SUMMARY
We have defined the molecular basis for association of the PH domain of the Arf GAP ASAP1 with phospholipid bilayers. Structures of the unliganded and dibutyryl PtdIns(4,5)P2-bound PH domain were solved. PtdIns(4,5)P2 made contact with both a canonical site (C site) and an atypical site (A site). We hypothesized cooperative binding of PtdIns(4,5)P2 to the C site and a nonspecific anionic phospholipid to the A site. PtdIns(4,5)P2 dependence of binding to large unilamellar vesicles and GAP activity was sigmoidal, consistent with cooperative sites. In contrast, PtdIns(4,5) P2 binding to the PH domain of PLC δ1 was hyperbolic. Mutation of amino acids in either the C or A site resulted in decreased PtdIns(4,5)P2-dependent binding to vesicles and decreased GAP activity. The results support the idea of cooperative phospholipid binding to the C and A sites of the PH domain of ASAP1. We propose that the mechanism underlies rapid switching between active and inactive ASAP1.
INTRODUCTION
PH domains are regulatory components of hundreds of human proteins involved in signaling, membrane traffic, and actin cytoskeleton remodeling (DiNitto and Lambright, 2006; Lemmon, 2008; Lemmon et al., 1996; Moravcevic et al., 2012). The domain is defined structurally as a sandwich of seven β strands capped at one end by an α helix. A subset of PH domains binds to phosphoinositides and proteins. Several mechanisms by which ligand binding to PH domains regulates protein activity have been proposed, but the kinetic and molecular basis for rapid switching between the ligand bound and unliganded forms of PH domains has not been explored.
A major function of PH domains is to localize proteins to specific membrane regions through binding to specific phosphoinositides (Moravcevic et al., 2012). For example, proteins containing phosphatidylinositol 3,4,5-triphosphate (PtdIns(3,4,5)P3)-binding PH domains are recruited to membranes in which PtdIns(3,4,5)P3 is produced. Membrane localization may be further specified by the coincidence of two signals, which was first described for two independent domains within a single protein, each domain having distinct ligand specificities (Moravcevic et al., 2012). Coincidence detection may also be mediated by a single PH domain binding two distinct ligands (Balla, 2005), as recently described for FAPP1, which simultaneously binds phosphoinositides and Arf1-GTP (Godi et al., 2004; He et al., 2011; Liu et al., 2014) and Grp1, which simultaneously binds PtdIns(3,4,5)P3 and Arf6-GTP (DiNitto et al., 2007; Malaby et al., 2013). The two ligands can also be different lipids, as seen with the yeast Slm1p PH domain (Gallego et al., 2010).
PH domains can function in capacities other than localization signals (DiNitto and Lambright, 2006). For example, in some Dbl-family Rho GTPase exchange factors (Rho GEFs), PH domains can form part of the substrate-binding site (Rossman et al., 2002) and, in other Rho GEFs, occlude the enzymatic site (He et al., 2013). PH domains of cytohesin family Arf guanine nucleotide exchange factors (Arf GEFs) position autoinhibitory motifs that are displaced on binding to Arf6-GTP (DiNitto et al., 2007; Malaby et al., 2013). In Akt the PH domain inhibits the associated catalytic domain, a serine/threonine kinase (Alessi et al., 1997; Chan et al., 1999; Milburn et al., 2003; Thomas et al., 2002).
Whether through localization or direct effects on catalytic sites, PH domain function in cellular signaling can be expected to involve rapid and complete switching between states. G proteins are another example of a protein motif used in signaling (Sprang, 1997; Vetter and Wittinghofer, 2001; Wittinghofer and Vetter, 2011). G proteins achieve rapid state transitions, necessary for function as molecular switches, through a cycle of binding and hydrolysis of GTP. PH domains, in contrast, do not have a catalytic function and depend on binding and dissociation of activating ligands.
Rapid and efficient switching driven by ligand binding and dissociation may be achieved by cooperativity. We propose that cooperative binding to two lipid ligands may be a function of a subset of PH domains. Indeed, two distinct lipid-binding sites have been described in PH domains: a canonical site between the β1/β2, β3/β4, and β6/β7 loops, and an “atypical” binding site on the opposite side of the β1/β2 loop, adjacent to the β5/β6 loop (Balla, 2005; DiNitto and Lambright, 2006; Moravcevic et al., 2012). The now atypical site was actually seen first in the structure of the Ins(1,4,5)P3-bound spectrin-β2 PH domain (Hyvonen et al., 1995), and has also been observed in Tiam1 and ArhGAP9 PH domains (Ceccarelli et al., 2007). Although in most cases examined only the canonical or non-canonical site is occupied, cooperative binding to both sites has been proposed for the PH domain of the yeast protein Slm1 (Anand et al., 2012; Gallego et al., 2010). Furthermore, biochemical evidence supports the existence of two lipid-binding sites in two other PH domains, those of Akt (Huang et al., 2011) and ASAP1 (Kam et al., 2000).
In the current study, we used the PH domain of ASAP1 to study the mechanism of cooperative lipid binding to two sites in a single PH domain (Brown et al., 1998; Che et al., 2005; Kam et al., 2000; Luo et al., 2008). ASAP1 is an Arf GTPase-activating protein (Arf GAP) that catalyzes the conversion of Arf-GTP to Arf-GDP by inducing the hydrolysis of GTP. ASAP1 is structurally complex, containing BAR, PH, Arf GAP, Ank repeat, Proline-rich, and SH3 domains. The SH3 and Proline-rich domains mediate targeting of ASAP1 to specific cellular sites by protein-protein interaction (Liu et al., 2002; Oda et al., 2003). The PH domain does not drive localization, but is nevertheless crucial for ASAP1 regulation. Phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) binds to the PH domain to increase enzymatic activity approximately 10,000-fold (Kam et al., 2000). Two cooperative lipid-binding sites have been biochemically defined for the ASAP1 PH domain. Mutagenesis was previously used to tentatively identify a canonical PtdIns(4,5)P2-binding site (Kam et al., 2000), but neither lipid-binding site has been unambiguously identified at atomic resolution, nor has the physical basis of apparent cooperativity been determined.
Here, we have solved the crystal structures of the unliganded PH domain of ASAP1 and the PH domain with dibutyryl PtdIns(4,5)P2 bound. The structures form the basis of our hypothesis of cooperative binding to canonical (C) and atypical (A) sites in the PH domain. Nuclear magnetic resonance (NMR) and mutagenesis results supported PtdIns(4,5)P2 binding to the C and A sites. We speculate that cooperativity enables rapid and reversible activation of ASAP1 through ligand binding and dissociation.
RESULTS
Structures of Unliganded and Dibutyryl PtdIns(4,5)P2-Bound ASAP1 PH Domain
We determined the structure of the PH domain (residues G325–D451 of ASAP1, Figure 1A) by X-ray crystallography. Crystals of purified ASAP1 PH domain were obtained and diffracted X-rays to 1.78 Å resolution using a synchrotron radiation source. The structure was determined by molecular replacement using the coordinates (PDB: 2DA0) from NMR studies and refined with a final Rwork and Rfree of 0.23 and 0.29, respectively (Table 1).
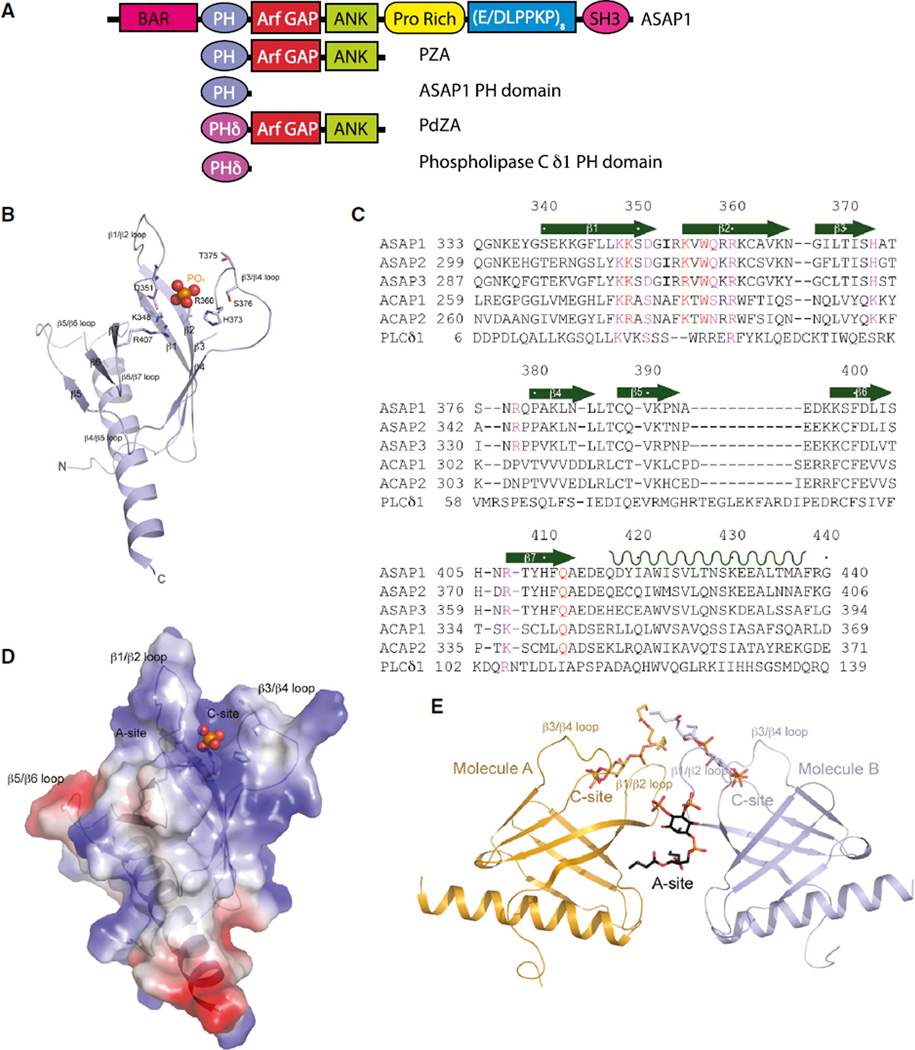
Figure 1. Structures of Unliganded PH Domain of ASAP1 and Its Complex with diC4-PtdIns(4,5)P2.
(A) Schematic representation of proteins used in this study.
(B) Ribbon representation of the structure of monomeric unliganded ASAP1 PH domain. Both N and C termini are labeled along with all β strands, as well as loops linking the β strands. The putative phosphate group is shown as a ball model with the phosphorus atom in orange and oxygen atoms in red. Residues that are interacting with the phosphate group are shown as stick models.
(C) Sequence of the ASAP1 PH domain aligned with other PH domains. Assignment of secondary structural elements defined by the crystal structure is shown. ASAP1, GI |4063616; ASAP2, GI |48474259; ASAP3, GI |221307473; ACAP1, GI |7661880; ACAP2, GI |39932727; PLCδ1, GI |130228.
(D) Electrostatic potential of monomeric ASAP1 PH domain. The surface representation of the electrostatic potential is superimposed on the ribbon structure of the ASAP1 PH domain. The phosphate group is shown as a ball model.
(E) One possible dimeric association of the two ASAP1 PH domains (A and B) in crystal. Ribbon representation of two molecules (A in coral and B in purple) in the asymmetric unit is shown. The two different PtdIns(4,5)P2 binding sites are illustrated as A site formed at the dimer interface and C site formed between β1/β2 and β3/β4 loops.
See also Figures S1, S2 and S3.
Table 1.
Statistics on Qualities of X-Ray Diffraction Datasets and Atomic Models
| ASAP1 PH Domain |
ASAP1 PH Domain:diC4- PtdIns(4,5)P2 |
|
|---|---|---|
| Diffraction Datasets | ||
| X-Ray wavelength (Å) | 1.0 | 1.54 |
| Space group | P43 | P21 |
| Cell dimensions (Å, °) | a = b = 48.12, c = 111.33 | a = 37.54, b = 64.70, c = 44.43, β = 95.63° |
| Resolution (Å) | 50–1.80 (1.86–1.80)a | 50–1.60 (1.66–1.60) |
| Merging R factors | 0.099 (0.515) | 0.075 (0.481) |
| Completeness (%) | 95.4 (75.9) | 97.6 (82.9) |
| No. of unique reflections | 22,383 | 27,403 |
| Redundancy | 4.6 (1.9) | (3.2) 1.9 |
| I/σI | 15.6 (1.35) | 15.3 (1.1) |
| Model Refinement | ||
| Resolution refined (Å) | 20–1.81 | 27–1.60 |
| Rwork | 0.21 (0.346) | 0.20 (0.474) |
| Rfree | 0.26 (0.376) | 0.23 (0.486) |
| No. of molecules/AU | 2 | 2 |
| No. of ligands/AU | 2 PO43− | 3 diC4-PtdIns(4,5)P2 |
| No. of protein atoms | 1,765 | 1,667 |
| No. of solvent molecules | 34 | 222 |
| No. of polymers | 3 | – |
| Ramachandran plot (%) | ||
| Preferred | 98.12 | 97.04 |
| Allowed | 0.47 | 2.96 |
| Outlier | 1.41b | 0 |
| PDB | 5C6R | 5C79 |
Numbers in parentheses are for the outermost-resolution shell.
Outliers are located in the disordered regions.
The ASAP1 PH domain displays a “stripped-down,” canonical PH domain fold of seven-stranded β sandwich consisting of N-terminal four-stranded (β1–β4) and C-terminal three-stranded (β5–β7) β sheets, and is terminated with a long α helix wedged between the two sheets of the β sandwich at one end (Figures 1B and 1C). Three of the loops connecting β strands are disordered in the unliganded structure, the β1/β2, β3/β4, and β5/β6 loops. Like other phospholipid-binding PH domains, the surface charge distribution is polarized (Figure 1D), consistent with its proposed function of membrane bilayer interaction. Strong, pyramid-shaped electron density was found in a surface cavity surrounded by positively charged residues from β1/β2, β3/β4, and β6/β7 loops, which corresponds to the canonical phosphoinositide (PtdIns)-binding site. Since sodium phosphate is present in the crystallization conditions, this density likely represents a phosphate molecule occupying the C site.
We also prepared the complex between ASAP1 PH domain and dibutyryl PtdIns(4,5)P2 (ASAP1 PH domain/diC4-PtdIns(4,5) P2) by incubating the two components in a molar ratio of 1:2.5. Crystals of ASAP1 PH domain/diC4-PtdIns(4,5)P2 complex diffracted X-rays to 1.6 Å resolution using an in-house X-ray source (Table 1). The structure was again determined by molecular replacement, using the coordinates of the unliganded ASAP1 PH domain as a starting model, and was refined to Rwork and Rfree values of 0.20 and 0.24, respectively.
In the ASAP1 PH domain/diC4- PtdIns(4,5)P2 complex crystal, there are two ASAP1 PH domain molecules (A and B) in a crystallographic asymmetric unit. Among the ways of selecting two monomers to form the asymmetric unit, one dimer pair is interesting due to its polarized surface charges and binding positions of the ligand (Figure 1E). The difference Fourier map showed a large peak of electron density in each of the canonical PtdIns(4,5)P2 binding sites, which matched the shape of the head group of diC4-PtdIns(4,5)P2. In addition, a large electron density peak was seen at the A site, present at the interface formed between the two monomers in the crystallographic asymmetric unit, which fits a diC4-PtdIns(4,5)P2 moiety (Figure 1E). This bound diC4-PtdIns(4,5)P2 is more ordered than those found in the canonical sites, and its chemical identity was verified by the presence of anomalous peaks for all three phosphate groups (Figure S1A), compared with the bound diC4-PtdIns(4,5)P2 at the C site that have weaker anomalous difference Fourier density (Figure S1B). Among the five residues that participate in interacting with this ligand, four come from molecule B (Q412, K349, K355, and W357), and are residues in the A site. NMR chemical-shift studies also argued that PtdIns(4,5)P2 interacts with both the C site and the A site in solution. In 15N heteronuclear single-quantum coherence experiments (Figure S1C), PtdIns(4,5)P2 titration resulted in chemical-shift perturbations for K349 and W357 in the A site and K348, R360, R378, and R407 in the C site, thus establishing that the interactions are not the result of crystal packing effects.
We found that the binding orientation of diC4-PtdIns(4,5)P2 in the C site of one molecule is different from that in the other, with the inositol ring flipped 180° (Figures S2A and S2B), although in both binding modes the same set of residues are employed for interactions. Consequently, residues in the two binding sites adopt slightly different conformations that best fit the bound ligand. This is not surprising because most of the protein-ligand binding energy appears to be derived from charge-charge interactions involving the two phosphate groups at the 4- and 5-positions. Indeed, the phosphates at 4- and 5-positions are super-imposable when molecule A is brought into alignment with B (Figure S3A).
Cooperative Dual Lipid-Binding-Dependent Membrane Bilayer Attachment and ASAP1 Activation
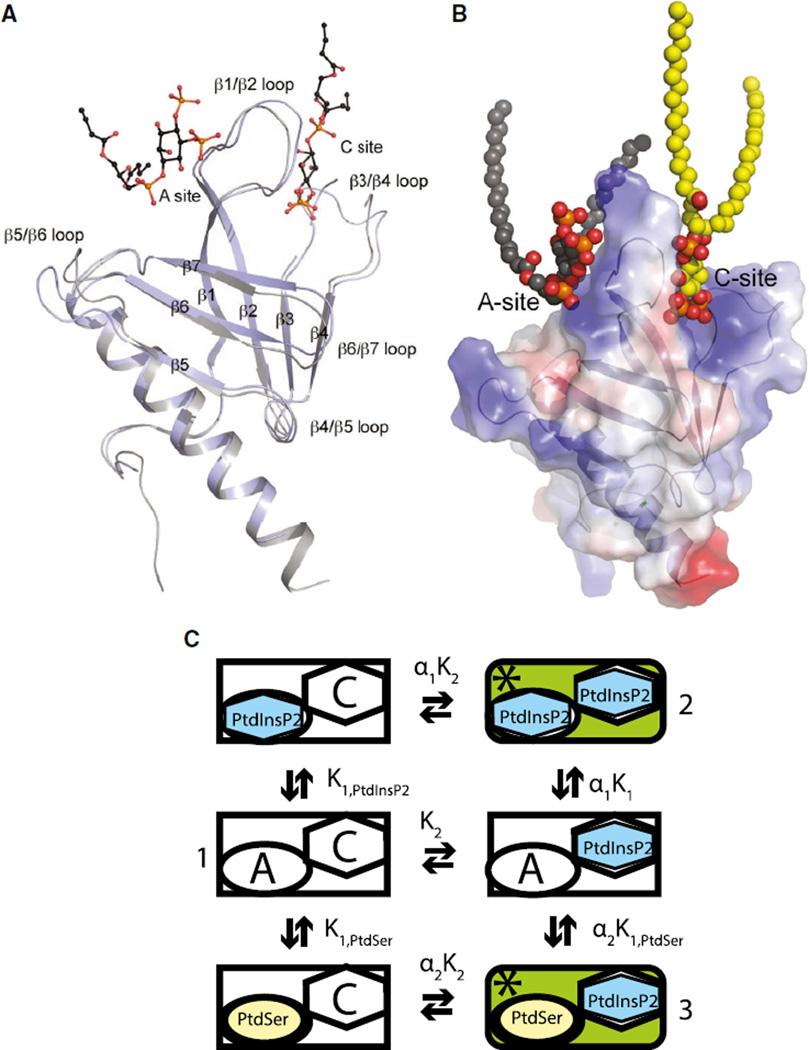
Based on the observed electron density, both 2Fo-Fc and anomalous difference Fourier maps (Figure S1), and the thermo-motion factor for bound ligands, the PtdIns(4,5)P2 bound at the A site is more ordered than that bound at the C site, suggesting the possibility of stronger binding of PtdIns(4,5)P2 at the A site, which may in turn suggest that this is the first binding site to be occupied when the ASAP1 PH domain binds to membranes. In the absence of the ligand, β1/β2, β3/β4, and β5/β6 loops are disordered. When the unliganded ASAP1 PH domain structure is aligned with the PtdIns(4,5)P2-bound structure (Figure 2A), several conformational changes are observed. (1) As expected upon ligand binding, the disordered loops, including the β1/β2 and β3/β4 loops, become better ordered; this transition is especially true for the β1/β2 loop that is sandwiched between the two lipid-binding sites. The lowered mobility for the β1/β2 and β3/β4 loops is to be expected, as both loops contribute to the formation of the A and C sites. (2) Binding of diC4-PtdIns(4,5)P2 alters the structure of ASAP1 PH domain locally, concentrating predominantly in the canonical phosphoinositide-binding site, inducing an outward movement of the β1/β2 and β3/β4 loop and thus a small expansion of the binding site compared with the unliganded ASAP1 PH domain (Figure 2A). (3) Binding of PtdIns(4,5)P2 also moves the second β sheet (made of β strands 5, 6, and 7) toward the C site (Figure 2A). Based on the identification of two inositol contact sites in the diC4-PtdIns(1,4,5)P3 crystal, we propose that the lipid bilayer-binding interface of the PH domain involves cooperation between the canonical phosphoinositide-binding site and the atypical phospholipid-binding site. We also propose, based on previous biochemical studies, that the atypical site can accommodate anionic phospholipid head groups, with little specificity for particular head groups. A representation of the hypothesis is schematized in Figure 2C. The atypical site is represented by an “A” and the canonical site by a “C” in the diagram. Binding is cooperative, and occupancy of both sites increases GAP activity. The active form of the enzyme is indicated by an asterisk. If binding is cooperative as proposed, forms 1, 2, and 3 are the primary species present at equilibrium.
Figure 2. Hypothesized Two Lipid-Binding Sites.
(A) Superposition of the structure of the unliganded ASAP1 PH domain with that bound with diC4-PtdIns(4,5)P2. The unliganded structure is in gray and the complex is shown in purple. The A- and C-site occupants are shown in the ball-and-stick models with carbon in black, phosphor in orange, and oxygen in red. Loops that show large conformational changes are labeled.
(B) Two lipid-binding sites model. The A site, C site, and the ordering of the β1/β2 basic loop upon simultaneous occupation of both A and C sites by lipids is shown. The ASAP1 PH domain is shown as transparent electrostatic potential surface with both sites taken by bound lipids.
(C) Schematic of interaction of two lipid-binding sites. Two binding sites are depicted. The A site can be occupied by any anionic phospholipid. The C site is specific for phosphoinositides. K1,PtdIns(4,5)P2 is the dissociation constant of PtdIns(4,5)P2 in the atypical site. K1,PtdSer is the dissociation constant for PtdSer in the atypical site. K2 is the dissociation constant for PtdIns(4,5)P2 binding to the canonical site. α is the interaction factor.
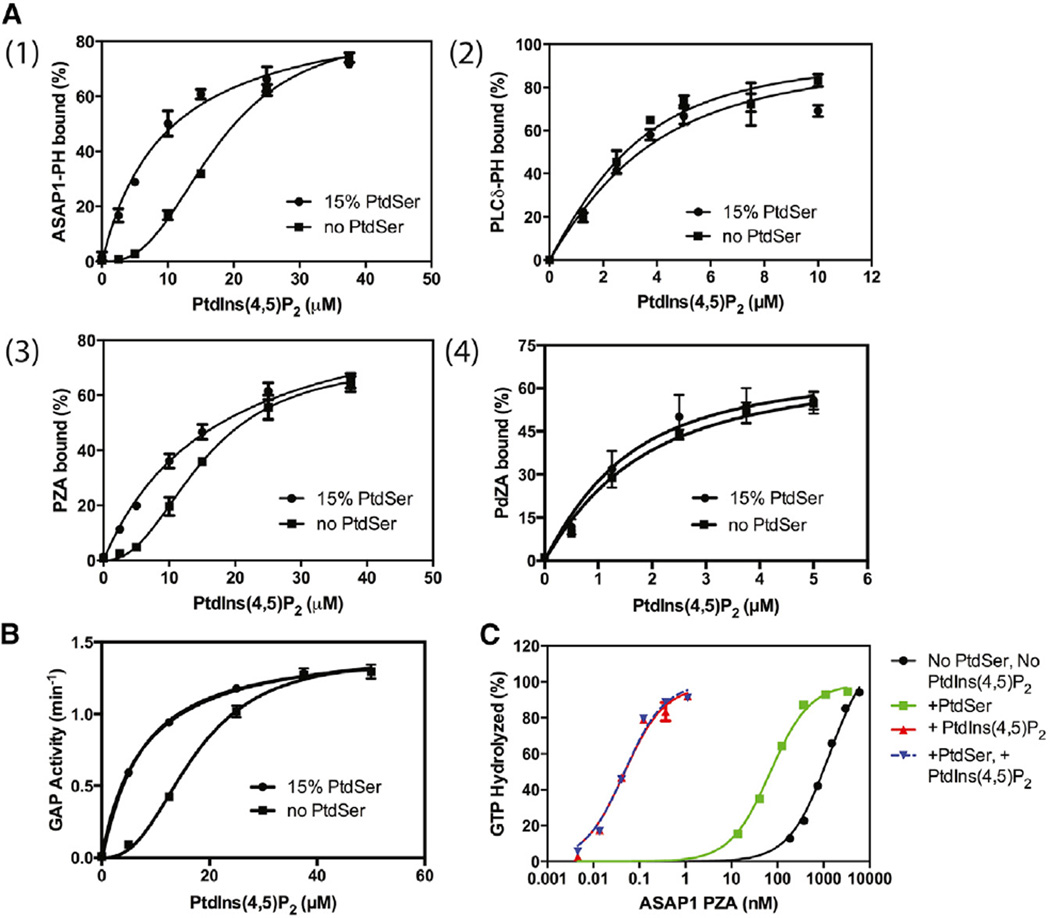
The hypothesis diagrammed in Figure 2 predicts that the PtdIns(4,5)P2 dependence, in the absence of other anionic lipids that might occupy the A site, is sigmoidal for both binding and activity (for equations, see Experimental Procedures). We titrated PtdIns(4,5)P2 in large unilamellar vesicles (LUVs) and measured binding of the isolated PH domain to the LUVs. As predicted, PtdIns(4,5)P2-dependent binding of the PH domain of ASAP1 to LUVs was sigmoidal (Figure 3A1). We propose that there are two sites, one that can accommodate any anionic phospholipid, including phosphatidylserine (PtdSer), and one specific for PtdIns(4,5)P2. With the second site occupied by PtdSer (with 15% PtdSer), the PtdIns(4,5)P2 dependence of binding is predicted to be hyperbolic, which was indeed observed (Figure 3A1). Results with a fragment of ASAP1 comprising the PH, Arf GAP, and Ankyrin repeat domains (henceforth called PZA), which has Arf GAP activity, were similar (Figure 3A3), with a sigmoidal PtdIns(4,5)P2 dependence for binding in the absence of PtdSer and a hyperbolic dependence in the presence of PtdSer. A recombinant protein comprising the PH domain of PLCδ1 either as an isolated protein or fused to the Arf GAP and Ank repeat domains of ASAP1 (henceforth called PdZA) had neither the sigmoidal PtdIns(4,5)P2 dependence for binding nor an effect of PtdSer (Figures 3A2 and A4, Table 2), and the PLCδ PH domain binds with significantly higher affinity. Thus, the sigmoidal PtdIns(4,5)P2-binding behavior was specific for the PH domain of ASAP1.
Figure 3. Anionic Phospholipid Dependence of ASAP1 and PLC δ1 PH Domain Binding to Lipid Vesicles and ASAP1 GAP Activity.
(A) PtdSer and PtdIns(4,5)P2 dependence for binding of ASAP1 and PLC δ1 PH domains to phospholipid vesicles. (1) Isolated PH domain of ASAP, (2) isolated PH domain of PLC δ1, (3) PZA (PH-ArfGAP-Ank repeats of ASAP1), and (4) PdZA (PH domain of PLC δ1 fused to the Arf GAP and Ank repeats of ASAP1) were incubated with large unilamellar vesicles (LUVs) containing the indicated concentration of PtdSer and PtdIns(4,5)P2. Protein binding to the LUVs was determined as described in Experimental Procedures.
(B) PtdSer and PtdIns(4,5)P2 dependence of GAP activity in PZA. GAP activity using myrArf1 · GTP as a substrate was estimated for PZA incubated with LUVs containing 15% PtdSer and PtdIns(4,5)P2 as indicated.
(C) Effect of PtdSer and PtdIns(4,5)P2 on the relative activity of PZA. PZA was titrated into a reaction containing myrArf1 · GTP as a substrate and LUVs containing 15% PtdSer, 10% PtdIns(4,5)P2, or both as indicated. The percentage of the GTP bound to Arf that was hydrolyzed in 3 min is plotted against PZA concentration. The data are one of three experiments used to estimate the concentration of PZA required for 50% hydrolysis (C50), which are summarized in Table 3.
Table 2.
PtdIns(4,5)P2-Dependent Binding of ASAP1 and PLC δ1 PH Domains to LUVs
| Protein | KD (µM) | |
|---|---|---|
| No PthSer | +PthSer | |
| ASAP1 PH | ND | 7 ± 1 |
| PLC δ1 PH | 1.3 ± 0.2 | 1.7 ± 0.2 |
| PZA | ND | 11 ± 2 |
| PdZA | 0.96 ± 0.15 | 0.73 ± 0.2 |
Binding of recombinant proteins to LUVs either lacking the anionic lipid PtdSer or containing 75 µM PtdSer and variable concentrations of PtdIns(4,5)P2 was determined as described in Experimental Procedures. Because the concentration of the PLC δ1 recombinant proteins was greater than the estimated KD, the data were fit to quadratic equations. The values presented are from two experiments. The data for ASAP1 PH and PZA were fit to a hyperbolic function to estimate of the KD for the PtdIns(4,5)P2-protein complexes. The values are the average ± SEM for six experiments. ND, not determined.
GAP activity of ASAP1 was similarly dependent on anionic phospholipids (Figure 3B). In the absence of a second anionic phospholipid, the PtdIns(4,5)P2 dependence of GAP activity was sigmoidal. In the presence of PtdSer, the PtdIns(4,5)P2 dependence was shifted from a half-maximal effect seen at 18 to 8 µM and was hyperbolic (Figure 3B).
In our hypothesis, GAP activity depends on two sites in the PH domain being occupied, with one site specific for PtdIns(4,5)P2 (Figure 2C). A prediction of the model is that saturating PtdIns(4,5)P2 (50 µM), or PtdIns(4,5)P2 together with PtdSer, should support similar levels of GAP activity, whereas ASAP1 would have less activity in the absence of PtdIns(4,5)P2 (since the canonical site is not occupied). To test this prediction, ASAP1 was titrated into reactions containing either no anionic phospholipid, PtdSer, PtdIns(4,5)P2, or PtdSer plus PtdIns(4,5) P2 (Figure 3C and Table 3). The amount of ASAP1 necessary to hydrolyze 50% of the GTP bound to Arf in the reaction (C50) was estimated. In the absence of anionic phospholipid, 1 µM ASAP1 was required. The presence of PtdSer increased activity by 15-fold. In contrast, PtdIns(4,5)P2 alone or PtdSer plus PtdIns(4,5)P2 increased activity approximately 20,000-fold compared with no anionic lipid and 1,500-fold compared with PtdSer alone. These data are similar to those previously obtained in the context of Triton X-100 mixed micelles (Kam et al., 2000) which, together with the comparison with the PLC δ1 PH domain, suggest that the differences are not the result of changes in lipid presentation, but rather are a consequence of two distinct contact surfaces for lipid head groups on the PH domain of ASAP1.
Table 3.
Effect of PtdIns(4,5)P2 and PtdSer on Activity of ASAP1 with Mutations in the PH Domain
| Protein | Site | C50 (nM) | |||
|---|---|---|---|---|---|
| No PthSer/no PthIns(4,5)P2 | +PthSer/no PthIns(4,5)P2 | No PthSer/+PthIns(4,5)P2 | +PthSer/+PthIns(4,5)P2 | ||
| Wild-type | 960 ± 350 | 63 ± 10 | 0.042 ± 0.007 | 0.044 ± 0.007 | |
| K365N | Neither | 860 ± 43 | 60 ± 5 | 0.033 ± 0.022 | |
| K348N | Canonical | 1,110 ± 260 | 170 ± 8 | 4.4 ± 1.8 | |
| R360Q | Canonical | 645 ± 31 | 110 ± 11 | 3.7 ± 0.8 | |
| R378Q | Canonical | 630 ± 10 | 145 ± 6 | 0.08 ± 0.03 | |
| R407Q | Canonical | 630 ± 60 | 92 ± 10 | 0.03 ± 0.01 | |
| K349N | Atypical | 2,180 ± 30 | 414 ± 20 | 1.0 ± 0.2 | |
| K355N | Atypical | 1,260 ± 62 | 270 ± 5 | 0.08 ± 0.02 | |
| I353D | Loop | >10,000 | 8,640 ± 560 | 11 ± 1 | |
A protein comprising the PH, Arf GAP, and Ankyrin repeat domains of ASAP1 with the indicated mutations were titrated into GAP reactions containing myrArf1·GTP and LUVs containing 75 µM PtdSer and 50 µM PtdIns(4,5)P2, as indicated. Reactions were stopped after 3 min and the fraction of GTP bound to Arf that was hydrolyzed was determined. GTP hydrolysis was plotted against protein concentration, from which the concentration of protein necessary to achieve 50% hydrolysis was estimated (C50). Estimates from three experiments with SD are provided for the wild-type PZA. The average from two experiments and the range are shown for the mutants.
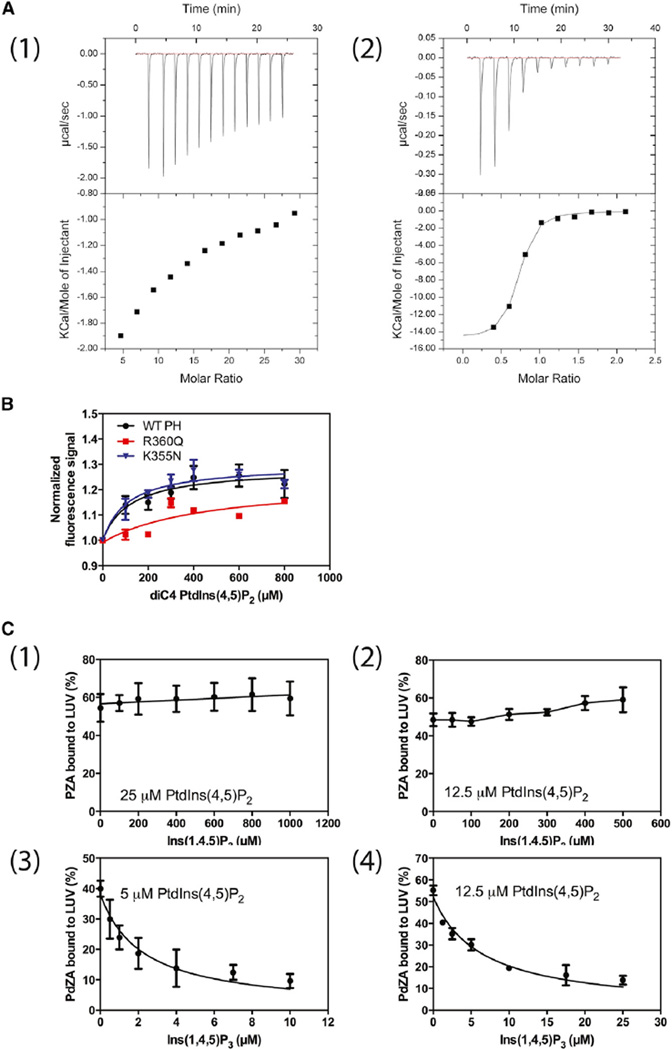
Our hypothetical binding mechanism also predicts that inositol 1,4,5-trisphosphate (Ins(1,4,5)P3), the head group of PtdIns(4,5)P2 that binds to the C site, should have lower affinity for the ASAP1 PH domain than does PtdIns(4,5)P2 in a bilayer containing PtdSer. The opposite has been reported for the PH domain of PLCδ1; the head group has higher affinity than does the lipid in a bilayer (Lemmon et al., 1995). We tested the prediction in three ways. First, we determined binding of Ins(1,4,5)P3 to the PH domain of ASAP1 and the PH domain of PLCδ1 by isothermal titration calorimetry (ITC) (Figure 4A1 and A2). We could not detect saturable binding for ASAP1; the dissociation constant (KD) was 100 µM or greater for Ins(1,4,5)P3, compared with 7 ± 1 µM (mean ± SEM, n = 6) for PtdIns(4,5)P2 in an LUV. The KD for Ins(1,4,5)P3 binding to the PLCδ1 PH domain, by contrast, was 0.18 ± 0.03 µM, compared with a KD of 1.7 ± 0.2 µM for PtdIns(4,5)P2 in an LUV, consistent with previous findings (Lemmon et al., 1995). We had previously found that binding of soluble analogs of PtdIns(4,5)P2 to the ASAP1 PH domain results in a change in tryptophan fluorescence. The effect of diC4-PtdIns(4,5)P2 was saturable with a half-maximal effect at 120 µM (Figure 4B). The third test of the prediction was to displace ASAP1 and PLCδ1 PH domains from PtdIns(4,5)P2 containing LUVs with Ins(1,4,5)P3 (Figure 4C1–4). The amount of Ins(1,4,5)P3, relative to PtdIns(4,5) P2, required to displace the PH domain is a measure of relative affinity of the PH domain for Ins(1,4,5)P3 and PtdIns(4,5)P2. To achieve similar saturation, more PtdIns(4,5)P2 was used for PZA, which has a KD of 11 µM, than for PdZA, which has a KD of 0.7 µM. PZA was not displaced by Ins(1,4,5)P3 at concentrations 40-fold greater than the concentration of PtdIns(4,5)P2, using vesicles with either 12.5 µM (Figure 4C2) or 25 µM PtdIns(4,5) P2 (Figure 4C1). In contrast, half of the PdZA was displaced from vesicles containing either 5 µM (Figure 4C3) or 12.5 µM (Figure 4C4) PtdIns(4,5)P2 at a concentration of Ins(1,4,5)P3 that was less than half the PtdIns(4,5)P2 concentration, indicating that the PLC δ1 PH domain has a greater affinity for Ins(1,4,5)P3 than for PtdIns(4,5)P2.
Figure 4. Affinity of PH Domains for the Inositol Phosphate Head Group and Soluble Forms of PtdIns.
(A) Ins(1,4,5)P3 binding to the PH domain of ASAP1 (1) and PLC δ1 (2) determined by ITC. A total of 13 injections of 3 µl of Ins(1,4,5)P3 (3 mM) were titrated into the ASAP1 PH domain (0.02 mM). Similarly, a total of 13 injections of 3 µl of Ins(1,4,5)P3 (0.1 mM) were titrated into the PLCδ1 PH domain protein (0.01 mM). The background heat of dilution generated during the experiment was subtracted in each experiment, which was determined by titrating the same amount of Ins(1,4,5)P3 into PBS alone.
(B) DiC4-PtdIns(4,5)P2 binding to the PH domain of ASAP1. Tryptophan fluorescence of the ASAP1 PH domain with the indicated amino acid changes in the presence of the indicated concentration of diC4-Ptdns(4,5)P2 was measured. Fluorescence normalized to fluorescence in the absence of diC4-PtdIns(4,5)P2 is plotted against diC4-PtdIns(4,5)P2 concentration. WT, wild-type.
(C) Displacement of PZA and PdZA from PtdSer/PtdIns(4,5)P2-containing vesicles by Ins(1,4,5)P3. PZA and PdZA were incubated with LUVs containing 15% PtdSer and the indicated concentrations of PtdIns(4,5)P2 and Ins(1,4,5)P3. The fraction of total protein in the incubation that adsorbed to the LUVs is plotted against Ins(1,4,5)P3 concentration.
Effects of Mutations on the Binding of PZA to a Membrane Bilayer
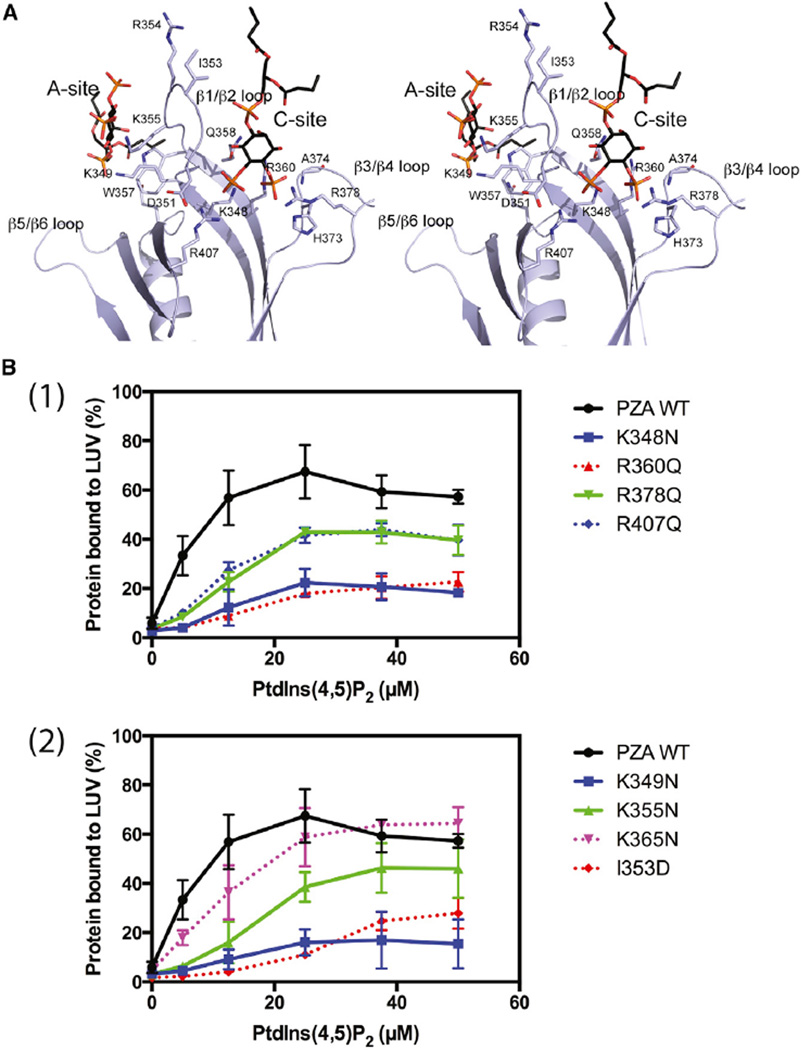
Four side chains from the A site interact with the bound PtdIns(4,5)P2: K349, K355, W357, and Q412 (Figure 5A). The two lysines (K349 and K355) form predicted salt bridges with phosphate groups at positions 1 and 5 of the inositol ring. The phosphate at the fourth position does not form any direct interactions with the protein. At the C site, the bound PtdIns(4,5)P2 contacts more than eight residues. Both the 4- and 5-phosphate interact with positively charged residues: phosphate-4 with R360, H373, R378, and the main-chain nitrogen of A374. The 5-phosphate interacts with two positively charged side chains, K348 and R407, with the latter stabilized by forming a salt bridge with D351 (Figure 5A).
Figure 5. Mutational Analysis of Phospholipid-Binding Sites.
(A) Stereo diagram showing detailed interactions of the two bound PtdIns(4,5)P2 with residues in the binding sites. Bound PtdIns(4,5)P2 molecules are shown as stick models with carbon atoms in black, oxygen in red, and phosphor in orange. Amino acid residues taking part in interactions with PtdIns(4,5)P2 are also displayed as stick models with carbons in purple, nitrogen in dark blue, and oxygen in red. The two lipid-binding sites are also labeled as A site and C site, respectively.
(B) Binding to vesicles. (1) Mutations in canonical site. (2) Mutations in the atypical site and β1/β2 loop. Wild-type (WT) PZA and PZA with the indicated changes in amino acids were incubated with LUVs containing 15% PtdSer and the indicated concentration of PtdIns(4,5)P2.
We changed amino acids identified as relevant in the crystal structure to further test the hypothesis of two distinct sites mediating binding to phospholipid bilayers (Figure 5A and Table 3). The prediction is that changing amino acids with positively charged side chains in either the A or C site to neutral side chains will reduce PtdIns(4,5)P2-dependent binding (Figure 5B1 and 2). The results of experiments examining PtdIns(4,5)P2-dependent binding to LUVs were consistent with the prediction. Figure 5B1 provides a comparison of wild-type protein with proteins with mutations in the canonical site. Figure 5B2 provides comparisons of wild-type with mutations in the atypical site and isoleucine 353, a residue in the β1/β2 loop that vacates the canonical site on PtdIns(4,5)P2 binding. All mutants affected PtdIns(4,5)P2-dependent binding, albeit to different extents. Mutation of arginine 360 and lysine 348, which are in the canonical site, lysine 349 in the atypical site, and isoleucine 353 had the largest effects, with 20% or less of the mutated protein bound at the highest concentration of PtdIns(4,5)P2 tested. Mutating arginine 378 in the canonical site and lysine 355 in the atypical site reduced binding but had less of an effect. Aspartate, a negatively charged amino acid, in position 353 would be predicted to be either trapped in the positively charged binding pocket for the inositol head group or be repelled by the negatively charged membrane surface. In either case, the affinity of [I353D]PZA for PtdIns(4,5)P2-containing vesicles would be lower than for wild-type, as was observed. Mutation of lysine 365, which is outside of the three elements, had little or no effect on binding to LUVs (Figure 5B1).
We determined the affinity of diC4-PtdIns(4,5)P2 for the ASAP1 PH domain and mutants using the change in tryptophan fluorescence as an indicator of binding (Che et al., 2005). Based on the hypothetical binding mechanism, mutating the canonical site should reduce affinity for diC4-PtdIns(4,5)P2 whereas mutation of residues in the atypical site should have no effect because, in the absence of a bilayer, binding is not cooperative. As predicted, mutating lysine 355 had no effect on binding compared with wild-type protein, whereas mutation of arginine 360 reduced binding (Figure 4B).
The effect of the mutations on PtdIns(4,5)P2-dependent GAP activity was examined (Table 3). In the absence of PtdSer and PtdIns(4,5)P2, all proteins with mutations in the canonical site had similar activity as wild-type ASAP1. Two mutants in the atypical site were examined. [K355N]PZA had activity that was similar to wild-type. [K349N]PZA had half the activity of wild-type. Mutation of isoleucine 353 to aspartate had more than a 10-fold reduction in activity. We determined the relative activity of the mutants using LUVs containing PtdSer as the sole anionic lipid. All mutants had reduced activity, with the largest effects observed for proteins containing mutations in the atypical site and isoleucine 353. We also determined the relative activity of each of the mutants at saturating PtdSer and PtdIns(4,5)P2. Effects of mutations on activity correlated with effects on binding, but in many cases were of greater magnitude. R360Q, K348N, and K349N bound to 10% the extent of wild-type but had 1%–4% the activity. [I353D]PZA bound to about 10% the extent of wild-type PZA but had ~0.25% the activity. In other cases, protein bound to the same extent as wild-type protein and had similar activity, including K355N, R378Q, and R407N. We did not detect an effect of mutating lysine 365, which is not in either binding site, to asparagine (Table 3).
DISCUSSION
We have tested the hypothesis that the PH domain of ASAP1 has cooperative canonical and atypical phosphoinositide-binding sites (Figure 3). The cooperative binding of phospholipids provides a means for efficient switching between active and inactive forms of ASAP1.
The complex regulation of the Arf GAPs may have evolved to provide site-specific function. Humans have five Arf isoforms, each with multiple sites of action (D’Souza-Schorey and Chavrier, 2006; Donaldson, 2003; Donaldson et al., 2005; Gillingham and Munro, 2007; Kahn et al., 2006). The Golgi in particular is enriched with several Arf isoforms, but the same isoforms function outside of the Golgi apparatus (Kahn et al., 2006; Volpicelli-Daley et al., 2005). The Arf GAPs are part of the regulatory machinery that ensures site-specific function. Humans have 31 genes encoding Arf GAPs (Kahn et al., 2008). Multiple Arf GAPs may engage each Arf isoform. If the Arf GAPs were not tightly controlled, they could be recruited by the substrate Arf-GTP to multiple sites. We propose that the Arf GAPs have recruitment mechanisms that are independent of the activation mechanism. ASAP1, for instance, appears to be recruited by its SH3 domains and PXXP motifs to sites containing FAK, Src, and Crk (Liu et al., 2002; Oda et al., 2003). Activity, on the other hand, is controlled by the PH domain. Once targeted to a specific membrane, cooperative binding of the activating lipid ligands ensures rapid and complete switching between active and inactive forms of the ASAP1. Because binding is cooperative, activation does not require a large change in the concentration of the activating ligand. Dependence on the coincidence of signals for both targeting and enzymatic activity may restrict ASAP1 function spatially and temporally.
Cooperative behavior is a means to efficiently switch a protein between two states (Koshland and Hamadani, 2002; Whitty, 2008). Cooperative binding of PtdIns(3,4,5)P3 and Arf6-GTP to the PH domain of Grp1 has been described (Malaby et al., 2013), but not for two lipid ligands to a PH domain as far as we are aware. ASAP1 has several possible sources of cooperative behavior. First, the PH domain by itself displays cooperative behavior as described here. Cooperative behavior may not require conformational changes in the protein. With two binding sites in a single protein that bind ligand spatially constrained to a surface, there is reduced entropic cost for binding the second site (Whitty, 2008). The difference in binding energy may result in a sigmoidal dependence for ligand binding. Second, ASAP1 has three properties of cooperative enzymes (Koshland and Hamadani, 2002). (1) Cooperative enzymes are composed of multiple identical subunits. ASAP1 is a homodimer. (2) Ligand binding in cooperative enzymes induces an intramolecular conformational change in the subunit to which it binds. In this study, we describe changes in the ASAP1 PH domain that occur on PtdIns binding. (3) The conformational change in one subunit affects the other subunit. Dimerization of ASAP1 is mediated by the BAR domain. Modeling indicates that the BAR domain interacts with the PH domain in trans (Jian et al., 2009), and therefore may be able to transmit conformational information between subunits. Cooperative behavior might also result from the hysteretic properties (Ainslie et al., 1972; Frieden, 1970) of ASAP1 (Luo et al., 2007).
The mechanism we propose for the ASAP1 PH domain may be generalized to a subset of other PH domains. Canonical and atypical binding sites for phosphoinositides have been described in PH domains. One site corresponds to the canonical site found in proteins such as PLC-δ, Akt, PDK, Btk, Grp1, FAPP1, and TAPP1 (DiNitto and Lambright, 2006; Moravcevic et al., 2012). The second site corresponds to the atypical site first described in spectrin and found in proteins such as TIAM and Slm1 (Hyvonen et al., 1995; Balla, 2005; DiNitto and Lambright, 2006; Moravcevic et al., 2012; Scheffzek and Welti, 2012). Some evidence also supports the idea that the sites are integrated. Simultaneous occupancy of the two sites has been proposed for Slm1 (Anand et al., 2012; Gallego et al., 2010). Akt has a patch of positive charge on its surface from side chains of amino acids in loop β1/β2 and loop β5/β6 (Bellacosa et al., 1998; Milburn et al., 2003; Thomas et al., 2002). These charged side chains could bind to anionic lipid head groups, and are in a position similar to that of the atypical site described in spectrin. A mutant Akt with one of the charged residues in this patch neutralized has less activity than wild-type Akt, consistent with the idea of an A site in Akt (Huang et al., 2011). Also, the dependence of Akt activity on PtdSer may be related (Huang et al., 2011). The specific interactions between binding sites will likely differ among PH domains. Furthermore, the hypothetical model likely applies to a subset of PH domains. Indeed, in this study we report differences between the ASAP1 and PLCδ1 PH domains.
The integration of structural elements involved in membrane binding has been reported for other phospholipid-binding domains. The PX domain of p47phox has two lipid-binding sites, one for PtdIns(3,4)P2 and the other for phosphatidic acid, separated by a hydrophobic ridge thought to insert into the membrane. Phospholipid binding is cooperative, as observed with ASAP1 (Karathanassis et al., 2002). Similarly, the C2 domain of PKCα binds cooperatively to PtdIns(4,5)P2 and PtdSer. Also similar to ASAP1, Ins(1,4,5)P3 binds with lower affinity than does PtdIns(4,5)P2 in a bilayer (Guerrero-Valero et al., 2009), which is different from what was reported for the PH domain of PLC δ1 (Lemmon et al., 1995). The multicontact protein-phospholipid bilayer interface may be a common feature of peripheral membrane proteins involved in signaling.
Association with a planar membrane likely has additional consequences for ASAP1 interaction with its substrate Arf-GTP. For some mutants we examined, the effect on activity was disproportionate to the effect on binding, e.g. I353, K349, and R360. The disproportion could indicate additional conformational changes in the PH domain that occur on binding a planar surface. diC4-PtdIns(4,5)P2 can bind but increases activity by only 2- to 4-fold, even when present at saturating concentrations (Che et al., 2005). Thus, the active conformation would not be apparent with dibutyryl PtdIns(4,5)P2 bound in the crystal.
Our crystal structure revealed plasticity in the canonical PtdIns(4,5)P2-binding site, which is able to accommodate the inositol phosphate ring in two orientations. This plasticity could account for the promiscuity for phosphoinositide isoforms that has been observed with ASAP1, which is similar to many PH domains. The PH domains that bind inositol lipids have been broadly classified into four groups (Maffucci and Falasca, 2001). From our biochemical and structural characterizations, the ASAP1 PH domain may belong to the group 2 PH domains that bind PtdIns(4,5)P2, PtdIns(3,4,5)P3, and others. A detailed comparison of the canonical PtdIns-binding sites for various PH domains shows a high similarity in the molecular interactions (Figure S3B), mostly involving charged interactions. However, bound PtdIns molecules display considerable flexibility in their interactions with their respective PH domains, e.g. symmetric rotation among PtdIns phosphates interacting with different residues in the binding pocket. Our observation of ASAP1 PH domain binding to diC4 PtdIns(4,5)P2 in two different configurations further supports this point and suggests that the binding pocket does not discriminate the configuration of the six-membered inositol ring.
There are numerous examples of phosphoinositide binding to the atypical binding site in PH domains (Figure S3C). In all these cases, the PH domain binds phospholipid between β1/β2 and β5/β6 loops. Superposition of ASAP1 PH domain to those with bound ligand in the atypical site shows that ASAP1 PH domain possesses the atypical site (Figure S3C).
In summary, we have tested the hypothesis that the PH domain of ASAP1 has two cooperative phospholipid-binding sites that control GAP activity, and we propose that this mechanism is combined with targeting mediated by coincident signals to spatially and temporally restrict ASAP1 function.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Bacterial expression vectors for His10-[325–724]ASAP1 (i.e. ASAP1 PZA), His10-[325–451]ASAP1 (i.e. ASAP1 PH), PLCδ-PH, PdZA, and myristoylated Arf1 (myrArf1) were described by Brown et al. (1998), Che et al. (2005), and Randazzo et al. (1995). [339–451]ASAP1 (used for NMR studies) was prepared as described for other ASAP1 PH domain constructs. Mutations in ASAP1 PZA and ASAP1 PH were generated using the QuikChange II site-directed mutagenesis kit (Agilent Technologies). ASAP1 PZA wild-type and mutant proteins, PdZA, as well as ASAP1 PH wild-type and mutant proteins were prepared as previously described (Jian et al., 2009). PLCδ-PH, corresponding to residues 11–140 of rat PLC-δ1, was expressed in bacteria and purified using cation-exchange column HiTrapSP (GE Healthcare) and size-exclusion chromatography as previously described (Lemmon et al., 1995). Bacterial expression and purification of myrArf1 has been previously described (Chen et al., 2012).
Preparation of LUVs
LUVs, prepared by extrusion with lipids purchased from Avanti Polar Lipids as described previously (Jian et al., 2009; Nie et al., 2006), contained molar ratios of 40% phosphatidylcholine (PtdCho), 25% phosphatidylethanolamine (PtdEtn), 15% phosphatidylserine (PtdSer), 10% PtdIns(4,5)P2, and 10% cholesterol (+PtdSer/+PtdIns(4,5)P2 LUV), or 40% PtdCho, 40% PtdEtn, 10% PtdIns(4,5)P2, and 10% cholesterol (no PtdSer/+PtdIns(4,5)P2 LUV), or 40% PtdCho, 25% PtdEtn, 15% PtdSer, 10% phosphatidylinositol (PtdIns), and 10% cholesterol (+PtdSer/no PtdIns(4,5)P2 LUV), or 40% PtdCho, 40% PtdEtn, 10% PtdIns, and 10% cholesterol (no PtdSer/no PtdIns(4,5)P2 LUV). For the PtdIns(4,5)P2 titration experiments, PtdIns(4,5)P2 amount varied from 0% to 10%, and the amount of phosphoinositide was changed to keep PtdIns plus PtdIns(4,5)P2 at 10%.
diC4-PtdIns(4,5)P2-Induced Change in PH Domain Tryptophan Fluorescence
The binding of diC4-PtdIns(4,5)P2 to the PH domain of ASAP1 wild-type or mutants increased tryptophan fluorescence. diC4-PtdIns(4,5)P2 was mixed with 1 µM of ASAP1 PH wild-type or mutants in a buffer containing 20 mM HEPES (pH 7.4) and 100 mM NaCl. Tryptophan fluorescence spectra were obtained using a FluoroMax-3 spectrophotometer (Jobin Yvon Horiba). The excitation wavelength was 280 nm, and the emission spectra from 310 to 450 nm were collected.
Lipid-Binding Assay
The assay for protein binding to lipid vesicles is described by Jian et al. (2009) and Nie et al. (2006). One µM for PZA wild-type, mutants, or PdZA or 3 µM ASAP1 PH or PLCδ-PH were incubated with sucrose-loaded LUVs containing 500 µM total phospholipids. The LUVs were precipitated by ultracentrifugation, and associated proteins were separated by SDS-PAGE and visualized by Coomassie blue staining. The signal was quantified by densitometry using ImageJ software.
GAP Assays
ASAP1-induced conversion of myrArf1·GTP to myrArf1·GDP was determined as described previously (Kam et al., 2000; Randazzo and Kahn, 1994; Randazzo et al., 2001, 2013). ASAP1 PZA was titrated into the reaction containing myrArf1·GTP as the substrate. The amount of ASAP1 required to hydrolyze 50% of the Arf bound GTP in 3 min (C50) is presented.
Isothermal Titration Calorimetry
ITC experiments were performed using an ITC200 calorimeter (Malvern). All samples were prepared in PBS. Ins(1,4,5)P3 was titrated into protein solutions at 25°C and the background heat of dilution was subtracted in each experiment. Data were fitted using a one-site model from Origin7 software (Origin-Lab). The averaged binding parameters were obtained from three independent experiments.
Crystallization Experiments of Apo PH Domain and Its Complex with Dibutyryl PtdIns(4,5)P2
Initial crystallization was performed utilizing the high-throughput crystallization screening service offered by the Hauptman-Woodward Medical Research Institute (www.hwi.buffalo.edu). ASAP1 PH domain (400 µM in 20 mM Tris-HCl [pH 7.4], 150 mM NaCl, and 0.01% NaN3) was crystallized for diffraction using a hanging-drop vapor diffusion setup with a one-to-one admixture of the protein and a well solution consisting of 100mMNa2HPO4, 100 mM Tris (pH 8.0) and 48% PEG400 (v/v) at 23°C. ASAP1 PH domain in the presence of dibutyryl PtdIns(4,5)P2 (400 µM ASAP1 PH domain, 1 mM diC4-PtdIns(4,5)P2, 20 mM Tris [pH 7.4], 150 mM NaCl, and 0.01% NaN3) was crystallized at 21°C by mixing the protein with equilibration solution (0.1 M HEPES [pH 7.5], 15% PEG3350) in a one-to-one ratio.
X-Ray Diffraction Experiments
Unliganded-ASAP1 PH domain crystals were flash-cooled directly in liquid nitrogen prior to X-ray diffraction. The PH domain crystals diffracted X-rays to 1.8 Å resolution at the insertion device magnet of SER-CAT at the Advanced Photon Source, Argonne National Laboratory. Diffraction images were recorded on a Marw225 CCD detector. Crystals of PH domain/diC4-PtdIns(4,5)P2 complex were cryoprotected by a quick soak in a cryoprotectant solution made of 7 µl of crystallization well solution supplemented with 3 µl of 50% PEG3350 followed by flash-cooling to 100 K of a nitrogen stream. A diffraction dataset was collected using X-rays from an in-house Rigaku X-ray generator operating at 100 mA and 50 kV and a Mar345 imaging plate scanner. Diffraction images from both types of crystals were processed with using the HKL2000 program suite (Otwinowski and Minor, 1997) (Table 1).
Structure Determination and Refinement
The crystal structure of unliganded-ASAP1 PH domain was determined by the molecular replacement method using coordinates from the solution structure of the PH domain of ASAP1 (PDB: 2DA0) (not yet published) as phasing template using the program Molrep (Vagin and Teplyakov, 2000). Subsequent refinement was performed with Refmac in the CCP4 package (CCP4, 1994) and molecular modeling was carried out in Coot (Emsley and Cowtan, 2004).
The unliganded PH domain structure was used as the search model for molecular replacement program Molrep to solve the structure of the PH domain/diC4-PtdIns(4,5)P2 complex. Subsequent refinement was performed with Refmac in the CCP4 package, and molecular modeling was carried out in Coot.
Equations
Equations based on the scheme presented in Figure 4 were derived as described by Cha (1968).
Assuming α1 = α2, the amount of PH domain-containing protein bound to vesicles is:
| (Equation 1) |
Further assuming that Vmax1 = Vmax2 and KM1 = KM2, the initial rate is related to phospholipid concentration by:
| (Equation 2) |
Assuming Arf•GTP ≪ KM activity determined as described by Randazzo et al. (2013) can be related to phospholipid concentration by:
| (Equation 3) |
In Equations 1 and 3, if PtdSer >> PtdInsP2, the square term of PtdInsP2 is negligible and the relationship of binding or activity to PtdInsP2 concentration is hyperbolic.
Supplementary Material
Highlights.
PH domain of ASAP1 has canonical and atypical anionic lipid-binding sites
Phospholipids bind cooperatively to the canonical and atypical sites
Occupancy of both sites is necessary for efficient ASAP1 enzymatic activity
Acknowledgments
We thank Mark Lemmon for advice, critical review of the manuscript, and providing the plasmid for expression of the isolated PH domain of PLC δ1, Tamas Balla for a plasmid for bacterial expression of GST fused to the PH domain of PLC δ1, and David Lambright for advice and review of the data. We thank Shangjin Sun for preliminary NMR experiments. The work was supported by the NIH Intramural Program (Project BC 007365).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes three figures and can be found with this article online at http://dx.doi.org/10.1016/j.str.2015.08.008.
AUTHOR CONTRIBUTIONS
X.J., P.Z., N.R., R.L., M.E.Y., and P.W.C. prepared proteins and performed biochemical experiments; P.Z. grew crystals; W.K.T. performed ITC experiments, X-ray diffraction experiments, and solved the structures; M.E.Y., J.M.G., R.A.B., D.X., and P.A.R. designed experiments; P.A.R. and D.X. wrote the manuscript.
REFERENCES
- Ainslie GR, Neet KE, Shill JP. Transients and cooperativity—slow transition model for relating transients and cooperative kinetics of enzymes. J. Biol. Chem. 1972;247:7088–7094. [PubMed] [Google Scholar]
- Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PRJ, Reese CB, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase B alpha. Curr. Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- Anand K, Maeda K, Gavin AC. Structural analyses of the Slm1-PH domain demonstrate ligand binding in the non-canonical site. PLoS One. 2012;7:e36526. doi: 10.1371/journal.pone.0036526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J. Cell Sci. 2005;118:2093–2104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- Bellacosa A, Chan TO, Ahmed NN, Datta K, Malstrom S, Stokoe D, McCormick F, Feng JN, Tsichlis P. Akt activation by growth factors is a multiple-step process: the role of the PH domain. Oncogene. 1998;17:313–325. doi: 10.1038/sj.onc.1201947. [DOI] [PubMed] [Google Scholar]
- Brown MT, Andrade J, Radhakrishna H, Donaldson JG, Cooper JA, Randazzo PA. ASAP1, a phospholipid-dependent Arf GTPase-activating protein that associates with and is phosphorylated by Src. Mol. Cell. Biol. 1998;18:7038–7051. doi: 10.1128/mcb.18.12.7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4. The CCP4 suit: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Ceccarelli DFJ, Blasutig IM, Goudreault M, Li ZQ, Ruston J, Pawson T, Sicheri F. Non-canonical interaction of phosphoinositides with pleckstrin homology domains of Tiam1 and ArhGAP9. J. Biol. Chem. 2007;282:13864–13874. doi: 10.1074/jbc.M700505200. [DOI] [PubMed] [Google Scholar]
- Cha S. A simple method for derivation of rate equations for enzyme-catalyzed reactions under rapid equilibrium assumption or combined assumptions of equilibrium and steady state. J. Biol. Chem. 1968;243:820–826. [PubMed] [Google Scholar]
- Chan TO, Rittenhouse SE, Tsichlis PN. AKT/PKB and other D3 phosphoinositide-regulated kinases: kinase activation by phosphoinositide-dependent phosphorylation. Annu. Rev. Biochem. 1999;68:965–1014. doi: 10.1146/annurev.biochem.68.1.965. [DOI] [PubMed] [Google Scholar]
- Che MM, Boja ES, Yoon HY, Gruschus J, Jaffe H, Stauffer S, Schuck P, Fales HM, Randazzo PA. Regulation of ASAP1 by phospholipids is dependent on the interface between the PH and Arf GAP domains. Cell. Signal. 2005;17:1276–1288. doi: 10.1016/j.cellsig.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Chen PW, Jian X, Luo R, Randazzo PA. Approaches to studying Arf GAPs in cells: in vitro assay with isolated focal adhesions. Curr. Protoc. Cell Biol. 2012;Chapter 17(Unit 17):13. doi: 10.1002/0471143030.cb1713s55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNitto JP, Lambright DG. Membrane and juxtamembrane targeting by PH and PTB domains. Biochim. Biophys. Acta. 2006;1761:850–867. doi: 10.1016/j.bbalip.2006.04.008. [DOI] [PubMed] [Google Scholar]
- DiNitto JP, Delprato A, Lee MTG, Cronin TC, Huang S, Guilherme A, Czech MP, Lambright DG. Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Mol. Cell. 2007;28:569–583. doi: 10.1016/j.molcel.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J. Biol. Chem. 2003;278:41573–41576. doi: 10.1074/jbc.R300026200. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Honda A, Weigert R. Multiple activities for Arf1 at the Golgi complex. Biochim. Biophys. Acta. 2005;1744:364–373. doi: 10.1016/j.bbamcr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Frieden C. Kinetic aspects of regulation of metabolic processes—hysteretic enzyme concept. J. Biol. Chem. 1970;245:5788–5794. [PubMed] [Google Scholar]
- Gallego O, Betts MJ, Gvozdenovic-Jeremic J, Maeda K, Matetzki C, Aguilar-Gurrieri C, Beltran-Alvarez P, Bonn S, Fernandez-Tornero C, Jensen LJ, et al. A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol. Sys. Biol. 2010;6:430. doi: 10.1038/msb.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4) P. Nat. Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Guerrero-Valero M, Ferrer-Orta C, Querol-Audi J, Marin-Vicente C, Fita I, Gomez-Fernandez JC, Verdaguer N, Corbalan-Garcia S. Structural and mechanistic insights into the association of PKC alpha-C2 domain to PtdIns(4,5)P-2. Proc. Natl. Acad. Sci. USA. 2009;106:6603–6607. doi: 10.1073/pnas.0813099106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Scott JL, Heroux A, Roy S, Lenoir M, Overduin M, Stahelin RV, Kutateladze TG. Molecular basis of phosphatidylinositol 4-phosphate and ARF1 GTPase recognition by the FAPP1 pleckstrin homology (PH) domain. J. Biol. Chem. 2011;286:18650–18657. doi: 10.1074/jbc.M111.233015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Kuo YC, Rosche TJ, Zhang XW. Structural basis for autoinhibition of the guanine nucleotide exchange factor FARP2. Structure. 2013;21:355–364. doi: 10.1016/j.str.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Akbar M, Kevala K, Kim H-Y. Phosphatidylserine is a critical modulator for Akt activation. J. Cell Biol. 2011;192:979–992. doi: 10.1083/jcb.201005100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvonen M, Macias MJ, Nilges M, Oschkinat H, Saraste M, Wilmanns M. Structure of the binding-site for inositol phosphates in a PH domain. EMBO J. 1995;14:4676–4685. doi: 10.1002/j.1460-2075.1995.tb00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian XY, Brown P, Schuck P, Gruschus JM, Balbo A, Hinshaw JE, Randazzo PA. Autoinhibition of Arf GTPase-activating protein activity by the BAR domain in ASAP1. J. Biol. Chem. 2009;284:1652–1663. doi: 10.1074/jbc.M804218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Cherfils J, Elias M, Lovering RC, Munro S, Schurmann A. Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J. Cell Biol. 2006;172:645–650. doi: 10.1083/jcb.200512057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Bruford E, Inoue H, Logsdon JM, Nie ZZ, Premont RT, Randazzo PA, Satake M, Theibert AB, Zapp ML, et al. Consensus nomenclature for the human ArfGAP domain-containing proteins. J. Cell Biol. 2008;182:1039–1044. doi: 10.1083/jcb.200806041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam JL, Miura K, Jackson TR, Gruschus J, Roller P, Stauffer S, Clark J, Aneja R, Randazzo PA. Phosphoinositide-dependent activation of the ADP-ribosylation factor GTPase-activating protein ASAP1—evidence for the pleckstrin homology domain functioning as an allosteric site. J. Biol. Chem. 2000;275:9653–9663. doi: 10.1074/jbc.275.13.9653. [DOI] [PubMed] [Google Scholar]
- Karathanassis D, Stahelin RV, Bravo J, Perisic O, Pacold CM, Cho WW, Williams RL. Binding of the PX domain of p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 2002;21:5057–5068. doi: 10.1093/emboj/cdf519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland DE, Hamadani K. Proteomics and models for enzyme cooperativity. J. Biol. Chem. 2002;277:46841–46844. doi: 10.1074/jbc.R200014200. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Obrien R, Sigler PB, Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. USA. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Schlessinger J. PH domains: diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell. 1996;85:621–624. doi: 10.1016/s0092-8674(00)81022-3. [DOI] [PubMed] [Google Scholar]
- Liu YH, Loijens JC, Martin KH, Karginov AV, Parsons JT. The association of ASAP1, an ADP ribosylation factor-GTPase activating protein, with focal adhesion kinase contributes to the process of focal adhesion assembly. Mol. Biol. Cell. 2002;13:2147–2156. doi: 10.1091/mbc.E02-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YZ, Kahn RA, Prestegard JH. Interaction of Fapp1 with Arf1 and PI4P at a membrane surface: an example of coincidence detection. Structure. 2014;22:421–430. doi: 10.1016/j.str.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo RB, Ahvazi B, Amariei D, Shroder D, Burrola B, Losert W, Randazzo PA. Kinetic analysis of GTP hydrolysis catalysed by the Arf1-GTP-ASAP1 complex. Biochem. J. 2007;402:439–447. doi: 10.1042/BJ20061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Jenkins LMM, Randazzo PA, Gruschus J. Dynamic interaction between Arf GAP and PH domains of ASAP1 in the regulation of GAP activity. Cell. Signal. 2008;20:1968–1977. doi: 10.1016/j.cellsig.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffucci T, Falasca M. Specificity in pleckstrin homology (PH) domain membrane targeting: a role for a phosphoinositide-protein co-operative mechanism. FEBS Lett. 2001;506:173–179. doi: 10.1016/s0014-5793(01)02909-x. [DOI] [PubMed] [Google Scholar]
- Malaby AW, van den Berg B, Lambright DG. Structural basis for membrane recruitment and allosteric activation of cytohesin family Arf GTPase exchange factors. Proc. Natl. Acad. Sci. USA. 2013;110:14213–14218. doi: 10.1073/pnas.1301883110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn CC, Deak M, Kelly SM, Price NC, Alessi DR, Van Aalten DMF. Binding of phosphatidylinositol 3,4,5-trisphosphate to the pleckstrin homology domain of protein kinase B induces a conformational change. Biochem. J. 2003;375:531–538. doi: 10.1042/BJ20031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins: facing up to limited specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie ZZ, Hirsch DS, Luo RB, Jian XY, Stauffer S, Cremesti A, Andrade J, Lebowitz J, Marino M, Ahvazi B, et al. A BAR domain in the N terminus of the Arf GAP ASAP1 affects membrane structure and trafficking of epidermal growth factor receptor. Curr. Biol. 2006;16:130–139. doi: 10.1016/j.cub.2005.11.069. [DOI] [PubMed] [Google Scholar]
- Oda A, Wada I, Miura K, Okawa K, Kadoya T, Kato T, Nishihara H, Maeda M, Tanaka S, Nagashima K, et al. CrkL directs ASAP1 to peripheral focal adhesions. J. Biol. Chem. 2003;278:6456–6460. doi: 10.1074/jbc.M210817200. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Randazzo PA, Kahn RA. GTP hydrolysis by ADP-ribosylation factor is dependent on both an ADP-ribosylation factor GTPase-activating protein and acid phospholipids. J. Biol. Chem. 1994;269:10758–10763. [PubMed] [Google Scholar]
- Randazzo PA, Terui T, Sturch S, Fales HM, Ferrige AG, Kahn RA. The myristoylated amino-terminus of ADP-ribosylation factor-1 is a phospholipid-sensitive and GTP-sensitive switch. J. Biol. Chem. 1995;270:14809–14815. doi: 10.1074/jbc.270.24.14809. [DOI] [PubMed] [Google Scholar]
- Randazzo PA, Miura K, Jackson TR. Assay and purification of phosphoinositide-dependent ADP-ribosylation factor (ARF) GTPase activating proteins. Method Enzymol. 2001;329:343–354. doi: 10.1016/s0076-6879(01)29096-x. [DOI] [PubMed] [Google Scholar]
- Randazzo PA, Jian X, Chen P-W, Zhai P, Soubias O, Northup JK. Quantitative analysis of guanine nucleotide exchange factors (GEFs) as enzymes. Cell. Log. 2013;3:e27609. doi: 10.4161/cl.27609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman KL, Worthylake DK, Snyder JT, Siderovski DP, Campbell SL, Sondek J. A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange. EMBO J. 2002;21:1315–1326. doi: 10.1093/emboj/21.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Welti S. Pleckstrin homology (PH) like domains—versatile modules in protein-protein interaction platforms. FEBS Lett. 2012;586:2662–2673. doi: 10.1016/j.febslet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Sprang SR. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- Thomas CC, Deak M, Alessi DR, van Aalten DMF. High-resolution structure of the pleckstrin homology domain of protein kinase B/Akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Curr. Biol. 2002;12:1256–1262. doi: 10.1016/s0960-9822(02)00972-7. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 2000;B56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A. Signal transduction—the guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- Volpicelli-Daley LA, Li YW, Zhang CJ, Kahn RA. Isoform-selective effects of the depletion of ADP-ribosylation factors 1–5 on membrane traffic. Mol. Biol. Cell. 2005;16:4495–4508. doi: 10.1091/mbc.E04-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitty A. Cooperativity and biological complexity. Nat. Chem. Biol. 2008;4:435–439. doi: 10.1038/nchembio0808-435. [DOI] [PubMed] [Google Scholar]
- Wittinghofer A, Vetter IR. Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 2011;80:943–971. doi: 10.1146/annurev-biochem-062708-134043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.