Abstract
The extracellular signal-regulated kinase 1 and 2 (ERK1/2) pathway plays a central role in defining various cellular fates. Scaffold proteins modulating ERK1/2 activity control growth factor signals transduced by the pathway. Here, we analyzed signals transduced by Shoc2, a critical positive modulator of ERK1/2 activity. We found that loss of Shoc2 results in impaired cell motility and delays cell attachment. As ERKs control cellular fates by stimulating transcriptional response, we hypothesized that the mechanisms underlying changes in cell adhesion could be revealed by assessing the changes in transcription of Shoc2-depleted cells. Using quantitative RNA-seq analysis, we identified 853 differentially expressed transcripts. Characterization of the differentially expressed genes showed that Shoc2 regulates the pathway at several levels, including expression of genes controlling cell motility, adhesion, crosstalk with the transforming growth factor beta (TGFβ) pathway, and expression of transcription factors. To understand the mechanisms underlying delayed attachment of cells depleted of Shoc2, changes in expression of the protein of extracellular matrix (lectin galactoside-binding soluble 3-binding protein; LGALS3BP) were functionally analyzed. We demonstrated that delayed adhesion of the Shoc2-depleted cells is a result of attenuated expression and secretion of LGALS3BP. Together our results suggest that Shoc2 regulates cell motility by modulating ERK1/2 signals to cell adhesion.
Keywords: Shoc2 scaffold, ERK1/2, motility, adhesion, transcription, LGALS3BP
1. Introduction
The mechanisms leading to activation of RAF, MEK and ERK kinases in the extracellular signal-regulated kinase 1 and 2 (ERK1/2) pathway have been studied extensively [1, 2]. However, what determines the activity of the pathway in the context of a specific set of downstream targets and how ERK signals result in distinct biological outcomes is not yet clear. Several studies suggest that divergent cell fates induced by the ERK pathway are the result of a tight spatio-temporal control of ERK1/2 targeting, sequestration and activation of the kinases and phosphatases, as well as modulation of the strength and duration of the ERK signaling [3, 4]. Scaffold proteins have been proposed to fulfill some of these requirements. Scaffolds have been implicated in controlling the spatial organization of signaling enzymes, insulation of active modules and prevention of a spurious cross-talk of signaling networks [5, 6]. Yet, detailed mechanisms allowing scaffolds to elicit specific cellular responses at the molecular level remain to be elucidated.
Scaffold proteins of the ERK1/2 pathway represent a diverse group of proteins [2]. A well-studied scaffold kinase suppressor of Ras 1 (KSR1) [7–9] is the multifunctional protein that binds to and accelerates activity of MEK and RAF kinases thereby stimulating expression of genes that drive cell proliferation and differentiation [10, 11]. Other ERK1/2 scaffolds, mitogen-activated protein 1 (MP1) and p14 (also called the LAMTOR2/3 complex), are believed to regulate cytoskeletal dynamics [12–15], and the MP1/p14 complex is involved in remodeling of focal adhesion and actin structures during cell spreading [16].
Ras-RAF-1-ERK1/2 signaling is accelerated by the scaffolding protein Shoc2 [17, 18]. This evolutionarily well-conserved protein is essential for normal development [19–21]. Loss of Shoc2 in mammalian cultured cells and C. elegans leads to a dramatic decrease in ERK1/2 activity [17, 22, 23]. As a scaffold protein, Shoc2 provides a molecular platform for multi-protein assemblies that modulate ERK1/2 activity [24, 25]. In addition to its signaling partners Ras and RAF-1, Shoc2 tethers the catalytic subunit of protein phosphatase 1c (PP1c) as well as proteins of the ubiquitin machinery HUWE1 and PSMC5 [23, 26, 27]. The ability of this non-catalytic scaffold to mediate ERK1/2 signaling is controlled through allosteric ubiquitination [24]. Alterations in the mechanisms controlling ubiquitination of the scaffold affect Shoc2-mediated ERK1/2 signals and cell motility [27].
Activation of the ERK1/2 pathway in response to epidermal growth factor (EGF) stimulation of the EGF receptor falls into three major regulatory loops: immediate, delayed, and late (secondary) [28–30]. The immediate regulatory loop induces phosphorylation of transcription factors such as FOS, Jun and EGR1 and does not require new protein synthesis for their transcription [30]. Expression of the genes of the immediate response induces transcription of delayed genes, such as the RNA-binding protein ZFP36 or dual specific phosphatases, which dephosphorylate ERK1/2 kinases that terminate the activity of the immediate loop [30]. Late (secondary) transcriptional response leads to expression of genes such as actin-binding proteins or genes encoding proteins that are involved in cell metabolism and biogenesis of membranes and appear to define cellular outcomes [31].
In the current study, we aimed to determine the specific ERK1/2 response elicited through the Shoc2 scaffolding module. Results of this study provide evidence that Shoc2-mediated ERK1/2 activity contributes to maintenance of the ERK1/2 feedback loop that regulates expression of genes of the TGFβ pathway. We also found that Shoc2-ERK1/2 signals control cell motility and adhesion, in part, through mechanisms that monitor expression of the protein of extracellular matrix- lectin galactoside-binding soluble 3-binding protein or LGALS3BP (also called Mac-2 binding protein) [32]. Deficient expression and secretion of this heavily glycosylated protein led to attenuated attachment of Shoc2-depleted cells. These results indicate that Shoc2 transduces signals to unique cellular responses and identifies novel molecular targets of the Shoc2-ERK1/2 signaling axis.
2. Materials and methods
2.1. Reagents and antibodies
EGF was obtained from BD Bioscience. U0126 and PD98059 were obtained from LC Laboratories. Respective proteins were detected using specific primary antibodies, including: GAPDH, phospho-ERK1/2, ERK1/2, MEK1/2, COL1A1 and EGFR (Santa Cruz Biotechnology); His, Shoc2 and LGALS3BP (Proteintech); phospho-AKT, KSR1, phospho-MEK1/2 (Cell Signaling).
2.2. Constructs
Shoc2-tRFP was described previously [25, 33]. The plasmid carrying full-length His-tagged LGAL3SBP was obtained from Dr. Enza Picollo (Chieti, Italy). The plasmid carrying shRNA specifically recognizing KSR1 was kindly provided by Dr. Tianyan Gao (University of Kentucky) and was obtained from the Sigma Mission collection. The shRNA sequence used to target the KSR1 transcripts was as follows: #1-5’-CCGGCAACAAGGAGTGGAATGATTTCTCGAGAAATCATTCCACTCCTTGTTGTTTTT G-3’; #2- 5’-CCGGTCGTACACAAAGATCTCAAATCTCGAGATTTGAGATCTTTGTGTACGATTTTT G-3’. Efficiency of the shRNA knockdown was validated by western blotting. Plasmid DNAs were purified using Zymo Research. All constructs were verified by dideoxynucleotide sequencing.
2.3. Cell culture and DNA transfections
Cos1 (ATCC), and stable cell lines (NT, LV1, SR) (derivative of Cos1 cells) were grown in Dulbecco Modified Eagle’s Medium (DMEM) containing 10% fetal bovine serum (FBS) supplemented with Sodium Pyruvate, MEM-NEAA, Penicillin, Streptomycin, and L-Glutamate (Invitrogen). MCF7, T47D and stable cell lines (NT, LV1, SR) (derivative of T47D cells) were grown in RPMI 1640 Medium containing 10% FBS. MCF7 and stable cell lines (NT, LV1, SR) (derivative of MCF7 cells) were grown in MEM containing 10% FBS. The transfections of DNA constructs were performed using PEI (Neo Transduction Laboratories, Lexington, KY) reagent.
2.4. Real-time quantitative polymerase chain reaction (qPCR)
Total RNA was isolated using PureZOL/Aurum Total RNA Isolation Kit (Bio-Rad) according to manufacturer instructions. Aliquots containing equal amounts of RNA were subjected to RT-PCR analysis. The RNA quality for RNA-seq was tested using Agilent Bioanalyzer 2100. Quantitative RT-PCR was performed using SoAdvanced™ SYBR® Green supermix and the Bio-Rad CFX detection system (Bio-Rad). Relative amounts of RNAs were calculated using the comparative CT method [34]. HPRT1 gene expression was used as a reference. Sequence-specific primer sets are presented in Supplemental Table 3.
2.5. Western blot analysis
Cells were placed on ice and washed with phosphate-buffered saline (PBS); proteins were then solubilized in lysis buffer containing 50 mM Tris pH 7.5, 150 mM NaCl, 1% Triton, 1 mM Na3OV4, 10 mM NaF, 0.5 mM phenylmethylsulfonyl fluoride, 10 µg/ml of leupeptin and 10 µg/ml of aprotinin for 15 min at 4°C. Lysates were centrifuged for 15 min to remove insoluble material. Cell lysates were denatured in the sample buffer at 95°C, resolved by electrophoresis, and probed with various antibodies, followed by chemiluminescence detection. Quantification was performed using the densitometry analysis mode of Image Lab software (Bio-Rad, Inc.).
2.6. Wound healing assays
Cells were seeded at a density of 1 × 105/mL in each well of IBIDI™ Culture-Inserts placed on a glass-bottom 33 mm culture dish. To create 500 µm cell-free gap, culture-inserts were removed 18h upon seeding. Cells were then treated with 10 nM EGF and observed with a 10× objective on a Zeiss Axiovert microscope. Images were acquired using Slidebook software (Intelligent Imaging Innovations, Denver, CO). Detection of GFP fluorescence was performed using a FITC filter channel every 12h up to 48h. The distance of closure was measured at three spots and the average from three images was shown as mean ± SD.
2.7. Migration assay
Cell migration assays were performed using 8.0 µm pore 24-well TC inserts (Greiner Bio-one, Monroe, NC, USA). Filters were coated with 15 µg/ml collagen at 37°C for 30 min. Cells were trypsinized (with 0.05% Trypsin-EDTA; Invitrogen), collected with serum-free medium containing soybean trypsin inhibitor (1 mg/ml), centrifuged (500 × g for 5 min), and then resuspended in serum-free medium. Cells (5 × 104) were then placed in the upper chamber and the lower chamber was filled with complete medium with 10% serum or 10nM EGF. Cells were migrated at 37°C for 4h. After removing non-migrated cells, membranes were fixed in methanol and stained with 1% crystal violet. Migrated cells were counted in three random fields per membrane under the microscope at × 20. Each assay was repeated more than three times. To test migration of T47D cells, plates were incubated for 24h at 37°C.
2.8. Cell attachment assay
Cells were trypsinized (with 0.05% Trypsin-EDTA; Invitrogen) and collected with serum-free medium containing soybean trypsin inhibitor (1 mg/ml) as described previously [35, 36]. Cells were centrifuged, washed once with serum-free medium, and resuspended gently in serum-free medium. After incubation for 90 min in a cell culture incubator at 37°C (5% CO2), cells were subsequently seeded at a density of 5 × 104 cells per well in a 96-well culture plate coated with collagen (10 ng/mL), fibronectin (5 ug/mL) or laminin (1 ug/mL). Cells were maintained at 37°C in a humidified 5% CO2 incubator. After incubation for indicated times, attached cells were fixed using 4% paraformaldehyde (Sigma-Aldrich) in PBS for 10 min, washed with PBS and stained with 1% crystal violet. Attached cells were then solubilized with 2% SDS, and the OD550 was measured by spectrophotometer. Each assay was repeated more than three times in duplicate.
2.9. Culture media concentration
Cells were grown to 90% confluency. Culture medium was collected 18–24h after seeding, then were filtered and concentrated in the presence of proteinase inhibitors using centrifugal filters (Amicon Ultra-15, >30KDa, Millipore, Germany).
2.10. Cell growth assay
Cells were seeded into a 96-well culture plate at a density of 1,000 cells/well and were grown in DMEM supplemented with 10% FBS. Cell viability was measured using CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (Promega) according to the manufacturer recommendations. The absorbance at 450 nm was measured on a 96-well plate reader. Each experiment was done in quadruplicate and repeated three times.
2.11. Immunoprecipitation and deglycosylation
The cells were transfected with LGALS3BP plasmid. 36h post-transfection, cells were placed on ice and washed with PBS, and the proteins were solubilized in lysis buffer containing 20 mM HEPES (Sigma) pH 7.6, 10 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA (Sigma), 1 mM EGTA (Sigma), 0.5 mM PMSF (Sigma), 10 µg/ml of leupeptin (Roche), 10 µg/ml of aprotinin (Roche), 5 µg/ml of pepstatin A (Sigma), and 50 mM β-glycrophosphate (Sigma) for 15 min at 4°C. Lysates were then centrifuged at 2,500 × g for 15 min to remove insoluble material. Lysates were incubated with anti-His antibody for 2h and the immuno-complexes were precipitated using Protein A Sepharose. Immunoprecipitates were digested with PNGase G or Endo H (New England BioLabs), denatured in the sample buffer at 95°C and resolved by electrophoresis.
2.12. RNA-seq analysis
Triplicates of RNA from Cos-NT and Cos-LV1 cells were purified as described above. For library preparation, mRNA was first extracted from total RNA using oligo (dT) magnetic beads and sheared into short fragments of about 200 bases. The cDNA library was sequenced using Illumina HiSeq 2500 sequencer. RNA-seq reads were trimmed using Trimmomatic-0.27 [37, 38]. Short reads were aligned to vervet monkey reference genome (Chlorocebus sabaeus) using TopHat [39] with the ChiSab gene description file from Ensembl. Aligned RNA-seq reads were assembled. Fragments per kilobase per million mapped (FPKM) reads were determined for all RefSeq genes using CuffLinks and CuffDiff (FDR <0.05) [39]. The RNA-seq data is publicly available as GEO series GSE67063. The data is MIAME compliant. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67063+
2.13. Gene ontology (GO) and pathway and network analysis
Differentially expressed genes determined by RNA-seq analysis were used for functional enrichment including the Ingenuity Pathways Analysis system (IPA, http://www.ingenuity.com) that predicts molecular and cellular functions using the Ingenuity Knowledge base as the background. Gene ontology terms within the data set were provided by Protein Analysis Through Evolutionary Relationships (PANTHER) [40].
2.14. Statistical analysis
Results are expressed as mean ± SD. Statistical significance of the differences between groups was determined using either Student's t test or one-way ANOVA (followed by the Tukey’s test). P-values <0.05 were considered statistically significant. All statistical analyses were carried out using SigmaStat 3.5 (Systat Software Inc., Point Richmond, CA, USA).
3. Results
3.1. Cells depleted of Shoc2 scaffold exhibit reduced cell motility
We and others have suggested that Shoc2 is involved in regulating cell polarity and movement [26, 27, 41]. To understand the cellular functions controlled by the Shoc2-mediated ERK1/2 signals, we utilized Cos1 cells stably depleted of Shoc2 (Cos-LV1) as well as cells stably depleted of Shoc2 and then rescued with Shoc2-tRFP (Cos-SR) [25, 42]. We also generated T47D and MCF7 cells constitutively depleted of Shoc2 (T47D-LV1, MCF7-LV1) and T47D and MCF7 cells depleted of Shoc2 and then rescued with Shoc2-tRFP (T47D-SR, MCF7-SR) (Suppl. Fig. 1). The effects of Shoc2 depletion or rescue in T47D and MCF7 cells were similar to that observed in Cos1 cells: silencing of Shoc2 resulted in decreased ERK1/2 activity that was rescued by the expression of Shoc2-tRFP [25].
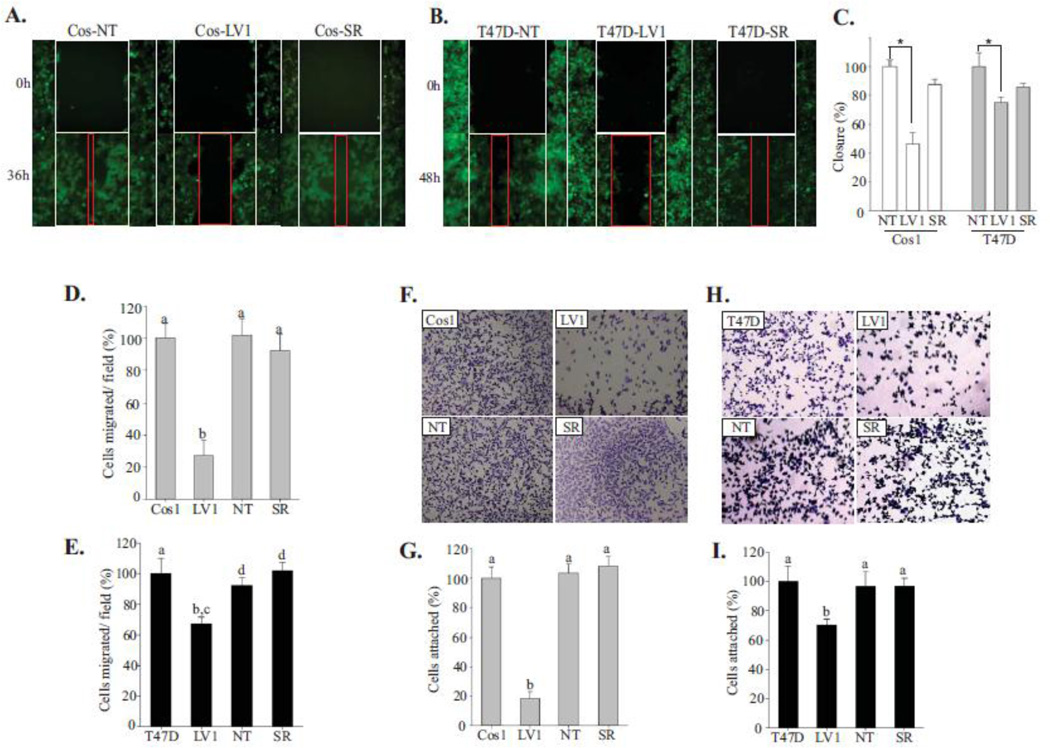
To analyze the motility of cells depleted of Shoc2 in more detail, we first monitored the ability of the cell monolayer to migrate into and across a nude area over time using time-lapse live-cell microscopy (Fig. 1A, B). The rate of gap closure was assessed over a 48h period. We found that control cells expressing Shoc2 (NT) as well as cells stably depleted of Shoc2 and then rescued with Shoc2-tRFP (Cos-SR) readily migrated into the wounded area and closed the wound after 36h (48h for T47D). In contrast, the lag in migration of the Shoc2-depleted Cos1 cells was clear even after 36h of continued culture observation (Fig. 1A–C). To extend the wound-closure analysis and to evaluate additional aspects of motility including chemotaxis, modified Boyden chamber migration assays were performed. Chemotaxis was evaluated in response to EGF. Consistent with the results of wound-healing analysis (Fig. 1A–C), the Shoc2-depleted cells migrated less efficiently than control cells (Fig. 1D, E). Together these data indicate that Shoc2 depletion inhibits cell motility.
Figure 1. Shoc2 knockdown affects cell motility.
(A, B) Wound healing assay was performed using Cos1 (A) and T47D (B) cells stably expressing Shoc2 shRNA (LV1), non-targeting shRNA (NT) or expressing Shoc2 shRNA and Shoc2-tRFP (Cos-SR). The white frames indicate the width of the wound at time 0; red frames show the width of the wound 36/48h post EGF treatment.
(C) Data from independent experiments in A and B were analyzed using a Tukey Test (mean ± SD, n = 3, * p<0.001). Bars represent the change in average wound closure rates at 36/48h. The closure was calculated by measuring the average width in open area, and normalized to control cells.
(D, E) Transwell migration assay was performed in Cos1 (D) and T47D (E) cells stably expressing Shoc2 shRNA (LV1), non-targeting shRNA (NT) and Shoc2-tRFP (SR). Cells in multiple independent fields for each well were counted and three independent experiments were analyzed. Data are shown as mean ± SD; a vs. b, c vs. d, p<0.05.
(F, H) 5×104 cells of Cos1 (F) and T47D (H) cells were seeded on collagen pre-coated 96-well plates for 10 min. Images of cells fixed and stained with crystal violet were obtained using a Nikon Eclipse E600 microscope.
(G, I) Cells from the experiments in F and H were solubilized with 2% SDS and subjected to colorimetric absorbance measurement (OD550). Data from three independent experiments was analyzed. Bars represent mean values ± SD, n = 3; a vs. b, p<0.05.
We next investigated the effect of Shoc2 ablation on the ability of cells to adhere to extracellular matrixes (i.e. collagen I, laminin, and fibronectin). We found that silencing of Shoc2 led to decreased cell attachment to collagen type I when seeded for 10 min (Fig. 1F–I). Once Shoc2-depleted cells were incubated on collagen I for longer times (30 min), the levels of cell attachment were comparable to those observed for the control cells (Suppl. Fig. 2A). Attachment rates of Cos1 and T47D cells in which Shoc2 was reconstituted with Shoc2-tRFP (SR) were similar to control cells (NT) (Fig. 1F–I). We have not detected changes in the attachment of Shoc2-depleted cells to fibronectin or laminin (Suppl. Fig. 2B–D). Growth assays to measure the proliferation of cells depleted of Shoc2 suggest that the reduced adhesion was not caused by cytotoxicity (Suppl. Fig. 1C, D). The compromised ability of Shoc2-depleted cells to adhere to collagen suggests that Shoc2 controls a pool of ERK1/2 signals that regulates cell attachment.
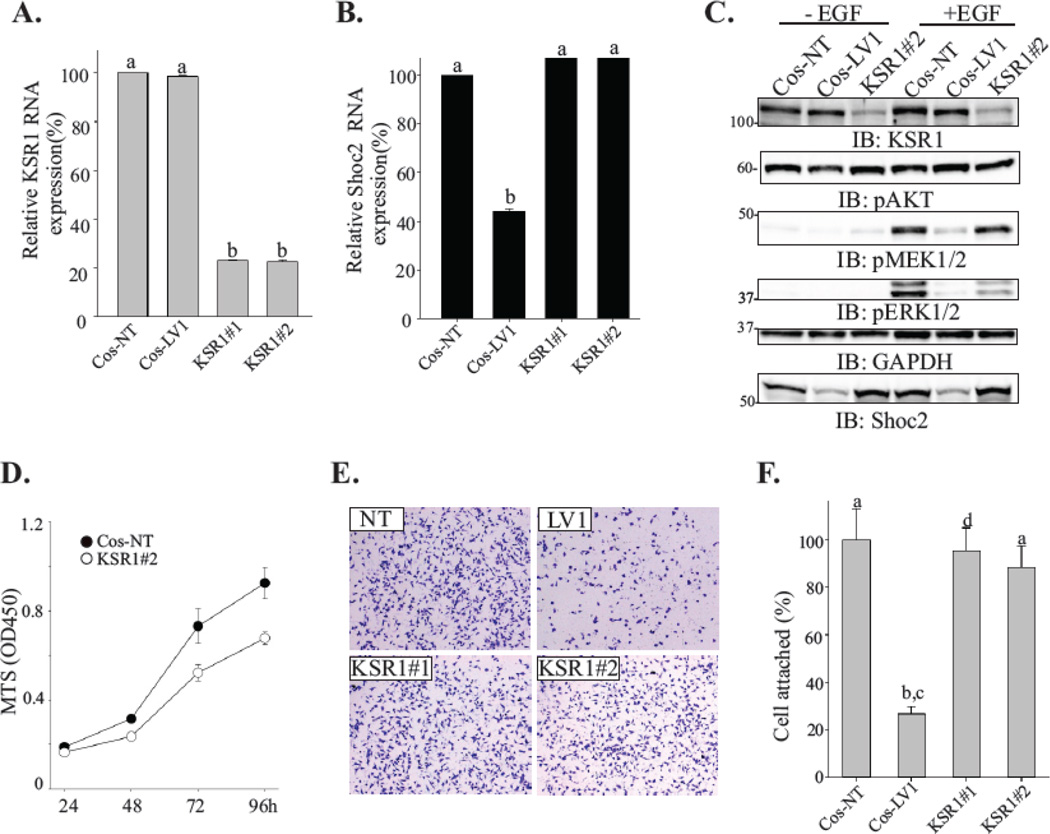
To test this notion, we examined whether depletion of another well-studied ERK1/2 scaffold, KSR1[8], affected cell attachment. Two different shRNA duplexes (#1 and #2) were utilized to generate cells constitutively depleted of KSR1. In these cells mRNA levels of KSR1 were reduced to about 20% of its endogenous expression (Cos-NT) (Fig. 2A). mRNA levels of Shoc2 were not affected (Fig. 2B). In order to prevent clonal variations due to different sites of viral genome incorporation, a pool population of shRNA-expressing Cos-KSR1 cells was used in subsequent experiments. Consistent with earlier reports, silencing of KSR1 attenuated ERK1/2 phosphorylation (Fig. 2C) leading to reduction in cell growth (Fig. 2D). We did not observe changes in AKT phosphorylation (Fig. 2C). Importantly, we found that attachment of KSR1-depleted cells was comparable to the control cells (Fig. 2E–F). Together, these data indicated that alterations in the ability of Shoc2-depleted cells to adhere were not due to the general attenuation of ERK1/2 activity but rather a result of a decrease in ERK1/2 signals mediated by the Shoc2 scaffold.
Figure 2. Depletion of KSR1 does not affect cell attachment.
(A, B) Total RNA was extracted from cells depleted of Shoc2 (LV1) or KSR1 (KSR#1 and KSR#2) and quantitative RT-PCR was performed using Shoc2 and KSR1-specific primers. Data are presented as the fold change of KSR1 (A) or Shoc2 (B) mRNA levels normalized to control (NT) (mean ± SD, n = 2; a vs. b, p<0.05).
(C) Cells were serum-starved for 18h and stimulated with 0.2 ng/mL EGF for 7 min. Expression of the indicated proteins was analyzed using specific antibodies. Representative blots are shown from multiple experiments.
(D) Equal numbers of cells constitutively expressing either KSR1 shRNA (open circle) or nontargeting shRNA (solid circle) were plated into a 96-well plate. The viability of cells was measured using a CellTiter 96 AQueous One solution cell proliferation assay. The mean number ± SD from triplicate experiments is shown.
(E) Cells were seeded on a collagen pre-coated 96-well plate for 10 min. Images of cells fixed and stained with crystal violet were obtained using a Nikon Eclipse E600 microscope.
(F) Cells from the experiments in E were solubilized with 2% SDS and subjected to the colorimetric absorbance measurement (OD550). Data from three independent experiments was analyzed and bars represent the mean values ± SD, n = 3; a vs. b, P < 0.05.
3.2. Transcriptome analysis identified differentially expressed genes controlling cell motility
Considerable effort devoted to characterization of genes of the immediate transcriptional response to EGF signaling identified a high redundancy of this response [43, 44]. Conversely, several other studies reported that the expression of numerous genes involved in regulation of cell motility, adhesion, and organization of actin cytoskeleton is induced during the late EGFmediated ERK1/2 transcriptional response [28, 31, 45]. Hence, to gain insight into the mechanisms responsible for the reduced motility and attachment of Shoc2-depleted cells, in the following studies we focused on the expression of the genes of the late (secondary) response.
To obtain a comprehensive understanding of the Shoc2-mediated ERK1/2 transcriptional response, we utilized comparative whole-genome profiling of cells depleted of Shoc2. RNA deep sequencing (RNA-seq) of Cos1 (NT vs. LV1) cells was performed. Cos1 cells were chosen as they presented the most dramatic changes in cell motility and attachment when Shoc2 was silenced (Fig. 1). mRNA isolated from the cells stimulated with EGF for 90 min were submitted for RNA-seq without RNA amplification to avoid any potential variances in transcripts. A flow chart of the experiment and RNA-seq analysis is shown in Suppl. Fig. 3A. As shown in Suppl. Fig. 3B, EGF induced a robust phospho-ERK1/2 response at 7 min of 0.2 ng/ml physiological response with only minimal phospho-ERK1/2 response in cells depleted of Shoc2. A summary of the RNA-seq data obtained in these experiments is shown in Suppl. Fig. 3C, D.
RNA-seq reads from Cos1 cells (African green monkey kidney) were aligned to the vervet reference genome (Chlorocebus sabaeus) ChiSab1.0 (GCA 000409795.1) (http://pre.ensembl.org/Chlorocebus_sabaeus/Info/Index) using TopHat (version 2.0.4) with the Ensembl pre-release gene annotation file. Interestingly, only 60% of the reads from each sample aligned to a transcript (Suppl. Fig. 3E), indicating that the genome transcript annotations are likely incomplete. Aligned RNA-seq reads were assembled and merged using TopHat/Cufflinks software package [46]. The number of mapped reads ranged from 32.3 million to 39.9 million per sample and resulted in 27,265 transcript identifiers from 23,709 unique gene regions. Taken together, this indicates both a depth and breadth of sequencing coverage allowing for comprehensive analysis of differentially regulated genes.
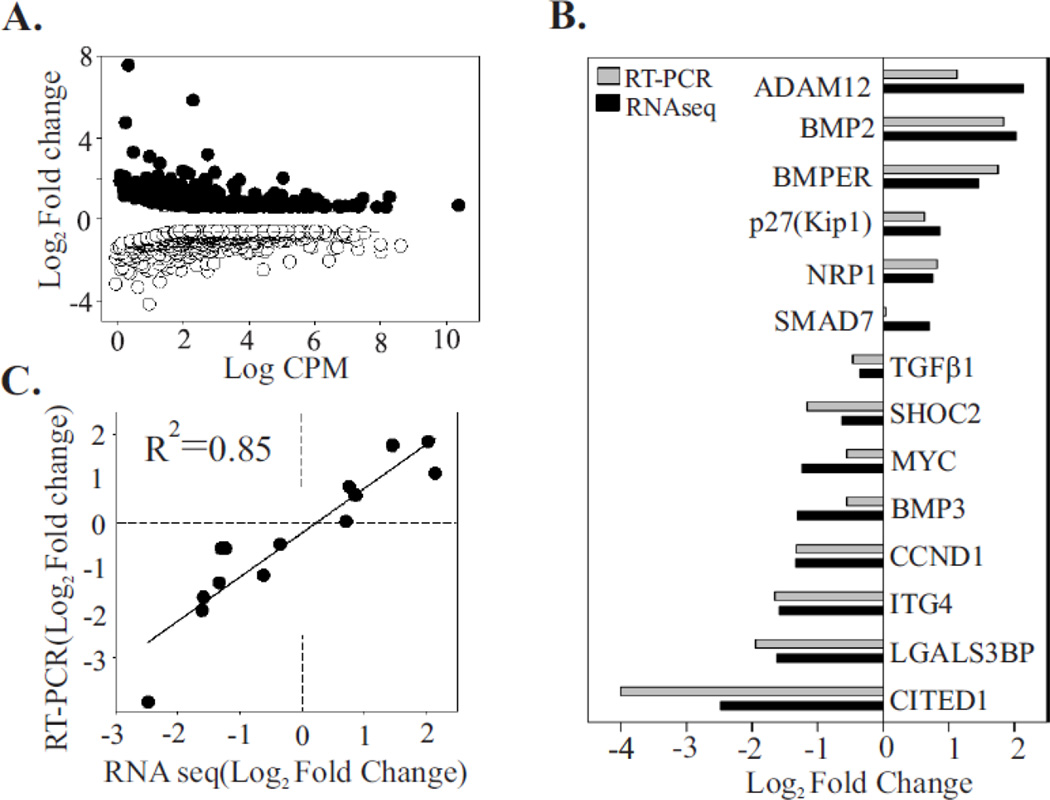
To identify differentially expressed genes, data was analyzed using Cufflinks and CuffDiff [47]. We found that ranking of the genes on the basis of a Log2fold change was somewhat misleading because no or very low transcript levels in Cos-LV1 led to very high rate of differential expression even if the transcript level in Cos-NT was very low. Thus, to rank differential expression in a more biologically meaningful way, we used a false discovery rate (FDR) < 0.05 and reads per kilobase of exon per million reads mapped (RPKM) ranking. 853 genes were ranked as differentially expressed, with 317 upregulated and 536 downregulated. The Log2fold changes for the obtained gene set are highlighted on an MA plot (Fig. 3A). A list of top 20 differentially expressed genes organized by intensity/RPKM can be found in [37]. Next, we selected 14 genes for additional confirmation by RT-PCR (Fig. 3B). The criteria for selection included >1.5-fold change and FDR < 0.05. These genes were also selected because of their relatively abundant gene expression levels. All of the analyzed genes showed consistent expression patterns (Log2fold change) between RNA-seq and RT-PCR analysis. A significant correlation (r2=0.85496, p<0.0001, Fig. 3C) between the Log2fold change in expression (relative to the control) of the RNA-seq and the RT-PCR data further confirmed accuracy of the results obtained with the RNA-seq data. Moreover, assessment of the gene expression in cells where endogenous Shoc2 was reconstituted with Shoc2-tRFP (Cos-SR) (Suppl. Fig. 4A) or in KSR1-depleted cells (KSR#2) (Suppl. Fig. 4B) showed no significant difference in mRNA levels of the target genes (relative to control), providing additional validation to our data and hypothesis.
Figure 3. Validation of the RNA-seq approach using RT-PCR.
(A) Differential gene expression between control sample (NT) and samples depleted of Shoc2 (LV1). The differential expression (log2fold change for LV1 samples compared to NT) is plotted against the log count per million for each gene. Each point represents a single ID of the reference transcriptome. Of 853 differentially expressed genes, 317 were upregulated (solid circle) and 536 were downregulated (open circle) (Fold change > 1.5, FDR <0.05).
(B) Fourteen differentially expressed genes were arbitrary selected from a range of upregulated and downregulated genes. Levels of expression were quantified by RT-PCR and the results were compared to those obtained by the RNA-seq approach. The log2fold change in gene expression of RT-PCR and RNA-seq approach were closely correlated (R2 = 0.85), indicating the accuracy of the RNA-seq analysis (C).
3.3. Depletion of Shoc2-mediated ERK1/2 signaling leading to the induction of alternate signaling pathways
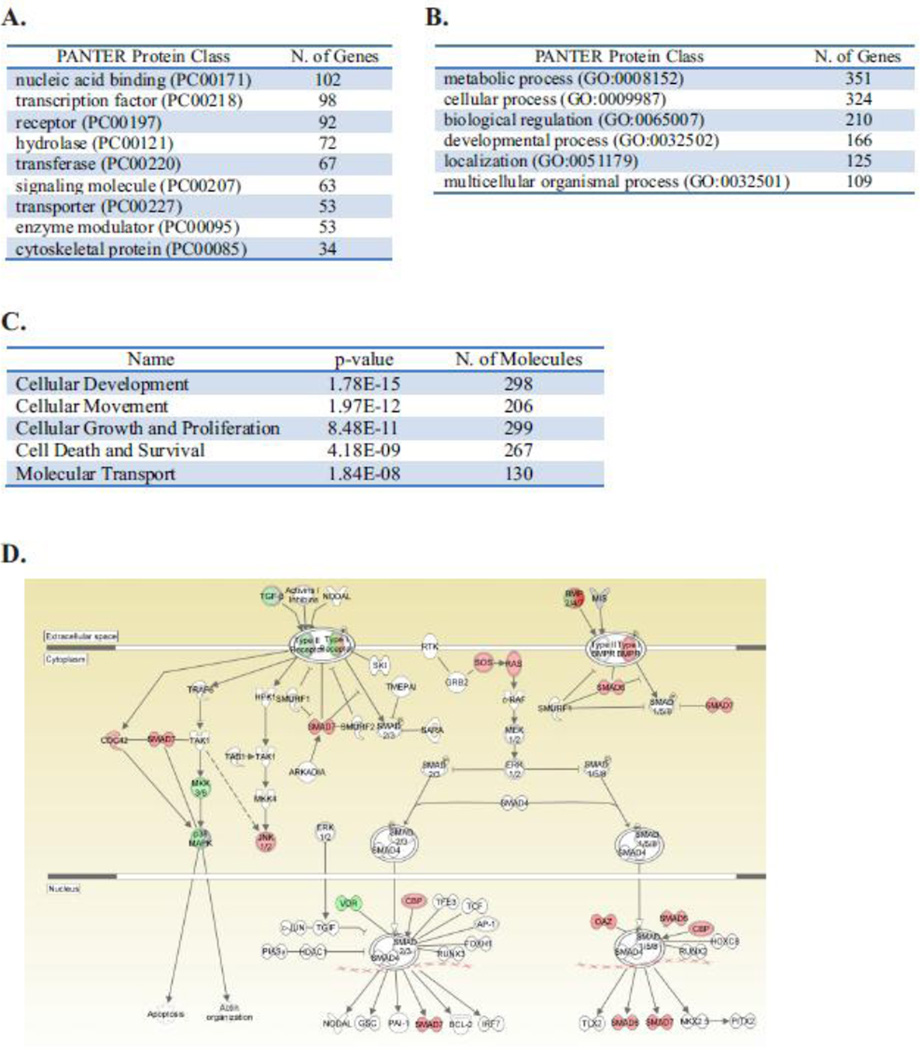
To allow for further identification of differentially expressed genes, data was annotated and analyzed by IPA Ingenuity™ and the Protein Analysis Through Evolutionary Relationships (PANTHER) resource [40]. Differentially expressed genes separated into 28 PANTHER protein classes with transcription factors, nucleic acid binders, signaling molecules and receptors comprising 32.3% of the differentially expressed genes (Fig. 4A and Table 12 in [37]). PANTHER analysis of significant gene ontology biological processes (GO BP) identified significant enrichment in several biological terms including “metabolic process” and “cellular process” (Fig. 4B and Table 13 in [37]). The comprehensive examination by IPA Ingenuity™ to analyze molecular and cellular functions also found that the differentially expressed genes are involved in cellular development, movement, cellular assembly and organization as well as cellular function and maintenance (Fig. 4C).
Figure 4. Biological processes associated with loss of Shoc2 in cells.
The top protein classes (A) and biological processes (B) of 853 differentially expressed genes (fold change >1.5, FDR <0.05) analyzed with PANTHER pathway analysis.
(C) Differentially expressed genes were analyzed by Ingenuity Pathway Analysis. The molecular and cellular functions identified by the analysis are presented.
(D) In-depth Ingenuity Pathway Analysis of differentially expressed genes revealed changes in regulators of the BMP/TGFβ pathway (red, upregulated expression; green, downregulated; gray, no-changed; white, not listed in A–C).
IPA pathway analysis of the differentially expressed transcripts was then used to examine changes in individual signaling modules. First, we analyzed the functional consequences of the loss of Shoc2 on transcriptional programs of ERK/MAPK signaling. Similarly to what described in our earlier studies [25, 33], no significant changes were found in the expression of genes of the ERK1/2 pathway upon loss of Shoc2 and no genes of the ERK/MAPK pathway showed a fold change ≥1.5 in expression (not shown). Further analysis of the genes exhibiting changes in transcription revealed that Shoc2 depletion altered the expression of genes of the BMP/TGFβ pathway such as SMAD6/SMAD7 (Log2fold =0.609), BMP2 (Log2fold =1.999), BMP3 (Log2fold = −1.038), BMP6 (Log2fold = −0.744) and chordin (Log2fold = −2.054) (Fig. 4D). This observed enrichment for the regulators of the TGFβ signaling pathway was consistent with reports of antagonism between ERK1/2 and TGFβ as regulating balance in the post-receptor phase of the pathway [48]. Interestingly, we also found alterations in the expression of growth factors such as TGFβ and BMP6/2/3, BDNF, FGF13, FGF5 and NRG3 in the Shoc2-knock down cells. These results indicate that loss of Shoc2 affected an important mechanism that balances signals through multiple signaling networks.
Next, we interrogated the response to a loss of Shoc2 in the expression of transcription factors and nucleic acid binding proteins. Differentially expressed transcription factors and proteins associated with transcription represented a large group of 171 genes (80 downregulated and 91 upregulated). Expression of transcription factors in the EGF-treated cells has been established by several studies [30, 44]. Therefore, we compared the results of our analysis to the data published by Amit et al., 2007 [30]. Surprisingly, very little overlap was found in identities of transcription factors in the two data sets. This may be due to the differences in the experimental and data analysis approaches of these studies (e.g. microarray analysis of HeLa cells vs. RNA-seq of Cos1 cells). Nevertheless, in addition to altered expression of the classic component of EGFR network v-Myc (Log2fold= −1.234), only four transcription factors were found in both sets of data (FOXC1, JUNB, KLF6, NAB2). All other transcription factors or proteins associated with transcription, in which expression changed in response to the loss of Shoc2, have not been previously reported to respond to EGFR activation (partial list is presented in Suppl. Table 1). Interestingly, IPA Ingenuity™ analysis of the upstream regulators found that similar changes in expression of transcription factors and proteins associated with transcription were observed when the TGFβ signaling pathway was affected. These results reveal novel pathway connectivity and may reflect on a role of Shoc2 in mediating ERK1/2 signals controlling activity of other signaling pathways.
3.4. Experimental validation of RNA-seq analysis predicts that expression of motility genes is affected by the Shoc2-mediated ERK1/2 signaling
Examination of cellular and molecular functions by IPA Ingenuity™ revealed an adhesion signature within the differentially expressed genes (ITG4, p= 3.60E-53, Neuropilin 1, p= 2.50E-37, CD97, p= 5.87E-15, and others; Suppl. Table 2), suggested a potential mechanism underlying changes in the motility of Shoc2-depelted cells. Therefore, we set to examine whether changes in gene expression identified by RNA-seq correlate with changes in protein synthesis using complementary biochemical methods.
For the experimental validation we chose the LGALS3BP due to its functional connection to focal adhesion assembly and cell migration [49]. LGALS3BP was top ranked using RPKM ranking with Log2fold= −1.613. Previous studies proposed that LGALS3BP docking of galectin 3 results in cross-linking and clustering of integrins on the cell surface [32]. In addition, LGALS3BP is a heavily glycosylated secreted molecule that has been implicated in the tumor metastatic process of breast cancers [50, 51]. LGALS3BP is associated with shorter survival in patients with breast carcinoma [52] and several other types of cancers [53].
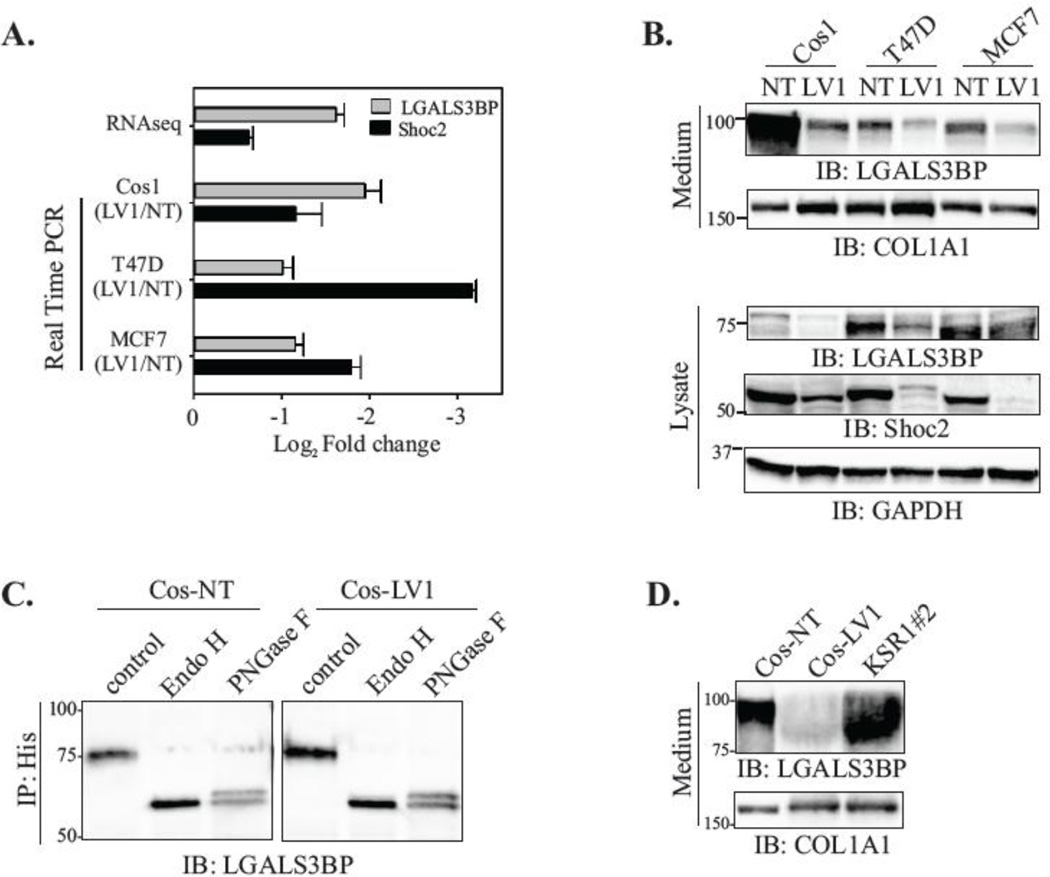
First, we utilized RT-PCR analysis to validate that loss of Shoc2 results in decreased RNA expression of LGALS3BP in T47D, Cos1 and MCF7 cells (Fig. 5A). We then examined whether protein expression of LGALS3BP was affected by silencing of Shoc2. The reduced LGALS3BP mRNA expression corresponded to the decreased intracellular protein expression (Fig. 5B, lysate). Moreover, we found a dramatic decrease in the LGALS3BP secretion to the culture media (Fig. 5B, medium). To validate that reduced secretion of LGALS3BP is not due to deficiencies in the glycosylation of LGALS3BP, we expressed His-tagged LGALS3BP in cells depleted of Shoc2 and then examined glycosylation of the intracellular His-LGALS3BP. His- LGALS3BP was immunoprecipitated from control and Shoc2-depleted cells and then digested with either Endoglycosidase H (Endo H) to remove high mannose N-glycans or with Peptide -N-Glycosidase F (PNGase F) to remove the high mannose, hybrid, and complex N-glycans from LGALS3BP [54]. We found that glycosylation of His-LGALS3BP expressed in Cos-LV1 cells was comparable to the glycosylation of His-LGALS3BP expressed in Cos-NT cells (Fig. 5C), indicating that deficiencies in secretion of LGALS3BP are not due to alterations in LGALS3BP post-translational processing. The secretion of LGALS3BP in KSR1-depleted cells was not affected (Fig. 5D). Together, these results suggest that ERK1/2 activity mediated by Shoc2 is necessary to induce transcription of LGALS3BP.
Figure 5. Shoc2 silencing results in reduced expression and secretion of LGALS3BP.
(A) The expression levels of LGALS3BP were quantified in Cos1, T47D and MCF7 cells depleted of Shoc2 using RT-PCR analysis and results were compared to those obtained by the RNA-seq approach. Data are presented as the log2fold change of the Shoc2 mRNA levels in Shoc2-depleted cells normalized to control (NT) (mean ± SD; n = 3).
(B) Cos1, T47D, and MCF7 cells depleted of Shoc2 (LV1) and control cells (NT) were harvested for immunoblotting. The expression of LGALS3BP in culture medium and cell lysate was analyzed using specific antibodies. Other indicated proteins were analyzed using specific antibodies.
(C) Cos-NT and Cos-LV1 cells were transiently transfected with His-LGALS3BP. At 48h post-transfection, His-LGALS3BP was immunoprecipitated using anti-His antibodies and then digested by Endoglycosidase H or Peptide -N-Glycosidase F (Endo H or PNGase F). Immunoprecipitates were analyzed by immunoblotting using anti-LGALS3BP antibody.
(D) Cos1 cells depleted of KSR1 (KSR1#2) and control cells (NT and LV1) were harvested for immunoblotting. LGALS3BP in culture medium was analyzed using specific anti-LGALS3BP antibodies.
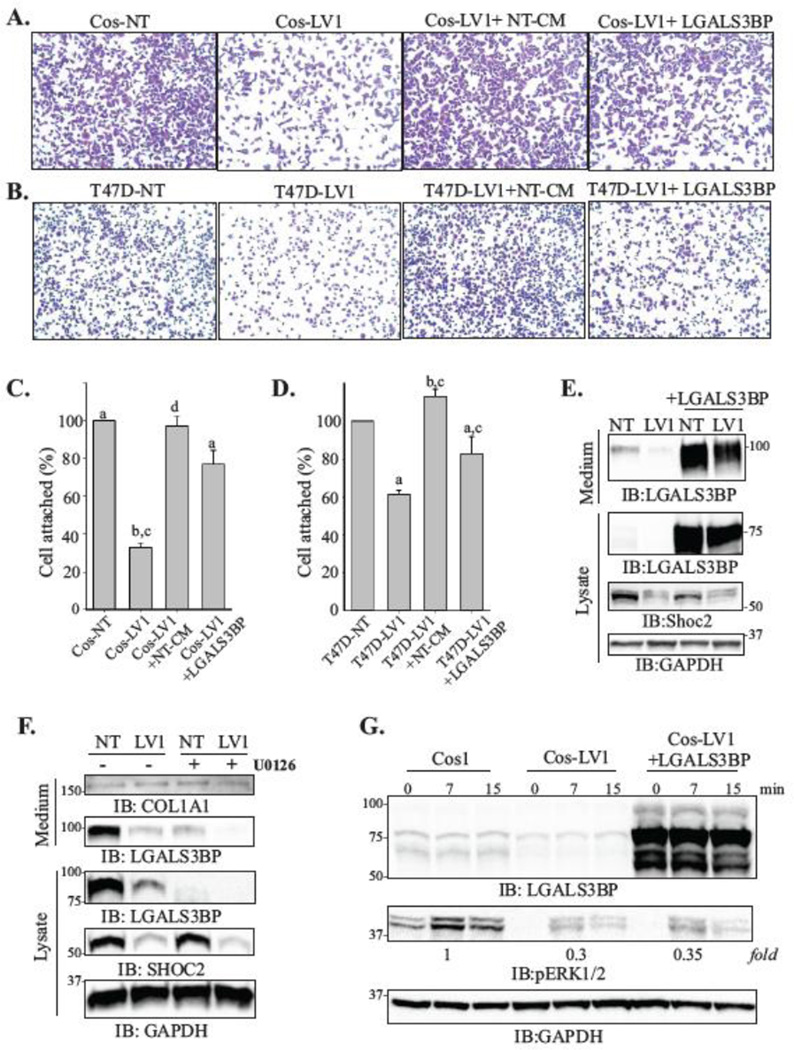
Next, to address whether deficient attachment of the Shoc2-depleted cells (LV1) is due to the decreased levels of LGALS3BP secretion, we examined attachment of Cos-LV1 and T47D-LV1 cells in the presence of the conditional media of control cells (LV1+NT-CM). We found that when the assay was performed in the presence of the culture media collected from NT cells, attachment of LV1 cells was restored to the rates comparable to those of cells expressing endogenous levels of Shoc2 (Fig. 6A–D, LV1+NT-CM). However, when cell attachment of the Shoc2 depleted (LV1) cells was examined in the presence of the culture media collected from cells lacking Shoc2 (LV1+ LV1-CM), we did not observe rescue in attachment of these cells (Suppl. Fig. 5A, B).
Figure 6. LGALS3BP rescues attachment of cells depleted of Shoc2.
(A, B) Cos1 (A) and T47D (B) cells (5×104 cells) depleted of Shoc2 (LV1) were seeded on collagen pre-coated 96-well for 10 min. LV1+NT-CM samples were seeded in presence of cultured media from the control cells. LV1+LGALS3BP cells were transiently transfected with His-LGALS3BP 48h prior to seeding. Images of cells fixed and stained with crystal violet were obtained using Nikon Eclipse E600 microscope.
(C, D) Cells from the experiments in A and B were then solubilized with 2% SDS and subjected to the colorimetric absorbance measurement (OD550). Data from three independent experiments exemplified in A and B was analyzed (bars represent mean values ± SD; n = 3; a vs. b; c vs. d, p<0.05).
(E) Cos-NT and Cos-LV1 were transiently transfected with His-LGALS3BP. At 48h post-transfection, expression of the indicated proteins in the lysate and secretion media was analyzed using specific antibodies.
(F) Cos-NT and Cos-LV1 cells were treated with UO126 (10 uM) or DMSO for 24h at 37°C. The expression of indicated proteins in the lysate and secretion media was analyzed using specific antibodies. The data are representative of three independent experiments.
(G) Cells were transiently transfected with His-LGALS3BP. At 48h post-transfection, the cells were serum starved for 18h and stimulated with EGF (0.2 ng/ml) for 7 or 15 min. Expression of the indicated proteins was analyzed using specific antibodies.
To directly examine whether changes in transcription of LGALS3BP led to the altered attachment of Shoc2-depleted cells, we transiently expressed His-LGALS3BP. As expected, ectopic expression of LGALS3BP restored secretion of LGALS3BP to the cell media (Fig. 6E) and the attachment of Cos-LV1 and T47D-LV1 cells (Fig. 6A–D, LV1+LGALS3BP).
To further validate the role of the ERK1/2 pathway in controlling expression of LGALS3BP, we used MEK1/2 inhibitors U0126 and PD98059. These inhibitors block the transfer of phosphate by MEK1/2 to ERK1/2, thus resulting in inhibition of the MEK kinase activity toward ERK [55]. Reduced phosphorylation of ERK1/2 observed in EGF-stimulated cells treated with UO126 and PD98059 (Suppl. Fig. 5C, E) affected the mRNA levels of LGALS3BP and other target genes (Suppl. Fig. 5D, F). Importantly, inhibition of MEK1/2 activity resulted in the dramatic reduction in the levels of the secreted LGALS3BP (Fig. 6F). These data indicate that the MEK-ERK kinase activity is required for the efficient LGALS3BP expression and secretion.
We next examined whether ectopic expression of LGALS3BP in cells affects ERK1/2 phosphorylation. No significant change in the levels of phospho-ERK1/2 in cells transiently expressing LGALS3BP was detected (Fig. 6G). Moreover, we did not observe changes in phosphorylation of FAK (not shown), suggesting restored adhesion of Cos-LV1 and T47D-LV1 cells expressing LGALS3BP is not due to the increased “inside-out” ERK1/2 signal, but is a consequence of restored LGALS3BP secretion. Together, this data not only validate the hypothesis that Shoc2-ERK1/2 signals control expression of LGALS3BP, but also suggest that deficiencies in adhesion of cells depleted of Shoc2 are the result of LGALS3BP decreased expression.
4. Discussion
Diverse cellular outcomes triggered by the ERK1/2 signaling cascade include modulation of cell cycle, proliferation, transcription, migration, and senescence, to name a few. Scaffold proteins have been suggested to offer conduits that convey a multipotent general ERK signal into a specific cellular outcome [6, 56]. However, our understanding of how signals are directed by the individual ERK1/2 scaffolds is still limited. The findings presented in this study provide sufficient evidence for the role of Shoc2 scaffold as a regulator of ERK1/2 signals to cell motility and attachment. This study shows that altered cell motility upon Shoc2 depletion is an effect of robust changes in transcription and establishes the existence of the ERK1/2/Shoc2-dependent signaling axis that regulates the expression of the extracellular matrix protein LGALS3BP. Our data support the exciting notion that the ERK1/2 signal generated by EGFR receptors is routed by the Shoc2 scaffold proteins to initiate cell attachment and motility. The notion that Shoc2 modulates ERK1/2 signals to cell motility is reinforced by the following findings: (i) constitutive depletion of Shoc2 reduced attachment and motility rates of multiple breast carcinoma cells to collagen I (Fig. 1); (ii) constitutive depletion of another ERK1/2 scaffold, KSR1, had no effect on cell motility attachment (Fig. 2); and (iii) expression of numerous genes controlling cell adhesion and motility was dramatically affected by Shoc2 silencing (Fig. 3).
Activation of an individual receptor tyrosine kinase controls multiple downstream outputs and transcriptional programs, and several studies have characterized total transcriptional ERK1/2 response in neurons, 3T3 fibroblasts or mouse embryonic palatal mesenchyme cells [43, 57, 58]. The RNA-seq analysis of cells depleted of Shoc2 provides insight into differences in gene expression following loss of the signaling scaffold, with the added benefit of producing a comprehensive list of total gene expression of Cos1 cells. Silencing of Shoc2 led to altered expression of a surprisingly large number of genes (Fig. 4). Validation of selected transcripts covering a wide range of expression by RT-PCR showed good agreement with RNA-seq data and demonstrated the feasibility of this approach. We found that Shoc2-ERK1/2 signals contribute to regulating the expression of several protein classes. Moreover, our data significantly expands a list of transcription factors and proteins associated with transcription stimulated by EGF-ERK1/2 activity (see supplemental data).
Finding altered expression of genes of the TGFβ pathway was somewhat surprising. While examples of ERK1/2 and TGFβ pathway cross-talk have been documented [59–61], there has been no direct evidence that ERK1/2 signals control expression of SMAD6 and SMAD7. Given the diverse molecular functions of the differentially expressed genes, some of the observed changes in gene expression may be due, at least in part, to the compensatory/indirect response of the signaling network or even alterations in mRNA stability. Nevertheless, our findings emphasize the significance of Shoc2 in controlling ERK1/2 signaling and open numerous questions that need to be addressed in future studies. Further studies are also needed to address whether Shoc2-ERK1/2 signals are involved in phosphorylation of genes in the immediate response and changes in the earlier waves of transcription. Our analysis, therefore, revealed an unanticipated complexity of kinase/scaffold-mediated regulation of the signaling pathway and uncovered a new link between EGFR signaling and transcription.
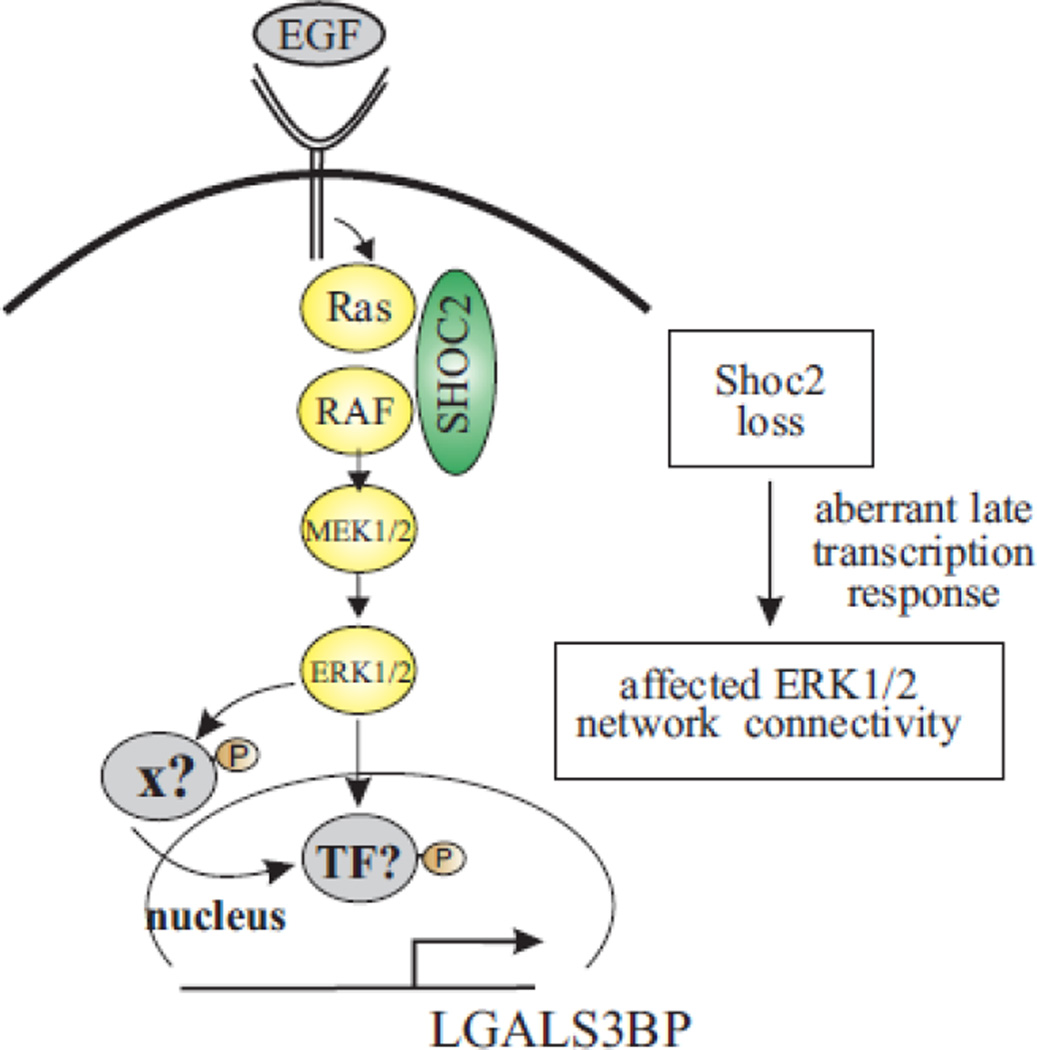
One important point worth mentioning is that loss of Shoc2 results in a dramatic decrease in expression of the protein of extracellular matrix, LGALS3BP. We found that the deficiencies in the attachment of cells depleted of Shoc2 are, to a large extent, due to aberrant expression and secretion of LGALS3BP (Fig. 5), and that ectopic expression of LGALS3BP rescues these deficiencies (Fig. 6). These results are in line with the notion that LGALS3BP induces clustering of integrins and recruitment of actin is needed for initiation of cell motility [49, 62]. Interestingly, ectopic expression of LGALS3BP rescued its secretion, but had no effect on the ERK1/2 phosphorylation in cells depleted of Shoc2 (Fig. 6), indicating that traditional “outsidein” activation of the ERK1/2 pathway was not affected by the transient expression of LGALS3BP. This result is also consistent with our observations of Shoc2 silencing having no effect on activity of FAK as well (not shown). Considering earlier reports of adhesion blockage as a result of the inhibited interaction of LGALS3BP-integrin β1 [49], it is possible that changes in cell attachment observed in our study are due to alterations in the integrin β1-LGALS3BPcollagen link. Further studies will decipher the molecular mechanism of this connection. Based on the currently available information, we propose the following model (Fig. 7). EGF-induced phosphorylation of the ERK1/2 pathway undergoes the check-point at the level of the Shoc2 scaffold. ERK1/2 then phosphorylates transcription factor that activates expression of LGALS3BP. It is also conceivable that ERK1/2 signals may be directed to inactivate a transcription repressor or intermediate co-transcriptional activator. Finding the factor controlling the expression of LGALS3BP mRNA would be the next logical step following the current study. Of note, the core promoter region of LGALS3BP was identified, and transcription factors AP1, AP2 and progesterone receptor were suggested to bind to the core promoter of LGALS3BP [63–65]. Our RNA-seq analyses, however, have not yielded changes in the expression of the transcription factors mentioned in these studies. Further functional analysis is needed to infer a plausible candidate. Additional research would also be necessary to address roles of other identified targets in controlling the Shoc2/ERK1/2-mediated cell motility. How various proteins identified in our RNA-seq screen are connected is yet to be defined and need to be studied.
Figure 7.
Schematic model recapitulating the role Shoc2 plays in controlling ERK1/2 activity. Shoc2 routes signals of the ERK1/2 pathway activated by EGFR for transcription of late response genes. Loss of Shoc2-mediated ERK1/2 signals affects crosstalk with other signaling pathways and transcription of various substrates, including LGALS3BP.
Elevated levels of LGALS3BP are found in many solid human cancers including breast, lung, prostatic, melanoma and colon cancers [52, 66–68]. These elevated levels are associated with poor prognosis or advanced stages of disease [53]. LGALS3BP has also been reported to have a proangiogenic effect in human breast cancers [51, 62]. Future studies, with respect to the Shoc2-ERK1/2 role in controlling expression and secretion of LGALS3BP, would be important for understanding the biology of solid tumors that present with increased levels of LGALS3BP. Such studies will lead to a better understanding of the pathology as well as to new possibilities for therapeutic interventions for aggressive cancers. It may also have implications for other type of cancers for which LGALS3BP is a prognostic marker [69, 70].
In this study we demonstrate that Shoc2 transduces signals specific to cellular programs including motility and attachment. Although scaffolds of the ERK1/2 pathway (i.e. calmodulin regulated actin binding protein with a GAP-like domain, IQGAP and mp1/p14 complex) have been suggested to regulate cell motility, their contributions to the process are distinct. IQGAP facilitates retention of Cdc42 and Rac in their GTP-bound states and cross-link the actin filament system with microtubules [15, 71]. Mp1/p14 was reported to be essential for focal adhesion dynamics [13]. Our work identifies that Shoc2 scaffold controls cell motility, in part, via regulating expression of the extracellular matrix protein, strongly supporting the model that scaffold proteins can function as gatekeepers of the multi-potent ERK1/2 signaling on the levels of the late transcriptional response.
In conclusion, these studies are the first report to identify LGALS3BP as a novel target of ERK1/2 signaling pathway and provide novel insights into the function of the scaffold protein Shoc2. Our data is also further supports the notion that scaffolds monitor the specificity of the cellular outcomes as well as the notion that ERK1/2 activity via separate scaffolding modules controls non-overlapping cellular responses.
Supplementary Material
Highlights.
Depletion of the Shoc2 scaffold attenuates cell motility and adhesion
Shoc2-mediated ERK1/2 signals control expression of multiple proteins, including LGALS3BP
Ectopic expression of LGALS3BP rescues adhesion deficiency of the Shoc2-depleted cells.
Acknowledgments
We thank Drs. Matthew Gentry, Tianyan Gao, Charles Waechter and Stacy Smith for providing reagents and critical reading of the manuscript; the Genetic Technologies Core at the Department of Molecular and Cellular Biochemistry (University of Kentucky) for assistance with the production of lentiviruses; the UK Flow Cytometry & Cell Sorting core facility for assistance in cell sorting.
The Genetic Technologies and Protein cores mentioned above are supported in part by a grant from the National Institute of General Medical Sciences (P20GM103486). The UK Flow Cytometry & Cell Sorting core facility is supported in part by the Office of the Vice President for Research, the Markey Cancer Center and an NCI Center Core Support Grant (P30CA177558) to the University of Kentucky Markey Cancer Center. Bioinformatics support for this work provided by National Institutes of Health (NIH) grants P20GM103436 (Nigel Cooper, PI).
This project was supported by grants from the National Cancer Institute (R00CA126161 to EG), the National Institute of General Medical Sciences (P20GM103486) (formally supported by the Center for Research Resources), the National Institute of General Medical Sciences (R01GM113087 to EG) and from the American Cancer Society (RSG-14-172-01-CSM to EG). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or the NIGMS.
Abbreviations
- EGF
epidermal growth factor
- ERK1/2
extracellular signal-regulated kinase 1 and 2
- KSR1
kinase suppressor of Ras (KSR1)
- MP1
mitogen-activated protein 1
- PNGase F
Peptide-N-Glycosidase F
- Endo H
Endoglycosidase H
- PP1c
protein phosphatase 1c
- qPCR
real-time quantitative polymerase chain reaction
- RPKM
reads per kilobase of exon per million reads mapped
- TGFβ
transforming growth factor beta
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
None
Author contributions
EG and MKJ planned the experiments, analyzed and interpreted data. MKJ and ERJ performed all biochemical assays. ER, JL, CW and WL performed Biostatistical analysis. EG and MKJ drafted the manuscript.
References
- 1.Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. The Biochemical journal. 2000;351(Pt 2):289–305. [PMC free article] [PubMed] [Google Scholar]
- 2.Pearson G, et al. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 3.Brown MD, Sacks DB. Protein scaffolds in MAP kinase signalling. Cellular signalling. 2009;21:462–469. doi: 10.1016/j.cellsig.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown MD, Sacks DB. Compartmentalised MAPK pathways. Handbook of experimental pharmacology. 2008:205–235. doi: 10.1007/978-3-540-72843-6_9. [DOI] [PubMed] [Google Scholar]
- 5.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nature reviews. Molecular cell biology. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 6.Langeberg LK, Scott JD. Signalling scaffolds and local organization of cellular behaviour. Nat Rev Mol Cell Biol. 2015;16:232–244. doi: 10.1038/nrm3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, et al. The dual function of KSR1: a pseudokinase and beyond. Biochem Soc Trans. 2013;41:1078–1082. doi: 10.1042/BST20130042. [DOI] [PubMed] [Google Scholar]
- 8.Pérez-Rivas LG, Prieto S, Lozano J. Modulation of Ras signaling by the KSR family of molecular scaffolds. 2010:10–23. [Google Scholar]
- 9.McKay MM, Ritt DA, Morrison DK. Signaling dynamics of the KSR1 scaffold complex. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11022–11027. doi: 10.1073/pnas.0901590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin J, et al. KSR1 modulates the sensitivity of mitogen-activated protein kinase pathway activation in T cells without altering fundamental system outputs. Molecular and cellular biology. 2009;29:2082–2091. doi: 10.1128/MCB.01634-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claperon A, Therrien M. KSR and CNK: two scaffolds regulating RAS-mediated RAF activation. Oncogene. 2007;26:3143–3158. doi: 10.1038/sj.onc.1210408. [DOI] [PubMed] [Google Scholar]
- 12.Sparber F, et al. The late endosomal adaptor molecule p14 (LAMTOR2) represents a novel regulator of Langerhans cell homeostasis. Blood. 2014;123:217–227. doi: 10.1182/blood-2013-08-518555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiefermeier N, et al. The late endosomal p14-MP1 (LAMTOR2/3) complex regulates focal adhesion dynamics during cell migration. J Cell Biol. 2014;205:525–540. doi: 10.1083/jcb.201310043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thauerer B, et al. LAMTOR2-mediated modulation of NGF/MAPK activation kinetics during differentiation of PC12 cells. PLoS One. 2014;9:e95863. doi: 10.1371/journal.pone.0095863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pullikuth AK, Catling AD. Scaffold mediated regulation of MAPK signaling and cytoskeletal dynamics: a perspective. Cellular signalling. 2007;19:1621–1632. doi: 10.1016/j.cellsig.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pullikuth A, et al. The MEK1 scaffolding protein MP1 regulates cell spreading by integrating PAK1 and Rho signals. Molecular and cellular biology. 2005;25:5119–5133. doi: 10.1128/MCB.25.12.5119-5133.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sieburth DS, Sun Q, Han M. SUR-8, a conserved Ras-binding protein with leucine-rich repeats, positively regulates Ras-mediated signaling in C. elegans. Cell. 1998;94:119–130. doi: 10.1016/s0092-8674(00)81227-1. [DOI] [PubMed] [Google Scholar]
- 18.Selfors LM, et al. soc-2 encodes a leucine-rich repeat protein implicated in fibroblast growth factor receptor signaling. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6903–6908. doi: 10.1073/pnas.95.12.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi J, et al. Endothelial SUR-8 acts in an ERK-independent pathway during atrioventricular cushion development. Developmental dynamics : an official publication of the American Association of Anatomists. 2010;239:2005–2013. doi: 10.1002/dvdy.22343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cordeddu V, et al. Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nature genetics. 2009;41:1022–1026. doi: 10.1038/ng.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hannig V, et al. A Novel SHOC2 Variant in Rasopathy. Hum Mutat. 2014 doi: 10.1002/humu.22634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshiki S, et al. Ras and calcium signaling pathways converge at Raf1 via the Shoc2 scaffold protein. Mol Biol Cell. 2010;21:1088–1096. doi: 10.1091/mbc.E09-06-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez-Viciana P, et al. A phosphatase holoenzyme comprised of Shoc2/Sur8 and the catalytic subunit of PP1 functions as an M-Ras effector to modulate Raf activity. Mol Cell. 2006;22:217–230. doi: 10.1016/j.molcel.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 24.Jang ER, et al. HUWE1 is a molecular link controlling RAF-1 activity supported by the Shoc2 scaffold. Mol Cell Biol. 2014 doi: 10.1128/MCB.00811-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeoung M, et al. Functional Integration of the Conserved Domains of Shoc2 Scaffold. PloS one. 2013;8:e66067. doi: 10.1371/journal.pone.0066067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young LC, et al. An MRAS, SHOC2, and SCRIB Complex Coordinates ERK Pathway Activation with Polarity and Tumorigenic Growth. Mol Cell. 2013;52:679–692. doi: 10.1016/j.molcel.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Jang E, et al. Spatial control of Shoc2 scaffold-mediated ERK1/2 signaling requires remodeling activity of the ATPase PSMC5. Journal of cell science. 2015 doi: 10.1242/jcs.177543. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nature reviews. Molecular cell biology. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 29.Katz M, Amit I, Yarden Y. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochimica et biophysica acta. 2007;1773:1161–1176. doi: 10.1016/j.bbamcr.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amit I, et al. A module of negative feedback regulators defines growth factor signaling. Nature genetics. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- 31.Katz M, et al. A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell migration. Nature cell biology. 2007;9:961–969. doi: 10.1038/ncb1622. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki T, et al. Mac-2 binding protein is a cell-adhesive protein of the extracellular matrix which self-assembles into ring-like structures and binds beta1 integrins, collagens and fibronectin. EMBO J. 1998;17:1606–1613. doi: 10.1093/emboj/17.6.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galperin E, Abdelmoti L, Sorkin A. Shoc2 is targeted to late endosomes and required for Erk1/2 activation in EGF-stimulated cells. PloS one. 2012;7:e36469. doi: 10.1371/journal.pone.0036469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 35.Chan BM, et al. Adhesion to vascular cell adhesion molecule 1 and fibronectin. Comparison of alpha 4 beta 1 (VLA-4) and alpha 4 beta 7 on the human B cell line JY. J Biol Chem. 1992;267:8366–8370. [PubMed] [Google Scholar]
- 36.John Wiley & Sons. Current protocols. New York, N.Y.: John Wiley & Sons; 1990. [Google Scholar]
- 37.Rouchka E, et al. Data set for transcriptional response to depletion of the Shoc2 scaffolding protein. Data in Brief. doi: 10.1016/j.dib.2016.03.012. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaduwal S, et al. Sur8/Shoc2 promotes cell motility and metastasis through activation of Ras-PI3K signaling. Oncotarget. 2015 doi: 10.18632/oncotarget.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeoung M, Galperin E. Visualizing of signaling proteins on endosomes utilizing knockdown and reconstitution approach. Methods Enzymol. 2014;534:47–63. doi: 10.1016/B978-0-12-397926-1.00003-2. [DOI] [PubMed] [Google Scholar]
- 43.Vasudevan HN, et al. Receptor tyrosine kinases modulate distinct transcriptional programs by differential usage of intracellular pathways. Elife. 2015;4 doi: 10.7554/eLife.07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fambrough D, et al. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell. 1999;97:727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 45.Kostler WJ, et al. Epidermal growth-factor-induced transcript isoform variation drives mammary cell migration. PLoS One. 2013;8:e80566. doi: 10.1371/journal.pone.0080566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 49.Stampolidis P, Ullrich A, Iacobelli S. LGALS3BP, lectin galactoside-binding soluble 3 binding protein, promotes oncogenic cellular events impeded by antibody intervention. Oncogene. 2015;34:39–52. doi: 10.1038/onc.2013.548. [DOI] [PubMed] [Google Scholar]
- 50.Tinari N, et al. High expression of 90K (Mac-2 BP) is associated with poor survival in node-negative breast cancer patients not receiving adjuvant systemic therapies. Int J Cancer. 2009;124:333–338. doi: 10.1002/ijc.23970. [DOI] [PubMed] [Google Scholar]
- 51.Piccolo E, et al. LGALS3BP, lectin galactoside-binding soluble 3 binding protein, induces vascular endothelial growth factor in human breast cancer cells and promotes angiogenesis. J Mol Med (Berl) 2013;91:83–94. doi: 10.1007/s00109-012-0936-6. [DOI] [PubMed] [Google Scholar]
- 52.Iacobelli S, et al. Prognostic value of a novel circulating serum 90K antigen in breast cancer. Br J Cancer. 1994;69:172–176. doi: 10.1038/bjc.1994.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grassadonia A, et al. 90K (Mac-2 BP) and galectins in tumor progression and metastasis. Glycoconj J. 2004;19:551–556. doi: 10.1023/B:GLYC.0000014085.00706.d4. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y, et al. Regulation of Mac-2BP secretion is mediated by its N-glycan binding to ERGIC-53. Glycobiology. 2013;23:904–916. doi: 10.1093/glycob/cwt027. [DOI] [PubMed] [Google Scholar]
- 55.Favata MF, et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 56.Ramos JW. The regulation of extracellular signal-regulated kinase (ERK) in mammalian cells. Int J Biochem Cell Biol. 2008;40:2707–2719. doi: 10.1016/j.biocel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 57.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 58.Yamamoto T, et al. Continuous ERK activation downregulates antiproliferative genes throughout G1 phase to allow cell-cycle progression. Curr Biol. 2006;16:1171–1182. doi: 10.1016/j.cub.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 59.Gui T, et al. The Roles of Mitogen-Activated Protein Kinase Pathways in TGF-beta- Induced Epithelial-Mesenchymal Transition. J Signal Transduct. 2012;2012:289243. doi: 10.1155/2012/289243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayashida T, Decaestecker M, Schnaper HW. Cross-talk between ERK MAP kinase and Smad signaling pathways enhances TGF-beta-dependent responses in human mesangial cells. FASEB J. 2003;17:1576–1578. doi: 10.1096/fj.03-0037fje. [DOI] [PubMed] [Google Scholar]
- 61.Miyazono K. Positive and negative regulation of TGF-beta signaling. J Cell Sci. 2000;113(Pt 7):1101–1109. doi: 10.1242/jcs.113.7.1101. [DOI] [PubMed] [Google Scholar]
- 62.Traini S, et al. Inhibition of tumor growth and angiogenesis by SP-2, an anti-lectin, galactoside-binding soluble 3 binding protein (LGALS3BP) antibody. Mol Cancer Ther. 2014;13:916–925. doi: 10.1158/1535-7163.MCT-12-1117. [DOI] [PubMed] [Google Scholar]
- 63.Brakebusch C, et al. Expression of the 90K immunostimulator gene is controlled by a promoter with unique features. J Biol Chem. 1997;272:3674–3682. doi: 10.1074/jbc.272.6.3674. [DOI] [PubMed] [Google Scholar]
- 64.Brakebusch C, et al. Isolation and functional characterization of the human 90K promoter. Genomics. 1999;57:268–278. doi: 10.1006/geno.1999.5760. [DOI] [PubMed] [Google Scholar]
- 65.Grassadonia A, et al. Upstream stimulatory factor regulates constitutive expression and hormonal suppression of the 90K (Mac-2BP) protein. Endocrinology. 2007;148:3507–3517. doi: 10.1210/en.2007-0024. [DOI] [PubMed] [Google Scholar]
- 66.Nonaka M, et al. Dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN) recognizes a novel ligand, Mac-2-binding protein, characteristically expressed on human colorectal carcinomas. J Biol Chem. 2011;286:22403–22413. doi: 10.1074/jbc.M110.215301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ulmer TA, et al. The tumor-associated antigen 90K/Mac-2-binding protein secreted by human colon carcinoma cells enhances extracellular levels of promatrilysin and is a novel substrate of matrix metalloproteinases-2, -7 (matrilysin) and-9: Implications of proteolytic cleavage. Biochim Biophys Acta. 2010;1800:336–343. doi: 10.1016/j.bbagen.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 68.Becker R, et al. Tumor stroma marker endosialin (Tem1) is a binding partner of metastasis-related protein Mac-2 BP/90K. FASEB J. 2008;22:3059–3067. doi: 10.1096/fj.07-101386. [DOI] [PubMed] [Google Scholar]
- 69.Piccolo E, et al. Prognostic relevance of LGALS3BP in human colorectal carcinoma. J Transl Med. 2015;13:248. doi: 10.1186/s12967-015-0606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Artini M, et al. Elevated serum levels of 90K/MAC-2 BP predict unresponsiveness to alpha-interferon therapy in chronic HCV hepatitis patients. J Hepatol. 1996;25:212–217. doi: 10.1016/s0168-8278(96)80076-6. [DOI] [PubMed] [Google Scholar]
- 71.Mataraza JM, et al. Multiple proteins mediate IQGAP1-stimulated cell migration. Cellular signalling. 2007;19:1857–1865. doi: 10.1016/j.cellsig.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.