Abstract
Ever since Stephen Paget’s 1889 hypothesis, metastatic organotropism has remained one of cancer’s greatest mysteries. Here we demonstrate that exosomes from mouse and human lung-, liver- and brain-tropic tumour cells fuse preferentially with resident cells at their predicted destination, namely lung fibroblasts and epithelial cells, liver Kupffer cells and brain endothelial cells. We show that tumour-derived exosomes uptaken by organ-specific cells prepare the pre-metastatic niche. Treatment with exosomes from lung-tropic models redirected the metastasis of bone-tropic tumour cells. Exosome proteomics revealed distinct integrin expression patterns, in which the exosomal integrins α6β4 and α6β1 were associated with lung metastasis, while exosomal integrin αvβ5 was linked to liver metastasis. Targeting the integrins α6β4 and αvβ5 decreased exosome uptake, as well as lung and liver metastasis, respectively. We demonstrate that exosome integrin uptake by resident cells activates Src phosphorylation and pro-inflammatory S100 gene expression. Finally, our clinical data indicate that exosomal integrins could be used to predict organ-specific metastasis.
Despite Stephen Paget’s 126-year-old “seed-and-soil” hypothesis1, insufficient progress has been made towards decoding the mechanisms governing organ-specific metastasis. In experimental metastasis assays, Fidler et al. demonstrated that cancer cells derived from a certain metastatic site displayed enhanced abilities to metastasize to that specific organ, providing support for Paget’s organ-specific metastasis theory2. Subsequent studies investigating organ-specific metastasis focused largely on the role of intrinsic cancer cell properties, such as genes and pathways regulating colonization, in directing organotropism3–8. Breast cancer cells express chemokine receptors, such as C-X-C motif receptor 4 (CXCR4) and C-C motif receptor 7 (CCR7), which partner with chemokine ligands expressed in lymph nodes (CXCL12) and lung (CCL21), thus guiding metastasis3,4. Tumour-secreted factors can also increase metastasis by inducing vascular leakiness5, promoting the recruitment of pro-angiogenic immune cells6, and influencing organotropism7. Furthermore, the ability of breast cancer to form osteolytic lesions depends on osteoclast-stimulating growth factors (for example, PTHRP and GM-CSF) released into the bone microenvironment4,8. Therefore, our previous observation that metastatic melanoma-derived factors dictate organotropism is not surprising9. We found that medium conditioned by highly metastatic murine B16-F10 melanoma cells was sufficient to expand the metastatic repertoire of Lewis lung carcinoma cells that would typically metastasize to the lung9. We also showed that pre-metastatic niche formation requires S100 protein and fibronectin upregulation by lung resident cells, and the recruitment of bone-marrow-derived myeloid cells in response to tumour-secreted factors9. These events establish a favourable microenvironment that promotes the growth of disseminated tumour cells upon their arrival9–11.
Recently, we demonstrated that exosomes are one of the tumour-derived factors inducing vascular leakiness, inflammation and bone marrow progenitor cell recruitment during pre-metastatic niche formation and metastasis11. Exosomes are small membrane vesicles (30–100 nm) containing functional biomolecules (that is, proteins, lipids, RNA and DNA) that can be horizontally transferred to recipient cells12–19. We showed that an ‘exosomal protein signature’ could identify melanoma patients at risk for metastasis to nonspecific distant sites11. Moreover, in the context of pancreatic cancer exosomes, we defined the sequential steps involved in liver pre-metastatic niche induction20.
Taken together, these findings led us to investigate whether molecules present on tumour-derived exosomes are ‘addressing’ them to specific organs. To test this idea, we profiled the exosomal proteome of several tumour models (osteosarcoma, rhabdomyosarcoma, Wilms tumour, skin and uveal melanoma, breast, colorectal, pancreatic and gastric cancers), all of which have a propensity to metastasize to specific sites (that is, brain, lung or liver). We subsequently analysed the biodistribution of tumour-secreted exosomes and found that exosomal integrins (ITGs) direct organ-specific colonization by fusing with target cells in a tissue-specific fashion, thereby initiating pre-metastatic niche formation. Remarkably, we found that tumour-secreted exosomes are sufficient to redirect metastasis of tumour cells that normally lack the capacity to metastasize to a specific organ. Finally, our clinical data indicate that integrin expression profiles of circulating plasma exosomes isolated from cancer patients could be used as prognostic factors to predict sites of future metastasis. Our findings pave the way for the development of diagnostic tests to predict organ-specific metastasis and therapies to halt metastatic spread.
Future metastatic sites uptake exosomes
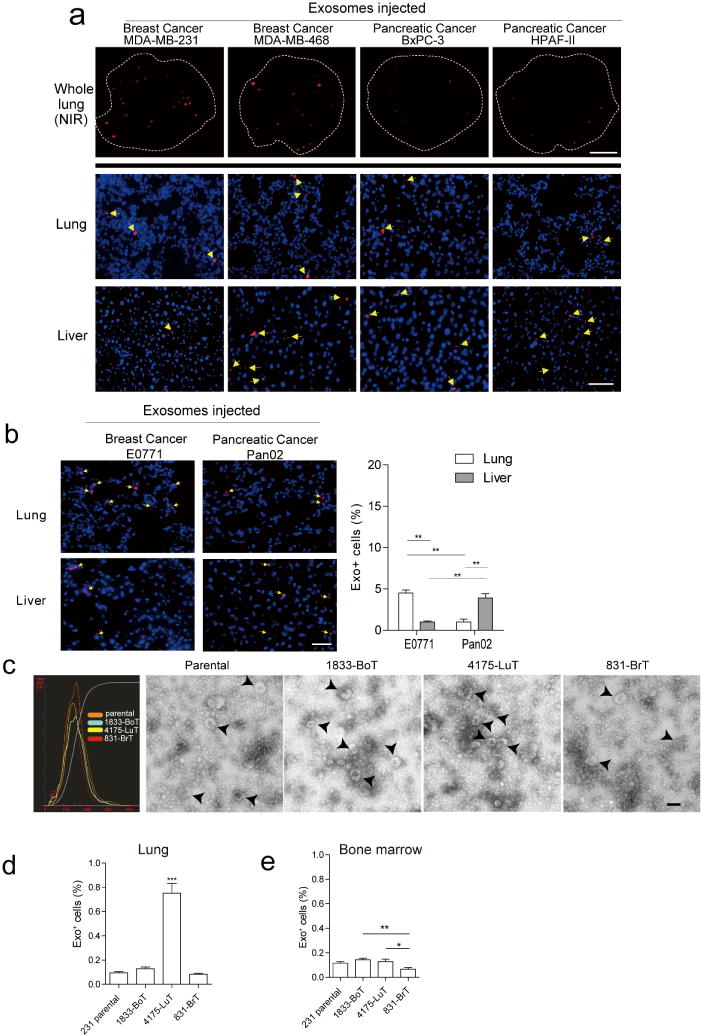
To examine whether tumour exosomes colonize specific organ sites, we isolated exosomes from organotropic human breast and pancreatic cancer cell lines that metastasize primarily to the lung (MDA-MB-231), liver (BxPC-3 and HPAF-II), or both (MDA-MB-468). We then retro-orbitally injected 10 μg of near infrared (NIR) or red fluorescently labelled exosomes into nude mice and, 24 h after injection, quantified exosome biodistribution and uptake in distant organs by NIR whole-lung imaging and confocal microscopy (Fig. 1a and Extended Data Fig. 1a). We observed a more than threefold increase in the uptake of MDA-MB-231 and/or 468- versus BxPC-3- and HPAF-II-derived exosomes in the lung (Fig. 1a, b). By contrast, liver uptake of BxPC-3 and HPAF-II exosomes was four times more efficient than that of MDA-MB-231 exosomes (Fig. 1a, b). Moreover, mouse E0771 breast cancer exosomes were four-to-fivefold more efficiently uptaken in lung, whereas mouse Pan02 pancreatic cancer exosomes were four times more efficiently uptaken in liver (Extended Data Fig. 1b). Therefore, the organ specificity of exosome biodistribution matched the organotropic distribution of the cell line of origin in both immune-compromised and immune-competent models.
Figure 1. Cancer-cell-derived exosomes localize to and dictate future metastatic organs.
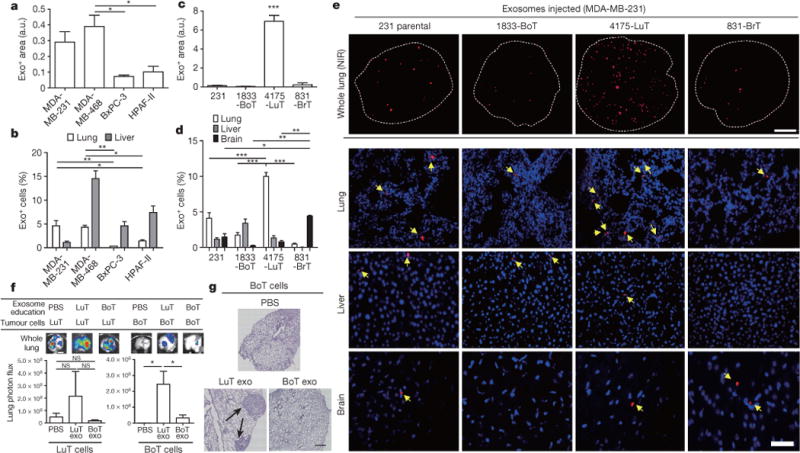
a, Biodistribution of human cancer-cell-line-derived exosomes in the lung and liver of naive mice. Quantification of exosome-positive (Exo+) areas by NIR imaging of whole lung, in arbitrary units (a.u.) (n = 3 per group). b, Immunofluorescence quantification of exosome-positive cells (n = 3, three independent experiments). c, MDA-MB-231- (parental), 1833-BoT-, 4175-LuT- and 831-BrT-derived exosome biodistribution. Quantification of exosome-positive areas by NIR imaging of whole lung (n = 3 for all, except 831-BrT, in which n = 4). d, Immunofluorescence quantification of exosome-positive cells (n = 5 animals pooled from two independent experiments). e, Top, NIR whole-lung imaging of MDA-MB-231 sublines. BoT, bone-tropic; BrT, brain-tropic; LuT, lung-tropic. Bottom, fluorescence microscopy of lung, liver and brain injected with MDA-MB-231 subline-derived exosomes. Arrows indicate exosome foci. All NIR and immunofluorescence images are representative of five random fields. f, Redirection of metastasis by education with organotropic exosomes. 4175-LuT or 1833-BoT cell metastasis in the lung after treatment with PBS, 4175-LuT or 1833-BoT exosomes. Top, quantitative bioluminescence of metastatic lesions. Bottom, graphs show quantification of luciferase activity (n = 5 for all, except for LuT exo/LuT cells, in which n = 4; data representative of two independent experiments). g, Lung haematoxylin/eosin staining for f. Arrows indicate lung metastasis. Scale bars, 5 mm (e, top, f), 50 μm (e, bottom) and 500 μm (g). Data are mean ± s.e.m. NS, not significant; * P < 0.05; ** P < 0.01; *** P < 0.001 by one-way analysis of variance (ANOVA).
These observations suggested that exosomes could promote organ-specific metastasis. We tested whether exosomes from the MDA-MB-231 sub-lines that colonize lung, bone or brain (4175-LuT, 1833-BoT or 831-BrT, respectively)21–24 would also exhibit organ tropism. Although exosomes from the MDA-MB-231 variants were similar in size and morphology (Extended Data Fig. 1c), their biodistribution varied 24 h after injection: lung-tropic 4175-LuT exosomes preferentially localized to the lung with a more than fourfold increase in exosome-positive cells compared to 1833-BoT and 831-BrT exosomes (Fig. 1c–e and Extended Data Fig. 1d), whereas 831-BrT exosomes efficiently localized to the brain with a more than fourfold increase compared to 1833-BoT and 4175-LuT exosomes (Fig. 1c–e). Liver and bone showed no significant differences in lung-, brain- or bone-tropic MDA-MB-231-derived exosome distribution, with the exception of 831-BrT exosomes that were uptaken less efficiently by bone marrow cells than exosomes isolated from other MDA-MB-231 sub-lines (Fig. 1d, e and Extended Data Fig. 1e). Taken together, our data suggest that exosomes from different cancer models recapitulate the organ specificity of their cell of origin.
Exosomes redirect metastatic distribution
We proposed that, in addition to cell-intrinsic genetic determinants of organotropism23,24, tumour exosomes could also facilitate organ-specific metastatic behaviour by preparing pre-metastatic niches.
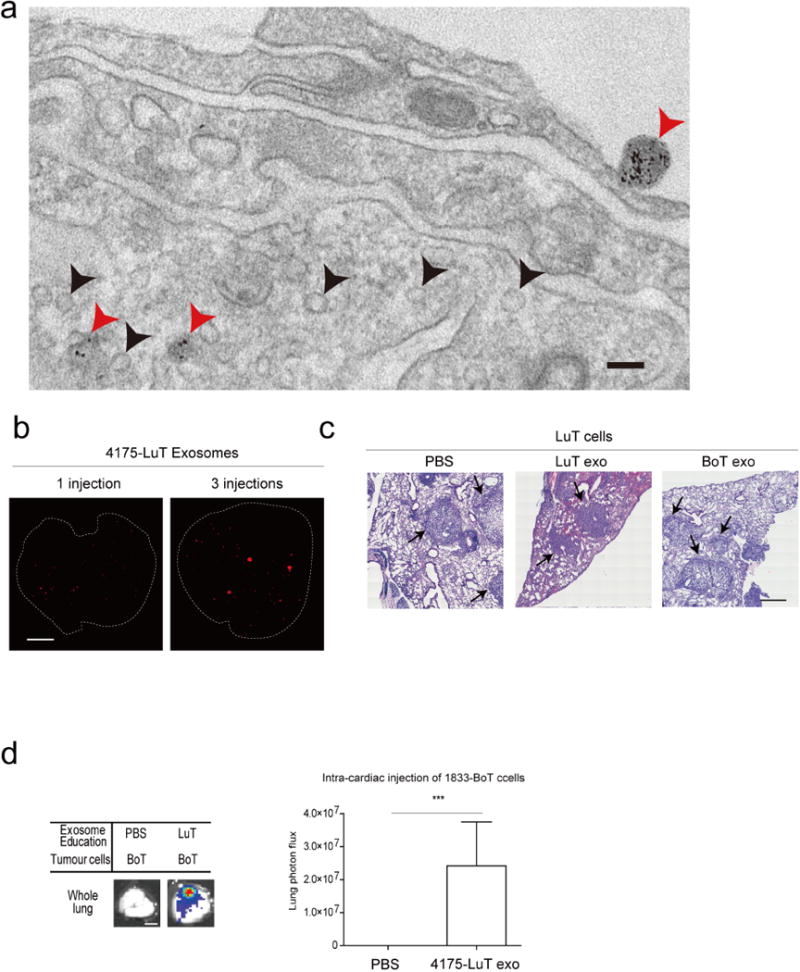
To gain insight into tumour exosome uptake at future metastatic sites, we intravenously injected 4175-LuT exosomes labelled with FM1-43 dye into naive animals, then used electron microscopy to distinguish endogenous from exogenous exosomes in lung sections. We detected tumour FM1-43-labelled exosomes in pre-metastatic cells (Extended Data Fig. 2a; red arrows, exogenous tumour-derived exosomes; black arrows, endogenous stromal exosomes). Moreover, NIR whole-mount lung imaging revealed that NIR-labelled 4175-LuT exosomes accumulated in the lungs of naive animals after three consecutive daily exosome injections (Extended Data Fig. 2b).
To condition or ‘educate’ cells in specific organs, we retro-orbitally injected 4175-LuT or 1833-BoT exosomes into mice every other day for three weeks11. To test exosome education of target organs functionally, luciferase-expressing 4175-LuT or 1833-BoT cells were injected into exosome-educated mice via the tail vein (Fig. 1f, g and Extended Data Fig. 2c) or intracardially (Extended Data Fig. 2d). Lung-tropic 4175 exosomes marginally increased the lung-metastatic capacity of 4175-LuT tumours. Remarkably, education with 4175-LuT-derived exosomes, but not with 1833-BoT exosomes or PBS, yielded a significant (sevenfold with intravenous and ten-thousand-fold with intracardiac injections) increase in lung metastatic capacity of 1833-BoT cells (Fig. 1f, g and Extended Data Fig. 2d). Conversely, 1833-BoT-derived exosomes did not affect 4175-LuT cell metastasis to the lung (Fig. 1f and Extended Data Fig. 2c). These data suggest that organotropic tumour exosomes prepare pre-metastatic niches potent enough to facilitate metastasis even for tumour cells poorly capable of colonizing these sites.
Exosomal ITGs determine organotropism
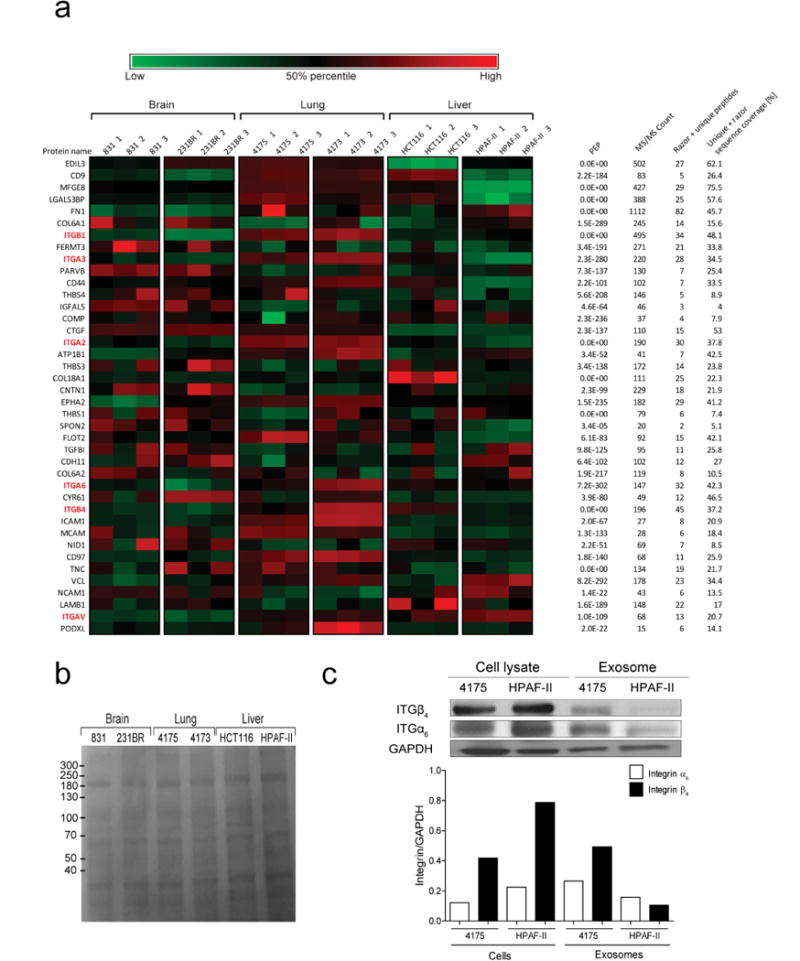
We then postulated that exosomal adhesion molecules could regulate local microenvironments within future metastatic organs. Quantitative mass spectrometry of brain-, lung- and liver-tropic metastatic exosomes identified six integrins among the top 40 most abundant adhesion molecules, making integrins the most highly represented protein family in this analysis. These data indicate a correlation between exosomal integrins and metastatic tropism (Extended Data Fig. 3a).
Interestingly, we found that integrin expression profiles correlated with tissue organotropism. Both quantitative mass spectrometry (Fig. 2a) and western blot analysis (Fig. 2b and Extended Data Fig. 3b) revealed that integrin alpha 6 (ITGα6), and its partners ITGβ4 and ITGβ1 (ref. 25), were present abundantly in lung-tropic exosomes. By contrast, ITGβ5, which associates only with ITGαv (ref. 25), was detected primarily in liver-tropic exosomes (Fig. 2a, b). We confirmed these findings by exosome proteomics for 28 organ-specific metastatic cell lines (Extended Data Tables 1 and 2). Qualitative mass spectrometry revealed that ITGα6 was present in lung-tropic exosomes, whereas ITGβ5 was found in liver-tropic exosomes (Extended Data Tables 1 and 2), consistent with our quantitative proteomics data. Exosomes from 4173, 4175 and 4180 lung-tropic MDA-MB-231 variants expressed ITGα6β4 (Extended Data Table 1). Meanwhile, ITGβ3 was present in exosomes isolated from brain-tropic cells (Extended Data Table 1). Notably, unlike non-cancerous lung fibroblast WI-38 or epithelial MCF10A exosomes, metastatic cell exosomes contained ITGα2β1, suggesting that this integrin could serve as a biomarker for metastasis (Extended Data Table 1). Importantly, exosomal integrin expression does not necessarily reflect cellular integrin expression, consistent with selective packaging of integrins in exosomes (Extended Data Fig. 3c). Taken together, our data suggest that exosomal integrin expression patterns underlie organotropism to the lung, liver and brain.
Figure 2. Organ-specific tumour exosomes interact with resident cells.
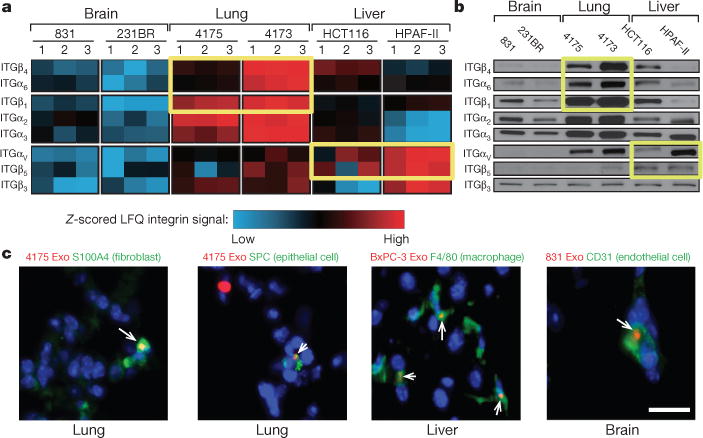
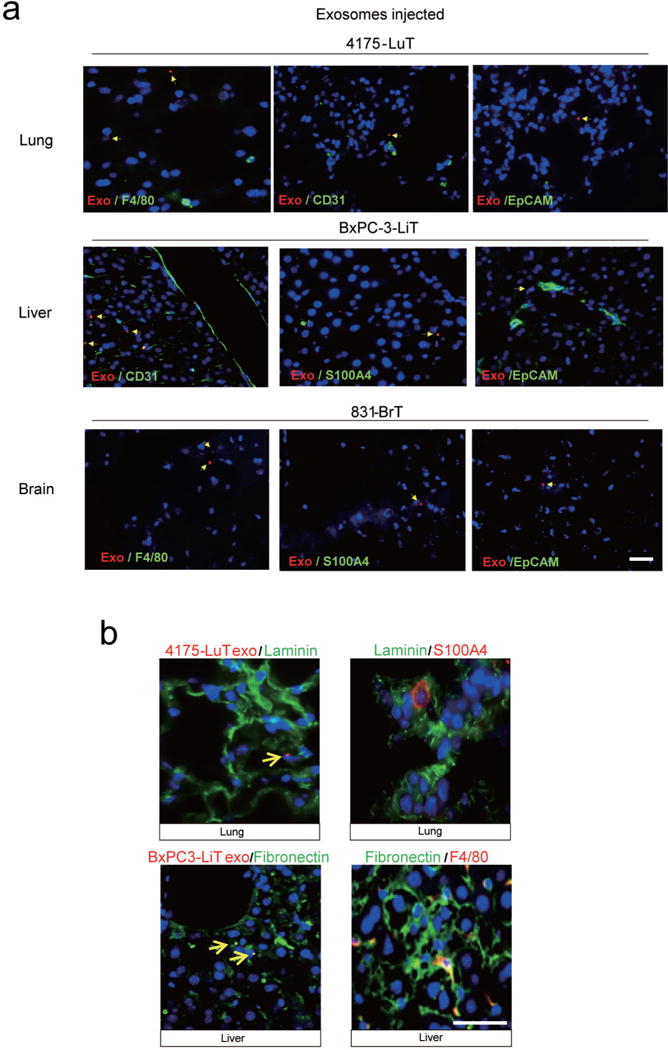
a, Heat map of integrin signals from quantitative mass spectrometry analysis, based on Z-scored label-free quantification (LFQ) values (technical triplicates). b, Western blot analysis of integrins from organotropic cell-line-derived exosomes, representative of three independent experiments. For western blot source data, see Supplementary Fig. 1a–h. c, Analysis by immunofluorescence of exosome distribution (red) and different resident cell types (green). Left to right: lung co-staining with 4175-LuT exosomes and S100A4 (fibroblasts) or SPC (epithelial cells), liver co-staining with F4/80 (macrophages) and BxPC-3-LiT exosomes, and brain co-staining with CD31 (endothelial cells) and 831-BrT exosomes. Scale bar, 30 μm. Immunofluorescence images are representative of five exosome-positive cells e ach, f rom n = 5 mice.
Distinct cells uptake tropic exosomes
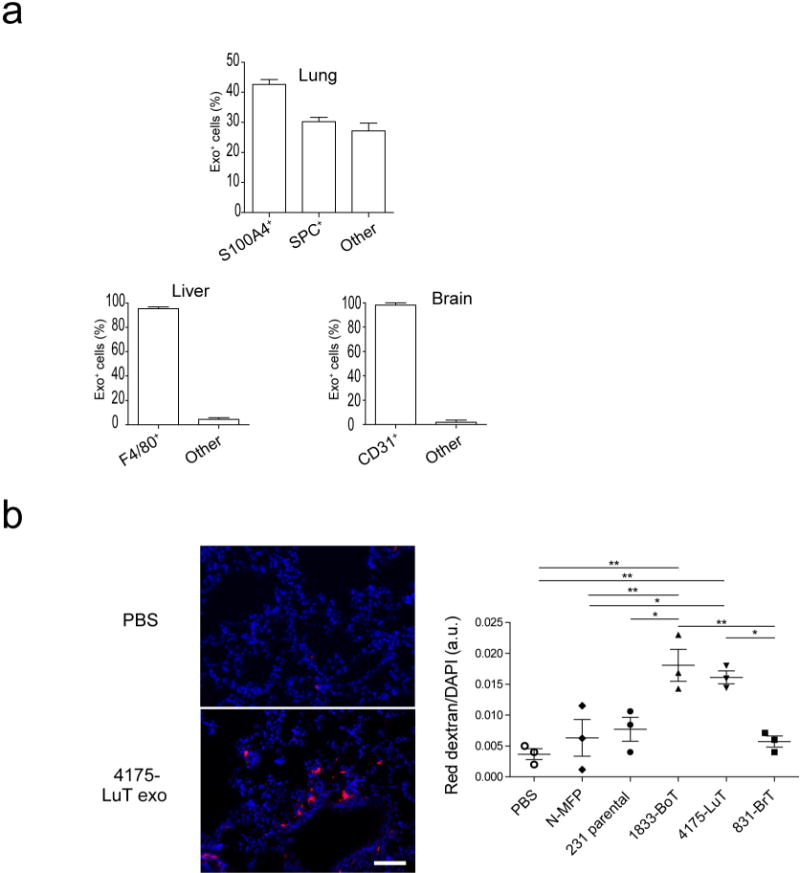
To identify the cells uptaking tumour exosomes in each organ, we intravenously injected red fluorescently labelled exosomes from 4175-LuT, 1833-BoT, BxPC-3-liver-tropic (BxPC-3-LiT) or 831-BrT cells into mice (Fig. 2c and Extended Data Fig. 4a, b). Both 1833-BoT and 4175-LuT exosomes promoted vascular leakiness 24 h after injection, before exosome uptake by specific lung cells (Extended Data Fig. 4b). These observations fit with our previous studies using melanoma exosomes11, suggesting that exosomes first permeabilize vessels, allowing for exosome diffusion before uptake by parenchymal cells. Unexpectedly, we found that the specific cell type responsible for exosome uptake varied depending on the metastatic organ. Lung-tropic 4175 exosomes mainly co-localized with S100A4-positive fibroblasts and surfactant protein C (SPC)-positive epithelial cells (40% and 30% of exosome-positive cells, respectively) in the lung (Fig. 2c and Extended Data Figs 4a and 5a, top). By contrast, pancreatic cancer exosomes derived from BxPC-3-LiT cells fused with Kupffer cells20 (90% of exosome-positive cells; Fig. 2c and Extended Data Figs 4a and 5a, middle). Finally, 831-BrT exosomes interacted mainly with CD31-positive brain endothelial cells (98% of exosome-positive cells; Fig. 2c and Extended Data Figs 4a and 5a, bottom). Collectively, these data demonstrate that specific tissue-resident stromal cells differentially uptake tumour exosomes in metastatic target organs.
We proposed that the unique exosomal integrins may interact with cell-associated extracellular matrix (ECM), mediating exosome uptake in specific target organs. We found that 4175-LuT exosomes expressing ITGα6β4 and ITGα6β1 co-localized with S100A4-positive cells in laminin-rich lung microenvironments (Extended Data Fig. 5b, top). Meanwhile, ITGαvβ5-expressing pancreatic BxPC-3-LiT exosomes co-localized with F4/80+ macrophages in fibronectin-rich liver microenvironments (Extended Data Fig. 5b, bottom). Therefore, specific exosomal integrins may selectively adhere to ECM-enriched cellular areas in the lung and liver.
Exosomal tropism requires ITGβ4 and ITGβ5
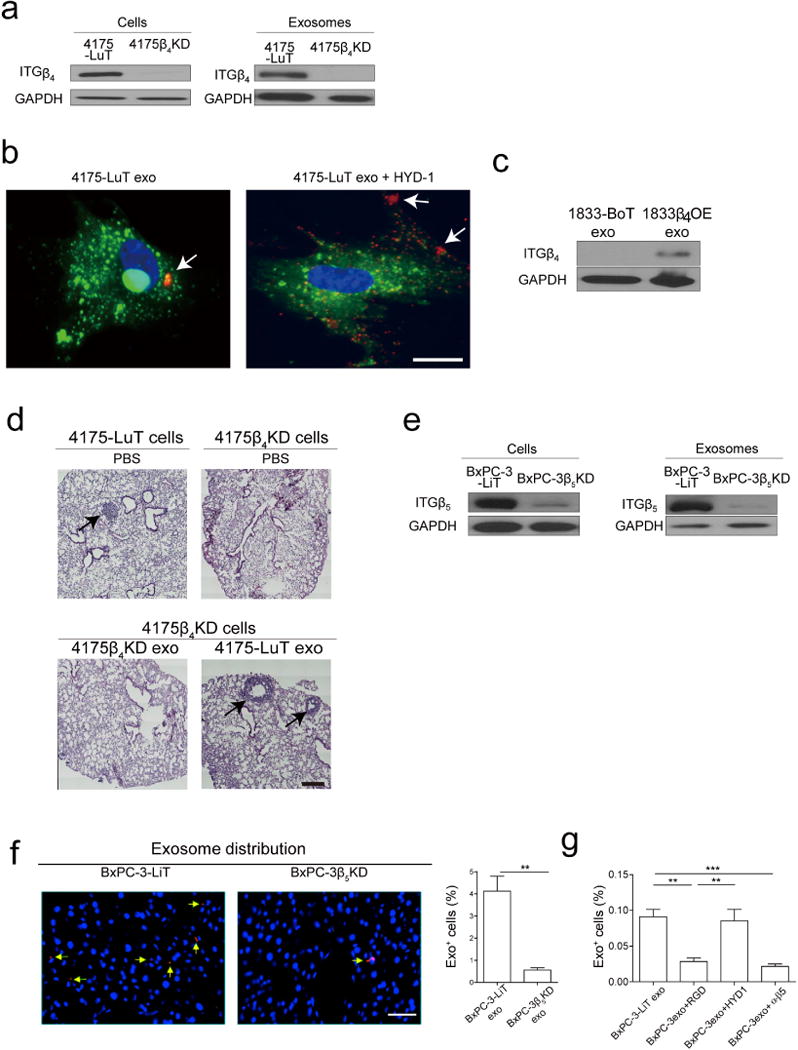
We next asked whether manipulating the integrin cargo packaged into exosomes could impact metastatic organotropism. To test the requirement for exosomal ITGβ4 in lung tropism, we knocked down ITGβ4 expression in 4175-LuT cells using short hairpin RNAs (shRNAs) (4175β4KD; Extended Data Fig. 6a). We found a more than threefold reduction in labelled ITGβ4KD exosomes in the lung compared with labelled control exosomes 24 h after injection (Fig. 3a–c). To test the requirement for exosomal ITGβ4 binding to laminin for lung tropism, we blocked integrin binding using RGD and HYD-1 peptides. Pre-incubation of 4175-LuT exosomes with HYD-1, which blocks laminin receptors26, markedly reduced exosome uptake in the lung, whereas RGD, which blocks fibronectin receptors but not ITGβ4 (ref. 27), did not significantly alter exosome uptake in the lung (Fig. 3a–c). Pre-treatment of 4175-LuT exosomes with HYD-1 peptide also prevented their uptake by WI-38 lung fibroblasts in vitro (Extended Data Fig. 6b). Conversely, ITGβ4 overexpression in 1833-BoT exosomes was sufficient to increase exosome uptake in lung (Extended Data Fig. 6c and Fig. 3d). These data demonstrate that integrins are responsible for organ-specific uptake of exosomes, and that ITGβ4 promotes tumour exosome adhesion within the lung.
Figure 3. Exosomal ITGβ4 expression functionally contributes to 4175-LuT exosome localization and mediates lung metastasis.
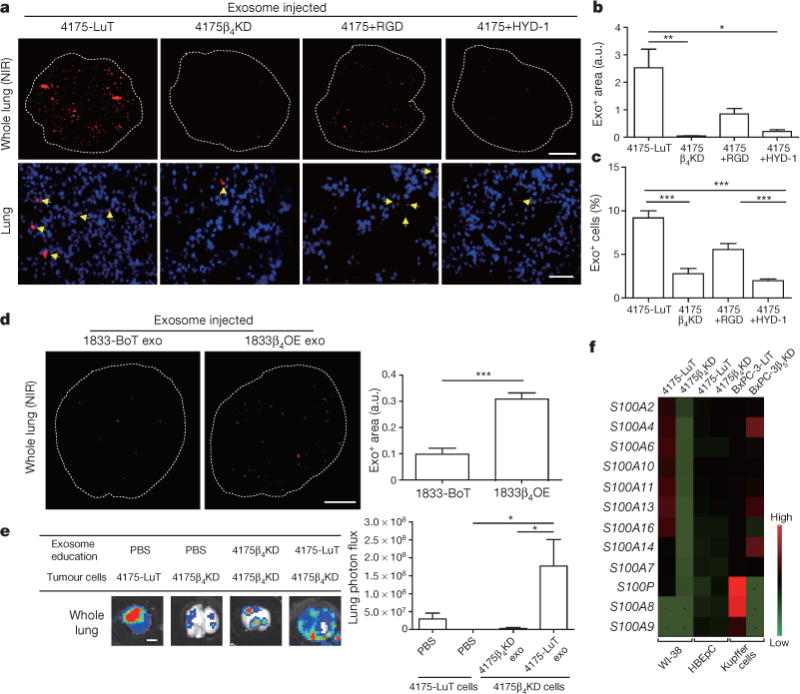
a, Top, NIR whole-lung imaging of 4175-LuT- or 4175β4KD-derived exosomes, or 4175-LuT-derived exosomes pre-incubated with RGD or HYD-1 peptides. Bottom, fluorescence microscopy. Arrowheads indicate exosome foci. b, Quantification of exosome-positive areas from the whole-lung images in a (top) (n = 4, except 4175, in which n = 6). c, Immunofluorescence quantification of exosome-positive cells from a (bottom) (n = 6 pooled from two independent experiments). d, Left, NIR whole-lung imaging of 1833-BoT (n = 5) or 1833-BoT overexpressing ITGβ4 (1833β4OE) (n = 4) exosomes. Right, quantification of the exosome-positive areas. e, Experimental lung metastasis of 4175β4KD cells after education with wild-type or 4175β4KD exosomes. Bioluminescence imaging of lung metastasis and quantification of luciferase activity (n = 6, data representative of two independent experiments). f, Heat map of S100 gene expression fold change by quantitative reverse transcription PCR (qRT–PCR) in 4175-LuT or 4175β4KD exosome-conditioned lung fibroblast (WI-38) or epithelial (HBEpC) cells, and liver-tropic BxPC-3 or BxPC-3β5KD exosome-conditioned Kupffer cells. Red represents high and green represents low expression (n = 3 in two independent experiments). Scale bars, 5 mm (a, top, d, e) and 50 μm (b, bottom). Data are mean ± s.e.m. * P < 0.05; ** P < 0.01; *** P < 0.001 by one-way ANOVA (b, c, e); *** P < 0.001 by two-tailed Student’s t-test (d).
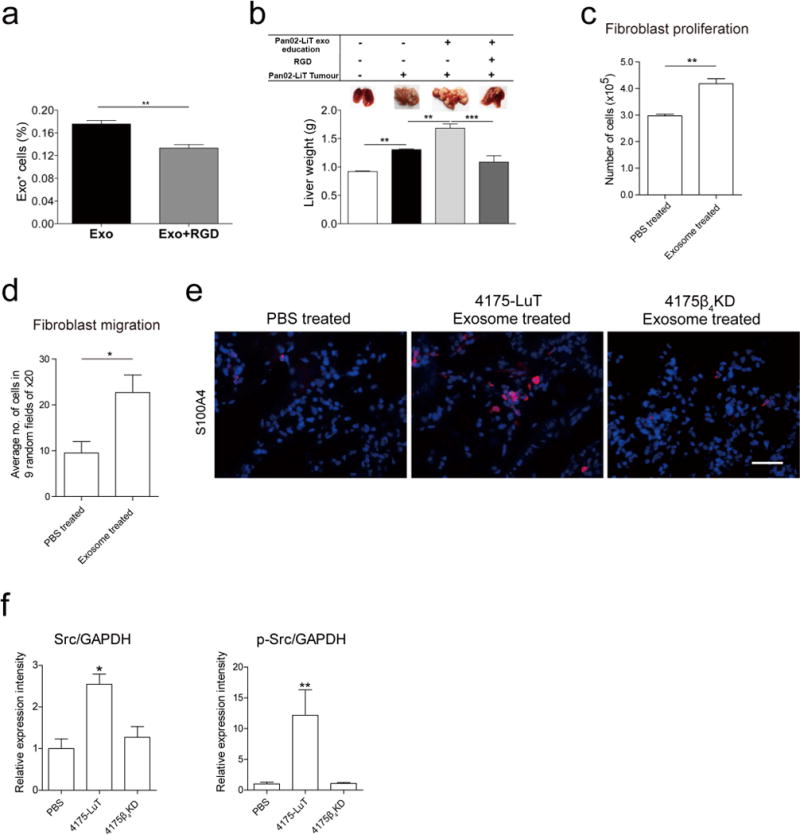
We next investigated whether ITGβ4KD exosomes could modulate the metastatic organotropism of 4175-LuT models. Knockdown of ITGβ4 was sufficient to reduce the lung metastatic capacity of 4175-LuT cells (Fig. 3e). Education with 4175-LuT exosomes, but not ITGβ4KD exosomes, rescued the metastatic ability of ITGβ4KD cells, yielding metastasis similar to 4175-LuT cells (Fig. 3e and Extended Data Fig. 6d). Therefore, ITGβ4-expressing exosomes can confer lung-metastatic behaviour to cells with limited capacity to colonize the lung. Similarly, ITGβ5 knockdown in BxPC-3-LiT exosomes decreased liver uptake by sevenfold compared with control BxPC-3-LiT exosomes (Extended Data Fig. 6e, f). Moreover, pre-incubation with RGD peptide or anti-ITGαvβ5 antibody, but not HYD-1 peptide, significantly reduced BxPC-3-LiT and Pan02-LiT exosome adhesion to the liver (Extended Data Figs 6g and 7a, respectively). Importantly, RGD peptides also inhibited the education effect of Pan02-LiT exosomes, subsequently blocking pre-metastatic niche formation and liver metastasis (Extended Data Fig. 7b). Our data support the hypothesis that local microenvironmental changes induced by specific exosomal cargo (that is, ITGβ4 or ITGβ5) can dictate metastatic organotropism.
Exosomal ITGs activate S100 genes
To identify the downstream effects of exosomal interaction with target cells, Kupffer cells were educated with either BxPC-3-LiT or BxPC-3-LiT ITGβ5KD exosomes every other day for two weeks. Unbiased analysis of gene expression by RNA sequencing in Kupffer cells identified 906 genes upregulated more than twofold after treatment with BxPC-3-LiT exosomes compared to BxPC-3-LiT ITGβ5KD exosomes. Cell migration genes were the most prominently upregulated (twofold for 221 genes; fourfold for 42 genes). Of these, S100A8 and S100P were upregulated more than fourfold (Fig. 3f; GEO accession GSE68919). Since pro-inflammatory S100 gene expression correlates with metastasis28,29, we analysed S100 genes in tumour exosome-educated lung WI-38 fibroblasts and in human bronchial epithelial cells (HBEpCs). Several S100 genes (S100A4, -A6, -A10, -A11, -A13 and -A16) were upregulated more than fivefold after WI-38 fibroblast treatment with 4175-LuT exosomes compared with 4175-LuT ITGβ4KD exosomes (Fig. 3f). Notably, S100 genes remained unchanged in HBEpCs treated with 4175-LuT exosomes (Fig. 3f). Moreover, exosome-treated lung fibroblasts proliferated and migrated more than controls (Extended Data Fig. 7c, d), which correlated with a higher frequency of S100A4+ cells in the lungs after three weeks of education with 4175-LuT, but not 4175β4KD, exosomes (Extended Data Fig. 7e). We then surveyed, by in-cell western blot analysis, ITGβ4 signalling proteins30–34 in WI-38 fibroblasts treated with 4175-LuT or 4175β4KD exosomes. Notably, only Src or phosphorylated Src (pSrc) levels increased in an exosomal ITGβ4-dependent manner (Extended Data Fig. 7f), consistent with the known roles of ITGα6β4 in Src activation and S100A4 expression30. Therefore, in addition to their adhesive properties, exosomal integrins can activate Src and upregulate pro-migratory and pro-inflammatory S100 molecules in specific resident cells within distant tissue microenvironments, influencing the expression of genes implicated in facilitating tumour metastasis.
Exosomal ITGs as organotropism biomarkers
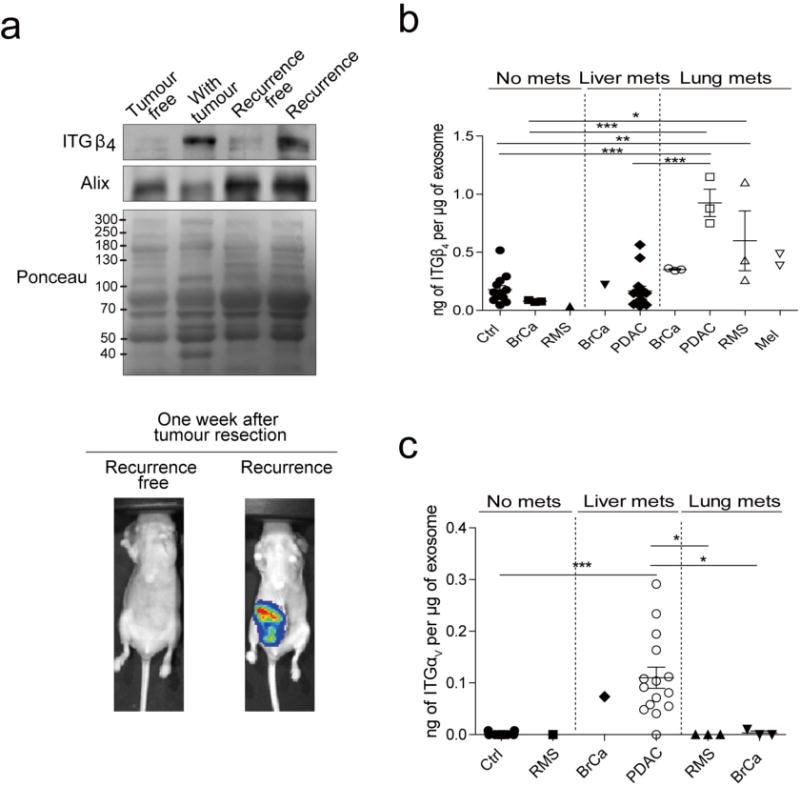
Next we investigated whether exosomal integrin content could predict tumour progression. ITGβ4 levels were increased in the plasma of mice six weeks after orthotopic 4175-LuT cell injection into the mammary fat pad, but were significantly reduced after successful tumour resection (Extended Data Fig. 8a). Furthermore, we performed ELISA assays for plasma-derived exosomal integrins in patients with lung (ITGβ4) or liver (ITGαV, the binding partner of ITGβ5) metastasis. We found increased ITGβ4 levels in exosomes from patients with lung metastasis (regardless of tumour type) compared with patients with no metastasis or liver metastasis (Extended Data Fig. 8b). Exosomes isolated before metastasis from patients with breast cancer that progressed to develop lung metastasis (POD) expressed the highest levels of exosomal ITGβ4 (Fig. 4a). ITGαv was significantly increased in exosomes isolated from cancer patients with liver metastasis compared with patients with no metastasis or lung metastasis (Extended Data Fig. 8c). Finally, exosomal ITGαv levels at diagnosis were higher in patients with pancreatic cancer who developed liver metastasis than in those without liver metastasis within three years of diagnosis or in control subjects (Fig. 4b). Taken together, our data indicate that the specific exosomal integrins in breast and pancreatic cancer patient plasma correlate with and predict likely sites of metastasis.
Figure 4. Exosomal integrin expression as a potential predictor of patient organ-specific metastasis.
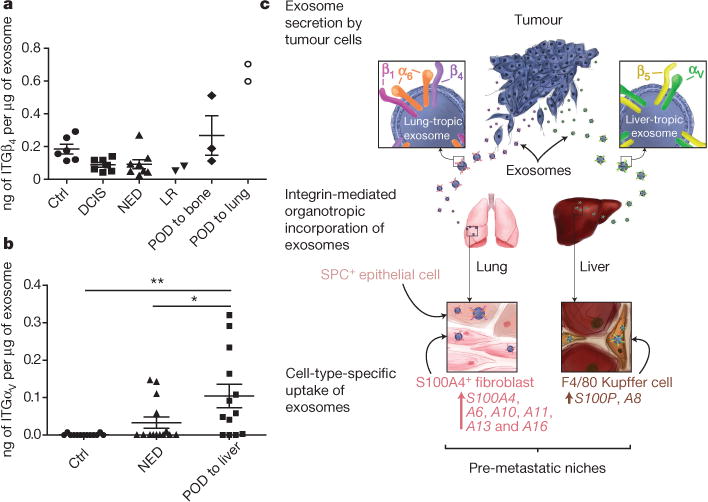
a, Exosomal ITGβ4 levels in breast cancer patients who were metastasis-free at the time of blood draw. Amount of ITGβ4 per microgram of exosome in healthy control (Ctrl) subjects (n = 6); patients with ductal carcinoma in situ (DCIS) (n = 7), invasive breast cancer without relapse within three years (NED, no evidence of disease) (n = 8), locoregional recurrence (LR) within three years (n = 2), bone metastasis within three years (n = 3), or lung metastasis within three years (n = 2). POD, progression of disease. b, Exosomal ITGαv in pancreatic cancer patients who were metastasis-free at the time of blood draw. Amount of ITGαv per microgram of exosome in healthy control subjects (n = 13); patients with pancreatic cancer without relapse within three years (n = 14), or liver metastasis within three years (n = 13). c, Model of exosome-mediated organotropic tumour dissemination. Tumour-derived exosomes are uptaken by organ-specific resident cells in future metastatic organs based on integrin expression. Data are mean ± s.e.m. * P < 0.05; ** P < 0.01 by one-way ANOVA.
Discussion
Since Stephen Paget’s hypothesis first emerged, many studies have focused on identifying cell-intrinsic determinants of organ-specific metastasis3,7,23,24,35. We now show that tumour-derived exosomes prepare a favourable microenvironment at future metastatic sites and mediate non-random patterns of metastasis. We identify determinants of exosome-mediated organ-specific conditioning that allow the redirection of metastasis. Previously, adhesion and ECM molecules, such as integrins, tenascin and periostin, were shown to promote metastasis of disseminating cancer cells36–39. We define a specific repertoire of integrins expressed on tumour-derived exosomes, distinct from tumour cells, which dictates exosome adhesion to specific cell types and ECM molecules in particular organs. Notably, exosomes expressing ITGαvβ5 specifically bind to Kupffer cells, mediating liver tropism, whereas exosomal ITGα6β4 and ITGα6β1 bind lung-resident fibroblasts and epithelial cells, governing lung tropism (Fig. 4c).
Interestingly, bone-tropic exosomes expressed a limited integrin repertoire, but were capable of inducing vascular leakiness in the lung despite lack of uptake in the lung parenchyma. These results suggest that whereas induction of vascular leakiness may be the first exosome-mediated step during the metastatic cascade, it is insufficient to promote metastasis. Thus, integrin-independent mechanisms may mediate vascular leakiness and exosome involvement in bone metastasis.
Cell-type-specific exosome integrin uptake promoted pro-migratory and pro-inflammatory S100 gene upregulation (S100A4, -A6, -A10, -A11, -A13 and -A16 in lung fibroblasts; S100P and -A8 in Kupffer cells). Notably, tumour exosomes failed to elicit S100 upregulation in lung epithelial cells, highlighting the cell-type specificity of exosomal education. Since S100A4 regulates lung metastasis40 and is controlled by ITGα6β4 (ref. 41), we conclude that exosomal ITGα6β4 activates the Src–S100A4 axis in lung fibroblasts during pre-metastatic niche formation. Therefore, we propose that exosomal integrins not only promote adhesion, but also trigger signalling pathways and inflammatory responses in target cells resulting in the education of that organ and rendering it permissive for the growth of metastatic cells.
We provide the proof-of-principle that integrin-blocking decoy peptides successfully ablate tumour exosome adhesion in an integrin-specific and organ-specific manner. Thus, it is no longer surprising that targeting ITGαv in breast cancer cells prevented metastasis to other organs but not to the lung42–44. However, strategies targeting exosomal integrins may effectively block organ-specific metastasis. Collectively, our data suggest that exosomal integrins and exosome-inducible S100 molecules in target cells represent candidates for anti-metastatic combination therapies.
Overall, our findings suggest that circulating tumour-derived exosomes may be useful not only to predict metastatic propensity7, but also to determine organ sites of future metastasis. We believe exosomes perform distinct roles during each of the sequential steps (that is, vascular leakiness, stromal cell education at organotropic sites, bone-marrow-derived cell education and recruitment) necessary to complete pre-metastatic niche evolution11,20,45.
Future studies will focus on identifying exosomal integrins and proteins that could dictate metastasis to other organs, as well as further exploring the potential of exosomal ITGα2β1 as a marker and driver of all cancer metastasis. Our findings demonstrate an important role for exosomes in dictating organ-specific metastasis, thus providing a basis for deciphering the mystery of organotropism.
METHODS
Cell lines and cell culture
The cell lines used in this study were provided as follows: human breast cancer MDA-MB-231 organotropic lines 4175, 1833 and 831 by J. Massagué; human breast cancer 4173 and 4180 cells by A. Minn; human breast cancer 231BR cells by P. Steeg; liver metastasis enriched uveal melanoma cells by V. Rajasekhar; human osteosarcoma 143B cells by A. Narendran; human melanoma 131/4-5B2 and 131/8-2L cells by R. Kerbel; human melanoma SB1B cells by C. F. Verschraegen; human rhabdomyosarcoma CT10 and RD cells by from R. Gladdy; and human Wilms tumour CCG9911 and CLS1 cells by A. Ketsis. Human breast cancer cell lines MDA-MB-231 and MDA-MB-468, human breast epithelial cells MCF10A, human pancreatic cancer cell lines, gastric cancer cell lines and colorectal cancer cell lines were purchased from American Type Culture Collection (ATCC). Although HT29 is commonly misidentified, we purchased this cell line directly from ATCC and the cell line was certified by this repository, therefore we are confident that it is indeed a colon cancer cell line. The C57BL/6 mouse pancreatic adenocarcinoma Pan02 was purchased from the National Cancer Institute Tumour Repository (DTP/DCTD, Frederick National Laboratory for Cancer Research). For in vitro education of human lung fibroblasts WI-38 (ATCC), human bronchial epithelial cells HBEpC (PromoCell), and human Kupffer cells (Life Technologies), cells were maintained in culture for 14 days, with media containing 0, 5 or 10 μg ml−1 of exosomes, replenished every other day. Kupffer cells were cultured in RPMI and WI-38 cells were cultured in alpha-MEM, both supplemented with 10% exosome-depleted FBS (Gibco, Thermo Fisher Scientific) and penicillin-streptomycin. HBEpC cells were cultured in airway epithelial cell growth medium (PromoCell). All cells were maintained in a humidified incubator with 5% CO2 at 37°C. FBS was depleted of bovine exosomes by ultracentrifugation at 100,000g for 70 min. All cells lines were routinely tested for mycoplasma and were found to be negative.
Exosome purification, characterization and analyses
Exosomes were purified by sequential centrifugation as previously described46. In brief, cells were removed from 3–4-day cell culture supernatant by centrifugation at 500g for 10 min to remove any cell contamination. To remove any possible apoptotic bodies and large cell debris, the supernatants were then spun at 12,000g for 20 min. Finally, exosomes were collected by spinning at 100,000g for 70 min. Exosomes were washed in 20 ml PBS and pelleted again by ultracentrifugation (Beckman 70Ti rotor). Exosome preparations were verified by electron microscopy. Exosome size and particle number were analysed using the LM10 or DS500 nanoparticle characterization system (NanoSight, Malvern Instruments) equipped with a blue laser (405 nm). Normal mammary fat pad tissue-derived exosomes were obtained by culturing five mammary fat pads isolated from healthy 4–6-week-old C57BL/6 mice in 3 ml of FBS-free RPMI for 12 h. The final exosome pellet was resuspended in PBS and protein concentration was measured by BCA (Pierce, Thermo Fisher Scientific).
Proteomics analysis
Mass spectrometry analyses of exosomes were performed at the Rockefeller University Proteomics Resource Center using 20 μg of exosomal protein. Samples were denatured using 8 M urea, reduced using 10 mM dithiothreitol (DTT), and alkylated using 100 mM iodoacetamide. This was followed by proteolytic digestion with endoproteinase LysC (Wako Chemicals) overnight at room temperature, and subsequent digestion with trypsin (Promega) for 5 h at 37°C. The digestion was quenched with formic acid and resulting peptide mixtures were desalted using in-house made C18 Empore (3M) StAGE tips47. Samples were dried and solubilized in the sample loading buffer containing 2% acetonitrile and 2% formic acid. Approximately 3–5 μg of each sample was analysed by reversed phase nano-liquid chromatography–tandem mass spectrometry (LC–MS/MS) (Ultimate 3000 coupled to QExactive, Thermo Scientific). After loading onto the C18 trap column (5 μm beads, Thermo Scientific) at a flow rate of 3 μl min−1, peptides were separated using a 75-μm inner diameter C18 column (3 μm beads, Nikkyo Technos Co.) at a flow rate of 200 nl min−1, with a gradient increasing from 5% buffer B (0.1% formic acid in acetonitrile)/95% buffer A (0.1% formic acid) to 40% buffer B/60% buffer A, over 140 min. All LC–MS/MS experiments were performed in data-dependent mode. Precursor mass spectra were recorded in a 300–1,400 m/z mass range at 70,000 resolution, and 17,500 resolution for fragment ions (lowest mass: m/z 100). Data were recorded in profile mode. Up to 20 precursors per cycle were selected for fragmentation and dynamic exclusion was set to 45 s. Normalized collision energy was set to 27.
Semi-quantitative data analysis
MS/MS spectra were extracted and searched against Uniprot complete human or mouse proteome databases (January 2013) concatenated with common contaminants48 using Proteome Discoverer 1.4 (Thermo Scientific) and Mascot 2.4 (Matrix Science). All cysteines were considered alkylated with acetamide. N-terminal glutamate to pyroglutamate conversion, oxidation of methionine, and protein N-terminal acetylation were allowed as variable modifications. Data were first searched using fully tryptic constraints. Matched peptides were filtered using a Percolator49 based 1% false discovery rate (FDR). Spectra not being matched at a FDR of 1% or better were re-searched allowing for semi-tryptic peptides. The average area of the three most abundant peptides for a matched protein50 was used to gauge protein amounts within and between samples.
Label-free quantitative mass spectrometry
LC–MS/MS data from three technical replicates of six organ-tropic samples were analysed using MaxQuant (version 1.5.0.30) and Perseus software (version 1.5.0.9)51, searching against a Uniprot human database (July 2014). Oxidation of methionine and protein N-terminal acetylation were allowed as variable modifications, and cysteine carbamidomethyl was set as a fixed modification. Two missed cleavages were allowed for specificity: trypsin/P. The ‘match between runs’ option was enabled. FDR values at the protein and peptide level were set to 1%. Protein abundance is expressed as LFQ values. Only proteins quantified in at least two out of three replicates in at least one group were retained, and missing values were imputed. A multiple sample ANOVA test was performed and corrected for multiple hypotheses testing using a permutation-based FDR threshold of 0.05.
Exosome treatment and labelling
To assess lung, liver and bone exosome distribution, exosomes were injected via the retro-orbital venous sinus, the tail vein or intracardially Exosome distribution patterns were consistent regardless of the route of injection. For brain distribution, exosomes were only observed in the brain after intracardiac injection. For 24-h exosome treatments, 10 μg of total exosomal protein were injected via the retro-orbital venous sinus, the tail vein, or intracardially in a total volume of 100 μl PBS. For exosome-tracking purposes, purified exosomes were fluorescently labelled using PKH67 (green) or PKH26 (red) membrane dye (Sigma-Aldrich) or FM1-43FX dye (Life Technologies) for the photo-conversion experiment. Labelled exosomes were washed in 20 ml of PBS, collected by ultracentrifugation and resuspended in PBS. When performing peptide blocking experiments, exosomes were incubated with 0.06 μM RGD or HYD-1 (peptide sequence: KIKMVISWKG) peptides for 30 min at 37°C before exosome injection. An average of five random fields was counted per sample at 20 × magnification, and representative pictures were taken at 40 × magnification. For education experiments, mice received 10 μg of exosomes retro-orbitally every other day for 3 weeks. To measure exosome uptake by specific cell types, labelled exosomes were injected 24 h before tissue collection and tissues were analysed for exosome-positive cells by immunofluorescence. Pictures were taken at 60 × magnification. For in vitro uptake assays, the membrane of WI-38 cells was labelled with PKH67 dye while 4175-LuT exosomes were labelled with PKH26 dye. Exosomes (10 μg ml−1) were first incubated with PBS or HYD-1 peptide for 30 min at 37°C, followed by an incubation for 1 h with WI-38 cells at 37°C. Excess exosomes were washed off and pictures were taken by Nikon confocal microscope (Eclipse TE2000U). The amount of exosomes localizing to the lung was analysed by immuno fluorescence or using the Odyssey imaging system (LI-COR Biosciences). In brief, NIR dye-labelled exosomes were injected 24 h before tissue collection and tissues were analysed for exosome-positive areas. Whole-lung images were analysed using image J software, quantifying red fluorescence area in arbitrary units.
Photoconversion and electron microscopy processing
Cryostat sections prepared at a 15-μm thickness were placed on glass slides and re-fixed in 0.075 M sodium cacodylate, pH 7.4, containing 2.5% glutaraldehyde. For photoconversion, slides were washed twice in 0.1 M sodium cacodylate buffer, pH 7.4. Autofluorescence was quenched using 100 mM NH4Cl in cacodylate buffer for 45 min. On the basis of optimization experiments, sections were photoconverted for 2 h by incubation in 5.4 mg ml−1 3,3′-diaminobenzidine in 0.1 M sodium cacodylate buffer, pH 7.4, and exposure to the light of an Intensilight C-HGFI 130-W mercury lamp and a 4×/0.1 NA objective (Nikon Inverted Microscope Eclipse Ti).
For electron microscopy processing, sections were post-fixed in 1% osmium tetroxide buffer for 15 min on ice. After washing with water, slides were placed in 1% aqueous uranyl acetate for 30 min. Sections were washed with water, dehydrated in a graded series of ethanol concentrations and subsequently in acetone for 10 min at room temperature. Samples were embedded in Eponate. Serial sections were cut at 70 nm in thickness and transferred to formvar-coated slot grids and imaged on a JEOL 100CX at 80 kV with an AMT XR41 digital imaging system.
Gene expression analysis
Cell lines were analysed for specific genes using pre-designed TaqMan assays (Applied Biosystems). In brief, RNA was extracted from tissues or cells using the RNeasy kit (Qiagen), and reverse transcribed using Superscript Vilo (Life Technologies). qRT–PCR was performed on a 7500 Fast Real Time PCR System (Applied Biosystems), using TaqMan Universal PCR Master Mix (Applied Biosystems). Relative expression was normalized to β-actin levels.
Knockdown and overexpression cell preparation
For shRNA-mediated knockdown of ITGβ4 and ITGβ5, specific interfering lentiviral vectors containing GFP reporter and puromycin resistance gene cassettes were used. In brief, oligonucleotide 5′-CCGGGAGGGTGTCATCACCATTGAACTCGAGTTCAATGGTGATGACACCCTCTTTTTG-3′ targeting the 5′-GAGGGTGTCATCACCATTGAA-3′ sequence in the human ITGB4 gene (EntrezGene ID: 3691) or oligonucleotide 5′-CCGGAGCTTGTTGTCCCAATGAAATCTCGAGATTTCATTGGGACAACAAGCTTTTTTG-3′ targeting the 5′-AGCTTGTTGTCCCAATGAAAT-3′ sequence in the human ITGB5 gene (EntrezGene ID: 3693) were cloned into the pLKO.1 vector. As a control, we used the empty pLKO.1 vector. For retrovirus production for integrin overexpression, the pWZL and pBabe vectors systems were used. pWZL-hygro-ITGB4 and pBabe-puro-ITGB4 were provided by F. Giancotti. Lentiviral and retroviral particles were packaged using 293T cells. Infected target cells were selected using 500 μg ml−1 hygromycin B or 2 μg ml−1 puromycin (Invitrogen).
Flow cytometry analysis
Bone marrow was prepared for flow cytometry as previously described1. For analysis of lung, tissues were minced and then digested at 37°C for 20 min with an enzyme cocktail (collagenase A, dispase and DNaseI, Roche Applied Science). Single-cell suspensions were prepared by filtering through a 70-μm strainer and passing through an 18G syringe. Lung fibroblasts were identified by flow cytometry using an anti-mouse rabbit polyclonal S100A4 (1:50, Abcam; ab27957), or SPC (1:100, Santa Cruz; FL-197), revealed by Alexa Fluor 568-conjugated goat anti-rabbit secondary (A-11011, Life Technologies, 1:400). For liver, tissues were mechanically dissociated, and single-cell suspensions were filtered through a 40-μm strainer. Allophycocyanin-conjugated F4/80 (1:100, eBioscience; clone BM8) was used to identify liver macrophages by flow cytometry. Cell fluorescence indicating fluorescently labelled exosome uptake was analysed using a FACSCalibur or a FACSCanto (Beckton Dickinson). FACS data was analysed with FlowJo software (TreeStar Inc.).
Migration assay
Twenty-thousand cells were plated in 24-well transwell plates with inserts (8-μm pore size, Corning) and were incubated at 37°C for 6 h. Cell inserts were fixed with 4% paraformaldehyde (PFA) for 10 min, followed by PBS wash and haematoxylin staining to allow visualization and counting. Nine random fields were counted per well at 20× magnification and the average number of migrated cells per field was calculated.
Human studies
Human peripheral blood samples were obtained from control healthy subjects and cancer patients with lung or liver metastasis, or from patients without distant metastasis at Weill Cornell Medical College, University Medical Center Hamburg-Eppendorf, Oslo University Hospital, Memorial Sloan Kettering Cancer Center and University of Nebraska Medical Center, all pathologically confirmed. All individuals provided informed consent for blood donation on approved institutional protocols (WCMC IRB 0604008488 (DL), MSKCC IRB 12-137A (JB)). Plasma or serum exosomes were isolated as previously described1. ITGβ4 and ITGαv levels in exosomes were measured by ELISA (ABIN417641 and ABIN417609 from Antibodies Online, and LS-F7188 from LifeSpan Biosciences), using 2 μg of exosomes per 100 μl of sample diluent, in duplicate reactions, according to the manufacturer’s instructions.
Mouse studies
All mouse work was performed in accordance with institutional, IACUC and AAALAS guidelines, by the animal protocol 0709-666A. All animals were monitored for abnormal tissue growth or ill effects according to AAALAS guidelines and euthanized if excessive deterioration of animal health was observed. No statistical method was used to pre-determine sample size. No method of randomization was used to allocate animals to experimental groups. The investigators were not blinded to allocation during experiments and outcome assessment. Mice that died before the predetermined end of the experiment were excluded from the analysis. In none of the experiments did tumours exceed the maximum volume allowed according to our IACUC protocol, specifically 2 cm3. For exosome localization, education and tumour implantation experiments for mouse cell lines, 6-week-old C57BL/6 Mus musculus females purchased from Jackson labs were used. For exosome localization, education and tumour implantation experiments for human cell lines, 6–8-week-old NCr nude (NCRNU-F sp./sp.) females purchased from Taconic were used. For lung metastasis studies using organotropic lines, 6–8-week-old nude female mice were pre-educated with exosomes for 3 weeks followed by tail vein injection of 2 × 105 or intracardiac injection of 1 × 105 luciferase-positive cancer cells resuspended in 100 μl PBS. Four weeks after intracardiac injection and eight weeks after tail vein injection, lung metastasis was measured using the IVIS 200 bioluminescence imaging system (Xenogen, Caliper Life Sciences), and tissues were cut in 6-μm sections and stained with haematoxylin and eosin for histology. To analyse the role of exosome education in tumour metastasis, 6–8-week-old C57BL/6 female mice pre-educated with pancreatic cancer-derived exosomes were injected intrasplenically with 1 × 106 Pan02 mCherry cells resuspende d in 30 μl of Matrigel (Corning). One or twenty-one days later, mice were euthanized, and livers were analysed for metastatic lesions by measuring liver weight.
To follow the levels of tumour-derived exosomes in plasma of tumour-bearing mice, 1 × 106 4175 lung-tropic cells were injected in the mammary fat pad of nude mice. Mouse blood (250 μl) was drawn from the retro-orbital sinus when tumour size was over 800 mm3, followed by tumour resection. One week after the tumour was resected, mice were analysed by bioluminescence IVIS imaging for luciferase activity and separated into two groups: recurrence/tumour-free and recurrent tumours. Mouse blood was drawn and the plasma of mice within the same group was pooled for exosome isolation. Western blot analysis with anti-human ITGβ4 antibodies was used to detect tumour-derived exosomes.
To assess exosome-induced vascular leakiness, 10 μg of total exosome protein were injected by retro-orbital injection. Then 20 h after exosome treatment, mice were injected with 2 mg of Texas Red-lysine fixable dextran 70,000 MW (Invitrogen) via retro-orbital injection. One hour after dextran injection, mice were euthanized and perfused with PBS. Lungs were dissected and fixed in a mix of 2% PFA and 20% sucrose overnight, then embedded in Tissue-tek O.C.T. embedding compound (Electron Microscopy Sciences) and frozen in a dry-ice/ethanol bath. O.C.T. blocks were sectioned and stained for DAPI, pictures were taken using a Nikon confocal microscope (Eclipse TE2000U). Images were analysed using image J software, quantifying red fluorescence area in arbitrary units.
Tissue processing and immunofluorescence
For histological analysis, tissues were dissected and fixed in a mix of 2% PFA and 20% sucrose in PBS overnight, then embedded in Tissue-tek O.C.T. embedding compound. Blocks were frozen in a dry-ice/ethanol bath. For immunofluorescence, 6 μm O.C.T tissue cryosections were stained with antibodies against F4/80 (1:100, eBioscience; BM8), fibronectin (1:50, Santa Cruz; IST-9), S100A4 (1:100, Abcam; ab27957), SPC (1:100, Santa Cruz; FL-197), laminin (1:50, abcam; ab11575), CD31 (1:100, Santa Cruz; MEC 13.3), EpCAM (1:50, Santa Cruz; HEA125). Secondary antibodies conjugated to Alexa Fluor 488 or 549 were used (A-11001 and A-11007, Life Technologies). Fluorescent images were obtained using a Nikon confocal microscope (Eclipse TE2000U) and analysed using Nikon software (EZ-C1 3.6).
Western blot analysis
Exosomes or cells were lysed with RIPA buffer containing a complete protease inhibitor tablet (Roche). Lysates were cleared by centrifugation at 14,000g for 20 min. Supernatant fractions were used for western blot. Samples were separated on a Novex 4–12% Bis-Tris Plus Gel (Life Technologies), and transferred onto a PVDF membrane (Millipore). Membranes were processed for Ponceau red staining followed by 1 h blocking and primary antibody incubation. The antibodies against the following proteins were used for western blot analysis: ITGβ 1 (1:1,000, Cell Signaling; 4706), ITGβ4 (1:500, Cell Signaling; 4707), ITGα6 (1:1,000, Cell Signaling; 3750), ITGα2 (1:10,000, abcam; ab133557), ITGα3 (1:1,000, abcam; ab190731), ITGαv (1:500, abcam; ab117611), ITGβ5 (1:500, Cell Signaling; 4708), ITGβ3 (1:500, Millipore; AB2984) Alix (1:1,000, Cell Signaling; 3A9), and GAPDH (1:10,000, Cell Signaling; 14C10). Anti-rabbit IgG, horseradish peroxidase (HRP)-linked antibody (1:3,000, Cell Signaling; 7074) and anti-mouse IgG, HRP-linked antibody (1:3,000, Cell Signaling; 7076) were used as secondary antibodies.
In situ protein expression analysis (in-cell western assay, LI-COR)
Cells were plated in a 96-well plate and treated with 10 μg ml−1 exosomes for 2 h and then processed according to the protocol provided by the manufacturer. In brief, cells were fixed with 4% PFA and washed with 0.1% TritonX-100/PBS. Cells were then blocked using Odyssey blocking buffer for 1 h and stained overnight at 4°C with primary antibody in Odyssey blocking buffer containing 0.1% Tween-20. The next day cells were washed again and incubated with LI-COR secondary antibodies for 1 h at room temperature followed by fluorescent imaging using Odyssey. Antibodies against the following proteins were used: Src (1:100, Cell Signaling; 2109), p-Src (1:100, Cell Signaling; 2101), AKT (1:100, Cell Signaling; 9272), p-AKT (1:100, Cell Signaling; 9271), p38 (1:100, Cell Signaling; 9212), p-p38 (1:100, Cell Signaling; 9211), NF-κB (1:100, Cell Signaling; 3034), p-NF-κB (1:100, Cell Signaling; 3033), NFAT (1:100, Thermo Scientific; PA1-023), ILK (1:100, abcam; ab52480), FAK (1:100, abcam; ab40794) and GAPDH (1:100, Cell Signaling; 14C10). IRDye 800CW anti-rabbit IgG (1:800, LI-COR) were used as secondary antibodies.
Statistical analysis
Error bars in graphical data represent mean ± s.e.m. Mouse experiments were performed in duplicate or triplicate, using 3–6 mice per treatment group. Statistical significance was determined using a two-tailed Student’s t-test and one-way ANOVA, in which P values of P < 0.05 were considered statistically significant. Variance was similar between the groups that were statistically compared.
Extended Data
Extended Data Figure 1. Characterization of organotropic exosome properties and biodistribution.
a, Human cancer exosome biodistribution in lung and liver. Exosomes (10 μg) derived from each cell line were labelled with lipophilic PKH26 dye (red) and injected retro-orbitally into nude mice 24 h before culling. Top, representative NIR whole-lung image by Odyssey imaging (n = 3). Middle and bottom, represent exosome biodistribution in the lung and liver as determined by immunofluorescence microscopy. Arrows indicate exosome foci (n = 3, three independent experiments). b, Biodistribution of exosomes isolated from mouse cell lines E0771 and Pan02. Mouse exosome biodistribution in the lung and liver was determined by immunofluorescence microscopy. Exosomes (10 μg) derived from each cell line were labelled with lipophilic PKH26 dye (red) and injected retro-orbitally into nude mice 24 h before culling. Top, lung at 40 × magnification. Bottom, liver at 40 × magnification. Arrows indicate exosome foci. Graph represents the quantification of exosome distribution by counting exosome-positive cells. An average of five random fields per sample were counted at 20 × magnification (three independent experiments, each with n = 3). ** P < 0.01 by two-tailed Student’s t-test. c, Analysis of organotropic cell-derived exosomes. MDA-MB-231 organotropic cell-line-derived exosomes were analysed for size distribution by NanoSight and phenotype (purity and shape) by electron microscopy; black arrows indicate representative exosomes. Technical triplicates were analysed, at least 10 images per sample. d, Flow cytometric analysis of exosome+ cells in lung. Exosomes (10 μg) derived from MDA-MB-231 organotropic cell lines were labelled with lipophilic PKH67 dye (green) and injected retro-orbitally into nude mice 24 h before culling. FITC-channel-positive cells were acquired on a FACS Calibur, and the percentage of exosome-positive cells was quantified (representing data pooled from two independent experiments, a total of n = 12). ***P < 0.001 by one-way ANOVA. e, Flow cytometric analysis of exosome-positive cells in the bone marrow. Exosomes (10 μm) derived from MDA-MB-231 organotropic cell lines were labelled with lipophilic PKH67 dye (green) and injected retro-orbitally into nude mice 24 h before culling. FITC-channel-positive cells were acquired on a FACS Calibur, and the percentage of exosome-positive cells was quantified (representative data pooled from two independent experiments, a total of n = 6). ** P < 0.01 and *P < 0.05 by one-way ANOVA for the 831-BrT to 1833-BoT and 4175-LuT comparisons, respectively. Data are mean ± s.e.m. Scale bars, 5 mm (a, top), 50 μm (a, middle and bottom, b) and 100 nm (c).
Extended Data Figure 2. 4175-LuT cell-derived exosomes localize to lung and dictate future metastatic sites.
a, Electron microscopy imaging of FM1-43-labelled 4175-LuT exosomes. Red arrows, FM1-43-positive exogenous exosomes; black arrows, endogenous exosomes. Two mice were tested, images were taken for several sections from each organ (n = 30 images in total). b, Representative NIR imaging of lung whole mount after daily exosome injections. Exosomes (10 μg) derived from 4175-LuT cells were injected daily for three consecutive days via the retro-orbital sinus and the whole lung was imaged by Odyssey imaging (n = 4). c, Representative haematoxylin/eosin staining of the lung from Fig. 1f at 20 × magnification; n = 5 for all, except for LuT exo/LuT cells, in which n = 4; data representative of two independent experiments. Arrows indicate lung metastasis. d, Analysis of 1833-BoT cell metastasis to the lung, after 3 weeks of continuous treatment with PBS or 4175-LuT exosomes, followed by intracardiac injection of 1 × 105 tumour cells. Mice were injected retro-orbitally with exosomes every other day for 3 weeks, before tumour cell injection. Quantitative bioluminescence imaging of luciferase activity by IVIS imaging. Metastasis was quantified 3 weeks after tumour cell injection (n = 4). Scale bars, 100 nm (a), 5 mm (b, d) and 500 μm (c). Data are mean ± s.e.m. *** P < 0.001 by two-tailed Student’s t-test.
Extended Data Figure 3. Characterization of organotropic exosome protein cargo.
a, Top 40 adhesion molecules packaged in exosomes isolated from organ tropic cell lines. Heat map of adhesion molecule signals based on Z-scored LFQ values obtained from quantitative mass spectrometry analysis. PEP (posterior error probability), MS/MS count is a number of fragmentation spectra (spectral counting), Razor + unique peptides refers to the number of peptides, and sequence coverage refers to percentage of peptide counts identified. b, Ponceau staining of exosome lysates isolated from organ tropic cell lines. Representative Ponceau staining of total protein from the organ tropic cell-line-derived exosomes. Exosomal protein (10 μg) was loaded in each well (n = 2, three independent experiments). c, Western blot analysis comparison of ITGα6 and ITGβ4 levels in cell lysates versus exosomes derived from organotropic breast cancer and pancreatic cancer cell lines. Graph represents the relative ratios of integrin to GAPDH signals as determined by densitometry. For western blot source data, see Supplementary Fig. 1i–k.
Extended Data Figure 4. Functional characterization of organotropic exosomes.
a, Quantification of organotropic exosome uptake by target cells in vivo. Top graph, flow cytometric quantification of the frequency of 4175-LuT exosome-positive fibroblasts and epithelial cells (n = 4). Left bottom graph, flow cytometric quantification of the frequency of BxPC-3 exosome-positive macrophages (n = 3). Right bottom graph, quantification of the frequency of 831-BrT exosome-positive endothelial cells by immunofluorescence microscopy (n = 5). b, Organotropic cell-line-derived exosomes induce vascular leakiness in the lung. Leakiness in the lung 24 h after retro-orbital injection of 10 μg of normal mammary fat pad or MDA-MB-231 organotropic cell-line-derived exosomes was quantified by imaging the presence of fluorescent dextran (red) outside of blood vessels, in the lung parenchyma. Left top panel, 40 × magnification of representative lung image after PBS injection. Left bottom panel, representative lung image after 4175-LuT exosome injection. Scale bar, 50 μm. Right graph depicts the quantification of five random areas at 20 × magnification in arbitrary units (data representative of two independent experiments; n = 3). Data are mean ± s.e.m. * P < 0.05; ** P < 0.01 by one-way ANOVA.
Extended Data Figure 5. Exosome co-localization with specific cell types within target tissues.
a, Immunofluorescence analysis of resident cells in lung, liver and brain after labelled exosome injection. Analysis of exosome (red) co-staining with markers (green) for tissue-specific stromal cell types. Top, representative images of immunofluorescence microscopy of 4175-LuT exosome co-staining with F4/80, CD31 and EpCAM. Middle, liver sections from mice injected with BxPC-3-LiT-derived exosomes were co-stained with CD31, S100A4 and EpCAM. Bottom, brain sections from mice injected with 831-BrT exosome were co-stained with F4/80, S100A4 and EpCAM (n = 3 per experiment for two independent experiments). b, Exosome biodistribution and co-localization with extracellular matrix proteins. Left top, representative immunofluorescence microscopy images of lung tissue, depicting 4175-LuT exosome (red) co-staining with laminin (green). Right top, laminin (green) co-staining with S100A4 (red). Left bottom, representative immunofluorescence microscopy of liver tissue co-stained for fibronectin (green) and BxPC-3-LiT exosomes (red). Right bottom, fibronectin (green) co-staining with F4/80 (red) (n = 3, two independent experiments). Scale bars, 30 μm.
Extended Data Figure 6. ITGs functionally regulate organotropic exosome uptake and exosome-mediated metastasis.
a, Representative western blot analysis of integrin expression in 4175-LuT and 4175β4KD cells and exosomes (representative of three independent experiments). For western blot source data, see Supplementary Fig. 1l. b, In vitro uptake of 4175-LuT exosomes by WI-38 lung fibroblasts. The WI-38 cell membrane was labelled with PKH67 green dye and 4175-LuT exosomes were labelled with PKH26 red dye. Exosomes (10 μg ml−1) were first incubated with PBS or HYD-1 peptide for 30 min at 37°C, followed by 1-h incubation with WI-38 cel ls at 37°C. Excess exosomes were washed and cells were imaged (n = 4 for two independent experiments). c, Representative western blot of ITGβ4 expression in exosomes isolated from wild-type or ITGβ4-overexpressing 1833-BoT cells (representative of two independent experiments). For western blot source data, see Supplementary Fig. 1m. d, Representative haematoxylin/eosin staining of lungs from Fig. 3e. Arrows indicate lung metastasis; n = 6, data representative of two independent experiments. e, Representative western blot analysis of integrin expression in BxPC-3-LiT and BxPC-3β5KD cells and exosomes. For western blot source data, see Supplementary Fig. 1n. f, Immunofluorescence analysis of BxPC-3-LiT control and BxPC-3β5KD-derived exosome biodistribution in the liver. Exosomes (10 μg) isolated from each cell line were labelled with lipophilic PKH26 dye (red) and injected retro-orbitally into nude mice 24 h before culling. Left, 40 × magnification. Arrows indicate exosome foci. Scale bar, 50 μm. Right, quantification of exosome distribution by exosome-positive cells. An average of five random fields were counted at 20 × magnification (data representative of two independent experiments; n = 3). g, Flow cytometry analysis of exosome-positive cells in the liver 24 h after exosome injection. Labelled BxPC-3-LiT exosomes (5 μg) per mouse were incubated with PBS, RGD, HYD-1 or ITGαvβ5 antibody for 30 min at 37°C before retro-orbital injection into nude mice. Livers were collected and analysed for exosome-positive cells by flow cytometry 24 h after injection (n = 4, except for the ITGαvβ5 antibody group, in which n = 5). Scale bars, 10 μm (b), 500 μm (d) and 500 μm (f). ** P < 0.01, *** P < 0.001 by two-tailed Students t-test (f) and one-way ANOVA (g). Data are mean ± s.e.m.
Extended Data Figure 7. Functional contribution of exosomes to metastasis.
a, Microscopic analysis of exosome-positive cells in the livers of mice injected with liver metastatic Pan02-LiT-derived exosomes. Before injection, Pan02-LiT exosomes were pre-incubated with RGD peptide for 30 min at 37°C. Pan02-LiT exosomes (10 μg) were labelled with lipophilic PKH67 green dye and injected retro-orbitally into C57BL/6 mice 24 h before culling. Livers were digested and exosome-positive cells were quantified by flow cytometr y (n = 3). b, Analysis of Pan02-LiT liver metastasis after 3 weeks of continuous treatment with PBS, Pan02-LiT-derived exosomes, or Pan02-LiT-derived exosomes pre-incubated with RGD peptide for 30 min at 37°C. Pan02-LiT cells were injected intrasplenically. Mice were injected retro-orbitally with 5 μg exosome every other day for 3 weeks. Top, representative liver images showing metastasis taken at culling. Bottom, liver weight quantification (n = 4 except for the control and peptide group for which n = 3 of one experiment). c, Functional analysis of lung fibroblasts educated with 4175-LuT-derived exosomes. Proliferation of lung fibroblasts educated with exosomes every other day for 2 weeks. Three days after cells were plated at equal density, cell numbers were counted using a haemocytometer (n = 3; three independent experiments). d, Migration of lung fibroblasts educated with exosomes every other day for 2 weeks was measured as follows. Fibroblasts were plated in 24-well transwell chamber inserts, and after 6 h the number of cells that migrated was counted using haematoxylin staining. Nine random fields were counted at 20 × magnification and the average number of cells per field was calculated (total of n = 4 from two independent experiments). e, Representative image of the lung stained for S100A4. Mice were treated every other day with PBS, 4175-LuT or 4175β4KD exosomes for 3 weeks. Scale bar, 50 μm; n = 4 mice. f, In situ (in-cell western) protein expression analysis of WI-38 fibroblasts treated with PBS, 4175-LuT or 4175ITGβ4KD exosomes. Relative expression levels of Src and phosphorylated (p-) Src (n = 3, three independent experiments). Data are mean ± s.e.m. * P < 0.05; ** P < 0.01; *** P < 0.001 by two-tailed Student’s t-test (a, c, d) and one-way ANOVA (b, f).
Extended Data Figure 8. Exosomal integrin expression as a potential metastatic site biomarker.
a, Exosomal ITGβ4 levels in the plasma of mice bearing orthotopic 4175-LuT tumours, as a function of tumour progression. Blood plasma was collected for exosome isolation 6 weeks after intra-mammary fat pad tumour injection, then again 1 week after tumour resection, from mice that were deemed to be either free of tumour or presenting with recurring tumours based on IVIS bioluminescence imaging (n = 5 were pooled for each group, based on one experiment). For western blot source data, see Supplementary Fig. 1o. b, Exosomal ITGβ4 in healthy control subjects (Ctrl) (n = 13); patients with breast cancer (BrCa) and no metastasis (n = 3), liver metastasis (n = 1), or lung metastasis (n = 3); patients with rhabdomyosarcoma (RMS) and no metastasis (n = 1) or lung metastasis (n = 3); patients with pancreatic cancer (PDAC) with liver metastasis (n = 14) and lung metastasis (n = 3); and patients with melanoma (Mel) with lung metastasis (n = 2). c, Exosomal ITGαV in healthy control subjects (n = 13); patients with rhabdomyosarcoma and no metastasis (n = 1) or lung metastasis (n = 3); patients with breast cancer and lung metastasis (n = 3) or liver metastasis (n = 1); and patients with pancreatic cancer and liver metastasis (n = 15). Data are mean ± s.e.m. * P < 0.05; *** P < 0.001 by one-way ANOVA.
Extended Data Table 1.
Integrin expression in human exosomes in multiple organotropic tumour models
| Sites of metastasis | None | Bone | Brain | Lung | Liver | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell type | Lung Fibroblast | Mammary Epithelial | Breast Cancer | Melanoma | Breast Cancer | Osteosarcoma | Rhabdomyosarcoma | Wilms’ Tumor | Melanoma | Uveal melanoma | Colorectal Cancer | Pancreatic Cancer | Gastric Cancer | |||||||||||||
| Cell line | WI-38 | MCF10A | 1833 | 831 | 231BR | 131/4-5B2 | SB1B | 4173 | 4175 | 4180 | 143B | RD | CT10 | CCG 9911 | CLS1 | 131/8-2L | Primary culture | HCT116 | HT29 | SW620 | BXPC-3 | HPAF-II | MiaPaca-2 | PANC-1 | SNU1 | SNU16 |
| ITGα1 | + | + | + | + | + | + | ||||||||||||||||||||
| ITGα2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||
| ITGα2b | + | + | ||||||||||||||||||||||||
| ITGα3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||
| ITGα4 | + | + | + | + | + | + | + | |||||||||||||||||||
| ITGα5 | + | + | + | + | + | + | + | + | + | + | ||||||||||||||||
| ITGα6 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||
| ITGα11 | + | |||||||||||||||||||||||||
| ITGαv | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||
| ITGβ1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| ITGβ3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||
| ITGβ4 | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||||
| ITGβ5 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||
| ITGβ6 | + | + | + | |||||||||||||||||||||||
Proteomic analysis of integrins in exosomes. ‘+’ indicates positive for integrin expression by qualitative mass spectrometry.
Extended Data Table 2.
Integrin expression in human and mouse cell-line-derived exosomes
| Human | Murine | ||||
|---|---|---|---|---|---|
| Sites of metastasis | Majority to lung | Lung and liver | Sites of metastasis | Lung | Liver |
| Cell type | Breast cancer | Cell type | Breast cancer | Pancreatic cancer | |
| Cell line | MDA-MB-231 | MDA-MB-468 | Cell line | E0771 | Pan02 |
| ITGα1 | ITGα1 | ||||
| ITGα2 | + | + | ITGα2 | ||
| ITGα2b | ITGα2b | + | |||
| ITGα3 | + | + | ITGα3 | + | + |
| ITGα4 | ITGα4 | ||||
| ITGα5 | + | ITGα5 | + | + | |
| ITGα6 | + | + | ITGα6 | + | + |
| ITGαv | + | ITGαv | + | + | |
| ITGβ1 | + | + | ITGβ1 | + | + |
| ITGβ3 | + | + | ITGβ3 | + | + |
| ITGβ4 | + | ITGβ4 | + | ||
| ITGβ5 | + | ITGβ5 | + | ||
| ITGβ6 | + | ITGβ6 | |||
Proteomic analysis of integrins in exosomes; ‘+’ indicates positive for integrin expression by qualitative mass spectrometry.
Supplementary Material
Acknowledgments
We thank S. Rudchenko at the Hospital for Special Surgery Flow Cytometry Core Facility. We acknowledge the MSK Cancer Center Support Grant/Core Grant (P30 CA008748). Our work is supported by grants from National Cancer Institute (U01-CA169538, D.L. and M.S.B.), National Institutes of Health (R01-CA169416, D.L. and H.P.), United States Department of Defense (W81XWH-13-10249, D.L.), W81XWH-13-1-0425 (D.L., J.B., B.A.G. and Y.K.), Melanoma Research Alliance (H.P.), Sohn Conference Foundation (H.P. and H.Z.), the Children’s Cancer and Blood Foundation (H.P. and D.L.), The Manning Foundation (D.L.), The Hartwell Foundation (D.L.), Fundação para a Ciência e a Técnologia (D.L.), The Nancy C. and Daniel P. Paduano Foundation (H.P. and D.L.), The Feldstein Foundation (H.P.), The Starr Cancer Consortium (H.P. and D.L.), The Mary Kay Foundation (D.L.), Pediatric Oncology Experimental Therapeutic Investigator Consortium (POETIC, D.L. and H.P.), James Paduano Foundation (D.L. and H.P.), Beth Tortolani Foundation (D.L. and J.B.), Malcolm Hewitt Weiner Foundation (D.L.), Theodore A. Rapp Foundation (D.L.), American Hellenic Educational Progressive Association 5th District Cancer Research Foundation (D.L., A.H.), Charles and Marjorie Holloway Foundation (J.B.), Sussman Family Fund (J.B.), Lerner Foundation (J.B.), Breast Cancer Alliance (J.B.), Manhasset Women’s Coalition Against Breast Cancer (J.B.), Ministry of Science and Technology Taiwan (101-2918-I-002-016, T.-L.S.), The JSPS Postdoctoral Fellowships for Research Abroad and Susan G. Komen Postdoctoral Fellowship (A.H.).
Footnotes
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary information is available in the online version of the paper.
Author Contributions
A.H. designed the experimental approach, performed the experimental work, analysed the data, coordinated the project and wrote the manuscript. B.C.-S. designed experiments investigating liver metastasis and performed the experimental work. T.-L.S. performed ECM studies. G.R. analysed brain tropic exosome distribution. A.H. performed western blot analysis. M.T.M. and H.M. performed and analysed exosome mass spectrometry. S.K. prepared overexpression vectors. S.S. and L.B. performed tissue processing and staining. S.C. designed and illustrated Fig. 4c. A.D.G., S.C., V.D.D.-C., Y.A. and C.W. received and processed human samples. N.S. and K.U. performed electron microscopy. A.E.D. performed animal surgeries and contributed to data interpretation and discussion. T.Z. performed RNA sequence analysis. B.A.G. performed initial proteomic analysis. V.K.R., G.K.S. and J.H.H. provided the uveal melanoma cell line. L.P., T.K., M.S.B., V.M., K.K., L.H.W., J.H., E.H.K., K.M., S.K.B., K.P., O.F., M.J., S.K., M.A.H., P.M.G., K.J.L., J.M.W., A.N. and W.R.J. provided and prepared human samples. H.Z., A.J.M. and P.S. read the manuscript and provided feedback. C.M.G., I.M. and H.P. discussed the hypothesis and contributed to data interpretation and wrote the manuscript. Y.K., M.d.S. and M.J.B. contributed to discussing the hypothesis, interpretation of data. J.B. coordinated the project, interpreted data and wrote the manuscript. D.L. conceived the hypothesis, led the project, interpreted the data and wrote the manuscript.
The raw data for quantitative mass spectrometry analysis of lung-tropic (4173 and 4175), liver-tropic (HPAF-II and HCT116) and brain-tropic (831 and 231BR) exosomes (Fig. 2a and Extended Data Fig. 3a) are available at http://dx.doi.org/10.6084/m9.figshare.1569781. The raw sequencing data for human Kupffer cells treated in vitro with BxPC-3 or BxPC-3 ITGβ5KD exosomes have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE68919.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Paget S. The distribution of secondary growths in cancer of the breast. Cancer Metastasis Rev. 1989;8:98–101. 1889. [PubMed] [Google Scholar]
- 2.Hart IR, Fidler IJ. Role of organ selectivity in the determination of metastatic patterns of B16 melanoma. Cancer Res. 1980;40:2281–2287. [PubMed] [Google Scholar]
- 3.Müller A, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 4.Weilbaecher KN, Guise TA, McCauley LK. Cancer to bone: a fatal attraction. Nature Rev Cancer. 2011;11:411–425. doi: 10.1038/nrc3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou W, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang Q, et al. The IL-6/JAK/Stat3 feed-forward loop drives tumorigenesis and metastasis. Neoplasia. 2013;15:848–862. doi: 10.1593/neo.13706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu X, Kang Y. Organotropism of breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2007;12:153–162. doi: 10.1007/s10911-007-9047-3. [DOI] [PubMed] [Google Scholar]
- 8.Cox TR, et al. The hypoxic cancer secretome induces pre-metastatic bone lesions through lysyl oxidase. Nature. 2015;522:106–110. doi: 10.1038/nature14492. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Kaplan RN, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiratsuka S, et al. MMP9 induction by vascular endothelial growth factor receptor-1 is involved in lung-specifc metastasis. Cancer Cell. 2002;2:289–300. doi: 10.1016/s1535-6108(02)00153-8. [DOI] [PubMed] [Google Scholar]
- 11.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nature Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balaj L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nature Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nature Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 15.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peinado H, Lavotshkin S, Lyden D. The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Semin Cancer Biol. 2011;21:139–146. doi: 10.1016/j.semcancer.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013;13:1554–1571. doi: 10.1002/pmic.201200329. [DOI] [PubMed] [Google Scholar]
- 18.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 19.Thakur BK, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa-Silva B, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nature Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang Y, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 22.Gupta GP, et al. Identifying site-specific metastasis genes and functions. Cold Spring Harb Symp Quant Biol. 2005;70:149–158. doi: 10.1101/sqb.2005.70.018. [DOI] [PubMed] [Google Scholar]
- 23.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bos PD, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nature Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sroka TC, Marik J, Pennington ME, Lam KS, Cress AE. The minimum element of a synthetic peptide required to block prostate tumor cell migration. Cancer Biol Ther. 2006;5:1556–1562. doi: 10.4161/cbt.5.11.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruoslahti E, Pierschbacher MD. Arg-Gly-Asp: a versatile cell recognition signal. Cell. 1986;44:517–518. doi: 10.1016/0092-8674(86)90259-x. [DOI] [PubMed] [Google Scholar]
- 28.Grum-Schwensen B, et al. Suppression of tumor development and metastasis formation in mice lacking the S100A4(mts1) gene. Cancer Res. 2005;65:3772–3780. doi: 10.1158/0008-5472.CAN-04-4510. [DOI] [PubMed] [Google Scholar]
- 29.Lukanidin E, Sleeman JP. Building the niche: the role of the S100 proteins in metastatic growth. Semin Cancer Biol. 2012;22:216–225. doi: 10.1016/j.semcancer.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Kim TH, Kim HI, Soung YH, Shaw LA, Chung J. Integrin (α6β4) signals through Src to increase expression of S100A4, a metastasis-promoting factor: implications for cancer cell invasion. Mol Cancer Res. 2009;7:1605–1612. doi: 10.1158/1541-7786.MCR-09-0102. [DOI] [PubMed] [Google Scholar]
- 31.Abdel-Ghany M, Cheng HC, Elble RC, Pauli BU. Focal adhesion kinase activated by β4 integrin ligation to mCLCA1 mediates early metastatic growth. J Biol Chem. 2002;277:34391–34400. doi: 10.1074/jbc.M205307200. [DOI] [PubMed] [Google Scholar]
- 32.Mainiero F, et al. p38 MAPK is a critical regulator of the constitutive and the beta4 integrin-regulated expression of IL-6 in human normal thymic epithelia cells. Eur J Immunol. 2003;33:3038–3048. doi: 10.1002/eji.200323931. [DOI] [PubMed] [Google Scholar]
- 33.Weaver VM, et al. β4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2002;2:205–216. doi: 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikolopoulos SN, et al. Targeted deletion of the integrin beta4 signaling domain suppresses laminin-5-dependent nuclear entry of mitogen-activated protein kinases and NF-κB, causing defects in epidermal growth and migration. Mol Cell Biol. 2005;25:6090–6102. doi: 10.1128/MCB.25.14.6090-6102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minn AJ, et al. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest. 2005;115:44–55. doi: 10.1172/JCI22320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oskarsson T, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nature Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukuda K, et al. Periostin is a key niche component for wound metastasis of melanoma. PLoS ONE. 2015;10:e0129704. doi: 10.1371/journal.pone.0129704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radisky D, Muschler J, Bissell MJ. Order and disorder: the role of extracellular matrix in epithelial cancer. Cancer Invest. 2002;20:139–153. doi: 10.1081/cnv-120000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weaver VM, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grum-Schwensen B, et al. Lung metastasis fails in MMTV-PyMT oncomice lacking S100A4 due to a T-cell deficiency in primary tumors. Cancer Res. 2010;70:936–947. doi: 10.1158/0008-5472.CAN-09-3220. [DOI] [PubMed] [Google Scholar]
- 41.Chen M, Sinha M, Luxon BA, Bresnick AR, O’Connor KL. Integrin α6β4 controls the expression of genes associated with cell motility, invasion, and metastasis, including S100A4/metastasin. J Biol Chem. 2009;284:1484–1494. doi: 10.1074/jbc.M803997200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bäuerle T, et al. Cilengitide inhibits progression of experimental breast cancer bone metastases as imaged noninvasively using VCT, MRI and DCE-MRI in a longitudinal in vivo study. Int J Cancer. 2011;128:2453–2462. doi: 10.1002/ijc.25563. [DOI] [PubMed] [Google Scholar]
- 43.Wu YJ, et al. Targeting αV-integrins decreased metastasis and increased survival in a nude rat breast cancer brain metastasis model. J Neurooncol. 2012;110:27–36. doi: 10.1007/s11060-012-0942-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y, et al. Tumor αvβ3 integrin is a therapeutic target for breast cancer bone metastases. Cancer Res. 2007;67:5821–5830. doi: 10.1158/0008-5472.CAN-06-4499. [DOI] [PubMed] [Google Scholar]
- 45.Tominaga N, et al. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nature Commun. 2015;6:6716. doi: 10.1038/ncomms7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nature Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 48.Bunkenborg J, Garcia GE, Paz MI, Andersen JS, Molina H. The minotaur proteome: avoiding cross-species identifications deriving from bovine serum in cell culture models. Proteomics. 2010;10:3040–3044. doi: 10.1002/pmic.201000103. [DOI] [PubMed] [Google Scholar]
- 49.Käll L, Canterbury JD, Weston J, Noble WS, Mac Coss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nature Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 50.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


