Abstract
Background
Ancestry informative markers (AIMs) measure genetic admixtures within an individual beyond self-reported racial/ethnic (SRR) groups. Here, we used genetically determined ancestry (GDA) across SRR groups and examine associations between GDA and HIV-1 RNA and CD4+ counts in HIV-positive children in the US.
Methods
41 AIMs, developed to distinguish 7 continental regions, were detected by real-time-PCR in 994 HIV-positive, antiretroviral naïve children. GDA was estimated comparing each individual’s genotypes to allele frequencies found in a large set of reference individuals originating from global populations using STRUCTURE. The means of GDA were calculated for each category of SRR. Linear regression was used to model GDA on CD4+ count and log10 RNA, adjusting for SRR and age.
Results
Subjects were 61% Black, 25% Hispanic, 13% White and 1.3% Unknown. The mean age was 2.3 years (45% male), mean CD4+ count 981 cells/mm3, and mean log10 RNA 5.11. Marked heterogeneity was found for all SRR groups with high admixture for Hispanics. In adjusted linear regression models, subjects with 100% European ancestry were estimated to have 0.33 higher log10 RNA levels (95% CI: (0.03, 0.62), p=0.028) and 253 CD4+ cells /mm3 lower (95% CI: (−517, 11), p = 0.06) in CD4+ count, compared to subjects with 100% African ancestry.
Conclusion
Marked continental admixture was found among this cohort of HIV-infected children from the US. GDA contributed to differences in RNA and CD4+ counts beyond SRR, and should be considered when outcomes associated with HIV infection are likely to have a genetic component.
Keywords: HIV, ancestry informative markers, CD4+ lymphocyte count, HIV RNA, continental ancestry
Introduction
Numerous studies have demonstrated that race/ethnicity can be an important factor in determining health and risk for specific diseases1–4. However, although most studies have used self -reported race/ethnicity (SRR), it is known that this approach to determining ancestry is often inaccurate, does not reveal the extent of genetic admixture and can mask the importance of race/ethnicity as a risk for disease or complications associated with specific drugs or drug combinations5–8. To measure the extent of admixture within a given individual, panels of ancestry informative markers (AIMs) are being used to characterize ancestry. Genetically determined ancestry (GDA) estimates are measurements, based on genomic variants, which are predictive of the regions (or ancestry) from which individuals inherited their genetic alleles. Due to population stratification the allele frequency for some single nucleotide polymorphisms (SNPs) can be extremely high within some regional populations, but low among other regional populations9. These differences can be extensive permitting the identification of panels of SNPs to identify continental ancestry9. Additionally, these same regional patterns suggest that AIMs may also be correlated with other SNPs which may be predictive or causal for a specific clinical phenotype10. Therefore, AIMs have been found to be useful for disease risk assessment and for control of confounding that may be due to population stratification.
As a result of the human diaspora that has occurred in the last 400–600 years, some regions in the world have populations with high admixtures of continental ancestry9. The result is that self-reported race and ethnicity groups can include a mixture of genetic backgrounds. Self-reported race and ethnicity can also reflect the historical and/or social context of the group with which the person identifies3. This implies that self-reported race measures a mixture of genetics and social factors leaving self-reported race as a poor measurement for the genetic composition of an individual.
Although human immunodeficiency virus type-1 (HIV) can infect all populations, in the United States and other countries, specific groups are at increased risk of infection. However, even among these risk groups, despite recurrent high risk exposures, some people do not become infected, while others who become infected may have substantially different rates of progression of HIV-associated diseases. Host genetic factors have been identified as important determinants for both risk of HIV infection and rate of disease progression11–14. Although racial and ethnic minorities, particularly those of African-American and Hispanic ancestry, are disproportionately infected with HIV, few studies have examined the impact of ancestry on HIV disease, and none to our knowledge in children. In a study of 91 HIV-infected adults, it was shown that the association with the CYP2B6 metabolizer phenotype and virologic response to an NNRTI-based (efavirenz or nevirapine) antiretroviral regimen was confounded by GDA and that self-reported race/ethnicity was insufficient15. A more nuanced conclusion was reached in a paper from the Multicenter AIDS Cohort that described dyslipidemia in 1,779 HIV-1 infected men16. They found a significant interaction between GDA and HIV/HAART status for all lipids tested, a low concordance between self-reported race and GDA in admixed populations, and better performance of GDA relative to self-reported race in statistical models. However, they still concluded that self-reported race remained a good clinical surrogate for GDA. In an additional study of 310 North American HIV-infected participants, the hazard ratios for the time to virologic suppression when comparing the CCR5-2459 GA and AA alleles to the AA alleles were stronger among participants with higher African ancestry, but no associations were found with GDA and the time to virologic suppression17.
In the research reported here, we have used a novel 41-SNP panel of AIMs18 to identify continental origin and admixture among HIV-infected children in the U.S. who predate the availability of effective combination antiretroviral therapy (cART). We have investigated the relationship between genetically determined ancestry (GDA) and self-reported race (SRR). Additionally, we have examined the associations of continental ancestry on CD4+ and HIV plasma RNA in children prior to the initiation of cART.
METHODS
Participants
The analysis participants were children who participated in the Pediatric AIDS Clinical Trial Group (PACTG) protocols P152 (n=431)19 and P300 (n=563)20. These trials were U.S. based, prospective, randomized, double blind, placebo controlled, multicenter protocols that assessed the efficacy of combination nucleoside reverse transcriptase inhibitor (NRTI) treatment regimens prior to the availability of effective cART. To be included in one of these trials a child needed to have symptomatic HIV infection, be between 3 months and 18 years of age for P15221, and between 42 days and 15 years of age for P300. For both protocols the children had to meet criteria for a diagnosis of HIV infection from Centers for Disease Control (CDC) classification system available at the time the protocols accrued. The vast majority of these children were HIV-infected through mother-to-child transmission as this cohort predates the routine administration of antiretrovirals to pregnant women.
Genetic Ancestry Determination
A recently described highly informative panel of 41 AIMs was used to determine continental ancestry18. Each of the 41 SNPs was detected using real-time PCR on DNA specimens obtained from peripheral blood mononuclear cells of 994 HIV-infected children in P152 and P300.
The continental origin of analysis participants was estimated by comparing each child’s genotypes to allele frequencies found in a large set of 3517 reference individuals originating from 107 populations around the world18. Reference populations were grouped into the 7 world-regions Europe, Africa, America, Central/South Asia, South/West Asia, East Asia and Oceania. Population structure and ancestry estimates were obtained in a trained clustering analysis using STRUCTURE v2.3.2.122,23. Five independent runs were performed at K=7, using 20,000 burn-in cycles and 20,000 MCMC replications under the admixture model, including prior population information of the reference set. Allele frequencies were updated using only individuals with population information at a migration prior of 0.05. Uniform priors were used for the degree of admixture (“infer α" option) and for the allele frequency (λ = 1 option). All other parameters were set at default. Continental ancestry calling was performed by assigning the predominant continental origin to each subject.
Three-way admixture of the analysis children determined to be from Africa, Europe, or America was further estimated using STRUCTURE with prior population information of reference populations from Africa (N=761), Europe (N=1011), and America (N=407) under an admixture model with correlated allele frequencies at K=3 groups and reported as percent GDA.
Outcome Measures
CD4+ lymphocyte count, CD4+ lymphocyte percentage, and HIV plasma RNA were used as the outcomes for this study. These were measured at entry prior to initiation of therapy. P152 used the NASBA HIV-1 RNA QT Amplification System21 and P300 used the Roche Amplicor quantitative RNA PCR assay20 to measure HIV plasma RNA.
Statistical Methods
Baseline characteristics are presented as percentages, means and standard deviations as appropriate. Linear regression with a robust variance estimator24 was used to measure the associations between GDA and CD4+ lymphocyte counts, CD4+ lymphocyte percent and log10 HIV RNA. Adjustment variables were all selected a-priori and included age, weight for age z-score, study (P152/P300) and where appropriate self-reported race/ethnicity. Genetically determined ancestry was used as a proportion in the regression analyses so that the regression slopes are interpreted for a 100% change in GDA. This parameterization was used so that a direct comparison to self-reported race could be made. Because GDA totaled to 100%, the region (e.g. African GDA) not in the model is interpreted as a reference group. In total, we considered six separate regression models to estimate the adjusted associations. The first model (Model 1) was used to estimate the effects for GDA without adjustment for SRR. The second model (Model 2) was used to estimate the effects of SRR without adjustment for GDA. The third model (Model 3) included both GDA and SRR so that GDA and SRR are adjusted for each other. The remaining models (Model 4 through Model 6) included GDA after subsetting based on SRR. All confidence intervals (CI) are 95% CI and p-values < 0.05 were considered to be statistically significant. R version 2.15.1 and SAS version 9.2 were used for the analyses.
RESULTS
Participants
Of the 994 participants for whom the complete panel of the 41 AIMs were determined, 61% self-reported as Black, 25% self-reported as Hispanic, 13% self-reported as White, and 1% reported as other races or without a self-reported race (Table 1). Fifty-five percent were female; the average age was 3.8 years; and the average weight for age z-score was −0.66. The mean CD4+ lymphocyte count was 981; the mean CD4+ percent was 24%; and the mean plasma log10 HIV RNA was 5.11. Of the 994 subjects with AIMs measurements, 826 (168 with missing data) had HIV-RNA data, and 987 (7 with missing data) had CD4+ counts and percentages. Missing HIV-RNA data was due to the availability of specimens.
Table 1.
Baseline Characteristics
| Study | ||||
|---|---|---|---|---|
| Characteristic | P152 (N=431) |
P300 (N=563) |
Total (N=994) |
|
| Self-reported race/ethnicity | Black | 227 (52.7%) | 378 (67.1%) | 605 (60.9%) |
| Hispanic | 135 (31.3%) | 111 (19.7%) | 246 (24.7%) | |
| White | 56 (13.0%) | 71 (12.6%) | 127 (12.8%) | |
| American | 0 (0.0%) | 1 (0.2%) | 1 (0.1%) | |
| Asian/Pacific | 1 (0.2%) | 1 (0.2%) | 2 (0.2%) | |
| Unknown | 12 (2.8%) | 1 (0.2%) | 13 (1.3%) | |
| Gender | Female | 216 (50.1%) | 326 (57.9%) | 542 (54.5%) |
| Male | 215 (49.9%) | 237 (42.1%) | 452 (45.5%) | |
| Age (years) | Mean (s.d.) | 3.90 (4.29) | 3.66 (3.46) | 3.77 (3.84) |
| Weight for age z-score | Mean (s.d.) | −0.84 (1.35) | −0.52 (1.36) | −0.66 (1.37) |
| Log10 HIV RNA (copies/mL) | Mean (s.d.) | 5.21 (1.03) | 5.04 (0.89) | 5.11 (0.94) |
| CD4 count (cell/mm3) | Mean (s.d.) | 1,047.94 (883.25) | 930.71 (799.73) | 981.19 (838.29) |
| CD4 Percent | Mean (s.d.) | 24% (11.7) | 23% (12.1) | 24% (12) |
HIV-1-infected children in the U.S. demonstrate a high degree of admixture
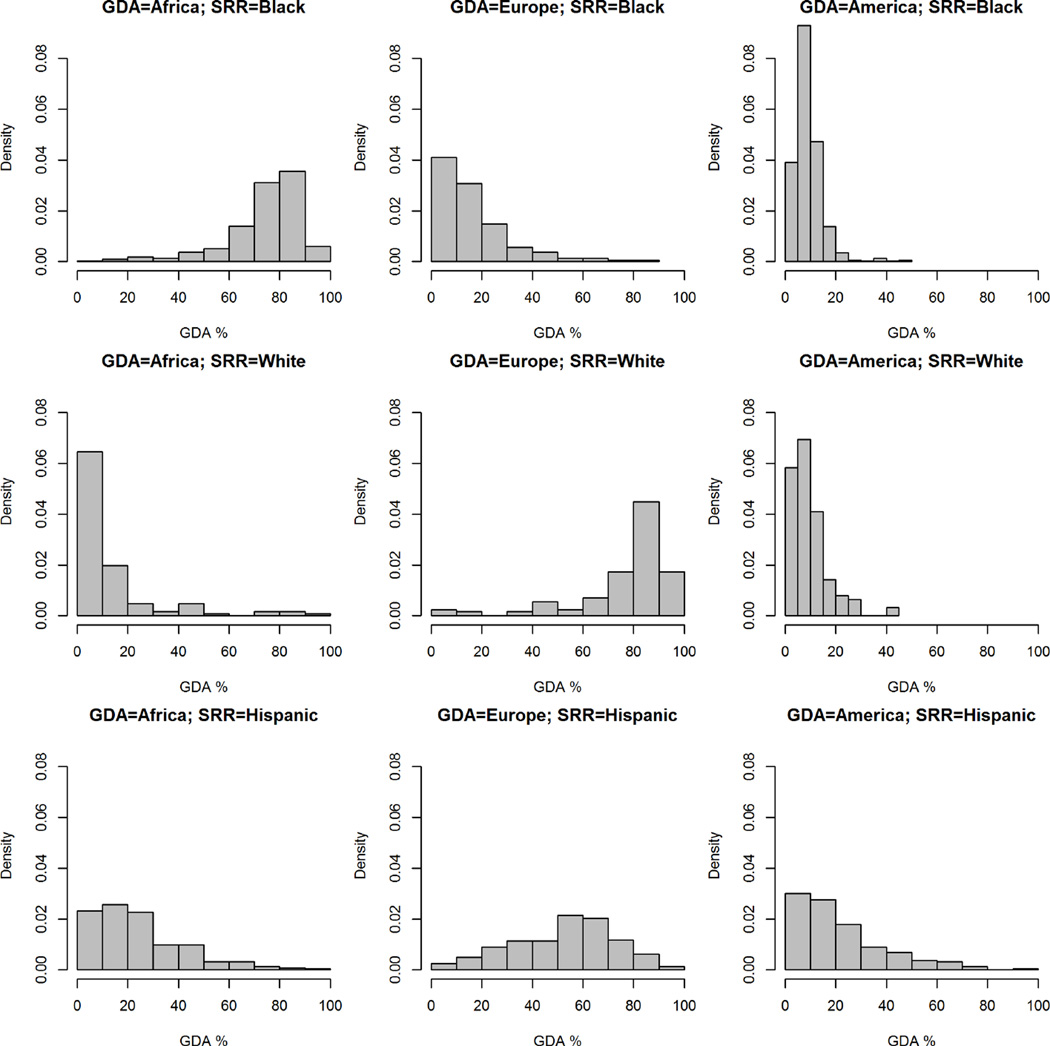
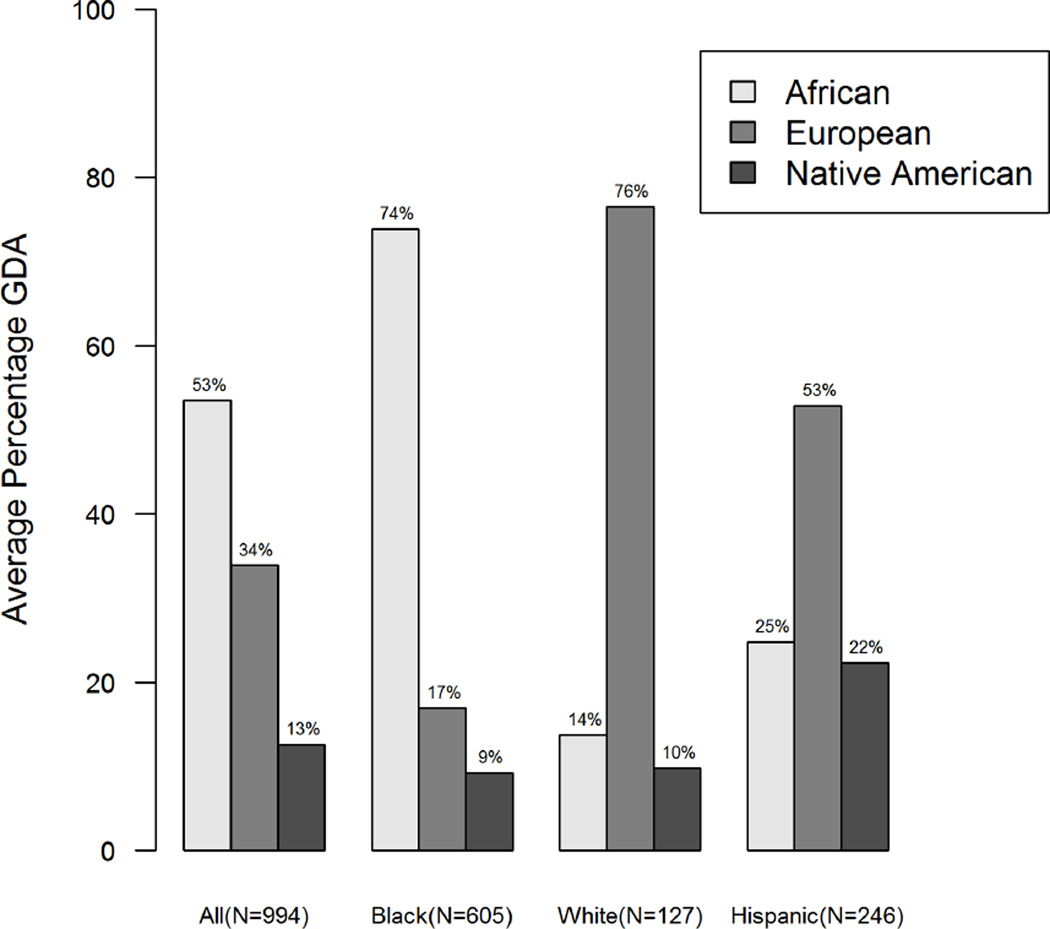
We first examined the association of SRR with GDA for each subject. Because the continental ancestry of the vast number of subjects clustered in three regions, Africa, Europe and the Americas, these continents were used to describe the GDA in these analyses. As seen in Figure 1, histograms for GDA for the three regions by self-reported race illustrate the relative skewness (departure from symmetry), kurtosis (degree of “peakedness”) and extensive variability among self-reported racial groups. All of the histograms display strong skewness, with the possible exception of the European GDA for those that self-report as Hispanic. Histograms with the most kurtosis include the Americas GDA for those that self-report as White or Black, and for the Africa GDA for those that self-report as White. Figure 2 displays the mean GDA by self-reported race and the overall continental ancestry distribution for the entire cohort. For those who self-reported as Black, the mean GDA was 74% for African ancestry, 17% for European ancestry and 9% for Native Americas ancestry. For those who self-reported as White, the mean GDA was 14% for African ancestry, 76% for European ancestry and 10% for Native Americas ancestry. For self-reported Hispanics, GDA was 25% for African ancestry, 53% for European ancestry and 22% for Native Americas ancestry.
Figure 1.
Histograms of genetically determined ancestry (GDA; i.e. % European, African, and Native American admixture) by self-reported racial groups (SRR).
The y-axis is scaled so that the area of each histogram totals to one.
Figure 2.
Bar plot of the average ancestry composition for genetically determined ancestry (GDA) estimates by self-reported race (SRR)
Knowledge of Genetic Ancestry and Admixture proportions adds Additional Information beyond Self-reported Race for pre-treatment HIV RNA
Because viral load is an important indicator of HIV replication and a predictor of disease progression, we examined the role of genetic ancestry in determining the quantity of virus detected in the plasma of subjects prior to their initiating antiretroviral therapy. In our initial analyses, we examined by regression analysis for all self-reported racial/ethnic groups, the pre-treatment HIV RNA comparing subjects with European or Americas ancestry to those with African ancestry (Table 2). In this analysis (Model 1) a higher percentage of European ancestry was associated with higher log10 RNA relative to children with more African ancestry (mean change in log10 RNA for a 100% change in GDA(slope) = 0.18, CI (0.01, 0.36), p-value = 0.041). In the same analysis, children with a higher percentage of Native American ancestry had a non-statistically significant lower log10 viral load compared to those with more African ancestry (slope = −0.29, CI (−0.79, 0.20), p-value = 0.25). Similarly, children with a higher percentage of Native American ancestry had marginally significant lower log10 viral load when compared to those with more European ancestry (slope = −0.47, CI (−1.00, 0.06), p-value =0.080). The higher log10 RNA for those with more European ancestry held up after controlling for self-reported race (slope = 0.33, CI (0.03, 0.62), p-value=0.028) (Model 3), and the directionality of the estimated slopes were similar after subsetting on self-reported race (Models 4 through 6). When the cohort was divided into subsets based on self-reported race, there was only one statistically significant result (self-reported Blacks: slope = 0.62, CI (0.18, 1.05), p-value=0.006). The interaction test for GDA and SRR was statistically significant (Model 3 plus an interaction term, p-value = 0.039), implying that the mean change of log10 HIV RNA as a function of GDA differed by SRR.
Table 2.
Associations of Self-reported Race (SRR) and Genetically Determined Ancestry (GDA) with CD4 and HIV RNA
| HIV RNA | CD4+ Count | CD4+ Percent | ||||||
|---|---|---|---|---|---|---|---|---|
| Model | Predictor | Comparison | Slope (LCI, UCI) |
P- value |
Slope (LCI, UCI) |
P- value |
Slope (LCI, UCI) |
P- value |
| 1 | GDA* | Eur vs. Afk(ref) | 0.18 (0.01, 0.36) | 0.041 | −4 (−163, 155) | 0.96 | 0.4 (−2.1, 3.0) | 0.74 |
| (All Racial Groups Included) | Amer vs. Afk(ref) | −0.29 (−0.79, 0.20) | 0.25 | −45 (−419, 330) | 0.82 | −0.6 (−6.7, 5.5) | 0.85 | |
| N=994 | Amer vs. Eur(ref) | −0.47 (−1.00, 0.06) | 0.080 | −40 (−448, 367) | 0.85 | −1.0 (−7.8, 5.8) | 0.77 | |
| 2 | SRR | Black (Ref) | ||||||
| (All Racial Groups Included) | White | 0.13 (−0.01, 0.28) | 0.065 | 92 (−53, 235) | 0.22 | 1.9 (−0.2, 4.1) | 0.079 | |
| Hispanic | −0.05 (−0.18, 0.08) | 0.45 | 16 (−90, 121) | 0.77 | −0.2 (−1.8, 1.5) | 0.85 | ||
| N=994 | Other | −0.28 (−0.89, 0.32) | 0.36 | 30 (−272, 333) | 0.84 | 0.2 (−3.1, 3.5) | 0.9 | |
| 3 | GDA* | Eur vs. Afk(ref) | 0.33 (0.03, 0.62) | 0.028 | −253 (−517, 11) | 0.060 | −2.5 (−6.7, 1.7) | 0.24 |
| (All Racial Groups Included) | Amer vs. Afk(ref) | 0.00 (−0.57, 0.58) | 1.00 | −195 (−653, 262) | 0.40 | −0.9 (−8.1, 6.3) | 0.81 | |
| N=994 | Amer vs. Eur(ref) | −0.32 (−0.90, 0.25) | 0.27 | 58 (−379, 495) | 0.80 | 1.6 (−5.8, 9.1) | 0.67 | |
| SRR | Black (Ref) | |||||||
| White | −0.07 (−0.29, 0.15) | 0.56 | 243 (31, 454) | 0.025 | 3.5 (0.2, 6.7) | 0.039 | ||
| Hispanic | −0.17 (−0.36, 0.02) | 0.081 | 132 (−39, 303) | 0.13 | 0.9 (−1.7, 3.4) | 0.51 | ||
| Other | −0.37 (−0.98, 0.24) | 0.24 | 111 (−211, 433) | 0.50 | 1.0 (−2.6, 4.6) | 0.60 | ||
| 4 | GDA* | Eur vs. Afk(ref) | 0.62 (0.18, 1.05) | 0.006 | −437 (−823, −52) | 0.026 | −5.1 (−11.2, 0.9) | 0.098 |
| (SRR Black only) | Amer vs. Afk(ref) | −0.13 (−1.06, 0.81) | 0.79 | 226 (−718, 1170) | 0.64 | −3.9 (−19.5, 11.7) | 0.63 | |
| N=605 | Amer vs. Eur(ref) | −0.74 (−1.78, 0.30) | 0.16 | 663 (−336, 1663) | 0.19 | 1.2 (−15.9, 18.4) | 0.89 | |
| 5 | GDA* | Eur vs. Afk(ref) | 0.40 (−0.09, 0.88) | 0.11 | 152 (−477, 781) | 0.64 | 0.8 (−9.7, 11.3) | 0.88 |
| (SRR White Only) | Amer vs. Afk(ref) | 0.43 (−1.41, 2.26) | 0.65 | 1048 (−758, 2854) | 0.26 | −9.0 (−38.8, 20.8) | 0.55 | |
| N=127 | Amer vs. Eur(ref) | 0.03 (−1.82, 1.88) | 0.97 | 897 (−779, 2572) | 0.29 | −9.8 (−38.2, 18.6) | 0.50 | |
| 6 | GDA* | Eur vs. Afk(ref) | 0.16 (−0.40, 0.73) | 0.57 | −308 (−800, 185) | 0.22 | −1.7 (−10.0, 6.6) | 0.69 |
| (SRR Hispanic only) | Amer vs. Afk(ref) | −0.21 (−1.04, 0.61) | 0.61 | −551 (−1169, 67) | 0.081 | 0.3 (−9.1, 9.8) | 0.94 | |
| N=246 | Amer vs. Eur(ref) | −0.38 (−1.13, 0.38) | 0.33 | −243 (−726, 240) | 0.32 | 2.0 (−6.8, 10.9) | 0.65 | |
All analyses are adjusted for age, weight for age z-score, study (P152/P300).
Linear regression models use a continuous GDA covariate as a proportion, and the estimates are therefore interpreted as a comparison for subjects with 100% GDA for a particular region relative to a reference group (ref) with 100% GDA. Negative values indicate lower average values than the reference. Eur=Europe; Afk=Africa; Amer=Americas, GDA=Genetically determined ancestry, SRR=Self-reported Race/Ethnicity; (LCI, UCI)=95% percent confidence interval.
Model 2 and Model 3 (Table 2) contains comparisons of self-reported race for the mean log10 HIV RNA without and with adjustment for GDA, respectively. When comparing those who self-report as White to those who self-report as Black, there was a marginally significant result (slope = 0.13, CI (−0.01, 0.28), p-value=0.065) (Model 2). This estimate is similar to the Europe estimate from Model 1. After controlling for GDA, no significant association was identified (Model 3, SRR based on self-report as Black as the reference) indicating that continental ancestry was a stronger predictor of viral set point than self-reported race/ethnicity.
Association of continental ancestry with CD4+ lymphocyte count and percentage
We performed similar regression analyses to those described above for the association of continental ancestry with CD4+ count and percentage (Table 2). When controlling for GDA (Model 3), in the analyses of self-reported race, those who self-identified as White had a higher CD4+ count compared to those who self-identified as Black (slope = 243, CI: (28,448), p-value = 0.025). Similarly, when CD4+ percentage was used in the analyses, subjects that self-reported as White had on average higher CD4+ percentage than those who self-identified as Black (slope = 3.5, CI: (0.2, 6.7), p-value=0.039). In adjusted linear regression models, subjects with 100% European ancestry were estimated to have 253 CD4+ cells /mm3 lower (95% CI: (−517, 11), p-value = 0.06) when compared to subjects with 100% African ancestry. When comparing the SRR estimates from Model 2 against Model 3 the estimates change due to the correlation of GDA and SRR.
When we adjusted for host genetic factors that were previously found to be related to HIV RNA, CD4+ Count and CD4+ percentage25 using the same participants the estimated regression slopes for the GDA were similar (Supplemental Table 1).
DISCUSSION
To our knowledge, this is the first report that has examined the importance of continental ancestry in a cohort of HIV-infected children from the U.S. Our findings indicate that AIMs provide information above and beyond self-reported race and ethnicity, and demonstrate that there is considerable ancestry variability within self-reported race for these U.S. based studies of HIV-infected children. Associations were identified between GDA and HIV disease severity markers, such as HIV RNA, CD4+ counts and CD4 percent, with effects remaining after adjusting for self-reported race; these associations were strongest among those who self-reported as Black. Additionally, the estimated associations between self-reported race and HIV RNA and CD4+ were stronger when adjusting for GDA. This implies that GDA may be a confounder for the socioeconomic effect of being a member of different racial groups, and that without adjustment for continental ancestry this socio-economic association could not be fully estimated. This argues for inclusion of AIMs in adjusted analyses when there is either a suspected strong genetic effect or a suspected strong socioeconomic effect on the outcome of interest.
Additionally, AIMs can be used to minimize bias associated with population stratification in case-control association studies of genetic markers26,27. Clinically, AIMs may be useful in disease classification and identification of genetic risk. For example, a study of European Americans would have misidentified genetic variants in LCT and IRF4 genes with rheumatoid arthritis without accounting for continental ancestry28. Moreover, in certain situations, differences may exist even within continental ancestry populations. For example, Menotti et al observed that when examining the risk for coronary heart disease applying a model of northern Europeans to southern Europeans overestimated the absolute risk and vice versa29. Because we also found similar results when controlling for some important genetic predictors, it is likely that additional genetic markers are related to CD4+ and HIV RNA, and are correlated with AIMs. This premise is supported by our findings that the associations with CD4+ and HIV RNA remain after adjustment for the genetic markers that we previously found associated with CD4+ and HIV RNA.
The previously reported confounding effect of GDA with the CYP2B6 metabolizer phenotype and virologic response to an NNRTI-based (efavirenz or nevirapine) antiretroviral regimen15 would not have played a role in our findings since HIV RNA and CD4+ were measured before study participants received any antiretrovirals. However, the association that we found with HIV RNA supports the plausibility that GDA is a confounder when investigating a virologic response to an NNRTI based antiretroviral regimen. In addition, the reported interaction with GDA and lipid levels16 poses an interesting question for pediatric research, particularly given that an increase in lipid levels have been reported in children30,31. This remains an area of open research. Lastly, the reported interaction with the CCR5-2459 genotype and GDA on the time to viral suppression17 might be congruent with our findings since it would be expected that there is a relationship with the interacting variables and the outcome under study32; nevertheless, viral suppression and GDA were not found to be statistically significant in the Cheruvu 2014 study. However, there are some important differences between the P152/P300 cohort examined in this study and these other HIV reports. P152/P300 consisted of children while the other studies included adults; therefore, extrapolation may not be valid. In addition, we did not study the time to virologic suppression, rather we studied pre-ART plasma HIV RNA, CD4+ counts and CD4+ percentages.
There are a few limitations to our study. The P152/P300 protocols did not collect information on socioeconomic status and thus, we were not able to more finely describe effects within varying socioeconomic levels. Also, since the P152/P300 studies were conducted in the 1990s it is possible that infants were born to women more likely to have difficulty with substance abuse than observed in more contemporary HIV-infected pregnant women33. Finally, children in this study had a median age of 3.77 years, and did not have access to cART. It would be unusual in the US for a child to reach this median age without having initiated antiretroviral therapy. Thus, the observed results might not generalize to children who have early access to cART.
In summary, we have found through the identification of continental ancestry that the population of children with HIV infection within the U.S. has considerable ancestral heterogeneity, and that self-reported race/ethnicity is often not truly reflective of a child’s genetic background. Moreover, identification of continental ancestry provides additional information with regards to HIV RNA and CD4+ cell count and percent beyond what is observed with self-reported race/ethnicity. Therefore, it is possible that many studies in the HIV literature that have included ancestry in the analysis based on self-reported race could have resulted in misleading conclusions. The utilization of AIMs to identify continental ancestry should be considered when outcomes associated with HIV infection are likely to have a genetic component.
Supplementary Material
Acknowledgements
This research was supported by R01 NS077874 (SAS), R01 MH085608(CNM, AXM), R03 MH103995 (CNM, AXM), and UM1AI068616(SSB, MF, MQ, and TF).
Overall support for the International Maternal Pediatric Adolescent AIDS Clinical Trials Group (IMPAACT) was provided by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under Award Numbers UM1AI068632 (IMPAACT LOC), UM1AI068616 (IMPAACT SDMC) and UM1AI106716 (IMPAACT LC), with co-funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and the National Institute of Mental Health (NIMH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Conflicts of Interest: None to declare
REFERENCES
- 1.Witzig R. The medicalization of race: scientific legitimization of a flawed social construct. Annals of Internal Medicine. 1996;125(8):675–679. doi: 10.7326/0003-4819-125-8-199610150-00008. [DOI] [PubMed] [Google Scholar]
- 2.Burchard EG, Ziv E, Coyle N, et al. The importance of race and ethnic background in biomedical research and clinical practice. New England Journal of Medicine. 2003;348(12):1170–1175. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- 3.Kaufman JS, Cooper RS. Commentary: considerations for use of racial/ethnic classification in etiologic research. American Journal of Epidemiology. 2001;154(4):291–298. doi: 10.1093/aje/154.4.291. [DOI] [PubMed] [Google Scholar]
- 4.Tang H, Quertermous T, Rodriguez B, et al. Genetic structure, self-identified race/ethnicity, and confounding in case-control association studies. The American Journal of Human Genetics. 2005;76(2):268–275. doi: 10.1086/427888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson J. Predictability of the effects of race or ethnicity on pharmacokinetics of drugs. International journal of clinical pharmacology and therapeutics. 2000;38(2):53–60. doi: 10.5414/cpp38053. [DOI] [PubMed] [Google Scholar]
- 6.Kappelhoff BS, Huitema AD, Yalvaç Z, et al. Population pharmacokinetics of efavirenz in an unselected cohort of HIV-1-infected individuals. Clinical pharmacokinetics. 2005;44(8):849–861. doi: 10.2165/00003088-200544080-00006. [DOI] [PubMed] [Google Scholar]
- 7.Bidulescu A, Choudhry S, Musani SK, et al. Associations of adiponectin with individual European ancestry in African Americans: the Jackson Heart Study. Frontiers in genetics. 2014;5:22. doi: 10.3389/fgene.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daya M, van der Merwe L, van Helden PD, Moller M, Hoal EG. The role of ancestry in TB susceptibility of an admixed South African population. Tuberculosis. 2014;94(4):413–420. doi: 10.1016/j.tube.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Winkler CA, Nelson GW, Smith MW. Admixture mapping comes of age. Annual review of genomics and human genetics. 2010;11:65–89. doi: 10.1146/annurev-genom-082509-141523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winkler C, An P, O'Brien SJ. Patterns of ethnic diversity among the genes that influence AIDS. Human molecular genetics. 2004;13(Spec No 1):R9–R19. doi: 10.1093/hmg/ddh075. [DOI] [PubMed] [Google Scholar]
- 11.Seldin MF, Price AL. Application of ancestry informative markers to association studies in European Americans. PLoS genetics. 2008;4(1):e5. doi: 10.1371/journal.pgen.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theodorou I, Capoulade C, Combadiere C, Debre P. Genetic control of HIV disease. TRENDS in Microbiology. 2003;11(8):392–397. doi: 10.1016/s0966-842x(03)00177-x. [DOI] [PubMed] [Google Scholar]
- 13.An P, Winkler CA. Host genes associated with HIV/AIDS: advances in gene discovery. Trends in genetics. 2010;26(3):119–131. doi: 10.1016/j.tig.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh KK, Spector SA. Host genetic determinants of human immunodeficiency virus infection and disease progression in children. Pediatric research. 2009;65:55R–63R. doi: 10.1203/PDR.0b013e31819dca03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frasco MA, Mack WJ, Van Den Berg D, et al. Underlying genetic structure impacts the association between CYP2B6 polymorphisms and response to efavirenz and nevirapine. AIDS. 2012;26(16):2097–2106. doi: 10.1097/QAD.0b013e3283593602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholaou MJ, Martinson JJ, Abraham AG, et al. HAART-associated dyslipidemia varies by biogeographical ancestry in the multicenter AIDS cohort study. AIDS Res Hum Retroviruses. 2013;29(6):871–879. doi: 10.1089/aid.2012.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheruvu VK, Igo RP, Jr, Jurevic RJ, et al. African ancestry influences CCR5-2459G>A genotype-associated virologic success of highly active antiretroviral therapy. Journal of acquired immune deficiency syndromes. 2014;66(1):102–107. doi: 10.1097/QAI.0000000000000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nievergelt CM, Maihofer AX, Shekhtman T, et al. Inference of human continental origin and admixture proportions using a highly discriminative ancestry informative 41-SNP panel. Investigative genetics. 2013;4(1):13. doi: 10.1186/2041-2223-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Englund JA, Baker CJ, Raskino C, et al. Zidovudine, didanosine, or both as the initial treatment for symptomatic HIV-infected children. AIDS Clinical Trials Group (ACTG) Study 152 Team. The New England journal of medicine. 1997;336(24):1704–1712. doi: 10.1056/NEJM199706123362403. [DOI] [PubMed] [Google Scholar]
- 20.McKinney RE, Jr, Johnson GM, Stanley K, et al. A randomized study of combined zidovudine-lamivudine versus didanosine monotherapy in children with symptomatic therapy-naive HIV-1 infection. The Pediatric AIDS Clinical Trials Group Protocol 300 Study Team. The Journal of pediatrics. 1998;133(4):500–508. doi: 10.1016/s0022-3476(98)70057-5. [DOI] [PubMed] [Google Scholar]
- 21.Palumbo PE, Raskino C, Fiscus S, et al. Virologic and immunologic response to nucleoside reverse-transcriptase inhibitor therapy among human immunodeficiency virus-infected infants and children. The Journal of infectious diseases. 1999;179(3):576–583. doi: 10.1086/314638. [DOI] [PubMed] [Google Scholar]
- 22.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White H. A heteroskedasticity-consistent covariance matrix estimator and a direct test for heteroskedasticity. Econometrica: Journal of the Econometric Society. 1980:817–838. [Google Scholar]
- 25.Qin M, Brummel S, Singh KK, Fenton T, Spector SA. Associations of Host Genetic Variants on CD4+ Lymphocyte Count and Plasma HIV-1 RNA in Antiretroviral Naive Children. The Pediatric infectious disease journal. 2014 doi: 10.1097/INF.0000000000000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin DY, Zeng D. Correcting for Population Stratification in Genomewide Association Studies. Journal of the American Statistical Association. 2011;106(495):997–1008. doi: 10.1198/jasa.2011.tm10294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nassir R, Kosoy R, Tian C, et al. An ancestry informative marker set for determining continental origin: validation and extension using human genome diversity panels. BMC genetics. 2009;10(1):39. doi: 10.1186/1471-2156-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian C, Plenge RM, Ransom M, et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS genetics. 2008;4(1):e4. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menotti A, Lanti M, Puddu P, Kromhout D. Coronary heart disease incidence in northern and southern European populations: a reanalysis of the seven countries study for a European coronary risk chart. Heart. 2000;84(3):238–244. doi: 10.1136/heart.84.3.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller TI, Borkowsky W, DiMeglio LA, et al. Metabolic abnormalities and viral replication are associated with biomarkers of vascular dysfunction in HIV-infected children. HIV medicine. 2012;13(5):264–275. doi: 10.1111/j.1468-1293.2011.00970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aldrovandi GM, Lindsey JC, Jacobson DL, et al. Morphologic and metabolic abnormalities in vertically HIV-infected children and youth. AIDS. 2009;23(6):661–672. doi: 10.1097/QAD.0b013e3283269dfb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.VanderWeele TJ, Robins JM. Four types of effect modification: a classification based on directed acyclic graphs. Epidemiology. 2007;18(5):561–568. doi: 10.1097/EDE.0b013e318127181b. [DOI] [PubMed] [Google Scholar]
- 33.Rough K, Tassiopoulos K, Kacanek D, et al. Dramatic decline in substance use by HIV-infected pregnant omen in the United States from 1990 to 2012. AIDS. 2015;29(1):117–123. doi: 10.1097/QAD.0000000000000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.