Abstract
Each amino acid has its intrinsic propensity for certain local backbone conformations, which can be further modulated by the physicochemical environment and post-translational modifications. In this work, we study the effects of phosphorylation on the intrinsic propensity for different local backbone conformations of serine/threonine by molecular dynamics simulations. We showed that phosphorylation has very different effects on the intrinsic propensity for certain local backbone conformations for the serine and threonine. The phosphorylation of serine increases the propensity of forming polyproline II, whereas that of threonine has the opposite effect. Detailed analysis showed that such different responses to phosphorylation mainly arise from their different perturbations to the backbone hydration and the geometrical constraints by forming side-chain–backbone hydrogen bonds due to phosphorylation. Such an effect of phosphorylation on backbone conformations can be crucial for understanding the molecular mechanism of phosphorylation-regulated protein structures/dynamics and functions.
Electronic supplementary material
The online version of this article (doi:10.1007/s10867-015-9405-0) contains supplementary material, which is available to authorized users.
Keywords: Phosphorylation, Intrinsic backbone conformation propensity, Proteins, Molecular dynamics
Introduction
The control of biological activity of proteins through phosphorylation plays important roles in many biochemical processes [1–5]. With the help of various kinases, phosphorylated proteins regularly adopt different conformations to promote or inhibit the functions of proteins, no matter that the proteins have well-folded native structures [6, 7] or are intrinsically disordered. One typical example is the tumor suppressor protein p53 [8]. In the N-terminal domain of p53, several residues could be phosphorylated, and the phosphorylation of different combinations of residues may produce diverse functions of p53. How does phosphorylation affect the protein structure/dynamics so as to generate different functional responses? The answer to this question is important for understanding the fundamental contributions of phosphorylation to protein structures/dynamics, and may help us to design or block phosphorylation sites with novel functions.
There are two possible mechanisms by which phosphorylation may modify protein structure/dynamics, including: (1) modifying the intrinsic propensity toward certain local backbone conformations and (2) modifying the global interactions due to the highly charged nature of the phosphate group. As a first step to this problem, it is necessary to elucidate possible effects of phosphorylation on the intrinsic propensity toward certain local backbone conformations of the central serine/threonine, although the final effects also rely on nonlocal interactions, which are the focus of many previous works [9, 10]. Such effect of phosphorylation on the intrinsic propensity toward certain local backbone conformations can be especially important for intrinsically disordered proteins, for which the long-range global interactions are less significant compared to those of well-structured proteins. In addition, the intrinsic propensity toward certain local backbone conformations of a certain amino acid is also an important input for some computational models of protein folding/aggregation and structure predictions [11–17]. For example, recent computational works showed that introducing amino acid-specific parameters to the intrinsic conformational propensity in the all-atom force field can drastically improve protein folding and structure predictions [18, 19].
Consistent with the above idea, there have recently been some experimental and computational works investigating the effects of phosphorylation on the backbone conformations for some short peptides. With FTIR and CD techniques, Kim and coworkers showed that phosphorylation has different effects on the backbone conformational preferences of serine and threonine peptides [20]. In Ref. [21], the effect of phosphorylation for a poly-serine peptide was studied by Brownian simulations with implicit solvent. In Ref. [22], by combining NMR measurement and molecular dynamics(MD) simulations for serine/arginine-rich proteins, the authors showed that phosphorylation of serine switches the serine/arginine-rich domain from a fully disordered state to a partially rigidified structure, demonstrating a dynamic switching mechanism. The effects of phosphorylation on protein–protein interactions, the cis-trans isomerization of peptidyl-prolyl bonds, and protein global structures have also been investigated [23–25]. These works provided valuable information about the effects of phosphorylation on protein conformational and dynamical features. However, the relationship between the structural/dynamics variations and the key interactions introduced by phosphorylation remains elusive. It is widely known that hydration plays an important role for polyproline II (PPII) conformational propensity of amino acids, especially through the interactions with the backbone groups (-NH-CO-) [26–28]. A possible reason is that the hydration of backbone groups in the PPII conformation produces minimal disruption of the nearby bulk water structure [28]. On the other hand, the local intra-peptide backbone hydrogen bonds favor the helical conformation. In addition, the hydrogen bonds formed between the side chain of a certain amino acid and its neighboring backbone groups may impose geometrical constraints, which can further modulate the intrinsic propensity toward certain backbone conformations [29]. How does phosphorylation modulate these interactions to induce structural variations? Facing such a question, an atomic-level understanding of the effects of phosphorylation on the structural propensity toward certain peptide backbone conformations is necessary, and requires further simulation studies.
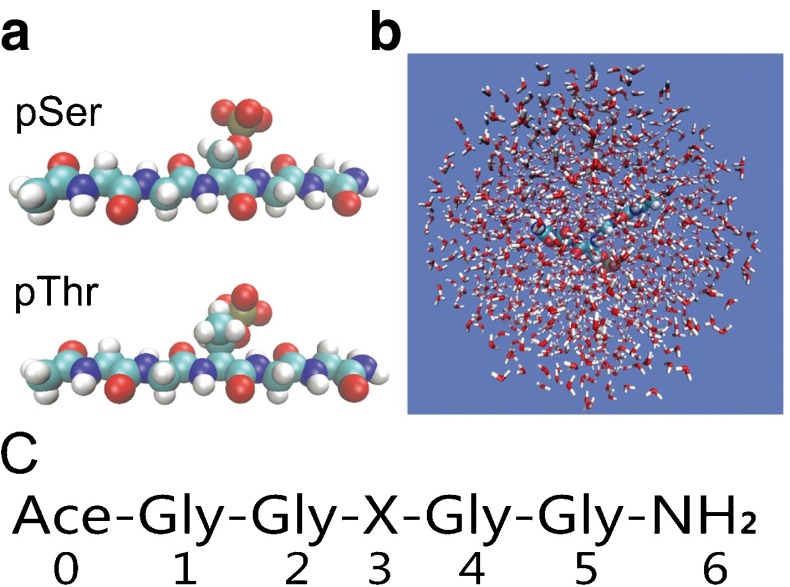
The intrinsic backbone conformation propensity of a certain amino acid is mainly determined by the interactions arising from the side-chain groups, the neighboring backbone peptide groups, and solvent [18, 29, 30]. To study the intrinsic propensity toward a given local backbone conformation for a certain amino acid, people often used a simple dipeptide (ACE-X-NME, with X denoting a certain amino acid) or a host-guest peptide (GXG) to parameterize/characterize the interactions both experimentally and computationally [31–33]. In addition, the host-guest peptide AcGGXGGNH 2 was also frequently used [29, 30, 34–38]. In the host-guest peptide AcGGXGGNH 2, the glycines flanking residue X act as a spacer, which alleviates the influence of the end groups. In this work, we study the effects of phosphorylation on the intrinsic propensity toward certain backbone conformations of serine and threonine by molecular dynamics simulations for the peptide AcGGSGGNH 2, AcGGTGGNH 2, and their phosphorylated counterparts (Fig. 1).
Fig. 1.
a Model peptides with the central serine (upper) and threonine (lower) phosphorylated; b Representative structure of the simulation system. c Sequences of the model peptides discussed in this work with X denoting the amino acids Ser, Thr, pSer, and pThr. The numbers represent the residue indices referred to in this work
We observed that the phosphorylation of serine enhances the propensity toward PPII conformation, whereas that of threonine decreases the PPII propensity, which is consistent with the experimental results by Kim and coworkers [20]. Further analysis showed that the phosphorylation of serine and threonine modifies the interactions between side-chain and backbone groups to different extents. Consequently, phosphorylation has distinct effects on backbone hydration and geometrical constraint, which leads to the opposite effects of phosphorylation on the PPII propensity for the central serine/threonine. Our results provide insights into the molecular mechanism for phosphorylation-regulated conformational changes of proteins.
Materials and methods
In the process of phosphorylation, the hydroxyl in the side chain of the serine/threonine residue is substituted by a phosphate group (Fig. 1a). The new side chain is negatively charged at neutral conditions and could establish strong electrostatic interactions with surrounding water molecules and neighboring residues [39]. In our simulation, the short peptides were generated with AmberTools [40]. The N- and C-termini of the short peptides were capped with acetyl (ACE) and amide (NHE), respectively, to keep them neutral. The molecular dynamics simulations were performed with the SANDER module of the AMBER11 package employing the Amber ff10 force field [40, 41]. The force-field parameters for the phosphorylated serine and threonine were taken from Ref. [42], in which a charge state of -2 was assigned to the phosphate groups. For comparison, similar simulations were conducted with force field charmm36 using GROMACS software [43–48]. Simulations were carried out for the four model peptides separately, i.e., AcGGSGGNH 2, AcGGTGGNH 2, and their phosphorylated counterparts. All the peptides were solvated in a truncated-octahedral TIP3P water box that extended at least 12 Å in each direction from the solute [49] (Fig. 1b). The size of the resulting water box is large enough to reduce the interactions between the peptide and its image as shown in Fig. S1. Sodium ions and chloride ions were added to neutralize the net charges of the simulation systems and to mimic the physiological salt concentration of 0.15 M. Non-bonded van der Waals interactions were truncated at 8 Å. All bonds involving hydrogen atoms were constrained using SHAKE algorithm [50]. Particle mesh Ewald (PME) was used to work with the electrostatic interactions with periodic boundary conditions [51]. After initial minimizations, the systems were gradually heated to 1000 K and then equilibrated for 1.0 ns. The obtained structures, which have minimum correlations, were used as the initial structures of the subsequent replica exchange molecular dynamics (REMD) simulations which have lasted for 100 ns for each replica [52, 53]. In total, 32 replicas were used for each REMD simulation with temperatures of 278.0, 282.2, 286.5, 290.9, 295.4, 300.0, 304.7, 309.5, 314.4, 319.4, 324.5, 329.8, 335.1, 340.6, 346.3, 352.1, 356.1, 362.0, 370.4, 376.8, 383.4, 390.2, 397.3, 404.5, 411.9, 419.7, 427.6, 435.9, 446.0, 455.3, 465.0, and 478.0 K. The first 10 ns of the REMD simulations were used for relaxing the initial unrealistic structures prepared by the high-temperature simulations, and were omitted during the subsequent analysis.
In all the model peptides, the host residue (Ser3, Thr3, pSer3, or pThr3) would generally adopt three major conformations, PPII, helix and β-strand. These conformations are predominantly even in phosphorylated states. In our analysis, the sampled snapshots were assigned as PPII (−125∘<Φ<0∘; Ψ>75∘ or Ψ<−125∘), helix (−125∘<Φ<0∘; −75∘<Ψ<25∘) and β-strand (Φ<−125∘; Ψ>75∘ or Ψ<−125∘) based on the corresponding areas in the Ramachandran plot. The hydrogen bond is assumed to be formed if the distance between the heavy atoms of donor and acceptor is less than 3.0 Å and the angle formed by donor-hydrogen-acceptor is larger than 160∘.
Results and discussion
Effects of phosphorylation on the conformational distribution of the serine and threonine peptides
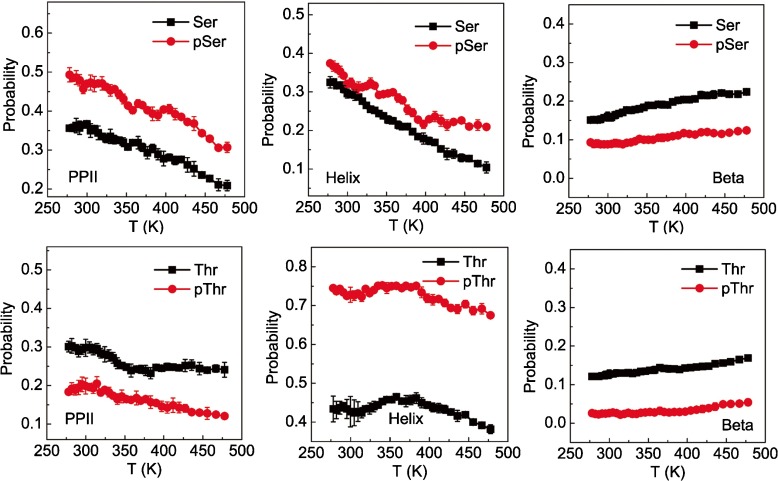
Based on our simulations, the backbone conformational propensities of the host residues at various temperatures were obtained for the peptides before and after phosphorylation (Fig. 2). It is worth noting that the number of conformations other than the above three conformations is rather small though they indeed could be observed. In consequence, these outlier conformations were not analyzed in the present study. As a result, the sum of the populations of the three major states is less than unity. To test possible convergence of the simulations, a jack-knife method was employed. That is, the full data sampled at each temperature were divided into ten groups with equal time duration; the free energy landscapes projected on the Φ- Ψ space constructed from the data of each group are similar (Fig. S2). In addition, we showed that the continuous trajectory reconstructed from the REMD simulations hops between different basins of the conformational space, suggesting high sampling quality (Fig. S3). The populations related to each group are calculated separately. Based on these populations, the standard errors were derived (as shown by the error bars in Fig. 2). It is clear that the errors are rather small compared to the variations of the populations, which indicates that the sampling in this work is reasonably converged and is reliable to be used in further structural comparisons. Actually, the populations converge to the saturation values even within the initial 10-ns relaxation simulations omitted in the data analysis (Fig. S4). The results in Fig. 2 show that the phosphorylation of serine enhances the propensity for the PPII conformation. Differently, for threonine, phosphorylation disfavors the formation of PPII. For both peptides, phosphorylation tends to decrease the propensity of β-strand formation and promotes the helical conformation. Clearly, phosphorylation has an opposite effect on the propensities of forming the PPII conformation for serine and threonine residues. Furthermore, when the temperature is higher, the proportion of amino acids adopting of helix and PPII conformations decrease and that of β-strand increases for both serine and threonine, which is consistent with the literature [54]. In this work, we used a cut-off of 8.0 Å for the non-bonded van der Waals interactions. To test the possible effect of cut-off the value on the results, we also conducted similar simulations for the peptide AcGGSGGNH 2, and the results do not change significantly (Fig. S5). In addition, we conducted similar simulations with the charmm36 force field, which shows similar qualitative trends as a result of phosphorylation, although the quantitative results can be different (Fig. S6).
Fig. 2.
Probability of the PPII (left), helix (middle) and β-strand (right) conformations of the host residues as a function of temperature for the model peptides AcGGSGGNH 2 (upper), AcGGTGGNH 2 (lower), and their phosphorylated counterparts
Molecular mechanism of phosphorylation-induced modulations of the intrinsic backbone conformation propensity
It is widely recognized that the backbone group (-CO-NH-) solvation on both sides of a certain amino acid is a major, if not the dominant, driving force in the formation of PPII [28, 55–58]. In addition, the hydrogen bonds between the side-chain and the neighboring backbone groups can impose geometrical constraints and therefore modify the local backbone structure. In this sense, it is necessary to evaluate the variations of the local interactions (often related to hydrogen bonds) during phosphorylation, which may help us to understand the variations of backbone conformational propensity of the model peptides.
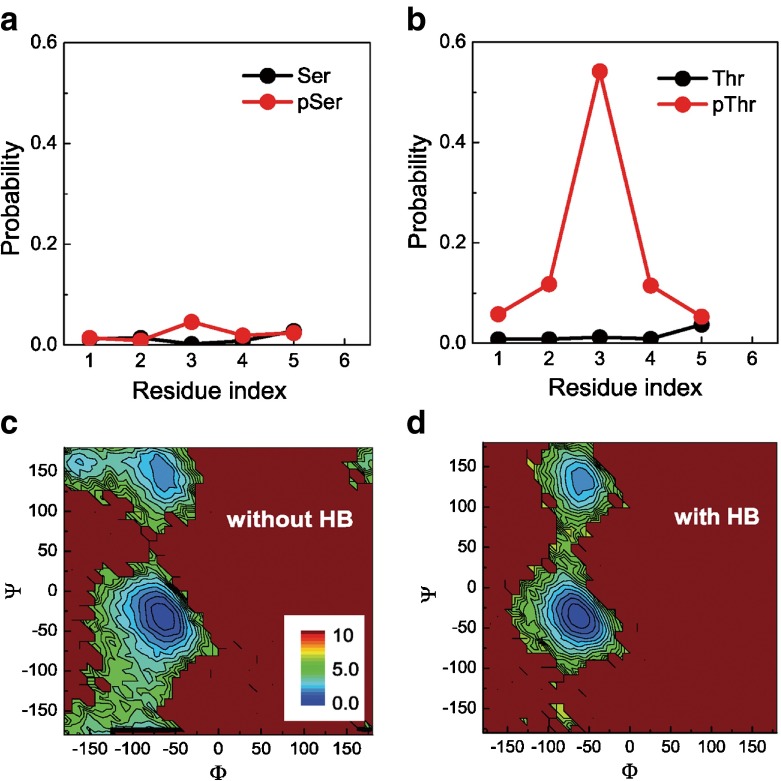
In the four model peptides, the main differences come from the side chains of the host residues. Upon phosphorylation, the side chains of serine and threonine are substituted with a long and negatively charged side chain. Compared to serine, threonine has an additional methyl on the Cβ atom, which may introduce differences in the interactions between the side-chain and the backbone groups and modulate backbone hydration and the propensity of backbone conformation. Following this line, we analyzed the interactions between the phosphate and the backbone groups. The probability of hydrogen bonds formed between the phosphate and each of the NH groups are shown in Fig. 3a, b. One can see that before phosphorylation, there are negligible interactions between the side-chain and backbone groups for either serine or threonine. After phosphorylation, the interactions between the side-chain and the backbone NH group increase drastically for the threonine-related peptide. In contrast, those of the serine-related peptide are much weaker. Particularly, the phosphate group has the highest probability to form hydrogen bonds with the NH group of the phosphorylated threonine/serine. Undoubtedly, such interaction between the side-chain phosphate and the backbone group can impose a geometrical constraint on the backbone’s local conformation and affect the intrinsic propensity of the backbone conformations.
Fig. 3.
a, b The distributions of hydrogen bonds between the side-chain and the NH group of serine (a) and threonine (b) before (black) and after (red) phosphorylation at 300 K; c, d free energy landscapes on the Φ- Ψ space of the phosphorylated threonine with (c) and without (d) the formation of hydrogen bonds between the phosphate and the NH group of the phosphorylated threonine residue at 300.0 K. The unit of free energy is k B T
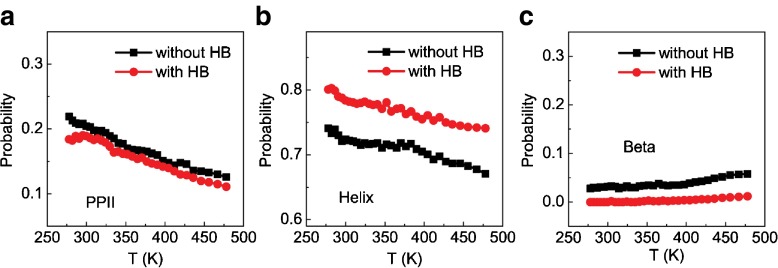
Figure 3c, d shows the free energy landscapes on the Φ- Ψ space of the phosphorylated central threonine with and without the formation of hydrogen bonds between the phosphate and the NH group of the phosphorylated threonine residue. One can see that the formation of the hydrogen bonds can largely affect the population of the major states, therefore imposing geometrical constraints to the backbone conformations of the central threonine. Such geometrical restraints will inevitably affect the propensity of the backbone conformations, as already shown in Fig. 3c, d. In Fig. 4, we show the probability of the PPII, helix and β-strand conformations of the phosphorylated threonine as a function of temperature with and without the formation of hydrogen bonds between the phosphate group and the NH group of the phosphorylated threonine. One can see that the phosphate–backbone interactions can drastically stabilize the helical conformation and moderately destabilize the β-strand conformation. In comparison, such interactions have negligible effect on the populations of the PPII conformation, implying that the geometrical constraint imposed by the interactions between side chain and backbone would be more compatible with the helical conformation.
Fig. 4.
Probability of the PPII (a), helix (b), and β-strand (c) conformations of the phosphorylated threonine as a function of temperature with (red) and without (black) the formation of the hydrogen bonds between the phosphate and the NH group of the same residue
Figure 5a, b shows the normalized histograms of the number of hydrogen bonds formed between water and the backbone groups neighboring the host residue at 300.0 K. The cases corresponding to the conditions before and after phosphorylation are represented with black and red colors, respectively. It is found that the backbone groups of the serine peptide can form more hydrogen bonds with water after phosphorylation. Therefore, the phosphorylated serine peptide has a higher ability to be hydrated, which tends to favor the PPII conformation. Such results are consistent with the above observations that the phosphorylation of serine could increase the content of PPII conformations. In comparison, the backbone groups of the threonine peptide becomes less accessible to water upon phosphorylation, which is consistent with the above observations that the PPII propensity is decreased by phosphorylation for the threonine-related peptide. Figure 5c, d shows detailed analysis of the hydrogen bond networks formed by water molecules and the backbone groups. The populations around the right-bottom corner represent the formation of backbone–water hydrogen bonds. One can see that the phosphorylation of the central serine increases the population of stable hydrogen bonds between water and the backbone carbonyl group, but almost does not change the hydrogen bonds between water and the backbone NH group (Fig. S7a). In comparison, the phosphorylation of the central threonine decreases the population of stable hydrogen bonds between water and the backbone NH group, and also decreases slightly the hydrogen bonds between water and the backbone carbonyl group (Fig. S7B). The correlation between hydration extent and PPII-forming propensity suggests that the modulations of the backbone hydration by phosphorylation can contribute to the observed effects of phosphorylation on the PPII-forming propensity.
Fig. 5.
a, b The probability distribution of the number of hydrogen bonds (HB) formed between water and the neighboring backbone groups of the host residues before (black) and after phosphorylation (red) for the serine (a) and threonine (b) related peptides at 300 K. c, d The free energy landscapes projected on the space formed by the distance and angle of the hydrogen bond acceptors and donors from the neighboring backbone groups and water molecules before and after phosphorylation for serine (c) and threonine (d) at 300.0 K. The distance is represented by the heavy atoms of the donor and acceptor. Only the results between backbone carbonyl (backbone NH) groups and water molecules were shown for serine (threonine), since these groups show the most significant changes by phosphorylation. The results for other groups are shown in Fig. ??. The unit of free energy is k B T
By combining the results shown in Figs. 2, 3, 4, and 5, we may discuss the molecular mechanism for the phosphorylation-modulated backbone conformation propsenity. For the phosphorylated threonine, the hydrogen bonds formed between the phosphate group and the backbone NH of the phosphorylated threonine impose a geometrical constraint, which tends to stabilize the helical conformation, as illustrated by Fig. 2b. Such enhanced helical propensity can equivalently contribute to the decreased PPII propensity observed in Fig. 2. On the other hand, the increased hydration of the backbone groups enhances the PPII-forming propensity for the phosphorylated serine. In comparison, the additional hydrophobic methyl group in the side chain disfavors the hydration of the nearby backbone group for the phosphorylated threonine, and therefore disfavors the PPII conformation as observed in Fig. 2.
In summary, the effects of phosphorylation on the intrinsic peptide backbone conformation propensity of serine and threonine have been studied by replica exchange molecular dynamics simulations. The phosphorylation of serine enhances backbone group hydration, which raises the likelihood of the PPII conformation. In comparison, the phosphorylation of threonine tends to decrease the PPII propensity and increase the likelihood of a helical conformation. Such different effects are attributed to the methyl group on the Cβ of the side chain, which leads to significant geometrical constraints favoring the helical conformation and decreased backbone hydration disfavoring the PPII conformation.
The intrinsic propensity of backbone local conformations is one of the most important inputs for the development of a protein force field. Recent computational work showed that considering the amino acid specificity of the intrinsic propensity of backbone conformations is very important in modeling protein structure and stability. Phosphorylation modifies the physiochemical properties of the serine and threonine, and therefore changes their intrinsic propensity to take certain local conformations as demonstrated in this work. Although there are a number of computational works focusing on the effects of phosphorylation on protein global structure and dynamics, phosphorylation effects on the intrinsic propensity of backbone local conformations of amino acids demonstrated in the current work can contribute to the understanding of the phosphorylation-induced conformational changes of functionally important proteins, especially intrinsically disordered proteins, from a different aspect.
Electronic supplementary material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant numbers 11574132, 11174133, 11174134, and 11274157) and Jiangsu Province (BK2011546). W.L. also acknowledges the support of the Program for New Century Excellent Talents in University and the PAPD project of Jiangsu higher education institutions. The numerical calculations in this paper have been done on the IBM Blade cluster system in the High Performance Computing Center of Nanjing University.
Contributor Information
Erbin He, Email: herbin@biophy.nju.edu.cn.
Guanghui Yan, Email: ghyan@njit.edu.cn.
Jian Zhang, Email: jzhang@nju.edu.cn.
Jun Wang, Email: wangj@nju.edu.cn.
Wenfei Li, Email: wfli@nju.edu.cn.
References
- 1.Walsh CT. Posttranslational Modification of Proteins: Expanding Nature’s Inventory. Englewood: Colorado: Roberts and Company Publishers; 2006. [Google Scholar]
- 2.Marks F. Protein Phosphorylation. Weinheim, Germany: VCH Verlagsgesellschaft mbH; 1996. [Google Scholar]
- 3.Chen HF. Molecular dynamics simulation of phosphorylated KID post-translational modification. PLoS ONE. 2009;4:e6516. doi: 10.1371/journal.pone.0006516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jho YS, Zhulina EB, Kim MW, Pincus PA. Monte Carlo simulations of tau proteins: effect of phosphorylation. Biophys. J. 2010;99:2387–2397. doi: 10.1016/j.bpj.2010.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valiev M, Yang J, Adams JA, Taylor SS, Weare JH. Phosphorylation reaction in cAPK protein kinase-free energy quantum mechanical/molecular mechanics simulations. J. Phys. Chem. B. 2007;111:13455–13464. doi: 10.1021/jp074853q. [DOI] [PubMed] [Google Scholar]
- 6.Mortishire-Smith RJ, Pitzenberger SM, Burke CJ, Middaugh CR, Garsky VM, Johnson RG. Solution structure of the cytoplasmic domain of phospholamban: phosphorylation leads to a local perturbation in secondary structure. Biochemistry. 1995;34:7603–7613. doi: 10.1021/bi00023a006. [DOI] [PubMed] [Google Scholar]
- 7.Quirk PG, Patchell VB, Colyer J, Drago GA, Gao Y. Conformational effects of serine phosphorylation in phospholamban peptides. Eur. J. Biochem. 1996;236:85–91. doi: 10.1111/j.1432-1033.1996.00085.x. [DOI] [PubMed] [Google Scholar]
- 8.Schon O, Friedler A, Bycroft M, Freund SMV, Fersht AR. Molecular mechanism of the interaction between MDM2 and p53. J. Mol. Biol. 2002;323:491–501. doi: 10.1016/S0022-2836(02)00852-5. [DOI] [PubMed] [Google Scholar]
- 9.Lee HJ, Srinivasan D, Coomber D, Lane DP, Verma CS. Modulation of the p53-MDM2 interaction by phosphorylation of Thr18: a computational study. Cell Cycle. 2007;6:2604–2611. doi: 10.4161/cc.6.21.4923. [DOI] [PubMed] [Google Scholar]
- 10.Fraser JA, Vojtesek B, Hupp TR. A novel p53 phosphorylation site within the MDM2 ubiquitination signal I. Phosphorylation at ser269 in vivo is linked to inactivation of p53 function. J. Biol. Chem. 2010;285:37762–37772. doi: 10.1074/jbc.M110.143099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li WF, Wolynes PG, Takada S. Frustration, specific sequence dependence, and nonlinearity in large-amplitude fluctuations of allosteric proteins. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3504–3509. doi: 10.1073/pnas.1018983108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li WF, Wang W, Takada S. Energy landscape views for interplays among folding, binding, and allostery of calmodulin domains. Proc. Natl. Acad. Sci. U.S.A. 2014;111:10550–10555. doi: 10.1073/pnas.1402768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terakawa T, Takada S. Multiscale ensemble modeling of intrinsically disordered proteins: p53 N-terminal domain. Biophys. J. 2011;101:1450–1458. doi: 10.1016/j.bpj.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li WF, Zhang Z, Wang J, Wang W. Metal-coupled folding of Cys2His2 zinc-finger. J. Am. Chem. Soc. 2008;130:892–900. doi: 10.1021/ja075302g. [DOI] [PubMed] [Google Scholar]
- 15.Mao XB, Guo YY, Luo Y, Niu L, Liu L, Ma XJ, Wang HB, Yang YL, Wei GH, Wang C. Sequence effects on peptide assembly characteristics observed by using scanning tunneling microscopy. J. Am. Chem. Soc. 2013;135:2181–2187. doi: 10.1021/ja307198u. [DOI] [PubMed] [Google Scholar]
- 16.Li WF, Zhang J, Su Y, Wang J, Qin M, Wang W. Effects of zinc binding on the conformational distribution of the amyloid-beta peptide based on molecular dynamics simulations. J. Phys. Chem. B. 2007;111:13814–13821. doi: 10.1021/jp076213t. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Chu XK, Longhi S, Roche P, Han W, Wang EK, Wang J. Multiscaled exploration of coupled folding and binding of an intrinsically disordered molecular recognition element in measles virus nucleoprotein. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E3743–E3752. doi: 10.1073/pnas.1308381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang F, Han W, Wu YD. The intrinsic conformational features of amino acids from a protein coil library and their applications in force field development. Phys. Chem. Chem. Phys. 2013;15:3413–3428. doi: 10.1039/c2cp43633g. [DOI] [PubMed] [Google Scholar]
- 19.Jiang F, Wu YD. Folding of fourteen small proteins with a residue-specific force field and replica-exchange molecular dynamics. J. Am. Chem. Soc. 2014;136:9536–9539. doi: 10.1021/ja502735c. [DOI] [PubMed] [Google Scholar]
- 20.Kim SY, Jung Y, Hwang GS, Han H, Cho M. Phosphorylation alters backbone conformational preferences of serine and threonine peptides. Proteins. 2011;79:3155–3165. doi: 10.1002/prot.23148. [DOI] [PubMed] [Google Scholar]
- 21.Shen TY, Wong CF, McCammon JA. Atomistic Brownian dynamics simulation of peptide phosphorylation. J. Am. Chem. Soc. 2001;123:9107–9111. doi: 10.1021/ja010190t. [DOI] [PubMed] [Google Scholar]
- 22.Xiang SQ, Gapsys V, Kim HY, Bessonov S, Hsiao HH, Mohlmann S, Klaukien V, Ficner R, Becker S, Urlaub H, Luhrmann R, De Groot B, Zweckstetter M. Phosphorylation drives a dynamic switch in serine/arginine-rich proteins. Structure. 2003;21:2162–2174. doi: 10.1016/j.str.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 23.Sellis D, Drosou V, Vlachakis D, Voukkalis N, Giannakouros T, Vlassi M. Phosphorylation of the arginine/serine repeats of lamin B receptor by SRPK1–Insights from molecular dynamics simulations. Biochim. Biophys. Acta. 2012;1820:44–55. doi: 10.1016/j.bbagen.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Vassall KA, Bessonov K, De Avila M, Polverini E, Harauz G. The effects of threonine phosphorylation on the stability and dynamics of the central molecular switch region of 18.5-kDa myelin basic protein. PLoS ONE. 2013;8:e68175. doi: 10.1371/journal.pone.0068175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Velazquez HA, Hamelberg D. Dynamical role of phosphorylation on serine/threonine-proline Pin1 substrates from constant force molecular dynamics simulations. J. Chem. Phys. 2015;142:075102. doi: 10.1063/1.4907884. [DOI] [PubMed] [Google Scholar]
- 26.Krimm S, Tiffany ML. Circular-dichroism spectrum and structure of unordered polypeptides and proteins. Isr. J. Chem. 1974;12:189–200. doi: 10.1002/ijch.197400018. [DOI] [Google Scholar]
- 27.Shi ZS, Olson CA, Rose GD, Baldwin RL, Kallenbach NR. Polyproline II structure in a sequence of seven alanine residues. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9190–9195. doi: 10.1073/pnas.112193999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kentsis A, Mezei M, Osman R. Origin of the sequence-dependent polyproline II structure in unfolded peptides. Proteins. 2005;61:769–776. doi: 10.1002/prot.20655. [DOI] [PubMed] [Google Scholar]
- 29.Li WF, Qin M, Tie ZX, Wang W. Effects of solvents on the intrinsic propensity of peptide backbone conformations. Phys. Rev. E. 2011;84:041933. doi: 10.1103/PhysRevE.84.041933. [DOI] [PubMed] [Google Scholar]
- 30.Liu ZG, Chen K, Ng A, Shi ZS, Woody RW, Kallenbach NR. Solvent dependence of PII conformation in model alanine peptides. J. Am. Chem. Soc. 2004;126:15141–15150. doi: 10.1021/ja047594g. [DOI] [PubMed] [Google Scholar]
- 31.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for Amber99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang F, Zhou CY, Wu YD. Residue-specific force field based on the protein coil library. RSFF1: modification of OPLS-AA/L. J. Phys. Chem. B. 2014;118:6983–6998. doi: 10.1021/jp5017449. [DOI] [PubMed] [Google Scholar]
- 33.Hagarman A, Measey TJ, Mathieu D, Schwalbe H, Schweitzer-Stenner R. Intrinsic propensities of amino acid residues in GxG peptides inferred from amide I’ band profiles and NMR scalar coupling constants. J. Am. Chem. Soc. 2010;132:540–551. doi: 10.1021/ja9058052. [DOI] [PubMed] [Google Scholar]
- 34.Shi ZS, Chen K, Liu ZG, Ng A, Bracken WC, Kallenbach NR. Polyproline II propensities from GGXGG peptides reveal an anticorrelation with beta-sheet scales. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17964–17968. doi: 10.1073/pnas.0507124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plaxco KW, Morton CJ, Grimshaw SB, Jones JA, Pitkeathly M, Campbell ID, Dobson CM. The effects of guanidine hydrochloride on the ‘random coil’ conformations and NMR chemical shifts of the peptide series GGXGG. J. Biomol. NMR. 1997;10:221–230. doi: 10.1023/A:1018340217891. [DOI] [PubMed] [Google Scholar]
- 36.van der Spoel D. The solution conformations of amino acids from molecular dynamics simulations of Gly-X-Gly peptides: comparison with NMR parameters. Biochem. Cell Biol. 1998;76:164–170. doi: 10.1139/o98-025. [DOI] [PubMed] [Google Scholar]
- 37.He L, Navarro AE, Shi ZS, Kallenbach NR. End effects influence short model peptide conformation. J. Am. Chem. Soc. 2012;134:1571–1576. doi: 10.1021/ja2070363. [DOI] [PubMed] [Google Scholar]
- 38.Toal S, Meral D, Verbaro D, Urbanc B, Schweitzer-Stenner R. pH-independence of trialanine and effects of termini blocking in short peptides: a combined vibrational, NMR, UVCD and molecular dynamics study. J. Phys. Chem. B. 2013;117:3689–3706. doi: 10.1021/jp310466b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dean AM, Koshland DE. Electrostatic and steric contributions to regulation at the active site of isocitrate dehydrogenase. Science. 1990;249:1044–1046. doi: 10.1126/science.2204110. [DOI] [PubMed] [Google Scholar]
- 40.Case, D A, Darden, T.A., Cheatham, III, T.E., Simmerling, C.L., Wang, J., Duke, R.E., Luo, R., Walker, R.C., Zhang, W., Merz, K.M., Roberts, B., Wang, B., Hayik, S., Roitberg, A., Seabra, G., Kolossvry, I., Wong, K.F., Paesani, F., Vanicek, J., Liu, J., Wu, X., Brozell, S.R., Steinbrecher, T., Gohlke, H., Cai, Q., Ye, X., Wang, J., Hsieh, M.-J., Cui, G., Roe, D.R., Mathews, D.H., Seetin, M.G., Sagui, C., Babin, V., Luchko, T., Gusarov, S., Kovalenko, A., Kollman, P.A.: AMBER 11. University of California, San Francisco (2010)
- 41.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong GM, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang JM, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 42.Craft JW, Legge GB. An AMBER/DYANA/MOLMOL phosphorylated amino acid library set and incorporation into NMR structure calculations. J. Biomol. NMR. 2005;33:15–24. doi: 10.1007/s10858-005-1199-0. [DOI] [PubMed] [Google Scholar]
- 43.Best RB, Zhu X, Shim J, Lopes PEM, Mittal J, Feig M, MacKerell AD. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ##φ##, ##ψ## and side-chain ##χ##(1) and ##χ##(2) dihedral angles. J. Chem. Theory. Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacKerell AD, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 45.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 46.Berendsen HJC, Van der Spoel D, Van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comp. Phys. Comm. 1995;91:43–56. doi: 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- 47.Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, Flexible and Free. J. Comp. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 48.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory. Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 49.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 50.Van Gunsteren WF, Berendsen HJC. Algorithms for macromolecular dynamics and constraint dynamics. Mol. Phys. 1977;34:1311–1327. doi: 10.1080/00268977700102571. [DOI] [Google Scholar]
- 51.Darden T, York D, Pedersen L. Particle mesh Ewald: an N∗log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 52.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999;314:141–151. doi: 10.1016/S0009-2614(99)01123-9. [DOI] [Google Scholar]
- 53.Walter N, Hansmann UHE. Dynamics and optimal number of replicas in parallel tempering simulations. Phys. Rev. E. 2007;76:065701. doi: 10.1103/PhysRevE.76.065701. [DOI] [PubMed] [Google Scholar]
- 54.Garcia AE. Characterization of non-alpha helical conformations in Ala peptides. Polymer. 2004;45:669–676. doi: 10.1016/j.polymer.2003.10.070. [DOI] [Google Scholar]
- 55.Avbelj F, Luo PZ, Baldwin RL. Energetics of the interaction between water and the helical peptide group and its role in determining helix propensities. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10786–10791. doi: 10.1073/pnas.200343197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Avbelj F, Baldwin RL. Origin of the neighboring residue effect on peptide backbone conformation. Proc. Natl. Acad. Sci. U.S.A. 2000;101:10967–10972. doi: 10.1073/pnas.0404050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poon CD, Samulski ET, Weise CF, Weisshaar JC. Do bridging water molecules dictate the structure of a model dipeptide in aqueous solution. J. Am. Chem. Soc. 2000;122:5642–5643. doi: 10.1021/ja993953+. [DOI] [Google Scholar]
- 58.Creamer TP, Campbell MN. Determinants of the polyproline II helix from modeling studies. Adv. Protein Chem. 2002;62:263–282. doi: 10.1016/S0065-3233(02)62010-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.