Have plants learned to forget stress? This review proposes benefits to forgetfulness and finds key roles for RNA turnover.
Keywords: RNA metabolism, gene silencing, epigenetics, DNA methylation, RNA turnover, abiotic stress, plants, arabidopsis, stress recovery, plant memory
Abstract
Plants grow in dynamic environments where they can be exposed to a multitude of stressful factors, all of which affect their development, yield, and, ultimately, reproductive success. Plants are adept at rapidly acclimating to stressful conditions and are able to further fortify their defenses by retaining memories of stress to enable stronger or more rapid responses should an environmental perturbation recur. Indeed, one mechanism that is often evoked regarding environmental memories is epigenetics. Yet, there are relatively few examples of such memories; neither is there a clear understanding of their duration, considering the plethora of stresses in nature. We propose that this field would benefit from investigations into the processes and mechanisms enabling recovery from stress. An understanding of stress recovery could provide fresh insights into when, how, and why environmental memories are created and regulated. Stress memories may be maladaptive, hindering recovery and affecting development and potential yield. In some circumstances, it may be advantageous for plants to learn to forget. Accordingly, the recovery process entails a balancing act between resetting and memory formation. During recovery, RNA metabolism, posttranscriptional gene silencing, and RNA-directed DNA methylation have the potential to play key roles in resetting the epigenome and transcriptome and in altering memory. Exploration of this emerging area of research is becoming ever more tractable with advances in genomics, phenomics, and high-throughput sequencing methodology that will enable unprecedented profiling of high-resolution stress recovery time series experiments and sampling of large natural populations.
INTRODUCTION
Abiotic stresses, such as temperature, moisture, nutrient limitation, and physical perturbations, require plants to continuously respond to the pressures of nature. Quite remarkably, plants are often able to acclimate their physiological response to challenges, allowing them to cope with these events. Abiotic stress extremes can persist for weeks or months or be part of a variable climate, promoting adaptation through resilience over subsequent generations. Yet, most abiotic stress is transient. By the same token, many transitory stresses are also recurring and indeed may progressively increase in severity. For instance, over the course of a day as the sun tracks through the sky, understory leaves may repeatedly be exposed to light flecks and/or to increasing dehydration. In fact, most environments are dynamic in this way, composed of many fluctuating variables; some may be cyclic and predictable because of diurnal and seasonal variation, whereas other variation may be random and unpredictable.
Prior mild exposure may prime a plant against future stress or promote an acclimated state that may persist until a subsequent exposure. In this way, it is widely accepted that plants have the capacity for what can be described as memory. Despite this capacity for priming or epigenetic memory, in many instances such memories are not formed, yet the reasons for this are poorly understood. Similarly, there is a significant gap in our knowledge regarding the length of memory and the mechanisms for memory dissipation, or “forgetfulness.”
In this review, we seek to emphasize that stress memory or stress priming is likely to be the exception rather than the rule. Dynamic environments characterized by predictable recurring stress could promote plant memory. Despite this, resetting and forgetfulness are likely the overriding strategies used by plants to maximize growth under favorable conditions as soon as they return (Fig. 1). Underpinning this process, RNA metabolism is a key regulatory point, either facilitating recovery to the original state or permitting memory formation in the acclimated state as RNA metabolism can swiftly clear the stress-responsive transcriptome or selectively stabilize transcripts. The RNA decay and quality control machinery are also gatekeepers for the gene silencing pathways and may antagonize this process during stress and recovery. In this review, we examine the growing body of literature describing stress priming and epigenetic mechanisms; we then focus on the potential roles for RNA metabolism as a key and novel avenue of regulation in this complex balancing act.
Fig. 1. Balancing act during recovery from a stress event.
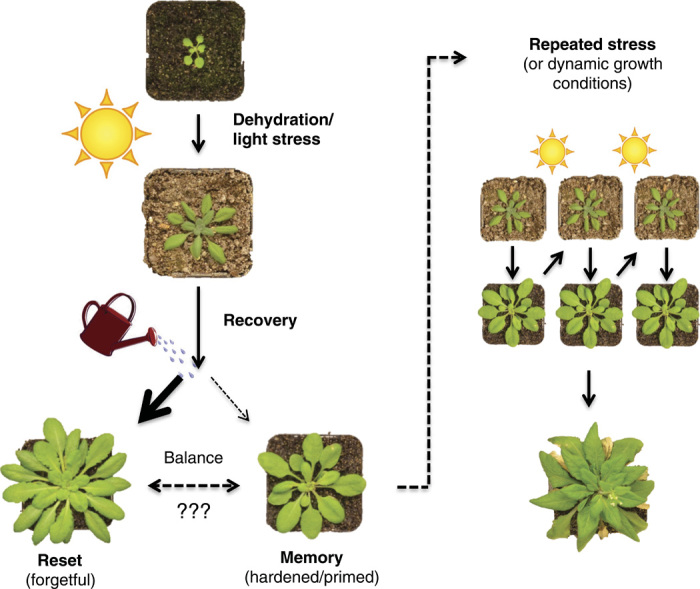
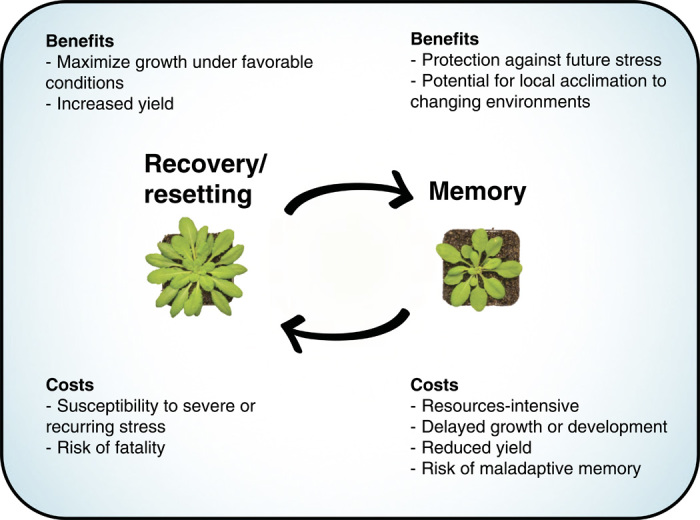
Abiotic stress, such as dehydration, heat stress, and light stress, imposed by the sun during a hot, dry spell activates plant defenses that are essential for survival. However, stress is transient and is followed by a period of recovery during which the plant must strike a balance between investing resources in continued priming versus resetting. We speculate that the predominant response is resetting (forgetfulness). Most transcripts, proteins, metabolites, and physiological responses return to a prestress state. This recovery is likely to be an important evolutionary strategy. Nevertheless, the degree of memory will likely be critical as well, particularly in dynamic environments characterized by a repetitive stress. Thus, plants must balance the potential protection from future stress by forming stress memories with the potential growth and yield advantages of resetting if favorable conditions persist.
PRIMING AND MEMORY IN PLANTS
One possible response of plants from exposure to stress is that they become more resistant to future exposure through the acquisition of memory, a response referred to as hardening, priming, conditioning, or acclimation. Multiple examples of memory in higher plants have been shown across multiple species and have been discussed in detail in past reviews (1–4). Table 1 highlights the potential for stress memory in response to stimuli including drought, salinity, abscisic acid (ABA), methyl jasmonate, excess light, oxidative stress, β-aminobutyric acid (BABA), and cold (2, 5–28).
Table 1. Examples of stress priming in plants.
| Species | Treatment | Outcome | Reference |
| Nicotiana sylvestris | Methyl jasmonate | Rapid nicotine accumulation | (5) |
| Arabidopsis thaliana | ABA | Sensitize light-triggered stomatal opening | (11) |
| Dehydration | Improved water retention | (9, 10) | |
| Excess light | Increased oxidative stress/excess light tolerance | (12, 18) | |
| Osmotic/oxidative stress | Altered Ca2+ signals under osmotic stress | (17, 19) | |
| BABA | Increased systemic acquired resistance; abiotic stress resistance | (15, 28) | |
| SA | Improved heat tolerance | (7) | |
| Cold | Vernalization response | (21) | |
| Zea mays | Dehydration | Improved water retention | (24, 25) |
| SA/BABA | Improved cold tolerance | (16) | |
| Triticum aestivum | Drought | Increased grain fill under drought | (26) |
| Salt | Improved resistance to salt stress | (13) | |
| SA | Increased salinity tolerance | (20) | |
| Conyza bonariensis | Paraquat | Increased oxidative stress recovery | (27) |
| Solanum lycopersicum | Salt | Improved resistance to salt stress | (6) |
| Cucumis sativus | SA/BABA | Improved cold tolerance | (16) |
| Oryza sativa | SA/BABA | Improved cold tolerance | (16) |
| Sinapis alba | SA | Improved heat tolerance | (8) |
There is also evidence that priming can persist between generations, a process referred to as adaptive transgenerational plasticity (4). Likewise, a selection of transgenerational memory events are summarized in Table 2, demonstrating transgenerational effects in relation to short-wavelength radiation (ultraviolet C), flagellin (an elicitor of plant defense), herbivory damage and treatment with jasmonic acid, BABA priming, day length, temperature, heat, and drought (4, 29–38). These results highlight the potential for memories to be passed down to offspring via transgenerational inheritance to increase offspring success.
Table 2. Examples of transgenerational stress priming in plants.
| Species | Priming treatment | Physiological response | Reference |
|
Polygonum persicaria |
Drought | Increased root growth and biomass, and improved drought tolerance (cumulative effect) in progeny |
(110) |
| Low light | Offspring produced more shoot tissue relative to root tissue | (37) | |
|
Arabidopsis thaliana |
BABA | Descendants exhibit biotic stress resistance | (36) |
| Mild heat (30.8°C) | Progeny (F3) displayed improved heat tolerance | (38) | |
| Herbivory damage (Pieris rapae); methyl jasmonate |
Progeny of treated parents displayed improved resistance to herbivory (reduced growth of P. rapae). This was shown to persist after a generation without herbivory |
(33) | |
| Short-wavelength radiation (ultraviolet C); flagellin |
Increased genomic flexibility in the form of increased homologous recombination |
(32) | |
|
Mimulus guttatus |
Herbivory damage (simulated) |
Increased trichome density in progeny | (34) |
| Picea abies | Day length and temperature during seed production |
Determines progeny developmental program | (30, 31, 35) |
|
Raphanus raphanistrum |
Herbivory damage (P. rapae); jasmonic acid |
Enhanced herbivory resistance in progeny of treated parents |
(29) |
|
Solanum lycopersicum |
Herbivory damage (Helicoverpa zea); methyl jasmonate |
Progeny of treated plants demonstrated improved resistance to herbivory |
(33) |
MEMORY MECHANISMS
Plant memory is often characterized by heightened molecular responses upon exposure to a subsequent stress, which can be composed of an enhanced response (for instance, hyperinduction of transcription; Fig. 2A, blue line), a more efficient response, or a more rapid response. Several molecular mechanisms underpinning plant memory have been elucidated to date. One mechanism of memory formation may be sustained alterations in levels of key signaling metabolites or transcription factors (Fig. 2A, red line), which provides an explanation for how plant metabolism is altered and maintained by exposure to various stresses (1, 2, 39–42). Another possible avenue could be alterations to chromatin states, such as histone tail modifications, DNA methylation, or paused RNA polymerase II (Pol II), which could play a further role in the coordinated changes in the patterns of gene expression that underpin memory responses (43–48). Accordingly, the underlying mechanisms of these phenomena are the subject of much research.
Fig. 2. Stress memory and the molecular pathways to recovery.
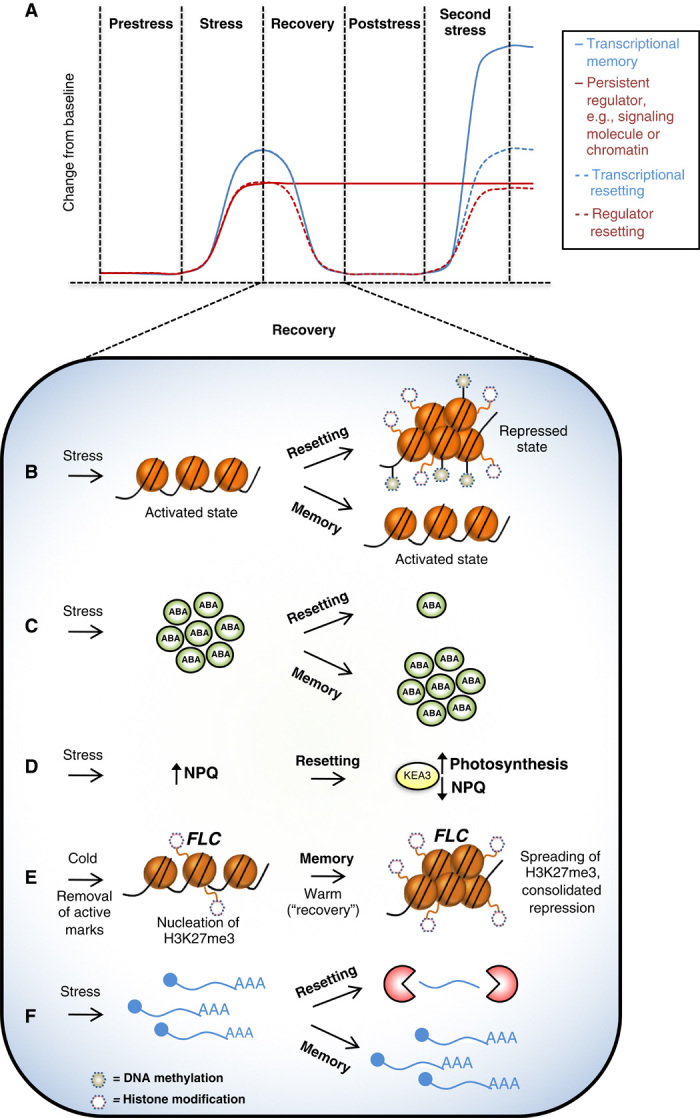
(A) A theoretical example of memory formation, where up to thousands of stress-inducible transcripts (blue lines) respond to the initial stress, concurrently with accumulation of signaling molecules and the release of repressive chromatin (red lines). Upon reexposure to a second stress, persistent signaling molecules and a retained accessible conformation of chromatin (solid lines) allow an enhanced stress response. The recovery period is a critical window where plant memory can be consolidated or resetting (dashed lines) can occur. (B) For instance, stress-induced changes in chromatin can be transient (possibly tied to regional accessibility for gene activation) or may persist, acting as a form of stress memory (90). (C) Similarly, signaling molecules may facilitate memory. In addition, signaling molecules can act during the recovery process; for instance, ABA may delay resumption of growth to enable embolism repair (113, 120). (D) KEA3 (potassium antiporter) activity accelerates recovery by relaxing non-photochemical quenching (NPQ) activity after dissipation of excess light stress (121). (E) Epigenetic silencing of FLC relies on the spreading of H3K27me3 specifically during transition to warm (recovery), consolidating repression and memory (66, 123, 124). (F) RNA decay reduces levels of stress-induced transcripts, resulting in resetting; impairment of decay may result in stress memory (161).
Memories can entail the persistence of signaling proteins or transcription factors after the initial stress; for example, sustained expression of microRNAs (miRNAs) to regulate SQUAMOSA PROMOTER BINDING PROTEIN–LIKE (SPL) transcription factors is critical for heat shock memory (49). The activation of signaling components and secondary messengers is required for BABA-induced priming of salicylate-dependent defense, because mutations in a cyclin-dependent kinase, the polyphosphoinositide phosphatase SUPPRESSOR OF ACTIN 1B (SAC1b) and the ABA biosynthetic enzyme ABA DEFICIENT 1 (ABA1), impair priming (50). Phosphorylation of MITOGEN-ACTIVATED PROTEIN KINASES (MPKs) (2) and accumulation of MPK3 and MPK6 and their messenger RNAs (mRNAs) after treatment with salicylic acid (SA) or the analog benzo(12,3)thiadiazole-7-carbothioic acid S-methyl ester have been associated with the priming of defense-related genes (51). Heat shock factors (Hsfs), including HsfB1, are also associated with systemic acquired resistance (52).
Another well-documented mechanism is seed provisioning, whereby environmental challenges, mostly to the maternal plant, influence the resources that are packaged into seeds, critical for germination and initial seedling growth (4). In particular, the quantity and composition of starch reserves, mRNAs, proteins, hormones, and other primary and secondary metabolites packaged into seeds are influenced by the environment (53–56). For instance, the ABA content of seeds can be increased by up to 44% by shading parental Amaranthus palmeri plants (57). These are examples of legacy maternal effects, a memory of the environment of the previous generation.
EPIGENETIC MEMORIES
Epigenetics describes heritable patterns of phenotypic variation that are not solely attributable to differences in DNA sequence (46). In plants, histone modifications and DNA methylation are known to be inherited through cell divisions (mitosis). In animals, epigenetic reprogramming and resetting occur during gamete formation and sexual reproduction, including the remodeling of histones and their modifications and demethylation of methylated DNA (58). Yet, many plants may circumvent such a process by proliferating partially or even exclusively by vegetative propagation, and plants are not known to undergo genome-wide demethylation in germ cells, as occurs in animals (58). Further, if such resetting does occur in plants it seems to be incomplete (48). Indeed, unlike animals, in Arabidopsis, DNA methylation can be inherited through meiotic cell divisions (59). Therefore, it is highly conceivable that mitotic and meiotic transmission of stress-induced epigenetic modifications could occur within higher plants.
Definitions of epigenetics.
The term “epigenetics” describes heritable patterns of phenotypic variation—that is, stable transmission of information through mitosis or meiosis—that are not solely attributable to differences in DNA sequence (46). “Epigenetics” is often used more broadly to describe changes in chromatin state, chromatin modification, and DNA methylation without regard for heritability. Indeed, these expanded definitions of epigenetics have been proposed to reflect popular usage of the term, to include long-term alterations in the transcriptional potential of a cell that are not necessarily heritable (60). However, others note that such definitions risk blurring important distinctions between the “proactive” and “responsive” roles of chromatin in transcription (43, 61). Delineating cause and effect in these situations remains challenging, and in the absence of established causality, labeling changes in chromatin structure as epigenetics is arguably misleading (62).
A diverse range of environmental stresses have been shown to alter chromatin and associated epigenetic marks (46). Of these, vernalization remains the best understood environmentally responsive epigenetic process, whereby FLOWERING LOCUS C (FLC) is transcriptionally repressed by cold exposure and repression is then epigenetically consolidated during subsequent development in warmer temperatures, facilitating a memory of the cold (63). Significantly, investigations of the FLC loci have demonstrated that memory is mitotically stable (64); memory is quantitative, with the degree of repression of FLC reflecting the length of the cold exposure (65); yet, the epigenetic state of FLC is cell-specific and likely digital, that is, ON/OFF (66, 67), and the chromatin state is inherited in cis, as opposed to trans or the inheritance of other diffusible factors (68).
DNA methylation changes can be induced by drought, flooding, nutrient limitation, temperature shock, pathogen infection, high salinity, heavy metal exposure, ultraviolet radiation, and herbivory (4, 69). Patterns of histone modification are likewise perturbed by environmental exposure and associate with altered expression at select loci (9, 19, 70–75). In fact, chromatin dynamics at drought-inducible genes during the transition from activation to inactivation have been shown to change within hours (72), similar to transcriptional responses.
Chromatin and small interfering RNA (siRNA) abundance also change in response to environmental conditions (76, 77). Several examples of environmentally responsive siRNAs have been reported (78, 79), although it should be noted that the vast majority of siRNAs in plants do not respond to the environmental perturbations profiled to date. Significantly, stress such as elevated temperature can transcriptionally activate chromosomal loci normally silenced by repressive chromatin (80–84). The transcriptional state of genes may also affect, and be affected by, nearby transposons that respond to changes in environmental conditions (80, 85–89). Indeed, recent evidence in rice (Oryza sativa) and Arabidopsis highlights that phosphate stress can trigger chromatin variation almost entirely targeting transposable elements near genes (90).
Naturally occurring epialleles such as the peloric variant of toadflax, Linaria vulgaris (91), and the colorless nonripening (CNR) locus from tomato (92) along with genome-wide studies (93, 94), natural variation (95, 96), and analysis of epigenetic recombinant inbred lines (97–99) demonstrate the stable inheritance of a range of DNA methylation variants that also affect fitness-related traits and confer differences in stress responses (100). Delineating pure epialleles from those linked to underlying genetic variation remains perhaps the biggest challenge in the field (46, 101). In fact, pure environment-induced epigenetic changes may only make minor contributions to durable genome-wide heritable chromatin structural variation, because this heritable chromatin variation largely reflects underlying genetic sequence variation (102). Numerous studies have also demonstrated that plants with compromised epigenetic machinery, mainly in the RNA-directed DNA methylation (RdDM) pathway, have reduced or eliminated capacity for priming or transgenerational transmission (33, 69, 103). Despite these links and correlations between epigenetic factors and acclimation, an ongoing challenge in the field is demonstrating causality (44, 46–48, 76, 102, 104). Recently, Secco et al. (90) provided one of the clearest examinations of chromatin-phenotype causality, in which phosphate stress in rice first leads to gene expression variation, which is then followed by changes in surrounding chromatin patterns.
Aside from issues of causality and pure epialleles, it is clear that the environment can and does induce epigenetic changes, which can be tightly correlated with transcriptional response. Nevertheless, examples of this phenomenon are rare and arguably represent the exception rather than the rule. As investigations continue to dissect epigenetic inheritance, it will be essential to also consider the trade-offs associated with the formation and duration of plant memories.
THE COST OF MEMORIES AND CHECKPOINTS
Plant memory, in particular epigenetic memory, is often hypothesized to complement genetic selection by providing a means to adapt, rapidly enabling local acclimation and even adaptation (46, 101). The potential instability and reversibility of memories could allow for adaptation to environmental variability and change, and opens the possibility of temporary adaptation or exploration of cryptic genomic information (4, 46). For instance, transgenerational plasticity contributes to biological invasions by invasive species (105). Yet, transgenerational memory could evolve specifically in cases where the parental environment more reliably predicts the offspring environment. Most often, it is the maternal environment that affects transgenerational plasticity, particularly for selfing species with a short seed dispersal range or for outcrossing species where pollen dispersal is wider than seed dispersal (106, 107). Nonetheless, many plants proliferate partially or even exclusively by vegetative propagation, in which case mitotic memories would be of greater significance. Yet, stress is often transitory, and in the same way that stress acclimation is balanced against stress avoidance, memories are counterbalanced by recovery and resetting.
There are numerous examples of maladaptive and adaptive memories with requisite trade-offs (108). Repeated stresses may result in increased sensitivity to deleterious effects (109), attenuation of photosynthesis, or perturbed growth and development (108). For instance, grape vines (Vitis vinifera) can become sensitized to ozone over successive years (109). Among closely related Polygonum species, drought-stressed Polygonum persicaria parents produce more well-provisioned seedlings, whereas Polygonum hydropiper parents transmit maladaptive traits leading to smaller seedlings with slower-extending root systems (110). There is also likely to be ascertainment bias in favor of adaptive traits in many studies; thus, the full extent to which plants form maladaptive memories is not known. Further, it remains an open question whether and when evolution would favor memory over forgetfulness. Analyses of natural populations are now just beginning to investigate such questions by looking for evidence of local nongenetic adaptations and so far they have served to reinforce the fact that genetic variation is a much more common influence on adaptation compared to epigenetics (95, 96, 102).
When memories turn out to be maladaptive, it is attractive to then consider possible mechanisms for resetting. One possibility proposed is a mechanism analogous to cell cycle checkpoints that could detect and remedy chromatin states in a manner analogous to DNA damage checkpoint mechanisms (76, 111). Iwasaki and Paszkowski (104) performed a screen to identify factors involved in the erasure of epigenetic stress memory. This revealed that Decrease in DNA Methylation 1 (DDM1) and Morpheus’ Molecule 1 (MOM1) are key to preventing transgenerational memory. In this case, environmental challenges such as elevated temperature can transcriptionally activate chromosomal loci normally silenced with repressive chromatin. This activation is only transient owing to the resilencing activity of DDM1 and MOM1, which reset the prior epigenetic state (80, 81, 83, 84).
STRESS RECOVERY
A critical window determining the degree of resetting versus the consolidation of memory occurs during the period of stress recovery, which we define to be the period of time following a stress (for example, rehydration following drought) until a new homeostasis is attained (Fig. 2). The poststress homeostasis may closely resemble the prestress state in cases where a stress is brief and resetting predominates. In other cases, a new distinct homeostasis may be reached due to priming and memory formation. In cases where a stress is prolonged, for instance, a drought, the poststress homeostasis may, in part, differ from the prestress state because of progression to a new developmental stage or permanently altered environment.
In contrast to the body of literature concerning acclimation and memory, there is a relative scarcity of reports on stress recovery. Several investigations have analyzed rehydration responses during drought stress recovery. In an early study, Oono et al. (112) used complementary DNA (cDNA) microarrays containing ~7000 Arabidopsis full-length cDNAs and identified 152 rehydration-inducible genes. Grapevine leaf petioles have been identified as a site of particular importance during rehydration following drought stress. Perrone et al. (113) analyzed transcriptomic responses during rehydration in petioles and found enrichment for genes involved in secondary metabolism, including genes linked to flavonoid biosynthesis; genes involved in sugar metabolism and transport; and several aquaporin genes. Zhang et al. (114) examined the global reprogramming of transcription and metabolism in Medicago truncatula during a progressive 14-day drought, including a sampling point 24 hours after rewatering. The authors observed that remarkably, most of the aboveground organs recovered fully within 24 hours of rewatering and most drought-responsive genes (~90%) reacted oppositely to the addition of water. In tobacco, recovery from drought stress was found to proceed by two pathways, restoration of photosynthesis and metabolism, or death, depending on the progress of senescence (115). Resetting was also observed in response to phosphate deprivation in Arabidopsis. Despite 21 days of starvation, resupplying phosphate for just 1 day reversed expression of 40% of induced genes, further increasing to 80% after 3 days and corresponding with a reestablished internal root phosphate concentration (90). Interestingly though, 80 genes remained differentially regulated even after 31 days of resupply. In addition, several studies have documented the reversion of chromatin states during recovery (Fig. 2B), including dehydration stress recovery (9, 72), phosphate starvation recovery (90), and recovery from oxygen depletion during submergence of rice seedlings (116).
Photosynthetic recovery is an essential step for the restoration of energy production and the resumption of growth. Rehydration in tobacco and olive revealed a relatively slow recovery of photosynthetic variables, particularly stomatal conductance (gs) (117, 118). The mechanism for this is not yet clear, although possibilities include both hydraulic limitations (119) and a residual ABA signal hindering stomatal opening (120). On the one hand, ABA may play a key role in facilitating embolism repair by controlling transpiration; however, such processes might also be manipulated to accelerate recovery and yield (Fig. 2C). In another example, Armbruster et al. (121) found that accelerating the deactivation of excess light defenses is critical for high photosynthetic efficiency under fluctuating light (Fig. 2D). In simulations, Zhu et al. (122) projected that slow down-regulation of these heat dissipation mechanisms can reduce photosynthetic efficiency in crop canopies by as much as 10%.
Mechanisms for memory formation may also operate specifically during stress recovery. In fact, in the case of FLC, although repressive chromatin marks are laid down at nucleation regions during the period of cold, it is not until returning to warm conditions (recovery) that the Polycomb Repressive Complex 2 (PHD-PRC2) is detected across the whole FLC locus and H3K27me3 increases substantially across the whole gene to facilitate effective epigenetic silencing (Fig. 2E) (66, 123, 124). Thus, the period of recovery is critical to vernalization and formation of memory of the cold even in the case of FLC.
Several key questions remain to be elucidated regarding the recovery period following stress. For instance, is recovery triggered by the alleviation of the stress stimulus, or the sensing of a return of favorable conditions? In this regard, “stress recovery” could alternatively be considered as a mere continuing response to new conditions. Whether this is the case requires further research. New investigations should also consider whether recovery is predominantly an active or passive process; for instance, in the case of the stress-responsive transcriptome, are transcripts switched off because of the dissipation of transcription factors and other activators or are they actively destabilized and targeted for decay during recovery? Through an examination of the processes of RNA decay, we may begin to address these questions.
ROLES FOR RNA DECAY IN STRESS RECOVERY
A pervasive theme in plant interactions with the environment is that dynamic remodeling of the transcriptome drives acclimation strategies; yet, gene expression studies often focus on transcription and overlook the critical contribution of RNA decay. RNA metabolism is presumably essential for clearing cells of stress-responsive transcripts (Fig. 3). Yet, RNA decay may also antagonize or circumvent the mechanisms that initiate epigenetic memories, for instance, by either generating or removing template RNA molecules that could be used by the posttranscriptional gene silencing (PTGS) or RdDM pathways (125–128).
Fig. 3. Roles of RNA metabolism during stress recovery leading to memory or resetting.
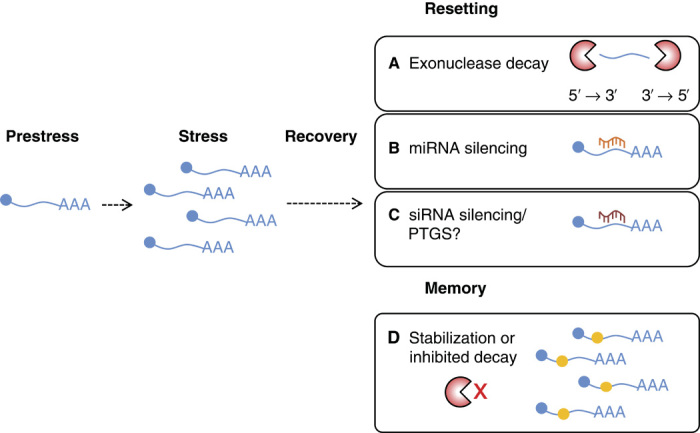
Stress is characterized by increased expression of many genes. (A to C) During the recovery period following a stress, RNA metabolism can facilitate resetting of the transcriptome by (A) exonuclease decay pathways, (B) miRNA decay pathways, and (we speculate) (C) siRNA pathways as well. (D) Alternatively, transcriptome memory may be created by inhibiting RNA decay or through stabilizing specific transcripts.
Increasing evidence also points to the specificity of particular RNA decay pathways (129–132). The implication of this specificity is that targeted decay of some transcripts and selected stabilization of others may underpin memory and resetting (Fig. 3). It is likely that the stabilities of specific mRNAs in plants are dynamic, for instance, by production of stabilization factors (133) or targeted decay (131); however, this is a relatively unexplored area. An as yet entirely unexplored possibility is the prospect that transcriptional memory might be underpinned by changes in mRNA stability rather than transcription.
RNA DECAY MECHANISMS AND REGULATION OF MRNA ABUNDANCE IN RESPONSE TO STRESS
In plants, mRNAs are stabilized by the 5′ cap, a 7-methyl guanine residue linked to the mRNA via a 5′-5′ triphosphate bond, and the 3′ poly(A) tail. RNA decay can proceed by removal of one or both of these structural stabilizers, or by endonuclease cleavage, which predominantly occurs in the cytoplasm and proceeds by either 5′-3′ or 3′-5′ exoribonuclease decay (125, 129, 134–136).
Steady-state transcript abundance is a product of the rates of transcription and decay; thus, RNA decay is clearly an essential component of gene regulation. RNA stability plays a major role in changing expression states; a role that is not necessarily obvious. As explained by Ross (137), consider two hypothetical transcripts—transcription factor 1 (TF1) and housekeeper 2 (HK2)—that are transcribed at the same rate as shown in Fig. 4. If TF1 is 10-fold less stable than HK2, then the TF1/HK2 ratio will be 1:10. If the transcription rate of both genes increases by 10-fold, ultimately the abundance of both transcripts will increase 10-fold, maintaining the 1:10 abundance ratio. However, TF1 will reach half its new steady state 10 times faster than the longer-lived HK2. This relationship holds for any system with zero-order input and first-order output as is the case for mRNA transcription and decay (137–139). Hence, there is logic for stress-responsive transcripts to also be highly unstable, enabling rapid responsiveness. No matter how sensitive and swift a transcriptional switch might be, mRNA transcripts that have prolonged half-lives will be unaffected by a change in transcriptional state as long as the transcripts remain stable (140). Thus, mRNA stability is a key component of transcript responsiveness and could be manipulated to modify recovery, priming, or memory.
Fig. 4. Relationship between mRNA stability and mRNA responsiveness [adapted from Ross (137)].
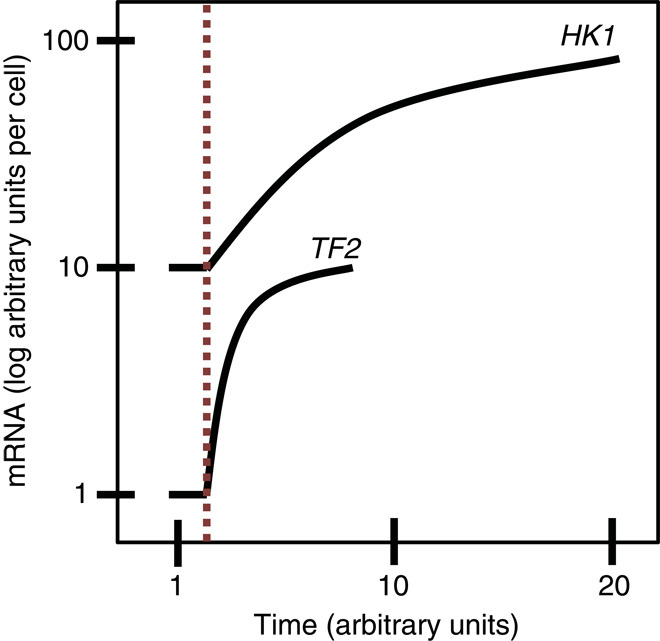
Following a change in transcription rate, an unstable transcript (TF2) can attain half its new steady state 10 times faster than the stable transcript (HK1). The dashed red line indicates the time point in which transcription increases 10-fold.
IMPRINTED MRNA RECOVERY DYNAMICS
Studies in yeast have discovered a fascinating coupling of transcription and decay that entails a “counteraction” between the two processes and underpins the enhanced responsiveness of some transcripts to stress (141). Several investigations found that many transcripts that are induced by stress are simultaneously destabilized in Saccharomyces cerevisiae (142, 143), Saccharomyces pombe (144), and mammals (138). Although cells expend valuable resources to increase the abundance of a transcript, in an outwardly fruitless waste of energy, these transcripts are also rendered more unstable: beneficially, this phenomenon enables fast, transient changes (145). An underlying mechanism is mRNA “imprinting” through the association of the RNA Pol II subunits, Rpb4 and Rpb7, which chaperone mRNAs into the cytoplasm and remain associated with the mRNA throughout its life cycle. These RNA-binding proteins promote degradation due to a higher affinity to the decay machinery. Stress triggers increased Rpb4/Rpb7 imprinting (146, 147), facilitating fast but transient changes in mRNA abundance (145). It is attractive to consider that other RNA binding proteins could imprint recovery or memory dynamics on mRNA molecules.
THE SPECIFICITY OF RNA DECAY
Although RNA decay may reset the transcriptome through nonspecific general RNA turnover of transcripts, a range of studies in plants point to specific and dynamic regulation of RNA stabilities and decay. Accordingly, the decay of specific transcripts may regulate and balance memory. Global investigations into RNA stability have reported a significant range of mRNA half-lives of 0.2 to >24 hours (mean, 5.9 hours; median, 3.8 hours) for transcripts in Arabidopsis (148). These analyses have also identified innate factors and motifs that affect mRNA stability, including conserved sequence elements in the 5′ and 3′ untranslated regions (UTRs) and the presence of introns and miRNA binding sites (148, 149). However, it is also clear that stability is dynamic and can change in response to developmental and environmental stimuli. The stability of mRNAs is known to change during the cell cycle (150–152), and multiple studies have demonstrated on a genome-wide scale that stress alters mRNA stability in yeast (142, 143, 153–156), although similar genome-wide investigations in plants are lacking. Xu and Chua (157) also found that dehydration stress activates Arabidopsis MPK6 to phosphorylate the decapping enzyme DECAPPING 1. Impairing this process led to stress hypersensitivity and the misregulation of dehydration-responsive transcripts. It has also been demonstrated that the RNA binding protein Tudor-SN stabilizes levels of stress-responsive mRNAs encoding secreted proteins in Arabidopsis and is thus required for stress tolerance (133). Cytoplasmic foci called stress granules and processing bodies also form during stress (158–160). These bodies sort, store, and likely decay mRNAs during stress and continue to be a focus of intense investigation.
EXORIBONUCLEASE 4 AND MRNA STRESS MEMORY
In Arabidopsis, several recent reports demonstrated specificity on the part of the general cytoplasmic RNA decay enzyme EXORIBONUCLEASE 4 (XRN4), uncovering roles in stress tolerance and recovery. Heat stress triggers the specific decay of many thousands of transcripts, a response that is critical for acclimation and tolerance of elevated temperatures (131). Here, LA RELATED PROTEIN 1A (LARP1) and XRN4 associate during heat stress (15 min at 38°C) to facilitate a massive heat-induced mRNA decay process targeting more than 4500 mRNAs. LARP1 is involved in associating XRN4 to polysomes, and xrn4 mutants are highly susceptible to prolonged moderate heat stress. In a recent investigation (161), it was reported that xrn4 mutants are more tolerant to transitory heat shock (44°C, 3 to 5 hours), in contrast to the earlier reported susceptibility to moderate prolonged temperature increases (35°C, 7 days). Increased thermotolerance was dependent in part on HEAT SHOCK TRANSCRIPTION FACTOR A2 (HSFA2) because xrn4 hsfa2 mutants are susceptible to heat stress. Significantly, following a mild heat stress, HSFA2 transcript levels return to prestress levels more slowly in xrn4 mutants. Thus, xrn4 mutants potentially retain a memory of stress for a longer period and XRN4 likely plays an important role during recovery to reset the transcriptome. Rymarquis et al. (132) also demonstrated substrate specificity for XRN4, related to conserved sequence motifs in target mRNAs.
THE FUNCTION OF SMALL RNAS AND PTGS DURING STRESS AND RECOVERY
Beyond the general RNA decay pathways, siRNAs and miRNAs provide the RNA decay machinery with exquisite specificity and have well-established roles during stress responses (77, 162), although their roles in recovery are largely unexplored. In one example, Stief et al. (49) uncovered a function for miRNAs in acquired thermotolerance. Using a typical heat stress time course, an initial moderate heat stress is followed by a more extreme stress designed to test acquired thermotolerance. Mutants impaired in small RNA (sRNA) biogenesis, specifically ago1, exhibited impaired thermotolerance. Furthermore, the induction of miRNA156, specifically during the recovery period, was found to be crucial for conveying this acclimation/priming through targeting the SPL family of transcription factors that are responsible for developmental transitions. An interesting follow-on from this study would be to see the effect of constitutive expression of miR156 on development once the stress has dissipated. Indeed, the authors did show enhanced thermotolerance when miR156 was constitutively overexpressed, as well as heat-inducible overexpression (49). Production of noncoding RNAs during stress and recovery could also provide substrates for siRNA production and mRNA regulation. For instance, naturally occurring antisense RNAs (natsiRNAs) that result from simultaneous expression of overlapping transcripts on opposite strands have the potential to affect mRNA expression during stress, generally in an inverse relationship through silencing of one of the transcripts. Numerous candidates (79, 163, 164) and several specific examples have been reported (165–168), although arguably a truly convincing reproducible example is lacking in the literature. Nevertheless, the possibility that natsiRNAs may be produced during stress recovery to facilitate transcriptional recovery and resetting is unexplored.
OUTLOOK: RNA DECAY ANTAGONIZES PRODUCTION OF SRNA THAT CAN DIRECT PTGS AND RDDM
The RNA decay machinery and the gene silencing machinery at times compete for the same RNA molecules (Fig. 5), establishing an antagonism between decay and silencing (125, 169). In the case of foreign genes, high expression of single-stranded sense transgenes from very active promoters results in susceptibility to S-PTGS and can cause cosuppression of endogenous genes (170, 171). This phenomenon is significantly exacerbated if cytoplasmic RNA decay is impaired (126, 128) and can lead to the production of sRNAs from endogenous transcripts (127). Mutations in an RNA splicing factor or several proteins acting in mRNA 3′ end formation can also lead to enhanced RNA silencing of a transgene (172). Two new studies further demonstrated that if the integrity of the decapping (173) or cytoplasmic RNA decay machinery (174) is impaired, endogenous transcripts can become susceptible to sRNA production and potentially silencing. All of these examples of gene silencing have been attributed to the availability of aberrant RNA molecules produced during transcription or decay, which serve as substrates for RNA-dependent RNA polymerases (RDRs), triggering double-stranded RNA (dsRNA) production and siRNA biogenesis. Indeed, some have proposed that such mechanisms may function to protect the genome against excessively expressed genes (175).
Fig. 5. RNA decay antagonizes sRNA production, PTGS, and RdDM.
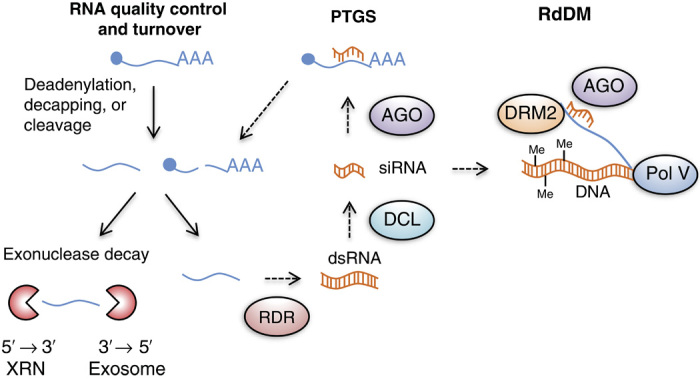
The RNA decay and gene silencing pathways use the same substrate RNA molecules, creating an antagonism between the RNA decay machinery and the gene silencing machinery. Transcripts are continuously turned over by the RNA decay machinery to achieve steady-state abundance and to ensure quality control; however, perturbations or defects in the RNA turnover or quality control pathway can cause transcripts to enter into the gene silencing pathways, leading to PTGS and potentially stable and heritable TGS via the RdDM pathway. DRM2, DOMAINS REARRANGED METHYLTRANSFERASE 2.
In addition to PTGS, siRNAs can also direct transcriptional gene silencing (TGS), notably through the RdDM pathway (176). In the canonical RdDM pathway, specialized plant-specific RNA Pol IV and Pol V transcribe “trigger” and “scaffold” RNAs, respectively, that facilitate sequence-specific DNA methylation via siRNAs (176). However, variations to the canonical RdDM pathway have also emerged. In rice, miRNA precursors transcribed by Pol II can be alternatively processed by DICER-LIKE 3 (DCL3) and, once bound to ARGONAUTE 4 (AGO4), can guide DNA methylation (177). Similarly, dsRNAs produced by the trans-acting siRNA (tasiRNA) pathway can also be processed by DCL1 to produce 21-nucleotide tasiRNAs that can act in Pol V–mediated RdDM following loading in AGO4 or AGO6 (178). RDR6 can also initiate a de novo DNA methylation from Pol II transcripts, a mechanism demonstrated to initiate canonical RdDM of young transposons (179). A transition from silencing by a PTGS mechanism to that by a TGS mechanism (which is a switch from posttranscriptional silencing to transcriptional silencing via DNA methylation) can also occur if high levels of dsRNA are produced by Pol II and RDR6 such that DCL2 and DCL4 become saturated and activate DCL3, which triggers RdDM (180).
Taking into account these findings, it is fascinating to consider how the RNA decay machinery might compete or collaborate with the epigenetic machinery to potentiate or circumvent either transcriptional recovery via PTGS or memory formation via RdDM (Fig. 5). For instance, stress is well known to induce very rapid production of transcripts (181), which conceivably could lead to an increase in aberrant mRNA that, in turn, would be potential substrates for RDRs. Indeed, the RNA decay inhibitor 3′-phosphoadenosine-5′-phosphate is also known to be produced during drought and light stress (182), potentially impairing RNA decay. In this scenario, mRNA molecules could be more readily susceptible to PTGS. During stress, this could in turn lead to RdDM and potentially heritable changes in gene expression (Fig. 5).
SUMMARY
In this review, we have highlighted recent advances in plant priming, memory, and epigenetics. These findings serve to demonstrate the capacity to confer acclimation and adaptive benefits within the life of a plant or future generations. Closer examination reveals that memory, in particular epigenetic memory, is likely a relatively rare event. The predominant strategy is resetting and recovery. Accordingly, a key regulatory step governing whether memories are formed or forgotten is the period of stress recovery. Within this period, plants balance resources allocated to sustained acclimation against the benefits of resetting and reallocation into growth and/or reproduction (Fig. 6). A key player in this process is RNA turnover, which may compete with the epigenetic machinery to circumvent memory formation. Future research into plant stress tolerance will be greatly aided by complementary analysis of stress recovery. In particular, the role of RNA metabolism in stress recovery has tremendous potential as a regulatory mechanism. A challenge is to consider epigenetic memory not only as a novelty in inheritance but also specifically in the context of the best adaptive strategies for the plant: Forgetfulness and resetting may be the more successful evolutionary strategies under unpredictable environmental conditions.
Fig. 6. Summary of the costs and benefits associated with recovery and resetting versus memory.
Acknowledgments
We thank G. Estavillo for his valuable review and discussion of the manuscript. Funding: P.A.C., D.G., S.R.E., J.O.B., and B.J.P. were funded by the Australian Research Council Centre of Excellence in Plant Energy Biology (CE140100008). P.A.C. and D.G. were supported by the Grains Research and Development Council (GRS184 and GRS10683) and Australian Postgraduate Awards. S.R.E. was funded by a Discovery Early Career Researcher Award (DE150101206). Author contributions: P.A.C. and B.J.P. conceived the review; P.A.C., D.G., and S.R.E. designed the figures; and all authors wrote and commented on the manuscript. Competing interests: The authors declare that they have no competing interests. Data availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
REFERENCES AND NOTES
- 1.Bruce T. J. A., Matthes M. C., Napier J. A., Pickett J. A., Stressful “memories” of plants: Evidence and possible mechanisms. Plant Sci. 173, 603–608 (2007). [Google Scholar]
- 2.Conrath U., Molecular aspects of defence priming. Trends Plant Sci. 16, 524–531 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Walter J., Nagy L., Hein R., Rascher U., Beierkuhnlein C., Willner E., Jentsch A., Do plants remember drought? Hints towards a drought-memory in grasses. Environ. Exp. Bot. 71, 34–40 (2011). [Google Scholar]
- 4.Herman J. J., Sultan S. E., Adaptive transgenerational plasticity in plants: Case studies, mechanisms, and implications for natural populations. Front. Plant Sci. 2, 102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldwin I. T., Schmelz E. A., Immunological “memory” in the induced accumulation of nicotine in wild tobacco. Ecology 77, 236–246 (1996). [Google Scholar]
- 6.Cayuela E., Pérez-Alfocea F., Caro M., Bolarín M. C., Priming of seeds with NaCl induces physiological changes in tomato plants grown under salt stress. Physiol. Plant. 96, 231–236 (1996). [Google Scholar]
- 7.Clarke S. M., Mur L. A. J., Wood J. E., Scott I. M., Salicylic acid dependent signaling promotes basal thermotolerance but is not essential for acquired thermotolerance in Arabidopsis thaliana. Plant J. 38, 432–447 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Dat J. F., Lopez-Delgado H., Foyer C. H., Scott I. M., Parallel changes in H2O2 and catalase during thermotolerance induced by salicylic acid or heat acclimation in mustard seedlings. Plant Physiol. 116, 1351–1357 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding Y., Fromm M., Avramova Z., Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat. Commun. 3, 740 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Ding Y., Virlouvet L., Liu N., Riethoven J.-J., Fromm M., Avramova Z., Dehydration stress memory genes of Zea mays; comparison with Arabidopsis thaliana. BMC Plant Biol. 14, 141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goh C.-H., Nam H. G., Park Y. S., Stress memory in plants: A negative regulation of stomatal response and transient induction of rd22 gene to light in abscisic acid-entrained Arabidopsis plants. Plant J. 36, 240–255 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Gordon M. J., Carmody M., Albrecht V., Pogson B., Systemic and local responses to repeated HL stress-induced retrograde signaling in Arabidopsis. Front. Plant Sci. 3, 303 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iqbal M., Ashraf M., Seed preconditioning modulates growth, ionic relations, and photosynthetic capacity in adult plants of hexaploid wheat under salt stress. J. Plant Nutr. 30, 381–396 (2007). [Google Scholar]
- 14.Jakab G., Cottier V., Toquin V., Rigoli G., Zimmerli L., Métraux J.-P., Mauch-Mani B., β-Aminobutyric acid-induced resistance in plants. Eur. J. Plant Pathol. 107, 29–37 (2001). [Google Scholar]
- 15.Jakab G., Ton J., Flors V., Zimmerli L., Métraux J.-P., Mauch-Mani B., Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol. 139, 267–274 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang H.-M., Saltveit M. E., Chilling tolerance of maize, cucumber and rice seedling leaves and roots are differentially affected by salicylic acid. Physiol. Plant. 115, 571–576 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Knight H., Brandt S., Knight M. R., A history of stress alters drought calcium signalling pathways in Arabidopsis. Plant J. 16, 681–687 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Rossel J. B., Wilson P. B., Hussain D., Woo N. S., Gordon M. J., Mewett O. P., Howell K. A., Whelan J., Kazan K., Pogson B. J., Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell 19, 4091–4110 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sani E., Herzyk P., Perrella G., Colot V., Amtmann A., Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 14, R59 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shakirova F. M., Sakhabutdinova A. R., Bezrukova M. V., Fatkhutdinova R. A., Fatkhutdinova D. R., Changes in the hormonal status of wheat seedlings induced by salicylic acid and salinity. Plant Sci. 164, 317–322 (2003). [Google Scholar]
- 21.Sung S., Amasino R. M., Vernalization and epigenetics: How plants remember winter. Curr. Opin. Plant Biol. 7, 4–10 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Sung D.-Y., Kaplan F., Lee K.-J., Guy C. L., Acquired tolerance to temperature extremes. Trends Plant Sci. 8, 179–187 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Ton J., Mauch-Mani B., β-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 38, 119–130 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Virlouvet L., Fromm M., Physiological and transcriptional memory in guard cells during repetitive dehydration stress. New Phytol. 205, 596–607 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Virlouvet L., Ding Y., Fujii H., Avramova Z., Fromm M., ABA signaling is necessary but not sufficient for RD29B transcriptional memory during successive dehydration stresses in Arabidopsis thaliana. Plant J. 79, 150–161 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Wang X., Vignjevic M., Jiang D., Jacobsen S., Wollenweber B., Improved tolerance to drought stress after anthesis due to priming before anthesis in wheat (Triticum aestivum L.) var. Vinjett. J. Exp. Bot. 65, 6441–6456 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye B., Gressel J., Transient, oxidant-induced antioxidant transcript and enzyme levels correlate with greater oxidant-resistance in paraquat-resistant Conyza bonariensis. Planta 211, 50–61 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Zimmerli L., Jakab G., Métraux J.-P., Mauch-Mani B., Potentiation of pathogen-specific defense mechanisms in Arabidopsis by β-aminobutyric acid. Proc. Natl. Acad. Sci. U.S.A. 97, 12920–12925 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agrawal A. A., Herbivory and maternal effects: Mechanisms and consequences of transgenerational induced plant resistance. Ecology 83, 3408–3415 (2002). [Google Scholar]
- 30.Johnsen Ø., Dæhlen O. G., Østreng G., Skrøppa T., Daylength and temperature during seed production interactively affect adaptive performance of Picea abies progenies. New Phytol. 168, 589–596 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Kvaalen H., Johnsen Ø., Timing of bud set in Picea abies is regulated by a memory of temperature during zygotic and somatic embryogenesis. New Phytol. 177, 49–59 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Molinier J., Ries G., Zipfel C., Hohn B., Transgeneration memory of stress in plants. Nature 442, 1046–1049 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Rasmann S., De Vos M., Casteel C. L., Tian D., Halitschke R., Sun J. Y., Agrawal A. A., Felton G. W., Jander G., Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol. 158, 854–863 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scoville A. G., Barnett L. L., Bodbyl-Roels S., Kelly J. K., Hileman L. C., Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytol. 191, 251–263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skrøppa T., Tollefsrud M. M., Sperisen C., Johnsen Ø., Rapid change in adaptive performance from one generation to the next in Picea abies—Central European trees in a Nordic environment. Tree Genet. Genomes 6, 93–99 (2009). [Google Scholar]
- 36.Slaughter A., Daniel X., Flors V., Luna E., Hohn B., Mauch-Mani B., Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 158, 835–843 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sultan S. E., Phenotypic plasticity for offspring traits in Polygonum persicaria. Ecology 77, 1791–1807 (1996). [Google Scholar]
- 38.Whittle C. A., Otto S. P., Johnston M. O., Krochko J. E., Adaptive epigenetic memory of ancestral temperature regime in Arabidopsis thaliana. Botany 87, 650–657 (2009). [Google Scholar]
- 39.Prime-A-Plant Group Conrath U., Beckers G. J. M., Flors V., García-Agustín P., Jakab G., Mauch F., Newman M.-A., Pieterse C. M. J., Poinssot B., Pozo M. J., Pugin A., Schaffrath U., Ton J., Wendehenne D., Zimmerli L., Mauch-Mani B., Priming: Getting ready for battle. Mol. Plant Microbe Interact. 19, 1062–1071 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Kinoshita T., Seki M., Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 55, 1859–1863 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Santos A. P., Serra T., Figueiredo D. D., Barros P., Lourenço T., Chander S., Oliveira M. M., Saibo N. J. M., Transcription regulation of abiotic stress responses in rice: A combined action of transcription factors and epigenetic mechanisms. OMICS 15, 839–857 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Vriet C., Hennig L., Laloi C., Stress-induced chromatin changes in plants: Of memories, metabolites and crop improvement. Cell. Mol. Life Sci. 72, 1261–1273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Avramova Z., Transcriptional ‘memory’ of a stress: Transient chromatin and memory (epigenetic) marks at stress-response genes. Plant J. 83, 149–159 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Boyko A., Kovalchuk I., Genome instability and epigenetic modification—Heritable responses to environmental stress? Curr. Opin. Plant Biol. 14, 260–266 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Chinnusamy V., Gong Z. Z., Zhu J.-K., Abscisic acid-mediated epigenetic processes in plant development and stress responses. J. Integr. Plant Biol. 50, 1187–1195 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eichten S. R., Schmitz R. J., Springer N. M., Epigenetics: Beyond chromatin modifications and complex genetic regulation. Plant Physiol. 165, 933–947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mirouze M., Paszkowski J., Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 14, 267–274 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Paszkowski J., Grossniklaus U., Selected aspects of transgenerational epigenetic inheritance and resetting in plants. Curr. Opin. Plant Biol. 14, 195–203 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Stief A., Altmann S., Hoffmann K., Pant B. D., Scheible W.-R., Bäurle I., Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26, 1792–1807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ton J., Jakab G., Toquin V., Flors V., Iavicoli A., Maeder M. N., Métraux J.-P., Mauch-Mani B., Dissecting the β-aminobutyric acid–induced priming phenomenon in Arabidopsis. Plant Cell 17, 987–999 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beckers G. J. M., Jaskiewicz M., Liu Y., Underwood W. R., He S. Y., Zhang S., Conrath U., Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21, 944–953 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pick T., Jaskiewicz M., Peterhäensel C., Conrath U., Heat shock factor HsfB1 primes gene transcription and systemic acquired resistance in Arabidopsis. Plant Physiol. 159, 52–55 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.M. Fenner, K. Thompson, The Ecology of Seeds (Cambridge Univ. Press, Cambridge, 2005). [Google Scholar]
- 54.M. Leishman, I. Wright, A. Moles, M. Westoby, in Seeds: The Ecology of Regeneration in Plant Communities (CAB International, Wallingford, UK, 2000), pp. 31–57. [Google Scholar]
- 55.A. Moles, M. Leishman, in Seedling Ecology and Evolution (Cambridge Univ. Press, New York, 2008), pp. 217–238. [Google Scholar]
- 56.Roach D. A., Wulff R. D., Maternal effects in plants. Annu. Rev. Ecol. Syst. 18, 209–235 (1987). [Google Scholar]
- 57.Jha P., Norsworthy J. K., Riley M. B., Bridges W. Jr, Shade and plant location effects on germination and hormone content of Palmer amaranth (Amaranthus palmeri) seed. Weed Sci. 58, 16–21 (2010). [Google Scholar]
- 58.Feng S., Jacobsen S. E., Reik W., Epigenetic reprogramming in plant and animal development. Science 330, 622–627 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calarco J. P., Borges F., Donoghue M. T. A., Van Ex F., Jullien P. E., Lopes T., Gardner R., Berger F., Feijó J. A., Becker J. D., Martienssen R. A., Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151, 194–205 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.NIH Roadmap for Medical Research: Epigenomics overview, http://www.roadmapepigenomics.org/overview [accessed 23 November 2015].
- 61.Bird A., Perceptions of epigenetics. Nature 447, 396–398 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Henikoff S., Shilatifard A., Histone modification: Cause or cog? Trends Genet. 27, 389–396 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Berry S., Dean C., Environmental perception and epigenetic memory: Mechanistic insight through FLC. Plant J. 83, 133–148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burn J. E., Bagnall D. J., Metzger J. D., Dennis E. S., Peacock W. J., DNA methylation, vernalization, and the initiation of flowering. Proc. Natl. Acad. Sci. U.S.A. 90, 287–291 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheldon C. C., Rouse D. T., Finnegan E. J., Peacock W. J., Dennis E. S., The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. U.S.A. 97, 3753–3758 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Angel A., Song J., Dean C., Howard M., A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476, 105–108 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Song J., Angel A., Howard M., Dean C., Vernalization—A cold-induced epigenetic switch. J. Cell Sci. 125, 3723–3731 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Berry S., Hartley M., Olsson T. S. G., Dean C., Howard M., Local chromatin environment of a Polycomb target gene instructs its own epigenetic inheritance. eLife 4, e07205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boyko A., Blevins T., Yao Y., Golubov A., Bilichak A., Ilnytskyy Y., Hollander J., Meins F. Jr, Kovalchuk I., Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of dicer-like proteins. PLOS One 5, e9514 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Dijk K., Ding Y., Malkaram S., Riethoven J.-J. M., Liu R., Yang J., Laczko P., Chen H., Xia Y., Ladunga I., Avramova Z., Fromm M., Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. BMC Plant Biol. 10, 238 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim J.-M., To T. K., Nishioka T., Seki M., Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 33, 604–611 (2010). [DOI] [PubMed] [Google Scholar]
- 72.Kim J.-M., To T. K., Ishida J., Matsui A., Kimura H., Seki M., Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 53, 847–856 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Kwon C. S., Lee D., Choi G., Chung W.-I., Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J. 60, 112–121 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Luo M., Liu X., Singh P., Cui Y., Zimmerli L., Wu K., Chromatin modifications and remodeling in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 129–136 (2012). [DOI] [PubMed] [Google Scholar]
- 75.Zong W., Zhong X., You J., Xiong L., Genome-wide profiling of histone H3K4-tri-methylation and gene expression in rice under drought stress. Plant Mol. Biol. 81, 175–188 (2013). [DOI] [PubMed] [Google Scholar]
- 76.Gutzat R., Mittelsten Scheid O., Epigenetic responses to stress: Triple defense? Curr. Opin. Plant Biol. 15, 568–573 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khraiwesh B., Zhu J.-K., Zhu J., Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819, 137–148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yao Y., Ni Z., Peng H., Sun F., Xin M., Sunkar R., Zhu J.-K., Sun Q., Non-coding small RNAs responsive to abiotic stress in wheat (Triticum aestivum L.). Funct. Integr. Genomics 10, 187–190 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moldovan D., Spriggs A., Yang J., Pogson B. J., Dennis E. S., Wilson I. W., Hypoxia-responsive microRNAs and trans-acting small interfering RNAs in Arabidopsis. J. Exp. Bot. 61, 165–177 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ito H., Gaubert H., Bucher E., Mirouze M., Vaillant I., Paszkowski J., An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119 (2011). [DOI] [PubMed] [Google Scholar]
- 81.Lang-Mladek C., Popova O., Kiok K., Berlinger M., Rakic B., Aufsatz W., Jonak C., Hauser M.-T., Luschnig C., Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in Arabidopsis. Mol. Plant 3, 594–602 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Makarevitch I., Waters A. J., West P. T., Stitzer M., Hirsch C. N., Ross-Ibarra J., Springer N. M., Transposable elements contribute to activation of maize genes in response to abiotic stress. PLOS Genet. 11, e1004915 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pecinka A., Dinh H. Q., Baubec T., Rosa M., Lettner N., Scheid O. M., Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22, 3118–3129 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tittel-Elmer M., Bucher E., Broger L., Mathieu O., Paszkowski J., Vaillant I., Stress-induced activation of heterochromatic transcription. PLOS Genet. 6, e1001175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bucher E., Reinders J., Mirouze M., Epigenetic control of transposon transcription and mobility in Arabidopsis. Curr. Opin. Plant Biol. 15, 503–510 (2012). [DOI] [PubMed] [Google Scholar]
- 86.Cavrak V. V., Lettner N., Jamge S., Kosarewicz A., Bayer L. M., Mittelsten Scheid O., How a retrotransposon exploits the plant’s heat stress response for its activation. PLOS Genet. 10, e1004115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Feschotte C., Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 9, 397–405 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Naito K., Zhang F., Tsukiyama T., Saito H., Hancock C. N., Richardson A. O., Okumoto Y., Tanisaka T., Wessler S. R., Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461, 1130–1134 (2009). [DOI] [PubMed] [Google Scholar]
- 89.Wheeler B. S., Small RNAs, big impact: Small RNA pathways in transposon control and their effect on the host stress response. Chromosome Res. 21, 587–600 (2013). [DOI] [PubMed] [Google Scholar]
- 90.Secco D., Wang C., Shou H., Schultz M. D., Chiarenza S., Nussaume L., Ecker J. R., Whelan J., Lister R., Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife 4, e09343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cubas P., Vincent C., Coen E., An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401, 157–161 (1999). [DOI] [PubMed] [Google Scholar]
- 92.Manning K., Tör M., Poole M., Hong Y., Thompson A. J., King G. J., Giovannoni J. J., Seymour G. B., A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38, 948–952 (2006). [DOI] [PubMed] [Google Scholar]
- 93.Becker C., Hagmann J., Müller J., Koenig D., Stegle O., Borgwardt K., Weigel D., Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480, 245–249 (2011). [DOI] [PubMed] [Google Scholar]
- 94.Schmitz R. J., Schultz M. D., Lewsey M. G., O’Malley R. C., Urich M. A., Libiger O., Schork N. J., Ecker J. R., Transgenerational epigenetic instability is a source of novel methylation variants. Science 334, 369–373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eichten S. R., Briskine R., Song J., Li Q., Swanson-Wagner R., Hermanson P. J., Waters A. J., Starr E., West P. T., Tiffin P., Myers C. L., Vaughn M. W., Springer N. M., Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 25, 2783–2797 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schmitz R. J., Schultz M. D., Urich M. A., Nery J. R., Pelizzola M., Libiger O., Alix A., McCosh R. B., Chen H., Schork N. J., Ecker J. R., Patterns of population epigenomic diversity. Nature 495, 193–198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Johannes F., Porcher E., Teixeira F. K., Saliba-Colombani V., Simon M., Agier N., Bulski A., Albuisson J., Heredia F., Audigier P., Bouchez D., Dillmann C., Guerche P., Hospital F., Colot V., Assessing the impact of transgenerational epigenetic variation on complex traits. PLOS Genet. 5, e1000530 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reinders J., Wulff B. B. H., Mirouze M., Marí-Ordóñez A., Dapp M., Rozhon W., Bucher E., Theiler G., Paszkowski J., Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 23, 939–950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Teixeira F. K., Heredia F., Sarazin A., Roudier F., Boccara M., Ciaudo C., Cruaud C., Poulain J., Berdasco M., Fraga M. F., Voinnet O., Wincker P., Esteller M., Colot V., A role for RNAi in the selective correction of DNA methylation defects. Science 323, 1600–1604 (2009). [DOI] [PubMed] [Google Scholar]
- 100.Kooke R., Johannes F., Wardenaar R., Becker F., Etcheverry M., Colot V., Vreugdenhil D., Keurentjes J. J. B., Epigenetic basis of morphological variation and phenotypic plasticity in Arabidopsis thaliana. Plant Cell 27, 337–348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weigel D., Colot V., Epialleles in plant evolution. Genome Biol. 13, 249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hagmann J., Becker C., Müller J., Stegle O., Meyer R. C., Wang G., Schneeberger K., Fitz J., Altmann T., Bergelson J., Borgwardt K., Weigel D., Century-scale methylome stability in a recently diverged Arabidopsis thaliana lineage. PLOS Genet. 11, e1004920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Luna E., Bruce T. J. A., Roberts M. R., Flors V., Ton J., Next-generation systemic acquired resistance. Plant Physiol. 158, 844–853 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Iwasaki M., Paszkowski J., Identification of genes preventing transgenerational transmission of stress-induced epigenetic states. Proc. Natl. Acad. Sci. U.S.A. 111, 8547–8552 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dyer A. R., Brown C. S., Espeland E. K., McKay J. K., Meimberg H., Rice K. J., SYNTHESIS: The role of adaptive trans-generational plasticity in biological invasions of plants. Evol. Appl. 3, 179–192 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Agrawal A. A., Laforsch C., Tollrian R., Transgenerational induction of defences in animals and plants. Nature 401, 60–63 (1999). [Google Scholar]
- 107.Galloway L. F., Maternal effects provide phenotypic adaptation to local environmental conditions. New Phytol. 166, 93–99 (2005). [DOI] [PubMed] [Google Scholar]
- 108.Skirycz A., Inzé D., More from less: Plant growth under limited water. Curr. Opin. Biotechnol. 21, 197–203 (2010). [DOI] [PubMed] [Google Scholar]
- 109.Soja G., Eid M., Gangl H., Redl H., Ozone sensitivity of grapevine (Vitis vinifera L.): Evidence for a memory effect in a perennial crop plant? Phys. Chem. Chem. Phys. 37, 265–270 (1997). [Google Scholar]
- 110.Sultan S. E., Barton K., Wilczek A. M., Contrasting patterns of transgenerational plasticity in ecologically distinct congeners. Ecology 90, 1831–1839 (2009). [DOI] [PubMed] [Google Scholar]
- 111.Cools T., De Veylder L., DNA stress checkpoint control and plant development. Curr. Opin. Plant Biol. 12, 23–28 (2009). [DOI] [PubMed] [Google Scholar]
- 112.Oono Y., Seki M., Nanjo T., Narusaka M., Fujita M., Satoh R., Satou M., Sakurai T., Ishida J., Akiyama K., Iida K., Maruyama K., Satoh S., Yamaguchi-Shinozaki K., Shinozaki K., Monitoring expression profiles of Arabidopsis gene expression during rehydration process after dehydration using ca. 7000 full-length cDNA microarray. Plant J. 34, 868–887 (2003). [DOI] [PubMed] [Google Scholar]
- 113.Perrone I., Pagliarani C., Lovisolo C., Chitarra W., Roman F., Schubert A., Recovery from water stress affects grape leaf petiole transcriptome. Planta 235, 1383–1396 (2012). [DOI] [PubMed] [Google Scholar]
- 114.Zhang J.-Y., Cruz De Carvalho M. H., Torres-Jerez I., Kang Y., Allen S. N., Huhman D. V., Tang Y., Murray J., Sumner L. W., Udvardi M. K., Global reprogramming of transcription and metabolism in Medicago truncatula during progressive drought and after rewatering. Plant Cell Environ. 37, 2553–2576 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vanková R., Dobrá J., Štorchová H., Recovery from drought stress in tobacco: An active process associated with the reversal of senescence in some plant parts and the sacrifice of others. Plant Signal. Behav. 7, 19–21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tsuji H., Saika H., Tsutsumi N., Hirai A., Nakazono M., Dynamic and reversible changes in histone H3-Lys4 methylation and H3 acetylation occurring at submergence-inducible genes in rice. Plant Cell Physiol. 47, 995–1003 (2006). [DOI] [PubMed] [Google Scholar]
- 117.Galle A., Florez-Sarasa I., Tomas M., Pou A., Medrano H., Ribas-Carbo M., Flexas J., The role of mesophyll conductance during water stress and recovery in tobacco (Nicotiana sylvestris): Acclimation or limitation? J. Exp. Bot. 60, 2379–2390 (2009). [DOI] [PubMed] [Google Scholar]
- 118.Perez-Martin A., Michelazzo C., Torres-Ruiz J. M., Flexas J., Fernández J. E., Sebastiani L., Diaz-Espejo A., Regulation of photosynthesis and stomatal and mesophyll conductance under water stress and recovery in olive trees: Correlation with gene expression of carbonic anhydrase and aquaporins. J. Exp. Bot. 65, 3143–3156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brodribb T. J., Cochard H., Hydraulic failure defines the recovery and point of death in water-stressed conifers. Plant Physiol. 149, 575–584 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lovisolo C., Perrone I., Hartung W., Schubert A., An abscisic acid-related reduced transpiration promotes gradual embolism repair when grapevines are rehydrated after drought. New Phytol. 180, 642–651 (2008). [DOI] [PubMed] [Google Scholar]
- 121.Armbruster U., Carrillo L. R., Venema K., Pavlovic L., Schmidtmann E., Kornfeld A., Jahns P., Berry J. A., Kramer D. M., Jonikas M. C., Ion antiport accelerates photosynthetic acclimation in fluctuating light environments. Nat. Commun. 5, 5439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhu X.-G., Ort D. R., Whitmarsh J., Long S. P., The slow reversibility of photosystem II thermal energy dissipation on transfer from high to low light may cause large losses in carbon gain by crop canopies: A theoretical analysis. J. Exp. Bot. 55, 1167–1175 (2004). [DOI] [PubMed] [Google Scholar]
- 123.Finnegan E. J., Dennis E. S., Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr. Biol. 17, 1978–1983 (2007). [DOI] [PubMed] [Google Scholar]
- 124.De Lucia F., Crevillen P., Jones A. M. E., Greb T., Dean C., A PHD-Polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. U.S.A. 105, 16831–16836 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Christie M., Brosnan C. A., Rothnagel J. A., Carroll B. J., RNA decay and RNA silencing in plants: Competition or collaboration? Front. Plant Sci. 2, 99 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gazzani S., Lawrenson T., Woodward C., Headon D., Sablowski R., A link between mRNA turnover and RNA interference in Arabidopsis. Science 306, 1046–1048 (2004). [DOI] [PubMed] [Google Scholar]
- 127.Gregory B. D., O’Malley R. C., Lister R., Urich M. A., Tonti-Filippini J., Chen H., Millar A. H., Ecker J. R., A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell 14, 854–866 (2008). [DOI] [PubMed] [Google Scholar]
- 128.Gy I., Gasciolli V., Lauressergues D., Morel J.-B., Gombert J., Proux F., Proux C., Vaucheret H., Mallory A. C., Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19, 3451–3461 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Belostotsky D. A., Sieburth L. E., Kill the messenger: mRNA decay and plant development. Curr. Opin. Plant Biol. 12, 96–102 (2009). [DOI] [PubMed] [Google Scholar]
- 130.Chekanova J. A., Gregory B. D., Reverdatto S. V., Chen H., Kumar R., Hooker T., Yazaki J., Li P., Skiba N., Peng Q., Alonso J., Brukhin V., Grossniklaus U., Ecker J. R., Belostotsky D. A., Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131, 1340–1353 (2007). [DOI] [PubMed] [Google Scholar]
- 131.Merret R., Descombin J., Juan Y.-t., Favory J.-J., Carpentier M.-C., Chaparro C., Charng Y.-y., Deragon J.-M., Bousquet-Antonelli C., XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress. Cell Rep. 5, 1279–1293 (2013). [DOI] [PubMed] [Google Scholar]
- 132.Rymarquis L. A., Souret F. F., Green P. J., Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories. RNA 17, 501–511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.dit Frey N. F., Muller P., Jammes F., Kizis D., Leung J., Perrot-Rechenmann C., Bianchi M. W., The RNA binding protein tudor-SN is essential for stress tolerance and stabilizes levels of stress-responsive mRNAs encoding secreted proteins in Arabidopsis. Plant Cell 22, 1575–1591 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chiba Y., Green P. J., mRNA degradation machinery in plants. J. Plant Biol. 52, 114–124 (2009). [Google Scholar]
- 135.Nagarajan V. K., Jones C. I., Newbury S. F., Green P. J., XRN 5′→3′ exoribonucleases: Structure, mechanisms and functions. Biochim. Biophys. Acta 1829, 590–603 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nakaminami K., Matsui A., Shinozaki K., Seki M., RNA regulation in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 149–153 (2012). [DOI] [PubMed] [Google Scholar]
- 137.Ross J., mRNA stability in mammalian cells. Microbiol. Rev. 59, 423–450 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Elkon R., Zlotorynski E., Zeller K. I., Agami R., Major role for mRNA stability in shaping the kinetics of gene induction. BMC Genomics 11, 259 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pérez-Ortín J. E., Alepuz P. M., Moreno J., Genomics and gene transcription kinetics in yeast. Trends Genet. 23, 250–257 (2007). [DOI] [PubMed] [Google Scholar]
- 140.Thomsen S., Anders S., Janga S. C., Huber W., Alonso C. R., Genome-wide analysis of mRNA decay patterns during early Drosophila development. Genome Biol. 11, R93 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Choder M., mRNA imprinting: Additional level in the regulation of gene expression. Cell Logist. 1, 37–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Molin C., Jauhiainen A., Warringer J., Nerman O., Sunnerhagen P., mRNA stability changes precede changes in steady-state mRNA amounts during hyperosmotic stress. RNA 15, 600–614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shalem O., Dahan O., Levo M., Martinez M. R., Furman I., Segal E., Pilpel Y., Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol. Syst. Biol. 4, 223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Amorim M. J., Cotobal C., Duncan C., Mata J., Global coordination of transcriptional control and mRNA decay during cellular differentiation. Mol. Syst. Biol. 6, 380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shalem O., Groisman B., Choder M., Dahan O., Pilpel Y., Transcriptome kinetics is governed by a genome-wide coupling of mRNA production and degradation: A role for RNA Pol II. PLOS Genet. 7, e1002273 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Farago M., Nahari T., Hammel C., Cole C. N., Choder M., Rpb4p, a subunit of RNA polymerase II, mediates mRNA export during stress. Mol. Biol. Cell 14, 2744–2755 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Selitrennik M., Duek L., Lotan R., Choder M., Nucleocytoplasmic shuttling of the Rpb4p and Rpb7p subunits of Saccharomyces cerevisiae RNA polymerase II by two pathways. Eukaryot. Cell 5, 2092–2103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Narsai R., Howell K. A., Millar A. H., O’Toole N., Small I., Whelan J., Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19, 3418–3436 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Christie M., Croft L. J., Carroll B. J., Intron splicing suppresses RNA silencing in Arabidopsis. Plant J. 68, 159–167 (2011). [DOI] [PubMed] [Google Scholar]
- 150.Alterman R. B., Ganguly S., Schulze D. H., Marzluff W. F., Schildkraut C. L., Skoultchi A. I., Cell cycle regulation of mouse H3 histone mRNA metabolism. Mol. Cell. Biol. 4, 123–132 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Goswami P. C., Roti Roti J. L., Hunt C. R., The cell cycle-coupled expression of topoisomerase IIalpha during S phase is regulated by mRNA stability and is disrupted by heat shock or ionizing radiation. Mol. Cell. Biol. 16, 1500–1508 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang W., Caldwell M. C., Lin S., Furneaux H., Gorospe M., HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 19, 2340–2350 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Castells-Roca L., García-Martínez J., Moreno J., Herrero E., Bellí G., Pérez-Ortín J. E., Heat shock response in yeast involves changes in both transcription rates and mRNA stabilities. PLOS One 6, e17272 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fan J., Yang X., Wang W., Wood W. H., Becker K. G., Gorospe M., Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc. Natl. Acad. Sci. U.S.A. 99, 10611–10616 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Molina-Navarro M. M., Castells-Roca L., Bellí G., García-Martínez J., Marín-Navarro J., Moreno J., Pérez-Ortín J. E., Herrero E., Comprehensive transcriptional analysis of the oxidative response in yeast. J. Biol. Chem. 283, 17908–17918 (2008). [DOI] [PubMed] [Google Scholar]
- 156.Romero-Santacreu L., Moreno J., Pérez-Ortín J. E., Alepuz P., Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA 15, 1110–1120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Xu J., Chua N.-H., Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J. 31, 1975–1984 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Xu J., Chua N.-H., Processing bodies and plant development. Curr. Opin. Plant Biol. 14, 88–93 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Maldonado-Bonilla L. D., Composition and function of P bodies in Arabidopsis thaliana. Front. Plant Sci. 5, 201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Weber C., Nover L., Fauth M., Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 56, 517–530 (2008). [DOI] [PubMed] [Google Scholar]
- 161.Nguyen A. H., Matsui A., Tanaka M., Mizunashi K., Nakaminami K., Hayashi M., Iida K., Toyoda T., Nguyen D. V., Seki M., Loss of Arabidopsis 5′–3′ exoribonuclease AtXRN4 function enhances heat stress tolerance of plants subjected to severe heat stress. Plant Cell Physiol. 56, 1762–1772 (2015). [DOI] [PubMed] [Google Scholar]
- 162.Sunkar R., Li Y.-F., Jagadeeswaran G., Functions of microRNAs in plant stress responses. Trends Plant Sci. 17, 196–203 (2012). [DOI] [PubMed] [Google Scholar]
- 163.Zhang X., Xia J., Lii Y. E., Barrera-Figueroa B. E., Zhou X., Gao S., Lu L., Niu D., Chen Z., Leung C., Wong T., Zhang H., Guo J., Li Y., Liu R., Liang W., Zhu J.-K., Zhang W., Jin H., Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol. 13, R20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang X.-J., Gaasterland T., Chua N.-H., Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 6, R30 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Katiyar-Agarwal S., Morgan R., Dahlbeck D., Borsani O., Villegas A. Jr, Zhu J.-K., Staskawicz B. J., Jin H., A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. U.S.A. 103, 18002–18007 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Held M. A., Penning B., Brandt A. S., Kessans S. A., Yong W., Scofield S. R., Carpita N. C., Small-interfering RNAs from natural antisense transcripts derived from a cellulose synthase gene modulate cell wall biosynthesis in barley. Proc. Natl. Acad. Sci. U.S.A. 105, 20534–20539 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zubko E., Meyer P., A natural antisense transcript of the Petunia hybrida Sho gene suggests a role for an antisense mechanism in cytokinin regulation. Plant J. 52, 1131–1139 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Iwamoto M., Higo K., Accumulation of sense–antisense transcripts of the rice catalase gene CatB under dark conditions requires signals from shoots. Gene 377, 186–194 (2006). [DOI] [PubMed] [Google Scholar]
- 169.Voinnet O., Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci. 13, 317–328 (2008). [DOI] [PubMed] [Google Scholar]
- 170.Que Q., Wang H. Y., English J. J., Jorgensen R. A., The frequency and degree of cosuppression by sense chalcone synthase transgenes are dependent on transgene promoter strength and are reduced by premature nonsense codons in the transgene coding sequence. Plant Cell 9, 1357–1368 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vaucheret H., Nussaume L., Palauqui J. C., Quillere I., Elmayan T., A transcriptionally active state is required for post-transcriptional silencing (cosuppression) of nitrate reductase host genes and transgenes. Plant Cell 9, 1495–1504 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Herr A. J., Baulcombe D. C., RNA silencing pathways in plants. Cold Spring Harb. Symp. Quant. Biol. 69, 363–370 (2004). [DOI] [PubMed] [Google Scholar]
- 173.Martínez de Alba A. E., Moreno A. B., Gabriel M., Mallory A. C., Christ A., Bounon R., Balzergue S., Aubourg S., Gautheret D., Crespi M. D., Vaucheret H., Maizel A., In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res. 43, 2902–2913 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang X., Zhu Y., Liu X., Hong X., Xu Y., Zhu P., Shen Y., Wu H., Ji Y., Wen X., Zhang C., Zhao Q., Wang Y., Lu J., Guo H., Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science 348, 120–123 (2015). [DOI] [PubMed] [Google Scholar]
- 175.Luo Z., Chen Z., Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell 19, 943–958 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Matzke M. A., Mosher R. A., RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394–408 (2014). [DOI] [PubMed] [Google Scholar]
- 177.Wu L., Zhou H., Zhang Q., Zhang J., Ni F., Liu C., Qi Y., DNA methylation mediated by a microRNA pathway. Mol. Cell 38, 465–475 (2010). [DOI] [PubMed] [Google Scholar]
- 178.Wu L., Mao L., Qi Y., Roles of DICER-LIKE and ARGONAUTE proteins in TAS-derived small interfering RNA-triggered DNA methylation. Plant Physiol. 160, 990–999 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nuthikattu S., McCue A. D., Panda K., Fultz D., DeFraia C., Thomas E. N., Slotkin R. K., The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162, 116–131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Marí-Ordóñez A., Marchais A., Etcheverry M., Martin A., Colot V., Voinnet O., Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 45, 1029–1039 (2013). [DOI] [PubMed] [Google Scholar]
- 181.Vogel M. O., Moore M., König K., Pecher P., Alsharafa K., Lee J., Dietz K.-J., Fast retrograde signaling in response to high light involves metabolite export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF transcription factors in Arabidopsis. Plant Cell 26, 1151–1165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Estavillo G. M., Crisp P. A., Pornsiriwong W., Wirtz M., Collinge D., Carrie C., Giraud E., Whelan J., David P., Javot H., Brearley C., Hell R., Marin E., Pogson B. J., Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23, 3992–4012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]


