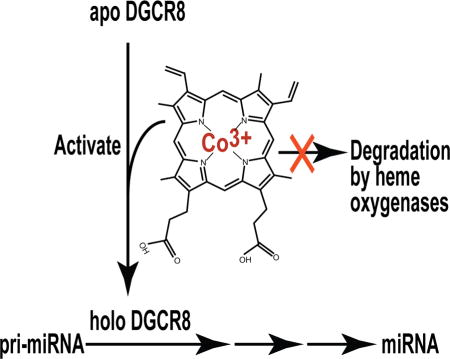
SUMMARY
Processing of microRNA primary transcripts (pri-miRNAs) is highly regulated and defects in the processing machinery play a key role in many human diseases. In 22q11.2 deletion syndrome (22q11.2DS), heterozygous deletion of DiGeorge critical region gene 8 (DGCR8) causes a processing deficiency, which contributes to abnormal brain development. The DGCR8 protein is the RNA-binding partner of Drosha ribonuclease, both essential for processing canonical pri-miRNAs. To identify an agent that can compensate reduced DGCR8 expression, we screened for metalloporphyrins that can mimic the natural DGCR8 heme cofactor. We found that Co(III) protoporphyrin IX (PPIX) stably binds DGCR8 and activates it for pri-miRNA processing in vitro and in HeLa cells. Importantly, treating cultured Dgcr8+/− mouse neurons with Co(III)PPIX can compensate the pri-miRNA processing defects. Co(III)PPIX is effective at concentrations as low as 0.2 μM and is not degraded by heme degradation enzymes, making it useful as a research tool and a potential therapeutic.
eTOC BLURB
miRNA processing defects have been reported in many human diseases including 22q11.2 deletion syndrome. Barr et al. show that Co(III) protoporphyrin activates miRNA processing by binding and activating the RNA-binding protein DGCR8 and compensates processing deficiency in Dgcr8+/− mouse neurons.
INTRODUCTION
pri-miRNA processing defects have been reported in a range of human diseases including cancer and schizophrenia (Beveridge and Cairns, 2012; Merritt, et al., 2008). Critical contribution of such defects to pathogenesis has been demonstrated (Kumar, et al., 2007; Mori, et al., 2014). In 22q11.2 deletion syndrome (also known as DiGeorge syndrome), the DGCR8 (called Pasha in Drosophila and Pash-1 in C. elegens) gene is heterozygously deleted along with 30–60 other genes in chromosome 22q11 (Karayiorgou, et al., 2010; Shiohama, et al., 2003). The haploinsufficiency of DGCR8 reduces the abundance of DGCR8 protein, resulting in lower pri-miRNA processing efficiency and abnormal expression of a subset of microRNAs (miRNAs) (Stark, et al., 2008). The miRNA deficiency has a pronounced effect on brain development and function, and contributes to the cognitive and behavioral deficits of 22q11.1DS (Fenelon, et al., 2011; Schofield, et al., 2011; Stark, et al., 2008). The DGCR8 protein and Drosha form the Microprocessor complex. Together they cooperate to recognize and cleave pri-miRNAs, thereby producing precursor miRNAs (pre-miRNAs) as intermediates (Denli, et al., 2004; Gregory, et al., 2004; Han, et al., 2004; Landthaler, et al., 2004). Following this initial step in the canonical miRNA maturation pathway, pre-miRNAs are exported to the cytoplasm and are further cleaved by the Dicer nuclease to generate mature miRNA strands, which are eventually incorporated into the miRNA-induced silencing complex (miRISC) and become functional in gene regulation (Ha and Kim, 2014).
To restore miRNA processing in 22q11.1DS and other diseases, it is highly desirable to enhance the activity of DGCR8 protein using small molecule agents, rather than engaging in risky gene therapies. DGCR8 has been shown to be regulated by cofactor binding, phosphorylation, acetylation and proteolytic cleavage (Gong, et al., 2012; Herbert, et al., 2013; Wada, et al., 2012; Weitz, et al., 2014). These posttranslational modifications offer opportunities for intervention. This study takes advantage of the fact that DGCR8 is activated by the cofactor heme b (Barr, et al., 2012; Barr, et al., 2011; Faller, et al., 2007). A purified recombinant DGCR8 construct NC1 (a.a. 276–751 of the 773-aa full-length protein) forms a stable complex with Fe(III) heme, but binds Fe(II) heme with much weaker affinity (Barr, et al., 2012; Barr, et al., 2011). Fe(III) heme in DGCR8 is coordinated by a unique two-cysteine configuration (Senturia, et al., 2010). DGCR8 binds heme using an RNA-binding heme domain (Rhed) that directly contacts pri-miRNAs and cooperates with two double-stranded RNA-binding domains to achieve high-affinity binding and specific recognition (Quick-Cleveland, et al., 2014). In cells, DGCR8 mutants that are unable to bind heme are unable to process pri-miRNAs (Weitz, et al., 2014). Additionally, DGCR8 activity can be modulated by removing heme from the cell culture media, adding back exogenous heme, or inhibiting endogenous heme biosynthesis (Weitz, et al., 2014).
It is well known that excess amounts of freely available heme in cells are degraded by heme oxygenases (Kikuchi, et al., 2005), making heme less useful as a sustainable activator for pri-miRNA processing. In this study, we set out to identify heme analogs that are capable of binding and activating DGCR8. We either removed the central Fe of heme or substituted it with a diverse set of other metals. We successfully identified one such derivative, Co(III)PPIX, and demonstrated that it can rescue miRNA expression in Dgcr8+/− mouse neurons. CoPPIX is not degraded by heme oxygenases and thereby should be able to activate pri-miRNA processing for a prolonged period of time.
RESULTS
A metal ion is necessary for porphyrin to stably associate with DGCR8
We examined potential interaction of porphyrin compounds with heme-free (apo) forms of DGCR8 via titration and electronic absorption spectroscopy. Size exclusion chromatography (SEC) was used to confirm the interaction and to estimate the stability of the complexes. Two apoDGCR8 proteins were used in the binding study: wild-type apoNC1 and the apoNC1-P351A mutant. NC1-P351A can be conveniently purified as a heme-free dimer (apoNC1-P351A) when recombinantly expressed in E. coli without the addition of δ-aminolevulinic acid (δ-ALA) to the media (Barr, et al., 2011). apoNC1-P351A can bind Fe(III) heme to form a stable complex similar to native Fe(III) heme-bound NC1. The porphyrin-binding assays using apoNC1-P351A were performed at pH 8 at which this protein is stable. This pH is close to physiological and is routinely used for our reconstituted pri-miRNA processing assays (Barr and Guo, 2014). In contrast, the porphyrin-binding assays using wild-type apoNC1 were performed at pH 6 for this protein is not very soluble at pH 8. Wild-type NC1 is expressed in E. coli as the Fe(III) heme-bound form and it associates with Fe(III) heme very tightly (Barr, et al., 2011). Thus, the preparation of apoNC1 requires several additional steps after purification of NC1, including buffer exchange to pH 6, heme reduction, incubation with apomyoglobin to scavenge the Fe(II) (ferrous) heme fast-dissociating from NC1 and SEC to separate apoNC1 from myoglobin(Barr, et al., 2012), thereby resulting in a lower yield. apoNC1 dimer prepared this way can bind Fe(III) heme and be activated for pri-miRNA processing (Barr, et al., 2012). Because of the comparative ease of preparation and handling, we chose to use apoNC1-P351A for initial binding assays with porphyrins. The binding or lack of binding for select porphyrins was confirmed using wild-type apoNC1.
We first tested protoporphyrin IX (PPIX), which does not contain a metal center (Figure 1A). Incubation of PPIX with apoNC1-P351A dimer resulted in a predominant Söret peak at 383 nm, which was slightly shifted from the 377 nm peak of PPIX without DGCR8 (Figure S1A). The peak wavelengths and shape of the spectra did not change as the molar ratio of PPIX and apoNC1-P351A increased above 1:1, indicating that the PPIX did not bind specifically. Consistent with this notion, SEC analysis of the 1:1 mixture revealed little 383 nm absorbance associated with the NC1-P351A elution peak (Figure S1B). Titration of PPIX to wild-type apoNC1 at pH 6.0 showed broadened split Söret peaks in the range of 300–500 nm with two maxima at 368 and 447 nm, respectively, and weaker Q bands at 535, 555, 592 and 643 nm (Figure 2A). These features were similar to those of free PPIX (Scolaro, et al., 2002) and no saturation behavior was observed (Figure 2B). Furthermore, the PPIX absorbance did not co-elute with apoNC1 in SEC analyses (Figure 2C). Therefore, we conclude that a metal ion is required for strong DGCR8-porphyrin interaction.
Figure 1.
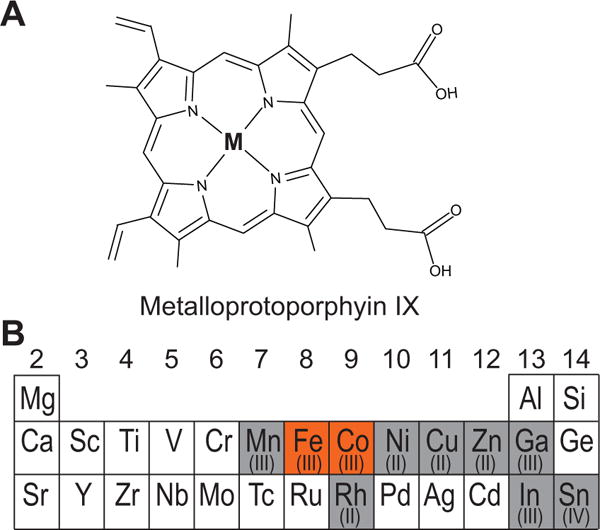
Metalloporphyrins that were tested for association with DGCR8 and for activation of pri-miRNA processing. (A) Schematic of a metalloprotoporphyrin IX. The M at the center represents a metal ion. (B) A section of the periodic table showing the elements used in this study, with the group numbers labeled. The protoporphyrin IX complexes with the metals shown in red background stably bind to DGCR8, whereas the complexes with the elements in gray do not.
Figure 2.

Protoporphyrin IX without a metal center does not stably associate with apoNC1. (A) Electronic absorption spectra of titration of PPIX into 1.25 μM apoNC1 dimer at 0.25 μM intervals, in 50 mM MES pH 6.0, 400 mM NaCl and 1 mM DTT The PPIX:apoNC1 ratios of the bold curves are indicated in italic. (B) Absorbance values at 368 nm and 447 nm obtained from the titration are plotted against PPIX:apoNC1 ratios. To avoid overlapping, the A368 values are nudged up by 0.01. In contrast to a similar titration experiment using Fe(III)PPIX (Barr, et al., 2011), no saturation point is observed at 1:1 PPIX:apoNC1 ratio, indicating a lack of specific binding. (C) Size exclusion chromatogram of a mixture of apoNC1 and PPIX, both at 4 μM concentration shows little co-elution. The low 368 nm absorption in the elution peak was contributed by the ~10% heme remaining bound to the apoNC1 preparation. The chromatogram is indistinguishable from that of the apoNC1 alone. See also Figure S1.
Co(III)PPIX stably associates with DGCR8
We screened nine metalloporphyrins (Figure 1) and found that at pH 8 only Co(III)PPIX is able to stably associate with apoNC1-P351A, that Zn(II)PPIX only weakly binds (Figure S2), and that the others do not seem to bind. In the titration of Co(III)PPIX, we observed hyperporphyrin (split Söret) spectra with peaks appearing at 365, 456, and 563 nm, and a non-specific peak at 430 nm became prominent as Co(III)PPIX was added in excess (Figure S3A). The 365 and 456 nm peaks are intense and of nearly equal heights, and the α/β band at 563 nm is much weaker. These features of the specific Co(III)PPIX-NC1-P351A complex are similar to those seen with Fe(III) heme titration (Barr, et al., 2011). Plotting of the specific and non-specific peak absorbance values over the Co(III)PPIX concentration revealed a common transition at 9 μM (Figure S3B), close to the 7.5 μM apoNC1-P351A concentration within experimental errors, indicating that the specific binding has a stoichiometry of one Co(III)PPIX per apoNC1-P351A dimer. SEC analyses showed that the Co(III)PPIX absorbance co-elutes with the protein, confirming that their association is stable (Figure S3C).
Titration of Co(III)PPIX to wild-type apoNC1 at pH 8 resulted in electronic absorption spectra remarkably similar to those from titration to apoNC1-P351A. The peaks of the specific complex are at 368, 456, 563 nm and a non-specific peak at 432 nm became prominent when Co(III)PPIX is in excess (Figure 3A). In SEC analyses, the A456 and slightly less intense A432 co-eluted with the 12.5 mL protein peak (Figure 3B), indicating that Co(III)PPIX remains bound to NC1. Therefore, we conclude that Co(III)PPIX stably binds DGCR8 at both pH 6 and 8.
Figure 3.
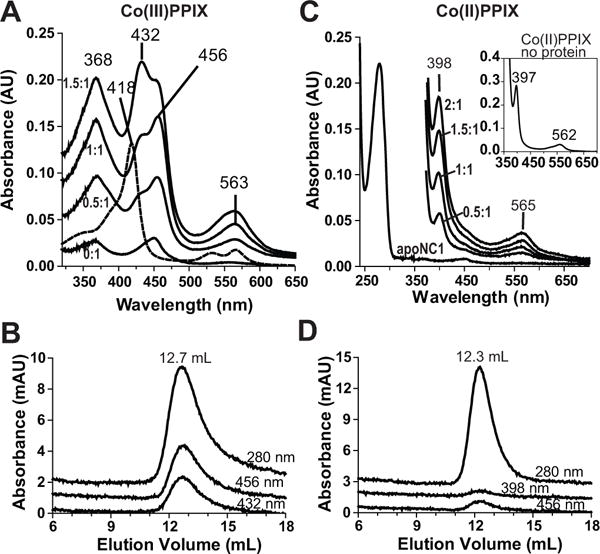
Co(III)PPIX, but not the Co(II) form, associates with apoNC1. (A) Electronic absorption spectra of Co(III)PPIX titrated into 2 μM apoNC1 dimer at 1 μM per step, in 50 mM MES pH 6.0, 400 mM NaCl and 1 mM DTT. The absorption peak at 432 nm results from a non-specific interaction. An absorbance spectrum of Co(III)PPK alone is shown as a dashed line. (B) Size exclusion chromatogram of the reconstituted Co(III)PPIX-NC1 complex. (C) Co(II)PPIX was titrated into 2.5 μM apoNC1 in 50 mM MES pH 6.0, 400 niM NaCl and 2 mM sodium dithionite at 1.25 μM per step. Sodium dithionite obscures peaks below 350 nm due to its high absorbance. The apoNC1 spectrum before addition of sodium dithionite is also shown. The absorption spectrum of Co(II)PPIX without proteins is shown in the inset. (D) Size exclusion chromatogram of apoNC1 with Co(II)PPIX. The SEC buffer contained 50 mM MES pH 6.0 and 400 mM NaCl, and was degassed to remove O2. The apoNC1 dimer elutes at an expected volume. However, there is no large accompanying absorbance at 398 nm (the Co(II)PPIX Söret wavelength) or 456 nm (potential re-oxidized Co(III)PPIX Söret wavelength). See also Figures S2 and S3 and Table S1.
The redox state of cobalt is important for DGCR8-CoPPIX interaction
The cobalt in Co(III)PPIX can be reduced to Co(II). We tested if the Co(II)PPIX can bind DGCR8. Co(II)PPIX was produced by reducing Co(III)PPIX using dithionite and was titrated to wild-type apoNC1. The electronic absorption spectra showed a prominent Söret peak at 398 nm and a relatively broad band at 565 nm, akin to the spectrum of free Co(II)PPIX (Figure 3C). SEC analyses of the Co(II)PPIX-apoNC1 mixture showed little 398 nm absorbance in the protein elution peak (Figure 3D). Therefore, we conclude that, in sharp contrast to Co(III)PPIX, Co(II)PPIX does not bind strongly to DGCR8. It is a common requirement for the metal ion in both CoPPIX and heme to be in a 3+ state to stably associate with DGCR8 (herein and published data (Barr, et al., 2012)).
Co(III)PPIX increases pri-miRNA processing activity of DGCR8
We next tested if Co(III)PPIX binding to apoNC1 can biochemically activate it for pri-miRNA processing. Uniformly 32P-labeled pri-miRNAs were incubated with purified recombinant Drosha and various forms of DGCR8. The latter included native Fe(III) heme-bound NC1, apoNC1, and in vitro reconstituted NC1-Co(III)PPIX and NC1-Fe(III)-heme complexes. The subsequent denaturing gel and autoradiography analyses showed that, compared to native NC1, apoNC1 displayed a deficiency in pri-miRNA processing, with nearly no pre-miRNA produced for pri-miR-23a, a >60% reduction of processing for pri-miR-21 and pri-miR-380, and a modest reduction (~30%) for pri-miR-30a (Figure 4, lanes 3 and 4 in all panels). Importantly, NC1 reconstituted with Co(III)PPIX and Fe(III) heme rescued pri-miRNA cleavage close to the levels of native NC1 for all four pri-miRNAs (Figure 4, lanes 4 and 5 in all panels). Our previous titration experiments showed that the activity of NC1 in the reconstituted assay coincides with its binding to the pri-miRNA substrate (Faller, et al., 2010). The NC1 concentration (25 nM) used in the current pri-miRNA processing assays is just above the K1/2 of binding (~10 nM) (Quick-Cleveland, et al., 2014), and this condition should be very sensitive to changes in activity of the heme-removed and reconstituted NC1 proteins. Therefore, our results clearly demonstrate that Co(III)PPIX is capable of activating DGCR8 for pri-miRNA processing, most likely by forming a complex similar to that containing Fe(III) heme.
Figure 4.
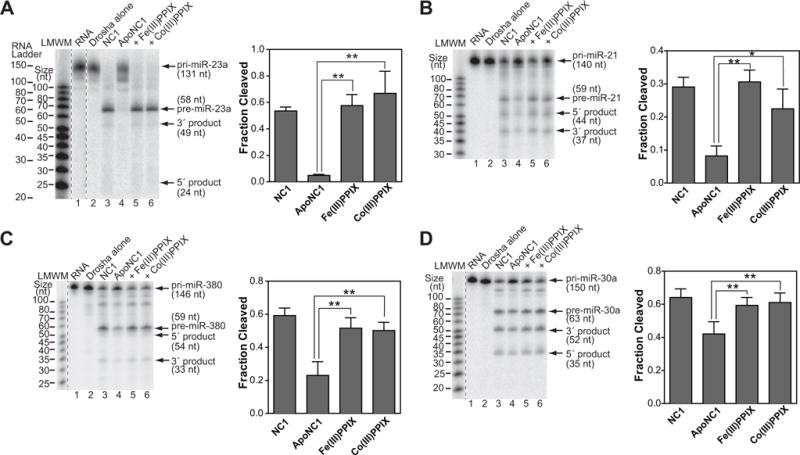
Co(III)PPIX activates DGCR8 for pri-miRNA processing in vitro. Denaturing gel analyses of cleavage assays of (A) pri-miR-23a, (B) pri-miR-21, (C) pri-miR-380 and (D) pri-miR-30a. The cleavage reactions contained trace amounts of uniformly 32P-labeled pri-miRNAs, recombinant His6-Drosha390–1374 and the various forms of DGCR8 proteins (25 nM dimer). NC1 is the native Fe(III) heme-bound dimer. apoNC1 is NC1 with the >90% of heme removed. The reactions labeled with “+Fe(III)PPIX” and “+Co(III)PPIX” contained apoNC1 incubated with equimolar of Fe(III) heme or Co(III)PPIX, respectively. Fractions of pri-miRNAs converted to pre-miRNAs are plotted as means ± SD (n = 3 for pri-miR-23a, pri-miR-21 and pri-miR-380; n = 4 for pri-miR-30a). Asterisks indicate statistically significant activation of apoNC1 (**, p ≤ 0.01; *, 0.01< p ≤ 0.05).
Co(III)PPIX activates pri-miRNA processing in cells
Unlike heme, CoPPIX is not degraded by heme oxygenases, the major heme degradation pathway in mammalian cells (Kappas and Drummond, 1986), making it potentially more useful as a pri-miRNA processing activator. We tested if Co(III)PPIX can activate pri-miRNA processing in human cells using a recently developed fluorescent live-cell reporter assay that specifically reflects changes in pri-miRNA processing efficiency (Weitz, et al., 2014). The reporter plasmid contains an mCherry expression cassette with a pri-miRNA sequence inserted in the 3′-untranslated region, so that cleavage of the pri-miRNA by Drosha/DGCR8 reduces the expression of this red fluorescent protein. The plasmid also expresses eYFP, which serves a normalization purpose. The total mCherry and eYFP fluorescent intensities of individual cells have a linear relationship over the wide range of expression levels usually seen in a transient transfection experiment. The eYFP vs mCherry slope indicates pri-miRNA processing efficiency. This assay has been extensively validated at both protein and RNA levels and was used to demonstrate that alternations in heme biosynthesis and availability modulate pri-miRNA processing efficiency (Weitz, et al., 2014).
We used our cellular assay to determine if Co(III)PPIX can activate pri-miRNA processing in mammalian cells. HeLa cells were adapted to grow in media with 50–80% of heme removed from the serum and then transfected with the pri-miR-9-1 reporter. Eighteen hours posttransfection, cells were treated with succinylacetone (SA) to block heme synthesis, either alone, with 0.2, 1 or 10 μM Co(III)PPIX, or with 10 μM hemin for 10 h. Cellular fluorescence signals indicated that, as expected, SA treatment decreased the slope (and thus the pri-miRNA processing efficiency) from 1.00 ± 0.02 (95% CI, which is shown for all fluorescence data) to 0.89 ± 0.03 and 10 μM hemin rescued it back to 1.07 ± 0.03 (Figure 5A). Importantly, Co(III)PPIX increased pri-miRNA processing efficiency in SA-treated cells in a dose-dependent manner. The fluorescence slope was increased by 8%, 19% and 25% in the presence of 0.2 μM, 1.0 μM and 10 μM of Co(III)PPIX, respectively (Figure 5A). Even the change cause by the lowest concentration (0.2 μM) of CoPPIX was highly significant (p = 0.0005). We subsequently tested if the boost of pri-miRNA processing efficiency resulted in increased production of mature miR-9 using qRT-PCR. With SA treatment, the abundance of mature miR-9 was reduced to 0.64-fold (± 0.05, SD) that of the no treatment control (Figure 5B). This defect was rescued by both Co(III)PPIX and hemin, with the miR-9 levels 1.15 ± 0.17 and 0.81 ± 0.25 folds that of the no treatment control, respectively. The effects of Co(III)PPIX and hemin on the reporter depend on the pri-miRNA, as the same treatments caused little changes in fluorescence slope when a reporter without pri-miRNA sequences was used (Figure S4). Treatment with any of these compounds is not toxic to cells, as indicated by cell viability measurements using an MTT assay (Figure 5C).
Figure 5.

Live-cell pri-miRNA processing assay shows that Co(III)PPIX activates pri-miRNA processing without inducing cytotoxicity. Hela cells were cultured in heme-depleted media, transfected with the pri-miR-9-1 reporter, treated for 10 h with succinylacetone (1 mM) either alone or together with Co(III)PPIX or hemin at the indicated concentrations. (A) Normalized eYFP/mCherry fluorescence slopes (± 95% CI). (B) Abundance of mature miR-9 normalized by that of β-actin mRNA (mean ± SD, n = 4). (C) MTT assays showed little cytotoxicity (mean ± SD, n = 4). (D–F) Reporter assays were performed similarly to described above, except that the N-flag-DGCR8 expression plasmid was cotransfected with the reporters, that the pri-miR-9-1 (D), pri-miR-185 (E), and pri-miR-30a (F) reporters were used, and that SA, Co(III)PPIX and hemin were added prior to transfection. See also Figure S4.
We performed the cellular assays in additional settings to further corroborate the conclusion. In the experiments just described, the changes in fluorescence slope caused by Co(III)PPIX and hemin (8–25%) are modest. This is due to low expression of DGCR8 from the endogenous loci in HeLa cells (Weitz et al., manuscript in preparation). With overexpression of N-terminally flag-tagged DGCR8 (N-flag-DGCR8), the fluorescence slope displayed by the pri-miR-9-1 reporter in cells treated with Co(III)PPIX increased by 29–73% relative to that of the SA-alone control (Figure 5D). We also tested two additional reporters that contain the pri-miR-185 and pri-miR-30a sequences, respectively. Upon Co(III)PPIX treatments, both reporters showed robust (23–51%) increases in fluorescence slope (Figure 5E–F), indicating that boost of pri-miRNA processing is general across difference pri-miRNA sequences. The dose responses vary among the pri-miRNA reporters. The increases for pri-miR-9-1 and pri-miR-185 were 29% and 23% respectively at 0.1 μM Co(III)PPIX, reached maxima of 73% and 51% at 1.0 μM, and dropped a little to 63% and 38% at 10 μM (Figure 5D–E). In contrast, the pri-miR-30a reporter showed similar increases (44–49%) at all three Co(III)PPIX concentrations (Figure 5F), which suggest that this compound may be effective at even lower concentrations for this pri-miRNA. The increases of fluorescence slope caused by 10 μM Co(III)PPIX are either similar to or slighter higher than that by 10 μM hemin (Figure 5D–F). We have evidence that with overexpression of DGCR8, pri-miRNA processing is no longer the rate limiting step in reporter miRNA production (Weitz et al. data not shown), thus mature miRNA abundance was not examined. Altogether, our data clearly demonstrate that Co(III)PPIX is able to generally enhance pri-miRNA processing in cells without obvious harmful side-effects.
Co(III)PPIX-mediated activation compensates miRNA processing deficiency caused by heterozygous deletion of Dgcr8 in mouse neurons
We investigated if Co(III)PPIX may be used to correct the miRNA processing deficiency in cultured neurons. Previous studies showed that heterozygous deletion of the Dgcr8 gene either alone or along with other genes in the Df(16)A region in mice (syntenic to the 1.5 Mb 22q11.2 microdeletion in humans) reduces both the mRNA and protein levels to ~50% those of the Dgcr8+/+ in wild-type mice (Stark, et al., 2008). This in turn reduces the maturation of certain miRNAs in the brain, including miR-185 and miR-134 that are important for dendritic spine development (Stark, et al., 2008; Xu, et al., 2013). We prepared primary cortical neurons from Dgcr8+/− pups and their wild-type littermate controls at embryonic day 17. These neurons were cultured in vitro and treated from 1 day in vitro (DIV) for 2 days with 0.2, 2 and 20 μM Co(III)PPIX. We then extracted total RNAs and measured the miRNA abundance. Similar to what has been reported previously (Xu, et al., 2013), the miR-185 levels in the Dgcr8+/− neurons decreased by 31% as compared to the Dgcr8+/+ neurons (Figure 6A). Treatment with Co(III)PPIX at all three concentrations elevated the miR-185 expression (Figure 6A). Even at the lowest concentration of 0.2 μM, the increase of miR-185 to 89% of wild type is statistically significant. At 20 μM, Co(III)PPIX restored miR-185 to a level indistinguishable to that of the wild type. Co(III)PPIX at 20 μM also significantly increased the expression of another miRNA, miR-134, that is negatively affected by Dgcr8 haploinsufficiency in cultured primary neurons (Figure 6B). The neurons all looked healthy with processes developing under all conditions (Figure 6C). Importantly, little activated (cleaved) caspase-3 was detected, indicating lack of apoptosis in the treated cells. These experiments demonstrate that Co(III)PPIX is capable of correcting the miRNA processing deficiency caused by reduced Dgcr8 gene dosage and thus pave the way to further explore the value of this metalloporphyrin in treating 22q11.2DS and other diseases linked to deficient miRNA processing.
Figure 6.
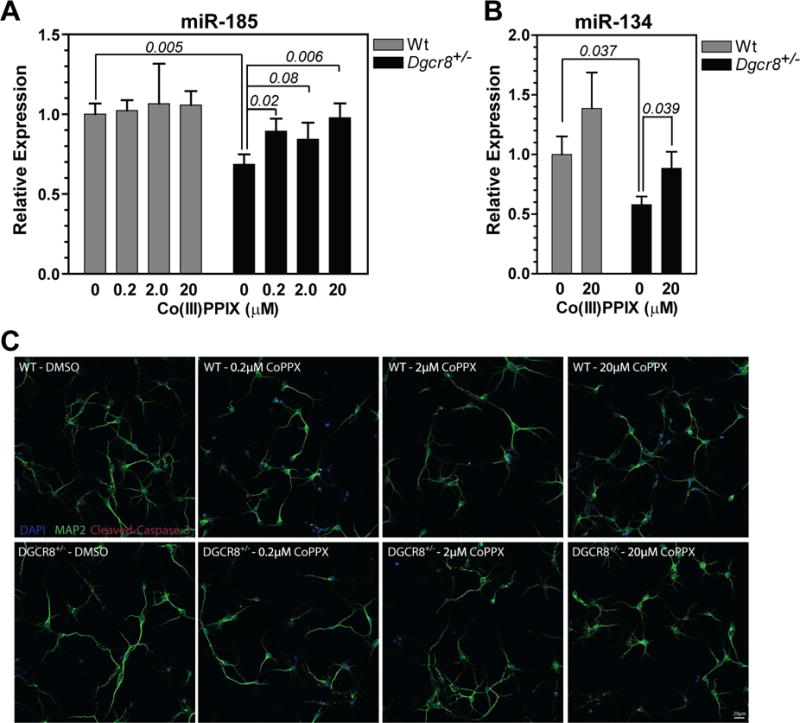
Co(III)PPIX restores deficient miRNA expression caused by heterozygous deletion of the Dgcr8 gene in mice. Primary cortical neurons dissected from Dgcr8+/− mouse embryos and their wild-type littermates were treated with Co(III)PPIX at the indicated concentrations between DIV 1 and 3, for 48 h. (A,B) Mature miR-185 (A) and miR-134 (B) levels after normalization to that of the Gapdh mRNA. The miR-134 data at lower Co(III)PPIX concentrations did not reach statistical significance and thus are not shown. Each plotted value is average ± SEM from seven independent experiments using seven animals for each genotype. Select P values are calculated using two-tailed Student’s t-test and are indicated on the graph. (C) Images of the treated and control neurons with processes visualized via MAP2 staining (shown in green).
DISCUSSION
Here we identify Co(III)PPIX as an activator of pri-miRNA processing. This activator mimics the required heme cofactor of DGCR8 but is not subjected to degradation by heme oxygenases. DGCR8 and its homologs constitute a unique class of heme protein in that they use two cysteine side chains (Cys352 from both subunits in a dimer) to ligate the heme iron (Barr, et al., 2012; Barr, et al., 2011). At least one (likely both) of the Cys352 side chains are in the deprotonated thiolate state. The electron-rich environment contributes to a tremendous preference for DGCR8 to bind Fe(III) heme over Fe(II) heme. The human DGCR8 does not dissociate with Fe(III) heme substantially even after incubation at room temperature for 4 days (koff < 3 × 10−6 s−1) with large excess of heme scavenger apomyoglobin (Barr, et al., 2011). In contrast, the dissociation rate of Fe(II) heme from DGCR8 cannot be measured using the stop flow method (koff > 100 s−1) (Barr, et al., 2012). Thus, a single electron (the difference between Fe(III) and Fe(II)) causes a >107-fold change in koff. The reduction of the heme iron causes loss of both cysteine ligands (Barr, et al., 2012), resulting in near elimination of DGCR8 binding and activation activities.
Among the metalloporphyrins with the central atoms highlighted in Figure 1B, only Co(III)PPIX and Fe(III)PPIX (heme) are capable of stably binding DGCR8 and activating its miRNA processing activity. Cobalt and iron in the 3+ state contain 5 and 6 electrons in their d shells respectively, leaving those orbitals approximately half empty and thereby favoring the octahedral coordination. Our previous EPR study indicated that the Fe(III) in DGCR8 is in a low-spin S = ½ state, with one unpaired electron (Barr, et al., 2011). The spin state of Co(III)PPIX in DGCR8 remains to be determined. Both Co and Fe are redox active with another most accessible state being 2+ when chelated by porphyrins. Similar to Fe(II)PPIX, the Co(II)PPIX does not stably bind DGCR8. The stringent metal ion and redox state requirements are likely to be demanded by the dual cysteine-ligation of DGCR8 and the electron-rich environment.
A key difference between CoPPIX and heme is that CoPPIX is not degraded by the heme degradation enzymes heme oxygenases (Kikuchi, et al., 2005). This property makes Co(III)PPIX much more stable in the body. Subcutaneous injection to rats results in retention of the compound in spleen, gonads, lung, and thymus for up to 4 weeks (Rosenberg, 1993). Furthermore, Kappa and colleagues treated rats with CoPPIX and observed a substantial decrease in body fat contents and overall bodyweight (Galbraith and Kappas, 1989). Either subcutaneous injection or intracerebroventricular injection into the brain was effective. The effects persisted over a long period of time, with those from a single intracerebroventricular dosage at 0.2 or 0.4 μmol per kg of body weight lasting strikingly for over 300 days. These effects were not produced by cobalt chloride, heme or other metalloporphyrins tested, therefore appear to be specific to CoPPIX. No histological abnormalities were observed in the brain, suggesting that administration of CoPPIX at low dosages may be safe for treating diseases such as 22q11.2DS. Additionally, there has been a substantial body of works trying to identify the in vivo targets of CoPPIX that contribute to the “slimming” effects, including heme oxygenase-1 (HO-1) (Csongradi, et al., 2012) and nitric oxide synthase (Li, et al., 2006). Our study suggests that boost of miRNA processing is another major effect of CoPPIX.
CoPPIX has been shown to potently induce the expression of HO-1, even though the compound itself is a competitive inhibitor of heme oxygenase enzymes (Dercho, et al., 2006; Shan, et al., 2006; Shan, et al., 2000). Previous studies showed that 5–10 μM of CoPPIX is needed to induce HO-1 expression (Dercho, et al., 2006; Shan, et al., 2006; Shan, et al., 2000). In the current study, we show that Co(III)PPIX is effective in activating pri-miRNA processing in human cells at concentrations as low as 0.2 μM. Thus, it should be possible to activate DGCR8 without substantially inducing HO-1. On the other hand, induction of HO-1 has major cytoprotective and other beneficial physiological effects (Motterlini and Foresti, 2014; Wu, et al., 2011). These effects are elicited by the products of the heme degradation reaction—biliverdin is a potent antioxidant and carbon monoxide has anti-inflammatory and anti-apoptotic activities. Therefore, activation of HO-1 by CoPPIX has been proposed as a potential therapeutic strategy for treating cardiovascular diseases and obesity (Csongradi, et al., 2012; Galbraith and Kappas, 1990). It may be beneficial to activate both pri-miRNA processing and HO-1 using modest dosages of Co(III)PPIX. Elevated heme oxygenase expression and activity would reduce the intracellular heme availability, which would in turn be expected to reduce pri-miRNA processing. At 10 or 20 μM of Co(III)PPIX, direct activation of DGCR8 appears to have a dominating effect on enhancing pri-miRNA processing efficiency over the potential indirect inhibition via HO-1 induction (Figures 5 and 6). The tight interaction between DGCR8 and Co(III)PPIX must have contributed to this overall positive effect of Co(III)PPIX on pri-miRNA processing.
CoPPIX shares major chemical properties with heme, and thus is able to interact with additional heme proteins and possibly affect their respective biological pathways. For example, CoPPIX has been shown to inhibit the rate-limiting heme synthesis enzyme δ-aminolevulenic acid synthase (Schoenfeld, et al., 1984). The combination of heme synthesis inhibition and degradation (the latter from induction of HO-1) causes a reduction of cytochrome P-450 in the liver (Drummond and Kappas, 1982). Cobalt-substituted cytochrome P-450cam, cystathionine β-synthase and horseradish peroxidase can be biochemically reconstituted using the corresponding apoproteins and CoPPIX (Smith, et al., 2011; Wagner, et al., 1981; Wang, et al., 1977), although it is not clear whether CoPPIX is incorporated to these heme proteins in vivo. Therefore, caution should be exercised when considering CoPPIX in long-term systemic treatments, especially at medium to high concentrations.
In summary, we found that Co(III)PPIX binds DGCR8 tightly and activates its pri-miRNA processing activity potently. We show that this compound is effective in correcting pri-miRNA processing defects by activating the DGCR8 protein in mouse neurons that mimic the DGCR8 haploinsufficiency in 22q11.2DS (Stark, et al., 2008; Xu, et al., 2013). Co(III)PPIX is not degraded by the heme degradation enzymes, thus has highly favorable pharmacokinetic half-lives (Galbraith and Kappas, 1989; Rosenberg, 1993). It works at low concentration, allowing the interaction with other heme proteins and potential undesired effects to be limited. These properties qualify Co(IIII)PPIX as a potential therapeutic for diseases in which pri-miRNA processing deficiency is involved. As pri-miRNA processing is highly regulated in broad biological contexts (Davis and Hata, 2009; Ha and Kim, 2014), Co(III)PPIX will also be useful as a research tool.
SIGNIFICANCE
miRNAs are extensively involved in development and cell physiology, and reduced miRNA production has been linked to many diseases. In cancers, decreased expression of miRNA processing factors has been extensively documented. Another disease, 22q11.2DS, is caused by the most common human chromosome microdeletion, with an incidence of 1 in 2,000–4,000 live births. Concrete evidence has demonstrated that these miRNA processing defects directly contribute to pathogenesis. However, it has not been possible to correct them pharmacologically. Here using a combined biochemical and cellular approach, we identify a miRNA processing activator, cobalt (III) protoporphyrin IX. This compound binds and activates DGCR8 by mimicking its essential heme cofactor, but is not degraded by heme degradation enzymes. Co(III)PPIX corrects the miRNA processing deficiency caused by heterozygous deletion of DGCR8 in cultured mouse neurons. Our study demonstrates that it is possible to compensate miRNA processing defects by activating the processing proteins.
EXPERIMENTAL PROCEDURES
Materials
Hemin and Co(III)PPIX were purchased from Sigma (St. Louis, MO, USA). All other metalloporphyrins were obtained from Frontier Scientific (Logan, UT, USA).
Expression and purification of recombinant DGCR8
Fe(III) NC1, apoNC1 and apoNC1 P351A proteins were purified as described previously (Barr, et al., 2012; Barr, et al., 2011). Briefly, the constructs were expressed in the pET-24a+ vector in E. coli and purified using cation exchange chromatography followed by size-exclusion chromatography (SEC). The SEC buffer contained 20 mM Tns pH 8.0, 400 mM NaCl, and 1 mM DTT. Reduction of the Fe(III) NC1 was accomplished using 2 mM sodium dithionite in 50 mM 2-(N-morpholino)ethanesulfonic acid (MES) pH 6.0, 400 mM NaCl, and 1 mM DTT.
DGCR8-porphyrin binding assay
All porphyrin solutions were made fresh on the day of use and stored at ≤ 4°C. Protoporphyrin IX was initially dissolved in 50% DMSO and 50% 0.1 M NaOH, concentration was measured (~1 mM), and diluted into the final binding buffer (pH 6 or 8). Metalloporphyrins were initially dissolved in 0.1 M NaOH, and then diluted to 100 μM stocks using binding buffer. The concentrations were determined either gravimetrically (Rh and In) or using published extinction coefficients. Several published absorbance peaks and extinction coefficient values are listed in Table S1.
The porphyrin-binding reaction buffer contained 1 mM DTT, 400 mM NaCl, and the buffer was either 50 mM MES pH 6.0 (for wild-type apoNC1) or 20 mM Tris pH 8.0 (for apoNC1 P351A). Absorbance spectra were collected using either a Varian Cary 300 Bio spectrophotometer with bandwidth set to 1 nm (Figures 2A and 3A,C), or a DU800 spectrophotometer (Beckman-Coulter, bandwidth ≤ 1.8 nm) (Figures S1, S2 and S3).
Reconstituted pri-miRNA processing assays
The assays were done as described previously (Barr, et al., 2012). The reactions were incubated at 37°C for either 30 min (pri-miR-23a and pri-miR-30a) or 45 min (pri-miR-21 and pri-miR-380), and were analyzed using 15% acrylamide, 7 M urea denaturing gels and autoradiography. The bands were quantified using the ImageQuant software (GE Healthcare). The intensities of pre-miRNAs were converted to the equivalents of pri-miRNAs by multiplying the ratio of the numbers of 32P-containing residues in the pri-miRNA and pre-miRNA p values were calculated using two-tailed Student’s t-test and Mann-Whitney U-test.
Cellular pri-miRNA processing assay
The cellular assays were performed as described previously (Mukherji, et al., 2011; Weitz, et al., 2014). Briefly, cells were grown in heme-depleted media for 24 h before splitting into smaller plates for both imaging and RNA analysis. For the experiments in Figure 5A–B, cells were transfected with the reporter. Eighteen hours posttransfection, 1 mM SA, either alone or with 0.2, 1 or 10 μM Co(III)PPIX, or 10 μM hemin, was added to the cells, and 1 μg/mL doxycycline was also added to induce expression of reporters. Co(III)PPIX and hemin were dissolved in DMSO shortly before addition to the cell cultures. After 10 h of treatment, cells were used either for imaging or for total RNA extraction. Imaging and data analysis were performed as described (Weitz, et al., 2014). Total RNAs were extracted using the miRNeasy mini kit (Qiagen). Taqman qRT-PCR assays (Life Technologies) were used to measure mature miR-9 and β-actin mRNA levels. A similar procedure was used for the experiments in Figures 5D–5F, except that the N-flag-DGCR8-expression plasmid or the pCMV-Tag2A vector (as a control) was included in the transfection together with the reporter. To assure that N-flag-DGCR8 was expressed under the heme-deficient and/or porphyrin-treated condition, SA, Co(III)PPIX, hemin were added 2 h prior to the transfection. Doxycycline was added shortly after the transfection. Imaging was done 16–18 h posttransfection.
Treatment of mouse Dgcr8+/− neurons using Co(III)PPIX
All animal protocols used in this study are approved by Columbia University IACUC. Dgcr8+/− mice have been described previously (Stark, et al., 2008) and have been backcrossed into C57BL/6J background for over ten generations. After embryonic dissection, dissociated neurons from individual pups were cultured at ~2 × 106 neurons per condition. At DIV 1, cells were treated with Co(III)PPIX or DMSO (no porphyrin control) as indicated in Figure 6. Co(III)PPIX was dissolved in DMSO. At DIV 3, total RNA was isolated using miRNeasy mini kit (Qiagen). Taqman qRT-PCR assays (Life Technologies) were used to measure mature miR-185, miR-134 and Gapdh mRNA levels, as described previously (Stark, et al., 2008; Xu, et al., 2013).
Supplementary Material
Table 1.
DGCR8 binding properties of metalloporphyrins.
| MPPIX | Soret peak, free (nm) |
Soret peak, bound (nm) |
Stably bind DGCR8 | Activate pri-miRNA processing |
|---|---|---|---|---|
| Fe(III) | 384 | 367, 450 | Yes | Yes |
| Fe(II) | ~390 | N.D. | No | No |
| Co(III) | 418 | 368, 456 | Yes | Yes |
| Co(II) | 397 | 398 | No | N.D. |
The properties of Fe(III) and Fe(II) heme were known from a previous study (Barr, et al., 2012) and are shown for comparison. N.D., not determined.
HIGHLIGHTS.
Co(III) protoporphyrin IX (PPIX) stably binds DGCR8 like its natural heme cofactor
Co(III)PPIX activates DGCR8 for pri-miRNA processing in vitro and in HeLa cells
Co(III)PPIX compensates the pri-miRNA processing defects in Dgcr8+/− mouse neurons
Co(III)PPIX is effective at low concentrations & is not degraded by heme oxygenases
Acknowledgments
We thank J. Valentine and K. Barnese for use of the anaerobic chamber and J. Feigon for use of spectrophotometer. This project was supported by National Institutes of Health Grants GM080563 (to F.G.), MH67068 (M.K. and J.A.G.), MH077235 (J.A.G.), T32GM008496 (S.H.W.). S.H.W. and I.B. were supported by UCLA Dissertation Year Fellowships. S.W. acknowledges support from Willard Chair funds.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
I.B. and F.G. conceived the project, I.B. performed all the biochemical experiments, S.H.W. and S.W. are responsible for the HeLa cells experiments, T.A., P.H., M.K. and J.A.G. are responsible for the mouse neuron experiments. I.B., S.H.W. and F.G. wrote the paper with input from other authors.
References
- Barr I, Guo F. Primary microRNA processing assay reconstituted using recombinant Drosha and DGCR8. Methods Mol Biol. 2014;1095:73–86. doi: 10.1007/978-1-62703-703-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr I, Smith AT, Chen Y, Senturia R, Burstyn JN, Guo F. Ferric, not ferrous, heme activates RNA-binding protein DGCR8 for primary microRNA processing. Proc Natl Acad Sci USA. 2012;109:1919–1924. doi: 10.1073/pnas.1114514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr I, Smith AT, Senturia R, Chen Y, Scheidemantle BD, Burstyn JN, Guo F. DiGeorge Critical Region 8 (DGCR8) is a double-cysteine-ligated heme protein. J Biol Chem. 2011;286:16716–16725. doi: 10.1074/jbc.M110.180844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge NJ, Cairns MJ. MicroRNA dysregulation in schizophrenia. Neurobiol Dis. 2012;46:263–271. doi: 10.1016/j.nbd.2011.12.029. [DOI] [PubMed] [Google Scholar]
- Csongradi E, Docarmo JM, Dubinion JH, Vera T, Stec DE. Chronic HO-1 induction with cobalt protoporphyrin (CoPP) treatment increases oxygen consumption, activity, heat production and lowers body weight in obese melanocortin-4 receptor-deficient mice. Int J Obes. 2012;36:244–253. doi: 10.1038/ijo.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hata A. Regulation of MicroRNA Biogenesis: A miRiad of mechanisms. Cell Commun Signal. 2009;7:18. doi: 10.1186/1478-811X-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Dercho RA, Nakatsu K, Wong RJ, Stevenson DK, Vreman HJ. Determination of in vivo carbon monoxide production in laboratory animals via exhaled air. J Pharmacol Toxicol Methods. 2006;54:288–295. doi: 10.1016/j.vascn.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Drummond GS, Kappas A. The cytochrome P-450-depleted animal: an experimental model for in vivo studies in chemical biology. Proc Natl Acad Sci U S A. 1982;79:2384–2388. doi: 10.1073/pnas.79.7.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller M, Matsunaga M, Yin S, Loo JA, Guo F. Heme is involved in microRNA processing. Nat Struct Mol Biol. 2007;14:23–29. doi: 10.1038/nsmb1182. [DOI] [PubMed] [Google Scholar]
- Faller M, Toso D, Matsunaga M, Atanasov I, Senturia R, Chen Y, Zhou ZH, Guo F. DGCR8 recognizes primary transcripts of microRNAs through highly cooperative binding and formation of higher-order structures. RNA. 2010;16:1570–1583. doi: 10.1261/rna.2111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenelon K, Mukai J, Xu B, Hsu PK, Drew LJ, Karayiorgou M, Fischbach GD, Macdermott AB, Gogos JA. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc Natl Acad Sci USA. 2011;108:4447–4452. doi: 10.1073/pnas.1101219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith RA, Kappas A. Cobalt-protoporphyrin suppresses expression of genetic obesity in homozygous (fa/fa) Zucker rats. Pharmacology. 1990;41:292–298. doi: 10.1159/000138738. [DOI] [PubMed] [Google Scholar]
- Galbraith RA, Kappas A. Regulation of food intake and body weight by cobalt porphyrins in animals. Proc Natl Acad Sci U S A. 1989;86:7653–7657. doi: 10.1073/pnas.86.19.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong M, Chen Y, Senturia R, Ulgherait M, Faller M, Guo F. Caspases cleave and inhibit the microRNA processing protein DiGeorge Critical Region 8. Protein Sci. 2012;21:797–808. doi: 10.1002/pro.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KM, Pimienta G, DeGregorio SJ, Alexandrov A, Steitz JA. Phosphorylation of DGCR8 increases its intracellular stability and induces a progrowth miRNA profile. Cell Rep. 2013;5:1070–1081. doi: 10.1016/j.celrep.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappas A, Drummond GS. Control of heme metabolism with synthetic metalloporphyrins. J Clin Invest. 1986;77:335–339. doi: 10.1172/JCI112309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Simon TJ, Gogos JA. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat Rev Neurosci. 2010;11:402–416. doi: 10.1038/nrn2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi G, Yoshida T, Noguchi M. Heme oxygenase and heme degradation. Biochem Biophys Res Commun. 2005;338:558–567. doi: 10.1016/j.bbrc.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Li MY, Vizzard MA, Jaworski DM, Galbraith RA. The weight loss elicited by cobalt protoporphyrin is related to decreased activity of nitric oxide synthase in the hypothalamus. J Appl Physiol. 2006;100:1983–1991. doi: 10.1152/japplphysiol.01169.2005. [DOI] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, Gregory RI. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motterlini R, Foresti R. Heme oxygenase-1 as a target for drug discovery. Antioxid Redox Signal. 2014;20:1810–1826. doi: 10.1089/ars.2013.5658. [DOI] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick-Cleveland J, Jacob JP, Weitz SH, Shoffner G, Senturia R, Guo F. The DGCR8 RNA-binding heme domain recognizes primary microRNAs by clamping the Hairpin. Cell Rep. 2014;7:1994–2005. doi: 10.1016/j.celrep.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg DW. Pharmacokinetics of cobalt chloride and cobalt-protoporphyrin. Drug Metab Dispos. 1993;21:846–849. [PubMed] [Google Scholar]
- Schoenfeld N, Wysenbeek AJ, Greenblat Y, Epstein O, Atsmon A, Tschudy DP. The effects of metalloporphyrins, porphyrins and metals on the activity of delta-aminolevulinic acid synthase in monolayers of chick embryo liver cells. Biochem Pharmacol. 1984;33:2783–2788. doi: 10.1016/0006-2952(84)90696-8. [DOI] [PubMed] [Google Scholar]
- Schofield CM, Hsu R, Barker AJ, Gertz CC, Blelloch R, Ullian EM. Monoallelic deletion of the microRNA biogenesis gene Dgcr8 produces deficits in the development of excitatory synaptic transmission in the prefrontal cortex. Neural Dev. 2011;6:11. doi: 10.1186/1749-8104-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolaro LM, Castriciano M, Romeo A, Patane S, Cefali E, Allegrini M. Aggregation behavior of protoporphyrin IX in aqueous solutions: Clear evidence of vesicle formation. J Phys Chem B. 2002;106:2453–2459. [Google Scholar]
- Senturia R, Faller M, Yin S, Loo JA, Cascio D, Sawaya MR, Hwang D, Clubb RT, Guo F. Structure of the dimerization domain of DiGeorge Critical Region 8. Protein Sci. 2010;19:1354–1365. doi: 10.1002/pro.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y, Lambrecht RW, Donohue SE, Bonkovsky HL. Role of Bach1 and Nrf2 in up-regulation of the heme oxygenase-1 gene by cobalt protoporphyrin. FASEB J. 2006;20:2651–2653. doi: 10.1096/fj.06-6346fje. [DOI] [PubMed] [Google Scholar]
- Shan Y, Pepe J, Lu TH, Elbirt KK, Lambrecht RW, Bonkovsky HL. Induction of the heme oxygenase-1 gene by metalloporphyrins. Arch Biochem Biophys. 2000;380:219–227. doi: 10.1006/abbi.2000.1921. [DOI] [PubMed] [Google Scholar]
- Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem Biophys Res Commun. 2003;304:184–190. doi: 10.1016/s0006-291x(03)00554-0. [DOI] [PubMed] [Google Scholar]
- Smith AT, Majtan T, Freeman KM, Su Y, Kraus JP, Burstyn JN. Cobalt cystathionine beta-synthase: a cobalt-substituted heme protein with a unique thiolate ligation motif. Inorg Chem. 2011;50:4417–4427. doi: 10.1021/ic102586b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- Wada T, Kikuchi J, Furukawa Y. Histone deacetylase 1 enhances microRNA processing via deacetylation of DGCR8. EMBO Rep. 2012;13:142–149. doi: 10.1038/embor.2011.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GC, Gunsalus IC, Wang MY, Hoffman BM. Cobalt-substituted cytochrome P-450cam. J Biol Chem. 1981;256:6266–6273. [PubMed] [Google Scholar]
- Wang MY, Hoffman BM, Hollenberg PF. Cobalt-substituted horseradish peroxidase. J Biol Chem. 1977;252:6268–6275. [PubMed] [Google Scholar]
- Weitz SH, Gong M, Barr I, Weiss S, Guo F. Processing of microRNA primary transcripts requires heme in mammalian cells. Proc Natl Acad Sci U S A. 2014;111:1861–1866. doi: 10.1073/pnas.1309915111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ML, Ho YC, Lin CY, Yet SF. Heme oxygenase-1 in inflammation and cardiovascular disease. Am J Cardiovasc Dis. 2011;1:150–158. [PMC free article] [PubMed] [Google Scholar]
- Xu B, Hsu PK, Stark KL, Karayiorgou M, Gogos JA. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell. 2013;152:262–275. doi: 10.1016/j.cell.2012.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


