Abstract
In the nitrogen-fixing heterocyst-forming cyanobacterium Anabaena sp. PCC 7120, the ferric uptake regulator FurA plays a global regulatory role. Failures to eliminate wild-type copies of furA gene from the polyploid genome suggest essential functions. In the present study, we developed a selectively regulated furA expression system by the replacement of furA promoter in the Anabaena sp. chromosomes with the Co2+/Zn2+ inducible coaT promoter from Synechocystis sp. PCC 6803. By removing Co2+ and Zn2+ from the medium and shutting off furA expression, we showed that FurA was absolutely required for cyanobacterial growth. RNA-seq based comparative transcriptome analyses of the furA-turning off strain and its parental wild-type in conjunction with subsequent electrophoretic mobility shift assays and semi-quantitative RT-PCR were carried out in order to identify direct transcriptional targets and unravel new biological roles of FurA. The results of such approaches led us to identify 15 novel direct iron-dependent transcriptional targets belonging to different functional categories including detoxification and defences against oxidative stress, phycobilisome degradation, chlorophyll catabolism and programmed cell death, light sensing and response, heterocyst differentiation, exopolysaccharide biosynthesis, among others. Our analyses evidence novel interactions in the complex regulatory network orchestrated by FurA in cyanobacteria.
Introduction
With only few exceptions, iron is absolutely essential for life of all forms since this metal participates as protein cofactor in major biological processes, such as photosynthesis, respiration, nitrogen fixation, and nucleic acids biosynthesis. Aerobic organisms must face a survival paradox: iron is required for growth, but on the other hand, it is also potentially toxic due to its ability to catalyze the formation of reactive oxygen species (ROS) by Fenton reactions [1]. Furthermore, iron became scarce and growth-limiting in most ecological niches, since the predominant form of this micronutrient in nature is extremely insoluble at neutral pH. To cope with this limitation, bacteria have evolved strategies to efficiently scavenge iron from the environment and thereby ensure its physiological demands, but at the same time they have developed molecular mechanisms to tightly regulate iron homeostasis in order to maintain its intracellular concentration within non-toxic levels [2].
In most bacterial species, the concerted expression of genes involved in iron scavenge, uptake, storage, and metabolism is transcriptionally regulated by the ferric uptake regulator (Fur) protein in response to iron availability [2, 3]. Fur is a homodimeric metalloprotein that complexes with ferrous iron under high intracellular Fe2+ concentrations and typically acts as a transcriptional repressor by binding to cis-acting regulatory elements known as Fur boxes, which are located in the promoters of target genes [4]. When intracellular iron is depleted, the Fur-Fe2+ complexes dissociate from the Fur boxes, allowing transcription of target genes. In less frequent cases, Fur has been reported to act positively rather than negatively on the expression of certain genes by the occurrence of different mechanisms of regulation [5–8]. To date, apo-regulation of Fur has been observed only in epsilon-proteobacteria like Helicobacter pylori [9] and Campylobacter jejuni [10].
Cyanobacteria constitute an abundant, widespread and diverse group of photoautotrophic microorganisms which possesses the unique ability among prokaryotes to perform oxygenic photosynthesis. In addition, many species of this group are able to fix atmospheric dinitrogen when combined nitrogen sources are shortly supplied, thereby contributing significantly to the Earth’s nitrogen cycle [11]. Since iron is an essential redox component for both, the photosynthetic electron transfer chain and the nitrogenase enzymatic complex, the requirements of iron in cyanobacteria far exceeds those of heterotrophic, non-diazotrophic bacteria [12, 13]. On the other hand, in addition to ROS produced by the respiratory machinery, cyanobacteria are overexposed to those ROS unavoidably generated during the photosynthetic electron flow [14]. Therefore, the challenge of balancing iron homeostasis and oxidative stress become particularly acute in these organisms.
In the filamentous, nitrogen-fixing, heterocyst forming cyanobacterium Anabaena sp. PCC 7120, iron homeostasis is orchestrated by the ferric uptake regulator FurA, which modulates the expression of siderophore biosynthesis machineries, TonB-dependent siderophore outer membrane transporters, siderophore periplasmic binding proteins, ABC inner membrane permeases, ferritin Dps family proteins, as well as several enzymes involved in the tetrapyrrole biosynthesis pathway [15–17]. FurA is a constitutive protein whose expression increases under iron limitation [18] and oxidative stress conditions [19]. This metalloregulator associates with DNA in an iron dependent manner, at a 19- to 23-bp consensus binding site with the sequence AAATAAATTCTCAATAAAT [20], which shares 58% homology to the canonical Escherichia coli Fur box consensus sequence [21]. Previous analyses have shown that FurA is a global transcriptional regulator, modulating not only the iron assimilation genes, but also a plethora of genes and operons involved in a variety of physiological processes including photosynthesis, respiration, oxidative stress defences, heterocyst differentiation, light-dependent signal transduction mechanisms, among others [17, 20, 22–24]. As other Fur orthologues, FurA displays a dual role as transcriptional modulator, acting both as repressor and as activator of gene expression [15, 20].
Although comparative global analyses of transcriptomes and proteomes for fur deletion mutants have been traditionally used to characterize Fur regulons [25–28], attempts to inactivate furA from the polyploid genome of Anabaena sp. have resulted in only a partial segregation of the mutated chromosomes, suggesting a putative essential role for this protein under standard culture conditions [29]. Similar results had been previously obtained with Fur from Synechococcus elongatus [30], which share 75% identity with FurA. Hence, most in vivo analyses of FurA regulatory roles have been carried out on furA-overexpressing backgrounds [15, 17, 20, 22, 24, 31]. To conclusively establish the essentiality of furA and go in-depth on the knowledge of its function, we developed a selectively regulating furA expression system by the replacement of furA promoter in the Anabaena sp. chromosomes with the Co2+ and Zn2+ inducible coaT promoter [32]. This approach allowed us to grow the cells with Co2+/Zn2+ so that furA was expressed and subsequently to remove Co2+/Zn2+ from the medium to shut off furA expression, mimicking the furA null phenotype. We showed that FurA was absolutely required for cyanobacterial growth. Comparative transcriptome analysis of the furA-turning off phenotype led us to identify 15 novel FurA direct targets involved in different physiological processes.
Results
FurA is an essential, well-conserved protein among cyanobacteria
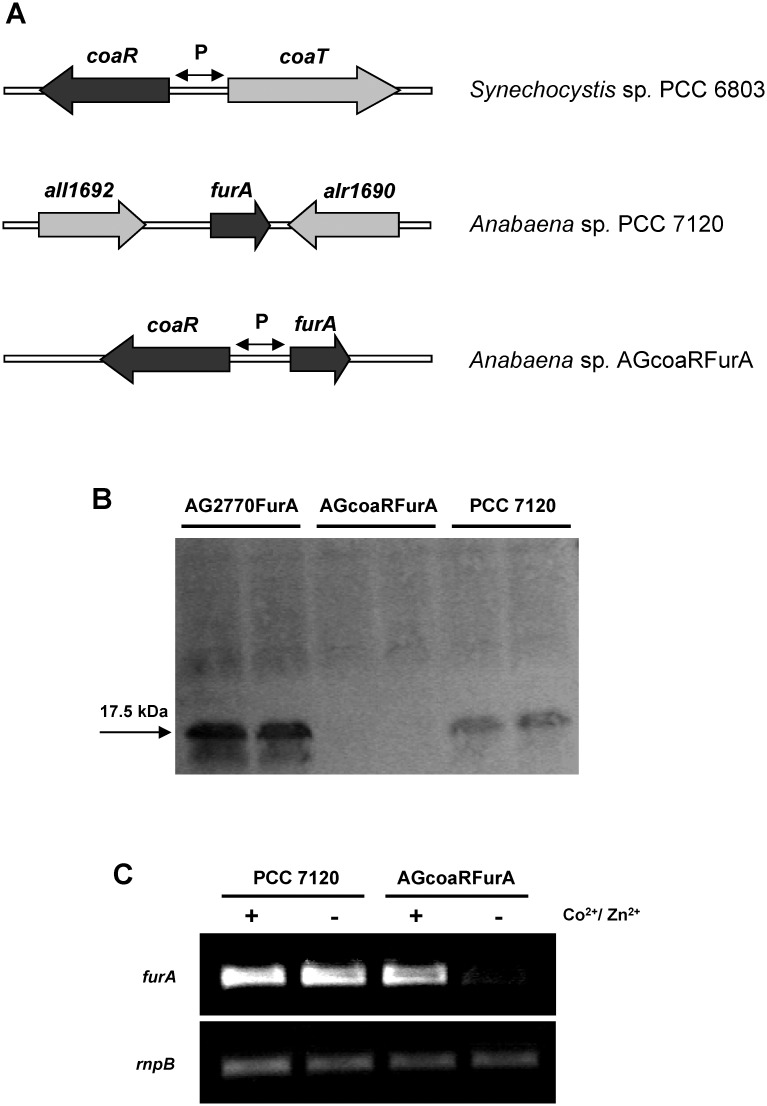
The native promoter region of the furA gene was replaced with the cobalt/zinc-inducible coaT promoter (PcoaT) to allow regulation of furA by controlling the level of Co2+ and Zn2+ in the medium. The divergently transcribed coaT and coaR genes (Fig 1A) mediate inducible resistance to Co2+ in Synechocystis sp. PCC 6803 [32]. In the coa operon, coaT encodes a Co2+ efflux P-type ATPase similarly induced by both Co2+ and Zn2+, while coaR encodes a transcriptional activator of coaT whose carboxyl-terminal Cys-His-Cys motif is required for cobalt sensing. Previous bioluminescence assays have shown that gene expression under the control of coaR-PcoaT responded to Co2+ and Zn2+ with a detection range of 0.3–6 μM and 1–3 μM, respectively. In mixed samples of Co2+ and Zn2+, the bioluminescence responded in an additive manner [32]. In the strain Anabaena sp. AGcoaRFurA, coaR-PcoaT and the coding region of furA were fused at their respective ATG translational start sites and were located immediately downstream of the predicted transcriptional terminator of all1692, the gene upstream of furA on the chromosome (Fig 1A). The region between furA and alr1690 remained the same as in wild-type Anabaena sp. PCC 7120.
Fig 1. Construction of the coaR-PcoaT::furA fusion strain AGcoaRFurA.
(A) The normal promoter region of the furA gene in the Anabaena sp. PCC 7120 chromosome was replaced with the divergent cobalt/zinc-inducible coaT promoter (PcoaT) and the coding region of the transcriptional activator CoaR from Synechocystis sp. PCC 6803. (B) Levels of FurA protein in wild-type Anabaena sp. strain PCC 7120, the furA-overexpressing strain AG2770FurA and the coaR-PcoaT::furA fusion strain AGcoaRFurA, revealed by Western blotting. Total cell extracts from filaments grown in Co2+/Zn2+ deprived medium (BG-11-Co/Zn) were separated in duplicate by SDS-PAGE, electrotransferred, and challenged with anti-FurA antiserum. Molecular weight is indicated. (C) furA expression in mid-log phase cultures of wild-type and AGcoaRFurA strains growing with and without Co2+/Zn2+, according to sqRT-PCR. Expression of the housekeeping gene rnpB is also shown.
To test the coaR-PcoaT::furA fusion strain, we grew it in standard BG-11 medium to mid-log phase (3–5 μg Chl/ml), the filaments were subsequently washed with BG-11 medium without Co2+/Zn2+ (BG-11-Co/Zn), and re-suspended in the same Co2+/Zn2+-free medium with further incubation for 48 h. Under those conditions, the furA expression in the AGcoaRFurA strain was completely suppressed according to western blotting (Fig 1B). Expression of furA by the strain AGcoaRFurA when grown in the standard BG-11 medium, which contains 0.4 μM of cobalt and 0.8 μM of zinc [33], was quite similar to the wild-type Anabaena sp. PCC 7120 (Fig 1C).
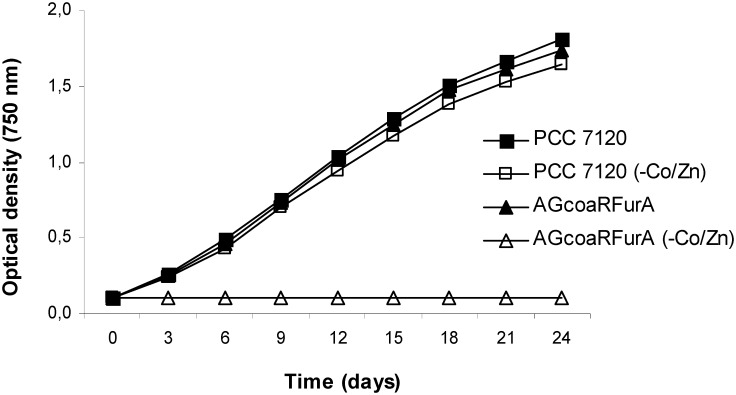
To evaluate the essentiality of FurA for the Anabaena sp. physiology, the strains wild-type Anabaena sp. PCC 7120 and Anabaena sp. AGcoaRFurA were photoautotrophically grown in BG-11 medium solidified with 1% Noble agar (Difco). Cultures were incubated under continuous white light illumination (20 μE/m2s) and controlled temperature of 30°C. Under these conditions, both strains grew abundantly, forming a confluent grow on the agar in 10–14 days (Figures A1 and A3 in S1 File). Both strains were subsequently subcultured in solidified BG-11 medium without Co2+ and Zn2+. As shown in Figures A2 and A4 in S1 File, elimination of both metals to the medium impaired growth of the coaR-PcoaT::furA fusion strain, suggesting that normal expression of FurA results essential for the growth of Anabaena sp. under standard culture conditions. Similar results were observed in liquid BG-11 medium (Fig 2). Since furA appears to have orthologs sharing ≥ 80% protein sequence identity in more than 10 cyanobacterial families and 25 genera (Table A in S1 File), this metalloregulator constitutes an essential well-conserved protein in cyanobacteria.
Fig 2. Expression of FurA results essential to the growth of Anabaena sp. under standard culture conditions.
Growth curves of the coaR-PcoaT::furA fusion strain Anabaena sp. AGcoaRFurA and the wild-type strain Anabaena sp. PCC 7120 in the presence or absence of Co2+/Zn2+.
Turning off furA induces substantial changes in the Anabaena sp. transcriptome
To examine the global impact of FurA in the Anabaena sp. physiology, comparative transcriptomic analysis was performed for the AGcoaRFurA and wild-type PCC 7120 strains in Co2+/Zn2+ restricted cultures. In total, 2089 genes (approximately 33% of the PCC 7120 genome) exhibited differential expression after turning off furA (at least 2-fold difference). The coaR-PcoaT::furA fusion strain showed a 4.4-fold decrease in expression of furA comparing with wild-type strain under the same culture conditions. Depletion of FurA levels resulted in the increased expression of 494 genes and the decreased expression of 1595 genes (Table B in S1 File). RPKM values and fold change for other ORF of the chromosome and six natural plasmids of Anabaena sp. are presented in an additional file (Table C in S1 File).
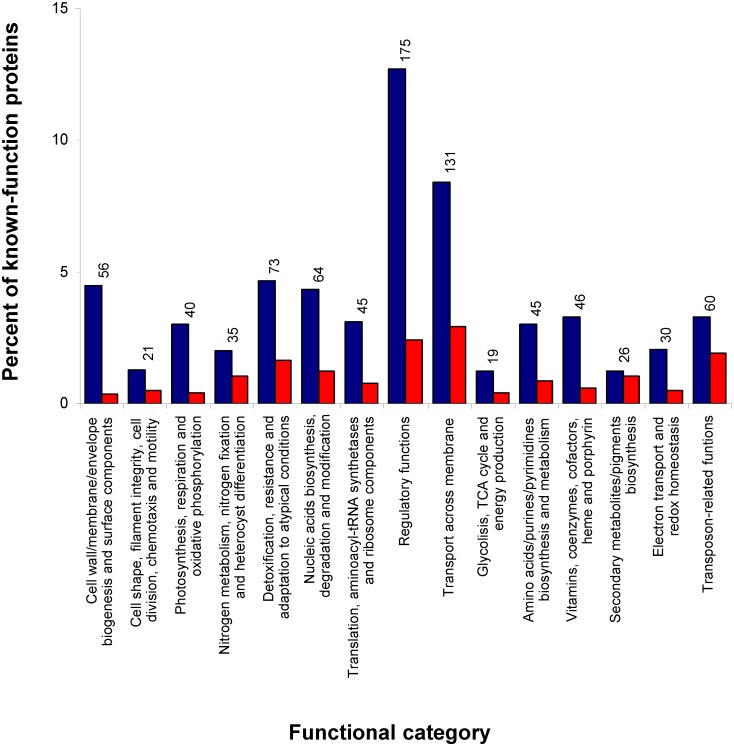
Among the 1155 genes with defined functions, two functional categories were highly enriched with differentially expressed products: (1) genes involved in regulatory functions, and (2) genes related to transport across membrane (Fig 3). Beyond those well-recognized FurA targets belonging to iron homeostasis, several genes involved in less studied Fur coordinated processes such as photosynthesis, detoxification, light sensing and response, exopolysaccharide biosynthesis, chlorophyll catabolism or transposon related functions appeared differentially expressed after depletion of FurA levels.
Fig 3. Distribution of known-function genes which exhibited differential expression after turning off furA in the coaR-PcoaT::furA fusion strain AGcoaRFurA.
Blue columns represent the percent of genes down-regulated of the total (n = 1155) genes with defined function, distributed according to their functional category. Red columns represent the percent of genes up-regulated. The numbers over the columns indicate the total amount of genes by functional category showing differential expression.
FurA controls the expression of novel direct targets
To determine whether the expression of genes identified by RNA-seq was directly regulated by FurA, we investigate the ability of recombinant FurA to bind in vitro the promoter regions of selected 30 up-regulated (Table 1) and 30 down-regulated genes (Table 2) in the coaR-PcoaT::furA fusion strain, belonging to more than 20 different functional categories. Since the analysis of those well-recognized FurA putative targets involved in iron homeostasis has been extensively studied in previous works [15, 17, 20], we made special emphasis in the identification of putative novel targets involved in distinctive processes of cyanobacteria as microbial group, such as oxygenic photosynthesis, biosynthesis and catabolism of chlorophyll as well as mechanisms of light sensing and response. We also investigate the implication of FurA in less studied cyanobacterial processes such as exopolysaccharide biosynthesis, transposon related function, biosynthesis of the Fe-S cluster, among others. Electrophoretic mobility shift assays (EMSA) were carried out using 300- to 400-bp-DNA fragments located immediately upstream of the translational start point, corresponding to the promoter regions of each selected gene. In the case of large promoters, the entire promoter region was analyzed as several fragments of 300- to 400-bp in independent experiments. To confirm the specificity of bindings, all assays included the promoter region of the nifJ gene as non-specific competitor DNA [23]. The impact of metal co-repressor and reducing conditions on the in vitro affinity of recombinant FurA to its putative targets was evaluated in all assays. The specific binding of FurA to the promoter region of its own gene was used as positive control [34], while promoters of Anabaena sp. superoxide dismutases genes sodA and sodB were included as negative controls [17].
Table 1. Subset of selected genes showing a ≥2-fold change increase of expression in the furA-turning off strain AGcoaRFurA.
| ORFa | Symbolb | Protein descriptionb | Functional category | Fold change | EMSAa |
|---|---|---|---|---|---|
| all1847 | CheB methylesterase | chemotaxis, cell motility | 6.31 | - | |
| all7348 | lethal leaf-spot 1 homolog | chlorophyll catabolism, developmentally programmed death | 4.73 | + | |
| alr5007 | similar to cell death suppressor protein, Lls1 | chlorophyll catabolism, developmentally programmed death | 3.15 | - | |
| all2097 | cell death suppressor protein | chlorophyll catabolism, developmentally programmed death | 3.15 | - | |
| alr3195 | glutathione S-transferase | detoxification, adaptations and atypical conditions | 6.31 | - | |
| alr0799 | glutaredoxin-related protein | detoxification, adaptations and atypical conditions | 3.94 | + | |
| alr7354 | glutathione S-transferase | detoxification, adaptations and atypical conditions | 2.60 | - | |
| all0177 | flv1b | heterocyst-specific flavodiiron protein, Flv1 | detoxification, adaptations and atypical conditions | 2.37 | - |
| asl4146 | srxA | sulfiredoxin | detoxification, adaptations and atypical conditions | 4.18 | + |
| all5185 | mercuric reductase | detoxification, adaptations and atypical conditions | 3.35 | + | |
| alr3090 | katB | Mn-catalase | detoxification, adaptations and atypical conditions | 2.37 | + |
| alr3100 | holliday junction resolvase, YqgF | DNA replication, recombination and repair | 4.73 | - | |
| all3135 | exoV | succinoglycan biosynthesis ketolase, ExoV | exopolysaccharide biosynthesis | 6.31 | + |
| alr3513 | Fe-S metabolism associated, SufE | Fe-S cluster biosynthesis | 7.88 | - | |
| all2566 | gap1 | glyceraldehyde-3-phosphate dehydrogenase | glycolisis | 4.73 | - |
| alr3356 | similar to phytochrome | light sensing and response | 3.94 | + | |
| alr0034 | menC | O-succinylbenzoic acid synthase, MenC | menaquinone biosynthesis | 6.31 | - |
| asr0941 | psbX | photosystem II protein, PsbX | photosynthesis | 4.60 | - |
| asr4517 | nblA | phycobilisome degradation protein, NblA | phycobilisome and phycobiliproteins | 2.37 | + |
| alr3707 | pcyA | phycocyanobilin:ferredoxin oxidoreductase | phytochromobilin biosynthesis | 2.01 | - |
| alr2945 | probable orotate phosphoribosyltransferase | pyrimidine biosynthesis | 3.68 | - | |
| all1681 | aspartate carbamoyltransferase | pyrimidine biosynthesis | 6.31 | - | |
| alr1010 | ccbP | calcium-binding protein, CcbP | regulatory functions | 2.37 | + |
| all1649 | similar to polyketide synthase | siderophore/cyanotoxin biosynthesis | 9.46 | - | |
| all1647 | peptide synthetase | siderophore/cyanotoxin biosynthesis | 3.94 | - | |
| alr2679 | polyketide synthase | siderophore/cyanotoxin biosynthesis | 3.15 | + | |
| alr2680 | polyketide synthase | siderophore/cyanotoxin biosynthesis | 2.15 | + | |
| all4026 | iacT | TonB-dependent receptor | transport across membrane | 3.15 | - |
| asr7385 | transposase | transposon related function | 3.17 | + | |
| alr7386 | transposase | transposon related function | 3.15 | + |
a FurA direct target genes according to EMSA results are indicated in bold letters.
b Gene symbol and protein description according to the cyanobacteria genome database CyanoBase (http://genome.microbedb.jp/cyanobase).
Table 2. Subset of selected genes showing a ≥2-fold change decrease of expression in the furA-turning off strain AGcoaRFurA.
| ORFa | Symbolb | Protein descriptionb | Functional category | Fold change | EMSAa |
|---|---|---|---|---|---|
| alr2495 | sufS | cysteine desulphurase, SufS | biosyntesis of Fe-S cluster | -2.78 | + |
| alr1358 | Mg-protoporphyrin IX monomethyl ester cyclase | chlorophyll biosynthesis | -2.22 | - | |
| alr1105 | arsenate reductase | detoxification, adaptations and atypical conditions | -7.08 | - | |
| all3033 | arsenical pump membrane protein | detoxification, adaptations and atypical conditions | -2.38 | - | |
| asr1102 | arsenical-resistance protein ACR3, efflux transporter | detoxification, adaptations and atypical conditions | -2.35 | - | |
| all1291 | cynS | cyanate lyase, CynS | detoxification, adaptations and atypical conditions | -5.56 | - |
| all4148 | petF | ferredoxin I | electron transfer agents in biological redox reactions | -2.87 | - |
| asl0884 | probable ferredoxin [2Fe-2S] | electron transfer agents in biological redox reactions | -5.90 | - | |
| asl2914 | similar to ferredoxin | electron transfer agents in biological redox reactions | -6.23 | - | |
| all3735 | fructose-bisphosphate aldolase class I | glycolisis, gluconeogenesis and the Calvin cycle | -4.15 | - | |
| alr2392 | fraC | filament integrity proteína, FraC | integrity of cell junctions | -2.75 | - |
| asr3846 | psbF | cytochrome b559 beta subunit | photosynthesis | -2.54 | - |
| alr3422 | petD | cytochrome b6/f-complex, apocytochrome subunit 4, PetD | photosynthesis | -4.60 | - |
| all1512 | petC | cytochrome b6/f-complex, iron-sulfur proteína, PetC | photosynthesis | -3.17 | - |
| all1365 | cytM | cytochrome, CytM | photosynthesis | -4.67 | - |
| asr1283 | psaX | photosystem I 4.8K protein, PsaX | photosynthesis | -2.54 | - |
| asr3848 | psbJ | photosystem II protein, PsbJ | photosynthesis | -2.85 | - |
| asl0846 | psbH | photosystem II protein, PsbH | photosynthesis | -2.83 | - |
| asl0885 | psbK | photosystem II protein, PsbK | photosynthesis | -3.07 | - |
| asr0847 | psbN | photosystem II protein, PsbN | photosynthesis | -5.31 | - |
| all0801 | psbW | photosystem II protein, PsbW | photosynthesis | -7.30 | - |
| all0450 | apcA | allophycocyanin alpha subunit, ApcA | phycobilisomes and phycobiliproteins | -5.08 | - |
| all3653 | apcD | allophycocyanin B alpha chain, ApcD | phycobilisomes and phycobiliproteins | -2.22 | - |
| asr8504 | nblA | phycobilisome degradation protein, NblA plasmidic | phycobilisomes and phycobiliproteins | -2.54 | - |
| alr3814 | nblB | phycocyanin alpha phycocyanobilin lyase, NblB | phycobilisomes and phycobiliproteins | -3.49 | - |
| all3549 | similar to phycoerythrobilin lyase subunit (cpeF) | phycobilisomes and phycobiliproteins | -2.19 | - | |
| alr3147 | phosphoenolpyruvate synthase | pyruvate metabolism and carboxylate cycle | -6.98 | - | |
| alr3397 | phosphoenolpyruvate synthase | pyruvate metabolism and carboxylate cycle | -5.00 | - | |
| alr4908 | lexA | SOS function regulatory protein, LexA | regulatory functions | -2.54 | - |
| alr4686 | cytochrome P450, germacrene A hydroxylase | sesquiterpene biosynthesis | -4.87 | + |
a FurA direct target genes according to EMSA results are indicated in bold letters.
b Gene symbol and protein description according to the cyanobacteria genome database CyanoBase (http://genome.microbedb.jp/cyanobase).
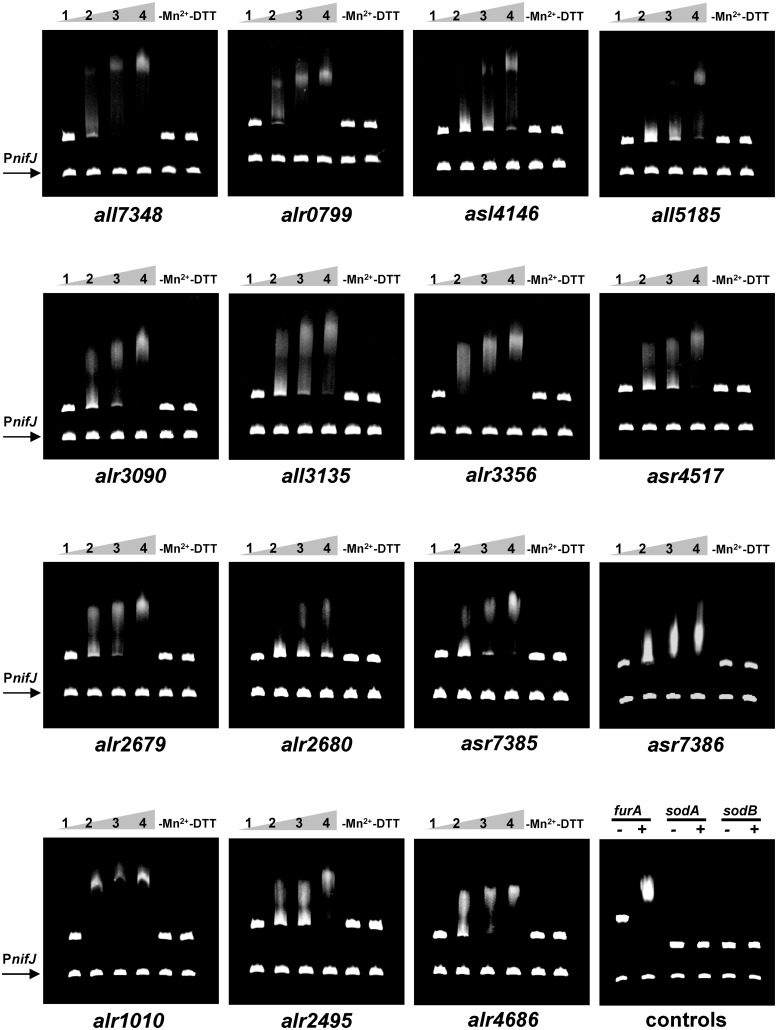
As shown in Fig 4, the EMSA experiments demonstrated that FurA specifically bound in vitro to the promoter regions of at least 15 of the selected 60 differently expressed genes. All the tested DNA fragments were shifted in the presence of up to 700 nM FurA in a dose-dependent manner, whereas the same concentrations of the regulator were unable to shift either the non-specific competitor or both negative controls. As occurs with all so far described FurA targets, the in vitro specific binding of the regulator to the operator regions of these novel target genes was strongly dependent on the presence of divalent metal ions and reducing conditions.
Fig 4. Electrophoretic mobility shift assays showing the ability of FurA to bind in vitro the promoter regions of novel direct target genes.
DNA fragments free (1) or mixed with recombinant FurA protein at concentration of 300 nM (2), 500 nM (3) and 700 nM (4) in the presence of Mn2+ and DTT were separated on a 4% PAGE. The impact of the metal co-regulator (removing Mn2+/adding EDTA) and reducing conditions (removing DTT) on the in vitro affinity of FurA (700 nM) to each target are also showed. The promoter region of nifJ gene was used as non-specific competitor DNA in all assays. Binding of FurA (700 nM) to its own promoter was included as positive controls, while promoter regions of superoxide dismutases genes sodA and sodB were used as negative controls.
Among the novel direct FurA targets, we found genes related to detoxification and protection against oxidative stress like the sulfiredoxin SrxA (Asl4146) and the Mn-catalase Alr3090, genes involved in siderophore/cyanotoxin biosynthesis like the polyketide synthases Alr2679 and Alr2680, and genes involved in transposon-related functions such as transposasas Asr7385 and Alr7386. Interestingly, FurA specifically bound to the promoter regions of the calcium-binding protein CcbP (Alr1010), the phycobilisome degradation protein NblA (Asr4517), and the cysteine desulphurase SufS (Alr2495). Thirteen of the 15 novel FurA direct targets corresponding to genes up-regulated in the coaR-PcoaT::furA fusion strain (Table 1).
FurA might function not only as a repressor, but also as an activator of gene expression
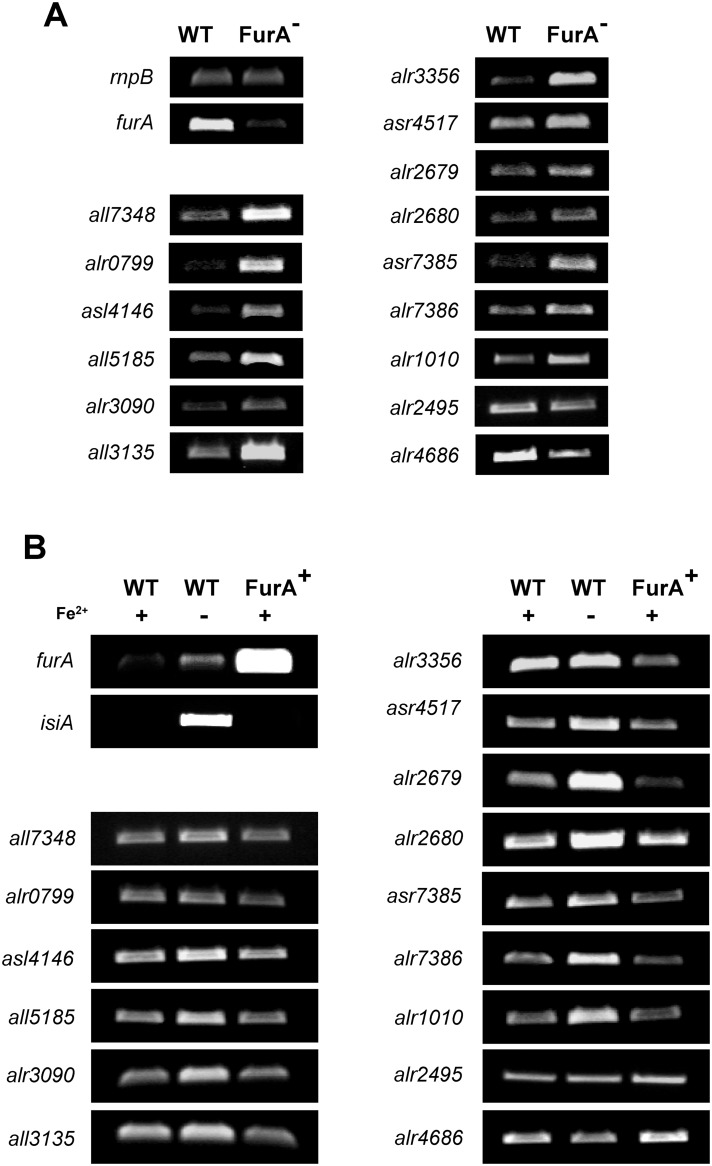
The RNA-seq data corresponding to the 15 novel FurA direct targets were validated in a second experiment by semi-quantitative reverse transcription-PCR (sqRT-PCR). In order to obtain accurate determinations, each measure was performed at the early exponential phase of PCR. The housekeeping gene rnpB was included in all RT-PCR analyses to ensure that equivalent amounts of total RNA were being used in all reactions. As shown in Fig 5A and Figure B in S1 File, the levels of transcripts of all selected genes were in correspondence with those previously obtained by RNA-seq. Thus, under a FurA depleted background, most of the novel FurA direct targets appeared up-regulated, suggesting a direct repression of FurA under the expression of those genes. By contrast, the genes alr2495 and alr4686 were down-regulated, which could suggest a direct activating role of FurA.
Fig 5. Semi-quantitative RT-PCR analyses showing the impact of FurA depletion, FurA overexpression and iron deprivation on the transcriptional pattern of novel FurA targets.
(A) Total RNA from the wild-type strain PCC 7120 (WT) and the coaR-PcoaT::furA fusion strain AGcoaRFurA (FurA-) were isolated from cells grown in Co2+/Zn2+ deprived medium (BG-11-Co/Zn). (B) Total RNA from the wild-type strain PCC 7120 (WT) and the furA overexpressing strain AG2770FurA (FurA+) were isolated from cells grown in standard BG-11 medium (+Fe2+) or iron deprived medium BG-11-Fe (-Fe2+). Housekeeping gene rnpB was used as control. Determinations for each gene were performed in the early exponential phase of PCR. Expression analyses of genes furA and isiA were included as controls of experimental conditions. All determinations were performed three times with independent biological samples, and the relevant portion of a representative gel is shown for each gene.
To further investigate the role of FurA on the expression of this novel targets, we analyse the transcriptional patterns observed to all of these genes under a FurA overexpression background. Wild-type strain PCC 7120 and the furA overexpressing strain AG2770FurA were grown in BG-11 medium under standard growth conditions to mid-log phase, in triplicate cultures. Total RNA was extracted and analysed by sqRT-PCR. The strain AG2770FurA contains a shuttle vector with a copy of gene furA under the control of the petE promoter [17]. This system allows increasing the expression of FurA in almost 30 times, respecting to the wild-type parental strain PCC 7120 [20]. As shown in Fig 5B and Figure C in S1 File, under a FurA overexpression background most of genes appeared down-regulated, while genes alr2495 and alr4686 showed and increase in transcript levels, corroborating a direct activating role of FurA on the expression of these two genes.
In addition, we evaluate the influence of metal co-regulator in the transcription of all novel direct targets. Mid-log growing filaments of the wild-type strain PCC 7120 were exposed to iron restricted conditions as described in Materials and Methods. Transcriptional patterns were compared with those obtained from both the wild-type and the furA-overexpressing strain under iron-replete conditions (Fig 5B and Figures D and E in S1 File). As previously observed in EMSA analyses, metal co-regulator appeared essential for a proper regulation of FurA in all of genes analysed. Under iron-restricted conditions, the regulatory activity of FurA was affected in all cases. In genes down-regulated, the transcript levels appeared slightly increased, while in FurA activated targets like alr2495 and alr4686 the restriction of the co-regulator led to a decrease in gene expression. The results demonstrated that the co-regulator appeared to be essential for FurA activity in both cases, as repressor and as activator of gene expression.
Discussion
FurA is the master regulator of iron homeostasis in Anabaena sp. PCC 7120 [15, 17, 20]. The goal of the present study was to discern novel regulatory roles of FurA in cyanobacteria beyond those well-recognized targets involved in iron metabolism. The essential role of FurA in the physiology of Anabaena sp. impairs classical genetic approaches to study Fur regulatory functions. Among others, several in vitro, in silico, and in vivo analyses on Anabaena sp. furA-overexpressing strains have been successfully used to expand our knowledge of the FurA regulon [15–17, 20, 22–24]. To date, the efforts to inactivate furA in Anabaena sp. and other cyanobacteria species had resulted in the creation of heteroallelic mutants expressing mostly the wild-type fur gene [29, 30]. Here, we constructed a coaR-PcoaT::furA fusion strain by the replacement of natural furA promoter in the Anabaena sp. chromosomes with the Co2+/Zn2+ inducible coaT promoter from Synechocystis sp. PCC 6803 [32]. This approach allowed us to selectively regulate the furA expression to effectively test the FurA essentiality. By growing the cells with Co2+/Zn2+ to allow the synthesis of FurA and subsequently removing Co2+/Zn2+ from the medium to shut off furA expression, we could simulate the furA null phenotype, or at least, allow a substantial depletion of FurA expression. Wild-type Anabaena sp. PCC 7120 was simultaneously grown under the same Co2+/Zn2+ restricted conditions in all analyses as control of the natural response of the cyanobacterium to this nutrient limitation.
Depletion of FurA expression in the coaR-PcoaT::furA fusion strain impaired photoautotrophic growth under standard culture conditions. Similar findings were observed in both solidified and liquid media. As we could note in a subsequent genome-wide transcriptome analysis, the depletion of FurA levels induced substantial changes in the pattern expression of a plethora of genes and operons involved in many essential physiological processes including photosynthesis, respiration, detoxification and gene regulation. The results led to conclude that the proper expression of this highly conserved global regulator appears to be essential for the cyanobacterial physiology under standard culture conditions.
Turning off furA induced differential expression in approximately 33% of the Anabaena sp. genome, which could indicate the total effect of both direct and indirect regulation of FurA. Curiously, the depletion of FurA levels, which mainly acts as a transcriptional repressor, mostly caused down-regulation of gene expression. This was the case of main genes involved in iron uptake (Table D in S1 File). Previous reports have shown that, as a general trend, the transcription levels of the main components of the iron transport system in Anabaena is repressed under iron sufficient condition, while iron limitation causes up-regulation of this machinery [17, 35–37]. In heterotrophic bacteria, fur mutants usually exhibit derepression of iron uptake mechanisms, even under iron sufficient condition [38–40]. Our results seem to suggest that in addition to FurA, other yet unidentified iron-responsive regulators are likely modulating iron homeostasis in Anabaena sp. PCC 7120, as occurs in Synechocystis sp. PCC 6803, where the PfsR regulator plays a crucial role in this process [41]. It is also remarkable that differential expressed products were mainly enriched by genes involved in regulatory functions, including dozens of other transcriptional regulators, two-component signal transduction systems, and other regulatory players. Hence, the effect of FurA depletion observed in the coaR-PcoaT::furA fusion strain goes beyond the direct transcripcional regulation exerted by FurA and reflects both the range and complexity of the FurA regulatory network.
The RNA-seq data showed differential expression of numerous genes that had not been previously identified as iron-responsive or FurA targets in cyanobacteria, including several genes related to important little studied processes such as chlorophyll catabolism and programmed death, phycobilisome degradation, exopolysaccharide biosynthesis, detoxification, light sensing and response, biosynthesis of Fe-S cluster, among others. Thus, among all differential expressed genes with known function, we selected 60 putative FurA targets and determine whether the expression of these genes was directly regulated by FurA according to EMSA. Fifteen of these further studied genes resulted FurA novel direct targets.
As expected, most of the novel direct targets described here appeared to be up-regulated under a FurA depletion background, while diminished their expression under a furA- overexpressing phenotype, suggesting a repressing function of FurA on the transcription of these genes. Among these FurA repressed targets, we found 4 genes directly involved in detoxification and defences against oxidative stress, including the sulfiredoxin SrxA (Asl4146) [42], the Mn-catalase KatB (Alr3090) [43], the glutaredoxin-related protein Alr0799, and the mercuric reductase All5185.
The intimate relationship between iron metabolism and oxidative stress has been extensively recognized in bacteria [2, 44, 45]. In E. coli, the expression of Fur is modulated by oxidative stress response regulators, and Fur directly or indirectly regulates the expression of antioxidant enzymes. Our findings further support the role of FurA as a key player in the regulation of defences against oxidative stress in Anabaena sp. PCC 7120. Previous analyses have identified other FurA targets belonging to the first line of defences against oxidative stress, including the peroxiredoxins Alr4641 and All1541 [24] and the flavodiiron protein Flv3 [20]. To date, all FurA targets involved in the response to oxidative stress appeared repressed by the regulator. In all cases, the modulation of gene expression depended of both, the presence of metal co-regulator and reducing conditions. Taking together, the results suggest that the antioxidant defences regulated by FurA in Anabaena sp. PCC 7120 are repressed by the regulator under iron-sufficient conditions, and those become up-regulated under iron deprivation. In fact, iron starvation leads to significant increase in ROS and induces oxidative stress in cyanobacteria [45], and this condition is the most abundant in nature. Other ferric uptake regulator paralogues described in Anabaena sp., like FurB or FurC, have also been associated to defences against oxidative stress [19, 36, 46]. Since the three Fur homologues resulted up-regulated in minor or major grade as a response to damage by ROS [19, 36], we could speculate the combined action of these regulators in the generation of a concerted response to oxidative stress in cyanobacteria. The implication of other Fur orthologues in modulation of gene expression as response of oxidative stress signals have been previously documented [47–50].
Our RNA-seq analysis and subsequent EMSA experiments led to identify an important novel FurA direct target, the phycobilisome degradation protein NblA (Asr4517) [51]. Phycobilisomes are the major light-harvesting complexes of the photosynthetic apparatus of cyanobacteria [52]. These multi-protein complexes, which can constitute up to 50% of the total cellular protein, are rapidly degraded when the microorganisms are starved for different nutrients, including combined nitrogen [53], sulphur, phosphorus [54], and iron [55], leading to a dramatic bleaching phenomenon known as chlorosis. NblA, a 59 amino acid polypeptide which is induced upon starvation, results essential for phycobilisome degradation [56]. Previous studies have shown that the nblA gene is differentially regulated by iron availability in Synechocystis sp. PCC 6803 [57]. Notably, Anabaena sp. PCC 7120 has two nblA homologues, one on the chromosome (asr4517) and one on plasmid delta (asr8504). Only the chromosomal nblA gene, which is up-regulated upon nitrogen starvation [53], was regulated by FurA.
We have previously documented that FurA influences heterocyst differentiation [20, 22]. Heterocyst development and its pattern formation are developmentally regulated processes, involving the coordinated action of several transcriptional regulators which orchestrated a complex regulatory cascade [58, 59]. Our previous analyses have shown that expression of furA is strongly induced by the global regulator of nitrogen metabolism NtcA in proheterocysts during the first 15 h after nitrogen step-down, remaining stably expressed in mature heterocysts [60]. On the other hand, in vitro, in silico and in vivo analyses have shown that FurA acts as a transcriptional modulator of ntcA [22], abp1 [24], hetC, patA, alr1728 and asr1734 [20]. Here, we identify another FurA direct target which plays a central role in the heterocyst development regulation, the calcium-binding protein CcbP [61]. A rapid increase of intracellular Ca2+ concentration due to the decreased expression of ccbP is observed in proheterocysts as soon as 4 hours after nitrogen deprivation. The ccbP message is down-regulated in differentiating cells and absent in mature heterocysts. It has been hypothesized that a regulatory pathway consisting of HetR, CcbP, and NtcA controls intracellular free calcium during heterocyst development [58]. The results presented here could suggest a role of FurA in repressing the ccbP expression in differentiating cells and mature heterocysts.
Our results underlined the role of FurA in controlling light-dependent signal transduction mechanisms. Sensing light signals and their subsequent transduction is essential for photosynthetic organisms, since it enables them to adapt to variable environmental conditions [62]. In previous studies [20], predicted and functionally validated FurA binding sites were determined in the promoter regions of the Anabaena sensory rhodopsin Asr [63] and the phytochrome-like protein AphC [64]. Here we identified the phytochrome-like protein Alr3356 as a novel FurA direct target. As AphC, the protein Alr3356 is a sensory photoreceptor characterized by the presence of a chromophore-binding GAF domain that is homologous but distinct to the tetrapyrrole-binding GAF domain of the phytochromes, and belongs to the family of cyanobacteriochromes [65, 66]. These findings could indicate that FurA might modulate certain light-dependent regulatory mechanisms based on iron availability or the redox status of cell.
New regulatory roles of FurA were also observed in less studied cyanobacterial processes such as chlorophyll catabolism and exopolysaccharide biosynthesis. Little is known about programmed death in cyanobacteria. In plants, leaf senescence is a genetically programmed process regulated by specific genes [67]. During this process, chlorophyll is degraded to colorless linear tetrapyrroles, termed nonfluorescent chlorophyll catabolites [68]. Exposition to large iron starvation periods could trigger chlorophyll catabolism and programmed death in cyanobacteria via FurA. Further studies will be required to verify this hypothesis.
Similarly, little is known regarding the cyanobacterial exopolysaccharides biosynthetic pathways and regulating factors. Many cyanobacteria strains have polysaccharidic structures surrounding their cells [69]. The ability to synthesize external polysaccharide layers confers the cells protection from unfavourable environmental conditions including dehydration, exposition to high UV radiation and biomineralization. In addition, the presence of negatively charged polysaccharidic layers surrounding cyanobacterial cells may play an important role in the sequestration of metal cations, and in creating a microenvironment enriched in those metals that are essential for cell growth but are present at very low concentrations in some environments. Recent studies demonstrated that cyanobacterial exopolysaccharides play a role in Fe-sorption, which protects Synechocystis sp. PCC 6803 against iron starvation [70]. Thus, in iron-limited environment FurA could modulate the expression of cyanobacterial exopolysaccharide in order to enhance metal sequestration.
Two genes, alr2495 and alr4686, decreased their transcript levels under a FurA depletion background, while appeared up-regulated in the furA-overexpressing strain. These results could indicate that two novel metabolic pathways appeared to be transcriptionally activated by FurA under iron-replete conditions: (1) the biosynthesis of the Fe-S cluster through the cysteine desulphurase SufS (Alr2495), and (2) the biosynthesis of sesquiterpene through the cytochrome P450 (Alr4686). These evidences further support the role of FurA as a dual regulator, acting both as repressor and activator of gene expression. Previous studies have shown other possible examples of direct positive modulation exerted by FurA [15, 20]. Despite Fur-mediated direct activation as been observed in several heterotrophic bacteria [5, 6, 71, 72], little is known about the mechanism or mechanisms by which Fur proteins function to directly activate gene transcription. In Neisseria gonorrhoeae [71], Fur-mediated activation occurs by competition to another repressor for overlapping binding sites, resulting in derepression of transcription. Interestingly, the locations of the Fur boxes in the promoters of these Fur-activated genes were close to the -10 and -35 motifs, which could suggest not only the competition with other repressors, but also the possibility of RNA polymerase recruitment to enhance transcription initiation. When Fur boxes are located far upstream of the transcription start site, Fur might activate gene expression by altering DNA morphology allowing or enhancing RNA polymerase binding [5].
In summary, the results presented here provide new insights into the complex regulatory network orchestrated by the essential cyanobacterial global regulator FurA. RNA-seq based transcriptome analysis employing a selectively regulating furA expression system led to define 15 novel direct iron-dependent transcriptional targets belonging to a variety of functional categories including detoxification and defences against oxidative stress, phycobilisome degradation, chlorophyll catabolism and programmed death, light sensing and response, exopolysaccharide biosynthesis, among others. Our analyses unravel the role of FurA as a global transcriptional regulator in cyanobacteria, acting both as repressor and activator of gene expression. In either case, the in vivo FurA-mediated regulation seems to be dependent of the environmental iron availability as well as the intracellular redox status.
Materials and Methods
Strains and culture conditions
Bacterial strains and plasmids used in this study are described in Table E in S1 File. Wild-type strain Anabaena sp. PCC 7120 and its derivatives were routinely grown in BG-11 medium as described previously [20]. Culture medium was supplemented with neomycin 50 μg ml-1 (Nm) in the case of strains AG2770FurA and AGcoaRFurA. Chlorophyll a (Chl) was determined in methanol extracts [73].
For cobalt/zinc restricted cultures, Anabaena sp. exponentially growing cells (3–5 μg Chl ml-1) from standard BG-11 were harvested by filtration, washed three times with BG-11 medium without Co2+/Zn2+ (BG-11-Co/Zn), re-suspended in the same Co2+/Zn2+-free medium and further grown for 48 h. For iron depletion experiments, exponentially growing filaments from standard BG-11 were collected by filtration, washed three times with BG-11 medium without iron (BG-11-Fe), re-suspended in the same iron-free medium and further grown for 72 h. All the glassware used in the experiments of metal restriction growth was soaked with 6M HCl and extensively rinsed with Milli-Q water to remove residual metal ions.
Escherichia coli strains carrying plasmids were grown in Luria broth supplemented, as appropriate, with 50 μg ml-1 kanamycin (Km), 50 μg ml-1 ampicillin (Amp) and/or 34 μg ml-1 chloramphenicol (Cm).
DNA manipulations
Cyanobacterial genomic DNA was isolated as described previously [74]. Plasmid preparations were performed using the GenElute™ Plasmid Miniprep Kit (Sigma). DNA purification of PCR products was performed using the GFX™ PCR DNA and the Gel Band Purification Kit (GE Healthcare). Standard protocols were used for cloning, E. coli transformation and PCR [75]. The sequences of all oligonucleotides used in plasmid constructions are described in Table F in S1 File.
The suicide plasmid pCoaRFurA, which contains a copy of the gene furA under the control of the Synechocystis sp. Co2+/Zn2+ inducible coaT promoter [32], was constructed in several steps (Figure F in S1 File): (i) The native 2-kb sequence from the upstream region of the furA gene was amplified from the Anabaena sp. PCC 7120 genomic DNA using primers PpetEfurA1 and PpetEfurA2, and cloned into pGEM-T vector (Promega) to generate pCoaR1. (ii) A sequence from upstream of the coaT gene containing the coaT promoter and the coaR regulator gene was amplified from the genomic DNA of Synechocystis sp. PCC 6803 using the primers 2770furA-coaR-P_up and 2770furA-coaR-P_dw. The resulting PCR product was digested with AvaI and NdeI, and inserted between the same restriction sites of vector pAM2770FurA [17] to create pCoaR2. (iii) Plasmid pCoaR3 was generated by moving the coaR-PcoaT::furA fusion fragment from pCoaR2 to pCoaR1. (iv) The PCR product generated with primers PpetEfurA3 and PpetEfurA4, which contained the native 2-kb sequence located immediately downstream of the furA gene in the Anabaena sp. PCC 7120 genome, was cloned into pGEM-T (pCoaR4), excised with BamHI-SacI and ligated into the same sites of pCoaR3 to create pCoaR5. (v) The Km/Nm-resistance transcription unit was amplified from pAM2770FurA [17] and cloned into the AvaI site of pCoaR5 to generate pCoaR6. (vi) Finally, the BglII-SacI fragment from pCoaR6 was ligated into the same site of pRL278 [76] to create the suicide vector pCoaRFurA. The final vector was partially sequenced to ensure that no modifications in the nucleotide sequence occurred during PCR and cloning.
Construction of Anabaena sp. strain AGcoaRFurA
Plasmid pCoaRFurA was transferred to Anabaena sp. PCC 7120 by conjugation according to a previously described method [77]. Triparental mating was carried out using the E. coli conjugal donor strain ED8654, which contains the plasmid pRL443, and the E. coli conjugal helper strain CPB1893 carrying the plasmid pRL623. Since replacement of native furA promoter by the Co2+/Zn2+ inducible coaT promoter in the Anabaena sp. chromosomes occurred by double homologous recombination events, double recombinants were selected by a sacB-mediated method of positive selection [74]. Briefly, several exconjugant clones resistant to neomycin were successively grown in liquid culture before the cyanobacterial filaments were fragmented to mostly single cells using a Branson 1200 ultrasonic cleaner. The cells were subsequently plated on BG-11 containing 5% sucrose to select for cells in which resolution of the plasmid resulted in loss of the conditional lethal gene sacB, which was part of the plasmid. Complete segregation of coaR-PcoaT::furA fusion was confirmed by PCR.
SDS-PAGE and Western blotting
Filaments of Anabaena sp. strains from 50 ml mid-log phase cultures were collected by centrifugation, washed with 50 mM Tris-HCl buffer (pH 8.0) and sonicated in an ice-water bath by five 30 s bursts with 30 s cooling intervals. The resulting crude extracts were centrifuged at 12 000 × g for 5 min at 4°C to remove cell debris, and protein concentration was determined using the BCA™ Protein Assay kit (Thermo Fisher Scientific). For each sample, 30 μg of total protein was loaded and separated by SDS-PAGE 17% polyacrylamide gel, and transferred to a polyvinylidine fluoride membrane (Millipore). Rabbit polyclonal antibodies raised against Anabaena sp. FurA recombinant protein were used, and the blot was visualized with an Universal Hood Image Analyser (Bio-Rad).
RNA sequencing and data analysis
Total RNA from Co2+/Zn2+ restricted cultures of Anabaena sp. wild-type and AGcoaRFurA treated as described previously (see Strains and culture conditions) were extracted in triplicate using the RNeasy Mini Kit (Qiagen). The triplicate samples of each strain were mixed for RNA sequencing. Ribosomal RNA was removed using the RiboMinus Transcriptome Isolation Kit (Thermo Fisher Scientific). RNA sequencing was performed on the Illumina Miseq desktop sequencer (Illumina) at STAB VIDA, Lda (Lisbon, Portugal), following the instructions of the manufacturer.
A total of 11,124,194 reads were obtained for both Anabaena sp. strains. The data were deposited in the NCBI Short Read Archive under accession SRP066361. Details about the mapping statistics of the individual libraries are provided in Table G in S1 File. Percents of Q30 bases were used as quality criteria. Short reads were aligned against the complete genome of Anabaena sp. PCC 7120 and plasmids pCC7120alpha, pCC7120beta, pCC7120gamma, pCC7120delta, pCC7120epsilon and pCC7120zeta (GenBank accession codes: NC_003272.1, NC_003276.1, NC_003240.1, NC_003267.1, NC_003273.1, NC_003270.1 and NC_003241.1, respectively) using Bow tie [78], allowing a maximum of three mismatches within the first 50 bases. The uniquely mapped reads were collected and analyzed with the DEG-seq package [79] to identify differentially expressed genes. The results of gene transcription were based in the fold-changes of RPKM (Reads Per Kilobase of transcript per Million mapped reads) [80], similarly to other previously published studies [81]. The significance of the expression was calculated using a proportions-based test [82], applicable in situations where the data sample consists of counts of a number of 'types' of data. Expression data were normalized against the housekeeping gene ilvD (alr2771) [83].
Electrophoretic mobility shift assays
Recombinant Anabaena sp. PCC 7120 FurA protein was expressed in E. coli BL21 (DE3) (EMD Biosciences) and purified according to previously described methods [84]. The promoter regions of each gene of interest were obtained by PCR using the primers described in Table F in S1 File. Electrophoretic mobility shift assays (EMSA) were performed as described previously [34]. Briefly, 120–140 ng of each DNA fragment were mixed with recombinant FurA protein at concentrations of 300, 500 and 700 nM in a 20 μl reaction volume containing 10 mM bis-Tris (pH 7.5), 40 mM potassium chloride, 100 μg/ml bovine serum albumin, 1 mM DTT, 100 μM manganese chloride and 5% glycerol. In some experiments, EDTA was added to a final concentration of 200 μM. To ensure the specificity of EMSA, the promoter region of Anabaena sp. nifJ (alr1911) gene was included as non-specific competitor DNA in all assays [17] The amount of competitor DNA (90–140 ng) was optimized in previous assays in order to detect unspecific binding of FurA without great molar excess. Mixtures were incubated at room temperature for 20 min, and subsequently separated on 4% non-denaturing polyacrylamide gels in running buffer (25 mM Tris, 190 mM glycine) at 90 V. Gels were stained with SYBR® Safe DNA gel stain (Invitrogen) and processed with a Gel Doc 2000 Image Analyzer (Bio-Rad).
Semi-quantitative reverse transcription-PCR
Samples of 1 μg RNA were heated at 85°C for 10 min and used as templates for the first-strand cDNA synthesis. Residual DNA in RNA preparations was eliminated by digestion with RNase-free DNase I (Roche). The absence of DNA was checked by PCR. Reverse transcription was carried out using SuperScript retrotranscriptase (Invitrogen) in a 20 μl reaction volume containing 150 ng of random primers (Invitrogen), 1 mM dNTP mix (GE Healthcare) and 10 mM DTT. The sequences of the specific primers used for semi-quantitative reverse transcription-PCR (sqRT-PCR) reactions are defined in Table F in S1 File. Housekeeping gene rnpB [17] was used as a control to compensate for variations in the input of RNA amounts and normalize the results. Exponential phase of PCR for each gene was determined by measuring the amount of PCR product at different time intervals. For the final results, 20–23 cycles at the early exponential phase were used in all genes analyzed. The PCR products were resolved by electrophoresis in 1% agarose gels, stained with ethidium bromide and analyzed using a Gel Doc 2000 Image Analyzer (Bio-Rad).
Relative induction ratio for each gene was calculated as the average of ratios between signals observed in two environmental conditions of interest in three independent determinations. Signal assigned to each gene corresponded to the intensity of its DNA band in the agarose gel stained with ethidium bromide normalized to the signal observed for the housekeeping gene rnpB in each strain.
Supporting Information
Figure A: Expression of FurA results essential to the growth of Anabaena sp. under standard culture conditions. Figure B: Fold changes in the expression of FurA direct targets in the furA-turning off strain Anabaena sp. AGcoaRFurA after cobalt/zinc deprivation as compared with the wild-type PCC 7120 strain under the same growth condition, as result of semi-quantitative RT-PCR analyses. Figure C: Fold changes in the expression of FurA direct targets in the furA-overexpressing strain Anabaena sp. AG2770FurA under iron-replete condition (BG-11 medium) as compared with the wild-type PCC 7120 strain under the same growth condition, as result of semi-quantitative RT-PCR analyses. Figure D: Fold changes in the expression of FurA direct targets in the wild-type strain Anabaena sp. PCC 7120 after iron deprivation as compared with the same strain under iron-replete condition (BG-11 medium), as result of semi-quantitative RT-PCR analyses. Figure E: Fold changes in the expression of FurA direct targets in the furA-overexpressing strain Anabaena sp. AG2770FurA under iron-replete condition (BG-11 medium) as compared with the wild-type PCC 7120 strain grown under iron deprivation, as result of semi-quantitative RT-PCR analyses. Figure F: Construction of Anabaena sp. strain AGcoaRFurA. Table A: Protein sequences producing significant alignments (≥80% identity) with FurA from Anabaena sp. PCC 7120. Table B: Complete list of genes showing ≥2-fold change in expression in the furA-turning off strain AGcoaRFurA. Table C: Genes showing <2-fold change in expression in the furA-turning off strain AGcoaRFurA. Table D: Main genes involved in iron uptake showing ≥2-fold change in expression in the furA-turning off strain AGcoaRFurA. Table E: Strains and plasmids used in this study. Table F: Oligonucleotides used in this study. Table G: Summary of sequencing run statistics
(PDF)
Acknowledgments
The authors thank Dr. Vladimir Espinosa Angarica and Dr. Jorge Cebolla for helpful comments, and Violeta Calvo for helping with cyanobacteria strains cultures.
Data Availability
All RNAseq files are available from the Sequence Read archive at NCBI under accession SRP066361 (Bioproject PRJNA301673).
Funding Statement
Support was provided by BFU2012-31458 Ministerio de Economía y Competitividad. Gobierno de España and Fondos FEDER [http://www.mineco.gob.es/portal/site/mineco/] to AG MTB MLP MFF; and B18, Grupo Consolidado Biología structural, Gobierno de Aragón [http://www.aragon.es/] to AG MTB MLP MFF. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Touati D. Iron and oxidative stress in bacteria. Archives of Biochemistry and Biophysics. 2000;373(1):1–6. . [DOI] [PubMed] [Google Scholar]
- 2.Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiology Reviews. 2003;27(2–3):215–37. . [DOI] [PubMed] [Google Scholar]
- 3.Fillat MF. The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Archives of Biochemistry and Biophysics. 2014;546:41–52. 10.1016/j.abb.2014.01.029 [DOI] [PubMed] [Google Scholar]
- 4.Escolar L, Pérez-Martín J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. Journal of Bacteriology. 1999;181(20):6223–9. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teixido L, Carrasco B, Alonso JC, Barbe J, Campoy S. Fur activates the expression of Salmonella enterica pathogenicity island 1 by directly interacting with the hilD operator in vivo and in vitro. PloS One. 2011;6(5):e19711 10.1371/journal.pone.0019711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbreath JJ, West AL, Pich OQ, Carpenter BM, Michel S, Merrell DS. Fur activates expression of the 2-oxoglutarate oxidoreductase genes (oorDABC) in Helicobacter pylori. Journal of Bacteriology. 2012;194(23):6490–7. 10.1128/JB.01226-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delany I, Rappuoli R, Scarlato V. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Molecular Microbiology. 2004;52(4):1081–90. . [DOI] [PubMed] [Google Scholar]
- 8.Dubrac S, Touati D. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. Journal of Bacteriology. 2000;182(13):3802–8. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delany I, Spohn G, Rappuoli R, Scarlato V. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Molecular Microbiology. 2001;42(5):1297–309. . [DOI] [PubMed] [Google Scholar]
- 10.Butcher J, Sarvan S, Brunzelle JS, Couture JF, Stintzi A. Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(25):10047–52. 10.1073/pnas.1118321109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitton BA, Potts M. The ecology of cyanobacteria: their diversity in time and space. Boston: Kluwer Academic; 2000. xvii, 669 p., 32 p. of col. plates p. [Google Scholar]
- 12.Shcolnick S, Summerfield TC, Reytman L, Sherman LA, Keren N. The mechanism of iron homeostasis in the unicellular cyanobacterium Synechocystis sp. PCC 6803 and its relationship to oxidative stress. Plant Physiology. 2009;150(4):2045–56. 10.1104/pp.109.141853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richier S, Macey AI, Pratt NJ, Honey DJ, Moore CM, Bibby TS. Abundances of iron-binding photosynthetic and nitrogen-fixing proteins of Trichodesmium both in culture and in situ from the North Atlantic. PloS One. 2012;7(5):e35571 10.1371/journal.pone.0035571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Latifi A, Ruiz M, Zhang CC. Oxidative stress in cyanobacteria. FEMS Microbiology Reviews. 2009;33(2):258–78. 10.1111/j.1574-6976.2008.00134.x [DOI] [PubMed] [Google Scholar]
- 15.González A, Bes MT, Valladares A, Peleato ML, Fillat MF. FurA is the master regulator of iron homeostasis and modulates the expression of tetrapyrrole biosynthesis genes in Anabaena sp. PCC 7120. Environmental Microbiology. 2012;14(12):3175–87. 10.1111/j.1462-2920.2012.02897.x [DOI] [PubMed] [Google Scholar]
- 16.Hernández JA, Pellicer S, Huang L, Peleato ML, Fillat MF. FurA modulates gene expression of alr3808, a DpsA homologue in Nostoc (Anabaena) sp. PCC 7120. FEBS Letters. 2007;581(7):1351–6. . [DOI] [PubMed] [Google Scholar]
- 17.González A, Bes MT, Barja F, Peleato ML, Fillat MF. Overexpression of FurA in Anabaena sp. PCC 7120 reveals new targets for this regulator involved in photosynthesis, iron uptake and cellular morphology. Plant & Cell Physiology. 2010;51(11):1900–14. . [DOI] [PubMed] [Google Scholar]
- 18.Hernández JA, Bes MT, Fillat MF, Neira JL, Peleato ML. Biochemical analysis of the recombinant Fur (ferric uptake regulator) protein from Anabaena PCC 7119: factors affecting its oligomerization state. Biochemical Journal. 2002;366(Pt 1):315–22. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.López-Gomollón S, Sevilla E, Bes MT, Peleato ML, Fillat MF. New insights into the role of Fur proteins: FurB (All2473) from Anabaena protects DNA and increases cell survival under oxidative stress. Biochemical Journal. 2009;418(1):201–7. 10.1042/BJ20081066 [DOI] [PubMed] [Google Scholar]
- 20.González A, Angarica VE, Sancho J, Fillat MF. The FurA regulon in Anabaena sp. PCC 7120: in silico prediction and experimental validation of novel target genes. Nucleic Acids Research. 2014;42(8):4833–46. 10.1093/nar/gku123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavrrar JL, McIntosh MA. Architecture of a fur binding site: a comparative analysis. Journal of Bacteriology. 2003;185(7):2194–202. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.González A, Valladares A, Peleato ML, Fillat MF. FurA influences heterocyst differentiation in Anabaena sp. PCC 7120. FEBS Letters. 2013;587(16):2682–90. 10.1016/j.febslet.2013.07.007 [DOI] [PubMed] [Google Scholar]
- 23.López-Gomollón S, Hernández JA, Pellicer S, Angarica VE, Peleato ML, Fillat MF. Cross-talk between iron and nitrogen regulatory networks in Anabaena (Nostoc) sp. PCC 7120: identification of overlapping genes in FurA and NtcA regulons. Journal of Molecular Biology. 2007;374(1):267–81. . [DOI] [PubMed] [Google Scholar]
- 24.González A, Bes MT, Peleato ML, Fillat MF. Unravelling the regulatory function of FurA in Anabaena sp. PCC 7120 through 2-D DIGE proteomic analysis. Journal of Proteomics. 2011;74(5):660–71. 10.1016/j.jprot.2011.02.001 [DOI] [PubMed] [Google Scholar]
- 25.Baichoo N, Wang T, Ye R, Helmann JD. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Molecular Microbiology. 2002;45(6):1613–29. . [DOI] [PubMed] [Google Scholar]
- 26.Gao H, Zhou D, Li Y, Guo Z, Han Y, Song Y, et al. The iron-responsive Fur regulon in Yersinia pestis. Journal of Bacteriology. 2008;190(8):3063–75. 10.1128/JB.01910-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McHugh JP, Rodríguez-Quiñones F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, et al. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. The Journal of Biological Chemistry. 2003;278(32):29478–86. . [DOI] [PubMed] [Google Scholar]
- 28.Wan XF, Verberkmoes NC, McCue LA, Stanek D, Connelly H, Hauser LJ, et al. Transcriptomic and proteomic characterization of the Fur modulon in the metal-reducing bacterium Shewanella oneidensis. Journal of Bacteriology. 2004;186(24):8385–400. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernández JA, Muro-Pastor AM, Flores E, Bes MT, Peleato ML, Fillat MF. Identification of a furA cis antisense RNA in the cyanobacterium Anabaena sp. PCC 7120. Journal of Molecular Biology. 2006;355(3):325–34. . [DOI] [PubMed] [Google Scholar]
- 30.Ghassemian M, Straus NA. Fur regulates the expression of iron-stress genes in the cyanobacterium Synechococcus sp. strain PCC 7942. Microbiology (Reading, England). 1996;142 (Pt 6):1469–76. . [DOI] [PubMed] [Google Scholar]
- 31.Hernández JA, Alonso I, Pellicer S, Luisa Peleato M, Cases R, Strasser RJ, et al. Mutants of Anabaena sp. PCC 7120 lacking alr1690 and alpha-furA antisense RNA show a pleiotropic phenotype and altered photosynthetic machinery. Journal of Plant Physiology. 2009. . [DOI] [PubMed] [Google Scholar]
- 32.Peca L, Kos PB, Mate Z, Farsang A, Vass I. Construction of bioluminescent cyanobacterial reporter strains for detection of nickel, cobalt and zinc. FEMS microbiology Letters. 2008;289(2):258–64. 10.1111/j.1574-6968.2008.01393.x [DOI] [PubMed] [Google Scholar]
- 33.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of Cyanobacteria. Journal of General Microbiology. 1979;111:1–61. [Google Scholar]
- 34.Hernández JA, López-Gomollón S, Muro-Pastor A, Valladares A, Bes MT, Peleato ML, et al. Interaction of FurA from Anabaena sp. PCC 7120 with DNA: a reducing environment and the presence of Mn(2+) are positive effectors in the binding to isiB and furA promoters. Biometals. 2006;19(3):259–68. . [DOI] [PubMed] [Google Scholar]
- 35.Stevanovic M, Hahn A, Nicolaisen K, Mirus O, Schleiff E. The components of the putative iron transport system in the cyanobacterium Anabaena sp. PCC 7120. Environmental Microbiology. 2012;14(7):1655–70. 10.1111/j.1462-2920.2011.02619.x [DOI] [PubMed] [Google Scholar]
- 36.Yingping F, Lemeille S, Talla E, Janicki A, Denis Y, Zhang CC, et al. Unravelling the cross-talk between iron starvation and oxidative stress responses highlights the key role of PerR (alr0957) in peroxide signalling in the cyanobacterium Nostoc PCC 7120. Environmental Microbiology Reports. 2014;6(5):468–75. . [DOI] [PubMed] [Google Scholar]
- 37.Stevanovic M, Lehmann C, Schleiff E. The response of the TonB-dependent transport network in Anabaena sp. PCC 7120 to cell density and metal availability. Biometals. 2013;26(4):549–60. 10.1007/s10534-013-9644-0 [DOI] [PubMed] [Google Scholar]
- 38.Ollinger J, Song KB, Antelmann H, Hecker M, Helmann JD. Role of the Fur regulon in iron transport in Bacillus subtilis. Journal of Bacteriology. 2006;188(10):3664–73. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bender KS, Yen HC, Hemme CL, Yang Z, He Z, He Q, et al. Analysis of a ferric uptake regulator (Fur) mutant of Desulfovibrio vulgaris Hildenborough. Applied and Environmental Microbiology. 2007;73(17):5389–400. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y, Harris DP, Luo F, Wu L, Parsons AB, Palumbo AV, et al. Characterization of the Shewanella oneidensis fur gene: roles in iron and acid tolerance response. BMC Genomics. 2008;9 Suppl 1:S11 10.1186/1471-2164-9-S1-S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng D, He Q. PfsR is a key regulator of iron homeostasis in Synechocystis PCC 6803. PloS One. 2014;9(7):e101743 10.1371/journal.pone.0101743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boileau C, Eme L, Brochier-Armanet C, Janicki A, Zhang CC, Latifi A. A eukaryotic-like sulfiredoxin involved in oxidative stress responses and in the reduction of the sulfinic form of 2-Cys peroxiredoxin in the cyanobacterium Anabaena PCC 7120. The New Phytologist. 2011;191(4):1108–18. 10.1111/j.1469-8137.2011.03774.x [DOI] [PubMed] [Google Scholar]
- 43.Bihani SC, Chakravarty D, Ballal A. Purification, crystallization and preliminary crystallographic analysis of KatB, a manganese catalase from Anabaena PCC 7120. Acta Crystallographica. 2013;69(Pt 11):1299–302. 10.1107/S1744309113028017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng M, Doan B, Schneider TD, Storz G. OxyR and SoxRS regulation of fur. Journal of Bacteriology. 1999;181(15):4639–43. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Latifi A, Jeanjean R, Lemeille S, Havaux M, Zhang CC. Iron starvation leads to oxidative stress in Anabaena sp. strain PCC 7120. Journal of Bacteriology. 2005;187(18):6596–8. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sein-Echaluce VC, Gonzalez A, Napolitano M, Luque I, Barja F, Peleato ML, et al. Zur (FurB) is a key factor in the control of the oxidative stress response in Anabaena sp. PCC 7120. Environmental Microbiology. 2015; 10.1111/1462-2920.12628 . [DOI] [PubMed] [Google Scholar]
- 47.Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440(7082):363–7. . [DOI] [PubMed] [Google Scholar]
- 48.Pelliciari S, Vannini A, Roncarati D, Danielli A. The allosteric behavior of Fur mediates oxidative stress signal transduction in Helicobacter pylori. Frontiers in Microbiology. 2015;6:840 10.3389/fmicb.2015.00840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infection & Immunity. 2001;69(6):3744–54. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Da Silva JF, Braz VS, Italiani VC, Marques MV. Fur controls iron homeostasis and oxidative stress defense in the oligotrophic alpha-proteobacterium Caulobacter crescentus. Nucleic Acids Research. 2009;37(14):4812–25. 10.1093/nar/gkp509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bienert R, Baier K, Volkmer R, Lockau W, Heinemann U. Crystal structure of NblA from Anabaena sp. PCC 7120, a small protein playing a key role in phycobilisome degradation. The Journal of Biological Chemistry. 2006;281(8):5216–23. . [DOI] [PubMed] [Google Scholar]
- 52.MacColl R. Cyanobacterial phycobilisomes. Journal of Structural Biology. 1998;124(2–3):311–34. . [DOI] [PubMed] [Google Scholar]
- 53.Baier K, Lehmann H, Stephan DP, Lockau W. NblA is essential for phycobilisome degradation in Anabaena sp. strain PCC 7120 but not for development of functional heterocysts. Microbiology (Reading, England). 2004;150(Pt 8):2739–49. . [DOI] [PubMed] [Google Scholar]
- 54.Richaud C, Zabulon G, Joder A, Thomas JC. Nitrogen or sulfur starvation differentially affects phycobilisome degradation and expression of the nblA gene in Synechocystis strain PCC 6803. Journal of Bacteriology. 2001;183(10):2989–94. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwarz R, Forchhammer K. Acclimation of unicellular cyanobacteria to macronutrient deficiency: emergence of a complex network of cellular responses. Microbiology (Reading, England). 2005;151(Pt 8):2503–14. . [DOI] [PubMed] [Google Scholar]
- 56.Collier JL, Grossman AR. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. The EMBO Journal. 1994;13(5):1039–47. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh AK, Sherman LA. Identification of iron-responsive, differential gene expression in the cyanobacterium Synechocystis sp. strain PCC 6803 with a customized amplification library. Journal of Bacteriology. 2000;182(12):3536–43. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar K, Mella-Herrera RA, Golden JW. Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol. 2010;2(4):a000315 10.1101/cshperspect.a000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flores E, Herrero A. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nature Reviews Microbiology. 2010;8(1):39–50. 10.1038/nrmicro2242 [DOI] [PubMed] [Google Scholar]
- 60.López-Gomollón S, Hernández JA, Wolk CP, Peleato ML, Fillat MF. Expression of furA is modulated by NtcA and strongly enhanced in heterocysts of Anabaena sp. PCC 7120. Microbiology (Reading, England). 2007;153(Pt 1):42–50. . [DOI] [PubMed] [Google Scholar]
- 61.Zhao Y, Shi Y, Zhao W, Huang X, Wang D, Brown N, et al. CcbP, a calcium-binding protein from Anabaena sp. PCC 7120, provides evidence that calcium ions regulate heterocyst differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(16):5744–8. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Montgomery BL. Sensing the light: photoreceptive systems and signal transduction in cyanobacteria. Molecular Microbiology. 2007;64(1):16–27. . [DOI] [PubMed] [Google Scholar]
- 63.Irieda H, Morita T, Maki K, Homma M, Aiba H, Sudo Y. Photo-induced regulation of the chromatic adaptive gene expression by Anabaena sensory rhodopsin. The Journal of Biological Chemistry. 2012;287(39):32485–93. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Okamoto S, Kasahara M, Kamiya A, Nakahira Y, Ohmori M. A phytochrome-like protein AphC triggers the cAMP signaling induced by far-red light in the cyanobacterium Anabaena sp. strain PCC7120. Photochemistry and Photobiology. 2004;80(3):429–33. . [DOI] [PubMed] [Google Scholar]
- 65.Ikeuchi M, Ishizuka T. Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochemical & Photobiological Sciences. 2008;7(10):1159–67. . [DOI] [PubMed] [Google Scholar]
- 66.Ma Q, Hua HH, Chen Y, Liu BB, Kramer AL, Scheer H, et al. A rising tide of blue-absorbing biliprotein photoreceptors: characterization of seven such bilin-binding GAF domains in Nostoc sp. PCC7120. The FEBS Journal. 2012;279(21):4095–108. 10.1111/febs.12003 [DOI] [PubMed] [Google Scholar]
- 67.Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MF, Yariv I, et al. Large-scale identification of leaf senescence-associated genes. Plant Journal. 2003;36(5):629–42. . [DOI] [PubMed] [Google Scholar]
- 68.Pruzinska A, Tanner G, Anders I, Roca M, Hortensteiner S. Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(25):15259–64. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pereira S, Zille A, Micheletti E, Moradas-Ferreira P, De Philippis R, Tamagnini P. Complexity of cyanobacterial exopolysaccharides: composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiology Reviews. 2009;33(5):917–41. 10.1111/j.1574-6976.2009.00183.x [DOI] [PubMed] [Google Scholar]
- 70.Jittawuttipoka T, Planchon M, Spalla O, Benzerara K, Guyot F, Cassier-Chauvat C, et al. Multidisciplinary evidences that Synechocystis PCC 6803 exopolysaccharides operate in cell sedimentation and protection against salt and metal stresses. PloS One. 2013;8(2):e55564 10.1371/journal.pone.0055564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu C, Genco CA. Fur-mediated global regulatory circuits in pathogenic Neisseria species. Journal of Bacteriology. 2012;194(23):6372–81. 10.1128/JB.00262-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Craig SA, Carpenter CD, Mey AR, Wyckoff EE, Payne SM. Positive regulation of the Vibrio cholerae porin OmpT by iron and Fur. Journal of Bacteriology. 2011;193(23):6505–11. 10.1128/JB.05681-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nicolaisen K, Moslavac S, Samborski A, Valdebenito M, Hantke K, Maldener I, et al. Alr0397 is an outer membrane transporter for the siderophore schizokinen in Anabaena sp. strain PCC 7120. Journal of Bacteriology. 2008;190(22):7500–7. 10.1128/JB.01062-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cai YP, Wolk CP. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. Journal of Bacteriology. 1990;172(6):3138–45. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd ed Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 76.Black TA, Cai Y, Wolk CP. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Molecular Microbiology. 1993;9(1):77–84. . [DOI] [PubMed] [Google Scholar]
- 77.Elhai J, Vepritskiy A, Muro-Pastor AM, Flores E, Wolk CP. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. Journal of Bacteriology. 1997;179(6):1998–2005. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10(3):R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11(10):R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5(7):621–8. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 81.Flaherty BL, Van Nieuwerburgh F, Head SR, Golden JW. Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genomics. 2011;12:332 10.1186/1471-2164-12-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baggerly KA, Deng L, Morris JS, Aldaz CM. Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics (Oxford, England). 2003;19(12):1477–83. . [DOI] [PubMed] [Google Scholar]
- 83.Pinto F, Pacheco CC, Ferreira D, Moradas-Ferreira P, Tamagnini P. Selection of suitable reference genes for RT-qPCR analyses in cyanobacteria. PloS One. 2012;7(4):e34983 10.1371/journal.pone.0034983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pellicer S, Bes MT, González A, Neira JL, Luisa Peleato M, Fillat MF. High-recovery one-step purification of the DNA-binding protein Fur by mild guanidinium chloride treatment. Process Biochemistry. 2010;45(2):292–6. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure A: Expression of FurA results essential to the growth of Anabaena sp. under standard culture conditions. Figure B: Fold changes in the expression of FurA direct targets in the furA-turning off strain Anabaena sp. AGcoaRFurA after cobalt/zinc deprivation as compared with the wild-type PCC 7120 strain under the same growth condition, as result of semi-quantitative RT-PCR analyses. Figure C: Fold changes in the expression of FurA direct targets in the furA-overexpressing strain Anabaena sp. AG2770FurA under iron-replete condition (BG-11 medium) as compared with the wild-type PCC 7120 strain under the same growth condition, as result of semi-quantitative RT-PCR analyses. Figure D: Fold changes in the expression of FurA direct targets in the wild-type strain Anabaena sp. PCC 7120 after iron deprivation as compared with the same strain under iron-replete condition (BG-11 medium), as result of semi-quantitative RT-PCR analyses. Figure E: Fold changes in the expression of FurA direct targets in the furA-overexpressing strain Anabaena sp. AG2770FurA under iron-replete condition (BG-11 medium) as compared with the wild-type PCC 7120 strain grown under iron deprivation, as result of semi-quantitative RT-PCR analyses. Figure F: Construction of Anabaena sp. strain AGcoaRFurA. Table A: Protein sequences producing significant alignments (≥80% identity) with FurA from Anabaena sp. PCC 7120. Table B: Complete list of genes showing ≥2-fold change in expression in the furA-turning off strain AGcoaRFurA. Table C: Genes showing <2-fold change in expression in the furA-turning off strain AGcoaRFurA. Table D: Main genes involved in iron uptake showing ≥2-fold change in expression in the furA-turning off strain AGcoaRFurA. Table E: Strains and plasmids used in this study. Table F: Oligonucleotides used in this study. Table G: Summary of sequencing run statistics
(PDF)
Data Availability Statement
All RNAseq files are available from the Sequence Read archive at NCBI under accession SRP066361 (Bioproject PRJNA301673).