Abstract
Viral infections remain a major threat to public health. The speed with which viruses are evolving drug-resistant mutations necessitates the further development of antiviral therapies with a large emphasis on drug discovery. To facilitate these efforts, there is a need for robust, high-throughput assays that allow the screening of large libraries of compounds, while enabling access to detailed kinetic data on their antiviral activity. We report here the development of a droplet-based microfluidic platform to probe viral fusion, an early critical step in infection by membrane-enveloped viruses such as HIV, Hepatitis C, and influenza. Using influenza A, we demonstrate the measurement of the kinetics of fusion of virions with target liposomes with sub-second temporal resolution. In analogy with acidification of the endosome that triggers fusion in a cellular context, we acidify the content of aqueous droplets containing virions and liposomes in situ by introducing acid from the dispersed phase and visualize the kinetics of fusion by using fluorescent probes.
I. INTRODUCTION
Droplet-based microfluidics, in which droplets of sub-nanoliter volumes are generated and used as individual chemical reactors, has allowed a wide range of biomedical applications1 such as digital polymerase chain reaction (PCR),2 rapid pathogen detection,3 and antibiotics screening.4 Droplet-based microfluidic approaches promise increased throughput, reduced usage of reactants, and the ability to rapidly execute complex workflows involving multiple steps. In droplet-based approaches, droplets are dispersed in an immiscible carrier fluid (an oil phase for aqueous droplets), which prevents evaporation of the droplet and enables easy transport and manipulation.5 The constant droplet size (reaction volume) and uniform concentration of oil-insoluble reactants within the droplet allow reactions to be performed with a high level of control and reproducibility. The development of technology to rapidly generate large numbers of droplets with uniform size has resulted in novel strategies to perform reactions in a high-throughput fashion.6
One field of research that has benefitted from such high-throughput approaches is the identification of biologically active compounds in disease mechanisms.7,8 Using microdroplets as reaction vessels and controlling them in microfluidic structures opens the possibility of developing assays that combine high-throughput approaches with the mechanistic insight that traditionally is reserved for biochemical studies.6 To increase the availability of methods that provide highly detailed kinetic and mechanistic information in a high-throughput fashion, we report here the development of a droplet-based microfluidic platform that allows the external triggering and real-time visualization of a pharmaceutically relevant process. In particular, we provide a proof of principle based on the visualization of the fusion kinetics of the influenza A virus with a target lipid vesicle.
Influenza, commonly called the flu, is a highly contagious infection of the upper respiratory tract and is estimated to be responsible for 250 000–500 000 deaths each year.9 The recent emergence of new, highly virulent porcine (H1N1) and avian (H5N1) strains underscores the difficulty we have as a society in addressing this public-health threat. Currently, there are two classes of antiviral drugs that are effective against influenza: M2 inhibitors, including amantadine and rimantadine, and the neuraminidase inhibitors Tamiflu and Relenza.10 Most circulating influenza strains are resistant to the former class of drugs, and Tamiflu-resistant strains of H5N1 have recently emerged. Additionally, no vaccine has been developed yet that provides lasting protection against influenza infection. Therefore, there is a great need for new antiviral drugs and vaccines and for experimental tools to develop these therapeutics.
The first key molecular event during the infection cycle of membrane-enveloped viruses, such as influenza, is the fusion of the viral lipid bilayer with the cellular membrane. The fusion process represents an attractive target for antiviral therapeutics, but currently only one antiviral drug that targets fusion—enfuvirtide, which targets human immunodeficiency virus (HIV) fusion—is commercially available.11 Treatment of HIV with combinations of drugs that target different steps in the viral lifecycle has been successful in slowing the emergence of drug resistance. Influenza antiviral treatments could similarly benefit from “drug cocktails,” which will require the development of new classes of therapeutics.
Among the barriers to the development of useful fusion inhibitors is the difficulty in designing a suitable assay that allows both parallel screening of multiple compounds and access to quantitative and mechanistic information, especially in cases where a structure-based molecular assay is not feasible. Conventional assays are generally based on single or multicycle virus growth curves that take multiple days to complete and provide little information on the mode of inhibition.12 Here, we report a method that combines highly quantitative fluorescence methods with the external control of fusion reactions inside microdroplets as a novel tool to screen for and characterize new antiviral compounds. We demonstrate our droplet-based assay for two strains of influenza virus, which differ in terms of their fusion proteins. In this study, we show that both viral particles remain functional within microdroplets. We then use one of these strains as a model system to extract fusion kinetics and demonstrate the action of a neutralizing antibody.
II. MATERIALS AND METHODS
A. Chip design and the pH switch
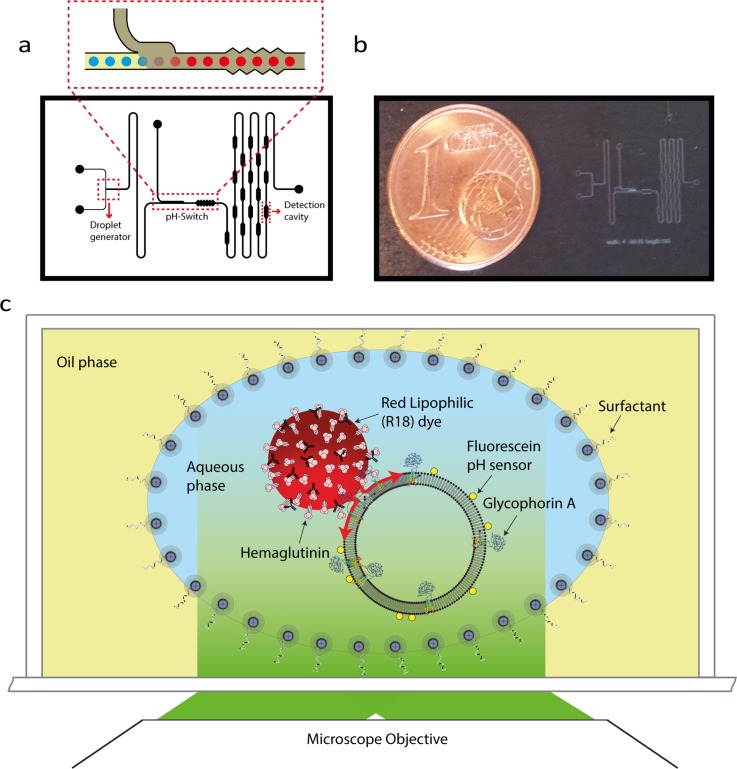
We use a 8 × 10 mm2 microfluidic chip fabricated from poly(dimethylsiloxane) (PDMS) using standard soft-lithography techniques. The width and height of the main channel are 45 and 100 μm, respectively. The oil used in this study is fluorinert-based oil with PEG-PE as the surfactant (See supplemental material at for more details).13 The device consists of three sections: a droplet generator, a pH switch, and observation cavities (Figure 1(a)). The droplet generator consists of a head-on structure to generate aqueous droplets in the oil phase. Fluid pressures of the aqueous and oil phase of droplet generation module are 10 and 8 mbar, respectively.
FIG. 1.
(a) Schematic of the microfluidic channel used in the droplet-based viral fusion assay. The components include a droplet generator, a pH switch, a micromixer, and detection cavities. The inset shows the pH switch, a structure that allows the rapid drop of droplet pH by introducing oil with an acid that is soluble in both the oil and aqueous phases. (b) The dimensions of the entire microfluidic device are 8 mm × 10 mm. (c) Schematic of the experimental assay. In this assay, droplets act as reaction chambers in which virus and receptor carrying liposome (host membrane mimic) fuse upon acidification. This process is visualized by fluorescence microscopy.
The microfluidic chip is equipped with a structure that introduces droplets to a new chemical environment, allowing for in situ adjustment of the pH of the aqueous droplets.14 The pH switch consists of a junction where a second oil phase containing an acid (acetic acid) that is both soluble in oil and water is mixed with the carrier oil in the main channel, allowing the acid to come into contact with the droplet and subsequently acidify its aqueous interior (Figure 2(a)). To achieve a pH of 5.9 in the droplets after the pH switch, acidic oil with an acid-to-oil ratio of 5.5 × 10−4% (by volume) was used at the switch with a positive pressure of 5 mbar at its inlet.
FIG. 2.
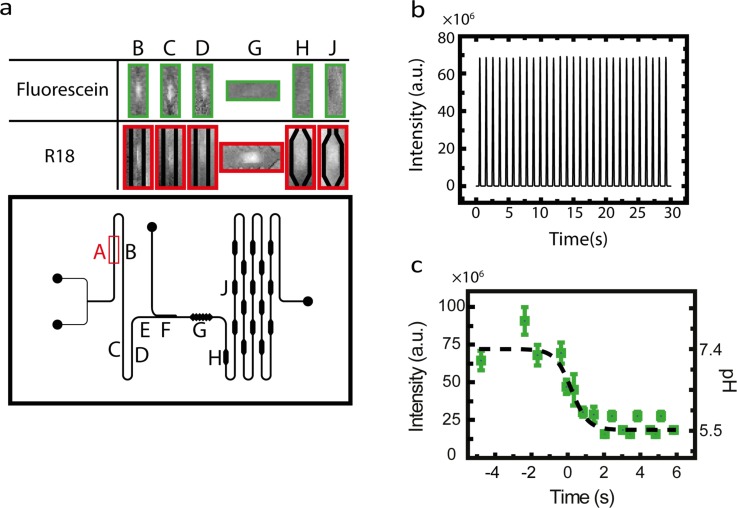
(a) Fluorescence images of droplets inside the microfluidic channel (top), obtained from positions as indicated in the lower panel. Position F shown in the channel layout is the point at which acidification is applied. The time corresponding to this position is set to t = 0. (b) The monitored fluorescence emission of selection (A), shown in panel (a), over time. Each peak represents a single droplet. The detected signal originates from fluorescein-labeled droplets. (c) Intensity of the fluorescein in the droplets at different time points (positions) in the microfluidic channel. The decrease in the fluorescence intensity at t = 0 s is due to the arrival of acidic oil from the pH switch in the main channel. Fluorescein at different known values of pH in the dispersed phase was used in the microfluidic channel with a neutral switch oil to generate the calibration curve. Given the initial pH of the aqueous phase and the calibration curve, the pH of the dispersed phase after the pH switch can be calculated.
The third section of the microfluidic chip contains a ∼48-mm long, serpentine-shaped flow channel that allows the droplets to stay within the confines of the chip for periods up to ∼8 s, allowing prolonged observation of the fusion reactions taking place within the droplets. The serpentine delay line contains periodically spaced cavities to transiently reduce the speed of a droplet to allow capturing of its fluorescence signal on a CCD camera and minimize blurring by motion.
A wide-field fluorescence microscope is used to visualize the droplet fluorescence in the microfluidic structure. The microscope is equipped with a 2×/0.08 NA objective, a custom-ordered microscope filter cube (Chroma Ltd. filters zet488/561 m and zt488/561rpc) and AVT Prosilica GX6600 camera. The home-built microscope is equipped with dual color lasers, 488 nm and 561 nm (Sapphire models, Coherent, Inc.) for visualization of both R18-labeled viruses and pH-sensitive fluorescein. The camera is controlled by Vimba 1.3 imaging software.
B. Virus, liposome, and proteo-liposome preparation
Two strains of influenza were used in this study, namely, A/Puerto Rico/8/34 (PR8) and A/HKX31 (X31). In our assay, functionalized liposomes were used as the host. To accommodate for the different surface proteins expressed at the viral surface (H3N2 for X31 and H1N1 for PR8), we engineered liposomes containing phosphatidylcholine (PC) and cholesterol supplemented with 1% Gd1A for X31 experiments and the sialoglycoprotein glycophorin A (GypA) for PR8 experiments. To prepare liposomes, lipids (DOPC:Cholesterol:gd1a/80:19:1 molar ratio) dissolved in chloroform were mixed and the chloroform was subsequently removed by argon flow followed by vacuum desiccation for 0.5–1.0 h. Subsequently, the lipid mixtures were suspended in HNE buffer (5 mM HEPES, 140 mM NaCl, 0.1 mM EDTA, pH 7.4). Next, the solution was exposed to repeated freeze/thawing cycles and extruded using polycarbonate membrane filters having a 100-nm pore diameter (Avanti Polar Lipids).
Proteoliposomes containing GypA (full-length with a GST tag, Abnova, Taipei City, Taiwan) were produced by mixing lipids solubilized in detergent with the GypA. Proteoliposomes were formed by removal of detergent with Bio-Beads (SM-2 absorbent, Bio-Rad Laboratories, Inc.).
C. Viral fusion reaction mixture
For the fusion assay, we mixed the liposome preparation with viral particles previously labeled with octadecyl rhodamine B chloride (R18). In order to label viruses, viral particles were first mixed with R18 (Figure S1) for 3 h in the dark. The viruses were then purified from the excess of dye by a PD10 size-exclusion column before use in the viral fusion experiments. We used a virus-liposome ratio of at least 1–10 to ensure an excess of liposomes compared to viruses in each droplet. After 30 min of incubation, which results in the virus binding to liposomes (Figure S2), the mixture (initial pH 7.4) was injected into the droplet-generating module. Before the injection of the mixture into the microfluidic chip, the pH-sensitive fluorescein (fluorescent above its pKa of 6.4 and non-fluorescent below this value) was added to the mixture, with a final concentration of 2 μg/ml. To perform a fusion inhibition experiment, we mixed inhibitory antibody with the labeled-virus solution at a final antibody concentration of 1 μM.15 The mixture was pre-incubated before addition of the liposomes and fluorescein.
D. Fluorescence detection of viral fusion
In this assay, the viral membrane is fluorescently labeled with a fluorophore, at a density sufficiently high to result in fluorescence quenching (Figure 1(c)). Fusion of the viral membrane with the target lipid bilayer allows the dye in the viral membrane to diffuse into the target bilayer resulting in a rapid dequenching of the fluorescence.16 For the liposome size used in our study (100 nm in diameter), fusion leads to an increase in the fluorescence signal with the signal intensity being larger than the intensity before acidification (Figure S3).
To detect fluorescence from the droplets, we measured the integrated intensity of a selected region of the main channel at a fixed position (such as selection A in Figure 2(a)) in the droplet path over time. Figure 2(b) shows an example of droplet detection on a fixed position on the chip as a function of time. Each peak corresponds to the passage of a droplet through the selected detection area. To follow the intensity of the droplet content over time, we recorded the fluorescence intensity of the droplets at different positions. Under the condition of constant fluid flow and droplet-generation rate, we could assign each position along the flow channel to a time corresponding to the travel time of the droplets in the channel in the microfluidic chip. We set the time corresponding to the passage of a droplet from pH-switch to zero. Knowing the exposure time of the collected data and the position of droplets at any given time, we assign each position in the channel to a time, with t = 0 being at the pH switch.
III. RESULTS
We initially used the X31 strain of influenza A as a model system to observe ganglioside-mediated viral fusion using our microfluidic droplet platform. We observed generation and passage of droplets through different modules of the microfluidic chip (Figure 2(a)). After generation, the droplets were moved to the first imaging section, where we recorded the initial fluorescence signal of the droplet (Figure 2(b)). Next, the droplets were transferred to the pH controller (switch), where we observed a sudden drop in the intensity of the fluorescein inside the droplets, consistent with a drop in the pH. Figure 2(c) shows that the fluorescence of fluorescein decreases at t = 0, which corresponds to the position of the pH switch. After pH adjustment, the droplets were then slowed down by their passage through detection cavities, where the fluorescence of droplets was monitored over time. Figure 3(a) illustrates the fusion of X31 with a liposome by monitoring the fluorescence of the R18 signal of the droplets (see also Figures S4 and S5). Upon viral fusion with liposomes, we detect an increase in the signal of viruses caused by the de-quenching of R18 dye.
FIG. 3.
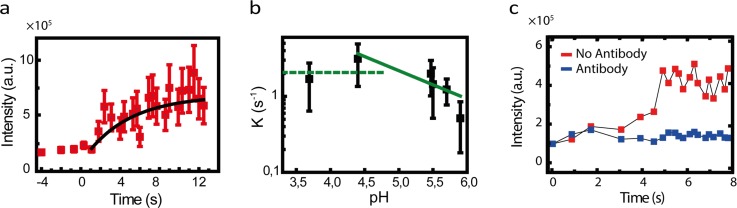
(a) Intensity of R18-labeled viruses in droplets over time. (b) Fusion rate as a function of pH. At low pH, the rate is nearly independent of pH. At high pH values, the fusion rate decreases with the increase in pH. Lines are a guide to the eye. (c) Time profiles of fluorescence intensity from R18 labeled viruses compared for viral fusion assays in the presence (red) and the absence (blue) of antibody.
Similarly, we performed the assay on the PR8 strain of influenza A, a virus that fuses more efficiently when using a glycophorin protein receptor.17 The proteoliposomes used as the host model in this assay were mixed with R18-labeled virus particles, and the mixture was used to generate aqueous droplets. The rest of the workflow was identical to the experiments described above using the X31 strain. A similar increase in fluorescence of the R18 dye was observed for PR8, as shown in Figure S6. These results show that the glycophorin protein receptor as well as PR8 virus are functional in the droplet-based platform.
Our setup enables a detailed kinetic characterization of the pH-dependent fusion of influenza with liposomes. To demonstrate this capability, we studied fusion of the X31 strain of influenza A for a number of pH values in the range of 3.5–6. Fusion of viral envelope and host membrane involves multiple kinetic barriers. The two membranes have to overcome the hydration force that prevents them from approaching each other. After coming into direct contact, the two bilayers have to deform to form a hemifusion stalk. Finally, there is a barrier associated with conversion of a hemifusion stalk to a progressively expanding pore.18,19 The envelope protein that mediates fusion, the hemagglutinin (HA) protein, is responsible for the catalysis of each of these three steps. Upon acidification, the protein complex undergoes a conformational change, which results in the formation of a long coiled-coil segment capable of penetrating a hydrophobic fusion peptide into host membrane. After binding, the protein complex folds back in a stochastic manner, a movement that forces the two membranes together.20–23
The timescale between acidification and hemifusion is determined by several factors, such as the size of the energy barrier to be overcome and the ability of the protein to coordinate its refolding with the collapse of neighboring HAs. To extract kinetic information, we measured the temporal profile of the fluorescence of the R18 of the X31 membrane for each pH value (Figure 3(b)). The probability distribution of the time intervals can be described by a gamma function. We extracted rate constants by fitting these time profiles with an integral of the gamma function (see supplemental material for more detail).13,24 The results are presented in Figure 3(b) in which multiple pH regimes can be identified. There is a pH regime in which the fusion kinetics of X31 are not affected. At lower pH values, acidity results in faster kinetics. The optimal pH is approximately 5, corresponding to the fastest hemifusion kinetics. The observed trends in Figure 3(b) are in excellent agreement with the previous reports.25
This platform can be readily adapted to study the effect of antiviral drugs on the kinetics of viral fusion. To illustrate this point, we studied inhibition of X31 viral fusion by a hemifusion-neutralizing antibody (Ab). The IgG antibody used in this study targets a conserved region in the stem region of HA and thereby inhibits influenza A HA-mediated membrane fusion. We first pre-incubated the antiviral antibody with the virus preparation and then added liposomes to the mixture. After incubation, the sample was used as the dispersed phase in our platform. When compared to our observations in the absence of Ab, setting the pH of the droplet to an acidic pH resulted in a less pronounced increase in the R18 fluorescence. We used a ratio of [virus]/[Ab] that completely prevents the rise in R18 fluorescence. Figure 3(c) compares the full inhibition of the X31 virus due to the presence of antibody with the X31 fusion experiment in the absence of the antibody.
IV. DISCUSSION
In this report, we describe the observation of viral fusion within emulsion droplets. The traditional in vitro approach to study viral fusion is by measuring lipid mixing, an approach that we adapted to our droplet-based platform. In our droplet-based assay, the observed increase in the R18 fluorescence can be attributed to fusion of the viral membrane with the liposome. Similarly, in traditional bulk liposome assays a rise in R18 fluorescence can be seen upon acidification. The fusion allows R18 (a self-quenching dye) to diffuse from the viral membrane to the host membrane. This dilution leads to dequenching and thus to an increased emission. We observed this pH-dependent dequenching for two strains of Influenza A (X31 and PR8), indicating that both viruses retain their integrity and fusogenicity in the oil/water emulsion. The measured fusion kinetics agree well with the previous reports based on single-particle and ensemble studies,25–30 where conventional oil-free systems were used.
Our droplet-based technique provides a number of advantages over bulk assays to monitor viral fusion, including increased throughput, reduced reagent use, and fast mixing needed for kinetic analysis. Viral fusion is a rapid process and fast pH change, as provided by our droplet-platform, is needed for reliable monitoring of the kinetics of the reaction. Mixing a large volume in a conventional dequenching assay may give rise to an asynchronous triggering, and the resulting poorly defined timing of the pH drop may mask the magnitude of the pH dependence of fusion. Therefore, to facilitate a rapid distribution of the acid throughout the cuvette, rapid mixing of contents is needed. However, rapid mixing results in shearing and may disrupt virus binding. We have overcome this limitation by using the on-chip pH switch module and small droplet volumes, providing a rapid pH change in our reaction chamber (droplet).
Introducing viral fusion assays into droplet microfluidics presents both challenges and opportunities. One challenge is to design the chemistry of the aqueous and oil phases in a way that maintains the functionality of the reactants. Care has to be taken when designing the experiments so that incorporated viruses and membranes do not penetrate the oil phase and preserve their native structure. Adsorption of biological material to the interface also affects droplet stability.31,32 Another important requirement is a detection technology that allows a sufficiently high signal-to-noise ratio to detect and analyze viral fusion at the level of individual droplets. In addition, the detection protocol has to be sufficiently rapid to monitor the relatively fast kinetics of viral fusion. Finally, additional controls may be needed when screening for previously uncharacterized therapeutics. For example, the antibody used in our study has been previously characterized,33 and optimal concentration for neutralization15 was known. This is not the case in screening applications; previously uncharacterized antibodies have to be compared with non-neutralizing antibodies as well as no-antibody controls.
The usage of Fluorinert FC-40 oil and a biocompatible surfactant (PEGylated-PE) in our assay ensured the integrity of the reactants. The amount of surfactant used in this assay was also optimized for droplet stability. A high amount of surfactant leads to the splitting of droplets as they travel along the chip, a process that is caused by roughness of the surface. If the amount of surfactant is too low, the droplets become prone to splitting at the chemical switch, a process that is the result of the interaction of biological material with the interface.
The speed of pH adjustment in our study can be increased by introducing a micro-switch and suppressing material exchange between consecutive droplets.34,35 In the experiments presented here, the pH of the chemical switch was typically set to a desired pH, followed by the collection of data from a train of droplets. After rigorous washing, the system was operated at a new desired pH. One can adjust the pH of the chemical switch with a much higher frequency using a micro-switch-equipped chemical switch. For certain applications, one might be interested in adjusting the pH of every single droplet independently to a different pH. The challenge would then be to make the pH of two consecutive droplets uncorrelated, particularly when the inter droplet distance approaches the droplet length. A solution to this problem could be the introduction of an air bubble before and after each aqueous droplet.
V. CONCLUSIONS AND OUTLOOK
Here, we report on the development of a droplet-based viral fusion assay that can be used for kinetic studies. We demonstrated this platform using two influenza viruses that exploit very different cell-surface receptors. This general approach can be used to study other similar viruses such as hepatitis C virus (HCV) and dengue. One can speculate on possible future extensions of the proposed technology beyond its application in antiviral drug studies. In view of recent developments in single-cell analysis and sorting, we envision that our assay can be further expanded to study single-cell interactions with single viral particles.36 This application will allow studies of not only fusion but also biological processes that follow the infection such as the changes in the gene expression profile. Furthermore, viruses are used as gene delivery vesicles for therapeutic purposes and engineering applications such as viral barcoding of cells. Our droplet-based viral fusion platform can be used for controlled viral barcoding of cells, which is needed, among others, to study cellular heterogeneity.37 Viral genetic barcoding can be used to track single cells in heterogeneous populations such as differentiating hematopoietic stem cells and growing tumors.38,39
ACKNOWLEDGMENTS
The authors would like to thank Dr. Alireza Mashaghi and Dr. Harshad Ghodke for critical reading of the manuscript. Funding of this research was from the European Research Council (ERC Starting Grant No. 281098) and the Dutch Science Foundation (NWO Vici Grant No. 680-47-607).
References
- 1. Villar G., Graham A. D., and Bayley H., Science 340(6128), 48–52 (2013). 10.1126/science.1229495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tewhey R., Warner J. B., Nakano M., Libby B., Medkova M., David P. H., Kotsopoulos S. K., Samuels M. L., Hutchison J. B., Larson J. W., Topol E. J., Weiner M. P., Harismendy O., Olson J., Link D. R., and Frazer K. A., Nat. Biotechnol. 27(11), 1025–1031 (2009). 10.1038/nbt.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang J., Morton M. J., Elliott C. T., Karoonuthaisiri N., Segatori L., and Biswal S. L., Anal. Chem. 86(3), 1671–1678 (2014). 10.1021/ac403437x [DOI] [PubMed] [Google Scholar]
- 4. Boedicker J. Q., Li L., Kline T. R., and Ismagilov R. F., Lab Chip 8(8), 1265–1272 (2008). 10.1039/b804911d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nightingale A. M., Phillips T. W., Bannock J. H., and de Mello J. C., Nat. Commun. 5, 3777 (2014). 10.1038/ncomms4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guo M. T., Rotem A., Heyman J. A., and Weitz D. A., Lab Chip 12(12), 2146–2155 (2012). 10.1039/c2lc21147e [DOI] [PubMed] [Google Scholar]
- 7. Dittrich P. S. and Manz A., Nat. Rev. Drug Discovery 5(3), 210–218 (2006). 10.1038/nrd1985 [DOI] [PubMed] [Google Scholar]
- 8. Wootton R. C. R. and deMello A. J., Nature 483(7387), 43–44 (2012). 10.1038/483043a [DOI] [PubMed] [Google Scholar]
- 9. McCullers J. A., Nat. Rev. Microbiol. 12(4), 252–262 (2014). 10.1038/nrmicro3231 [DOI] [PubMed] [Google Scholar]
- 10. Yang J., Li M., Shen X., and Liu S., Viruses 5(1), 352–373 (2013). 10.3390/v5010352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arts E. J. and Hazuda D. J., Cold Spring Harbor Perspect. Med. 2(4), a007161 (2012) 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zuckerman A. J., Banatvala J. E., Griffiths P., Schoub B., and Mortimer P., Principles and Practice of Clinical Virology ( Wiley, 2009). [Google Scholar]
- 13.See supplementary material at http://dx.doi.org/10.1063/1.4943126E-BIOMGB-10-011602 for more details of Materials and Methods.
- 14. Mashaghi S. and van Oijen A. M., Sci. Rep. 5, 11837 (2015). 10.1038/srep11837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Otterstrom J. J., Brandenburg B., Koldijk M. H., Juraszek J., Tang C., Mashaghi S., Kwaks T., Goudsmit J., Vogels R., Friesen R. H. E., and van Oijen A. M., Proc. Natl. Acad. Sci. U.S.A. 111(48), E5143–E5148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoekstra D. B. T., Klappe K., and Wilschut J., Biochemistry 20(23(24)), 5675–5681 (1984) 10.1021/bi00319a002. [DOI] [PubMed] [Google Scholar]
- 17. Gambaryan A., Yamnikova S., Lvov D., Tuzikov A., Chinarev A., Pazynina G., Webster R., Matrosovich M., and Bovin N., Virology 334(2), 276–283 (2005). 10.1016/j.virol.2005.02.003 [DOI] [PubMed] [Google Scholar]
- 18. Chernomordik L. V. and Kozlov M. M., Nat. Struct. Mol. Biol. 15(7), 675–683 (2008). 10.1038/nsmb.1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marsden H. R., Tomatsu I., and Kros A., Chem. Soc. Rev. 40(3), 1572–1585 (2011). 10.1039/C0CS00115E [DOI] [PubMed] [Google Scholar]
- 20. Skehel J. J. and Wiley D. C., Annu. Rev. Biochem. 69(1), 531–569 (2000). 10.1146/annurev.biochem.69.1.531 [DOI] [PubMed] [Google Scholar]
- 21. Harrison S. C., Virology 479–480(0), 498–507 (2015). 10.1016/j.virol.2015.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kielian M., Annu. Rev. Virol. 1(1), 171–189 (2014). 10.1146/annurev-virology-031413-085521 [DOI] [PubMed] [Google Scholar]
- 23. Harrison S. C., Nat. Struct. Mol. Biol. 15(7), 690–698 (2008). 10.1038/nsmb.1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feller W., An Introduction to Probability Theory and its Applications ( John Wiley & Sons, 2008). [Google Scholar]
- 25. Floyd D. L., Ragains J. R., Skehel J. J., Harrison S. C., and van Oijen A. M., Proc. Natl. Acad. Sci. 105(40), 15382–15387 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mashaghi S. and van Oijen A. M., Biotechnol. Bioeng. 111(10), 2076–2081 (2014) 10.1002/bit.25273. [DOI] [PubMed] [Google Scholar]
- 27. Hoekstra D., Klappe K., De Boer T., and Wilschut J., Biochemistry 24(18), 4739–4745 (1985). 10.1021/bi00339a005 [DOI] [PubMed] [Google Scholar]
- 28. Kemble G. W., Danieli T., and White J. M., Cell 76(2), 383–391 (1994). 10.1016/0092-8674(94)90344-1 [DOI] [PubMed] [Google Scholar]
- 29. Danieli T., Pelletier S. L., Henis Y. I., and White J. M., J. Cell Biol. 133(3), 559–569 (1996). 10.1083/jcb.133.3.559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chernomordik L. V., Leikina E., Frolov V., Bronk P., and Zimmerberg J., J. Cell Biol. 136(1), 81–93 (1997). 10.1083/jcb.136.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Retzinger G. S., DeAnglis A. P., and Patuto S. J., Arterioscler. Thromb. Vasc. Biol. 18(12), 1948–1957 (1998). 10.1161/01.ATV.18.12.1948 [DOI] [PubMed] [Google Scholar]
- 32. Yeung A., Dabros T., Czarnecki J., and Masliyah J., Proc. R. Soc. London, A: Math. Phys. Eng. Sci. 455(1990), 3709–3723 (1999). 10.1098/rspa.1999.0473 [DOI] [Google Scholar]
- 33. Ekiert D. C., Friesen R. H. E., Bhabha G., Kwaks T., Jongeneelen M., Yu W., Ophorst C., Cox F., Korse H. J. W. M., Brandenburg B., Vogels R., Brakenhoff J. P. J., Kompier R., Koldijk M. H., Cornelissen L. A. H. M., Poon L. L. M., Peiris M., Koudstaal W., Wilson I. A., and Goudsmit J., Science 333(6044), 843–850 (2011). 10.1126/science.1204839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang Z. and Chakrabarty K., IEEE Trans. Comput. Aided Des. Integr. Circuits Syst. 31(6), 817–830 (2012). 10.1109/TCAD.2012.2183369 [DOI] [Google Scholar]
- 35. Courtois F., Olguin L. F., Whyte G., Theberge A. B., Huck W. T. S., Hollfelder F., and Abell C., Anal. Chem. 81(8), 3008–3016 (2009). 10.1021/ac802658n [DOI] [PubMed] [Google Scholar]
- 36. Mazutis L., Gilbert J., Ung W. L., Weitz D. A., Griffiths A. D., and Heyman J. A., Nat. Protoc. 8(5), 870–891 (2013). 10.1038/nprot.2013.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu R., Neff N. F., Quake S. R., and Weissman I. L., Nat. Biotechnol. 29(10), 928–933 (2011). 10.1038/nbt.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nolan-Stevaux O., Tedesco D., Ragan S., Makhanov M., Chenchik A., Ruefli-Brasse A., Quon K., and Kassner P. D., PLoS One 8(6), e67316 (2013). 10.1371/journal.pone.0067316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Porter S., Baker L., Mittelman D., and Porteus M., Genome Biol. 15(5), R75 (2014). 10.1186/gb-2014-15-5-r75 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4943126E-BIOMGB-10-011602 for more details of Materials and Methods.