Abstract
Visceral leishmaniasis (VL) is a debilitating, often fatal disease caused by Leishmania donovani complex; however, it is a neglected tropical disease. L. donovani complex comprises two closely related species, L. donovani that is mostly anthroponotic and L. infantum that is zoonotic. Differentiation between these two species is critical due to the differences in their epidemiology and pathology. However, they cannot be differentiated morphologically, and their speciation using isoenzyme-based methods poses a difficult task and may be unreliable. Molecular characterization is now the most reliable method to differentiate between them and to determine their phylogenetic relationships. The present study aims to characterize Leishmania species isolated from bone marrows of Yemeni pediatric patients using sequence analysis of the ribosomal internal transcribed spacer-1 (ITS1) gene. Out of 41 isolates from Giemsa-stained bone marrow smears, 25 isolates were successfully amplified by nested polymerase chain reaction and sequenced in both directions. Phylogenetic analysis using neighbor joining method placed all study isolates in one cluster with L. donovani complex (99% bootstrap). The analysis of ITS1 for microsatellite repeat numbers identified L. infantum in 11 isolates and L. donovani in 14 isolates. These data suggest the possibility of both anthroponotic and zoonotic transmission of VL-causing Leishmania species in Yemen. Exploring the possible animal reservoir hosts is therefore needed for effective control to be achieved.
Introduction
Human leishmaniasis is a vector-borne disease caused by more than 20 Leishmania species in 98 countries on five continents [1]. The disease is categorized as one of the most “neglected tropical diseases'' and has a strong and complex association with poverty [2,3]. Approximately 350 million people are at risk of leishmaniasis, with a yearly incidence of about 0.5 million cases of visceral leishmaniasis (VL), which is responsible for 59,000 deaths annually and losses of 2,357,000 disability-adjusted life years (DALYs), ranking leishmaniasis the ninth in a global analysis of infectious diseases [2]. The disease is caused by two closely related species of the Leishmania donovani complex: L. donovani that is mainly considered to be anthroponotic and L. infantum that is considered to be zoonotic, with dogs being the main reservoir hosts [4–6]. In the Eastern Mediterranean Region (EMR) of the World Health Organization (WHO), L. donovani causes anthroponotic VL in Somalia and Sudan [7,8] and is a probable cause of anthroponotic VL in Yemen and Saudi Arabia [6]. On the other hand, L. infantum causes zoonotic VL in several countries of the EMR, including Yemen [6]. VL is the most severe systemic form of leishmaniasis that is characterized by persistent fever, pancytopenia and organomegaly and is usually fatal if left untreated, particularly among children. Moreover, anti-leishmanial drugs are toxic and given parenterally for long periods [4,9].
In Yemen, leishmaniasis is a major public health problem with a nationwide distribution and is responsible for about 60% of DALYs lost due to tropical diseases prevalent in the country [10]. However, a few studies have been published on VL among pediatric patients from endemic areas of the country. Diagnosis of VL is routinely based on the microscopic examination of bone marrow aspirates together with clinical diagnosis and hematologic investigations [11], which does not enable species identification. This in turn, limits the possibility of developing an effective VL control strategy. A high seroprevalence rate (34.7%; 99/285) of antibodies to Leishmania species was reported among schoolchildren from areas endemic with infantile VL in Sana’a and Hajjah governorates [12]. In addition, natural infection of feral dogs from the study areas with Leishmania species (50%; 8/16) was also documented for the first time [12]. Haidar et al. [13] reported that the clinical presentation of VL among children in Hajjah governorate, north of Yemen, is similar to that of the Mediterranean type. In Aden, south of Yemen, Abdul Hamid and Gobah [11] reported fever, hepatosplenomegaly and pancytopenia as the most common clinical and hematologic manifestations of VL among pediatric patients. In central Yemen, VL was reported to be the least prevalent form of leishmaniasis, being responsible for 3.3% of cases [14]. However, children and women are at highest risk for contracting the disease [14].
Except for a single study on the molecular characterization of Leishmania species causing cutaneous leishmaniasis (CL) in Yemen [15], characterization of parasite isolates was based on isoenzyme analysis. Eight human isolates of Leishmania species causing VL in Yemen were analyzed using the isoenzyme method, where seven were identified as L. donovani and one as L. infantum [16]. Later, a case of CL in a French visitor to Yemen was also reported to be caused by L. donovani [17]. However, the isoenzyme method has been recently indicated to be non-robust for the distinction between L. donovani and L. infantum compared to molecular discrimination using sequence analysis of ribosomal internal transcribed spacer gene [18]. Therefore, this is the first study conducted to characterize Leishmania species causing VL among Yemeni children using the sequence analysis of ITS1.
Materials and Methods
Ethical clearance and samples
This study used archived bone marrow smears of 41 pediatric patients admitted in 2010 to Al-Sabeen Hospital, the main referral hospital for women and children in Sana’a, and diagnosed with VL (30 males and 11 females with a mean age of 3.5 years old). The study protocol was approved by the Ethical Committee of University of Science and Technology, Yemen. Permission to use the archived samples was obtained from the hospital management. No informed consent was acquired and patients’ information was maintained confidentially and analyzed anonymously. It should be noted that Al-Sabeen Hospital is a teaching hospital and patients understand that samples may be used in future research. Confirmation of VL was made by the examination of Giemsa-stained smears of bone marrow aspirates, and amastigote density was then determined [19]. Leishmania isolates were coded according to the international code system for Leishmania isolates following the recommendations of the UNDP/ WHO meeting in Washington, D.C., in 1980 [20].
DNA extraction, amplification and sequencing
Parasite DNA was extracted from Giemsa-stained bone marrow smears positive for Leishmania as previously described [15]. A nested polymerase chain reaction (PCR) was then used to amplify the ITS1 region [15, 21]. PCR products were purified using the QIAquick PCR Purification Kit (QIAgen, Hilden, Germany) according to the manufacturer’s instructions and sequenced using the ABI PRISM® BigDyeTM Terminator v3.0 Ready Reaction Cycle Sequencing Kit (Applied Biosystems, USA) in ABI PRISM® 3700 DNA Analyzer (Applied Biosystems, USA).
Sequence analysis
Consensus ITS1 sequences of the study isolates were created and multiple-aligned with reference sequences of L. donovani, L. infantum, L. major, L. aethiopica and L. tropica retrieved from GenBank using BioEdit software (www.mbio.ncsu.edu/BioEdit/BioEdit.html). Phylogenetic analysis was conducted in MEGA 6 software [22] using neighbor-joining (NJ) method [23], with evolutionary distance calculated by Kimura 2-parameter model [24] and 1000-replicate bootstrap test [25]. For species identification of L. donovani complex, ITS1 sequences of study isolates were multiple-aligned using ClustalW [26], with ITS1 sequences of previously defined L. infantum and L. donovani, representing the different types of ITS1 sequences, and manually analyzed for microsatellite repeats [18]. Sequences from this study have been deposited in the GenBank database under the accession numbers KT751245 to KT751269.
Results
All Giemsa-stained bone marrow smears were positive for Leishmania species amastigotes by light microscopy. The densities of amastigotes were 1+, 2+, 3+ and 4+ in 5, 13, 7 and 16 samples, respectively. One patient had a VL co-infection with malaria. Twenty five (61%; 25/41) isolates were successfully amplified by nested PCR producing the 350 bp amplicon after gel electrophoresis and directly sequenced in both directions. Multiple alignment and analysis of four polymorphic microsatellites in these sequences divided these isolates into two alleles with a single nucleotide polymorphism.
Phylogenetic analysis using NJ method with reference strains of L. infantum, L. donovani, L. tropica, L. major and L. aethiopica placed all Leishmania isolates causing VL among Yemeni pediatric patients in one cluster with L. donovani complex (L. donovani and L. infantum) originating from different geographical regions with 99% bootstrap support (Fig 1). Based on the analysis of four polymorphic microsatellite of ITS1 sequences of L. donovani complex, including the study isolates and reference sequences representing different types of L. infantum and L. donovani sequences, Yemeni isolates showed two sequence types, named TDRC1 and TDRC2, differing in a single nucleotide polymorphism. TDRC1 sequence type included 11 isolates that have microsatellite repeat numbers identical to those of L. infantum isolates from China and Mediterranean region. TDRC2 sequence type included 14 isolates that have microsatellite repeat numbers identical to those of L. donovani isolates from China and East Africa (Table 1).
Fig 1. Phylogenetic study of L. donovani complex causing VL among Yemeni children.
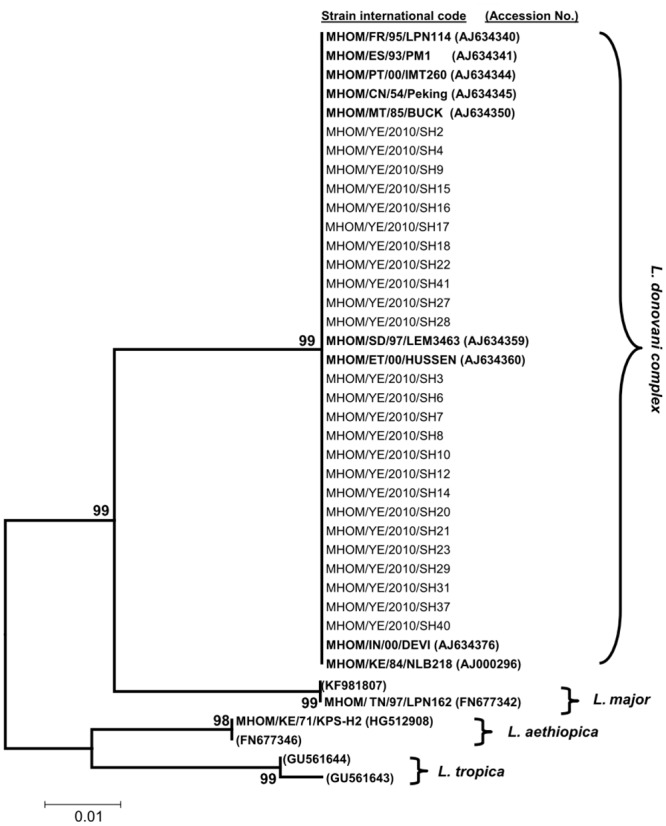
Unrooted NJ phylogenetic tree showing the relationships of 25 ITS1 sequences of Leishmania species isolates from Yemeni children with VL and sequences representing L. infantum, L. donovani, L. major, L. tropica and L. aethiopica. Bold-type represents reference sequences from GenBank. Abbreviations of countries of origib: FR, France; ES, Spain; PT, Portugal; CN, China; MT, Malta; SD, Sudan; ET, Ethiopia; KE, Kenya; IN, India; TN, Tunisia, YE, Yemen.
Table 1. Microsatellite repeat numbers of ITS1 gene found for L. donovani complex isolates from Yemeni children infected with VL compared to reference strains.a.
| Strain/ isolate international code | Species | Origin | Poly C | Poly A | Poly TA | Poly A | GenBank accession No. |
|---|---|---|---|---|---|---|---|
| MHOM/FR/95/LPN114 | L. infantum | France | 3 | 6 | 4 | 8 | AJ634340 |
| MHOM/ES/93/PM1 | L. infantum | Spain | 3 | 6 | 4 | 8 | AJ634341 |
| MHOM/PT/00/IMT260 | L. infantum | Portugal | 3 | 6 | 4 | 8 | AJ634344 |
| MHOM/CN/54/Peking | L. infantum | China | 3 | 6 | 4 | 8 | AJ634345 |
| MHOM/TN/80/IPT1 | L. infantum | Tunisia | 3 | 6 | 4 | 8 | AJ000289 |
| MHOM/IT/94/ISS1036 | L. infantum | Italy | 3 | 6 | 4 | 8 | AJ634353 |
| MHOM/YE/10/SH2, 4, 9, 15, 16, 17, 18, 22, 27, 28, 41b | L. infantum | Yemen | 3 | 6 | 4 | 8 | KT751245 to KT751255 |
| MHOM/CN/00/Wangjie1 | L. donovani | China | 3 | 6 | 4 | 7 | AJ000294 |
| MCAN/SD/00/LEM3946 | L. donovani | Sudan | 3 | 6 | 4 | 7 | AJ634356 |
| MHOM/ET/00/HUSSEN | L. donovani | Ethiopia | 3 | 6 | 4 | 7 | AJ634360 |
| MHOM/YE/10/SH3, 6, 7, 8, 10, 12, 14, 20, 21, 23, 29, 31, 37, 40c | L. donovani | Yemen | 3 | 6 | 4 | 7 | KT751256 to KT751269 |
| MHOM/SD/62/LRC-L61 | L. donovani | Sudan | 2 | 8 | 6 | 8 | AJ634365 |
| MHOM/SD/68/1S | L. donovani | Sudan | 2 | 8 | 6 | 8 | AJ000293 |
| MHOM/SD/75/LV139 | L. donovani | Sudan | 2 | 8 | 6 | 8 | AJ000291 |
| MHOM/IN/71/LRC-L51a | L. donovani | India | 2 | 8 | 5 | 7 | AJ000290 |
| MHOM/IN/80/DD8 | L. donovani | India | 2 | 8 | 5 | 7 | AJ000292 |
| MHOM/KE/84/NLB218 | L. donovani | Kenya | 2 | 8 | 5 | 7 | AJ000296 |
| MHOM/SD/93/9S | L. donovani | Sudan | 2 | 9 | 5 | 7 | AJ634372 |
| MHOM/ET/67/HU3 | L. donovani | Ethiopia | 2 | 9 | 5 | 7 | AJ634373 |
a Reference strains are published by Kuhls et al. [18];
b isolates with TDRC1 sequence type in the present study;
c isolates with TDRC2 sequence type in the present study
Discussion
VL is the most severe form of leishmaniasis that is caused by the species of L. donovani complex (L. donovani and L. infantum). Although differentiation between these two species is important because of the differences in their epidemiology and pathology, this had been a difficult task. Typing this parasite using isoenzyme analysis is difficult and requires a large amount of the parasite [27,28] or may sometimes be unreliable [29]. The advent of molecular tools provided methods which are robust in discriminating these two species and to study their phylogenetic relationships [18,30–32]. Although pediatric VL among Yemeni children was clinically described [12,14], the molecular characterization and phylogenetic relationship among VL-causing Leishmania isolates had not been studied before. The present study deploys molecular approaches to characterize VL-causing Leishmania species from clinical isolates collected from bone marrows of Yemeni children based on the analysis of ribosomal ITS1 sequences.
L. donovani complex causing VL among Yemeni children exhibited a monophyletic cluster that is consistent with L. infantum isolates from Europe and China as well as L. donovani isolates from Africa. This finding is in agreement with that of Mauricio et al. [33], where phylogenetic trees generated by the combined data of five genes of L. donovani complex could not give a clear division between L. infantum and L. donovani. Hence, this finding strongly supports early hypotheses of the monophyletic origin of Leishmania species from the Old World and the New World based on zymodeme analysis [34] and DNA-based characterization [35]. The finding is also consistent with the monophyletic relationship among Turkish isolates of L. infantum recently reported in comparison to L. tropica that showed different phylogenetic groups based on ITS1 sequence [36]. In contrast, Yang et al. [37] reported the heterogeneity of Chinese Leishmania isolates and suggested that they do not form a monophyletic group based on the sequence analysis of ITS1, which was then confirmed by sequencing of kinetoplast cytochrome oxidase II gene [38]. The phylogenetic tree constructed from the sequences of the present study isolates and sequences from L. donovani complex from other parts of the world shows that Yemeni isolates are genetically similar to them.
Based on the analysis of the four polymorphic microsatellites in the ITS1 sequences of L. donovani complex in the present study, isolates showed two distinct alleles or sequence types (TDRC1 and TDRC2) differing in a single nucleotide polymorphism. The ITS1 sequence types of 11 isolates were similar to those of L. infantum strain zymodemes originating from different geographical regions, with the exception of L. infantum zymodemes from Sudan (MON-30, 81, 267) [18]. However, these Sudanese zymodemes were later regarded as L. donovani rather than L. infantum based on microsatellite analysis and that L. donovani is the only agent causing VL in East Africa [18,29,39,40]. In general, phylogenetic trees generated from DNA-based analyses might not correlate with isoenzyme-based taxonomy.
These 11 isolates with TDRC1 sequence type showed the same sequence types at the four polymorphic microsatellites as those reported by Kuhls et al. [18] from France (MON-1, 11, 29, 108), Spain (MON-1, 77, 183, 198, 199), Portugal (MON-1), Italy (MON-188, 228), Malta (MON-78), China (MON-1 and LON-49) and Tunisia (MON-1). Sequence similarity also exists with MON-1 zymodeme of L. chagasi from Brazil [18]. It is important to consider that L. chagasi has been regarded as a synonym of L. infantum based on DNA analysis by a variety of techniques [18,41,42]. It is noteworthy to highlight that L. infantum differs from L. donovani with regards to its ITS1 microsatellite repeat numbers as shown in the study by Kuhls et al. [18]. This strongly supports the existence of L. infantum as a causative agent of VL among Yemeni children based on molecular sequencing. Moreover, reporting canine leishmaniasis and that dogs possibly act as reservoir hosts for VL [12] further supports the possible role of L. infantum as a causative agent of VL in Yemen.
On the other hand, 14 of the studied isolates with TDRC2 sequence type are similar to those of L. donovani zymodemes originating from Old World countries; namely, Sudan (MON-274), Ethiopia (MON-31) and China (MON-35) as well as those of L. archibaldi zymodemes from Sudan (MON-257, 258) [18]. L. archibaldi is a disputed species that has been suggested to be a synonym of L. donovani based on DNA analysis by a variety of techniques [31,40,41]. However, ITS1 sequences of Yemeni isolates differ from those of L. donovani zymodemes from India (MON-2, 38), Kenya (MON-274), Sudan and Ethiopia (MON-18) [18]. Sequence differences of the latter 14 isolates from sequences of well-characterized L. infantum and their similarity to well-characterized L. donovani strongly suggest that these isolates are L. donovani. In addition to its small sample size, another limitation of this study is that it did not consider ITS2 sequencing. However, according to Schonian et al. [32], sequencing of the ITS1 allows the differentiation of the Old World L. donovani complex, with a clear distinction of L. infantum from L. donovani. Moreover, beyond species identification, ITS1 sequencing enables the assignment of L. donovani complex strains to different phylogenetic groups supported by their biological characteristics and clinical outcomes [18]. With the presence of L. donovani and L. infantum in the country, there is a need to determine the relative contribution of both parasite species to the anthroponotic and/or zoonotic epidemiology of VL.
Questions remain unanswered include whether dogs and other animals are possible reservoir hosts and whether L. donovani has an anthroponotic transmission similar to that in the Indian subcontinent. The detection of circulating antibodies against Leishmania species in sera of dogs in Yemen [12] raises concerns about the possible role of zoonotic transmission from dogs to humans. Postigo [6] reported the occurrence of zoonotic VL caused by L. infantum in most countries of the EMR of the WHO, including Yemen. In contrast to the anthroponotic CL in Yemen, the anthroponotic VL caused by L. donovani remains unclear yet probable in the country [6]. The presence of L. donovani and L. infantum necessitates further investigation for the nature of transmission and the possible involvement of animal reservoir hosts in their transmission to tailor appropriate prevention and control strategies.
In conclusion, the present study reveals that both L. infantum and L. donovani are causative agents of pediatric VL in Yemen in line with previous characterization using isoenzyme analysis. Therefore, both anthroponotic and zoonotic epidemiological patterns of transmission could not be ruled out. For effective control, determination of possible animal reservoir hosts should be explored using molecular methods.
Acknowledgments
The authors thank the management and personnel of Al-Sabeen Hospital for their cooperation and for giving the permission to use the archived samples. This work was supported by University of Malaya (RG302/11HTM).
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the University of Malaya (RG302/11HTM).
References
- 1.Alvar J, Velez ID, Bern C, Herrero M, Desjeux P, Cano J, et al. (2012) Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7: e35671 10.1371/journal.pone.0035671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotez PJ, Remme JH, Buss P, Alleyne G, Morel C, Breman JG (2004) Combating tropical infectious diseases: report of the Disease Control Priorities in Developing Countries Project. Clin Infect Dis 38: 871–878. [DOI] [PubMed] [Google Scholar]
- 3.Alvar J, Yactayo S, Bern C (2006) Leishmaniasis and poverty. Trends Parasitol 22: 552–557. [DOI] [PubMed] [Google Scholar]
- 4.Chappuis F, Sundar S, Hailu A, Ghalib H, Rijal S, Peeling RW, et al. (2007) Visceral leishmaniasis: what are the needs for diagnosis, treatment and control? Nat Rev Microbiol 5: 873–882. [DOI] [PubMed] [Google Scholar]
- 5.Quinnell RJ, Courtenay O (2009) Transmission, reservoir hosts and control of zoonotic visceral leishmaniasis. Parasitology 136: 1915–1934. 10.1017/S0031182009991156 [DOI] [PubMed] [Google Scholar]
- 6.Postigo JA (2011) Leishmaniasis in the World Health Organization Eastern Mediterranean Region. Int J Antimicrob Agents 36: S62–S65. [DOI] [PubMed] [Google Scholar]
- 7.Raguenaud ME, Jansson A, Vanlerberghe V, Deborggraeve S, Dujardin JC, Orfanos G, et al. (2007) Epidemiology and clinical features of patients with visceral leishmaniasis treated by an MSF clinic in Bakool region, Somalia, 2004–2006. PLoS Negl Trop Dis 1: e85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolaczinski JH, Hope A, Ruiz JA, Rumunu J, Richer M, Seaman J (2008) Kala-azar epidemiology and control, southern Sudan. Emerg Infect Dis 14: 664–666. 10.3201/eid1404.071099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palumbo E (2010) Visceral leishmaniasis in children: a review. Minerva Pediatr 62: 389–395. [PubMed] [Google Scholar]
- 10.WHO (2008) The global burden of disease: 2004 update. Geneva: World Health Organization. [Google Scholar]
- 11.Abdul Hamid G, Gobah G (2009) Clinical and hematological manifestations of visceral leishmaniasis in Yemeni children. Turk J Hematol 26: 25–28. [PubMed] [Google Scholar]
- 12.Al-Shamahy HA (1998) Seroprevalence of kala-azar among humans and dogs in Yemen. Ann Saudi Med 18: 66–68. [DOI] [PubMed] [Google Scholar]
- 13.Haidar NA, Diab AL, El-Sheikh AM (2001) Visceral leishmaniasis in children in the Yemen. Saudi Med J 22: 516–519. [PubMed] [Google Scholar]
- 14.Al-Kamel MA (2015) Leishmaniasis in Yemen: a clinicoepidemiological study of leishmaniasis in central Yemen. Int J Dermatol [in press]. [DOI] [PubMed] [Google Scholar]
- 15.Mahdy MA, Al-Mekhlafi HM, Al-Mekhlafi AM, Lim YA, Bin Shuaib NO, Azazy AA, et al. (2010) Molecular characterization of Leishmania species isolated from cutaneous leishmaniasis in Yemen. PLoS One 5: e12879 10.1371/journal.pone.0012879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rioux JA, Dereure J, Daoud W, el Kubati Y, Rageh HA, Moreno G, et al. (1989) Eco-epidemiology of visceral and cutaneous leishmaniasis in the Yemen Arab Republic. I. Presence, in sympatric condition, of Leishmania infantum and Leishmania donovani complexes]. Bull Soc Pathol Exot Filiales 82: 658–664. [PubMed] [Google Scholar]
- 17.Pratlong F, Bastien P, Perello R, Lami P, Dedet JP (1995) Human cutaneous leishmaniasis caused by Leishmania donovani sensu stricto in Yemen. Trans R Soc Trop Med Hyg 89: 398–399. [DOI] [PubMed] [Google Scholar]
- 18.Kuhls K, Mauricio IL, Pratlong F, Presber W, Schonian G (2005) Analysis of ribosomal DNA internal transcribed spacer sequences of the Leishmania donovani complex. Microbes Infect 7: 1224–1234. [DOI] [PubMed] [Google Scholar]
- 19.Ramirez JR, Agudelo S, Muskus C, Alzate JF, Berberich C, Barker D, et al. (2000) Diagnosis of cutaneous leishmaniasis in Colombia: the sampling site within lesions influences the sensitivity of parasitologic diagnosis. J Clin Microbiol 38: 3768–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chance ML, Walton BC (1982) Biochemical characterization of Leishmania: Proceedings of a workshop held at the Pan American Health Organization, Washington, D.C., 9–11 December 1980. Geneva: UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases. [Google Scholar]
- 21.Schonian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HD, Presber W, et al. (2003) PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis 47: 349–358. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 24.Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 25.Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 26.Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rioux JA, Lanotte G, Serres E, Pratlong F, Bastien P, Perieres J (1990) Taxonomy of Leishmania. Use of isoenzymes. Suggestions for a new classification. Ann Parasitol Hum Comp 65: 111–125. [DOI] [PubMed] [Google Scholar]
- 28.Hide M, Banuls AL, Tibayrenc M (2001) Genetic heterogeneity and phylogenetic status of Leishmania (Leishmania) infantum zymodeme MON-1: epidemiological implications. Parasitology 123: 425–432. [DOI] [PubMed] [Google Scholar]
- 29.Jamjoom MB, Ashford RW, Bates PA, Chance ML, Kemp SJ, Watts PC, et al. (2004) Leishmania donovani is the only cause of visceral leishmaniasis in East Africa; previous descriptions of L. infantum and "L. archibaldi" from this region are a consequence of convergent evolution in the isoenzyme data. Parasitology 129: 399–409. [DOI] [PubMed] [Google Scholar]
- 30.Hide M, Banuls AL (2006) Species-specific PCR assay for L. infantum/L. donovani discrimination. Acta Trop 100: 241–245. [DOI] [PubMed] [Google Scholar]
- 31.Lukes J, Mauricio IL, Schonian G, Dujardin JC, Soteriadou K, Dedet JP, et al. (2007) Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc Natl Acad Sci U S A 104: 9375–9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schonian G, Kuhls K, Mauricio IL (2011) Molecular approaches for a better understanding of the epidemiology and population genetics of Leishmania. Parasitology 138: 405–425. 10.1017/S0031182010001538 [DOI] [PubMed] [Google Scholar]
- 33.Mauricio IL, Yeo M, Baghaei M, Doto D, Pratlong F, Zemanova E, et al. (2006) Towards multilocus sequence typing of the Leishmania donovani complex: resolving genotypes and haplotypes for five polymorphic metabolic enzymes (ASAT, GPI, NH1, NH2, PGD). Int J Parasitol 36: 757–769. [DOI] [PubMed] [Google Scholar]
- 34.Thomaz-Soccol V, Lanotte G, Rioux JA, Pratlong F, Martini-Dumas A, Serres E (1993) Monophyletic origin of the genus Leishmania Ross, 1903. Ann Parasitol Hum Comp 68: 107–108. [PubMed] [Google Scholar]
- 35.Mauricio IL, Howard MK, Stothard JR, Miles MA (1999) Genomic diversity in the Leishmania donovani complex. Parasitology 119: 237–246. [DOI] [PubMed] [Google Scholar]
- 36.Toz SO, Culha G, Zeyrek FY, Ertabaklar H, Alkan MZ, Vardarli AT, et al. (2013) A real-time ITS1-PCR based method in the diagnosis and species identification of Leishmania parasite from human and dog clinical samples in Turkey. PLoS Negl Trop Dis 7: e2205 10.1371/journal.pntd.0002205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang BB, Guo XG, Hu XS, Zhang JG, Liao L, Chen DL, et al. (2010) Species discrimination and phylogenetic inference of 17 Chinese Leishmania isolates based on internal transcribed spacer 1 (ITS1) sequences. Parasitol Res 107: 1049–1065. 10.1007/s00436-010-1969-9 [DOI] [PubMed] [Google Scholar]
- 38.Cao DP, Guo XG, Chen DL, Chen JP (2011) Species delimitation and phylogenetic relationships of Chinese Leishmania isolates reexamined using kinetoplast cytochrome oxidase II gene sequences. Parasitol Res 109: 163–173. 10.1007/s00436-010-2239-6 [DOI] [PubMed] [Google Scholar]
- 39.Oskam L, Pratlong F, Zijlstra EE, Kroon CC, Dedet IP, Kager PA, et al. (1998) Biochemical and molecular characterization of Leishmania parasites isolated from an endemic focus in eastern Sudan. Trans R Soc Trop Med Hyg 92: 120–122. [DOI] [PubMed] [Google Scholar]
- 40.Zemanova E, Jirku M, Mauricio IL, Miles MA, Lukes J (2004) Genetic polymorphism within the Leishmania donovani complex: correlation with geographic origin. Am J Trop Med Hyg 70: 613–617. [PubMed] [Google Scholar]
- 41.Mauricio IL, Gaunt MW, Stothard JR, Miles MA (2001) Genetic typing and phylogeny of the Leishmania donovani complex by restriction analysis of PCR amplified gp63 intergenic regions. Parasitology 122: 393–403. [DOI] [PubMed] [Google Scholar]
- 42.Kuhls K, Alam MZ, Cupolillo E, Ferreira GE, Mauricio IL, Oddone R, et al. (2011) Comparative microsatellite typing of new world Leishmania infantum reveals low heterogeneity among populations and its recent old world origin. PLoS Negl Trop Dis 5: e1155 10.1371/journal.pntd.0001155 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.


