Abstract
Transposable elements (TEs) are both a boon and a bane to eukaryotic organisms, depending on where they integrate into the genome and how their sequences function once integrated. We focus on two types of TEs: long interspersed elements (LINEs) and short interspersed elements (SINEs). LINEs and SINEs are retrotransposons; that is, they transpose via an RNA intermediate. We discuss how LINEs and SINEs have expanded in eukaryotic genomes and contribute to genome evolution. An emerging body of evidence indicates that LINEs and SINEs function to regulate gene expression by affecting chromatin structure, gene transcription, pre-mRNA processing, or aspects of mRNA metabolism. We also describe how adenosine-to-inosine editing influences SINE function and how ongoing retrotransposition is countered by the body’s defense mechanisms.
Transposable elements (TEs) are DNA sequences that have the ability to be integrated elsewhere in a genome. With few exceptions, TEs have been identified in all eukaryotic genomes sequenced to date (1). There are two main classes of TEs: Retrotransposons (class I) transpose via an RNA intermediate, whereas DNA transposons (class II) transpose directly without an RNA intermediate (2). The three major retrotransposon orders are long terminal repeat (LTR) retrotransposons, long interspersed elements (LINEs), and short interspersed elements (SINEs). Retrotransposons propagate via a copy-and-paste amplification mechanism that has allowed them to accumulate in DNA, giving rise to the bulk of repeats in eukaryotic genomes.
Mobile LINEs are RNA polymerase II (Pol II)–transcribed autonomous retrotransposons of several thousand base pairs (bp) (3). In the copy step, their internal Pol II promoter generates an mRNA-like capped and polyadenylated transcript (4). The transcript of LINE-1 (L1), which is the only active class of autonomous retrotransposons in humans, contains two open reading frames (ORFs) that are crucial for retrotransposition: ORF1 encodes an RNA-binding protein, and ORF2 encodes a protein with reverse transcriptase (RT) and endonuclease activities (Fig. 1A) (5). In the subsequent paste step, these proteins recognize a specific sequence in the 3′ end of the LINE transcript that encodes them, create two staggered nicks at specific sequences in the genome and, by using the genomic sequence as a primer, reverse-transcribe the LINE RNA into cDNA that is simultaneously incorporated into the genome (Fig. 1B) (5, 6). Acquisition of an additional L1 ORF 5′ to ORF1 (ORF0) was recently demonstrated in the primate lineage (7).
Fig. 1. LINE and SINE transposition.
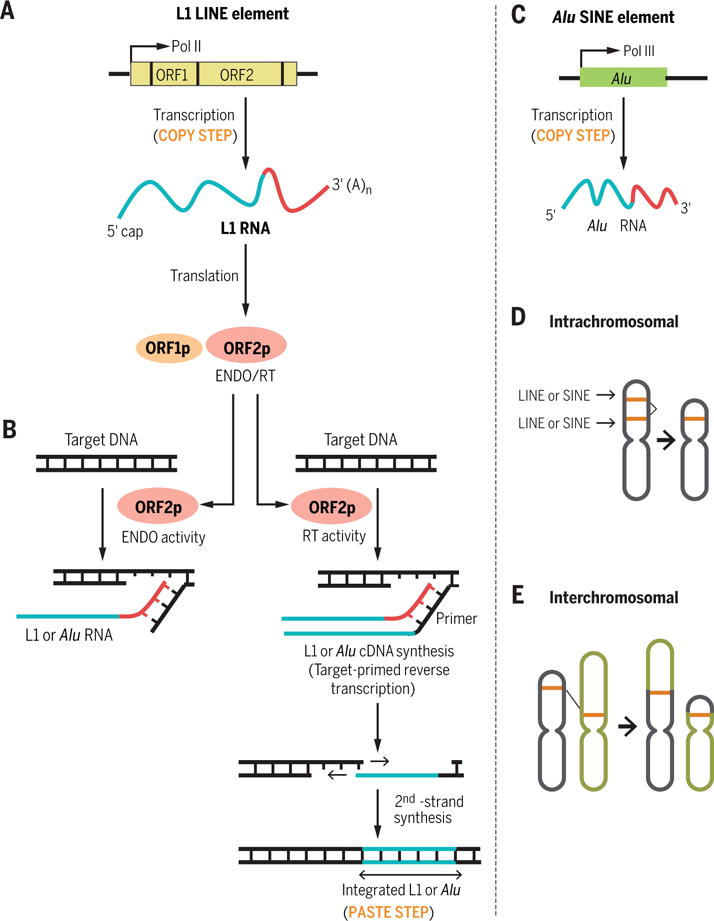
(A) Copy step of a LINE: L1 gene transcription by Pol II followed by L1 RNA translation. (B) Paste steps of L1 and Alu element transposition using the endonuclease (ENDO) and reverse transcriptase (RT) activities of ORF2p. (C) Copy step of a SINE: Alu element transcription by Pol III. (D) Intrachromosomal recombination between related LINEs or SINEs resulting in genomic deletion. (E) Interchromosomal recombination between related LINEs or SINEs resulting in genomic rearrangements.
Mobile SINEs are RNA polymerase III (Pol III)–transcribed nonautonomous retrotransposons that do not encode any proteins (Fig. 1C) but retrotranspose by hijacking the RT and endonuclease activities of a partner LINE-encoded protein (Fig. 1B). In most cases, LINE-encoded proteins recognize SINE RNAs with 3′ sequences that are similar to the 3′ sequence of the LINE RNA from which these proteins were synthesized; subsequently, they generate and integrate a cDNA copy of the SINE RNA into the genome (Fig. 1, B and C) (8).
The lengths of SINE family members generally range from 85 to 500 bp (9). A SINE typically has three parts: a 5′ head, a body, and a 3′ tail. Head sequences, which harbor the internal Pol III promoter, have been used to categorize SINEs into three superfamilies according to their derivation from, and thus similarity to, cellular Pol III genes encoding tRNA (such as mouse B2 or ID elements), 7SL RNA (such as mouse B1 and human Alu elements), or 5S rRNA (SINE3) (2, 9, 10). Most LINEs and SINEs in mammalian genomes have lost their functional promoters and thus lack the ability to retrotranspose (5).
LINEs and SINEs constitute ~30% of the human genome sequence and show a nonrandom genomic distribution (11). SINEs are generally localized in gene-rich regions, whereas LINEs are enriched in intergenic regions (12). The relative sparsity of LINEs in genic regions likely reflects negative selection against insertion of their large sequence (several thousand bp) in or near genes. In contrast, the smaller SINEs are more apt to be tolerated, and some SINEs in genic regions have assumed regulatory roles that control gene expression. The expansion of LINEs and SINEs has drastically shaped the genomes of multicellular organisms by providing regions of similarity that act as hotspots for nonallelic homologous recombination (Fig. 1, D and E) and acting as reservoirs of potential coding, regulatory, or disruptive sequences (13, 14). In addition to their own retrotransposition and that of SINEs, LINEs have supported the retrotransposition of mRNAs (15, 16). The resulting “retrogenes,” in the presence of their functional counterpart, are free from selective pressure and thus can accumulate mutations and acquire novel functions (16). Thus, retrotransposition contributes to genetic diversity within a species and among different species in many ways. Additionally, retrotransposition appears to be active in some somatic tissues, including early in development (17), in developing neurons (18, 19), and in the adult brain (20), leading to mosaicism whereby different cells within an individual have different genetic sequences. Most insertion events are neutral or detrimental to the host; here, we describe instances whereby inserted LINEs and SINEs have been harnessed to regulate gene expression.
Regulation of chromatin structure and transcription
Primate LINEs and SINEs have a high GC content, making them hotspots for DNA methylation, which is used by cells to suppress transcription (21). The methylation of LINE- and SINE-embedded CpG islands has the potential to silence the expression of nearby genes (22). LINEs and SINEs can demarcate the boundary between heterochromatin and euchromatin. For example, one mouse B2 element functions as a boundary element to prevent cis-residing heterochromatin from silencing developmental expression of the five genes located in the mouse growth hormone locus (Fig. 2A) (23). LINEs also participate in X-chromosome inactivation (XCI) via one of two mechanisms: Transcriptionally silent L1 elements contribute to the formation of a silent nuclear compartment during XCI, whereas L1 RNAs that derive from young LINE elements (which are enriched in the X chromosome) participate in inactivating X-chromosome loci that would otherwise escape XCI (24).
Fig. 2. LINE- and SINE-mediated gene regulation.
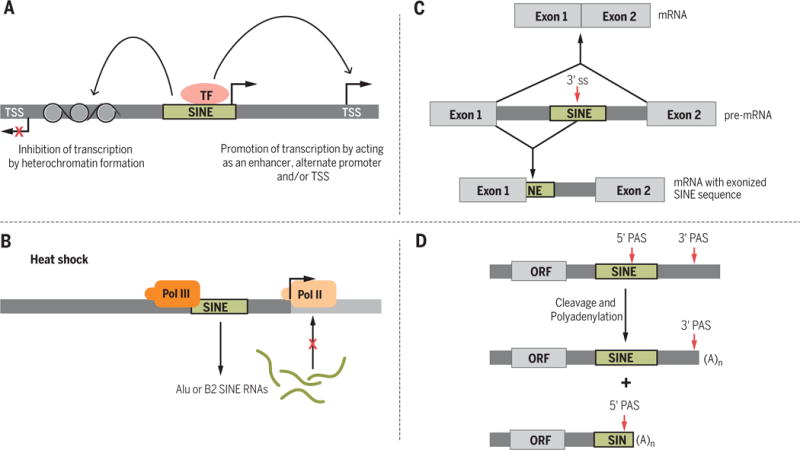
(A) SINEs (and LINEs) can promote or inhibit the transcription of nearby genes. TSS, transcription start site; TF, transcription factor. (B) Upon heat shock, increased expression of Alu and B2 RNAs inhibits Pol II. (C) SINEs contain potential splice sites (ss) that, if used, can lead to mRNAs with intronic sequences. (D) SINEs (in particular, Alu elements) can contain a polyadenylation signal (PAS).
SINEs, and Alu elements in particular (25), can function as transcriptional enhancers, as exemplified by two members of the ancient SINE family Amniota SINE1 (AmnSINE1), which act as enhancers for the genes encoding fibroblast growth factor 8 (Fgf8) and special AT-rich sequence–binding protein 2 (Satb2) in the developing brain (Fig. 2A) (25, 26). Retrotransposons located immediately upstream of protein-coding genes may function as promoters because putative binding sites for many transcription factors have been identified in SINEs (Fig. 2A) (27). However, it is unclear whether the majority of SINE-embedded transcription factor binding sites act to modulate gene transcription or simply act as sinks that titrate transcription factors away from their active binding sites. LINEs and SINEs can also introduce a new transcription start site (TSS); 6 to 30% of human and mouse 5′-capped transcripts use repetitive sequence-associated TSSs (27).
The functional influences of LINEs and SINEs on transcription are mediated not only as DNA elements but also via the RNAs that they encode. SINEs normally are transcriptionally silenced in somatic tissues; however, in response to stressors such as heat shock, SINE Pol III promoters are activated and SINE RNAs are massively up-regulated. Stress-mediated up-regulation of human Alu and mouse B2 RNAs inhibits the transcription of most genes, excluding those up-regulated during heat shock, by binding to Pol II (Fig. 2B) (28, 29).
Regulation of RNA processing
Some SINE insertions, in particular Alu elements, can influence gene expression by altering pre-mRNA splicing. In humans, 66% of Alu elements and 65% of mammalian-wide interspersed repeats (MIRs) are found in introns (30). In the antisense orientation, the consensus Alu sequence contains seven potential 5′ splice sites and 12 potential 3′ splice sites, whereas sense Alu elements contain three potential 5′ splice sites and one potential 3′ splice site (31, 32). Alu-derived splice sites are usually cryptic, requiring few mutations to become functional and to promote exonization (i.e., inclusion of a intronic sequence within the resulting spliced mRNA) (Fig. 2C) (31). It is estimated that 5% of alternatively spliced exons in humans derive from Alu sequences and that most Alu-containing exons are alternatively spliced (32). Although the majority of exonized Alu elements form cassette exons that are included in one or more minor splice isoforms (30), in the human brain a substantial portion of Alu-containing exons reside in major splice isoforms (33). RNA-binding proteins are able to regulate the availability of splice signals within SINEs to associate with the splicing machinery (34).
When embedded within Pol II transcripts, the length of Alu elements (~300 bp) and their high (>70%) similarity (14) allow two elements that coexist in opposite orientation within the same transcript (inverted-repeat Alus or IRAlus) to form intramolecular imperfect duplexes of >100 bp (35, 36). Recently, intronic IRAlus have been shown to promote pre-mRNA “backsplicing” so as to facilitate the formation of circular noncoding RNAs (circRNAs) that may have biological functions (37–39).
The vast majority of protein-coding mRNAs, as well as many long noncoding RNAs (lncRNAs), are polyadenylated at their 3′ ends. Most polyadenylation occurs upon recognition of a polyadenylation signal (PAS) that consists of the conical AAUAAA sequence or the closely related AUUAAA sequence (40). Most human genes harbor more than one PAS that, when used, generate mRNA isoforms with alternative 3′ ends (Fig. 2D) (41). New PAS sequences are commonly created via spontaneous mutations within the A-rich tails of LINEs and SINEs (42–44). Retrotransposon-associated PASs are largely not conserved between different species, which suggests that retrotransposons have contributed to interspecies differences in transcript 3′ ends (43). Alu-derived PASs, 99% of which derive from sense Alu elements, are efficiently used (43, 44). Some of these putative Alu-embedded PASs are intronic and result in shortened transcripts, presumably explaining the observed low abundance of sense Alu elements relative to antisense Alu elements in intronic regions (44).
When transcribed as part of mRNA 3′ untranslated regions (3′UTRs), SINEs have the potential to act in cis and/or in trans to influence mRNA turnover. The poly(T) sequence that exists in antisense Alu elements is the source of ~40% of identified 3′UTR AU-rich elements (AREs), which regulate mRNA half-life through the competitive binding of proteins that stabilize or destabilize the transcript (Fig. 3A) (45). Additionally, LINEs and SINEs can activate the function of microRNAs (miRNAs) by acting as promoters for miRNA synthesis or as miRNA-binding sites in target mRNAs (46, 47) (Fig. 3B); miRNAs are ~22-nucleotide noncoding RNAs that mediate decay and/or translational repression of transcripts to which they bind.
Fig. 3. Effects on mRNA stability by SINE insertions.
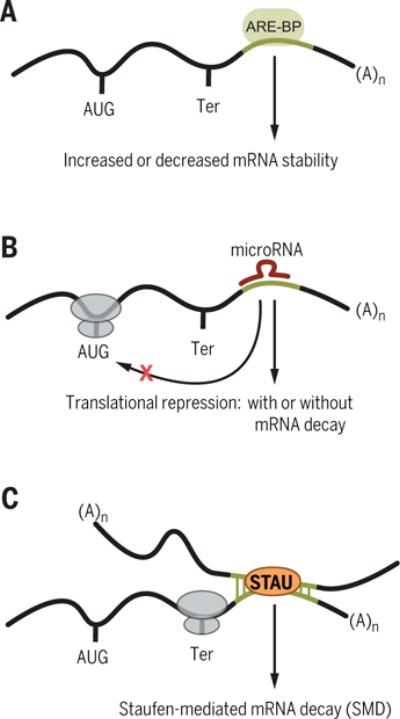
(A) AU-rich element–binding proteins (ARE-BPs) may bind a 3′UTR Alu element–derived ARE and either stabilize or destabilize the mRNA. (B) Alu element–derived microRNA-binding sites within an mRNA can promote mRNA decay and/or inhibit mRNA translation. (C) Intermolecular base pairing via partially complementary SINEs can create Staufen-binding sites that trigger Staufen-mediated mRNA decay.
As a consequence of their high similarity and presence in 5.7% of human mRNA 3′UTRs (27), Alu elements can also mediate intermolecular base pairing between two RNA molecules. For example, Alu elements in some human mRNA 3′UTRs can form base pairs with partially complementary, reverse-oriented Alu elements in lncRNAs (48) or in the 3′UTRs of other mRNAs (Fig. 3C) (49). The intermolecular double-stranded RNA (dsRNA) formed, if bound by the dsRNA-binding protein (dsRBP) Staufen 1 (STAU1) and/or its paralog STAU2, can result in mRNA decay in a mechanism that depends on translation (Fig. 3C) (50, 51). This STAU-mediated mRNA decay (SMD) contributes to cell motility, cell invasion, and other processes (48, 49, 52). B SINEs and identifier (ID) SINEs in mouse mRNA 3′UTRs can also form duplexes with, respectively, partially complementary B and ID SINEs in lncRNAs (or, most likely, in the 3′UTRs of mRNAs) and likewise trigger SMD so as to regulate cellular processes (53). The presence of a 3′UTR Alu or B element is not always predictive of SMD (54). Both CUB domain–containing protein 1 (CDCP1) mRNA and BCL2-associated athanogene 5 (BAG5) mRNA have a 3′UTR Alu element that is predicted to bind the same lncRNA Alu element, but only CDCP1 mRNA is an SMD target in HeLa cells (48). The features that distinguish a SINE-directed SMD target remain to be determined until more is understood about transcript folding, what defines a STAU-binding site, and how other dsRNA-binding proteins compete with STAU for binding to duplexed SINEs (48).
Messenger RNA localization and translation
Not all mRNAs are efficiently exported from the nucleus to the cytoplasm for translation. Some nuclear-retained transcripts contain 3′UTR IRAlus (IRAlus mRNAs; Fig. 4A) (55–60) and are localized in paraspeckles (Fig. 4A), which are subnuclear bodies containing the lncRNA NEAT1 and multiple RNA-binding proteins (61). Localization of IRAlus mRNAs can be determined by use of alternative PASs, which could exclude the 3′UTR IRAlus from product mRNA. STAU1 binding to the 3′UTR IRAlus of a subset of IRAlus mRNAs precludes the binding of p54nrb (a protein component of paraspeckles), thereby permitting their nuclear export (Fig. 4A) (57, 58). Furthermore, the affinity of dsRBPs for IRAlus can be altered by posttranslational modifications. For example, the methylation of p54nrb by the coactivator-associated arginine methyltransferase 1 (CARM1) reduces the binding of p54nrb to the 3′UTR IRAlus of particular IRAlus mRNAs, promoting their nuclear export (Fig. 4A) (59, 60). Because different mRNAs with apparently similar 3′UTR IRAlus manifest distinct subcellular localizations, the regulation of IRAlus mRNAs is substantially more complicated than depicted in current models.
Fig. 4. 3′UTR IRAlus regulate mRNA localization and translation.
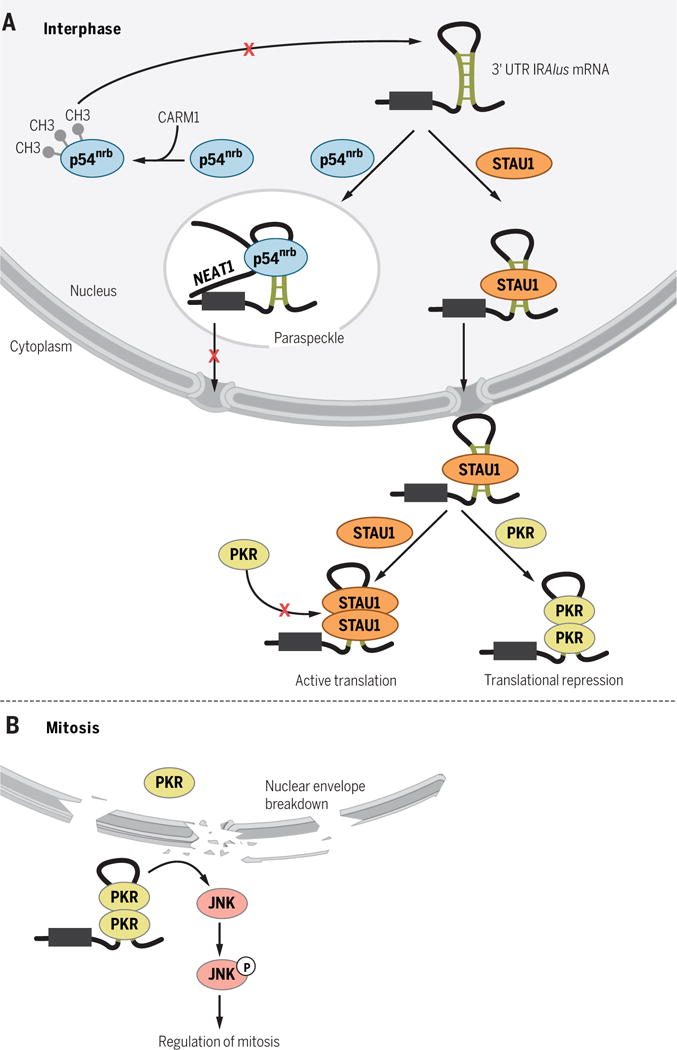
(A) During cellular interphase, 3′UTR IRAlus localize many newly synthesized mRNAs to nuclear paraspeckles by binding p54nrb, which is relieved by Staufen binding or by CARM1-mediated methylation of p54nrb. In the cytoplasm, 3′UTR IRAlus can inhibit mRNA translation in cis and in trans by binding PKR, and this inhibition is relieved by STAU1 binding. (B) During mitosis, breakdown of the nuclear envelope allows mixing of nuclear-retained 3′UTR IRAlus and cytoplasmic PKR, resulting in PKR binding to 3′UTR IRAlus and PKR-mediated phosphorylation of JNK.
In the cytoplasm, 3′UTR IRAlus can also repress the translation of their host mRNAs and accelerate their accumulation in stress granules (57, 58, 62–64). A subset of 3′UTR IRAlus mediate translational repression by binding and activating dsRNA-dependent protein kinase (PKR) (Fig. 4A), which is activated by autophosphorylation once dimerized on dsRNA. PKR activation results in phosphorylation of eukaryotic translation initiation factor 2α, which in turn inhibits the bulk of cellular translation (57, 58, 64). Thus, 3′UTR IRAlus can act as translational repressors not only in cis but also in trans. STAU1 binding to 3′UTR IRAlus in the cytoplasm precludes PKR binding, alleviating translational repression of STAU1-bound IRAlus mRNAs and, to a lesser extent, the bulk of cellular mRNAs (Fig. 4A) (57, 58, 64). During interphase, nuclear-retained IRAlus mRNAs are physically segregated away from cytoplasmic PKR, thereby preventing PKR activation (Fig. 4A) (57, 58, 64). However, after breakdown of the nuclear envelope during mitosis, the boundary between nuclear-retained IRAlus mRNAs and cytoplasmic PKR is removed, resulting in PKR activation (Fig. 4B) (64). Activated PKR is necessary for the regulation of mitosis because it acts as an upstream kinase for c-Jun N-terminal kinase (JNK), which controls the abundance of multiple mitotic factors (Fig. 4B) (64).
SINEs can also enhance mRNA translation in trans. The normally low cellular abundance of Pol III–synthesized Alu RNAs transiently increases under stressful conditions that include viral infection, heat shock, and the inhibition of protein synthesis (65). Other mammalian SINEs, such as mouse B1 and B2 elements and the rabbit C element, exhibit a similar response, suggesting a common mode of regulation during the stress response (65). During heat shock, Alu RNAs enhance translation, presumably by sequestering PKR to discrete loci so as to inhibit its activation (66). Additionally, transiently introduced Alu RNAs in human cells, and B1 and B2 RNAs in mouse cells, selectively enhance the translation of reporter mRNAs independently of PKR without affecting global cell translation (67). In mouse cells, lncRNAs called SINEUPs stimulate the translation of mRNAs with which they form base pairs via 5′-end complementary sequences. Stimulation depends on a B2 element embedded within the SINEUP (68).
Influence of A-to-I editing on SINE function
Adenosine-to-inosine (A-to-I) RNA editing is a tissue-specific posttranscriptional process whereby adenosine residues located within dsRNAs are deaminated to inosines by the dsRNA-specific adenosine deaminase (ADAR) proteins (69). In primates, IRAlus are the main binding site for ADARs and are subject to editing at multiple sites (Fig. 5) (35, 36, 70). More than 90% of A-to-I editing in humans occurs within Alu elements (71–75). Multisite A-to-I editing within exonized Alu elements are predicted to result in amino acid recoding, because inosines are recognized as guanosines by translating ribosomes (55, 76, 77). A-to-I editing within intronic IRAlus can generate new splice sites that lead to the exonization of Alu elements (Fig. 5A) (78). For instance, exonization of the alternatively spliced exon 8 of human nuclear prelamin A recognition factor pre-mRNA results from the A-to-I editing–dependent generation of a functional 3′ splice site within an intronic Alu (78, 79). The editing of IRAlus embedded in UTRs is not site-specific and the biological significance is not known. One possible function of UTR-embedded IRAlus is to act as “sponges” that titrate ADAR away from site-specific editing sites within ORFs to prevent amino acid recoding (55, 76, 77). Because inosine forms base pairs with cytosine, A-to-I editing influences the stability of the IRAlus double-stranded structure by creating mismatches or, less likely, matches (Fig. 5B). Thus, A-to-I editing might change the repertoire of proteins that bind IRAlus and thereby have an impact on the metabolism of transcripts within which IRAlus reside.
Fig. 5. The roles of A-to-I editing of IRAlus.
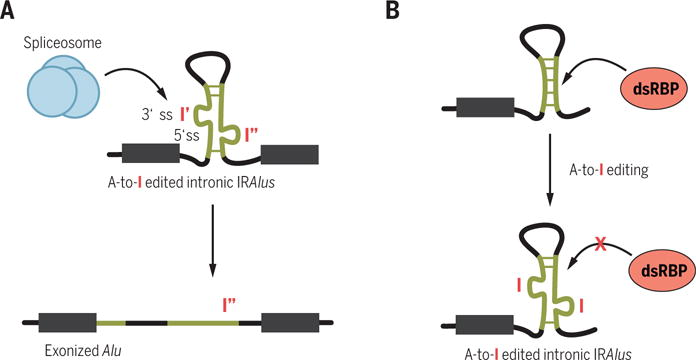
(A) Edited intronic IRAlus can create a new splice site. (B) Editing in IRAlus might destabilize their dsRNA structure and reduce dsRBP binding.
A-to-I editing within Alu elements might also reprogram the interaction network between miRNAs and their Alu-embedded target sites via deactivating or, possibly creating miRNA-binding sites (80). Recent profiling of mRNAs that bind the miRNA machinery indicates that the majority of miRNA targets within Alu elements are used less than those residing outside of Alu elements, presumably because Alu-element A-to-I editing and tight secondary structures prevent access to computationally predicted miRNA-binding sites within Alu elements (81).
Host defense against retrotransposition
Whereas some LINE and SINE insertions regulate gene expression, retrotransposition is necessarily mutagenic with the potential to cause disease. A small minority of L1, Alu, and SVA (SINE–variable number of tandem repeats–Alu) elements retain functional promoters that enable them to be transcribed and to retrotranspose. Alu elements are currently the most active retrotransposon in the human germ line, manifesting an estimated insertion rate of 1 in 20 live births (82). The estimated L1 insertion rate is 1 in 20 to 1 in 200 live births; the estimated SVA insertion rate is 1 in 900 live births (3). Germline insertions have been implicated in ~100 genetic diseases (Table 1) (83, 84) and insertion events in somatic tissues, although not heritable, also have the potential to cause disease (20, 85). Indeed, ongoing retrotransposition that results from the removal of inhibitory methylation marks on LINE and SINE promoters is a hallmark of many cancers (86) and also typifies neurological disorders, including schizophrenia (87) and Rhett syndrome (88).
Table 1. Some human diseases linked to LINE and SINE insertions.
The extensive role of LINEs and SINEs in the regulation of human gene expression suggests that they contribute to disease in as yet undiscovered ways.
| Effect of LINE or SINE insertion | Possible mechanism(s) of pathogenesis | Examples of associated diseases | Reference |
|---|---|---|---|
| Genomic deletions and rearrangements | LINE/SINE-mediated homologous recombination: DNA sequence loss; genomic instability | Prostate cancer, pyruvate dehydrogenase complex deficiency, leukemia, Alport syndrome, breast cancer | (83) |
| Hereditary nonpolyposis colorectal cancer, Von Hippel–Lindau disease | (86) | ||
| Disruption of protein-coding sequences | Aberrant protein production; nonsense-mediated mRNA decay (NMD) | Hemophilia B, breast cancer, colon cancer, neurofibromatosis type 1 | (83) |
| Altered DNA methylation | Increased expression of LINE and SINE RNA | Early event in many cancers | (86) |
| Altered pre-mRNA splicing | Aberrant protein production; NMD | Fukuyama-type congenital muscular dystrophy, neurofibromatosis type 1, hemophilia A | (83) |
| Neurofibromatosis type 1, hemophilia A, breast cancer, Coffin-Lowry syndrome | (84) | ||
| Altered 3′-end formation | Premature transcription termination; altered protein production; NMD; altered mRNA stability, localization, or translatability | X-linked retinitis pigmentosa | (83) |
| Altered mRNA stability | Reduced protein production; altered temporal and/or spatial gene expression | X-linked dilated cardiomyopathy | (83) |
| Hemophilia A, hereditary nonpolyposis colorectal cancer, hyper–immunoglobulin M syndrome | (84) | ||
| Sites of A-to-I editing | Loss of ADAR editing of target sites, possibly at Alu elements | Amyotrophic lateral sclerosis (ALS), astrocytoma, metastatic melanoma, Aicardi-Goutières syndrome, hepatocellular carcinoma | (100) |
Organisms have developed various mechanisms to protect their genomes from the deleterious effects of retrotransposon insertions [reviewed in (89, 90)]. Transcriptional silencing of retrotransposons by DNA methylation has been described as a major host defense mechanism in mammals (89, 90). However, recent evidence suggests that histone methylation, rather than DNA methylation, is the predominant suppressor of SINE transcription in human and mouse cells (91). Mammalian cells have also developed an arsenal of sequence-specific RNA degradation mechanisms to eliminate retrotransposon transcripts once produced. Endogenous small interfering RNAs (endo-siRNAs) or PIWI-interacting RNAs (piRNAs) can initiate degradation of LINE and SINE RNAs (92–96). Microprocessor, a nuclear complex of miRNA-processing enzymes, also recognizes and cleaves L1, Alu, and SVA transcripts, at least in vitro (97). The finding that L1 and Alu RNAs are enclosed within human cell autophagosomes has implicated autophagy (i.e., the delivery of cytosolic constituents to the lysosome) as another host defense mechanism that degrades retrotransposon RNAs (98). Indeed, mice lacking the critical autophagy gene, Atg6/Beclin1, are characterized by higher levels of retrotransposon RNAs and increased rates of genomic insertions (98). Additionally, retrotransposition is restricted in both somatic and germ cells by members of the apolipoprotein B mRNA-editing enzyme 3 (APOBEC3) family (89, 99).
Conclusions
Here we have focused on the functions of human and mouse LINEs and SINEs. However, the prevalence of LINEs and SINEs in other organisms, and known examples whereby evolutionarily unrelated human and mouse SINEs have been exapted for similar functions, leads us to propose that at least some of the LINEs and SINEs found in many organisms are likely to be used analogously as regulatory elements. In addition to their exaptation as functional sequences, the recent discovery that LINEs and SINEs are actively retrotransposing in somatic tissues (including brain) and the myriad of potential consequences of LINE and SINE insertions implicate LINEs and SINEs in the molecular pathogenesis of acquired diseases, including diseases of aging.
Acknowledgments
We thank T. Eickbush, X. Li, and M. Popp for critically reading the manuscript. We apologize to our colleagues whose work we were unable to include because of space restrictions. Research on SINEs in the Maquat lab is supported by NIH grant R37 GM074593 (L.E.M.). R.A.E. is supported by NIH Pilot Award P30 AR061307.
REFERENCES AND NOTES
- 1.Huang CRL, Burns KH, Boeke JD. Active transposition in genomes. Annu Rev Genet. 2012;46:651–675. doi: 10.1146/annurev-genet-110711-155616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 3.Beck CR, García-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroutter EN, Belancio VP, Wagstaff BJ, Roy-Engel AM. The RNA polymerase dictates ORF1 requirement and timing of LINE and SINE retrotransposition. PLOS Genet. 2009;5:e1000458. doi: 10.1371/journal.pgen.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 6.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-86749390078-5. [DOI] [PubMed] [Google Scholar]
- 7.Denli AM, et al. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell. 2015;163:583–593. doi: 10.1016/j.cell.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 8.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 9.Kramerov DA, Vassetzky NS. Origin and evolution of SINEs in eukaryotic genomes. Heredity. 2011;107:487–495. doi: 10.1038/hdy.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapitonov VV, Jurka J. A novel class of SINE elements derived from 5S rRNA. Mol Biol Evol. 2003;20:694–702. doi: 10.1093/molbev/msg075. [DOI] [PubMed] [Google Scholar]
- 11.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 12.Medstrand P, van de Lagemaat LN, Mager DL. Retroelement distributions in the human genome: Variations associated with age and proximity to genes. Genome Res. 2002;12:1483–1495. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sen SK, et al. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 16.Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: Mechanistic and evolutionary insights. Nat Rev Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kano H, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 19.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichiyanagi K. Epigenetic regulation of transcription and possible functions of mammalian short interspersed elements, SINEs. Genes Genet Syst. 2013;88:19–29. doi: 10.1266/ggs.88.19. [DOI] [PubMed] [Google Scholar]
- 22.Estécio MRH, et al. SINE retrotransposons cause epigenetic reprogramming of adjacent gene promoters. Mol Cancer Res. 2012;10:1332–1342. doi: 10.1158/1541-7786.MCR-12-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lunyak VV, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 24.Chow JC, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 25.Sasaki T, et al. Possible involvement of SINEs in mammalian-specific brain formation. Proc Natl Acad Sci USA. 2008;105:4220–4225. doi: 10.1073/pnas.0709398105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tashiro K, et al. A mammalian conserved element derived from SINE displays enhancer properties recapitulating Satb2 expression in early-born callosal projection neurons. PLOS ONE. 2011;6:e28497. doi: 10.1371/journal.pone.0028497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faulkner GJ, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 28.Mariner PD, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 29.Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 30.Sela N, et al. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu’s unique role in shaping the human transcriptome. Genome Biol. 2007;8:R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lev-Maor G, Sorek R, Shomron N, Ast G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 32.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen S, et al. Widespread establishment and regulatory impact of Alu exons in human genes. Proc Natl Acad Sci USA. 2011;108:2837–2842. doi: 10.1073/pnas.1012834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zarnack K, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deininger P. Alu elements: Know the SINEs. Genome Biol. 2011;12:236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang XO, et al. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 39.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: Extent, regulation and function. Nat Rev Genet. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 42.Roy-Engel AM, et al. Human retroelements may introduce intragenic polyadenylation signals. Cytogenet. Genome Res. 2005;110:365–371. doi: 10.1159/000084968. [DOI] [PubMed] [Google Scholar]
- 43.Lee JY, Ji Z, Tian B. Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3′-end of genes. Nucleic Acids Res. 2008;36:5581–5590. doi: 10.1093/nar/gkn540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C, Ara T, Gautheret D. Using Alu elements as polyadenylation sites: A case of retroposon exaptation. Mol Biol Evol. 2009;26:327–334. doi: 10.1093/molbev/msn249. [DOI] [PubMed] [Google Scholar]
- 45.An HJ, Lee D, Lee KH, Bhak J. The association of Alu repeats with the generation of potential AU-rich elements (ARE) at 3′ untranslated regions. BMC Genomics. 2004;5:97. doi: 10.1186/1471-2164-5-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piriyapongsa J, Mariño-Ramírez L, Jordan IK. Origin and evolution of human microRNAs from transposable elements. Genetics. 2007;176:1323–1337. doi: 10.1534/genetics.107.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehnert S, et al. Evidence for co-evolution between human microRNAs and Alu-repeats. PLOS ONE. 2009;4:e4456. doi: 10.1371/journal.pone.0004456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gong C, Tang Y, Maquat LE. mRNA-mRNA duplexes that autoelicit Staufen1-mediated mRNA decay. Nat Struct Mol Biol. 2013;20:1214–1220. doi: 10.1038/nsmb.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gleghorn ML, Gong C, Kielkopf CL, Maquat LE. Staufen1 dimerizes through a conserved motif and a degenerate dsRNA-binding domain to promote mRNA decay. Nat Struct Mol Biol. 2013;20:515–524. doi: 10.1038/nsmb.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park E, Gleghorn ML, Maquat LE. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci USA. 2013;110:405–412. doi: 10.1073/pnas.1213508110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gong C, Maquat LE. “Alu”strious long ncRNAs and their role in shortening mRNA half-lives. Cell Cycle. 2011;10:1882–1883. doi: 10.4161/cc.10.12.15589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J, Gong C, Maquat LE. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 2013;27:793–804. doi: 10.1101/gad.212639.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gong C, Popp MWL, Maquat LE. Biochemical analysis of long non-coding RNA-containing ribonucleoprotein complexes. Methods. 2012;58:88–93. doi: 10.1016/j.ymeth.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen LL, Carmichael GG. Gene regulation by SINES and inosines: Biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7:3294–3301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- 56.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elbarbary RA, Li W, Tian B, Maquat LE. STAU1 binding 3′ UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes Dev. 2013;27:1495–1510. doi: 10.1101/gad.220962.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elbarbary RA, Maquat LE. Dodging two bullets with one dsRNA-binding protein. Cell Cycle. 2014;13:345–346. doi: 10.4161/cc.27512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu SB, et al. Protein arginine methyltransferase CARM1 attenuates the paraspeckle-mediated nuclear retention of mRNAs containing IRAlus. Genes Dev. 2015;29:630–645. doi: 10.1101/gad.257048.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elbarbary RA, Maquat LE. CARMing down the SINEs of anarchy: Two paths to freedom from paraspeckle detention. Genes Dev. 2015;29:687–689. doi: 10.1101/gad.261438.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fox AH, et al. Paraspeckles: A novel nuclear domain. Curr Biol. 2002;12:13–25. doi: 10.1016/S0960-98220100632-7. [DOI] [PubMed] [Google Scholar]
- 62.Capshew CR, Dusenbury KL, Hundley HA. Inverted Alu dsRNA structures do not affect localization but can alter translation efficiency of human mRNAs independent of RNA editing. Nucleic Acids Res. 2012;40:8637–8645. doi: 10.1093/nar/gks590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fitzpatrick T, Huang S. 3′-UTR-located inverted Alu repeats facilitate mRNA translational repression and stress granule accumulation. Nucleus. 2012;3:359–369. doi: 10.4161/nucl.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim Y, et al. PKR is activated by cellular dsRNAs during mitosis and acts as a mitotic regulator. Genes Dev. 2014;28:1310–1322. doi: 10.1101/gad.242644.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu WM, Chu WM, Choudary PV, Schmid CW. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chu WM, Ballard R, Carpick BW, Williams BR, Schmid CW. Potential Alu function: Regulation of the activity of double-stranded RNA-activated kinase PKR. Mol Cell Biol. 1998;18:58–68. doi: 10.1128/MCB.18.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rubin CM, Kimura RH, Schmid CW. Selective stimulation of translational expression by Alu RNA. Nucleic Acids Res. 2002;30:3253–3261. doi: 10.1093/nar/gkf419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carrieri C, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 69.Savva YA, Rieder LE, Reenan RA. The ADAR protein family. Genome Biol. 2012;13:252. doi: 10.1186/gb-2012-13-12-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eisenberg E, et al. Is abundant A-to-I RNA editing primate-specific? Trends Genet. 2005;21:77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 71.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLOS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim DDY, et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Levanon EY, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 75.Morse DP, Aruscavage PJ, Bass BL. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc Natl Acad Sci USA. 2002;99:7906–7911. doi: 10.1038/nn.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li JB, Church GM. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat Neurosci. 2013;16:1518–1522. doi: 10.1038/nn.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mallela A, Nishikura K. A-to-I editing of protein coding and noncoding RNAs. Crit Rev Biochem Mol Biol. 2012;47:493–501. doi: 10.3109/10409238.2012.714350. [DOI] [PubMed] [Google Scholar]
- 78.Lev-Maor G, et al. RNA-editing-mediated exon evolution. Genome Biol. 2007;8:R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Möller-Krull M, Zemann A, Roos C, Brosius J, Schmitz J. Beyond DNA: RNA editing and steps toward Alu exonization in primates. J Mol Biol. 2008;382:601–609. doi: 10.1016/j.jmb.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 80.Borchert GM, et al. Adenosine deamination in human transcripts generates novel microRNA binding sites. Hum. Mol. Genet. 2009;18:4801–4807. doi: 10.1093/hmg/ddp443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hoffman Y, Dahary D, Bublik DR, Oren M, Pilpel Y. The majority of endogenous microRNA targets within Alu elements avoid the microRNA machinery. Bioinformatics. 2013;29:894–902. doi: 10.1093/bioinformatics/btt044. [DOI] [PubMed] [Google Scholar]
- 82.Cordaux R, Hedges DJ, Herke SW, Batzer MA. Estimating the retrotransposition rate of human Alu elements. Gene. 2006;373:134–137. doi: 10.1016/j.gene.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 83.Hancks DC, Kazazian HH., Jr Active human retrotransposons: Variation and disease. Curr Opin Genet Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kaer K, Speek M. Retroelements in human disease. Gene. 2013;518:231–241. doi: 10.1016/j.gene.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 85.Helman E, et al. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014;24:1053–1063. doi: 10.1101/gr.163659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Belancio VP, Roy-Engel AM, Deininger PL. All y’all need to know ’bout retroelements in cancer. Semin Cancer Biol. 2010;20:200–210. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bundo M, et al. Increased L1 retrotransposition in the neuronal genome in schizophrenia. Neuron. 2014;81:306–313. doi: 10.1016/j.neuron.2013.10.053. [DOI] [PubMed] [Google Scholar]
- 88.Muotri AR, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12:615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Crichton JH, Dunican DS, Maclennan M, Meehan RR, Adams IR. Defending the genome from the enemy within: Mechanisms of retrotransposon suppression in the mouse germline. Cell Mol Life Sci. 2014;71:1581–1605. doi: 10.1007/s00018-013-1468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Varshney D, et al. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat Commun. 2015;6:6569. doi: 10.1007/s00018-013-1468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: New insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012;26:2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weick EM, Miska EA. piRNAs: From biogenesis to function. Development. 2014;141:3458–3471. doi: 10.1242/dev.094037. [DOI] [PubMed] [Google Scholar]
- 94.Claycomb JM. Ancient endo-siRNA pathways reveal new tricks. Curr Biol. 2014;24:R703–R715. doi: 10.1016/j.cub.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 95.Piatek MJ, Werner A. Endogenous siRNAs: Regulators of internal affairs. Biochem Soc Trans. 2014;42:1174–1179. doi: 10.1042/BST20140068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aravin AA, Hannon GJ. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:283–290. doi: 10.1101/sqb.2008.73.058. [DOI] [PubMed] [Google Scholar]
- 97.Heras SR, et al. The Microprocessor controls the activity of mammalian retrotransposons. Nat Struct Mol Biol. 2013;20:1173–1181. doi: 10.1038/nsmb.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guo H, et al. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat Commun. 2014;5:5276. doi: 10.1038/ncomms6276. [DOI] [PubMed] [Google Scholar]
- 99.Arias JF, Koyama T, Kinomoto M, Tokunaga K. Retroelements versus APOBEC3 family members: No great escape from the magnificent seven. Front Microbiol. 2012;3:275. doi: 10.3389/fmicb.2012.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Slotkin W, Nishikura K. Adenosine-to-inosine RNA editing and human disease. Genome Med. 2013;5:105. doi: 10.1186/gm508. [DOI] [PMC free article] [PubMed] [Google Scholar]


