Abstract
Osteogenesis imperfecta (OI) is a group of genetic disorders characterized by bone fragility and deformity. OI type VI is unique owing to the mineralization defects observed in patient biopsies. Furthermore, it has been reported to respond less well to standard therapy with bisphosphonates (1). Others and we have previously identified SERPINF1 mutations in patients with OI type VI. SERPINF1 encodes pigment epithelium derived factor (PEDF), a secreted collagen-binding glycoprotein that is absent in the sera of patients with OI type VI. Serpinf1 null mice show increased osteoid and decreased bone mass, and thus recapitulate the OI type VI phenotype. We tested whether restoration of circulating PEDF in the blood could correct the phenotype of OI type VI in the context of protein replacement. To do so, we utilized a helper-dependent adenoviral vector (HDAd) to express human SERPINF1 in the mouse liver and assessed whether PEDF secreted from the liver was able to rescue the bone phenotype observed in Serpinf1−/− mice. We confirmed that expression of SERPINF1 in the liver restored the serum level of PEDF. We also demonstrated that PEDF secreted from the liver was biologically active by showing the expected metabolic effects of increased adiposity and impaired glucose tolerance in Serpinf1−/− mice. Interestingly, overexpression of PEDF in vitro increased mineralization with a concomitant increase in the expression of bone gamma-carboxyglutamate protein, alkaline phosphatase and collagen, type I, alpha I, but the increased serum PEDF level did not improve the bone phenotype of Serpinf1−/− mice. These results suggest that PEDF may function in a context-dependent and paracrine fashion in bone homeostasis.
Keywords: PEDF, SerpinF1, gene transfer, Osteogenesis Imperfecta, Helper-dependent adenovirus
1. Introduction
Osteogenesis Imperfecta (OI) is a group of heterogeneous genetic disorders largely characterized by bone fragility and deformity. Among the different types of OI, the recessively inherited OI type VI is unique owing to the mineralization defects observed in patient biopsies. Individuals with OI type VI display a dramatic increase in osteoid and prolonged mineralization lag time, and show variable response to standard treatment with bisphosphonates (2). Whole exome studies have identified loss of function mutations in SERPINF1 as causative of OI type VI (3-5). SERPINF1 encodes pigment epithelium derived factor (PEDF), a 50kDA collagen-bin(2)ding glycoprotein that belongs to the serine protease inhibitor superfamily. PEDF was first isolated from the retina and was described as having potent neurotropic and antiangiogenic activity (6); recent reports support a potential anti-tumor role for the protein in many forms of cancers (7,8).
Serpinf1−/− mice recapitulate the hypomineralization and low bone mass phenotypes observed in patients with OI type VI, and are thus an excellent model for therapeutic studies (9). In this study, we examined whether restoration of PEDF into the serum using a helper dependent adenoviral (HDAd) system was able to correct the skeletal defects observed in the Serpinf1−/− mice. Helper-dependent adenoviral vectors are devoid of viral genes, and hence, exhibit dramatically lower host adaptive immune response. In multiple animal models including nonhuman primates, a single injection can lead to life-long sustained expression from the liver (10). SERPINF1 expression was mediated using the HDAd approach owing to the ability of the vector system to transduce hepatocytes and secrete PEDF into the serum (11).
2. Materials and Methods
2.1 Ethics Statement
All research involving animals was approved by the Center for Comparative Medicine, in conjunction with the Institutional Animal Care and Use Committee, and conducted according to the relevant national and international guidelines. Veterinarians supervised animal care according to the standards of the Baylor College of Medicine Center for Comparative Medicine, a program fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
2.2 Animals
The Serpinf1−/− mice were a generous gift from S. Crawford and were generated as described previously (12). The Serpinf1−/− mouse colony was maintained in a C57Black/6J genetic background and housed in the Baylor College of Medicine Animal Vivarium. All of the mice were housed under pathogen-free conditions; food and water were provided ad libitum. Mice used in the experiments were males and between 7–9 weeks of age. All experimental procedures were conducted in accordance with institutional guidelines for animal care and use.
2.3 Cell culture, virus production and stable cell generation
HEK293T and MC3T3-E1 cells were cultured in DMEM or α-MEM medium with 10% FBS, 1% glutamine and 1% penicillin-streptomycin. The SERPINF1 cDNA was cloned into the expression plasmid pLenti CMV Puro DEST (w118-1), a gift from Eric Campeau (Addgene plasmid #17452). 293T cells were used to generate a lentivirus constitutively expressing SERPINF1, as previously described (13). Briefly, pLentiSERPINF1PuroDEST was cotransfected with the psPAX2 and pMD2.G packaging vectors using Superfect (Qiagen). The supernatant was collected at 36 and 48h post transfection. After centrifugation, viral particles were concentrated overnight at 4°C using PEG5000 and resuspended in 10mM Tris after centrifugation at 4°C. MC3T3-E1 cells were transduced with the virus; after 48h, cells were selected with medium containing 5μg/ml puromycin for a period of 2 weeks to obtain clones that stably expressed SERPINF1.
2.4 Western blotting and mineralization assay
Alizarin red staining assays were performed as previously described on MC3T3 cells stably expressing SERPINF1 (13). A total of 15μl of conditioned media was electrophoresed and transferred onto PVDF membranes (Millipore). The membranes were blocked with 5% milk in Tris buffered saline/0.1% Tween (TBST) and incubated with a 1:1000 dilution of primary antibodies for PEDF (Millipore). Primary antibodies were detected with appropriate HRP-conjugated secondary antibodies, and the signal was detected using a chemiluminescence detection kit (Pierce).
2.5 RNA extraction and real time RT-PCR
Total RNA from MC3T3 cells overexpressing SERPINF1 was extracted using Trizol (Life Technologies). Samples were treated with DNase (Roche), and the Superscript III First Strand RT-PCR kit (Life Technologies) was used to synthesize cDNA. qRT-PCR was performed on a LightCycler instrument (Roche), with Gapdh as the internal control to normalize gene expression. Three independent replicates and one technical replicate were included for each analysis. Two-tailed, unpaired t-tests with unequal variances were used for statistical analysis.
Viral vector construction, production and injection in animals
HDAd HDΔ25.3E4PEPCK-SERPINF1 constructs containing the human SERPINF1 cDNA transgene driven by the liver-restricted phosphoenolpyruvate carboxykinase (PEPCK) promoter (HDAd-SERPINF1) were produced as described elsewhere (14,15). Purified HDAds were diluted in phosphate buffered saline (PBS) and injected into tail veins, with a total volume of 200 μL. All injections were done in the mornings. Blood was collected retro-orbitally, and plasma and serum samples were frozen immediately and stored at −80°C until analysis. Once the animals were sacrificed, the livers were harvested and kept at −80°C until analysis.
2.6 ELISA
Concentrations of serum PEDF were determined per manufacturer's protocol using the commercial human PEDF ELISA kit (Biovendor).
2.7 Intraperitoneal glucose tolerance test
Mice were fasted overnight, with access to plenty of water. After measuring the baseline glucose level, 10% glucose solution per gram bodyweight was injected intraperitoneally. Glucose levels were measured using a glucose meter at 15, 30, 60 and 120 minutes post injection.
2.8 Animal tissue collection, processing and histology
Serpinf1−/− mice and wild-type littermates were sacrificed at 6 months of age. Liver, femurs and spines were dissected, fixed, embedded, and sectioned according to methods previously described (13). Routine histologic analysis of the tissues was performed according to standard protocols. ANOVA was used to analyze fat pad weights (Graphpad Prism).
2.9 Micro-computed tomography (μCT)
For μCT, spine and femur samples were placed into a 16mm tube filled with 70% ethanol and scanned, at a resolution of 16μm, using a ScanCo μct40 scanner (N=4–5, males). Trabecular and cortical bone analysis was performed using the ScanCo software, as previously described (13). Two-tailed, unpaired t-tests with unequal variances were used for statistical analysis.
3. Results
3.1 Overexpression of SERPINF1 in MC3T3 cells results in enhanced osteoblast differentiation and mineralization
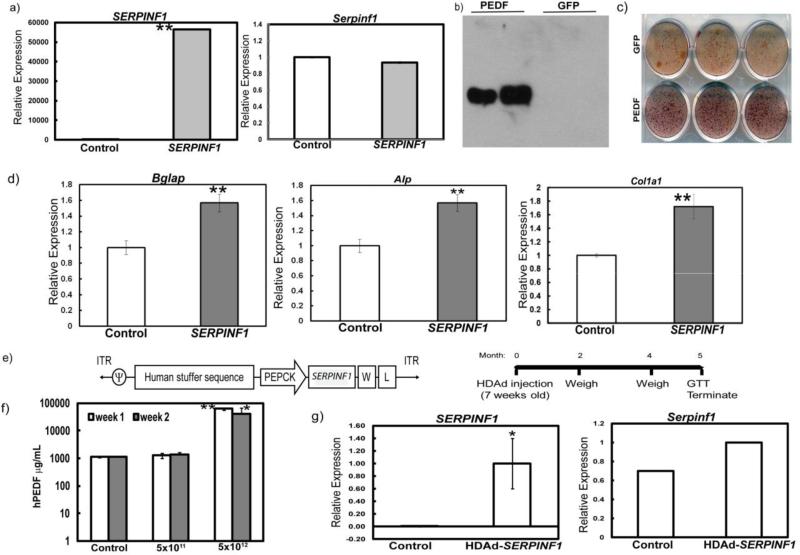
Since loss of Serpinf1 in mice results in a mineralization defect, we first assessed whether overexpression of SERPINF1 would have an effect on mineralization. MC3T3 cells were transduced with a lentivirus overexpressing SERPINF1. Real-time PCR data showed increased expression of human SERPINF1, but not of mouse Serpinf1 (Fig. 1a). Western blot analysis using conditioned medium from MC3T3 cells overexpressing SERPINF1 shows a band of approximately 50kDa, which suggests that cells secreted PEDF (Fig. 1b). In comparison to control MC3T3 cells, cells overexpressing SERPINF1 exhibited a high degree of mineralization as shown by alizarin red staining (Fig. 1c). Real-time PCR analysis revealed a concomitant increase in the expression of osteoblast differentiation markers, bone gamma-carboxyglutamate protein (Bglap), alkaline phosphatase (Alp), and collagen, type I, alpha I (Col1a1) (Fig. 1d). Together, these results suggest that the overexpression of SERPINF1 increases mineralization and is bioactive in this in vitro context.
Figure 1. Overexpression of SERPINF1 in MC3T3 cells and mouse liver.
a) Real-time PCR analysis for SERPINF1 and Serpinf1 in cells overexpressing SERPINF1 (hPEDF) and a vector containing GFP showing relative expression to control (**p<0.005); b) Western blotting using anti-PEDF antibody of media from cells overexpressing SERPINF1 (hPEDF) and a vector containing GFP; c) Alizarin Red staining of MC3T3 cells transduced with SERPINF1 vs. GFP control vector; d) Real-time PCR analysis for Bglap, Alp and Col1a1 (**p<0.005; unpaired t-test) relative expression to GFP control infected cells; e) Schematics of the HDAd-mediated SERPINF1 expression construct (left panel) and experimental time course (right panel); f) Serum ELISA of mice injected with PBS (Control), 5×1011 and 5×1012 viral particles per kilogram (VP/kg) one and two weeks after injection; N=5 per group (*p< 0.05; **p<0.005; unpaired t-test compared to the PBS injected controls); g) Real-time PCR for human SERPINF1 and mouse Serpinf1 in the livers of mice injected with PBS (Control, N=4) or human PEDF virus, N=3 (*p<0.05; unpaired t-test) showing relative expression compared to control.
3.2 SERPINF1 transduction produces biologically active PEDF in the liver of Serpinf1−/− mice
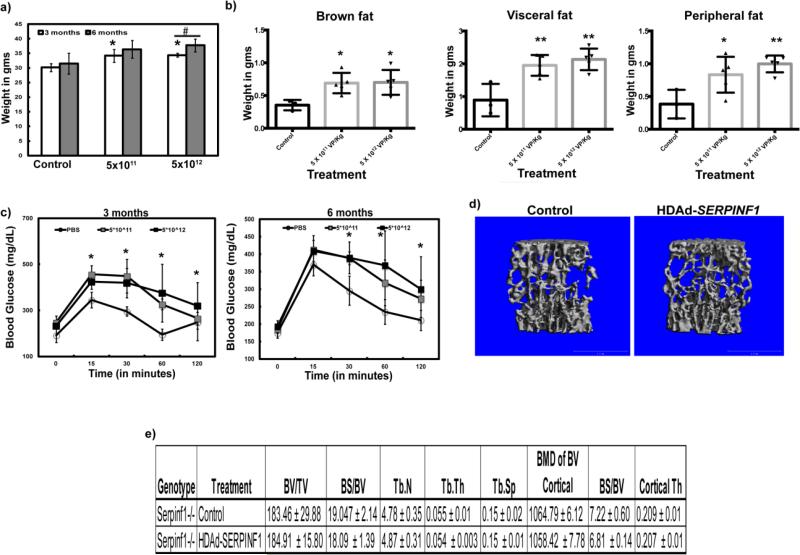
To test whether restoration of PEDF in the serum is sufficient to rescue the mineralization defect present in the Serpinf1−/− mice, we generated a helper-dependent adenoviral (HDAd) vector expressing SERPINF1 specifically in the liver (Fig. 1e). We injected different dosages of viral particles (5×1011VP/kg or 5×1012VP/kg of HDAd-SERPINF1) into Serpinf1−/− mice to evaluate the dose-dependent secretion of PEDF from the liver. We confirmed the presence of PEDF in the blood by ELISA after one and two weeks of injection (Fig 1f). We observed SERPINF1 gene expression by real time PCR analysis in the livers 5 months after injection of the virus (Fig. 1g). We observed a significant increase in serum PEDF in the Serpinf1−/− mice injected with 5×1012VP/kg of HDAd-SERPINF1. Previous work by others showed that overexpression of Serpinf1 led to metabolic dysfunction, with increased body weight and impaired glucose tolerance (16-18). To determine whether the PEDF produced by the liver was biologically active, we measured the body weights of mice injected with control (PBS) and HDAd-SERPINF1 at 3 and 6 months of age and performed an intraperitoneal glucose tolerance test (IP GTT). Serpinf1−/− mice injected with 5×1011VP/kg and 5×1012VP/kg weighed significantly more than Serpinf1−/− mice injected with control virus at 3 months of age. Additionally, Serpinf1−/− animals injected with 5×1012VP/kg weighed significantly more at 6 months of age (5 months after injection) than at 3 months (2 months after injection) of age (Fig. 2a). We also observed an increase in fat pad weights in mice injected with both 5×1011VP/kg and 5×1012VP/kg of HDAd-SERPINF1 (Fig. 2b). Blood glucose levels in mice injected with 5×1012VP/kg HDAd-SERPINF1 remained significantly higher than in PBS-injected control mice, suggesting that these mice have impaired glucose tolerance (Fig. 2c). The increase in body weight, fat mass and glucose intolerance in the Serpinf1−/− mice injected with either 5X1011VP/kg or 5×1012VP/kg of HD-Ad SERPINF1 suggests that PEDF produced in the transduced liver was biologically active.
Figure 2. Biological activity of human PEDF expressed from mouse liver.
a) Weight (g) of Serpinf1−/− mice injected with PBS (Control), 5×1011 and 5×1012 HDAd-SERPINF1 viral particles per kilogram (VP/kg) at 3 and 6 months of age (*p<0.05, compared to control; #p<0.05 comparing the 3 and 6 month time points); b) Fat pads were weighed at 6 months (*p<0.05; **p<0.005; one-way ANOVA); c) Blood glucose levels at 3 and 6 months of age; N=4-5 per group (*p<0.05, compared to PBS injected controls); d) μCT analysis of bones from Serpinf1−/− mice injected with PBS (Control) and 5×1012 HDAd-SERPINF1 VP/kg at 6 months; e) Bone parameters obtained from the μCT analysis of bones from Serpinf1−/− mice injected with PBS (Control) and 5×1012 HDAd-SERPINF1 VP/kg at 6 months.
3.3 Restoration of PEDF in the serum is not sufficient to rescue the bone phenotype in Serpinf1−/− mice
To determine whether restoration of serum PEDF is able to rescue the skeletal phenotype observed in Serpinf1−/− mice, we analyzed bones collected from Serpinf1−/− mice treated with either control or HDAd-SERPINF1 virus by micro-computed tomography (μCT). Surprisingly, we did not observe any improvement in the low bone mass phenotype of Serpinf1−/− mice injected with either 5×1011VP/kg or 5×1012VP/kg of HDAd-SERPINF1 (Fig. 2d, data not shown for the lower viral dose). Other bone parameters such as trabecular thickness and separation were also unchanged (Fig. 2e, data not shown for the lower viral dose). These data suggest that serum restoration of PEDF produced by the liver does not rescue the bone phenotype observed in the Serpinf1−/− mice.
4. Discussion
Here we show that in vitro overexpression of SERPINF1 results in increased mineralization, accompanied with increased osteoblast differentiation. We succeeded in constitutively secreting PEDF from the liver of mice using a liver-directed helper-dependent adenovirus gene transfer system. We have shown that while overexpression of SERPINF1 results in metabolic phenotypes, including impaired glucose tolerance and increased adiposity measured by increase fat pad weights in Serpinf1−/− mice, the high amount of SERPINF1 in the blood did not correct the low bone mass phenotype observed in these mice.
These results are surprising because local treatment with recombinant PEDF has been successfully used to treat eye diseases in mouse models of degenerative eye diseases, which may be attributed to the presence of previously described PEDF receptors in the eye (19-22). PEDF protein interactions and binding partners have not been well characterized in bone, and it is possible that lack of PEDF in bone cells results in a reduction in either the expression or the binding capacity of its interacting partner.
The amount of PEDF secreted into the serum in treated Serpinf1−/− mice was well above the normal range observed in control animals. Recent findings by Rauch et al. found that SERPINF1 mutation heterozygous carriers have reduced levels of serum PEDF when compared to controls (23). A clinical analysis of the carriers found that they have normal bone density parameters, suggesting that partial restoration of PEDF serum levels may be sufficient to rescue the bone phenotype though the impact of source of PEDF production on the skeletal system is unknown. The lack of correction of PEDF from the liver on bone may suggest that PEDF needs to be produced locally to act in either paracrine or autocrine fashion. Alternatively, the requirements for bone homeostasis may require a narrow range of expression.
The heterologous system used for overexpressing the human protein in a Pedf-deficient mouse model might be the reason for the lack of improvement in the bone phenotype, although there is nearly 90% homology between human and mouse PEDF at the protein level and overexpression of SERPINF1 did result in weight gain and impaired glucose tolerance in the mutant mice. Interestingly, expression of SERPINF1 in the liver resulting in increased serum PEDF is not sufficient to overcome the deleterious bone phenotype in Serpinf1−/− mice. Taken together, our results raise the possibility of tissue-specific requirements, and perhaps even tissue-specific modifications, for PEDF.
Highlights.
The expression of SERPINF1 in the liver restored the serum levels of PEDF.
PEDF secreted from the liver results in a metabolic phenotype.
Increased serum PEDF levels did not improve the bone phenotype of Serpinf1−/− mice.
Acknowledgements
Author roles: Study design: EPH, AR, KSJ, MS and BL. Study conduct: EPH, AR, KSJ and MS. Data collection: EPH, AR, KSJ and MS. Data analysis: EPH, AR, KSJ and MS. Data interpretation: EPH, AR, KSJ and MS. Drafting manuscript: EPH and AR. Revising manuscript: KSJ and MS. Approving final version of manuscript: BHL. EPH, AR, KSJ and MS take responsibility for data integrity.
We thank Addgene for the destination vector (pLenti CMV Puro DEST) to clone the SERPINF1 cDNA into the expression vector (https://www.addgene.org/17452/).
Grants and fellowships that supported this work include the BCM Intellectual and Developmental Disabilities Research Center (HD024064) grant from the Eunice Kennedy Shriver National Institute of Child Health & Human Development, funding from the NIH to the BCM Advanced Technology Cores (AI036211, CA125123, and RR024574), funding from the Rolanette and Berdon Lawrence Bone Disease Program of Texas and the BCM Center for Skeletal Medicine and Biology, and NIH grants PO1 HD22657 (BHL), PO1 HD070394 (BHL), R00HL098692 (MS), AR063616 (KSJ), F31 DE020954 (EPH), and NIDDK 5T32DK060445-10 (AR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure page
The authors have nothing to disclose.
References
- 1.Land C, Rauch F, Travers R, Glorieux FH. Osteogenesis imperfecta type VI in childhood and adolescence: effects of cyclical intravenous pamidronate treatment. Bone. Mar. 2007;40(3):638–644. doi: 10.1016/j.bone.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Shaheen R, Alazami AM, Alshammari MJ, Faqeih E, Alhashmi N, Mousa N, et al. Study of autosomal recessive osteogenesis imperfecta in Arabia reveals a novel locus defined by TMEM38B mutation. J Med Genet. Oct. 2012;49(10):630–635. doi: 10.1136/jmedgenet-2012-101142. [DOI] [PubMed] [Google Scholar]
- 3.Becker J, Semler O, Gilissen C, Li Y, Bolz HJ, Giunta C, et al. Exome sequencing identifies truncating mutations in human SERPINF1 in autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. Mar 11. 2011;88(3):362–371. doi: 10.1016/j.ajhg.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Homan EP, Rauch F, Grafe I, Lietman C, Doll JA, Dawson B, et al. Mutations in SERPINF1 cause osteogenesis imperfecta type VI. J Bone Miner Res. Dec. 2011;26(12):2798–2803. doi: 10.1002/jbmr.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rauch F, Husseini A, Roughley P, Glorieux FH, Moffatt P. Lack of circulating pigment epithelium-derived factor is a marker of osteogenesis imperfecta type VI. J Clin Endocrinol Metab. Aug. 2012;97(8):E1550–6. doi: 10.1210/jc.2012-1827. [DOI] [PubMed] [Google Scholar]
- 6.Tombran-Tink J, Chader GG, Johnson LV. PEDF: a pigment epithelium-derived factor with potent neuronal differentiative activity. Exp Eye Res. Sep. 1991;53(3):411–414. doi: 10.1016/0014-4835(91)90248-d. [DOI] [PubMed] [Google Scholar]
- 7.Hong H, Zhou T, Fang S, Jia M, Xu Z, Dai Z, et al. Pigment epithelium-derived factor (PEDF) inhibits breast cancer metastasis by down-regulating fibronectin. Breast Cancer Res Treat. Nov. 2014;148(1):61–72. doi: 10.1007/s10549-014-3154-9. [DOI] [PubMed] [Google Scholar]
- 8.Nelius T, Martinez-Marin D, Hirsch J, Miller B, Rinard K, Lopez J, et al. Pigment epithelium-derived factor expression prolongs survival and enhances the cytotoxicity of low-dose chemotherapy in castration-refractory prostate cancer. Cell Death Dis. May 8. 2014;5:e1210. doi: 10.1038/cddis.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogan R, Riddle RC, Li Z, Kumar S, Nandal A, Faugere MC, et al. A mouse model for human osteogenesis imperfecta type VI. J Bone Miner Res. Jul. 2013;28(7):1531–1536. doi: 10.1002/jbmr.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunetti-Pierri N, Ng T, Iannitti D, Cioffi W, Stapleton G, Law M, et al. Transgene expression up to 7 years in nonhuman primates following hepatic transduction with helper-dependent adenoviral vectors. Hum Gene Ther. Aug. 2013;24(8):761–765. doi: 10.1089/hum.2013.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vetrini F, Ng P. Liver-directed gene therapy with helper-dependent adenoviral vectors: current state of the art and future challenges. Curr Pharm Des. 2011;17(24):2488–2499. doi: 10.2174/138161211797247532. [DOI] [PubMed] [Google Scholar]
- 12.Doll JA, Stellmach VM, Bouck NP, Bergh AR, Lee C, Abramson LP, et al. Pigment epithelium-derived factor regulates the vasculature and mass of the prostate and pancreas. Nat Med. Jun. 2003;9(6):774–780. doi: 10.1038/nm870. [DOI] [PubMed] [Google Scholar]
- 13.Homan EP, Lietman C, Grafe I, Lennington J, Morello R, Napierala D, et al. Differential effects of collagen prolyl 3-hydroxylation on skeletal tissues. PLoS Genet. Jan. 2014;10(1):e1004121. doi: 10.1371/journal.pgen.1004121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunetti-Pierri N, Ng T, Iannitti DA, Palmer DJ, Beaudet AL, Finegold MJ, et al. Improved hepatic transduction, reduced systemic vector dissemination, and long-term transgene expression by delivering helper-dependent adenoviral vectors into the surgically isolated liver of nonhuman primates. Hum Gene Ther. Apr. 2006;17(4):391–404. doi: 10.1089/hum.2006.17.391. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki M, Cela R, Clarke C, Bertin TK, Mourino S, Lee B. Large-scale production of high- quality helper-dependent adenoviral vectors using adherent cells in cell factories. Hum Gene Ther. Jan. 2010;21(1):120–126. doi: 10.1089/hum.2009.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crowe S, Wu LE, Economou C, Turpin SM, Matzaris M, Hoehn KL, et al. Pigment epithelium-derived factor contributes to insulin resistance in obesity. Cell Metab. Jul. 2009;10(1):40–47. doi: 10.1016/j.cmet.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Bohm A, Ordelheide AM, Machann J, Heni M, Ketterer C, Machicao F, et al. Common genetic variation in the SERPINF1 locus determines overall adiposity, obesity-related insulin resistance, and circulating leptin levels. PLoS One. 2012;7(3):e34035. doi: 10.1371/journal.pone.0034035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carnagarin R, Dharmarajan AM, Dass CR. PEDF-induced alteration of metabolism leading to insulin resistance. Mol Cell Endocrinol. Feb 5. 2015;401C:98–104. doi: 10.1016/j.mce.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Subramanian P, Shen D, Tuo J, Becerra SP, Chan CC. Pigment epithelium-derived factor reduces apoptosis and pro-inflammatory cytokine gene expression in a murine model of focal retinal degeneration. ASN Neuro. Nov 26. 2013;5(5):e00126. doi: 10.1042/AN20130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amaral J, Becerra SP. Effects of human recombinant PEDF protein and PEDF-derived peptide 34-mer on choroidal neovascularization. Invest Ophthalmol Vis Sci. Mar. 2010;51(3):1318–1326. doi: 10.1167/iovs.09-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian P, Locatelli-Hoops S, Kenealey J, DesJardin J, Notari L, Becerra SP. Pigment epithelium-derived factor (PEDF) prevents retinal cell death via PEDF Receptor (PEDF-R): identification of a functional ligand binding site. J Biol Chem. Aug 16. 2013;288(33):23928–23942. doi: 10.1074/jbc.M113.487884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian P, Notario PM, Becerra SP. Pigment epithelium-derived factor receptor (PEDF-R): a plasma membrane-linked phospholipase with PEDF binding affinity. Adv Exp Med Biol. 2010;664:29–37. doi: 10.1007/978-1-4419-1399-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Jallad H, Palomo T, Moffatt P, Roughley P, Glorieux FH, Rauch F. Normal bone density and fat mass in heterozygous SERPINF1 mutation carriers. J Clin Endocrinol Metab. Nov. 2014;99(11):E2446–50. doi: 10.1210/jc.2014-2505. [DOI] [PubMed] [Google Scholar]