracy of using a multitrait model when genetic correlations between the two training populations and the predicted population were different can be explained as follows. When the genetic correlations are different, it is beneficial to take into account that estimated SNP effects from one training population are more related to SNP effects in the predicted population than estimated SNP effects from the other training population. When the genetic correlation was the same, the use of a multitrait model was not beneficial, even when the genetic correlation among the training populations was different from 1. This can be explained by the fact that estimated SNP effects in each of the training populations are equally related to SNP effects in the predicted population. In the single-trait model, averages of the SNP effects in both training populations are estimated, which have the same correlation with the SNP effects in the predicted population as the SNP effects in each of the training populations. Therefore, taking the genetic correlation between the training populations into account had no effect on the obtained accuracy for those scenarios.
Conclusion
A deterministic equation is derived to predict the accuracy of genomic values when the training population comprises individuals of different populations, such as populations from different lines or environments or populations measured for different traits. In this study, the equation was validated for different multienvironment and multitrait scenarios. Results showed that the accuracy of estimating genomic values can be accurately predicted for these scenarios, provided that the effective number of chromosome segments across predicted and training populations, the heritability of the trait in each of the training populations, the genetic correlations between the populations, and the proportion of the genetic variance in the predicted population captured by the SNPs in the training population are known. Therefore, the derived equation can be used to investigate the potential accuracy of different multipopulation genomic prediction scenarios and to decide on the most optimal design of training populations.
Acknowledgments
The authors are thankful for useful comments from Chris Schrooten and Henk Bovenhuis. The RobustMilk project and the National Institute of Food and Agriculture are acknowledged for providing the 50k genotypes of the Holstein–Friesian cows, and the global Dry Matter Initiative (gDMI) is acknowledged for imputing those to 777k genotypes. This study was financially supported by Breed4Food (KB-12-006.03-005-ASG-LR), a public–private partnership in the domain of animal breeding and genomics, and CRV BV (Arnhem, The Netherlands).
Appendix A
Derivation Based on a Random-Effects Model
In the main text, Equation 2 and others were derived by analogy, based on the idea that the accuracy is the square root of the proportion of variance explained by a locus. In Appendix A, we provide a proof based on first principles for estimating a random effect.
Consider an additive trait determined by M independently segregating loci, where each locus explains an equal amount of additive genetic variance. The total additive genetic variance equals where pi is the allele frequency at the ith locus, and is the variance of the average effect at that locus [this expression is valid, since is the same for all loci]. Thus the variance of the average effect at a locus can be written as
| (A1) |
Since loci are independent, the effects at each of the loci can be estimated one at a time. Thus, the average effect at the ith locus can be estimated using a random-effects model,
| (A2) |
in which y is an N × 1 vector with phenotypes corrected for fixed effects for N individuals, ai is a random genetic effect for locus i, and zi is an N × 1 incidence vector with genotypes for all N individuals at locus i. Elements of zi are 0 − 2pi, 1 − 2pi, and 2 − 2pi for the three genotype classes, and e is a vector of residuals. Since each locus explains only a small part of the variance, the residual variance can be approximated as where is the total phenotypic variance.
The variance of y follows from
| (A3) |
in which I is an N × N identity matrix, and h2 is the heritability.
Following the mixed-model equations, the effect of one locus is estimated as
| (A4) |
Thus the variance of the estimated effect for one locus equals
| (A5) |
With best linear prediction, the accuracy of an estimated random effect follows from the variances of the estimated and true effects (Falconer and Mackay 1996),
| (A6) |
where is the variance explained by a single locus. This result is equivalent to Equation 3 from the main text and shows that the accuracy of an estimated gene effect follows from the proportion of variance explained by the locus.
The estimated effects can be used to calculate an estimated genomic value for individual j,
| (A7) |
in which zj is an M × 1 vector with genotypes for individual j for all M loci (modeled similarly to zi above), and is an M × 1 vector with estimated effects for all loci.
The true genomic value of an individual equals
| (A8) |
in which a is a vector with true effects for all loci.
The accuracy of the EGV equals
| (A9) |
This result shows that, when all loci explain an equal amount of the genetic variance, the accuracy of the EGV is equal to the accuracy of estimating a single-locus effect.
The above represents an alternative derivation of the result of Daetwyler et al. (2008) and is conceptually simpler than the original derivation that treats estimated gene effects as both fixed and random.
Appendix B
Deriving the Accuracy of Estimating SNP Effects in a Combined Training Population
The accuracy of the selection index, representing the accuracy of estimating the effect of one locus, can be calculated as
| (B1) |
For simplicity, we start by referring to the first element of this inversed P matrix as A, to the off-diagonal elements as B, and to the last element as C. Hence, Equation B1 can be written as
| (B2) |
The inverse of the P matrix can be written as
| (B3) |
Hence, Equation B2 can be written as
| (B4) |
Dividing both the numerator and the denominator by and results in
| (B5) |
Since each locus is assumed to explain an equal amount of the genetic variance, the accuracy of estimating the effect of one SNP is the same for each of the SNPs and represents the overall accuracy of estimating SNP effects (reffect).
Appendix C
Alternative Way of Deriving the Prediction Equation
In this section, an alternative derivation of the prediction equation is presented. In this derivation, the estimated genomic values for population C based on two different training populations (population A and population B) are combined in a selection index to calculate the estimated genomic values for population C when the two populations are combined in one training population. The estimated genomic value for individual i from population C () can be calculated using the estimated marker effects in a training population of population A, following
| (C1) |
in which is the genetic correlation between populations A and C, is the genotype of individual i from population C for marker j, and is the estimated effect of marker j in population A. In an equivalent way, the estimated genomic value for individual i from population C can be calculated using the estimated marker effects in a training population of population B, i.e.,
Both estimated genomic values, and can be combined in a selection index to estimate the genomic value for individual i from population C when both populations A and B are combined in the training population (), following
| (C2) |
in which bA and bB are the regression coefficients on and to predict the estimated genomic value for individual i from population C for the combined training population ().
The regression coefficients on and that would maximize the estimation of the genomic value for individual i from population C can be calculated as
| (C3) |
in which P is the (co)variance matrix between the information sources and and g is a vector with covariances between the information sources, and and the true genomic value for individual i from population C ():
| (C4) |
and
| (C5) |
In the following part, we assume that the variances of the estimated and true genomic values are scaled, such that the true genomic values in population C have a variance of 1. The variance of the estimated genomic values for population C using population A in the training population is then equal to the reliability of predicting genomic values for population C:
| (C6) |
The covariance between and can be written as
| (C7) |
The covariance between the marker effects estimated in population A and B can be written as
| (C8) |
Using the path coefficient method as described by Dekkers (2007), it can be shown that the correlation between the estimated marker effects is equal to
| (C9) |
in which is the genetic correlation between populations A and B, and and are the accuracies of estimating the marker effects in, respectively, populations A and B. The square root of the variance of the estimated marker effects in each of the populations is equal to the accuracy of the estimated marker effects; i.e., therefore
| (C10) |
and
| (C11) |
The accuracy of estimating marker effects in population A multiplied by the genetic correlation between populations A and C equals the accuracy of the estimated genomic values, i.e., under the assumption that all genetic variance of the predicted population is captured by the training populations. Hence, the covariance can be written as
| (C12) |
Hence, P can be written as
| (C13) |
The covariance between the estimated genomic values for individual i from population C using population A as the training population is also equal to the reliability of predicting genomic values for population C; i.e., Hence, g can be written as
| (C14) |
Since it is assumed that the variance of the true genomic values in population C is scaled to 1, the accuracy of this selection index, representing the accuracy of estimating genomic values for population C based on a training population of population A and B, can be calculated as
| (C15) |
For simplicity, we start by referring to the first element of matrix P−1 as A, to the off-diagonal elements as B, and to the last element as C. Hence, Equation C15 can be written as
| (C16) |
The matrix P−1 can be written as
| (C17) |
Hence, Equation C16 can be written as
| (C18) |
If we assume that all genetic variance in population C can be captured by the SNPs in the training population, the accuracies for each of the populations can be replaced by the corresponding equation to predict the accuracy of genomic prediction (Daetwyler et al. 2008, 2010; Wientjes et al. 2015b):
| (C19) |
and
| (C20) |
Using this in Equation C18 results in
| (C21) |
Multiplying both the numerator and the denominator by and results in
| (C22) |
This last equation is equivalent to the equation derived before, using the same assumption that all genetic variance of the predicted population is captured by the SNPs in the training populations.
Footnotes
Communicating editor: D. J. de Koning
Literature Cited
- Andreescu C., Avendano S., Brown S. R., Hassen A., Lamont S. J., et al. , 2007. Linkage disequilibrium in related breeding lines of chickens. Genetics 177: 2161–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brard S., Ricard A., 2015. Is the use of formulae a reliable way to predict the accuracy of genomic selection? J. Anim. Breed. Genet. 132: 207–217. [DOI] [PubMed] [Google Scholar]
- Calus M. P. L., De Haas Y., Veerkamp R. F., 2013. Combining cow and bull reference populations to increase accuracy of genomic prediction and genome-wide association studies. J. Dairy Sci. 96: 6703–6715. [DOI] [PubMed] [Google Scholar]
- Calus M. P. L., Huang H., Vereijken A., Visscher J., Ten Napel J., et al. , 2014. Genomic prediction based on data from three layer lines: a comparison between linear methods. Genet. Sel. Evol. 46: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. A., Wiggans G. R., VanRaden P. M., 2015. Short communication: analysis of genomic predictor population for Holstein dairy cattle in the United States—effects of sex and age. J. Dairy Sci. 98: 2785–2788. [DOI] [PubMed] [Google Scholar]
- Daetwyler, H. D., 2009 Genome-wide evaluation of populations. Ph.D. Thesis, Animal Breeding and Genomics Centre, Wageningen University, Wageningen, The Netherlands. [Google Scholar]
- Daetwyler H. D., Villanueva B., Woolliams J. A., 2008. Accuracy of predicting the genetic risk of disease using a genome-wide approach. PLoS One 3: e3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Pong-Wong R., Villanueva B., Woolliams J. A., 2010. The impact of genetic architecture on genome-wide evaluation methods. Genetics 185: 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Calus M. P. L., Pong-Wong R., De los Campos G., Hickey J. M., 2013. Genomic prediction in animals and plants: simulation of data, validation, reporting, and benchmarking. Genetics 193: 347–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Candia T. R., Lee S. H., Yang J., Browning B. L., Gejman P. V., et al. , 2013. Additive genetic variation in schizophrenia risk is shared by populations of African and European descent. Am. J. Hum. Genet. 93: 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haas Y., Calus M. P. L., Veerkamp R. F., Wall E., Coffey M. P., et al. , 2012. Improved accuracy of genomic prediction for dry matter intake of dairy cattle from combined European and Australian data sets. J. Dairy Sci. 95: 6103–6112. [DOI] [PubMed] [Google Scholar]
- De Los Campos G., Gianola D., Allison D. B., 2010. Predicting genetic predisposition in humans: the promise of whole-genome markers. Nat. Rev. Genet. 11: 880–886. [DOI] [PubMed] [Google Scholar]
- De Los Campos G., Klimentidis Y. C., Vazquez A. I., Allison D. B., 2012. Prediction of expected years of life using whole-genome markers. PLoS One 7: e40964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Los Campos G., Vazquez A. I., Fernando R., Klimentidis Y. C., Sorensen D., 2013. Prediction of complex human traits using the genomic best linear unbiased predictor. PLoS Genet. 9: e1003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roos A. P. W., Hayes B. J., Spelman R. J., Goddard M. E., 2008. Linkage disequilibrium and persistence of phase in Holstein-Friesian, Jersey and Angus cattle. Genetics 179: 1503–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roos A. P. W., Schrooten C., Veerkamp R. F., Van Arendonk J. A. M., 2011. Effects of genomic selection on genetic improvement, inbreeding, and merit of young vs. proven bulls. J. Dairy Sci. 94: 1559–1567. [DOI] [PubMed] [Google Scholar]
- Dekkers J. C. M., 2007. Prediction of response to marker-assisted and genomic selection using selection index theory. J. Anim. Breed. Genet. 124: 331–341. [DOI] [PubMed] [Google Scholar]
- Erbe M., Gredler B., Seefried F. R., Bapst B., Simianer H., 2013. A function accounting for training set size and marker density to model the average accuracy of genomic prediction. PLoS One 8: e81046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics. Pearson Education, Harlow, UK. [Google Scholar]
- Garrick D. J., Taylor J. F., Fernando R. L., 2009. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet. Sel. Evol. 41: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour A. R., Gogel B., Cullis B., Thompson R., Butler D., et al. , 2009. ASReml User Guide Release 3.0. VSN International, Hemel Hempstead, UK. [Google Scholar]
- Goddard M. E., 2009. Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 136: 245–257. [DOI] [PubMed] [Google Scholar]
- Goddard M. E., Hayes B. J., 2009. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat. Rev. Genet. 10: 381–391. [DOI] [PubMed] [Google Scholar]
- Goddard M. E., Hayes B. J., Meuwissen T. H. E., 2011. Using the genomic relationship matrix to predict the accuracy of genomic selection. J. Anim. Breed. Genet. 128: 409–421. [DOI] [PubMed] [Google Scholar]
- Haile-Mariam M., Pryce J. E., Schrooten C., Hayes B. J., 2015. Including overseas performance information in genomic evaluations of Australian dairy cattle. J. Dairy Sci. 98: 3443–3459. [DOI] [PubMed] [Google Scholar]
- Harris B. L., Johnson D. L., 2010. Genomic predictions for New Zealand dairy bulls and integration with national genetic evaluation. J. Dairy Sci. 93: 1243–1252. [DOI] [PubMed] [Google Scholar]
- Hayes B. J., Bowman P. J., Chamberlain A. J., Verbyla K., Goddard M. E., 2009a Accuracy of genomic breeding values in multi-breed dairy cattle populations. Genet. Sel. Evol. 41: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B. J., Visscher P. M., Goddard M. E., 2009b Increased accuracy of artificial selection by using the realized relationship matrix. Genet. Res. 91: 47–60. [DOI] [PubMed] [Google Scholar]
- Hazel L. N., 1943. The genetic basis for constructing selection indexes. Genetics 28: 476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffner E. L., Sorrells M. E., Jannink J. L., 2009. Genomic selection for crop improvement. Crop Sci. 49: 1–12. [Google Scholar]
- Huang H., Windig J. J., Vereijken A., Calus M. P., 2014. Genomic prediction based on data from three layer lines using non-linear regression models. Genet. Sel. Evol. 46: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannink J. L., Lorenz A. J., Iwata H., 2010. Genomic selection in plant breeding: from theory to practice. Brief. Funct. Genomics 9: 166–177. [DOI] [PubMed] [Google Scholar]
- Karoui S., Carabaño M., Díaz C., Legarra A., 2012. Joint genomic evaluation of French dairy cattle breeds using multiple-trait models. Genet. Sel. Evol. 44: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper K. E., Goddard M. E., 2012. Understanding and predicting complex traits: knowledge from cattle. Hum. Mol. Genet. 21: R45–R51. [DOI] [PubMed] [Google Scholar]
- Kemper K. E., Hayes B. J., Daetwyler H. D., Goddard M. E., 2015. How old are quantitative trait loci and how widely do they segregate? J. Anim. Breed. Genet. 132: 121–134. [DOI] [PubMed] [Google Scholar]
- Khatkar M. S., Nicholas F. W., Collins A. R., Zenger K. R., Cavanagh J. A. L., et al. , 2008. Extent of genome-wide linkage disequilibrium in Australian Holstein-Friesian cattle based on a high-density SNP panel. BMC Genomics 9: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., DeCandia T. R., Ripke S., Yang J., Sullivan P. F., et al. , 2012. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat. Genet. 44: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehermeier C., Krämer N., Bauer E., Bauland C., Camisan C., et al. , 2014. Usefulness of multiparental populations of maize (Zea mays L.) for genome-based prediction. Genetics 198: 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund M. S., De Roos S. P. W., De Vries A. G., Druet T., Ducrocq V., et al. , 2011. A common reference population from four European Holstein populations increases reliability of genomic predictions. Genet. Sel. Evol. 43: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier R., Moser G., Chen G.-B., Ripke S., Coryell W., et al. , 2015. Joint analysis of psychiatric disorders increases accuracy of risk prediction for schizophrenia, bipolar disorder, and major depressive disorder. Am. J. Hum. Genet. 96: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matukumalli L. K., Lawley C. T., Schnabel R. D., Taylor J. F., Allan M. F., et al. , 2009. Development and characterization of a high density SNP genotyping assay for cattle. PLoS One 4: e5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy B. P., Powell J. E., Goddard M. E., Visscher P. M., 2011. Human population dispersal “Out of Africa” estimated from linkage disequilibrium and allele frequencies of SNPs. Genome Res. 21: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. E., Visscher P. M., Goddard M. E., 2010. Reconciling the analysis of IBD and IBS in complex trait studies. Nat. Rev. Genet. 11: 800–805. [DOI] [PubMed] [Google Scholar]
- Pryce J. E., Johnston J., Hayes B. J., Sahana G., Weigel K. A., et al. , 2014. Imputation of genotypes from low density (50,000 markers) to high density (700,000 markers) of cows from research herds in Europe, North America, and Australasia using 2 reference populations. J. Dairy Sci. 97: 1799–1811. [DOI] [PubMed] [Google Scholar]
- R Development Core Team , 2011. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Román-Ponce S. I., Samoré A. B., Dolezal M. A., Bagnato A., Meuwissen T. H. E., 2014. Estimates of missing heritability for complex traits in Brown Swiss cattle. Genet. Sel. Evol. 46: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S. L., Mukherjee N., Pakstis A. J., Feuk L., Kidd J. R., et al. , 2005. Linkage disequilibrium patterns vary substantially among populations. Eur. J. Hum. Genet. 13: 677–686. [DOI] [PubMed] [Google Scholar]
- Schaeffer L. R., 1994. Multiple-country comparison of dairy sires. J. Dairy Sci. 77: 2671–2678. [DOI] [PubMed] [Google Scholar]
- Spelman R. J., Ford C. A., McElhinney P., Gregory G. C., Snell R. G., 2002. Characterization of the DGAT1 gene in the New Zealand dairy population. J. Dairy Sci. 85: 3514–3517. [DOI] [PubMed] [Google Scholar]
- Thaller G., Krämer W., Winter A., Kaupe B., Erhardt G., et al. , 2003. Effects of DGAT1 variants on milk production traits in German cattle breeds. J. Anim. Sci. 81: 1911–1918. [DOI] [PubMed] [Google Scholar]
- Van den Berg, S., M. P. L. Calus, T. H. E. Meuwissen and Y. C. J. Wientjes, 2015 Across population genomic prediction scenarios in which Bayesian variable selection outperforms GBLUP. BMC Genet. (in press). [DOI] [PMC free article] [PubMed]
- VanRaden P. M., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423. [DOI] [PubMed] [Google Scholar]
- Venter J. C., Adams M. D., Myers E. W., Li P. W., Mural R. J., et al. , 2001. The sequence of the human genome. Science 291: 1304–1351. [DOI] [PubMed] [Google Scholar]
- Veroneze R., Lopes P. S., Guimarães S. E. F., Silva F. F., Lopes M. S., et al. , 2013. Linkage disequilibrium and haplotype block structure in six commercial pig lines. J. Anim. Sci. 91: 3493–3501. [DOI] [PubMed] [Google Scholar]
- Wientjes Y. C. J., Veerkamp R. F., Calus M. P. L., 2013. The effect of linkage disequilibrium and family relationships on the reliability of genomic prediction. Genetics 193: 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wientjes Y. C. J., Calus M. P. L., Goddard M. E., Hayes B. J., 2015a Impact of QTL properties on the accuracy of multi-breed genomic prediction. Genet. Sel. Evol. 47: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wientjes Y. C. J., Veerkamp R. F., Bijma P., Bovenhuis H., Schrooten C., et al. , 2015b Empirical and deterministic accuracies of across-population genomic prediction. Genet. Sel. Evol. 47: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wientjes Y. C. J., Veerkamp R. F., Calus M. P. L., 2015c Using selection index theory to estimate consistency of multi-locus linkage disequilibrium across populations. BMC Genet. 16: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray N. R., Goddard M. E., Visscher P. M., 2007. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 17: 1520–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Shah T., Warburton M. L., Buckler E. S., McMullen M. D., et al. , 2009. Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS One 4: e8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Dekkers J. C. M., Fernando R. L., Jannink J.-L., 2009. Factors affecting accuracy from genomic selection in populations derived from multiple inbred lines: a barley case study. Genetics 182: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]