Abstract
The emergence of drug resistance has devastating economic and social consequences, a testimonial of which is the rise and fall of inhibitors against the respiratory component cytochrome bc1 complex, a time tested and highly effective target for disease control. Unfortunately, the mechanism of resistance is a multivariate problem, including primarily mutations in the gene of the cytochrome b subunit but also activation of alternative pathways of ubiquinol oxidation and pharmacokinetic effects. There is a considerable interest in designing new bc1 inhibitors with novel modes of binding and lower propensity to induce the development of resistance. The accumulation of crystallographic data of bc1 complexes with and without inhibitors bound provides the structural basis for rational drug design. In particular, the cytochrome b subunit offers two distinct active sites that can be targeted for inhibition - the quinol oxidation site and the quinone reduction site. This review brings together available structural information of inhibited bc1 by various quinol oxidation- and reduction-site inhibitors, the inhibitor binding modes, conformational changes upon inhibitor binding of side chains in the active site and large scale domain movements of the iron-sulfur protein subunit. Structural data analysis provides a clear understanding of where and why existing inhibitors fail and points towards promising alternatives.
Keywords: cyt bc1 complex, mechanism of inhibition, crystal structure, resistance, inhibitors
1. Introduction
In the efforts to obtaining ever larger crops needed to sustain a growing world population and to improve the quality of life, humanity has not only been quite successful by using crop protection agents, but on the downside experiences a most dramatic demonstration of the principle described by the words: "that what does not kill me, makes me stronger" [F. Nietzsche, Götzen-Dämmerung: Sprüche und Pfeile] [1]. This refers to the crop pests that need to be kept in check but show the ability to become resistant against any pesticide we can dish out. Similar phenomena have been extensively documented in modern medicine in treatment of pathogenic microbial infections and in cancer chemotherapy.
Cellular respiration is an essential housekeeping process that yields large amounts of ATP on which the cell depends for growth, division, maintenance and survival. A number of large integral membrane protein complexes located in the inner membrane of mitochondria in eukaryotic cells, or plasma membrane in bacteria are crucial for the conversion of energy. These membrane protein complexes take part in the process of oxidative phosphorylation (Oxphos), where electrons from chemical reducing equivalents are transferred sequentially through a series of complexes, maintaining a proton motive force across the membrane for ATP synthesis. Inhibition of Oxphos has evolved as an efficient strategy in nature to gain competitive advantage in survival and is used extensively in disease control. A similar evolutionary pattern has emerged from the development of crop-protection agents. Historically, in the 1940s crop protection using effective but environmentally toxic inorganic, organo-metallic or simple organic compounds shifted to targeted, pest-specific fungicides [2]. Even though some 70 years later, pesticides with nine different modes of action (and several of unknown modes) exist, the two largest groups in total market value are those that inhibit sterol biosynthesis and respiration, respectively.
As a central component of the cellular respiratory chain, the bc1 complex became an easy target for numerous natural antibiotics. Examples of natural compounds that specifically target the bc1 complex are antimycin from Streptomyces bacteria, strobilurin from Strobilurus tenacellus fungus, myxothiazol from Myxococcus fulvus bacteria, and stigmatellin from Stigmatella aurantica bacteria, etc. However, targeting housekeeping enzymes such as the bc1 complex can be a double-edged sword due to its ubiquitous presence in vast number of organisms. By manipulating differences in uptake or taking advantage of structural differences at the active sites between the host and pathogens, some of the inhibitors or their derivatives have been developed into anti-microbial and anti-parasitic agents and are used in agriculture to control fungal or bacterial diseases and in medicine to treat infections. In particular, the wide spread success in the use of anti-complex III agents in control of fungal disease in crops is considered a remarkable achievement in history, resulting in an annual sale of pesticides worth $2.7 billion [3]. They exhibit protective, systemic and eradicative action.
As observed in the application of most chemical interventions, development of resistance is almost inevitable, rendering the chemical agents useless and causing the recurrence of diseases. In agriculture, the development of resistant pathogens means hefty losses of crops and investments in agrochemicals, and in medicine it means failed therapy, patient suffering and potential loss of lives. Resistance is a cellular stress response to a particular cytotoxic agent, which can occur at either cellular or target level. At a cellular level, the use of cytotoxic chemicals can induce up-regulated expression of transporters and metabolic pathways that effectively reduce intracellular accumulation of the toxic compound. In some organisms, mechanisms exist that bypass the drug-targeted molecules, leading to resistance. At the target level, nature will take advantage of variations in the population (mutants) to select out those variants that are resistant to the treatment.
In this article, we will first review the current view of the mechanism of function of bc1 in coupling electron transfer to proton translocation, which is followed by an overview on experiments detailing the structural basis of inhibitor binding and the mechanism of resistance against bc1 inhibitors. Strategies on how to overcome this resistance will be discussed.
2. Importance of Ubiquinol cytochrome C oxidoreductase
Cellular respiration depends on the functions of the membrane embedded protein complexes NADH dehydrogenase (I), succinate dehydrogenase (II), ubiquinol cyt c oxido reductase (III), cyt c oxidase (IV) and ATP synthase (V) (Fig. 1). In eukaryotic cells, these respiratory chain complexes are located in the inner mitochondrial membrane. The complex III or bc1 complex is the mid-segment of the respiratory chain. Structurally, it is a homo-dimeric complex, containing 10 to 11 subunits per monomer, of which only three subunits are essential for the electron transfer (ET) and proton translocation function: cyt b, cyt c1 and the iron-sulfur-protein. In bacteria, bc1 is embedded in the plasma membrane and may contain one supernumerary subunit. Although the main focus of this review is on mitochondrial bc1, it should be pointed out that in the photosynthetic apparatus of plant chloroplasts, the analogous cyt b6f complex is used, which operates in the thylakoid membrane (Fig. 2a). Out of the 10 or 11 subunits of mitochondrial bc1, only the cyt b subunit is encoded by the mitochondrial DNA and synthesized in the mitochondrion; the remaining subunits are coded in the nucleus. Containing two b-type hemes bL and bH, the cyt b subunit contains two quinol/quinone binding sites that are referred to as quinol oxidation (outside QO) and quinone reduction (inside Qi) pockets, respectively (Fig. 2b). The attribute outside or inside indicates the physical location of the active site, but since the orientations of the mitochondrial inner membrane and their related chloroplast thylakoid membrane are reversed, confusion can be avoided by labeling them with respect to the constant, relative membrane potential. Thus, the QO and the QI sites being close to either the positive or negative side of the membrane are referred to as QP and QN site, respectively.
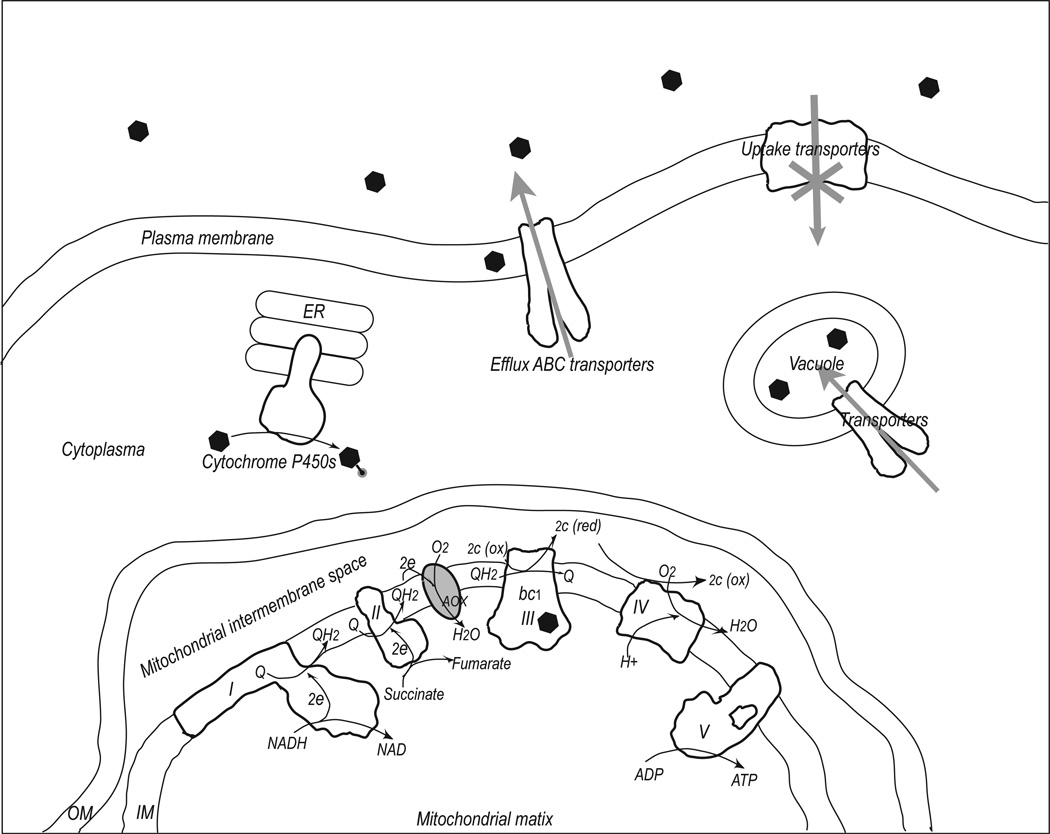
Figure 1. Mitochondrial respiratory chain and mechanisms of resistance to inhibitors of cyt bc1 complex.
Cellular components shown include mitochondria, plasma membrane, ER and vacuole. On the mitochondrial inner membrane (IM), the five respiratory chain components are shown and labeled sequentially as I – V. Their basic functions in Oxphos are also illustrated. Additionally, the alternative oxidase (AOX) is shown, which bypasses the bc1 by sending electrons from ubiquinol directly to oxygen. Inhibitors of cyt bc1 are shown as filled hexagons, whose uptake may require assistance of uptake transporters. Resistance to bc1 inhibitors can arise if the cellular accumulation of the inhibitors is reduced, which includes blockage of uptake pathway, active transport by efflux ABC transporters, getting pumped into vacuoles, or chemically modified by cyt P450s. Additionally, mutations in bc1 can lead to resistance.
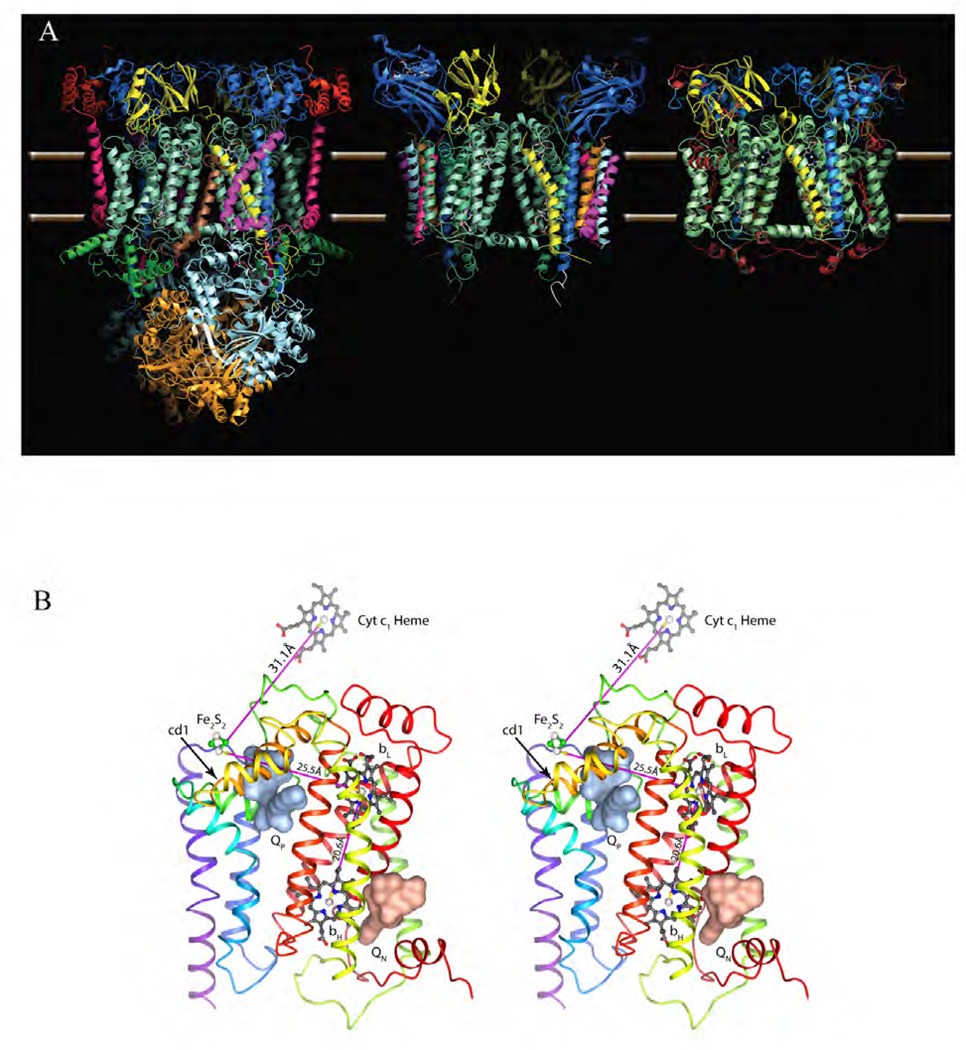
Figure 2. Atomic model of cyt bc1/b6f complexes.
(A) Comparison of the dimeric complexes with their respective 11, 8 and 3 subunits per monomer found in the crystal structures: bovine mitochondrial bc1, Chlamydomonas Reinhardtii algal b6f and Rhodobacter Sphaeroides bc1 with the three essential subunits: cyt b (green-cyan), cyt c1 (dark blue) and the iron-sulfur-protein (yellow). Subunit IV of R.S. bc1 was not found in the crystal structure. (B) Stereo diagram depicting cyt b with its eight TM helices and two b-type heme group bL and bH. The QP and QN site are occupied by stigmatellin (pale red surface) and antimycin (pale blue surface), respectively. Also shown are the iron-sulfur cluster Fe2S2 and the heme group of cyt c1. The magenta arrows indicate the shortest connection between the redox centers.
The reaction catalyzed by bc1 is shown in equations (a) and (b). In this reaction, ubiquinol is the electron donor and cyt c is the acceptor; the ET is coupled to proton translocation across the membrane. In the effective summary (Eq. b) of the reaction, only molecules of QH2 can be cancelled but not protons. This is because we can only cancel molecules that stay in the same phase like the quinone or quinol molecules, which are only soluble in the membrane phase and freely diffuse between sites. But we cannot cancel the protons. Only under highly regulated conditions is it possible for protons to exit or enter the membrane phase as in the case of the four protons leaving the QP site or the two protons entering at the QN site. This leads to the stoichiometry of four protons that are released for each molecule of oxidized quinol.
| (a) |
| (b) |
The stoichiometry of the reaction can be explained by the “Q cycle hypothesis” [4, 5]. In this hypothesis, ubiquinol (QH2) enters a large hydrophobic pocket in cyt b near the positive side of the membrane. There it is flanked by, and, through space electronically connected to, the cyt b-bound heme group (bL) and the mobile Fe2S2 cluster of the ISP. When the two-electron oxidation of QH2 is completed, only one electron will have entered the high potential chain, which consists of ISP, cyt c1 and cyt c subunits, yet both hydrogen atoms of QH2, having been stripped of their electrons, enter, as protons, the aqueous phase on the positive side of the membrane. While the electron that entered the Fe2S2 cluster has been removed from the membrane, the second electron stays in it, moving through the components (bL and bH) of the low potential chain, and is directed towards the negative side some 20 Å onto the bH heme. Here it reduces a pre-bound quinone molecule (Fig. 3). This process of a one-electron reduction has to be completed twice and must be coupled to the protonation of the otherwise negatively charged (reduced) reaction product. These protons stem necessarily from the negative side of the membrane. The elegance of this two-step forwards and one-step back process lies in the ability of bc1 to split two electrons of a quinol molecule at the quinol oxidation site (electron bifurcation), thus double the ratio of transported protons per oxidized quinol molecule compared to a simple oxidation with its 1:1 ratio.
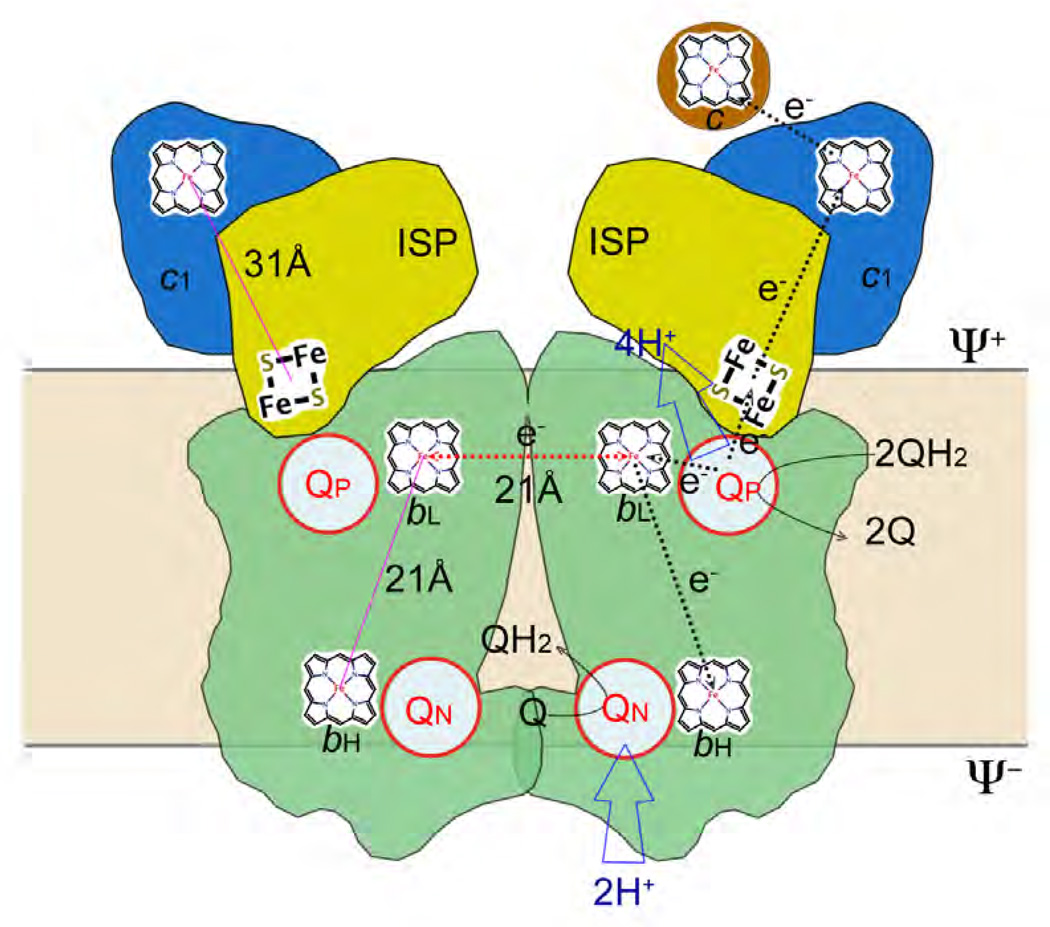
Figure 3. The Q-cycle hypothesis.
The Q cycle mechanism defines two reaction sites: quinol oxidation (Center P or QP) and quinone reduction (Center N or QN). The 1st QH2 moves into the QP site and undergoes bifurcated oxidation with one electron going to cyt c via ISP and cyt c1 (high potential chain), and another ending in the QN via hemes bL and bH (low potential chain) to form a semiubiquinone, and releasing its two protons to the Ψ+ site of the membrane. The 2nd QH2 is oxidized in the same way at the QP site but its low potential chain electron ends up reducing the semiubiquinone and releasing a QH2 upon picking up two protons from the Ψ− side. As a result of the Q cycle, 4 protons are transferred to the Ψ+ side, 2 protons are picked up from the Ψ− side and one QH2 oxidized.
3. Structural characterization of Complex III with and without bound inhibitors
3.1 Structure determination of bc1 and b6f complexes
Structural insights into the bovine mitochondrial enzyme at atomic resolution were gained first by single crystal X-ray diffraction techniques. The crystallization of bovine heart mitochondrial bc1 was reported as early as 1980 [6]. While the early trials did not produce high quality crystals, they at least suggested feasibility and initiated further trials in a number of laboratories [7–10]. Crystallization trials were focused on mitochondrial bc1 as opposed to bacterial, yeast or plant analogues because of its stability: the prevailing literature pointed out that mitochondrial bc1 is more stable against disintegration and is enzymatically more active than its plant and bacterial counterparts [11]. Additionally, the success in crystallizing mitochondrial bc1 was attributed to the availability of large amounts of purified proteins from natural sources, a factor particularly important in an era before the wide spread use of recombinant technology for membrane protein production. Crystals of high quality of bovine bc1 were eventually obtained by rigorously optimizing purification and crystallization procedures leading to the publication of the 11-subunit homo dimeric bc1 structure in 1997 [12, 13]. Since then, 54 structures of bc1 or b6f complexes from nine different species have been deposited into the Protein Data Bank (PDB, Table 1). The high-resolution limit of the X-ray data ranges from 3.80 Å to 1.9 Å, and is sufficiently high to provide detailed atomic information. Historically, structures of unaltered mammalian bc1 were determined first [13–15] followed by the ten-subunit bc1 complex of S. cerevisiae in 2000 [16], which required both the attachment of the Fv fragment of a monoclonal antibody and the respiratory inhibitor stigmatellin for crystal formation. In 2003, another milestone was reached when the structures of two cyt b6f complexes with eight subunits from the thermophilic cyanobacterium M. laminosus and from algae C. reinhardtii were solved [17, 18]. Although bacterial complexes of bc1 were extensively studied biochemically [19], they were the last to be crystallized and the three or four subunit bc1 complexes from anoxygenic photosynthetic bacteria R. capsulatus, R. sphaeroides and P. denitrificans were reported in 2004 (3.8 Å), 2006 (2.3 Å) and 2011 (2.7 Å), respectively [20–22].
Table 1.
Cytochrome bc1/b6f structures in the protein data bank (PDB).
| PDB1 | Organism2 | QP Site occupant |
Inhibitor Type |
QN Site occupant |
Resolution3 [Å] |
R (Free)4 |
No. Subunits: Found/In vivo |
Reference |
|---|---|---|---|---|---|---|---|---|
| 1SQB | B.T. | Azoxystrobin | Pm | - | 2.69 | 0.288 | 11 / 11 | Xia [112] |
| 3L71 | G.G. | Azoxystrobin | Pm | Coenzyme Q10 | 2.84 | 0.281 | 10 / 11 | Berry TBP5 |
| 3L70 | G.G. | Trifloxystrobin | Pm | Coenzyme Q10 | 2.75 | 0.297 | 10 / 11 | Berry TBP |
| 1SQP | B.T. | Myxothiazol | Pm | - | 2.7 | 0.314 | 11 / 11 | Xia [112] |
| 1SQQ | B.T. | MOA Stilbene | Pm | Ubiquinone 2 | 3.0 | 0.295 | 11 / 11 | Xia [112] |
| 3L72 | G.G. | Iodo-Kresoxim-dimethyl | Pm | Coenzyme Q10 | 3.06 | 0.294 | 10 / 11 | Berry TBP |
| 3H1K | G.G. | Iodo-Kresoxim-dimethyl | Pm | Coenzyme Q10 | 3.48 | 0.284 | 10 / 11 | Berry [113] |
| 3TGU | G.G. | WF3 | Pm | Coenzyme Q10 | 2.70 | 0.289 | Berry[47] | |
| 3L73 | G.G. | Triazolone | Pm | Coenzyme Q10 | 3.04 | 0.293 | 10 / 11 | Berry TBP |
| 1L0L | B.T. | Famoxadone | Pf | - | 2.35 | 0.306 | 11 / 11 | Xia [88] |
| 3L74 | G.G. | Famoxadone | Pf | Coenzyme Q10 | 2.76 | 0.286 | 10 / 11 | Berry TBP |
| 2FYU | B.T. | JG144 | Pf | - | 2.26 | 0.283 | 11 / 11 | Xia [27] |
| 3L75 | G.G. | Fenamidone | Pf | Coenzyme Q10 | 2.79 | 0.275 | 10 / 11 | Berry TBP |
| 3H1L | G.G. | Ascochlorin | Pf, PN | Ascochlorin | 3.21 | 0.295 | 10 / 11 | Berry [93] |
| 3CWB | G.G. | Iodo-Crocacin-D | Pf | Coenzyme Q10 | 3.51 | 0.319 | 10 / 11 | Berry [114] |
| 1NU1 | B.T. | NQNO | Pf, PN | NQNO | 3.2 | 0.296 | 11 / 11 | Xia [88] |
| 2E75 | M.L. | - | / | NQNO | 3.55 | 0.267 | 8 / | Cramer [36] |
| 1SQV | B.T. | UHDBT | Pf | Ubiquinone 2 | 2.85 | 0.285 | 11 / 11 | Xia [112] |
| 1P84 | S.C. | HDBT | Pf | Ubiquinone 6 | 2.5 | 0.252 | Hunte [43] | |
| 1SQX | B.T. | Stigmatellin | Pf | Ubiquinone 2 | 2.6 | 0.281 | 11 / 11 | Xia [112] |
| 2A06 | B.T. | Stigmatellin | Pf | Coenzyme Q10 | 2.1 | 0.258 | 10 / 11 | Berry [89] |
| 1PP9 | B.T. | Stigmatellin | Pf | Coenzyme Q10 | 2.1 | 0.287 | 10 / 11 | Berry [89] |
| 1PPJ | B.T. | Stigmatellin | Pf | Antimycin | 2.1 | 0.26 | 10 / 11 | Berry [89] |
| 2BCC | G.G. | Stigmatellin | Pf | Coenzyme Q10 | 3.5 | 0.317 | 10 / 11 | Berry [89] |
| 3BCC | G.G. | Stigmatellin | Pf | Antimycin | 3.7 | 0.321 | 10 / 11 | Berry [25] |
| 3H1I | G.G. | Stigmatellin | Pf | Antimycin | 3.53 | 0.306 | 10 / 11 | Berry [93] |
| 3H1J | G.G. | Stigmatellin | Pf | Coenzyme Q10 | 3.0 | 0.277 | 10 / 11 | Berry [25] |
| 1EZV | S.C. | Stigmatellin | Pf | Coenzyme Q6 | 2.3 | 0.254 | 9 / 10? | Hunte [115] |
| 1KB9 | S.C. | Stigmatellin | Pf | Coenzyme Q6 | 2.3 | 0.249 | 9 / 10? | Hunte [116] |
| 1KYO | S.C. | Stigmatellin | Pf | - | 2.97 | 0.268 | 9 / 10? | Hunte [117] |
| 3CX5 | S.C. | Stigmatellin | Pf | - | 1.9 | 0.263 | 9 / 10? | Hunte [118] |
| 2IBZ | S.C. | Stigmatellin | Pf | Ubiquinone Q6 | 2.3 | 0.256 | 9 / 10? | Hunte [92] |
| 3CXH | S.C. | Stigmatellin | Pf | - | 2.5 | 0.256 | 9 / 10? | Hunte [118] |
| 1ZRT | R.C. | Stigmatellin | Pf | - | 3.5 | 0.358 | 3 / 3 | Berry [119] |
| 2FYN | R.S. | Stigmatellin | Pf | - | 3.2 | 0.254 | 3 / 4 | Xia [27] |
| 2QJP | R.S. | Stigmatellin | Pf | Antimycin | 2.6 | 0.277 | 3 / 4 | Xia [90] |
| 2QJK | R.S. | Stigmatellin | Pf | Antimycin | 3.1 | 0.266 | 3 / 4 | Xia [90] |
| 2QJY | R.S. | Stigmatellin | Pf | Ubiquinone 2 | 2.4 | 0.251 | 3 / 4 | Xia [90] |
| 2YIU | P.C. | Stigmatellin | Pf | - | 2.7 | 0.29 | 3 / 3 | Carola Hunte |
| 1Q90 | C.R. | Tridecyl Stigmatellin | Pf | - | 3.1 | 0.261 | 9 / 9 | Picot [82] |
| 1VF5 | M.L. | Tridecyl Stigmatellin | Pf | Plastoquinone | 3.0 | 0.346 | 8 / 8 | Cramer [81] |
| 2E76 | M.L. | Tridecyl Stigmatellin | Pf | Tridecyl Stigmatellin | 3.41 | 0.256 | 8 / 8 | Cramer [36] |
| 2YBB | TH.TH. | Stigmatellin | Pf | Ubiquinone Q1 | 19.0 | N/A | - | Kühlbrandt [120] |
| 1QCR | B.T. | - | - | - | 2.7 | 0.375 | 11 / 11 | Xia [84] |
| 1BE3 | B.T. | - | - | - | 3.0 | 0.32 | 11 / 11 | Iwata [121] |
| 1BGY | B.T. | - | - | - | 3.0 | 0.36 | 11 /11 | Iwata [121] |
| 1L0N | B.T. | - | - | - | 2.6 | 0.297 | 11 / 11 | Xia [51] |
| 1NTM | B.T. | - | - | - | 2.4 | 0.285 | 11 / 11 | Xia [88] |
| 1NTZ | B.T. | Ubiquinone Q2 | - | Ubiquinone Q2 | 2.6 | 0.283 | 11 / 11 | Xia [88] |
| 1NTK | B.T. | - | - | Antimycin | 2.6 | 0.27 | 11 / 11 | Xia [88] |
| 1BCC | G.G. | - | - | Coenzyme Q10 | 3.16 | 0.31 | 10 / 11 | Berry [25] |
| 3H1H | G.G. | - | - | Coenzyme Q10 | 3.16 | 0.291 | Berry [25] | |
| 2E74 | M.L. | - | - | - | 3.0 | 0.268 | Cramer [36] | |
| 2D2C | M.L. | Dibromo-benzoquinone5 | P5 | - | 3.8 | 0.378 | Cramer, TBP | |
| 2ZT9 | NOS. | - | - | - | 3.0 | 0.259 | Cramer, TBP |
PDB – protein data bank
B.T., B. taurus; G.G., G. gallus; M.L., M. laminosus; NOS, Nostoc sp. PCC7120; T.H., T. thermophilus; C.R., C. reinhardtii; P.D.; P. denitrificans; R.S., R. sphaeroides; R.C., R. capsulatus; S.C., S. cerevisiae
Diffraction limit of the crystal in units of Å: 1Å = 10−10 m.
Quality index; predictive power of model. A lower value of R (free) is better.
TBP, to be published
As much as they differ in the subunit compositions, at the core all bc1 complexes contain the three essential subunits: cyt b, cyt c1 and the iron-sulfur-protein (ISP) as shown in Figure 2a (RS bc1). Dimeric cyt b with its 16 transmembrane (TM) helices forms the core of the bc1 and sits entirely inside the membrane bilayer. By contrast, both cyt c1 and ISP subunits have each only one TM helix, which is largely supported by cyt b and both feature large extra membrane domains which are exposed to the positive side of the membrane (intermembrane space in mitochondria or periplasmic space in bacteria). While the cyt c1 subunit is associated with one cyt b monomer, the ISP subunit has its TM helix associated with one cyt b monomer and its functional domain interacting with the other, providing the first structural evidence for a functional dimer [13]. The eight TM helices of a monomeric cyt b subunit organize into two helical bundles. Helices A–E form the first bundle and enclose the two b-type hemes. The second bundle consists of helices F–H. With the structures of all subunits known, distances between positions of iron atoms for all prosthetic groups were resolved [13]. Interestingly, the extrinsic domain of ISP subunit (ISP-ED) demonstrated significant conformation flexibility and was found in at least three different positions. In the majority of crystal forms, ISP-ED was found docked at cyt b subunit (the b-state), but in a few reported crystal structures, it was either near cyt c1 (c1-state) or in a position between c1- and b-states. As predicted by the Q-cycle mechanism, there are two quinol/quinone reaction sites in the bc1 complex. The quinone reduction site (QN) is close to the bH heme near the N-side of the membrane, whereas the quinol oxidation site (QP) is a relatively elongated hydrophobic pathway located between the bL heme and Fe2S2 cluster of ISP subunit near P-side of the membrane. The portion of the QP site close to the bL heme is referred to as the proximal site and that close to the Fe2S2 cluster is called the distal site.
3.2. Structures of bc1 in complex with inhibitors and the discovery of the controlled ISP mobility
The establishment of the Q-cycle mechanism can be attributed to the use of specific cyt bc1 inhibitors, some of which had the ability to stall the enzymatic reaction halfway through allowing the discovery of the "oxidant-induced" reduction of cyt b [23, 24]. The structure determination of native mitochondrial bc1 was followed and/or accompanied by structures with QP and QN inhibitors bound to cyt b [13, 25, 26]. It should be mentioned that bovine heart mitochondrial bc1 crystallized in the space group I4122, which turned out to be particularly useful in the study of the motion of the ISP extrinsic domain (ISP-ED) since this crystal form did not involve ISP-ED in packing interactions. Table 1 lists all currently downloadable bc1 models with their respective QP and QN site occupants. The importance of an unrestrained ISP-ED (~14.6 kDa domain of ISP, i.e. ISP minus the TM helix) became clear when it was discovered that its conformational state depended on the inhibitor or inhibitor type bound at the QP site [26–28]. Furthermore, a controlled, mobile ISP-ED proved instrumental in explaining how a ~31 Å gap (Fig. 2b) between the redox centers Fe2S2 and heme cyt c1 could be bridged, how the oxidation and de-protonation of the substrate could be initiated and how the bifurcated electron transport could be accomplished. It is important to emphasize that a flexible ISP-ED is indeed necessary, though not sufficient, for the function of bc1. Through a series of elegant experiments based on suitably designed mutants of bacterial bc1 it was finally established that restricting the mobility of ISP-ED yielded an enzyme with poor or abolished activities, both in vivo and in vitro [29–31].
3.3. Classification of bc1 inhibitors
Historically, bc1 inhibitors were classified based on their effects on bL-heme spectrum and impact on the redox potential of ISP, which suffered from a lack of understanding of the underlying process [32]. On the basis of structural information, bc1 inhibitors are currently divided into QP site inhibitors (Fig. 4), the QN site inhibitors (Fig. 5) (N) and the dual site inhibitors (PN) [28]. Although a classification of bc1 inhibitors that goes beyond the obvious of discriminating solely between P and N types may appear useful only in Q-cycle research, the additional information contained in inhibitor sub-types (P sub-types: Pf, Pm), may impact the design of future generations of pesticides with an emphasis on reducing resistance risk. Initial concepts of whether a QP inhibitor increases the redox potential of Fe2S2 and/or changes the optical spectrum of the bL-heme were useful for instance in drawing attention to a connection between two prosthetic groups through the QP site, it could not reveal nor make predictions about the motion of ISP-ED that depends on the inhibitor type [32]. For instance, famoxadone was assigned to the group of strobilurin and myxothiazol-like inhibitors [33] but in fact rather belongs to the group of stigmatellin-like inhibitors [27, 28, 34]. An increasing body of structural evidence starting with the first structures of bc1 [13] followed by a number of high resolution inhibitor bound bc1 complexes suggested that the influence of the inhibitor on the mobility of the ISP-ED was systematic. Accordingly, all known QP site inhibitors (QoI) can be divided into two subgroups [27, 28], namely those that fix the conformation of the ISP-ED in the b-state (Pf-type) and those that mobilize it (Pm-type). In the latter case, the ISP-ED may be found in the c1-state (pdb entries 3L70, 3L71, 3L72, 3TGU) but could also be anywhere between the b and c1-state (pdb entries 1SQP, 1SQQ). The division into Pf and Pm type became clear after superposition of all available inhibitor bound structures. As it turns out, the formation of the narrow channel in the QP site between Pro270 and Gly142 is such that an inhibitor (and by implication the natural substrate quinol) must interact with the cd1 helix, of which Ile146 (Ile147 in S. cerevisiae numbering, Fig. 6) is the most prominent feature. Mechanistically, this ISP-motion switch is achieved by modulating the ISP-binding affinity of cyt b, onto which ISP-ED docks, from high to low. The binding surface itself is made up of the cd1–cd2 helix pair, which is capable of undergoing a bi-directional movement, and portions of E-ef-loop (~260–268), F1 helix (~282–287) and the G-H loop (~342–344). An alternative explanation as to the origin of the fixation of ISP-ED was recently proposed by Berry & Huang [35] pointing to the highly conserved residue Tyr278 (bovine numbering), which replaces the lost (stigmatellin) O4…H-Nε2 hydrogen bond in the complex. However, the proposal lacked any mechanism by which the inhibitor could interact with Tyr278 to make it form the hydrogen bond that it is notably absent from the native (apo enzyme) structure, because Tyr278 by itself has not been seen to undergo conformational changes in response to the presence or absence of inhibitors.
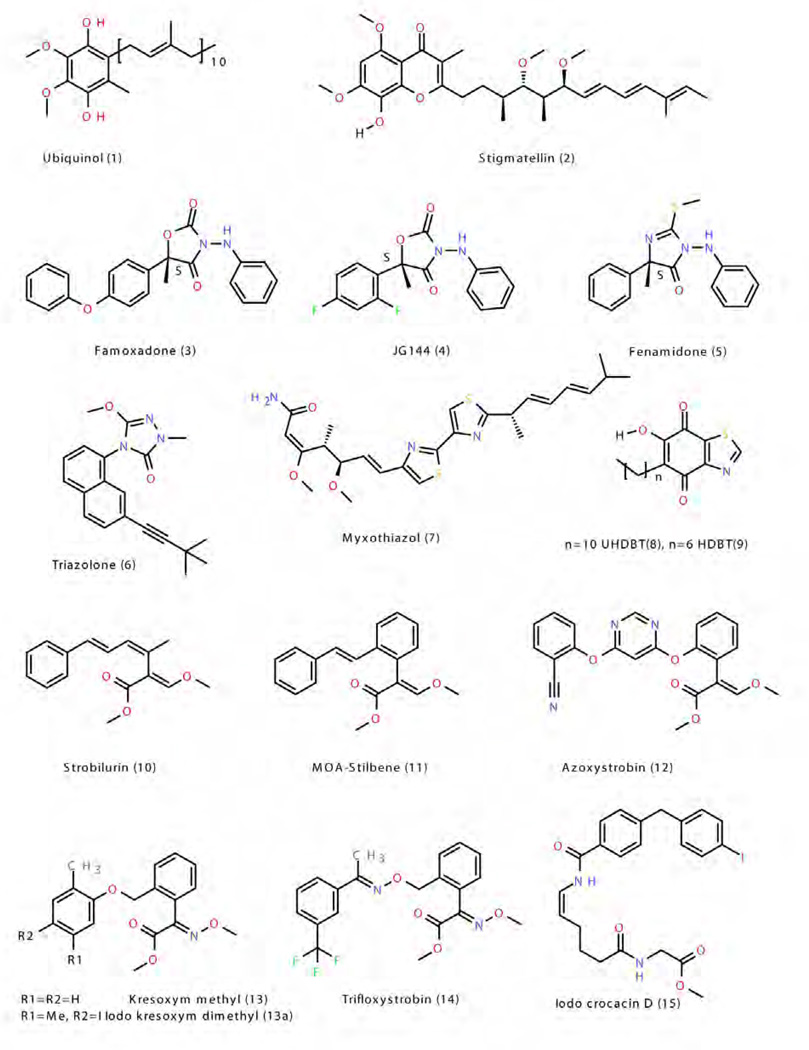
Figure 4. Molecular structures of quinol and QP site inhibitors.
Figure 5. Molecular structures of quinone and QN site inhibitors.
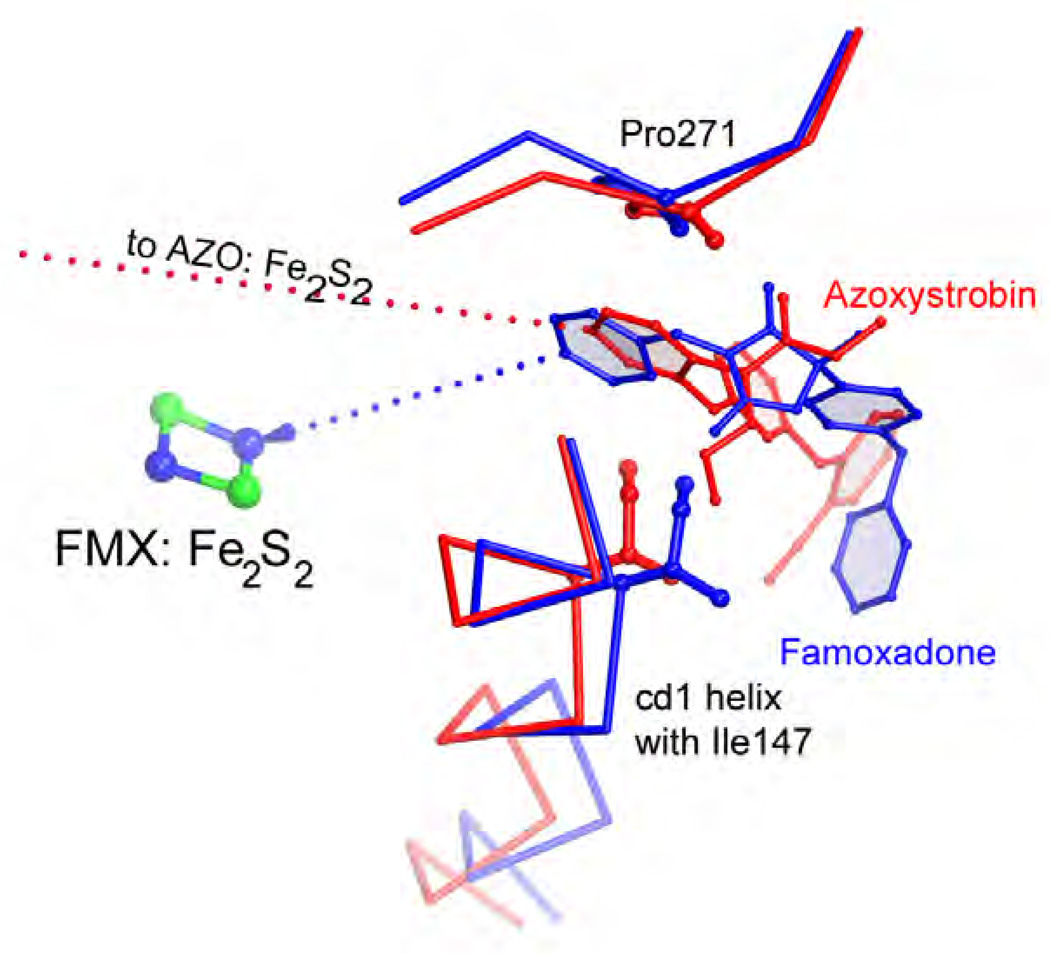
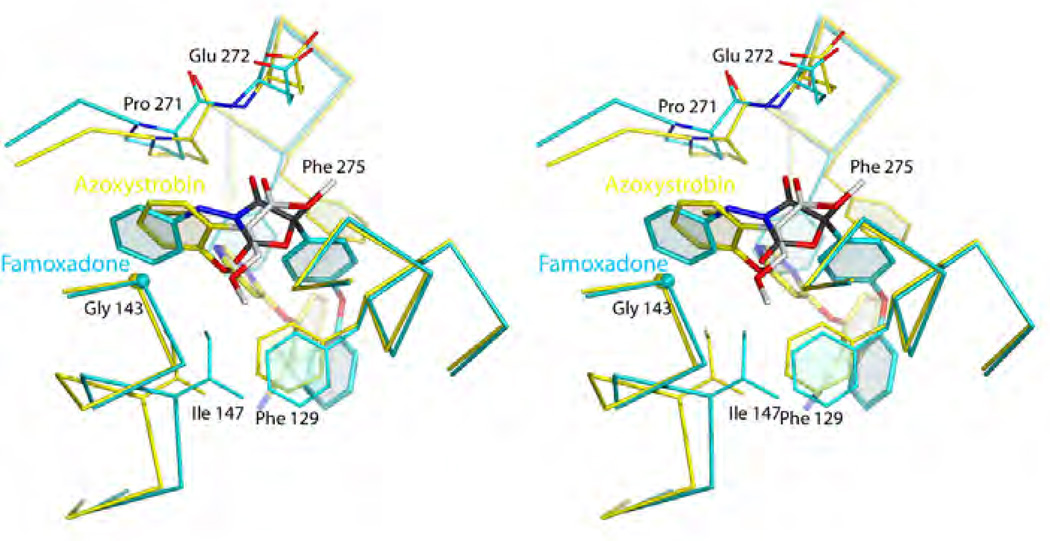
Figure 6. Comparison of the Pm and Pf type inhibitors azoxystrobin (red) and famoxadone (blue).
Note that the iron-sulfur protein (here represented by the Fe2S2 cluster) is docked at cyt b even though no direct inhibitor-ISP interaction occurs.
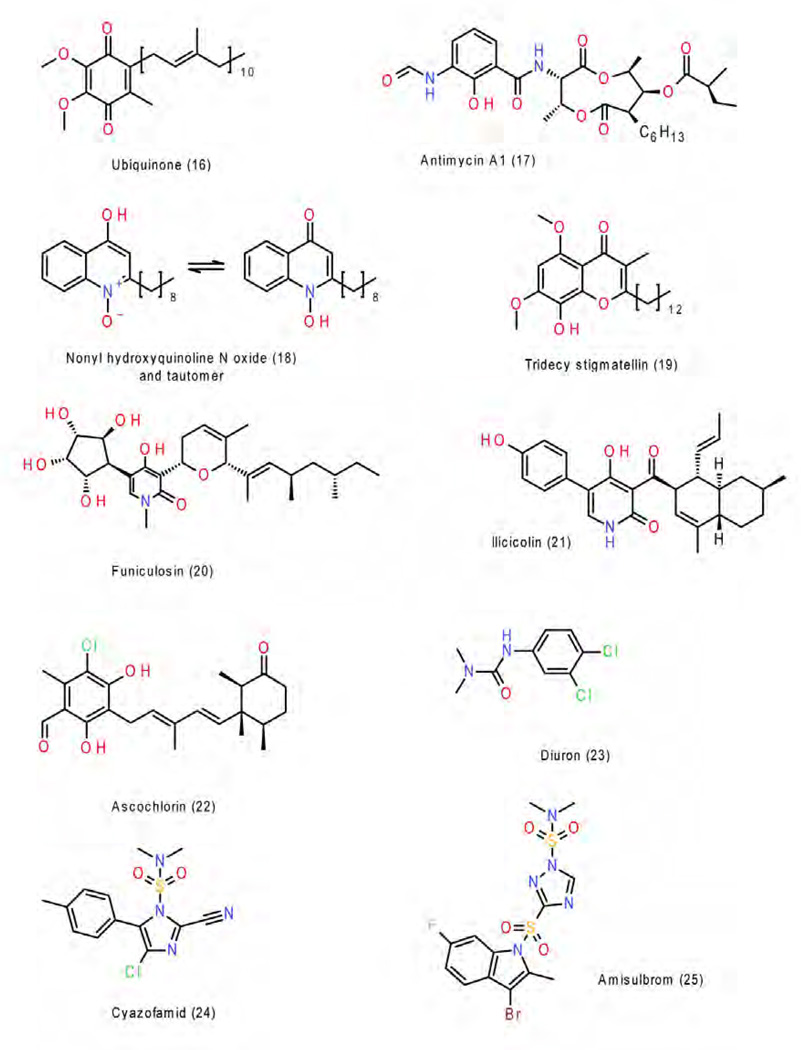
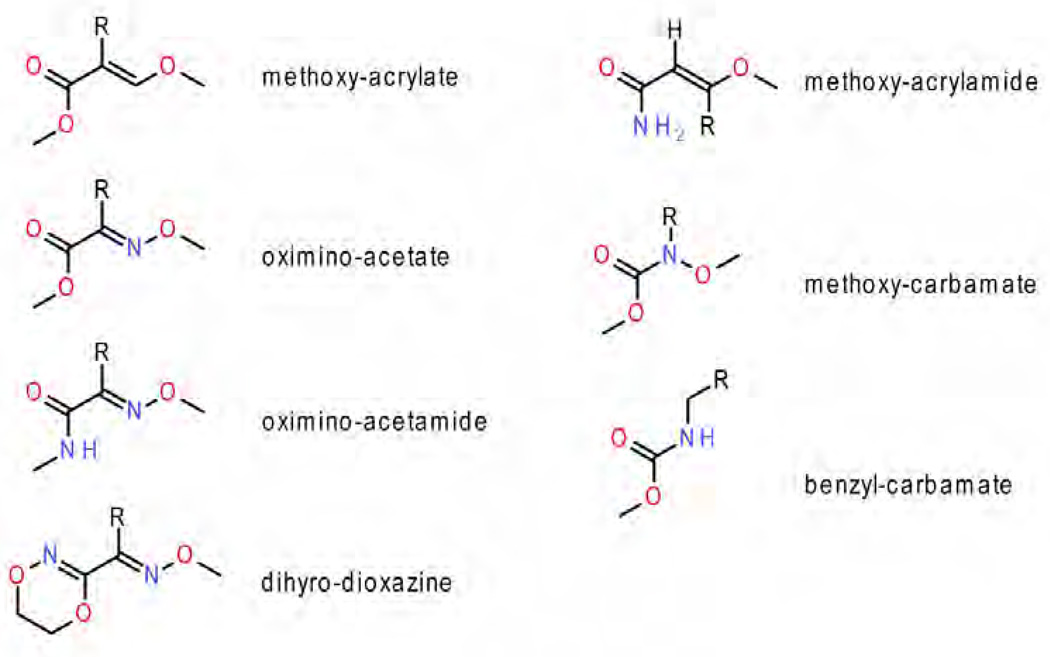
The Pf class contains the classic inhibitors stigmatellin 2, n-undecyl hydroxy dibenzothiazole (UHDBT) 8, HDBT 9 and n-nonyl quinoline N-oxide (NQNO) 18, modern pesticides like famoxadone 3, 3-anilino-5-(2,4-difluorophenyl)-5-methyl-oxazolidine-2,4-dione (jg144) 4, fenamidone 5 and the natural inhibitor ascochlorin 22 as well as iodo-crocacin-D 15 (a compound optimized from natural crocacin D). It was the structure of bc1 with famoxadone bound that first demonstrated that ISP-ED can be stabilized without direct inhibitor contact. This required a re-evaluation of the mode of binding of the QP inhibitors and led to a re-organization of the class of QP inhibitors. Based on structural similarity with an emphasis on the effects of substitution patterns and steric bulk of side chains, compounds belonging to the oxazolidinediones, and imidazolinones are expected to be Pf-type inhibitors. However care must be taken not to rush to conclusions based on the shape of the toxophore alone: triazolone 6 [35] features a 5-membered ring just as 3, 4 and 5 but it deviates from the side chain pattern that is found in methoxy-acrylates and therefore does not belong to the Pf type but rather is unambiguously of type Pm.
The class of Pm inhibitors contains the well-known natural inhibitor myxothiazol 7 but also the synthetic compounds methoxy-acrylate stilbene and the widely used pesticides azoxystrobin 11, kresoxim methyl 13, trifloxystrobin 14, and triazolone 6. This is the group that contains derivatives of strobilurin whose structure-activity relationship advanced the research in respiratory chain complex III inhibitors. Given the consistency of the mode of binding in this group, it can be expected that all methoxy-acrylates, methoxy-carbamates, oximino-acetates, oximino-acetamides and benzyl-carbamates will belong to this group.
The class of N inhibitors is comprised of compounds that occupy the QN site and covers the category of QiI pesticides. This group is still rather small and starts with the natural inhibitor antimycin but also contains NQNO 18 and ascochlorin 22, for which crystal structures were determined. No structural information is currently available that would illuminate how a number of natural inhibitors like funiculosin 20 or the important pesticides cyano-imidazole (cyazofamid 24) and sulfamoyl-triazole (amisulbrom 25) might bind.
The class of PN inhibitors is essentially not a new class as members of this group are listed in both, the P and N class. Its presence only underlines the fact that some inhibitors have features (just as the quinol/quinone pair) that allow them to bind to both active sites. This group includes NQNO, ascochlorin and tridecyl-stigmatellin 19 (observed in b6f) [36].
4. binding mode of QP site Inhibitors
Much biochemical, spectroscopic and mutational work on isolated bc1 and in vivo studies on organisms bearing bc1 mutants, including many pathogens had been done before the crystal structure of bc1 was determined. For instance, the entire lead optimization that led to azoxystrobin, kresoxym-methyl [37] and other pesticides from strobilurin was accomplished without the benefit of 3D structural information: Not even the native bc1 structure was known, much less any atomic details on how and where QOIs might bind. Nowadays however, with the increasing number of high-resolution structures, drug design can take advantage of fairly detailed information across a diverse group of inhibitors that in fact illuminates a rather complex and dynamic quinol oxidation site. Recent attempts employing rational drug design aim for instance at the development of new medicines against the human scourge of malaria [38, 39]. Structural information will play also an increasingly important role in the design of new QoI-type inhibitors, which will continue to be developed despite the recognition by FRAC (Fungicides Resistance Action Committee) [40] that respiration inhibitors (codes C3 / C4) bear a high risk of causing the emergence of resistant pathogens.
4.1. Binding mode of the classic inhibitor Stigmatellin and related compounds at QP site
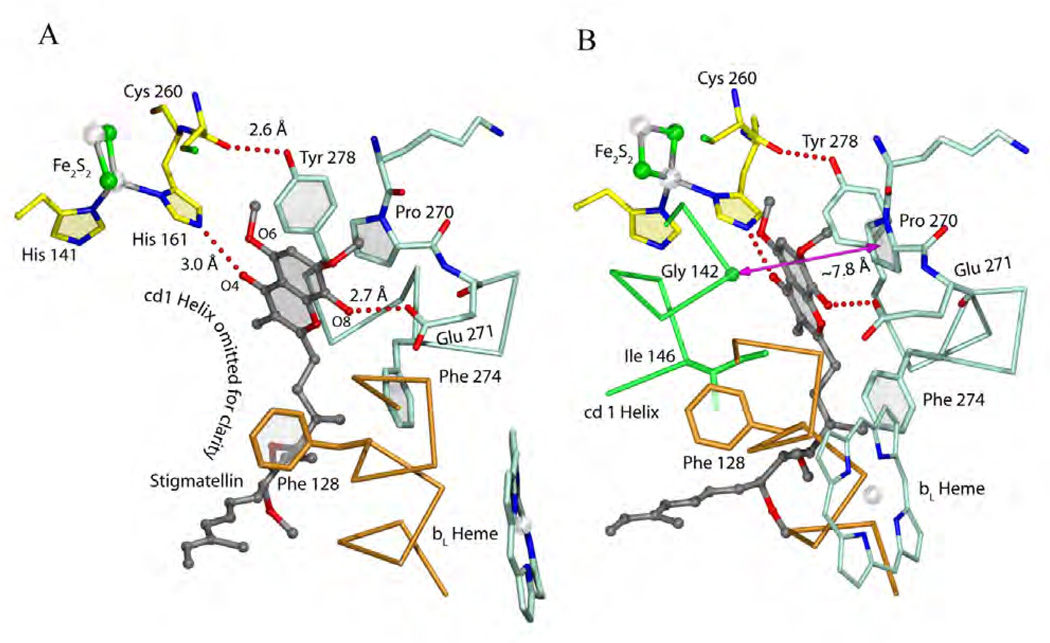
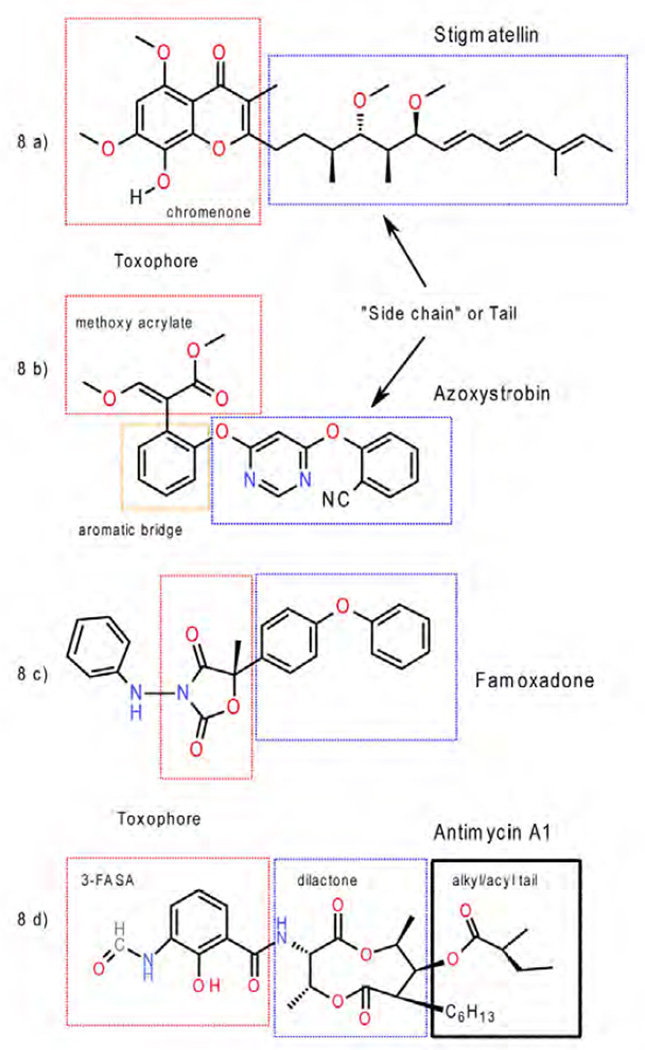
Stigmatellin 2 from the myxobacterium Stigmatella aurantica is not only a QP-site inhibitor with one of the lowest dissociation constants (Kd < 10−11M) [41, 42] but is also one of the largest inhibitors in terms of atom count / molecular formula: C30H42O7 (Mw = 514.56 g/mol). Not surprisingly, in crystal structures of bc1 complexed with this inhibitor (Table 1), stigmatellin (Fig. 7a, b) was found to take advantage of a large portion of the QP-site [16, 21, 28]. It consists of a chromone moiety that resembles two fused quinol/quinone units and a long olefinic tail. It is a reoccurring theme that inhibitors themselves can be divided into a pharmacophore (or toxophore) and a supporting "side chain" (Fig. 8a). While the pharmacophore is providing the essential binding capacity and specificity, the side chain plays a role in support and solubilization. In the case of stigmatellin it was found that the chromone ring is the toxophore, interacting with the iron-sulfur protein in a way that is believed to be accessible to the natural substrate as well (Fig. 7). Furthermore, even relatively subtle changes in the composition and/or degree of saturation of the hydrophobic tail fragment were shown to lower the binding affinity by up to five orders of magnitude [42].
Figure 7. Quinol oxidation site and binding of stigmatellin.
(A) Stigmatellin is bound at the QP site and forms hydrogen bonds with ISPHis161 and the side chain of Glu271. (B) Stigmatellin is inserted in the ~7.8 Å gap (magenta arrow) between Gly142 and Pro270.
Figure 8. Illustration of the location of the toxophores and side chains in inhibitors.
a) stigmatellin, b) azoxystrobin, c) famoxadone, and d) antimycin.
It is no accident that crystallographers prefer stigmatellin as an inhibitor over all others as can be seen in the disproportionally large number of bc1/b6f structures that contain it (24 out of 54, see Table 1). There is even an argument to be made that without it, crystal structures of bc1 with fewer subunits (for instance bacterial bc1 with 3 or 4 subunits) could not have been obtained without stigmatellin arresting the motion of ISP-ED or at least would have been significantly delayed. Stigmatellin sits in the distal portion of the QP site (Fig. 7a, b) and forms two hydrogen bonds: carbonyl oxygen atom O4 accepts a hydrogen bond from the Fe2S2-cluster ligand His161 of ISP and on the opposite side, the phenolic hydroxyl group (O8) donates a hydrogen bond to the carboxylate side chain of Glu271. Compared to its "native" position in which Glu271 is pointing towards the aqueous phase, the side chain of Glu271 has swung over to accommodate this special inhibitor. Apart from very specific interactions, the flat toxophore of stigmatellin inserts halfway between the cd1 helix on one side and the Pro270 residue of the classical PEWY motif (ef-loop) (Fig 7b). The gap between Gly142 (cd1 helix) and Pro270 indicated by the magenta arrow of about 7.8 Å is of an ideal size for inserting flat aromatic ring systems with a ~3.5 Å van-der-Waals contact distance. Indeed, all QoIs take advantage of this characteristic QP-site feature, which unfortunately makes them vulnerable to changes by the target molecule when a Gly142Ala mutation occurs. The binding of stigmatellin exerts pressure on Ile146 causing a horizontal shift of the cd1/cd2 helix pair with respect to the membrane surface, which was correlated with an increased binding of ISP-ED [27]. Despite the highly optimized structure of stigmatellin's tail there are no detectable specific interactions with the QP pocket. The function of the tail in its complexity may simply rest in the need to maintain the overall distinct shape of stigmatellin resembling a straight edge.
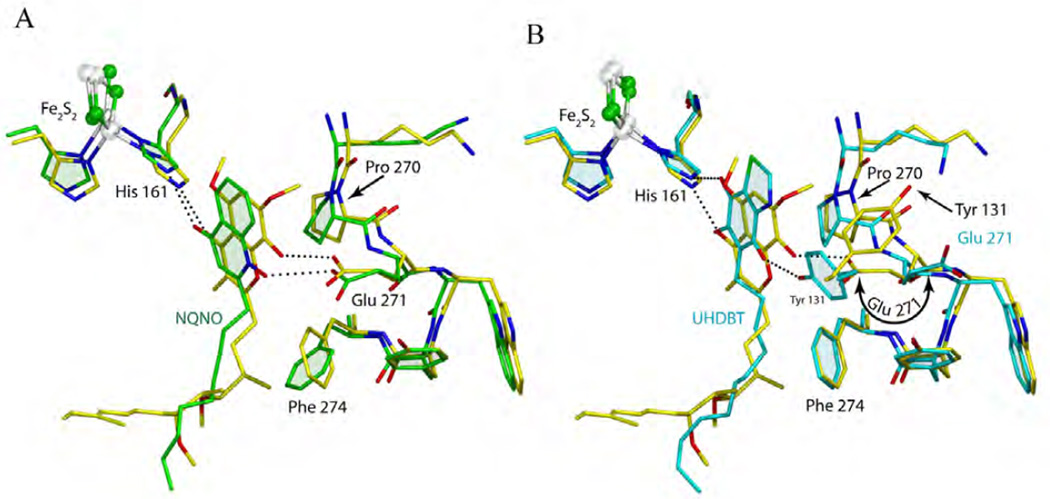
The inhibitors that come closest to stigmatellin in their binding modes are 2-nonyl-4-hydroxyl quinoline N-oxide (NQNO) and 5-n-alkyl-6-hydroxyl-4,7-dioxobenzothiazole (alkyl = n-heptyl for HDBT and n-undecyl for UHDBT). In fact, NQNO 18 in form of the 1-hydroxyl (amine)-4 oxo tautomer (Fig. 5) binds to ISPHis161 of the ISP-ED and Glu271 of cyt b in exactly the same manner as stigmatellin. Figure 9a shows that the pharmacophores of stigmatellin and NQNO overlap after superposition. The n-nonyl chain of NQNO follows approximately the tail of stigmatellin, but being unrestrained, it adopts a far more variable conformation possibly leading to a lower affinity. The unusual features of the tail of stigmatellin may also prevent it from being a QiI inhibitor. This is in contrast to the observation that both tridecyl stigmatellin (with a much more flexible tail) and NQNO could be detected in the quinone reduction site (1NU1, 2E78). Clearly, NQNO has a binding mode very similar to that of stigmatellin (Fig. 9a); the same cannot be said for UHDBT or HDBT. While the pharmacophores of UHDBT and HDBT coincide very well with that of stigmatellin and while both inhibitors form comparable hydrogen bonds with His161 of ISP-ED (Fig. 9b), they do not induce the rotation of Glu271 towards the inner part of the QP site. An increase in distance towards Glu271 (the DBT moiety is smaller than a chromone) may prevent H-bond formation but it could also be due to the absence of an H-bond donor: at neutral pH (pH 7, in the intermembrane space) Glu271 is ionized and the 4-hydroxy-6,7-dioxo tautomer of the 6-hydroxy-4,7-dioxo benzothiazole that could act as a donor may not be accessible because of the energy cost involved in losing an intra molecular hydrogen bond. A striking difference between the binding modes of the HDBT and UHDBT is that in the latter case the 'lost' H-bond to Glu271 is replaced by a newly formed bond to the phenolic group of Tyr131, which requires this residue to rotate by ~140° (from X1 = −150° to X1 = 71°) from its native position (Fig. 9b) [28]. Furthermore, HDBT ends up interacting only with a surrogate water molecule (not shown, see [43]).
Figure 9. Comparison of NQNO and UHDBT to stigmatellin in their binding to the QP pocket.
(A) The superposition of stigmatellin (yellow) and NQNO (green) reveals their similarities in position and in binding to ISP-ED and Glu271. (B) The superposition of stigmatellin and UHDBT shows different cyt. b side chain conformations when UHDBT is bound. Specifically, Glu271 rotates back to the 'native' state while Tyr131 rotates to form an H-bond with the inhibitor - in a way taking over from Glu271.
4.2. Binding mode of the "strobilurins" at QP site
A series of natural strobilurin derivatives were identified as powerful antibiotics in the late 1970s. Their toxophores were identified as the relatively small E-β-methoxy methyl acrylate moiety [44]. Ultimately, by lead optimization, which at times included major deviations from the basic methoxy acrylate toxophore due to patent restrictions, the current set of some seven fundamental groups of compounds (Fig. 10) were developed. For reviews see [37, 45].
Figure 10. The current set of seven types of QP inhibitor toxophores.
The group of methoxy methyl acrylates contains the strobilurins, oudemansins, methoxy acrylate stilbene (MOA-stilbene 11), myxothiazol, a very large number of trial compounds that were used in lead optimizations, and the ultimately successful azoxystrobin and the more recent picoxystrobin. Based on similarities, the structure of strobilurin and strobilurin-like compounds (collectively referred as the "strobilurins") was divided into three regions that are shown in Fig. 8b as the toxophore (red), aromatic bridge (red-orange) and side chain (blue) [45].
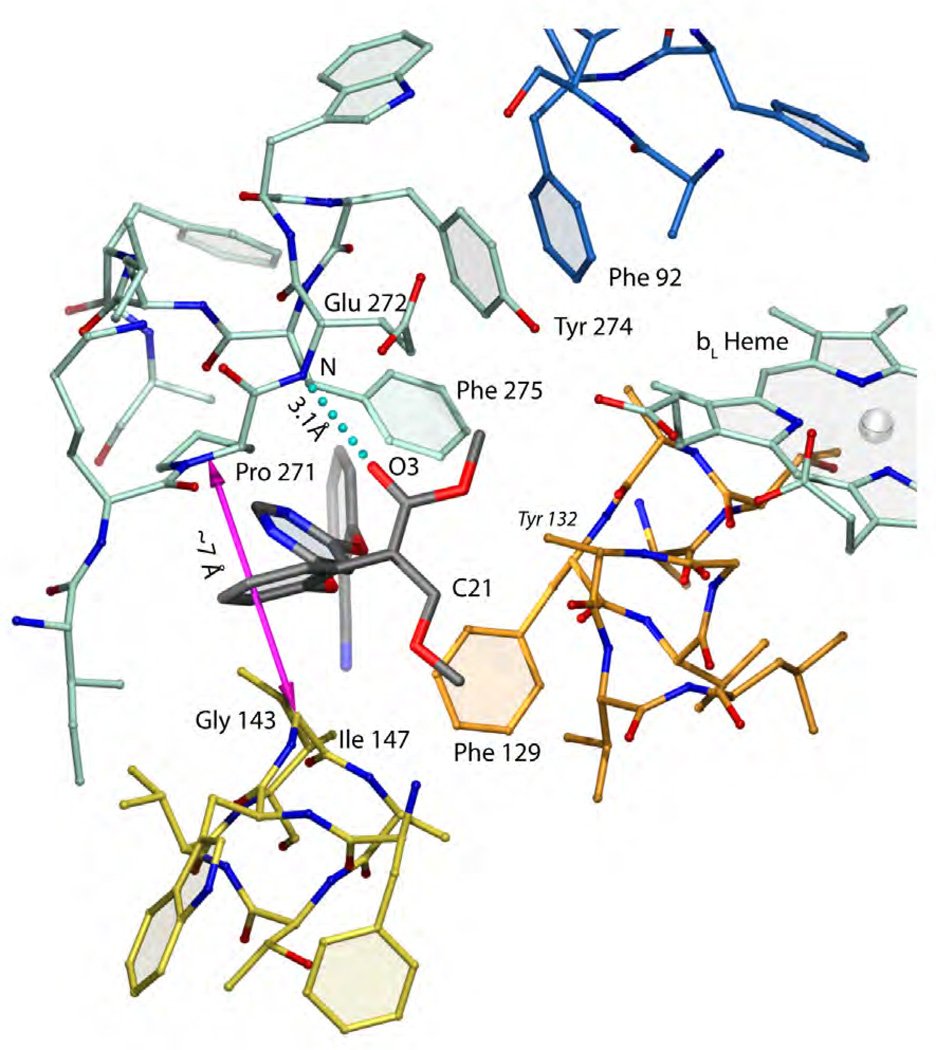
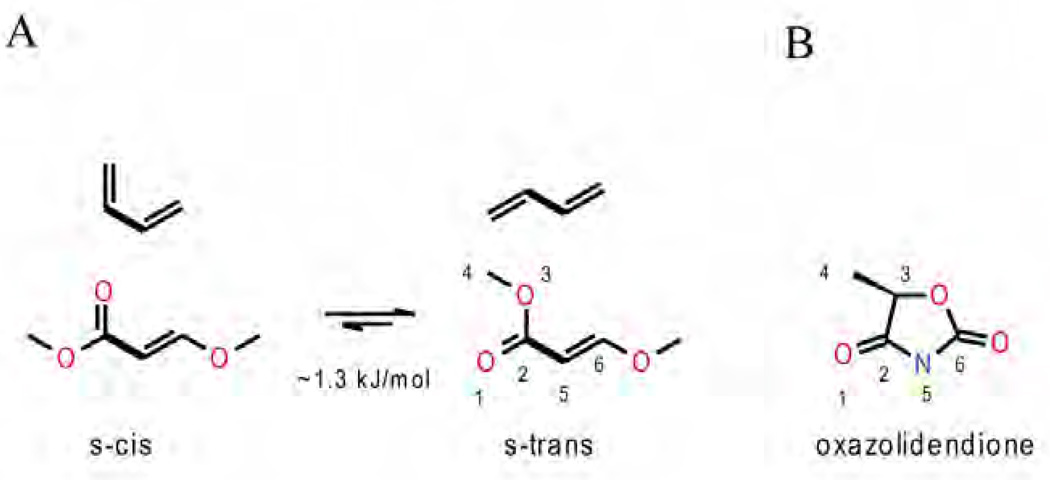
To date, two structures of bc1 in complex with azoxystrobin have been solved from bovine heart (1SQB) and chicken (3L71) mitochondria bc1, respectively. Interestingly, neither species is the intended target of the fungicide. There are several bc1 structures from S. cerevisiae known, but unfortunately none contains azoxystrobin or any other Pm inhibitors. In Figure 11, azoxystrobin (black) is shown bound to the QP site in chicken bc1. Since in the field of drug design considerable interest is focused on the yeast structure, it is important to point out that the sequence conservation of cyt b is very high and the sequence numbering of chicken cyt b in the proximal QP site is identical to that of yeast so that no adjustments with respect to the residue numbers have to be made. The choice of azoxystrobin as a representative of the strobilurins is a good one as it turns out that the toxophores of MOA-stilbene (1SQQ) and even the chemically different moieties of myxothiazol (1SQP), trifloxystrobin (3l70) and iodo-kresoxym-dimethyl (3l72 and 3h1k) superimpose well. Unlike stigmatellin, azoxystrobin is bound in the proximal position close to the bL-heme within a distance of about 11 Å to the heme iron ion. The methoxy acrylate group adopts here the s-trans or antiperiplanar conformation while it was found to be s-cis in the bovine heart mitochondrial bc1 structure (1sqb). Energetically the two alternatives (Fig. 12a) can interconvert as the barrier is only of the order of 1.3 kJ/mol for methyl acrylates [46]. Importantly, both conformers are capable of accepting a hydrogen bond. In fact, azoxystrobin (and by extension all strobilurins) forms a critical hydrogen bond to the backbone of Glu272: N-H … O3 of about 3.1 Å as shown in Fig. 11. A carbonyl or imino functionality in a position that can H-bond to the backbone of Glu272 was found to be an essential feature of the toxophore by toxicity analysis in yeast mitochondria of suitably modified compounds [45]. The enol-methyl ether of the toxophore cannot engage in accepting hydrogen bonds, as it is flanked by portions of the cd1 helix (no polar side chains, all amide H-atoms are engaged in helix formation) and Phe129 of the C-helix. The mechanistically important residue Tyr132, also of the C-helix, covers the methoxy-acrylate group but without major interactions. The "aromatic bridge" intercalates between Gly143 of the cd1 helix and Pro271 of the highly conserved PEWY motif (ef-loop). This opening of about 7 Å is not induced by azoxystrobin, as it is also present in the apo form (1ntm). While the pyrimidine portion of the "side chain" forms an energetically favorable π–π stacking interaction with Phe275, the degree to which the terminal 2-cyano phenyl group contributes to the binding energy is less certain judging from visual inspection. It has been pointed out, however, that the pyrimidine moiety was introduced to replace the photolabile diene in strobilurin for both increased stability and water solubility. This was done in view of allowing azoxystrobin to be absorbed by the leaves and distributed to the entire plant (systemic fungicide). On the other side of the stacking interaction, the pyrimidine ring is flanked by Ile147 - a key residue in a model for the mechanism of quinol oxidation [27].
Figure 11. Binding of azoxystrobin in cyt b as revealed by x-ray crystallography.
Figure 12. Chemical configurations of various toxophores.
(A) Illustration of s-cis and s-trans conformation of conjugated double bonds. (B) The oxazolidendione ring is oriented and its atoms are numbered (1–5) to optimally match the methoxy methyl acrylate (MOA) group to the left. The five-membered ring resembles a 'tied' back version of the MOA group.
Structural information for the oximino-acetates (iodo-kresoxim-dimethyl and trifloxystrobin) has become available and their toxophores are expectedly isostructural with the methoxy acrylate group having the methin carbon atom (labeled C21) replaced by an imine (nitrogen) atom. In both cases, the aromatic bridge [45], consisting of an ortho-substituted phenyl ring whose side chains exhibit considerable flexibility and chemical diversity, intercalates between Gly143 and Pro271. Again the closest contact partners with the remainder of the fungicides' side chains are Ile147 of the cd1 helix and Phe275, which engages in π–π stacking with the 4-iodo-5-methyl substituted kresole moiety of "kresoxim-methyl". However, in trifloxystrobin, the longer chain connecting the aromatic bridge with the terminal aromatic ring bearing the trifluoro-methyl group misaligns the two π electron systems. Since no adjustment is made by Phe275, trifloxystrobin may just miss this energetically favorable interaction. By consequently taking advantage of π-π stacking interactions with Phe275 combined with the stabilizing effect of filling cavities, the first pico molar methoxy-acrylate based inhibitors were developed and structurally characterized[47].
4.3. Binding mode of oxazolidine-diones and imidazolinones at QP site
In this group, famoxadone is the best known representative. It was discovered through a chemical scouting program based on the lead compound of 5-methyl-5-phenyl-3-phenyl-amino-2-thioxo-4-oxazolidinone [33, 48, 49] and its new structure that included a chiral toxophore with a more active S(−) enantiomer [49] suggested a novel mode of action [50]. The crystal structure of bc1 bound with famoxadone [51] revealed that famoxadone (Fig. 13) binds in the proximal site of the QP pocket overlapping partially with azoxystrobin (Fig. 14). Unlike azoxystrobin, famoxadone causes ISP-ED to adopt a fixed conformation that resembles that found in the complex with stigmatellin. Thus, the oxazolidine-diones with structures resembling famoxadone, are classified as Pf inhibitors.
Figure 13. Famoxadone binding to cyt b.
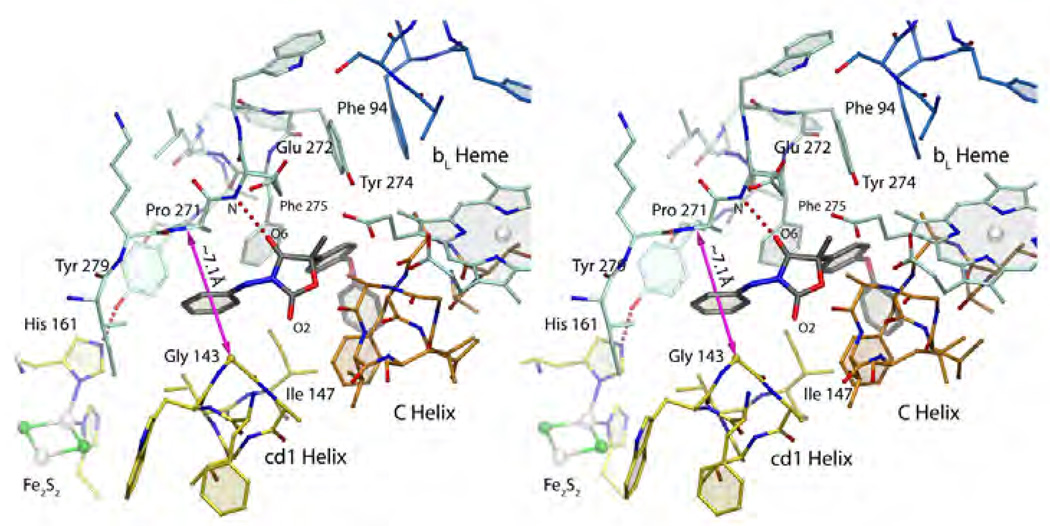
Stereo figure of famoxadone bound to the QP site. The magenta arrow spans the width of the Gly143-Pro271 constriction.
Figure 14. Stereo diagram of the superimposed structures of azoxystrobin (yellow) and famoxadone (blue) using yeast numbering.
Their toxophores share remarkable similarities Phe275 undergoes a conformational change to accommodate famoxadone's large aromatic side chain.
In this class of Pf compounds, the complex structures of famoxadone (1L0L BT, 3L74 GG), jg144 (2FYU BT) and fenamidone (3L75 GG) have been determined. As shown in Figure 14, the flat oxazolidendione ring assumes a central position between the PEWY motif and portions of the cyt b helices B and C. Oxygen atom O6 of famoxadone forms a hydrogen bond of 2.86 Å with the backbone amide of Glu272 whereas the opposing oxygen atom O3 (of the pseudo-symmetric ring) faces directly a fairly featureless portion of the C-helix around residues 140–144. In agreement with experimental studies [52], crystallographic fitting of the S-(−) enantiomer can be accomplished with much greater ease than with R-(+)-famoxadone. Not visible in the depicted chicken bc1 complex is a water molecule that was found in the bovine heart mitochondrial structure. This water molecule bridges the phenylamine atom N1 with Tyr131 (bovine sequence) and carbonyl oxygen atom of Lys269. The importance of this interaction in stabilizing famoxadone may be seen in the loss of 3-fold inhibitory action when N1-methylated famoxadone is used [51, 52]. Whether it is an integral part of the toxophore or rather a second side chain, the aniline moiety of famoxadone inserts into the narrow cleft between Gly143 and Pro271. The aromatic ring plane is essentially normal to a vector connecting the cyt b residues at a distance of about 7.1 Å. The binding of famoxadone is further characterized by a number of Ar-Ar interactions. The highly aromatic side chain of famoxadone and residues of cyt b (in particular, Phe129, Phe275 but also Tyr274 and Phe151) cooperate in forming an extended aromatic network reminiscent of that in solid benzene. Details of these interactions that lead to enhanced binding have been described by Gao et al [51]. The terminal phenoxy moiety of famoxadone appears to provide the least amount of binding energy as it has the fewest contacts with aromatic residues. That it is indeed not needed can be seen in the structures of the pesticides jg144 and fenamidone that exhibit the same binding mode as famoxadone but lack any large additions. However changing the side chain of the toxophore to a bromo-pyridyl moiety causes a loss in inhibitory activity [34].
5. Comparison of QoIs binding modes and mechanisms of pathogen resistance
The effectiveness of QoIs in disease control combined with their generally favorable properties, is the driving force behind the continued design of new quinol oxidation site inhibitors despite the occurrence of resistance. Finding different types of compounds, i.e. inhibitors that are non-methoxy methyl acrylate based promise new modes of actions and a means by which resistance to existing fungicides can be overcome. Crystal structures reveal that the shape of QP pocket is rather elongated, of which "Mother Nature" has taken full advantage by devising both Pf and Pm inhibitors such as stigmatellin and strobilurin. Compounds with an oxazolidendione toxophore, originally thought to be non-competitive QP-site inhibitors [52], were developed by man as alternatives to the classical strobilurins. As crystal structures were solved, it became clear that the 'toxophoric' cores of these competing classes are based on the same principles but differ significantly in their support for side chains resulting in inhibitors with different spatial use of the QP site. For example, by superimposing the structures of famoxadone and azoxystrobin, it becomes clear that the toxophores, despite their chemical differences, fulfill the same function: they are both flat systems that have one carbonyl oxygen atom poised to accept a hydrogen bond from the Glu271 amide backbone. Comparing methoxy methyl acrylates and oxazolidendione ring (Fig. 12b), Zheng pointed out that the latter appears to be just a "tied-back" version of the open toxophore originally found in nature [50]. An illustration of the similarity of the two types of toxophore can be seen in Fig. 14 that shows a superposition of the azoxystrobin and famoxadone bound chicken bc1. Furthermore, key residues that are mutated in nature to impart drug resistance by pathogens are shown.
5.1. Mutations that confer resistance
There are more than a dozen sites [53, 54] within the QP pocket, whose mutations confer at least some resistance to QP site inhibitors and facilitated mechanistic studies of bc1 complexes, but we will limit the discussion to those that greatly diminish the efficacy of current pesticides and drugs. The most frequently found mutation in field isolates is Gly143Ala [55] and to a lesser extent Gly143Ser. The effectiveness of these mutations is substantial as evidenced by a resistance factor of >1000 [56]. This can be understood on the basis of the structure that shows that the methyl side chain of Ala is ideally placed to sterically clash with an aromatic ring of the inhibitor. Since an aromatic side chain is common to all current fungicides regardless of the toxophore a high level of cross resistance [57] is inevitable. Given the importance of this narrow passageway in ubiquinol binding, it is rather remarkable that not all species with the Gly143Ala mutation incur a severe performance penalty in ubiquinol oxidation. In fact, strobilurins not only originated from natural compounds, nature also outfitted the fungi that produce strobilurin (and strobilurin-like compounds) with a Gly143Ala mutation for self-protection. Organisms with the Gly143Ser mutation have a reduced capacity for Oxphors and have compromised reproduction [56].
Changing Phe129 to Leu or Gly137 to Arg are other mutations that reduce the affinity of cyt b for strobilurins [58]. This is in the former case possible because the methoxy acrylate moiety rests on the aromatic ring and a change in geometry eliminates this stabilization. Famoxadone with the cyclic toxophore is less affected by a Phe129Leu mutation but loses favorable aromatic interactions when Phe275 is mutated to Leu. None of the Phe to Leu mutations is known to interfere with ubiquinol oxidation. The more spatially remote residue Gly137, when mutated to Arg, causes resistance but mutants with this change are observed with very low frequency. How this Arg substitution lowers the binding of an inhibitor is more difficult to rationalize except to note, that the very long side chain of Arg can potentially reach and H-bond with Glu272 and Tyr132. A pull on Glu272 might impact the backbone as well and with it possibly weaken the important Glu272 N-H … O=C (toxophore) hydrogen bond.
A comparison of the structures of azoxystrobin and famoxadone is instructive, as it demonstrates that the differential branching off of the respective inhibitor side chains influences the binding mode. In the azoxystrobin structure Phe275 and Ile147 are spatially close to the native position but change significantly in response to a bound famoxadone molecule. These changes, especially the shift in the position of Ile147 (switch residue) can be correlated with a dramatic change in the conformation of the ISP-ED [27] from a state characterized by heightened mobility to firmly docked at cyt b.
The malaria parasite P. falciparum treated with atovaquone rapidly becomes resistant after undergoing various mutations in cyt b with the most prevalent mutation of Tyr268Ser/Cys (Tyr278 in bovine or Tyr279 in yeast). Mutations at this position cause an increase in IC50 values in the range between 1,700 and 10,000 fold for atovaquone [59, 60]. Atovaquone (2-{trans-4-[4’-chlorophenyl]cyclohexyl}-3-hydroxy-1,4-hydroxynaphthoquinone) is a quinone derivative and given its similarity to NQNO and stigmatellin, it could be modeled into cyt b using stigmatellin as a reference [61]. In the modeling study, atovaquone forms a hydrogen bond with His181, the ligand of the iron-sulfur cluster of ISP, and residue Tyr268 appears to form a critical part of the binding pocket for atovaquone. Conceivably, the mutation Tyr268Ser/Cys would result in a loss of a large portion of this interaction, leading to resistance, even though it comes at a cost of the parasite’s overall fitness [62]. Mitochondrial and bacterial bc1 bearing mutations of this conserved tyrosine have elevated reactive oxygen species (ROS) production and reduced activity [63].
5.2. Implications for structure-based QoI design
Much as researchers in structural biology would like to "read off" the next successful drug from coordinates derived from crystal structures (for instance by intuition or virtual screening), in reality the complexity of the whole process in creating a new, successful fungicide/drug involves a large number of factors not even associated with the drug target, such as drug uptake and efflux, metabolic modification and degradation, and the use of alternate pathways. Thus, the straightforward, combinatorial screening against pathogens and the subsequent lead optimization processes has met with more successes, though it is not even fathomable how many variations on strobilurins have been tried by now (conservative estimate of 30,000 in 1998 [45]) and what their performances were as this information hardly ever appears in print. Despite the complexity, improvement in affinity toward target molecules is still fundamentally important and it is possible to suggest a number of restraints on the next generation of molecules that should be tested.
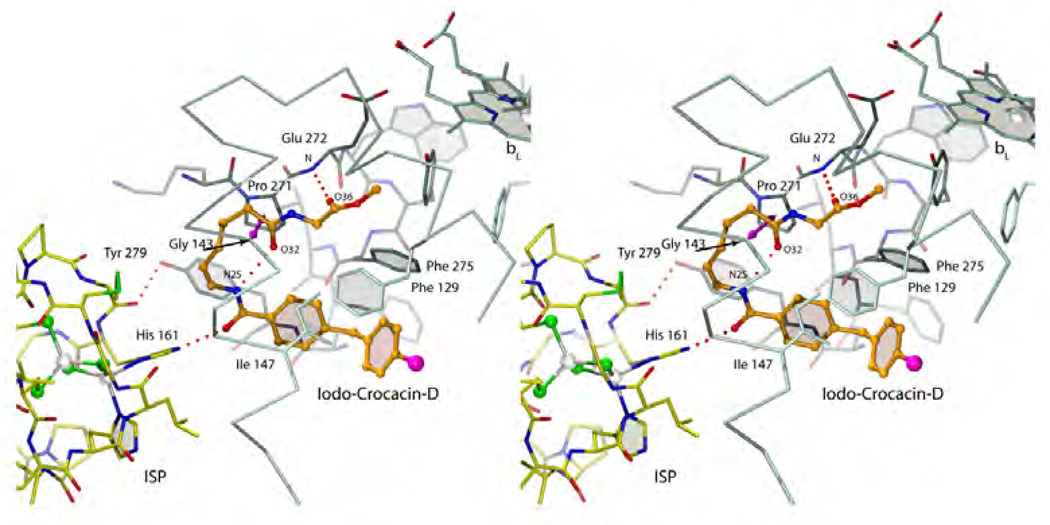
It has become amply clear that the methoxy acrylate backbone in its various incarnations is very useful and amenable to even significant modification ranging from simple CH to N substitutions (oxime) via truncations (carbamate) to additions (dihyro-oxazine). As some additional space can be explored in this pocket, the toxophore only needs to maintain a degree of flatness and its ability to accept a hydrogen bond from the non-mutable Glu272 backbone. The position of the inhibitor side chain branching off the toxophore differs significantly between the methoxy acrylates and the oxazolidendiones. As was shown, only the latter class of compounds is able to recruit ISP-ED to its cyt b docking site sealing off the distal portion of the QP site. A docked ISP-ED offers additional possibilities of interactions especially with His161 as seen in the stigmatellin complex. Both types of commercial inhibitors rely (presumably for affinity reasons) heavily on aromatic fragments or aromatic bridge [45] that intercalate between Gly143 and Pro271. Unfortunately, this turned out to be one of their major weaknesses as pathogens emerged with a Gly143Ala mutation for all practical purposes abolishing the binding and toxicity of these fungicides. Future generations of fungicides in the QP pocket may start with any of the established (and still modifiable) toxophores but should then avoid the Gly143-Pro271 constriction. Furthermore, they should trigger the capture of ISP-ED for additional protection and polar interactions with non-mutable residues. For instance His161 is an integral part of the Fe2S2 complex involved in the deprotonation [64–66] of the substrate and is certainly a determinant of the redox potential of ISP. It is inconceivable that this residue will undergo mutation and any inhibitor interacting with this residue can be expected to have an increase lifespan as fungicide. As an example of a compound that comes close to exploiting all the features of the QP site is a product derived from the (+)-crocacins [67] - a natural fungicide from C. crocatus. The synthetic analogue of crocacin binds to the QP site (Fig. 15, chicken bc1 complex, 3CWB [68]) using a minimalistic glycine ester in place of the "methoxy acrylate" toxophore but still manages to form the characteristic Glu272 NH…O(36) hydrogen bond. The linker traverses the narrow Gly-Pro passageway but may incur less interference by a potential Gly143Ala mutation than compounds with aromatic bridges. This was however achieved by introducing an internal hydrogen bond whose perturbation by a Gly143Ala mutation may weaken the inhibitor's binding again. The terminal moiety accomplishes two things: its large aromatic end helps push Ile147 switch to the "on" position that leads to ISP-ED docking and secondly, the amide oxygen atom (O24) accepts a hydrogen bond from His161 in much the same way stigmatellin does. While this novel compound was clearly successful as a bc1 inhibitor, its overall biological efficacy was deemed not optimizable for commercial purposes [68, 69].
Figure 15. Stereo diagram of a crocasin-D analogue bound to cyt b.
6. Inhibition of quinone reduction site
6.1. Binding mode of QiI inhibitors
Quinone reduction site inhibitors are increasingly of interest as pathogen resistance to QP site inhibitors develops. The QN site is characterized by one prosthetic group, namely the bH heme, and is passive compared to the quinol oxidation site. At the QN site, the quinone substrate only awaits the sequential arrival of two electrons from the bH heme and two protons from the negative side of the membrane. While the QN site binds inhibitors that resemble quinone like tridecyl stigmatellin [70, 71] and NQNO, the majority of the well-known QN site inhibitors that have been used for countless mechanistic investigations show only remote resemblance to 1,4-benzoquinone.
Figure 5 compares the structure of the natural substrate ubiquinone-10 10 with that of the natural antifungal agents, antimycin 17, funiculosin 20 [72, 73], ilicicolin 21 [74] and ascochlorin 22 [75, 76] and the synthetic compounds diuron 23 [77], cyazofamid 24 [78] and amisulbrom 25 (see for instance [79]). Crystal structure information regarding tridecyl stigmatellin 19, NQNO 18, ascochlorin 22 and antimycin 17 are available [80]. Disregarding for now the possibility of accidentally inhibiting plant mitochondrial bc1, for the design of fungicides that cannot bind to and thus inhibit b6f which is critically important in photosynthesis, it is advantageous to target the QN site of bc1 as the plastoquinone reduction site in b6f carries an additional and covalently bound heme group (heme-X) exactly in the place where the quinone substrate in a superimposed bc1 molecule binds [81, 82]. Thus, the QN sites of bc1 and b6f can hardly be more different. The observation that tridecyl stigmatellin binds near a QN center has so far only been made in b6f of Mastigocladus laminosus [70].
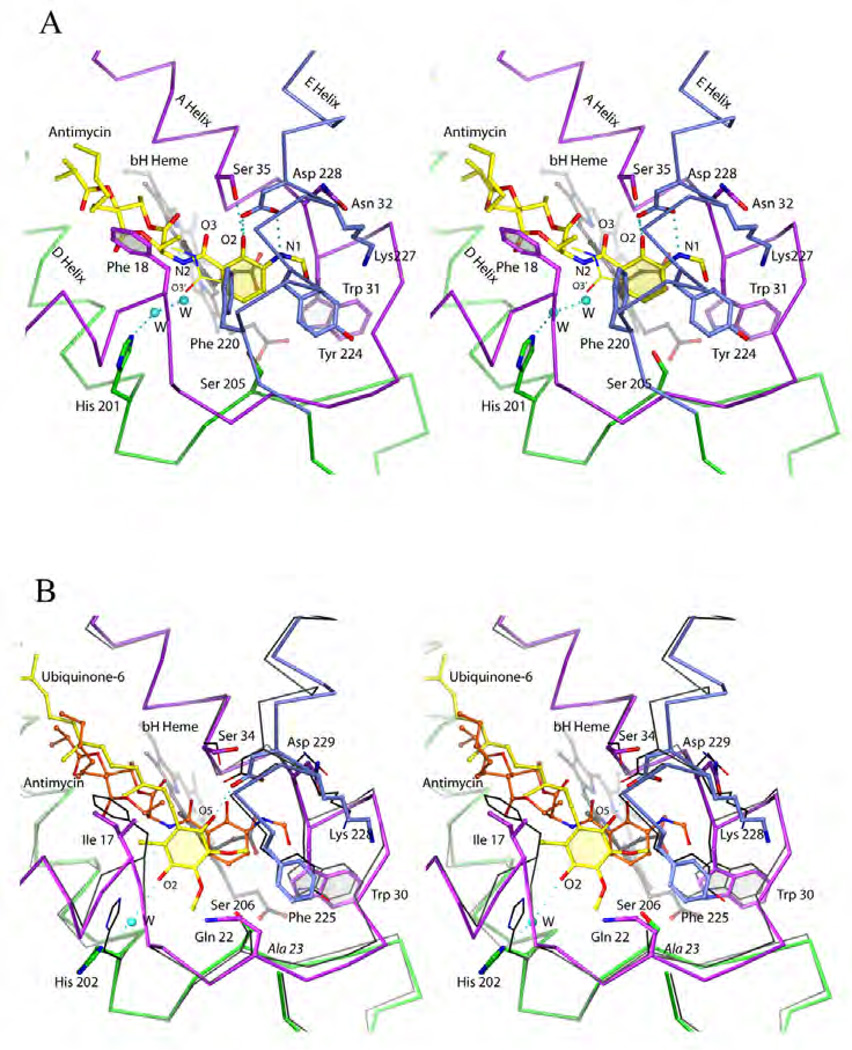
It will be instructive to compare structural determinants that are involved in binding of natural ubiquinone and the natural inhibitor antimycin for which high resolution data are available (see Table 1). Antimycin [83] was isolated from Streptomyces as an antifungal agent. By spectroscopic methods it was discovered that it binds to the QN site and was instrumental in unraveling the Q-cycle hypothesis. In the first crystal structure of bc1 antimycin was used to locate the QN site [84], which is located at the matrix side in the first helical bundle flanked by helices A, D and E and allows direct access to the bH heme. In recent times antimycin has been extensively used to investigate the mechanism of ROS production by bc1, which increases dramatically when the electron flow through bH is inhibited by a potent inhibitor like antimycin. The molecular structure [85] of antimycin (A1) can be divided into three moieties (Fig. 8d), namely the toxophore (3-formyl amino salicylic acid or 3-FASA [86]), a central dilactone and terminal alkyl and acyl chains of different lengths that may indeed differ among the various subtypes (for a review see [87]). The current view based on three crystal structures (Fig. 16a) [88–90] describes the 3-FASA toxophore to be bound by at least three hydrogen bonds to the highly conserved residues Ser35 and Asp228 inside an amphiphilic cavity of 7 × 10 × 13 Å3. Residues that further stabilize the aromatic system are Phe220, Tyr224, Trp31 and to some extend Lys227 and a propionate chain of heme bH. The lack of formation of an intramolecular hydrogen-bond between phenolic OH and amide carbonyl groups resulted in a remarkable loss of the activity by four orders of magnitude, indicating that this hydrogen-bond is essential for the inhibition [91]. However, this intramolecular H-bond was destroyed upon association of antimycin with the protein. The amide linkage to the dilactone was described as in syn conformation in the 2003 structure but has since been found to be better described in its anti form [89, 90]. The alkyl/acyl substituted dilactone does not engage in specific interactions and can in fact be replaced entirely by alkyl chains albeit with some loss of inhibitory action [86]. An overlay with the high resolution crystal structure of yeast bc1 with ubiquinone (Fig. 16b) in the QN site [92] reveals that the natural substrate is engaged in only one hydrogen bond with Asp229 and none with Ser34. One other hydrogen bond is accepted from water as is the case of antimycin. That the position of the benzophenone group is off-set from antimycin's salicylic acid may hint at the differences in purpose: as an inhibitor, antimycin needs to bind as tightly as possible to the site whereas ubiquinone has to dissociate after reduction. However, the shape of the dilactone including its (S)-methyl butanoic acyl chain coincides nicely with the isoprenyl chain of ubiquinone suggesting that their interactions with the hydrophobic cavity are equivalent. Separate superpositions using the other two structurally known inhibitors, NQNO [88] and ascochlorin [93] reveal that the former inserts ~2Å deeper into the QN site than antimycin whereas ascochlorin stays closer to the position of ubiquinone.
Figure 16. Binding of ubiquinol and antimycin to the QN site.
(A) Stereo diagram of antimycin bound to the QN site. The available crystal structures differ only in the conformation of the amide linkage (indicated by O3 vs. O3'). (B) Stereo diagram of quinone (yellow) superimposed onto antimycin (orange, cyt b in black). The secondary structure is color-coded: magenta, green or blue for a/A-helix, D-helix and E-Helix respectively.
6.2. Comparison of QiI binding and mechanisms of pathogen resistance
Antimycin [94, 95], diuron [96], funiculosin [97] and HQNO resistance has been investigated in some details. Resistance can be built up by two mechanisms involving a relatively small number of amino acids: elimination (or redirection) of stabilizing hydrogen bonds or steric exclusion. For the former mechanism, typical examples include polar to non-polar mutations (Ser35Ile, Asp228Ala, Lys227Met/Ile) or geometry alterations (Asp228Asn/His/Glu). Considering that conserved residues Ser35, Asp228 and Lys227 are implicated in proton transport, it is surprising that some of the mutations are tolerated.
In contrast, no mutations at residue His201 have been observed in any QiI resistant mutants, which may indicate that His201 does not significantly contribute to the inhibitor binding or it fulfils an indispensible function [54] in quinone reduction/protonation. As observed in the crystal structure with ubiquinone bound at the QN site the pathway is flat, permitting access only to planar molecules or planar portions of molecules such as the substrate ubiquionone, antimycin, and NQNO. The bH heme and opposing aromatic residues (Phe220 and Phe18) seem to be important for orienting the substrate molecule with their edges contacting the aromatic ring of the substrate. The ubiquinone molecule inserted into the site interacts, mediated by a water molecule W, with His201 at one side and with Ser34 and Asp228 at the other [80].
Cases of steric exclusions frequently involving not only Phe220 but also Trp31, Gly38, Ser205 and Tyr224 are understandable, given the binding characteristics of the inhibitors and roles of these residues in shaping the binding site, and explain the considerable cross resistance for antimycin and HQNO (and by extension for NQNO).
6.3. Implications for new QiIs design
Given the available structural information of several native and inhibitor complexes, it becomes clear that the QN site offers a variety of exploitable polar, aromatic and non-polar interactions for drug binding. Unfortunately, the flexibility of the QN site in being able to tolerate even drastic changes while maintaining function puts it in the 'high risk' category for drug resistance development [40]. To increase the lifespan of an inhibitor, a new design should reverse the preference of existing N inhibitors in binding towards residues Ser34 and Asp228 and exploit residues (like His201) that so far have shown no propensity for change, backbone carbonyl functions (Ala17) and/or possibly one of the propionate chains (A-branch) of the bH heme. Furthermore, size may matter i.e. smaller inhibitors may have an edge over large compounds like antimycin which could be affected by a simple Gly38Val mutation. Although no structural data is available that would show how the new pesticides cyazofamid and amisulbrom interact with the QN site, their small sizes and novel structural features might delay the onset of pathogen resistance against them.
7. Other mechanisms of acquired resistance against bc1 inhibitors
Conceivably, drugs/fungicides aimed at bc1 must reach their target in order to be effective. Resistance can arise if drugs fail to overcome barriers that reduce their cellular accumulation. While animals and plants are not intended targets of the fungicides designed to inhibit cyt bc1, we and others have shown that mammalian cyt bc1 complexes such as those from bovine and chicken mitochondria are indeed inhibited by these compounds. Clearly, different mechanisms exist that protect animals and plants but not pathogenic fungi. Alarmingly, reports on resistant field strains that do not bear mutations in bc1 have been accumulating, indicating the emergence of other mechanisms of resistance [98, 99].
7.1. Resistance by alternate pathways
Although less efficient in energy metabolism, many prokaryotic organisms are capable of bypassing cyt bc1 under certain growth conditions. Such a mechanism can be activated when bc1 is inhibited. Alternate ubiquinol oxidases (AOX) catalyze the reaction of transferring electron from quinol directly to oxygen and in this way maintain Oxphos by bypassing cyt bc1 and cyt c oxidase (Fig. 1). However this comes at the price of reduced proton pumping activity since AOX does not pump protons across the membrane and two complexes that did are shut down. In general, it has been observed that organism using AOX cannot support ATP demanding activities such as spore germination of fungus [100]. Therefore, it is believed that the expression of AOX is not going to impact the efficacy of bc1 inhibitors. Contrary to this belief, it was reported that the mixture of QoIs with salicylhydroxamic acid (SHAM), an inhibitor of the alternative oxidase, exerted synergistic effects in inhibiting the spore germination of the fungus V. inaequalis [99]. Induction of alternative oxidase (AOX) by trifloxystrobin was observed in mycelium cells at the molecular level for the sensitive but not the resistant isolate. Thus, the bc1 and the alternative respiration pathways are assumed to play different roles, depending on the developmental stage of the fungus [99].
8.2. Resistance by reduced uptake and increased efflux
Over expression of efflux pumps in bacteria, fungi and cancer cells is known to be one of the major mechanisms for drug resistance. For example, a broad spectrum of resistance to the treatment with azoles achieved by up-regulating the efflux ABC transporter cdr1 has been known for the wild type pathogenic fungi C. albicans [101]. Furthermore, a laboratory strain of S. cerevisiae was made highly sensitive to the bc1 inhibitor atovaquone when nine plasma membrane ABC transporters were genetically deleted [102]. In agriculture, however, there are few clear-cut cases of plant pathogens showing resistance to bc1 inhibitors due to active transporters. An efflux transporter mediated mechanism of resistance to bc1 fungicides in field isolates of the wheat pathogen P. triticirepentis was reported; the resistance was acquired after repeated applications of low doses of strobilurin fungicides [103]. The involvement of efflux pumps in fungicide resistance was demonstrated by using inhibitors of these membrane transporters such as the hydroxyflavone derivative 2-(4-ethoxy-phenyl)-chromen-4-one in combination with fungicides: such inhibitors restored normal sensitivity levels of resistant isolates, preventing infection of wheat leaves [103]. The first gene to be identified as that of an efflux transporter imparting QoI insensitivity to a plant pathogen was MgMfs1, a major facilitator superfamily (MFS) transporter gene of M. graminicola [104]. However, most field strobilurin resistant isolates of M. graminicola not only overexpress MgMfs1 but also contain the Gly143Ala substitution in bc1, suggesting that MgMfs1 plays only a supporting role.
8. Overcoming drug resistance
8.1. Challenges in structure-based development of novel drugs
Structure-based ab initio design of novel drugs has proven to be challenge. Historically, structure-based drug design [105] has a less than stellar record of finding new drugs or new lead compounds. A number of factors hamper virtual screening efforts starting with the fact that the crystal structure of the target may be incomplete or incorrect. Even a model that is considered good by today's standards is likely incomplete with respect to bound water molecule, ions of light elements like Na+, Mg2+, Cl− and bound small molecules. While it is sensible and necessary to remove water to make way for a ligand to bind, care should be taken not to remove water that is essential to maintain protein structure. The problem of having to deal with molecules that are present but incompletely seen by x-ray crystallography is greatly compounded in the case of membrane proteins. Here the presence of lipid and detergent molecules is often only hinted at in the electron density and remains unaccounted for by the crystallographer due to the difficulties in modeling partially recognized or recognizable molecules. This may inadvertently cause the identification of lead compounds for sites that are not available in reality.
Another problem in virtual screening approaches arises when the receptor is flexible. As we have seen, the quinol oxidation site has a very high degree of plasticity evidenced not only in changes of side chain rotamers (for instance Glu272, Phe129), and shifts in backbone positions (for instance Pro271 and the entire cd1/cd2 helix) but also in dramatic domain movements (ISP-ED). In a recent review on structure-based drug design, Ivetac and McCammon [106] pointed out that each crystal structure of a complex is only a static snapshot of a conformation that may not even be the one with the highest binding affinity. To improve on the "rigid body-protein flexible-ligand" model, modern programs and protocols such as the Dynamic Pharmacophore Model [107, 108] and the Relaxed Complex Scheme [109] incorporate molecular dynamics simulations to diversify the receptor conformations. These methods hold the promise to finding novel leads provided that the MD simulations needed in the process are carefully planned and properly deal with the interactions between cyt b and ISP-ED. In contrast, the QN site poses virtually no discernable challenge with respect to any plasticity beyond changes in side chain conformation.
Despite all the challenges, structural information has been routinely used as an indispensible tool for understanding the precise mechanism of binding and as a guide for lead improvement. In the field of drug design for bc1, a recent example where a combination of traditional and modern ideas named pharmacophore-like fragment virtual screening (PFVS) resulted in the first pico molar methoxyacrylate-based inhibitors was recently introduced by the Yang group[47, 110].
8.2. Combining protein targets
The use of chloroquine in conventional treatment of malaria often leads to development of resistance. The role of quine-type drugs is to prevent the formation of hemozoin crystals in the digestive vacuoles of the parasites, leading to toxic levels of heme molecules. The wide spread use of chloroquine led to the emergence of malaria strains that are resistant to chloroquine such as P. falciparum. Atovaquone is a hydroxynapthoquinone that targets specifically the bc1 complex, causing the collapse of the mitochondrial membrane potential, the disruption of pyrimidine biosynthesis and the subsequent death of the parasite. It is currently most widely applied in the treatment of opportunistic infections in immuno-suppressed patients and against the chloroquine-resistant malaria strain P. falciparum.
It has also been shown that the use of high doses of atovaquone against malaria is a guaranteed recipe for developing resistance. By combining atovaquone with proguanil in a ratio of 59 to 24 (malarone) or with other antibiotics such as tetracycline, it has been shown that such an approach prolongs the period of developing resistance [111]. In agriculture, it is a common practice switching among fungicides that have different modes of action: from ergosterol biosynthesis inhibitors such as triazoles to respiratory chain inhibitors as reviewed here.
8.3 Combinatorial use of chemicals targeting multiple sites
In perfect analogy to the structurally similar quinol/quinone pair, it should be possible to design "double kill" inhibitors that block both sites. Since no single point mutation can reverse the inhibition of both sites by the same molecule, the occurrence of resistance may be substantially delayed. While examples of "double kill-" compounds already exist (NQNO, ascochlorin), finding new ones may still be an insurmountable task by rational drug design due to the increase in complexity given the structurally and chemically different sites. An alternative approach would be to develop new “double-kill” compounds by making use of existing QoIs and QiIs linking them chemically. As shown in Figure 17, the distance between tails of QP site inhibitor myxothiazol and QN site inhibitor antimycin is ~19 Å or about the distance of decaethylenglycol linker, making such an approach feasible.
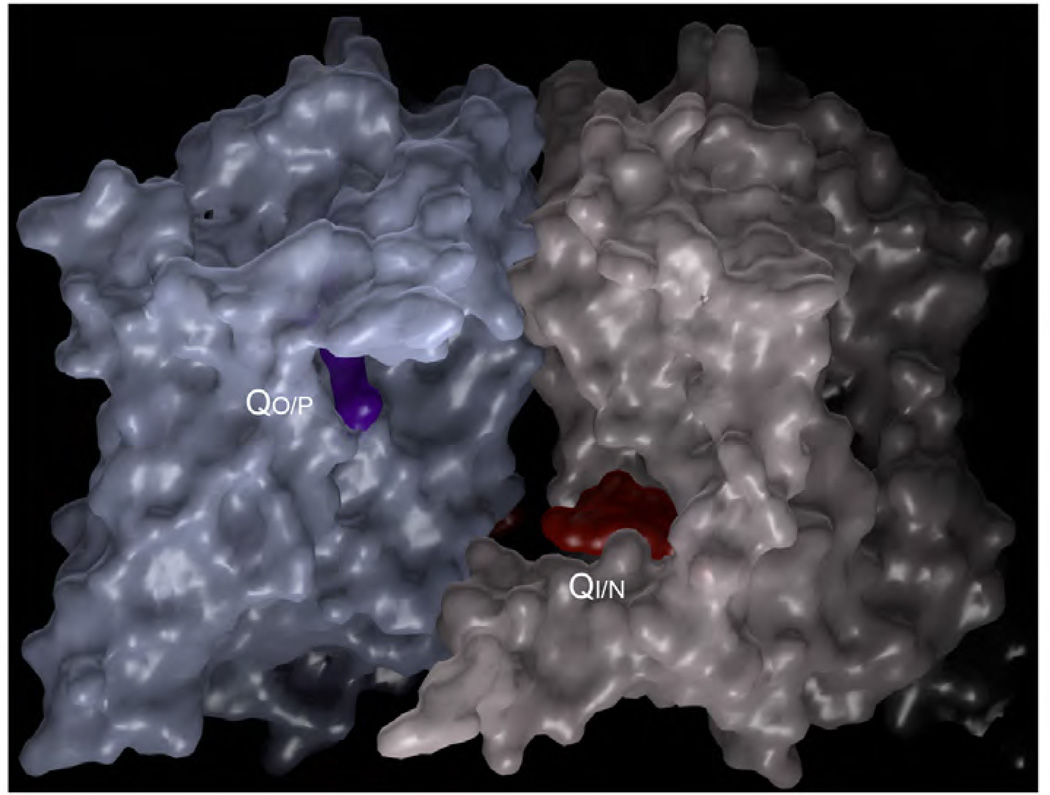
Figure 17. Dimer of bc1 showing the molecular surface as well as inhibitors bound at the QP and QN site and their spatial relationship.
The inhibitor bound at the QP site is stigmatellin and that occupies the QN site is antimycin A.
In the end the best way of dealing with the pathogen resistance may be by adhering to a rigorous disease-control protocol that includes applications of fungicides/drugs but alternating between QP and QN inhibitors, which might be called 'inhibitor ping-pong'. As resistance to one inhibitor builds up, the pathogen has time to revert inhibition to the currently uninhibited site from the previous round. Success with this method however may mean international coordination to minimize the occurrence of strains that are permanently resistant to both types of inhibitors.
Abbreviations
- cyt
cytochrome
- cyt bc1
cytochrome bc1 complex
- bL
low potential heme of cytochrome b
- bH
high potential heme of cytochrome b
- ET
electron transfer
- FRAC
fungicides resistance action committee
- ISP
iron-sulfur protein subunit of bc1 complex
- ISP-ED
ISP extrinsic domain
- Fe2S2
iron-sulfur-cluster
- Oxphos
oxidative phosphorylation
- QiI
Qi site inhibitor
- QoI
Qo site inhibitor
- QP
quinol oxidation site at the positive side of the membrane
- QN
quinone reduction site at the negative side of the membrane
- TM
transmembrane
- MD
molecular dynamics.
References
- 1.Nietzsche FW. Nietzsche's Werke. Leipzig: C.G. Naumann; 1899. [Google Scholar]
- 2.Morton V, Staub T. A Short History of Fungicides. The American Phytopathological Society. 2008 [Google Scholar]
- 3.Leadbeater A. Resistance Risk to QoI Fungicides and Anti-Resistance Strategies. Fungicide Resistance in Crop Protection Risk and Management: CABI. 2008:141–152. [Google Scholar]
- 4.Mitchell P. Possible molecular mechanisms of the protonmotive function of cytochrome systems. Journal of Theoretical Biology. 1976;62:327–367. doi: 10.1016/0022-5193(76)90124-7. [DOI] [PubMed] [Google Scholar]
- 5.Trumpower BL. The protonmotive Q cycle. Energy transduction by coupling of proton translocation to electron transfer by the cytochrome bc1 complex. Journal of Biological Chemistry. 1990;265:11409–11412. [PubMed] [Google Scholar]
- 6.Ozawa T, Tanaka M, Shimomura Y. Crystallization of the middle part of the mitochondrial electron transfer chain: cytochrome bc1-cytochrome c complex. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5084–5086. doi: 10.1073/pnas.77.9.5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yue WH, Zou YP, Yu L, Yu CA. Crystallization of Mitochondrial Ubiquinol-Cytochrome c Reductase. Biochemistry. 1991;30:2303–2306. doi: 10.1021/bi00223a002. [DOI] [PubMed] [Google Scholar]
- 8.Kubota T, Kawamoto M, Fukuyama K, Shinzawa-Itoh K, Yoshikawa S, Matsubara H. Crystallization and preliminary X-ray crystallographic studies of bovine heart mitochondrial cytochrome bc1 complex. J Mol Biol. 1991 Sep 20;221(2):379–382. doi: 10.1016/0022-2836(91)80059-4. [DOI] [PubMed] [Google Scholar]
- 9.Berry EA, Huang L, Earnest T, Jap BK. X-ray diffraction by crystals of beef heart ubiquinol: cytochrome c oxidoreductase. Journal of Molecular Biology. 1992;224:1161–1166. doi: 10.1016/0022-2836(92)90476-z. [DOI] [PubMed] [Google Scholar]
- 10.Lee JW, Chan M, Law TV, Kwon HJ, Jap BK. Preliminary cryocrystallographic study of the mitochondrial cytochrome bc1 Complex:improved crystallization and flash-cooling of a large membrane protein. Journal of Molecular Biology. 1995;252:15–19. doi: 10.1006/jmbi.1994.0470. [DOI] [PubMed] [Google Scholar]
- 11.Ljungdahl PO, Pennoyer JD, Robertson DE, Trumpower BL. Purification of highly active cytochrome bc1 complexes from phylogenetically diverse species by a single chromatophaphic procedure. Biochimica et Biophysica Acta. 1987;891:227–241. doi: 10.1016/0005-2728(87)90218-0. [DOI] [PubMed] [Google Scholar]
- 12.Yu CA, Xia JZ, Kachurin AM, Yu L, Xia D, Kim H, et al. Crystallization and preliminary structure of beef heart mitochondrial cytochrome-bc1 complex. Biochim Biophys Acta. 1996;1275:47–53. doi: 10.1016/0005-2728(96)00049-7. [DOI] [PubMed] [Google Scholar]
- 13.Xia D, Yu CA, Kim H, Xia JZ, Kachurin AM, Zhang L, et al. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z, Huang L, Shulmeister VM, Chi YI, Kim KK, Hung LW, et al. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 15.Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, et al. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex [see comments] Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 16.Hunte C, Koepke J, Lange C, Rossmanith T, Michel H. Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 2000;15:669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 17.Kurisu G, Zhang H, Smith JL, Cramer WA. Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science. 2003;302:1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- 18.Stroebel D, Choquet Y, Popot JL, Picot D. An atypical haem in the cytochrome b(6)f complex. Nature. 2003;426:413–418. doi: 10.1038/nature02155. [DOI] [PubMed] [Google Scholar]
- 19.Crofts AR, Wang Z. How rapid are the internal reactions of the ubiquinol:cytochrome c2 oxidoreductase? Photosynthesis Research. 1989;22:69–87. doi: 10.1007/BF00114768. [DOI] [PubMed] [Google Scholar]
- 20.Kleinschroth T, Castellani M, Trinh CH, Morgner N, Brutschy B, Ludwig B, et al. X-ray structure of the dimeric cytochrome bc(1) complex from the soil bacterium Paracoccus denitrificans at 2.7-A resolution. Biochim Biophys Acta. 2011 Dec;1807(12):1606–1615. doi: 10.1016/j.bbabio.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Esser L, Elberry M, Zhou F, Yu CA, Yu L, Xia D. Inhibitor complexed structures of the cytochrome bc1 from the photosynthetic bacterium Rhodobacter sphaeroides at 2.40 Å resolution. J Biol Chem. 2008;283:2846–2857. doi: 10.1074/jbc.M708608200. [DOI] [PubMed] [Google Scholar]
- 22.Berry EA, Huang L, Saechao LK, Pon NG, Valkova-Valchanova M, Daldal F. X-ray structure of Rhodobacter capsulatus cytochrome bc1: comparison with its mitochondrial and chloroplast counterparts. Photosynthesis Research. 2004;81:251–275. doi: 10.1023/B:PRES.0000036888.18223.0e. [DOI] [PubMed] [Google Scholar]
- 23.Chance B, Wilson DF, Dutton PL, Erecinska M. Energy-coupling mechanisms in mitochondria: kinetic, spectroscopic, and thermodynamic properties of an energy-transducing form of cytochrome b. Proc Natl Acad Sci U S A. 1970 Aug;66(4):1175–1182. doi: 10.1073/pnas.66.4.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wikstrom MK, Berden JA. Oxidoreduction of cytochrome b in the presence of antimycin. Biochim Biophys Acta. 1972;283:403–420. doi: 10.1016/0005-2728(72)90258-7. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Huang L, Shulmeister VM, Chi Y-I, Kim KK, Hung L-W, et al. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392(6677):677–684. doi: 10.1038/33612. [10.1038/33612] [DOI] [PubMed] [Google Scholar]
- 26.Kim H, Xia D, Yu CA, Xia JZ, Kachurin AM, Zhang L, et al. Inhibitor binding changes domain mobility in the iron-sulfur protein of the mitochondrial bc1 complex from bovine heart. Proceedings of National Academy of Sciences U S A. 1998;95:8026–8033. doi: 10.1073/pnas.95.14.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esser L, Gong X, Yang S, Yu L, Yu CA, Xia D. Surface-modulated motion switch: capture and release of iron-sulfur protein in the cytochrome bc1 complex. Proc Natl Acad Sci U S A. 2006 Aug 29;103(35):13045–13050. doi: 10.1073/pnas.0601149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esser L, Quinn B, Li Y, Zhang M, Elberry M, Yu L, et al. Crystallographic studies of quinol oxidation site inhibitors: A modified classification of inhibitors for the cytochrome bc1 complex. Journal of Molecular Biology. 2004;341:281–302. doi: 10.1016/j.jmb.2004.05.065. [DOI] [PubMed] [Google Scholar]
- 29.Tian H, Yu L, Mather MW, Yu CA. Flexibility of the neck region of the rieske iron-sulfur protein is functionally important in the cytochrome bc1 complex. Journal of Biological Chemistry. 1998;273:27953–27959. doi: 10.1074/jbc.273.43.27953. [DOI] [PubMed] [Google Scholar]
- 30.Tian H, White S, Yu L, Yu CA. Evidence for the head domain movement of the Rieske iron-sulfur protein in electron transfer reaction of the cytochrome bc1 complex. Journal of Biological Chemistry. 1999;274:7146–7152. doi: 10.1074/jbc.274.11.7146. [DOI] [PubMed] [Google Scholar]
- 31.Darrouzet E, Moser CC, Dutton PL, Daldal F. Large scale domain movement in cytochrome bc(1): a new device for electron transfer in proteins. Trends in Biochemical Sciences. 2001;26:445–451. doi: 10.1016/s0968-0004(01)01897-7. [DOI] [PubMed] [Google Scholar]
- 32.Link TA, Haase U, Brandt U, von Jagow G. What information do inhibitors provide about the structure of the hydroquinone oxidation site of ubihydroquinone: cytochrome c oxidoreductase? Journal of Bioenergetics and Biomembrane. 1993;25:221–232. doi: 10.1007/BF00762584. [DOI] [PubMed] [Google Scholar]
- 33.Jordan DB, Livingston RS, Bisaha JJ, Duncan KE, Pember SO, Picollelli MA, et al. Mode of action of famoxadone. Pesticide Science. 1999 Feb;55(2):105–118. [Google Scholar]
- 34.Wang F, Li H, Wang L, Yang WC, Wu JW, Yang GF. Design, syntheses, and kinetic evaluation of 3-(phenylamino)oxazolidine-2,4-diones as potent cytochrome bc complex inhibitors. Bioorg Med Chem. 2011 Aug 1;19(15):4608–4615. doi: 10.1016/j.bmc.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Berry EA, Huang LS. Conformationally linked interaction in the cytochrome bc(1) complex between inhibitors of the Q(o) site and the Rieske iron-sulfur protein. Biochim Biophys Acta. 2011 Oct;1807(10):1349–1363. doi: 10.1016/j.bbabio.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita E, Zhang H, Cramer WA. Structure of the Cytochrome b6f Complex: Quinone Analogue Inhibitors as Ligands of Heme cn. Journal of Molecular Biology. 2007;370(1):39–52. doi: 10.1016/j.jmb.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ypema HL, Gold RE. Modification of a naturally occurring compound to produce a new fungicide. Plant Disease. 1999 Jan;83(1):4–19. doi: 10.1094/PDIS.1999.83.1.4. [DOI] [PubMed] [Google Scholar]
- 38.El Hage S, Ane M, Stigliani JL, Marjorie M, Vial H, Baziard-Mouysset G, et al. Synthesis and antimalarial activity of new atovaquone derivatives. Eur J Med Chem. 2009 Nov;44(11):4778–4782. doi: 10.1016/j.ejmech.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 39.Hughes LM, Covian R, Gribble GW, Trumpower BL. Probing binding determinants in center P of the cytochrome bc(1) complex using novel hydroxy-naphthoquinones. Biochim Biophys Acta. 2010 Jan;1797(1):38–43. doi: 10.1016/j.bbabio.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leadbeater A. Fungicide Resistance Action Committee. Basel, Switzerland: 2011. [updated 2005]. Available from: www.frac.info. [Google Scholar]
- 41.Thierbach G, Kunze B, Reichenbach H, Hofle G. On Antibiotics from Gliding Bacteria .20. The Mode of Action of Stigmatellin, a New Inhibitor of the Cytochrome-B-C1 Segment of the Respiratory-Chain. Biochimica Et Biophysica Acta. 1984;765(2):227–235. [Google Scholar]
- 42.Ohnishi T, Brandt U, Vonjagow G. Studies on the Effect of Stigmatellin Derivatives on Cytochrome-B and the Rieske Iron-Sulfur Cluster of Cytochrome-C Reductase from Bovine Heart-Mitochondria. European Journal of Biochemistry. 1988 Sep 15;176(2):385–389. doi: 10.1111/j.1432-1033.1988.tb14293.x. [DOI] [PubMed] [Google Scholar]
- 43.Palsdottir H, Lojero CG, Trumpower BL, Hunte C. Structure of the yeast cytochrome bc1 complex with a hydroxyquinone anion Qo site inhibitor bound. J Biol Chem. 2003 Aug 15;278(33):31303–31311. doi: 10.1074/jbc.M302195200. [DOI] [PubMed] [Google Scholar]
- 44.Becker WF, von Jagow G, Anke T, Steglich W. Oudemansin, strobilurin A, strobilurin B and myxothizaol: new inhibitors of the bc1 segment of the respiratory chain with an E-b-methoxyacrylate system as common structural element. FEBS Letters. 1981;132:329–333. doi: 10.1016/0014-5793(81)81190-8. [DOI] [PubMed] [Google Scholar]
- 45.Sauter H, Steglich W, Anke T. Strobilurins: Evolution of a new class of active substances. Angewandte Chemie-International Edition. 1999;38(10):1329–1349. doi: 10.1002/(SICI)1521-3773(19990517)38:10<1328::AID-ANIE1328>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 46.George WO, Hassid DV, Maddams WF. Conformations of Some Alpha-Beta-Unsaturated Carbonyl-Compounds .3. Infrared Solution Spectra of Methyl, [H-2-3]Methyl, Ethyl, and [H-2-5]Ethyl Acrylates and Trans-Crotonates. Journal of the Chemical Society-Perkin Transactions 2. 1972;(4):400-&. [Google Scholar]
- 47.Hao GF, Wang F, Li H, Zhu XL, Yang WC, Huang LS, et al. Computational discovery of picomolar Q(o) site inhibitors of cytochrome bc1 complex. J Am Chem Soc. 2012 Jul 11;134(27):11168–11176. doi: 10.1021/ja3001908. [DOI] [PubMed] [Google Scholar]
- 48.Sternberg JA, Geffken D, Adams JB, Jr, Jordan DB, Poestages R, Sternberg CG, Campbell CL, Moberg WK, Livingston RS. Oxazolidinones: a new class of agricultural fungicides. In: Baker DR, Feynes JG, Basarab GS, Hunt DA, editors. Synthesis and Chemistry of Agrochemicals. Washington DC: American Chemical Society; 1998. pp. 216–227. [Google Scholar]
- 49.Sternberg JA, Geffken D, Adams JB, Jr, Postages R, Sternberg CG, Campbell CL, et al. Famoxadone: the discovery and optimisation of a new agricultural fungicide. Pest Manag Sci. 2001 Feb;57(2):143–152. doi: 10.1002/1526-4998(200102)57:2<143::AID-PS282>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 50.Zheng YJ. Molecular basis for the enantioselective binding of a novel class of cytochrome bc1 complex inhibitors. J Mol Graph Model. 2006 Sep;25(1):71–76. doi: 10.1016/j.jmgm.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Gao X, Wen X, Yu C, Esser L, Tsao S, Quinn B, et al. The crystal structure of mitochondrial cytochrome bc1 in complex with famoxadone: the role of aromatic-aromatic interaction in inhibition. Biochemistry. 2002 Oct 1;41(39):11692–11702. doi: 10.1021/bi026252p. [DOI] [PubMed] [Google Scholar]
- 52.Zheng YJ, Shapiro R, Marshall WJ, Jordan DB. Synthesis and structural analysis of the active enantiomer of famoxadone, a potent inhibitor of cytochrome bc1. Bioorg Med Chem Lett. 2000 May 15;10(10):1059–1062. doi: 10.1016/s0960-894x(00)00164-5. [DOI] [PubMed] [Google Scholar]
- 53.Esposti MD, De Vries S, Crimi M, Ghelli A, Patarnello T, Meyer A. Mitochondrial cytochrome b: evolution and structure of the protein. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1993;1143(3):243–271. doi: 10.1016/0005-2728(93)90197-n. [DOI] [PubMed] [Google Scholar]
- 54.Brasseur G, Saribas AS, Daldal F. A compilation of mutations located in the cytochrome b subunit of the bacterial and mitochondrial bc(1) complex. Biochimica Et Biophysica Acta-Bioenergetics. 1996 Jul 18;1275(1–2):61–69. doi: 10.1016/0005-2728(96)00051-5. [DOI] [PubMed] [Google Scholar]
- 55.Samuel S, Papayiannis LC, Leroch M, Veloukas T, Hahn M, Karaoglanidis GS. Evaluation of the incidence of the G143A mutation and cytb intron presence in the cytochrome bc-1 gene conferring QoI resistance in Botrytis cinerea populations from several hosts. Pest Manag Sci. 2011 Aug;67(8):1029–1036. doi: 10.1002/ps.2226. [DOI] [PubMed] [Google Scholar]
- 56.Avila-Adame C, Koller W. Characterization of spontaneous mutants of Magnaporthe grisea expressing stable resistance to the Qo-inhibiting fungicide azoxystrobin. Curr Genet. 2003 Mar;42(6):332–338. doi: 10.1007/s00294-002-0356-1. [DOI] [PubMed] [Google Scholar]
- 57.Sierotzki H, Parisi S, Steinfeld U, Tenzer I, Poirey S, Gisi U. Mode of resistance to respiration inhibitors at the cytochrome bc(1) enzyme complex of Mycosphaerella fijiensis field isolates. Pest Management Science. 2000 Oct;56(10):833–841. [Google Scholar]
- 58.Sierotzki H, Frey R, Wullschleger J, Palermo S, Karlin S, Godwin J, et al. Cytochrome b gene sequence and structure of Pyrenophora teres and P. tritici-repentis and implications for QoI resistance. Pest Manag Sci. 2007 Mar;63(3):225–233. doi: 10.1002/ps.1330. [DOI] [PubMed] [Google Scholar]
- 59.Korsinczky M, Chen N, Kotecka B, Saul A, Rieckmann K, Cheng Q. Mutations in Plasmodium falciparum cytochrome b that are associated with atovaquone resistance are located at a putative drug-binding site. Antimicrob Agents Chemother. 2000 Aug;44(8):2100–2108. doi: 10.1128/aac.44.8.2100-2108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Srivastava IK, Morrisey JM, Darrouzet E, Daldal F, Vaidya AB. Resistance mutations reveal the atovaquone-binding domain of cytochrome b in malaria parasites. Mol Microbiol. 1999 Aug;33(4):704–711. doi: 10.1046/j.1365-2958.1999.01515.x. [DOI] [PubMed] [Google Scholar]
- 61.Fisher N, Abd Majid R, Antoine T, Al-Helal M, Warman AJ, Johnson DJ, et al. Cytochrome b mutation Y268S conferring the atovaquone resistance phenotype in the malaria parasite results in reduced parasite bc1 catalytic turnover and protein expression. J Biol Chem. 2012 Jan 26; doi: 10.1074/jbc.M111.324319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peters JM, Chen N, Gatton M, Korsinczky M, Fowler EV, Manzetti S, et al. Mutations in cytochrome b resulting in atovaquone resistance are associated with loss of fitness in Plasmodium falciparum. Antimicrob Agents Chemother. 2002 Aug;46(8):2435–2441. doi: 10.1128/AAC.46.8.2435-2441.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee DW, Selamoglu N, Lanciano P, Cooley JW, Forquer I, Kramer DM, et al. Loss of a conserved tyrosine residue of cytochrome b induces reactive oxygen species production by cytochrome BC1. J Biol Chem. 2011 Mar 23; doi: 10.1074/jbc.M110.214460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crofts AR. Proton-coupled electron transfer at the Q(o)-site of the bc(1) complex controls the rate of ubihydroquinone oxidation. Biochimica Et Biophysica Acta-Bioenergetics. 2004 Apr 12;1655(1–3):77–92. doi: 10.1016/j.bbabio.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 65.Hsueh KL, Westler WM, Markley JL. NMR investigations of the Rieske protein from Thermus thermophilus support a coupled proton and electron transfer mechanism. J Am Chem Soc. 2010 Jun 16;132(23):7908–7918. doi: 10.1021/ja1026387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin IJ, Chen Y, Fee JA, Song JK, Westler WM, Markley JL. Rieske protein from Thermus thermophilus: N-15 NMR titration study demonstrates the role of iron-ligated histidines in the pH dependence of the reduction potential. Journal of the American Chemical Society. 2006 Aug 23;128(33):10672–10673. doi: 10.1021/ja0627388. [DOI] [PubMed] [Google Scholar]
- 67.Kunze B, Jansen R, Hofle G, Reichenbach H. Crocacin, a new electron transport inhibitor from Chondromyces crocatus (myxobacteria). Production, isolation, physico-chemical and biological properties. J Antibiot (Tokyo) 1994 Aug;47(8):881–886. doi: 10.7164/antibiotics.47.881. [DOI] [PubMed] [Google Scholar]
- 68.Crowley PJ, Berry EA, Cromartie T, Daldal F, Godfrey CR, Lee DW, et al. The role of molecular modeling in the design of analogues of the fungicidal natural products crocacins A and D. Bioorg Med Chem. 2008 Dec 15;16(24):10345–10355. doi: 10.1016/j.bmc.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nising CF, Hillebrand S, Rodefeld L. Recent developments in the total synthesis of fungicidal natural products--a crop protection perspective. Chem Commun (Camb) 2011 Apr 14;47(14):4062–4073. doi: 10.1039/c0cc05178k. [DOI] [PubMed] [Google Scholar]
- 70.Yamashita E, Zhang H, Cramer WA. Structure of the cytochrome b6f complex: quinone analogue inhibitors as ligands of heme cn. J Mol Biol. 2007 Jun 29;370(1):39–52. doi: 10.1016/j.jmb.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Degli Esposti M, Ghelli A, Crimi M, Estornell E, Fato R, Lenaz G. Complex I and complex III of mitochondria have common inhibitors acting as ubiquinone antagonists. Biochem Biophys Res Commun. 1993 Feb 15;190(3):1090–1096. doi: 10.1006/bbrc.1993.1161. [DOI] [PubMed] [Google Scholar]
- 72.Ando K, Suzuki S, Saeki T, Tamura G, Arima K. Funiculosin, a new antibiotic. I. Isolation, biological and chemical properties (studies on antiviral and antitumor antibiotics. 8) J Antibiot (Tokyo) 1969 May;22(5):189–194. doi: 10.7164/antibiotics.22.189. [DOI] [PubMed] [Google Scholar]
- 73.Nelson BD, Walter P, Ernster L. Funiculosin: an antibiotic with antimycin-like inhibitory properties. Biochim Biophys Acta. 1977 Apr 11;460(1):157–162. doi: 10.1016/0005-2728(77)90162-1. [DOI] [PubMed] [Google Scholar]
- 74.Gutierrez-Cirlos EB, Merbitz-Zahradnik T, Trumpower BL. Inhibition of the yeast cytochrome bc1 complex by ilicicolin H, a novel inhibitor that acts at the Qn site of the bc1 complex. J Biol Chem. 2004 Mar 5;279(10):8708–8714. doi: 10.1074/jbc.M311805200. [DOI] [PubMed] [Google Scholar]
- 75.Tamura G, Suzuki S, Takatsuki A, Ando K, Arima K. Ascochlorin, a new antibiotic, found by the paper-disc agar-diffusion method. I. Isolation, biological and chemical properties of ascochlorin. (Studies on antiviral and antitumor antibiotics. I) J Antibiot (Tokyo) 1968 Sep;21(9):539–544. doi: 10.7164/antibiotics.21.539. [DOI] [PubMed] [Google Scholar]
- 76.Nawata Y, Ando K, Tamura G, Arima K, Iitaka Y. The molecular structure of ascochlorin. J Antibiot (Tokyo) 1969 Oct;22(10):511–512. doi: 10.7164/antibiotics.22.511. [DOI] [PubMed] [Google Scholar]
- 77.Convent B, Briquet M. Properties of 3-(3, -4-dichlorophenyl)-1, 1-dimethylurea and other inhibitors of the cytochrome bc1 segment of the mitochondrial respiratory chain in Saccharomyces cerevisiae. Eur J Biochem. 1978 Jan 16;82(2):473–481. doi: 10.1111/j.1432-1033.1978.tb12041.x. [DOI] [PubMed] [Google Scholar]
- 78.Mitani S, Araki S, Yamaguchi T, Takii Y, Ohshima T, Matsuo N. Biological properties of the novel fungicide cyazofamid against Phytophthora infestans on tomato and Pseudoperonospora cubensis on cucumber. Pest Manag Sci. 2002 Feb;58(2):139–145. doi: 10.1002/ps.430. [DOI] [PubMed] [Google Scholar]
- 79.Aliferis KA, Jabaji S. Metabolomics – A robust bioanalytical approach for the discovery of the modes-of-action of pesticides: A review. Pesticide Biochemistry and Physiology. 2011;100(2):105–117. [Google Scholar]
- 80.Gao X, Wen X, Esser L, Yu L, Yu CA, Xia D. Structural basis for the quinone reduction in bc1 complex: a comparative analysis of crystal structures of mitochondrial cytochrome bc1 with bound substrate and inhibitors. Biochemistry. 2003;42:9067–9080. doi: 10.1021/bi0341814. [DOI] [PubMed] [Google Scholar]
- 81.Kurisu G, Zhang H, Smith JL, Cramer WA. Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science. 2003 Nov 7;302(5647):1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- 82.Stroebel D, Choquet Y, Popot J-L, Picot D. An atypical haem in the cytochrome b6f complex. Nature. 2003;426(6965):413–418. doi: 10.1038/nature02155. [10.1038/nature02155] [DOI] [PubMed] [Google Scholar]
- 83.Leben C, Keitt GW. An Antibiotic Substance Active against Certain Phytopathogens. Phytopathology. 1948;38(11):899–906. [Google Scholar]
- 84.Xia D, Yu CA, Kim H, Xia JZ, Kachurin AM, Zhang L, et al. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997 Jul 4;277(5322):60–66. doi: 10.1126/science.277.5322.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim H, Esser L, Hossain MB, Xia D, Yu CA, Riso J, et al. Structure of antimycin A1, a specific electron transfer inhibitor of ubiquinol-cytochrome c oxidoreductase. Journal of the American Chemical Society. 1999 May 26;121(20):4902–4903. [Google Scholar]
- 86.Dickie JP, Loomans ME, Farley TM, Strong FM. The Chemistry of Antimycin A. XI. N-Substituted 3-Formamidosalicylic Amides1. Journal of Medicinal Chemistry. 1963 Jul 01;6(4):424–427. doi: 10.1021/jm00340a018. 1963. [DOI] [PubMed] [Google Scholar]
- 87.Rieske JS. Inhibitors of respiration at energy-coupling site 2 of the respiratory chain. Pharmacol Ther. 1980;11(2):415–450. doi: 10.1016/0163-7258(80)90036-4. [DOI] [PubMed] [Google Scholar]
- 88.Gao X, Wen X, Esser L, Quinn B, Yu L, Yu CA, et al. Structural basis for the quinone reduction in the bc1 complex: a comparative analysis of crystal structures of mitochondrial cytochrome bc1 with bound substrate and inhibitors at the Qi site. Biochemistry. 2003 Aug 5;42(30):9067–9080. doi: 10.1021/bi0341814. [DOI] [PubMed] [Google Scholar]
- 89.Huang L-s, Cobessi D, Tung EY, Berry EA. Binding of the Respiratory Chain Inhibitor Antimycin to the Mitochondrial bc1 Complex: A New Crystal Structure Reveals an Altered Intramolecular Hydrogen-bonding Pattern. Journal of Molecular Biology. 2005;351(3):573–597. doi: 10.1016/j.jmb.2005.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Esser L, Elberry M, Zhou F, Yu CA, Yu L, Xia D. Inhibitor-complexed structures of the cytochrome bc1 from the photosynthetic bacterium Rhodobacter sphaeroides. J Biol Chem. 2008 Feb 1;283(5):2846–2857. doi: 10.1074/jbc.M708608200. [DOI] [PubMed] [Google Scholar]
- 91.Miyoshi H, Tokutake N, Imaeda Y, Akagi T, Iwamura H. A model of antimycin A binding based on structure-activity studies of synthetic antimycin A analogues. Biochimica et Biophysica Acta. 1995;1229:149–154. doi: 10.1016/0005-2728(94)00185-8. [DOI] [PubMed] [Google Scholar]
- 92.Lancaster CR, Hunte C, Kelley J, 3rd, Trumpower BL, Ditchfield R. A comparison of stigmatellin conformations, free and bound to the photosynthetic reaction center and the cytochrome bc1 complex. J Mol Biol. 2007 Apr 20;368(1):197–208. doi: 10.1016/j.jmb.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 93.Berry EA, Huang L-s, Lee D-W, Daldal F, Nagai K, Minagawa N. Ascochlorin is a novel, specific inhibitor of the mitochondrial cytochrome bc1 complex. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2010;1797(3):360–370. doi: 10.1016/j.bbabio.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.di Rago JP, Colson AM. Molecular basis for resistance to antimycin and diuron, Q-cycle inhibitors acting at the Qi site in the mitochondrial ubiquinol-cytochrome c reductase in Saccharomyces cerevisiae. Journal of Biological Chemistry. 1988;263(25):12564–12570. [PubMed] [Google Scholar]
- 95.Schnaufer A, Sbicego S, Blum B. Antimycin A resistance in a mutant Leishmania tarentolae strain is correlated to a point mutation in the mitochondrial apocytochrome b gene. Curr Genet. 2000 Apr;37(4):234–241. doi: 10.1007/s002940050525. [DOI] [PubMed] [Google Scholar]
- 96.Colson AM, Thevan L, Convent B, Briquet M, Goffeau A. Mitochondrial Heredity of Resistance to 3-(3,4-Dichlorophenyl)-1,1-Dimethylurea, an Inhibitor of Cytochrome-B Oxidation, in Saccharomyces-Cerevisiae. Eur J Biochem. 1977;74(3):521–526. doi: 10.1111/j.1432-1033.1977.tb11419.x. [DOI] [PubMed] [Google Scholar]
- 97.Pratje E, Michaelis G. Allelism Studies of Mitochondrial Mutants Resistant to Antimycin a or Funiculosin in Saccharomyces-Cerevisiae. Mol Gen Genet. 1977;152(2):167–174. doi: 10.1007/BF00267422. [DOI] [PubMed] [Google Scholar]
- 98.Fernandez-Ortuno D, Tores JA, de Vicente A, Perez-Garcia A. Field resistance to QoI fungicides in Podosphaera fusca is not supported by typical mutations in the mitochondrial cytochrome b gene. Pest Manag Sci. 2008 Jul;64(7):694–702. doi: 10.1002/ps.1544. [DOI] [PubMed] [Google Scholar]
- 99.Steinfeld U, Sierotzki H, Parisi S, Poirey S, Gisi U. Sensitivity of mitochondrial respiration to different inhibitors in Venturia inaequalis. Pesticide Mangement Science. 2001;57:787–796. doi: 10.1002/ps.356. [DOI] [PubMed] [Google Scholar]
- 100.Wood PM, Hollomon DW. A critical evaluation of the role of alternative oxidase in the performance of strobilurin and related fungicides acting at the Qo site of complex III. Pest Manag Sci. 2003 May;59(5):499–511. doi: 10.1002/ps.655. [DOI] [PubMed] [Google Scholar]
- 101.Gaur M, Choudhury D, Prasad R. Complete inventory of ABC proteins in human pathogenic yeast, Candida albicans. J Mol Microbiol Biotechnol. 2005;9(1):3–15. doi: 10.1159/000088141. [DOI] [PubMed] [Google Scholar]
- 102.Hill P, Kessl J, Fisher N, Meshnick S, Trumpower BL, Meunier B. Recapitulation in Saccharomyces cerevisiae of cytochrome b mutations conferring resistance to atovaquone in Pneumocystis jiroveci. Antimicrob Agents Chemother. 2003 Sep;47(9):2725–2731. doi: 10.1128/AAC.47.9.2725-2731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Reimann S, Deising HB. Inhibition of efflux transporter-mediated fungicide resistance in Pyrenophora tritici-repentis by a derivative of 4'-hydroxyflavone and enhancement of fungicide activity. Appl Environ Microbiol. 2005 Jun;71(6):3269–3275. doi: 10.1128/AEM.71.6.3269-3275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roohparvar R, De Waard MA, Kema GH, Zwiers LH. MgMfs1, a major facilitator superfamily transporter from the fungal wheat pathogen Mycosphaerella graminicola, is a strong protectant against natural toxic compounds and fungicides. Fungal Genet Biol. 2007 May;44(5):378–388. doi: 10.1016/j.fgb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 105.Schneider G. Virtual screening: an endless staircase? Nat Rev Drug Discov. 2010;9(4):273–276. doi: 10.1038/nrd3139. [10.1038/nrd3139] [DOI] [PubMed] [Google Scholar]
- 106.Ivetac A, McCammon JA. Molecular recognition in the case of flexible targets. Curr Pharm Des. 2011;17(17):1663–1671. doi: 10.2174/138161211796355056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Carlson HA, Masukawa KM, Rubins K, Bushman FD, Jorgensen WL, Lins RD, et al. Developing a dynamic pharmacophore model for HIV-1 integrase. J Med Chem. 2000 Jun 1;43(11):2100–2114. doi: 10.1021/jm990322h. [DOI] [PubMed] [Google Scholar]
- 108.Carlson HA, McCammon JA. Accommodating protein flexibility in computational drug design. Mol Pharmacol. 2000 Feb;57(2):213–218. [PubMed] [Google Scholar]
- 109.McCammon JA. Target flexibility in molecular recognition. Biochim Biophys Acta. 2005 Dec 30;1754(1–2):221–224. doi: 10.1016/j.bbapap.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 110.Zhao PL, Wang L, Zhu XL, Huang X, Zhan CG, Wu JW, et al. Subnanomolar inhibitor of cytochrome bc1 complex designed by optimizing interaction with conformationally flexible residues. J Am Chem Soc. 2010 Jan 13;132(1):185–194. doi: 10.1021/ja905756c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bloland PB. In: Drug resistance in malaria. Organization WH, editor. 2001. [Google Scholar]
- 112.Esser L, Quinn B, Li YF, Zhang M, Elberry M, Yu L, et al. Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex. J Mol Biol. 2004 Jul 30;341(1):281–302. doi: 10.1016/j.jmb.2004.05.065. [DOI] [PubMed] [Google Scholar]
- 113.Berry EA, Zhang Z, Bellamy HD, Huang L. Crystallographic location of two Zn2+-binding sites in the avian cytochrome bc1 complex. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2000;1459(2–3):440–448. doi: 10.1016/s0005-2728(00)00182-1. [DOI] [PubMed] [Google Scholar]
- 114.Crowley PJ, Berry EA, Cromartie T, Daldal F, Godfrey CRA, Lee D-W, et al. The role of molecular modeling in the design of analogues of the fungicidal natural products crocacins A and D. Bioorganic & Medicinal Chemistry. 2008;16(24):10345–10355. doi: 10.1016/j.bmc.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hunte C, Koepke J, Lange C, Rossmanith T, Michel H. Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 2000 Jun 15;8(6):669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 116.Lange C, Nett JH, Trumpower BL, Hunte C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 2001 Dec 3;20(23):6591–6600. doi: 10.1093/emboj/20.23.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lange C, Hunte C. Crystal structure of the yeast cytochrome bc1 complex with its bound substrate cytochrome c. Proc Natl Acad Sci U S A. 2002 Mar 5;99(5):2800–2805. doi: 10.1073/pnas.052704699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Solmaz SR, Hunte C. Structure of complex III with bound cytochrome c in reduced state and definition of a minimal core interface for electron transfer. J Biol Chem. 2008 Jun 20;283(25):17542–17549. doi: 10.1074/jbc.M710126200. [DOI] [PubMed] [Google Scholar]
- 119.Berry EA, Huang LS, Saechao LK, Pon NG, Valkova-Valchanova M, Daldal F. X-Ray Structure of Rhodobacter Capsulatus Cytochrome bc (1): Comparison with its Mitochondrial and Chloroplast Counterparts. Photosynth Res. 2004;81(3):251–275. doi: 10.1023/B:PRES.0000036888.18223.0e. [DOI] [PubMed] [Google Scholar]
- 120.Althoff T, Mills DJ, Popot J-L, Kuhlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 2011;30(22):4652–4664. doi: 10.1038/emboj.2011.324. [10.1038/emboj.2011.324] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, et al. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998 Jul 3;281(5373):64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]