Abstract
The S100A1 protein mediates a wide variety of physiological processes through its binding of calcium (Ca2+) and endogenous target proteins. S100A1 presents two Ca2+-binding domains: a high-affinity “canonical” EF (cEF) hand and a low-affinity “pseudo” EF (pEF) hand. Accumulating evidence suggests that both Ca2+-binding sites must be saturated to stabilize an open state conducive to peptide recognition, yet the pEF hand’s low affinity limits Ca2+ binding at normal physiological concentrations. To understand the molecular basis of Ca2+ binding and open-state stabilization, we performed 100 ns molecular dynamics simulations of S100A1 in the apo/holo (Ca2+-free/bound) states and a half-saturated state, for which only the cEF sites are Ca2+-bound. Our simulations indicate that the pattern of oxygen coordination about Ca2+ in the cEF relative to the pEF site contributes to the former’s higher affinity, whereas Ca2+ binding strongly reshapes the protein’s conformational dynamics by disrupting β-sheet coupling between EF hands. Moreover, modeling of the half-saturated configuration suggests that the open state is unstable and reverts toward a closed state in the absence of the pEF Ca2+ ion. These findings indicate that Ca2+ binding at the cEF site alone is insufficient to stabilize opening; thus, posttranslational modification of the protein may be required for target peptide binding at subsaturating intracellular Ca2+ levels.
Introduction
S100A1 is a Ca2+-binding protein commonly found in neuronal, renal (1), and cardiac (2, 3) tissue, in the latter of which it is implicated in cardiomyopathy (for reviews, see (4, 5)). S100A1 contributes to a variety of protein-protein interaction (PPI) events after Ca2+-dependent activation, including those of the sarcoplasmic reticulum (ryanodine receptor (6, 7), SERCA2a and phospholamban (8, 9), and a cationic channel (TRPM3 (3)) to regulate Ca2+ homeostasis (10). The S100A1 protein forms a globular homodimer that contains two Ca2+-binding sites per monomer, as shown in Fig. 1. Each S100A1 monomer presents two Ca2+-binding sites consisting of a helix-loop-helix secondary-structure motif that is commonly called the “EF hand” (11). The canonical form of the EF hand, which we refer to as the cEF hand, consists of 12 residues and is highly conserved in many Ca2+-binding proteins, such as calmodulin (CaM) and troponin C (TnC) (12, 13).
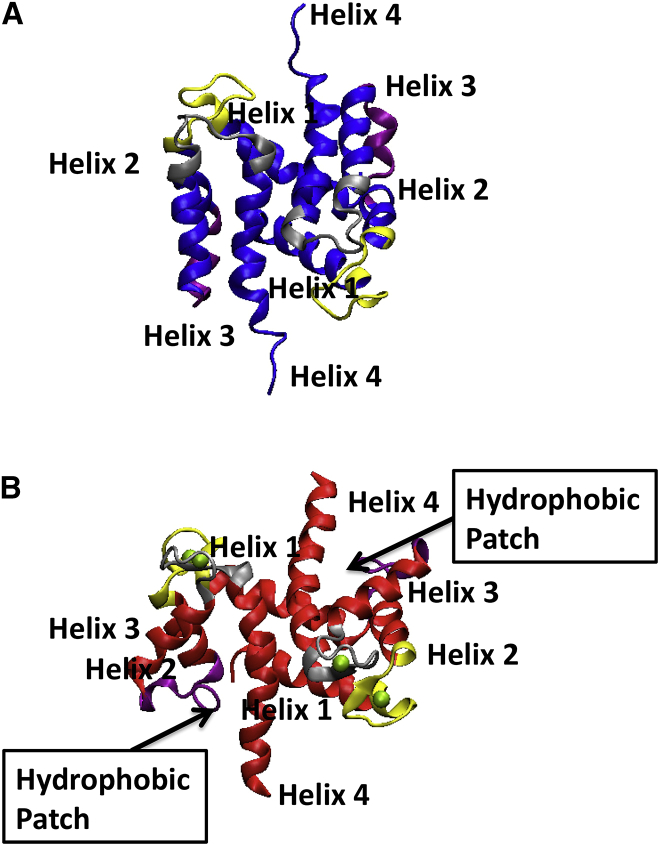
Figure 1.
NMR structures of the human apo (PDB: 2L0P) (A) and holo (PDB: 2LP3) S100A1 (B) states (26, 29). The pEF (yellow), linker (purple), cEF (gray), and helical bundle (red) are represented in the structures, and the Ca2+ ions (green) are shown for the holo form. The binding of Ca2+ ions reveals a hydrophobic protein recognition patch near H3. To see this figure in color, go online.
The S100A1 cEF region spans 12 residues between D62 and E73, whereas the second, “pseudo” EF hand (pEF), is unique to the S100 family and related proteins, with 14 residues bridging S19 to E32 (11, 14). Experiments show that the first Ca2+ ion binds to the cEF with a dissociation constant, Kd, of 27–250 μM compared to a Kd of 250–16,700 μM for the pEF domain (15, 16)). It is further believed that Ca2+ binding to pEF proceeds in a noncooperative fashion (15), but the structural basis of consecutive binding remains unclear.
Upon saturation of the S100A1 monomer with two Ca2+ ions, the protein undergoes a conformational change from a closed to an open state that is characterized by the exposure of hydrophobic residues between helices 3 (H3) and 4 (H4) (15, 16) (see Fig. 1). It is the presentation of this open hydrophobic patch that promotes PPIs (17, 18), in a fashion similar to PPI formation by the TnC (19, 20, 21, 22) and CaM proteins (23, 24, 25). Nuclear magnetic resonance (NMR) studies have revealed atomic-resolution details of the S100A1 conformations in the apo (Ca2+-free) (26, 27, 28), holo (Ca2+-saturated) (16, 29) (Fig. 1, A and B), and target-protein-bound states (6, 17, 18). An open question, however, is how S100A1 participates in peptide-binding events given that saturation of the cEF and pEF hands requires Ca2+ ion concentrations of 100–10,000 μM, whereas physiological intracellular Ca2+ rarely exceeds tens of micromolar Ca2+ in localized regions (30, 31).
To answer this question, we characterized key physiochemical drivers of high-affinity Ca2+ binding and the stabilization of the open state in the apo and holo states using molecular simulations. We complemented these analyses with simulations of the “half-saturated” configuration, for which only the cEF site is Ca2+ bound, to investigate the relative stability of the open and closed states when the pEF hand is devoid of Ca2+ ions. To this end, we performed 100 ns molecular dynamics (MD) simulations of the apo and holo states, as well as extended (>120 ns) runs on the half-saturated holo states to model the S100A1 structure and dynamics under physiological binding configurations. We relate the protein’s Ca2+ and target-protein-binding mechanisms to structural dynamics local to the binding domains as well as to allosterically driven global conformational motions. First, we analyzed the binding site by estimating NMR amide order parameters as indicators of protein flexibility, for Ca2+-binding domains in the apo and holo states. Second, we compared the relative positioning and coordination of EF-hand oxygens in the presence and absence of Ca2+ ion in the pEF and cEF hands. Third, we examined the impact of Ca2+-ion binding on the global conformation of the protein through cross correlation, principal-component analysis (PCA), and measuring bundle helix displacements. Our results support prior experimental studies (15) suggesting that full EF-hand saturation is required for open-state stabilization. Hence, posttranslational modification of the protein may be required to form the open state for peptide recognition at physiological Ca2+ concentration levels. Moreover, our simulations provide, to our knowledge, new insight into 1) how chelating oxygen distributions and charges within the EF hands may tune Ca2+ binding affinities, and 2) the allosteric control of open-state stabilization via correlative motions and disruption of β-sheet formation between the cEF and pEF domains.
Materials and Methods
MD simulations
NMR-derived homodimeric structures were used as starting conformations for the apo state (PDB: 2L0P (26) and 2LLU (28)), and holo state (PDB: 2LP3) (29). Using TLeap (32, 33), all systems were neutralized, explicitly solvated in a 20.0-Å-margin TIP3P water box in 0.15 M KCl solution and parameterized with the AMBER-12SB force field (FF) (34). The protein consists of 186 residues in a system of ∼20,000 water molecules and 130 neutralizing ions, for a total of ∼60,000 atoms. Ca2+ parameters were based on a recent formulation for divalent ions (Li-Merz (35)) shown to reproduce experimental coordination numbers and hydration free energies through optimizing the cations’ 12-6 Lennard Jones interactions (35). In the Results and Discussion section, we verify that the Li-Merz cation reasonably reproduces experimentally estimated coordination distances and EF-hand amide order parameters. Generally speaking, we have found that standard Ca2+ parameters, such as those from Marchand et al. (36), perform reasonably well for recapitulating coordinating structures and order parameters in EF-hand-based Ca2+-binding proteins, including TnC (37, 38). The PMEMD module within the AMBER 14 package (32, 33) was used for minimization and equilibration, whereas the pmemd.cuda engines were used for the initial 100 ns production MD runs performed in quadruplicate. MD simulations in part utilized resources provided by the XSEDE high performance computing facilities (39). Our model of the half-saturated S100A1 was based on removing Ca2+ ions from the pEF hand of the holo structure (PDB: 2LP3); attempts to add Ca2+ ions to the apo state did not lead to an adequately coordinated structure. We recognize that the unresolved equilibrium conformations of the half-saturated state may entail localized reorganization of the protein; we approached this challenge by first performing normal equilibration routines to relax the EF-hand regions, followed by long-timescale MD simulations performed in succession. As discussed in the Results and Discussion section, significant relaxation of the protein toward an apo-like state was observed within 60 ns. We also performed four additional runs for snapshots from two of the half-saturated trajectories (see Section S1.3.6 in the Supporting Material). Further details of the MD simulation configurations are summarized in Section S1.1.1 in the Supporting Material).
Simulation trajectory analysis
Order parameters
Backbone N-H order parameters were calculated using the isotropic reorientational eigenmode dynamics approach (40) via the ptraj and mat2s2.py programs. Twenty scans consisting of window sizes of 5 ns (2500 frames) from the 100 ns (50,000 frames) saved for each simulation were used, which is comparable to the window sizes suggested by Genheden et al. (40).
Helix angles
Interhlx (K. Yap, University of Toronto, Toronto, Ontario, Canada) was used to calculate the interhelical angles. Helical lengths matched those from the respective NMR references and are outlined in Table S1. The helical angles were calculated for 500 snapshots saved every 0.2 ns during the 100 ns simulations.
Trajectory analysis and Ca2+/oxygen radial distributions
Standard trajectory analyses including root mean square fluctuations (RMSFs), hydrogen-bonding patterns, and oxygen/Ca2+ distances were computed using the Lightweight Object-Oriented Structure library (LOOS) ((41) and T. D. Romo and A. Grossfield, 2013, Biophys. J., abstract). Common reference positions were obtained by aligning the α-carbon atoms to the holo-state configuration (PDB: 2LP3 (29)). For Ca2+ coordination comparisons with the apo cases, the Ca2+ ion position was approximated by the corresponding location in the aligned holo state. Except where noted, all custom analyses were written in python and are available at https://bitbucket.org/pkhlab/pkh-lab-analyses/.
Electrostatic score
Two-dimensional histograms of the number of Ca2+-oxygen contacts with respect to time and distance were computed for all side-chain oxygens and backbone oxygens to illustrate interactions between the Ca2+ ion and its oxygen ligands. We defined an electrostatic score (Eq. 1) to assess the interactions indicated in these histograms:
| (1) |
where is the electrostatic score, is the charge on the Ca2+ ion (+2), and are based on CHARMM partial charges for the backbone and side-chain oxygen atoms (−0.51 and −0.76, respectively) (42, 43, 44), and ρ is the density of oxygen atoms at distance (Å). Although this formula resembles the Poisson model of electrostatic energy for a cloud of point charges, we opted for a simplified score given the difficulty of estimating a position-dependent dielectric constant in the neighborhood of the bound ion. The advantage of this approach is that we can qualitatively assess how the electrostatic field in the EF-hand domain changes between the apo and holo states. Conventional procedures for assessing interaction energies, such as molecular mechanics/Poisson Boltzmann surface area analysis (MM/PBSA) (45, 46, 47, 48), are facilitated when an explicit Ca2+ ion is included in all simulations, which is unfeasible for the apo state. Nevertheless, to ensure that our electrostatic score was consistent with conventional interaction energy methods, we determined the MM electrostatic energies from the MM/PBSA program for the holo and half-saturated cases, as discussed in Section S1.2 in the Supporting Material.
PCA
The Bio3D package (49) was used to quantify the large-scale protein conformational changes shared by proteins homologous to S100A1, according to procedures we outlined for two related Ca2+-binding proteins (see Kekenes-Huskey et al. (37) and Lindert et al. (38)). A BLAST search (50) on the human S100A1 sequence was performed with the NMR structure (PDB: 2L0P) (26) as input. Seven hundred and seventy-eight sequences were found, and the top 56 chains from 28 homodimer proteins with sequence identities ranging above 92.5% were used for subsequent sequence alignment and PCA. From these homologous sequences, Bio3D identified an invariant core of atoms with the least positional variation across all available protein structures. The invariant core most strongly consisted of those atoms buried within the S100A1 helical bundle. All structural sequences were aligned to this core, after which PCA was performed to identify bases presenting the largest modes of structural variation. The largest principal components (PCs) represent the most significant conformational differences between the apo and holo proteins; PCs 1 and 2 accounted for 74.4% of the variance, with PC1 making up 57.2% of the variance.
Residue cross correlation analysis
The four trajectories for each case were aligned using LOOS to both chains in the first frame of the respective NMR conformations. In Bio3D, residue cross correlation analysis was conducted with the dynamical cross correlation map (49, 51, 52) on the α-carbons of chain A (residues 1–93) for each protein.
Results and Discussion
Predicted Ca2+-coordination and N-H order parameters are consistent with NMR data
We first verified that our predicted coordination distances in the holo state are consistent with NMR-derived NOESY inter-proton distance constraints (29) (Table S2). The coordination distances in the simulations were measured between the specified atoms for every time step (2 fs) over the course of 100 ns, from which means and standard deviations were reported based on the averages for each independent simulation. The simulated data were compared against coordinating distances reflected in the ensemble of holo NMR structures (PDB: 2LP3 (29)). Nearly all average coordinating distances shown for the NMR structures were within 2.91 Å, compared to 2.77 Å in our simulations. One exception was the Ca2+-N64 Oδ1 distance, for which we report an average distance of 4.26 ± 2.28 Å versus the NMR measured distances of 2.55 ± 0.04 Å. The larger standard deviation in our data reflects that the Ca2+/N64 Oδ1 atoms were bound for a significant fraction of the simulation, although the Ca2+ ion would frequently exchange binding partners with water in the EF-hand domain. These data confirm that the Li-Merz Ca2+ parameter set reasonably reproduce experimentally determined coordination numbers and distances, despite their optimization for reproducing solvation structure and energies (35).
We next compared protein conformational dynamics reflected in our simulations with appropriate experimentally derived observables. Specifically, amide nitrogen/hydrogen order parameters have been widely used to probe backbone mobility in EF-hand-based proteins, particularly in the presence and absence of Ca2+ (29, 53). By definition, order parameters approaching 1.0 signify greater protein rigidity, whereas order parameters approaching 0.0 imply greater flexibility. Order-parameter calculations thereby enable assessment of the Ca2+-binding site and linker region stability, which can further be validated against experimental data. Specifically, prior NMR studies (29) of S100A1 in its apo and holo states indicated significant changes in backbone mobility, especially near the Ca2+-binding region, where the presence of Ca2+ suppressed EF-hand fluctuations (leading to larger order parameters) relative to the apo state.
In Fig. 2, we compare prior NMR-derived order parameters with computed estimates averaged from four 100 ns simulations. In the three regions presented, the pEF hand (residues S19–E32; see Table S1), the linker (residues L41–D50), and the cEF hand (residues D62–E73), the estimates for the apo state are in good agreement with experiment (0.80 ± 0.01 theoretical versus 0.79 ± 0.02 experimental for pEF, 0.81 ± 0.01 versus 0.78 ± 0.02 for cEF, and 0.83 ± 0.02 versus 0.85 ± 0.03 for the linker). For the holo state, we note that the calculated order parameters were greater for the Ca2+-binding sites relative to those calculated for the the apo state (0.85 ± 0.01 calculated holo versus 0.80 ± 0.01 calculated apo for the pEF and 0.84 ± 0.02 calculated holo versus 0.81 ± 0.01 calculated apo for the cEF), which signifies more rigid EF hands in the Ca2+-bound state. The decreased mobility in the EF holo state undoubtedly arises in part because the backbone and side-chain oxygens involved in coordinating Ca2+ are strongly bound and thus restrict backbone movement. We note, however, that our computed order parameters for the holo cases were similar to, but somewhat smaller than, the experimental holo measurements. This modest difference may suggest that our simulations moderately overestimated the lability of Ca2+-binding regions. If this is the case, further refined Ca2+/oxygen parameters (54) or use of polarizable force fields (55) may better stabilize coordination and recapitulate the order parameters derived from NMR studies. We nevertheless find agreement with the experimental trends for the linker region, residues 41–50, which suggest smaller order parameters for the holo state (calculated 0.78 ± 0.02) than for the apo state (calculated 0.83 ± 0.02). The holo state’s smaller order parameters indicate greater linker mobility compared to the apo state, although the underlying mechanism is unclear.
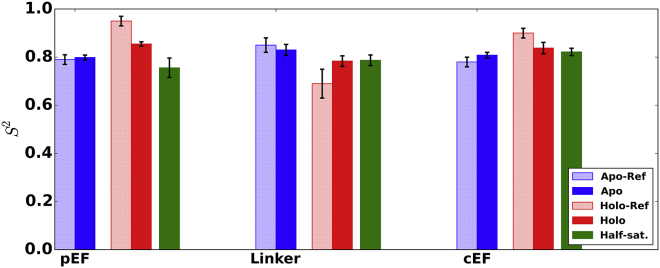
Figure 2.
Experimental order parameters (29), S2, for the apo (light blue) and holo (light red) cases versus the respective calculated order parameters (dark blue and dark red, respectively) and the calculated half-saturated cEF states (green). Vertical error bars represent standard deviations of the four averages of the two chains in each simulation. Predictions for the apo and holo states were consistent with experiment. To see this figure in color, go online.
The concurrence of our predicted order parameters and coordination distances with experiment suggest that the Li-Merz parameters are sufficiently reliable for predicting conformational and dynamic properties of small Ca2+-binding proteins. These findings are also consistent with those of prior studies of similar globular Ca2+-binding proteins, including TnC (37, 38, 56, 57), CaM (58, 59), and calbindin (36, 60), which demonstrated good agreement with experimental measurements such as chelation distances, order parameters, and global conformational dynamics, using conventional MD protocols without polarization effects. Also remarkable were the results from Jiang et al. for Ca2+ binding to calbindin (60), for which the free energy of binding was within 2.1 kcal/mol of the experimental measurements. We note, however, that explicit consideration of polarization and charge transfer effects via ab initio methods (61) or polarizable models (45, 55, 62) are likely required for obtaining further refined estimates of protein/cation affinity. Validation of such methodologies and parameters may consider excellent experimental protein/cation affinity data such as those reported for CaM (63), for which site-specific affinity data were precisely measured. We nevertheless anticipate that subtle variation in chelation energetics due to these nonclassical phenomena would have limited impact on the chelation shell structure and global protein conformational dynamics, given the consistency of classical force-field MD simulations of Ca2+-binding proteins with experimental probes of protein dynamics.
It has been reported (15) that at physiological Ca2+ concentrations, the pEF hand is not Ca2+ bound, and thus, the S100A1 open state is inaccessible. To provide insight into the backbone dynamics of the most probable binding configuration in physiological systems, we present order parameters for the half-saturated (cEF-only) S100A1 protein (see Fig. 2). Our calculated results indicate similar order parameters in the cEF region for the fully saturated holo and half-saturated systems (0.84 ± 0.02 and 0.82 ± 0.02, respectively), with modestly smaller order parameters for the latter configuration. Order parameters for the pEF region of the half-saturated state (0.76 ± 0.04) were comparable to and somewhat smaller than those reported for the unsaturated (apo) state (calculated as 0.80 ± 0.01) as opposed to the fully saturated holo state (calculated as 0.85 ± 0.01). The similarity of the pEF dynamics in the apo and half-saturated cases suggests that the presence of cEF-hand Ca2+ has little impact on the structure and mobility of the pEF region, which may support experimental observations of minimal Ca2+-binding cooperativity (15). We find that the linker region is equally mobile in the half- and fully saturated configurations, which may indicate that the cEF-bound Ca2+ ion is sufficient to induce linker mobility.
In Section S1.3.2 in the Supporting Material, we present backbone amide nitrogen root mean-square fluctuations (RMSFs) for the apo, holo, and half-saturated states that quantify the relative lability of the EF-hand domains in a manner similar, but not equivalent, to the amide order parameters (Fig. S1). We find that in the pEF hand, the reported RMSFs were significantly greater for the apo and half-saturated proteins than for the holo states. This behavior was not unexpected, given that strong Ca2+/oxygen interactions in the holo state should be expected to dampen pEF hand mobility. In the cEF hand, we note that the holo and half-saturated RMSFs were statistically comparable. Since the RMSF values merely indicate the relative fluctuations of the residues and not their positional decorrelation implied in order parameter analysis (64), these data indicate that the amides continued to fluctuate, although they were strongly constrained by the complexed Ca2+.
Oxygen radial distribution about the bound Ca2+ ion
We further investigated the Ca2+ coordination to understand the molecular basis for higher Ca2+ affinity at the cEF relative to the pEF hand (15). Canonical high-affinity EF-hand Ca2+ binding is typically afforded by short Ca2+/oxygen distances (1.8–3.5 Å) stemming from seven oxygens arranged in a pentagonal bipyramidal fashion (12, 13, 65). In the cEF hand (D62–E73), the Ca2+ ion coordinated with seven oxygens, six of which were side-chain carboxylate oxygens and one backbone amide oxygen. The pEF hand (S19–E32) presented two additional residues relative to the cEF hand, which still promoted seven coordinate oxygens, although in this case, four arose from backbone oxygens, two from side-chain oxygens, and one from a bound water molecule (14). In Fig. 3, we report radial distributions of EF-hand oxygens within 12 Å of the coordinated Ca2+ position for the half-saturated and fully saturated holo states. For comparison, we also present data for the oxygen radial distribution in the apo state to demonstrate the substantial reorganization of coordinating ligands upon presentation of the Ca2+ ion. Fig. 3 A shows the oxygen population with respect to distance and time for the pEF and cEF hands, for which 7.0 is the optimal coordination number (11, 14). For both pEF and cEF hands in the holo case, nearly optimal coordination at 2.5 Å is evidenced by a distinct red line that persists throughout the simulations. However, the cEF hand presents a higher degree of coordination, 6.45 ± 0.47, relative to the pEF hand, 5.32 ± 0.06. The nonintegral coordination number reflects the transient nature of Ca2+/S100A1 oxygen binding owing to the rotation of side-chain carboxylate groups and exchange with bound solvent waters (not shown). Overall, the higher radial probability of coordinating oxygens for the cEF hand relative to pEF is consistent with the former’s greater Ca2+ affinity.
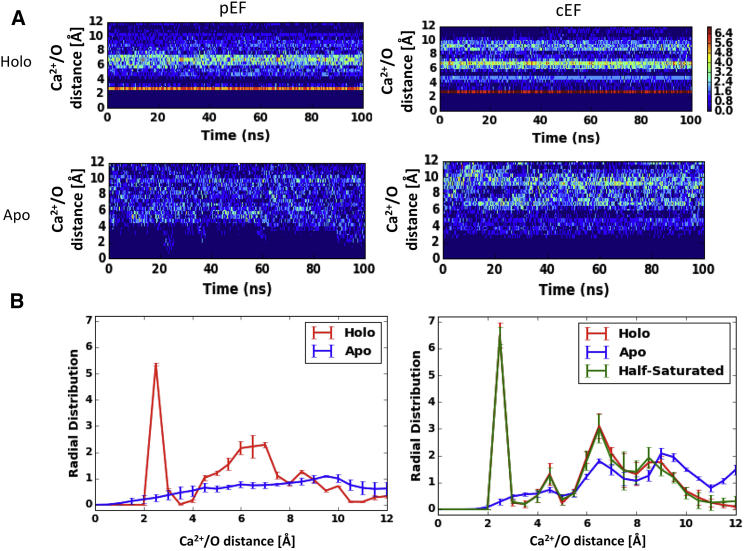
Figure 3.
Time-dependent Ca2+/oxygen distances (A) and averaged radial distribution of oxygen/Ca2+ (B) for the pEF (left) and cEF (right) hands. Vertical error bars indicate the mean ± SD of all four runs. Ca2+ binding localizes chelating oxygens to an ∼2.5-Å-radius coordination shell. To see this figure in color, go online.
The extent of EF-hand reorganization upon binding Ca2+ ions can be inferred from our apo-state data, for which the absence of Ca2+ ions permitted broadly and sparsely distributed chelating oxygens (see Fig. 3, A and B). Although this point of reference is biased toward the Ca2+-bound configuration, it nevertheless illuminates the considerable mobility of the ligating oxygens. This behavior is consistent with the small order parameters reported in the EF-hand regions by Nowakowski et al. (29). For the half-saturated states, the reported cEF distribution is nearly coincident with the statistics observed in the holo configuration. In other words, the loss of the pEF Ca2+ ions has no apparent effect on the cEF coordinating oxygens, which may support prior experimental evidence suggesting that pEF and cEF bind Ca2+ noncooperatively (15). We note, however, that the half-saturated pEF-hand conformation significantly differed from that of the Ca2+-free and fully saturated states, so we are unable to unambiguously define an approximate Ca2+ ion position for computing oxygen radial distributions. In the next section, we assess the approximate energetic stabilization due to these chelation patterns.
Calculated electrostatic scores based on chelating oxygens
The Ca2+ cation is anchored in the EF-hand binding site via electrostatic interactions with negatively charged oxygens, the resulting enthalpy of which likely dominates the binding free energy (66). To relate our simulated coordination geometries to an approximate electrostatic contribution to binding enthalpies, we introduce an “electrostatic score” that resembles Coulomb’s law (Eq. 1). Similar to techniques applied to Ca2+-binding channels, this score provides a simple metric for assessing the electrostatic potential from the chelating ions for both the Ca2+-free- and Ca2+-bound-state protein/cation interactions (67). We note that the higher negative partial charge in the coordinating side-chain carboxylic acid oxygens relative to backbone amide oxygens should lead to more favorable scores for the cEF versus pEF coordination. Indeed, the holo state cEF hand reports a more favorable average electrostatic score than the pEF hand by 1.46 units (pEF, −5.87 ± 0.04; cEF: −7.33 ± 0.29), as shown in Fig. 4 A, which is consistent with experimental binding trends. In Fig. 4 B and Section S1.3.3 in the Supporting Material, we show that the difference in electrostatic scores narrowed when all oxygens were assumed to share the same partial charge, as is assumed in some models (68). Hence, although it is understood that the positions of coordinating ligands are important determinants of Ca2+ affinity (12, 13, 65), these data indicate that oxygen partial charges and potentially mobilities further tune affinity. Although this electrostatic score assessment is simplistic, it captures the predominant electrostatic enthalpic contribution to the binding free energy and has a consistent trend with MM electrostatic interaction energies determined directly from AMBER MM/PBSA analysis (see Section S1.3.4 in the Supporting Material).
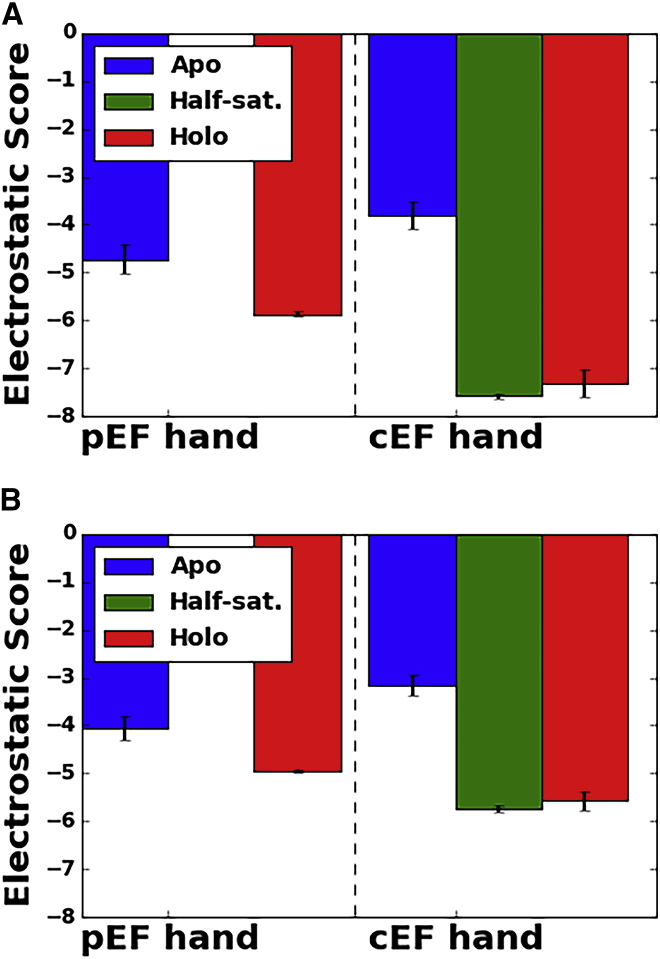
Figure 4.
Electrostatic scores for the apo (blue), half-saturated (green), and fully saturated holo (red) states averaged over quadruplicate simulations. Results are given assuming partial charges of −0.51 and −0.76 on side-chain and backbone oxygens (A) versus uniform charges of −0.51 for both oxygen types (B). Vertical error bars indicate the mean ± SD of the eight electrostatic scores for two protein chains and four simulation runs. The higher density of coordinating oxygens and more negative partial charges in the cEF hand yield more stabilizing scores than for the pEF hand. To see this figure in color, go online.
We additionally computed the electrostatic score based on the apo state conformation to assess the gain in favorable enthalpic interactions as the protein relaxes in the presence of Ca2+ ions to form optimal binding interactions. As in the previous section, we present data for the apo state centered about the approximate position the Ca2+ ion would assume upon binding, to demonstrate the considerable change in the electrostatic environment upon chelating Ca2+. Given comparable electrostatic scores for the apo states of the pEF and cEF hands (−4.72 ± 0.32 and −3.80 ± 0.30, respectively), Ca2+ ion binding yields greater reorganization energy for the latter, as holo state scores were 1.15 and 3.53 units more negative, respectively. Similar to our analysis of oxygen radial distributions in the previous section, we find that the electrostatic score for the Ca2+-bound cEF hand in the half-saturated state (−7.58 ± 0.07) is nearly indistinguishable from that in the fully saturated holo state and further supports lack of cooperativity in pEF Ca2+ binding (15). However, there remains the possibility that the Ca2+-bound cEF site preorganizes the pEF hand to facilitate rapid Ca2+ ion association rates, as we speculated previously for the EF hand in TnC mutants (37). Moreover, significant entropic contributions from coordinating oxygen desolvation and conformational reorganization will understandably contribute to the overall observed affinity (14). Although it is beyond the scope of this study, rigorous approaches for deriving binding free energies, such as free-energy perturbation or thermodynamic integration, could potentially help quantify these effects (47, 69, 70, 71).
Stability of hydrogen bonds within the EF-hand β-sheets
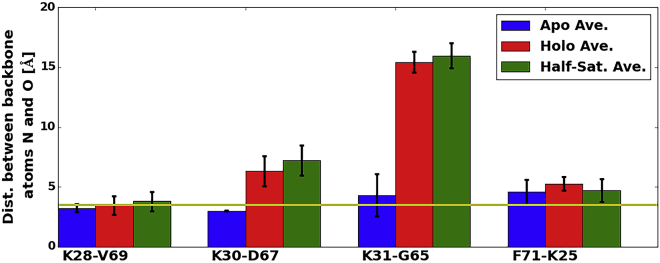
To examine stabilizing contributions to the EF-hand regions, we calculated the average distance between the backbone amide nitrogen and oxygen atoms comprising β-sheets linking the pEF and cEF hands (Fig. 5). We observe that four hydrogen bonds, including K25-F71, K28-V69, K30-D67, and K31-G65 (polar heavy atom distances <3.5 Å), form within the β-sheets of the apo case, compared with only one in the holo case between K28 and V69. The larger degree of hydrogen bonding in the apo configuration likely helps stabilize the EF hand, which may help preorganize the binding site for Ca2+-ion recognition (14). Meanwhile, the half-saturated case presents an intermediate degree of hydrogen bonding (two bonds, L28-V69 and K25-F71), which correlates with its order parameters. Notably, the largest change in the distance between two potential hydrogen-bond partners occurred between K31 and G65. In the apo state, they are separated by 4.33 ± 1.77 Å in the intact β-sheet secondary structure, but in the holo and half-saturated states, that distance increases to 15.43 ± 0.85 Å and 15.98 ± 1.04 Å, respectively, and thereby disrupt the β-sheet. As discussed further in Section S1.3.5 in the Supporting Material, we attribute this deviation to Ca2+ ion binding at the cEF hand. Specifically, N64 and D66 reorient to directly coordinate the Ca2+ ion, thereby pulling G65 away from active hydrogen bonding with residue K31 and compromising the β-sheet interaction. We will later present evidence that the disrupted β-sheet coupling upon Ca2+ binding directly correlates with increased mobility of helix 4 (H4), which together suggests that β-sheet hydrogen-bond breaking may trigger open-state formation.
Figure 5.
Average distances (Å) between backbone amide nitrogen and oxygen atoms of the residues in the EF hands forming the β-sheets in the apo case (blue), holo case (red), and half-saturated case (green). Standard deviations are shown. The yellow line indicates the 3.5 Å mark, which we define as the hydrogen-bond cutoff. Ca2+ ion binding disrupts the β-sheet hydrogen-bond network in both the half-saturated and holo states. To see this figure in color, go online.
Configuration and variability of the helix 3 and helix 4 angles
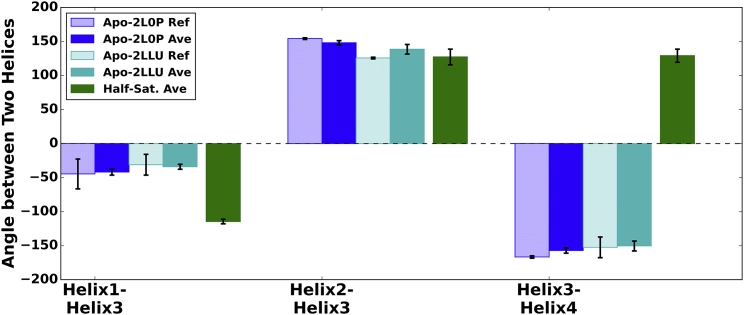
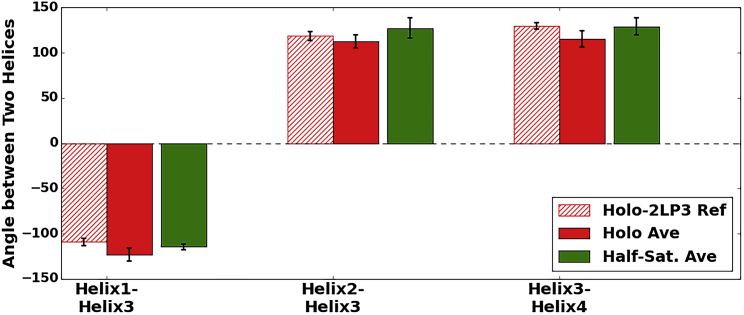
Ca2+ ion binding at the cEF and pEF hands is known to expose a hydrophobic PPI site between H3 and H4 (6, 15, 16, 17, 18). Interhelical angles have thus been commonly used (6, 16, 17, 28, 29, 72) to assess the transition between open and closed states. Experimental studies (26, 28, 29) of the apo versus holo conformations have indicated significant interhelical angle changes between H1 and H3 (−44.6 ± 21.9° versus −109.1 ± 4.1° for the 2L0P apo and 2LP3 holo cases, respectively) and between H3 and H4 (−166.6 ± 1.7° versus 129.9 ± 3.6° for the 2L0P apo and 2LP3 holo cases, respectively), culminating from the progression of H3 from closed to open states. Our helical angle measurements for the apo (Fig. 6) and holo states (Fig. 7) show excellent agreement between the simulation and the experiment. Namely, we observe for H3 and H4 that average values for the apo simulations are −157.2 ± 3.9° and −150.4 ± 7.3° versus 115.3 ± 8.9° in the holo state. For the half-saturated case, the H3 and H4 angle approached 129.1 ± 9.7°, which is consistent with the holo state configuration. We will note in the subsequent section, however, that the half-saturated state fleetingly progresses toward an apo-like configuration characterized by closing of the H3/H4 angle. The comparatively sparse numbers and short durations of these excursions are apparently insufficient to significantly shift the interhelical angle average.
Figure 6.
Computed versus experimental mean interhelical angles between H1 and H3, H2 and H3, and H3 and H4 for two apo NMR cases (PDB: 2L0P (blue) (26), 2LLU (cyan) (28)), and the half-saturated state (green). On average, the H1/H3 and H3/H4 angles in the half-saturated state are inconsistent with those in the apo state. To see this figure in color, go online.
Figure 7.
Computed (solid) versus experimental (hatched) mean interhelical angles between H1 and H3, H2 and H3, and H3 and H4 for one holo NMR case (PDB: 2LP3 (red) (29)) and the half-saturated state (green). On average, H1/H3 and H3/H4 angles in the half-saturated state are consistent with those in the holo state. To see this figure in color, go online.
PCA of protein-recognition-site exposure
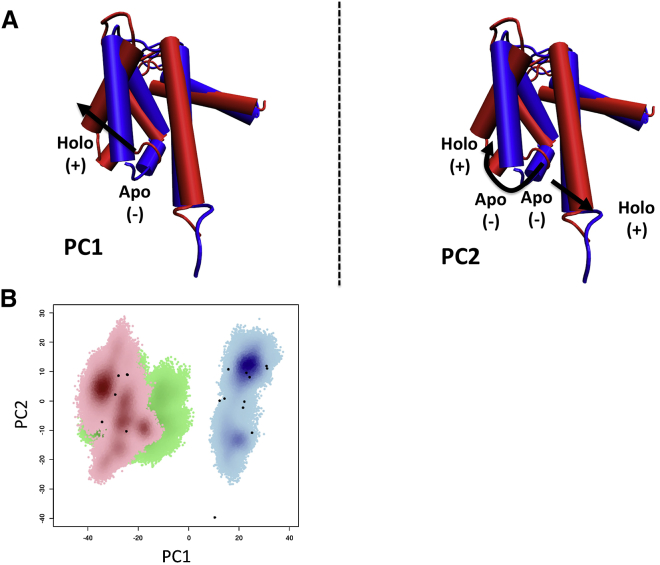
Ca2+ ion binding is known to expose the hydrophobic PPI site via migration of H3 toward an open-state configuration (17, 18), which is accompanied by significant structural reorganization of the protein. To simplify interpretation of our simulations, we used PCA to project the protein’s predominant conformational changes into a basis that clearly distinguished the apo from the holo states. The first two (largest eigenvalue) PCs for all S100A1 homologous structures available in the Protein Data Bank are shown in Fig. 8. In Fig. 8 A, we relate the PC bases to the conformational motions of the protein. PC1 is characterized by the swinging movement of H3 as it tilts outward from the apo state (26) toward the holo state (29) to allow room for binding peptides (6, 18). PC2 is characterized by the unwinding of the H4 C-terminus (data not shown), though it is not clear whether this loss of helical secondary structure plays a role in tuning Ca2+ affinity in S100A1 variants. In Fig. 8 B, we demonstrate that the apo and holo structures are clearly separated by the first PC (PC1); specifically, the holo structures are localized to PC1 = −10 to −45 and the apo structures to PC1 = 5–40. We further find that S100A1 variants with higher Ca2+ ion affinity (for example, the holo rat S100A1 bound to the TRTK12 peptide) tend to have more positive PC2 values than those with weaker affinity. Similarly, two peptide-bound S100A1 rat protein structures with high Ca2+ ion affinity exhibit more positive PC2 values than the human holo state examined in this study.
Figure 8.
(A) PC1 (left) and PC2 (right) compared against the NMR structures of the human apo protein (blue; PDB: 2L0P) (26) and the human holo protein (red; PDB: 2LP3) (29). Movement of the respective PCs is in the direction from apo, indicated by (−), to holo, indicated by (+), as shown by the black arrows. PC1 is characterized by the swinging of the third helix away from the helical bundle. PC2 is characterized by the rotation of the third helix and the tilting of the fourth helix, both away from the helical bundle. (B) PCA for the four MD trajectories of the apo (blue), holo (red), and half-saturated (green) proteins, with NMR structures indicated as black dots. The half-saturated system samples intermediate regions along PC1 between the apo and holo states. To see this figure in color, go online.
Using these PCs, we show in Fig. 8 B the projections of the four MD trajectories of the apo, fully saturated holo, and half-saturated cases. For the apo case, the protein conformation samples PC2 extensively −27 to 27), but not PC1 (5–40), which suggests that the apo state remained in the closed configuration. Similarly, for the holo case, the trajectory is restricted to more negative PC1 values, indicating that the protein remains open. In contrast, the half-saturated state trajectories span a larger range of PC1 values, suggesting occasional excursions toward an apo-like closed state.
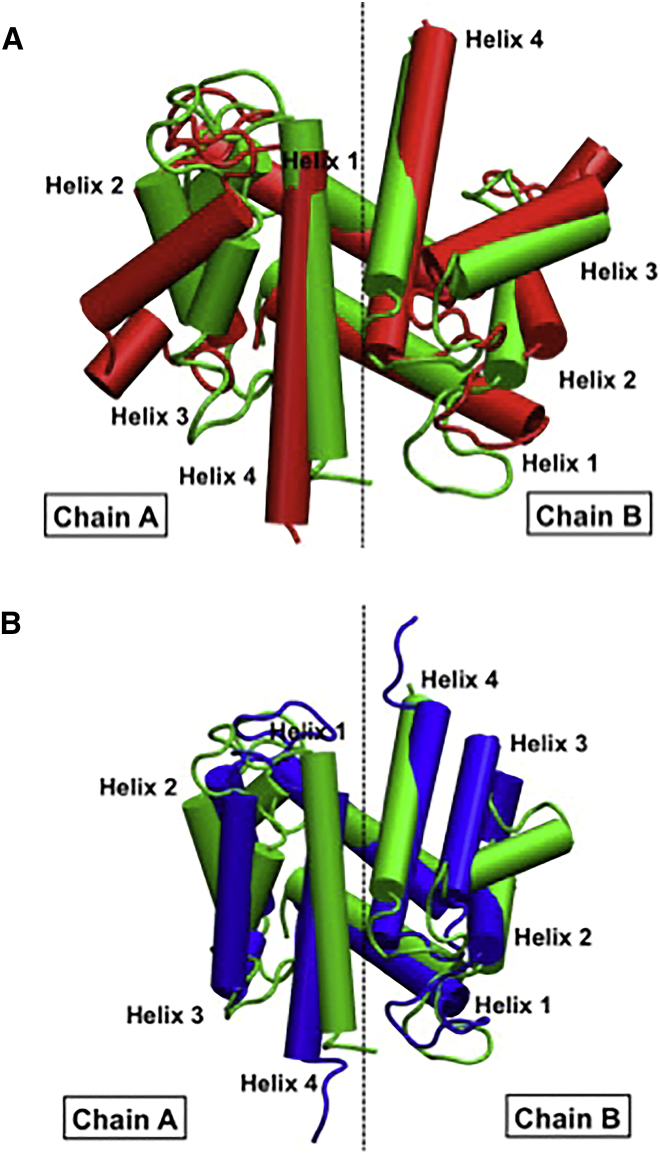
To investigate the hypothesis that the removal of the pEF Ca2+ ions may prompt relaxation from the open toward the closed state, we selected four snapshots from intermediate PC1 values for extended MD simulations. In Fig. 9, we compare a snapshot from one 100 ns extended simulation with apo and holo S100A1 NMR structures (26, 29). The snapshot indicates that the chain A H3 unwinds by several turns and tilts inward to closely resemble the apo NMR structure, whereas the chain B H3 maintains its original open position. Thus, it is apparent that in the absence of pEF Ca2+ions, the open state may relax back to the closed state. Similar trends are reported in the other extended trajectories in Fig. S4. These results agree with data from Goch et al. (15) indicating that the hydrophobic patch was less solvent-exposed in subsaturating levels of Ca2+. Overall, our data imply that the Ca2+ ion-bound pEF plays a critical role in stabilizing the holo-like state, despite its lesser affinity for Ca2+ ions relative to the cEF hand. Nevertheless, longer-duration MD simulations are likely required to fully repack H3 into the S100A1 helical bundle to more closely resemble the apo state. It is interesting to note that thiolation, glutathionylation, and nitrosylation of C85 on H4 has been suggested to increase Ca2+ binding affinity and induce significant conformational changes in the protein (15, 28, 72). Although few chemically modified C85 structures have been resolved, an attractive hypothesis is that covalently modified C85 could prevent H3 and H4 from fully closing, thereby producing a semiopen state that facilitates target peptide binding in the absence of pEF Ca2+ ions.
Figure 9.
Comparison of a half-saturated S100A1 snapshot (green) extended for 100 ns beyond the initial 60 ns MD run against holo (A, red) and apo (B, blue) S100A1 structures (26, 29). These half-saturated snapshots show a closer resemblance to the closed apo state; similar trends are reported for three other extended half-saturated simulations. To see this figure in color, go online.
Cross-correlation analysis of interresidue coupling
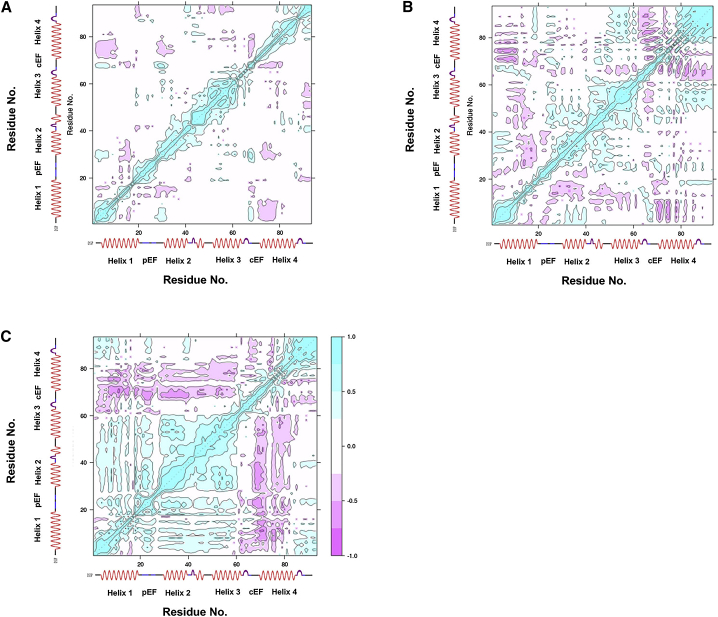
Ca2+ ion binding at the pEF site (formed from H1 and H2) appears to modulate the H3 position, despite the fact that H3 and H4 are bridged by the cEF hand. This behavior is a hallmark of allostery, for which substrate binding induces protein conformational changes distal to the recognition region. A prominent hypothesis for allostery evokes the notion that long-range correlations between a binding site and distal regions of the protein (73) are involved in transferring the binding signal. We therefore report in Fig. 10 the cross correlation of Cα atom positions to uncover potential trends in this allosteric mechanism.
Figure 10.
Residue cross correlation analysis for the human S100A1 protein in the apo (A), half-saturated (B), and holo (C) states. Cyan regions represent positive correlation, and magenta regions represent negative or anticorrelation. The half-saturated case presents correlation structure intermediate to the apo and holo cases. To see this figure in color, go online.
For the apo state (Fig. 10 A), we observe a small number of anticorrelated residues arising from breathing (opening) motions of H4 with H1 and H2, as well as with the EF hands. We note, however, a distinct positive correlation approximately between residues K25–K30 and E68–F71. This region corresponds to the β-sheet that tightly couples the EF hands in the apo state (14). Beyond these correlations, minimal coupling between distal regions is apparent. In contrast, the holo state (Fig. 10 C) presents a rich variety of off-diagonal (distal) correlations that evidence strong coupling between distal regions of the protein. We observe strong positive correlations, for instance, between H2, the linker region, and H3, as well as between H1, the pEF, the linker, and H3. Moreover, the negative correlations involving cEF and H4 appear to be intensified in the holo state relative to the apo site, which is suggestive of a breathing motion near the hydrophobic patch. Our earlier observations of EF-hand β-sheet decoupling upon Ca2+ binding appear to abolish the positive correlation in the K25–K30/E68–F71 region reported in the apo state with anticorrelative motions. Since these negative correlations extend from the β-sheet over the entirety of H4, we speculate that the β-sheet decoupling is the allosteric driver of the open-state formation. We additionally note in Fig. 10 C that C85 was anticorrelated with H3 residues 55–60 in the holo state, which may serve as additional evidence that C85 functionalization could enhance decoupling of H3 from H4. Hence, the barrier to exposing the hydrophobic region may be reduced and thus obviate the binding of Ca2+ ions at the pEF site, in support of findings from Zhukov et al (72). Similar analysis applied to the half-saturated state (Fig. 10 B) yields correlations intermediate between those of the apo and holo states that are suggestive of partial progression toward those configurations. More sophisticated methodologies for analyzing allostery in proteins, such as the mutual-information (74) or transmission-pathway (75) approach, could potentially provide greater insight into the Ca2+-driven allostery of open-state formation.
Conclusions
Our MD simulations of the human S100A1 protein reveal structural and dynamic details of Ca2+ binding at the EF hands and its influence in forming the protein’s functionally active open state. Using a primitive electrostatic scoring function, we find that the higher density and more negative partial charges of the coordinating oxygens in the cEF hand relative to the pEF region correlate with the former’s stronger affinity for Ca2+ ions. In a separate manuscript, we are extending our primitive electrostatic score to include enthalpic and entropic terms to better quantify these affinities. We further find that in the absence of Ca2+ ion binding at the pEF hand, S100A1 appears to revert to an apo-like closed configuration whereby anticorrelation between H4 and the helical bundle in the holo state is diminished. We believe this transition is afforded by decoupling β-sheets linking the EF hands upon Ca2+ ion binding. Finally, the evident role of H4 in the transition between the apo and holo states appears consistent with reports of enhanced signaling propensity for chemically modified H4 C85 variants at subsaturating levels of Ca2+. This insight into the molecular basis of high-affinity Ca2+ binding and its impact on allostery in S100A1 will be invaluable for understanding the function of the broad family of S100A-class proteins.
Author Contributions
C.E.S. designed the research, performed the research, analyzed the data, and wrote the article. P.K.H. designed the research, analyzed the data, and wrote the article.
Acknowledgments
P.K.H. thanks Jeff Wereszczynski for many helpful discussions. C.E.S. and P.K.H. thank Jason Swails and Kenneth Merz for their advice and help.
This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1053575.
Editor: Michael Feig.
Footnotes
Supporting Materials and Methods, Supporting Results, four figures, and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)00050-3.
Supporting Citations
References (76, 77, 78, 79, 80) appear in the Supporting Material.
Supporting Material
References
- 1.Zimmer D.B., Chessher J., Zimmer W.E. S100A1 and S100B expression and target proteins in type I diabetes. Endocrinology. 1997;138:5176–5183. doi: 10.1210/endo.138.12.5579. [DOI] [PubMed] [Google Scholar]
- 2.Kato K., Kimura S. S100ao (α-α) protein is mainly located in the heart and striated muscles. Biochim. Biophys. Acta. 1985;842:146–150. doi: 10.1016/0304-4165(85)90196-5. [DOI] [PubMed] [Google Scholar]
- 3.Haimoto H., Kato K. S100A0 (α-α) protein in cardiac muscle: isolation from human cardiac muscle and ultrastructural localization. Eur. J. Biochem. 1988;171:409–415. doi: 10.1111/j.1432-1033.1988.tb13805.x. [DOI] [PubMed] [Google Scholar]
- 4.Kraus C., Rohde D., Most P. S100A1 in cardiovascular health and disease: closing the gap between basic science and clinical therapy. J. Mol. Cell. Cardiol. 2009;47:445–455. doi: 10.1016/j.yjmcc.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rohde D., Ritterhoff J., Most P. S100A1: a multifaceted therapeutic target in cardiovascular disease. J. Cardiovasc. Transl. Res. 2010;3:525–537. doi: 10.1007/s12265-010-9211-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright N.T., Prosser B.L., Weber D.J. S100A1 and calmodulin compete for the same binding site on ryanodine receptor. J. Biol. Chem. 2008;283:26676–26683. doi: 10.1074/jbc.M804432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prosser B.L., Hernandez-Ochoa E.O., Schneider M.F. S100A1 and calmodulin regulation of ryanodine receptor in striated muscle. Cell Calcium. 2011;50:323–331. doi: 10.1016/j.ceca.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiewitz R., Acklin C., Heizmann C.W. Ca2+-dependent interaction of S100A1 with the sarcoplasmic reticulum Ca2+-ATPase2a and phospholamban in the human heart. Biochem. Biophys. Res. Commun. 2003;306:550–557. doi: 10.1016/s0006-291x(03)00987-2. [DOI] [PubMed] [Google Scholar]
- 9.Most P., Remppis A., Fink R.H.A. The C terminus (amino acids 75–94) and the linker region (amino acids 42–54) of the Ca2+-binding protein S100A1 differentially enhance sarcoplasmic Ca2+ release in murine skinned skeletal muscle fibers. J. Biol. Chem. 2003;278:26356–26364. doi: 10.1074/jbc.M303338200. [DOI] [PubMed] [Google Scholar]
- 10.Duarte-Costa S., Castro-Ferreira R., Leite-Moreira A.F. S100A1: a major player in cardiovascular performance. Physiol. Res. 2014;63:669–681. doi: 10.33549/physiolres.932712. [DOI] [PubMed] [Google Scholar]
- 11.Grabarek Z. Structural basis for diversity of the EF-hand calcium-binding proteins. J. Mol. Biol. 2006;359:509–525. doi: 10.1016/j.jmb.2006.03.066. [DOI] [PubMed] [Google Scholar]
- 12.Yang J.J., Gawthrop A., Ye Y.Y. Obtaining site-specific calcium-binding affinities of calmodulin. Protein Pept. Lett. 2003;10:331–345. doi: 10.2174/0929866033478852. [DOI] [PubMed] [Google Scholar]
- 13.Kirberger M., Wang X., Yang J.J. Statistical analysis of structural characteristics of protein Ca2+-binding sites. J. Biol. Inorg. Chem. 2008;13:1169–1181. doi: 10.1007/s00775-008-0402-7. [DOI] [PubMed] [Google Scholar]
- 14.Gifford J.L., Walsh M.P., Vogel H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- 15.Goch G., Vdovenko S., Bierzynski A. Affinity of S100A1 protein for calcium increases dramatically upon glutathionylation. FEBS J. 2005;272:2557–2565. doi: 10.1111/j.1742-4658.2005.04680.x. [DOI] [PubMed] [Google Scholar]
- 16.Wright N.T., Varney K.M., Weber D.J. The three-dimensional solution structure of Ca2+-bound S100A1 as determined by NMR spectroscopy. J. Mol. Biol. 2005;353:410–426. doi: 10.1016/j.jmb.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 17.Wright N.T., Cannon B.R., Weber D.J. Solution structure of S100A1 bound to the CapZ peptide (TRTK12) J. Mol. Biol. 2009;386:1265–1277. doi: 10.1016/j.jmb.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright, N. T., Morgan, M. T., Cannon, B. R., K. M. Varney, and D. J. Weber. 2008. Solution structure of Ca-S100A1-TRTK12. http://www.rcsb.org/pdb/explore.do?structureId=2kbm. Accessed February 4, 2016.
- 19.Herzberg O.J.M., Moult J. A model for the Ca2+-induced conformational transition of troponin C. A trigger for muscle contraction. J. Biol. Chem. 1986;261:2638–2644. [PubMed] [Google Scholar]
- 20.Houdusse A., Love M.L., Cohen C. Structures of four Ca2+-bound troponin C at 2.0 Å resolution: further insights into the Ca2+-switch in the calmodulin superfamily. Structure. 1997;5:1695–1711. doi: 10.1016/s0969-2126(97)00315-8. [DOI] [PubMed] [Google Scholar]
- 21.Strynadka N.C.J., Cherney M., James M.N.G. Structural details of a calcium-induced molecular switch: x-ray crystallographic analysis of the calcium-saturated N-terminal domain of troponin C at 1.75 Å resolution. J. Mol. Biol. 1997;273:238–255. doi: 10.1006/jmbi.1997.1257. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z.Y., Gergely J., Tao T. Characterization of the Ca2+-triggered conformational transition in troponin C. Proc. Natl. Acad. Sci. USA. 1992;89:11814–11817. doi: 10.1073/pnas.89.24.11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn B.E., Drakenberg T., Forsen S. The structure of apo-calmodulin. A 1H NMR examination of the carboxy-terminal domain. FEBS Lett. 1993;336:368–374. doi: 10.1016/0014-5793(93)80839-m. [DOI] [PubMed] [Google Scholar]
- 24.Finn B.E., Evenas J., Forsen S. Calcium-induced structural changes and domain autonomy in calmodulin. Nat. Struct. Biol. 1995;2:777–783. doi: 10.1038/nsb0995-777. [DOI] [PubMed] [Google Scholar]
- 25.Tan R.Y., Mabuchi Y., Grabarek Z. Blocking the Ca2+-induced conformational transitions in calmodulin with disulfide bonds. J. Biol. Chem. 1996;271:7479–7483. doi: 10.1074/jbc.271.13.7479. [DOI] [PubMed] [Google Scholar]
- 26.Nowakowski M., Jaremko L., Ejchart A. Solution NMR structure and dynamics of human apo-S100A1 protein. J. Struct. Biol. 2011;174:391–399. doi: 10.1016/j.jsb.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Rustandi R.R., Baldisseri D.M., Weber D.J. Three-dimensional solution structure of the calcium-signaling protein Apo-S100Al as determined by NMR. Biochemistry. 2002;41:788–796. doi: 10.1021/bi0118308. [DOI] [PubMed] [Google Scholar]
- 28.Zivkovic M.L., Zareba-Koziol M., Wyslouch-Cieszynska A. Post-translational S-nitrosylation is an endogenous factor fine tuning the properties of human S100A1 protein. J. Biol. Chem. 2012;287:40457–40470. doi: 10.1074/jbc.M112.418392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nowakowski M., Ruszczynska-Bartnik K., Ejchart A. Impact of calcium binding and thionylation of S100A1 protein on its nuclear magnetic resonance-derived structure and backbone dynamics. Biochemistry. 2013;52:1149–1159. doi: 10.1021/bi3015407. [DOI] [PubMed] [Google Scholar]
- 30.Berridge M.J., Lipp P., Bootman M.D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 31.Shannon T., Wang F., Bers D. A mathematical treatment of integrated Ca dynamics within the ventricular myocyte. Biophys. J. 2004;87:3351–3371. doi: 10.1529/biophysj.104.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearlman D.A., Case D.A., Kollman P. AMBER, A package of computer-programs for applying molecular mechanics, normal-mode analysis, molecular dynamics, and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 33.Case D.A., Cheatham T.E., Woods R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cornell W.D., Cieplak P., Kollman P.A. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 35.Li P.F., Roberts B.P., Merz K.M. Rational design of particle mesh Ewald compatible Lennard-Jones parameters for +2 metal cations in explicit solvent. J. Chem. Theory Comput. 2013;9:2733–2748. doi: 10.1021/ct400146w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchand S., Roux B. Molecular dynamics study of calbindin D9k in the apo and singly and doubly calcium-loaded states. Proteins. 1998;33:265–284. [PubMed] [Google Scholar]
- 37.Kekenes-Huskey P., Lindert S., McCammon J. Molecular basis of calcium-sensitizing and desensitizing mutations of the human cardiac troponin C regulatory domain: a multi-scale simulation study. PLOS Comput. Biol. 2012;8:e1002777. doi: 10.1371/journal.pcbi.1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindert S., Kekenes-Huskey P., McCammon J. Dynamics and calcium association to the N-terminal regulatory domain of human cardiac troponin C: a multiscale computational study. J. Phys. Chem. B. 2012;116:8449–8459. doi: 10.1021/jp212173f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Towns J., Cockerill T., Wilkens-Diehr N. XSEDE: accelerating scientific discovery. Comput. Sci. Eng. 2014;16:62–74. [Google Scholar]
- 40.Genheden S., Diehl C., Ryde U. Starting-condition dependence of order parameters derived from molecular dynamics simulations. J. Chem. Theory Comput. 2010;6:2176–2190. doi: 10.1021/ct900696z. [DOI] [PubMed] [Google Scholar]
- 41.Romo T., Grossfield A. LOOS: An extensible platform for the structural analysis of simulations. Conf. Proc. IEEE Eng. Med. Biol. Sci. 2009;2009:2332–2335. doi: 10.1109/IEMBS.2009.5335065. [DOI] [PubMed] [Google Scholar]
- 42.Best R.B., Zhu X., MacKerell A.D. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacKerell A.D., Feig M., Brooks C.L. Improved treatment of the protein backbone in empirical force fields. J. Am. Chem. Soc. 2004;126:698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 44.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 45.Kollman P.A., Massova I., Cheatham T.E. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 46.Gohlke H., Case D.A. Converging free energy estimates: MM-PB(GB)SA studies on the protein-protein complex Ras-Raf. J. Comput. Chem. 2004;25:238–250. doi: 10.1002/jcc.10379. [DOI] [PubMed] [Google Scholar]
- 47.Hou T., Wang J., Wang W. Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated from docking. J. Comput. Chem. 2011;32:866–877. doi: 10.1002/jcc.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller I., Bill R., Roitberg A.E. MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 49.Grant B.J., Rodrigues A.P.C., Caves L.S.D. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 50.Altschul S.F., Madden T.L., Lipman D.D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCammon A.J., Harvey S.C. Cambridge University Press; Cambridge, United Kingdom: 1986. Dynamics of Proteins and Nucleic Acids. [Google Scholar]
- 52.Lange O., Grubmuller H. Generalized correlation for biomolecular dynamics. Proteins. 2006;62:1053–1061. doi: 10.1002/prot.20784. [DOI] [PubMed] [Google Scholar]
- 53.Spyracopoulos L., Lavigne P., Sykes B.D. Temperature dependence of dynamics and thermodynamics of the regulatory domain of human cardiac troponin C. Biochemistry. 2001;40:12541–12551. doi: 10.1021/bi010903k. [DOI] [PubMed] [Google Scholar]
- 54.Saxena A., Sept D. Multisite ion models that improve coordination and free energy calculations in molecular dynamics simulations. J. Chem. Theory Comput. 2013;9:3538–3542. doi: 10.1021/ct400177g. [DOI] [PubMed] [Google Scholar]
- 55.Kohagen M., Lepšík M., Jungwirth P. Calcium binding to calmodulin by molecular dynamics with effective polarization. J. Phys. Chem. Lett. 2014;5:3964–3969. doi: 10.1021/jz502099g. [DOI] [PubMed] [Google Scholar]
- 56.Wang D., Robertson I.M., Regnier M. Structural and functional consequences of the cardiac troponin C L48Q Ca2+-sensitizing mutation. Biochemistry. 2012;51:4473–4487. doi: 10.1021/bi3003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lindert S., Kekenes-Huskey P., McCammon J. Long-timescale molecular dynamics simulations elucidate the dynamics and kinetics of exposure of the hydrophobic patch in troponin C. Biophys. J. 2012;103:1784–1789. doi: 10.1016/j.bpj.2012.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang C., Jas G., Kuczera K. Structure, dynamics and interaction with kinase targets: computer simulations of calmodulin. Biochim. Biophys. Acta. 2004;1697:289–300. doi: 10.1016/j.bbapap.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 59.Aykut A.O., Atilgan A.R., Atilgan C. Designing molecular dynamics simulations to shift populations of the conformational states of calmodulin. PLOS Comput. Biol. 2013;9:e1003366. doi: 10.1371/journal.pcbi.1003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang W., Luo Y., Roux B. Calculation of free energy landscape in multi-dimensions with Hamiltonian-exchange umbrella sampling on petascale supercomputer. J. Chem. Theory Comput. 2012;8:4672–4680. doi: 10.1021/ct300468g. [DOI] [PubMed] [Google Scholar]
- 61.Biekofsky R.R., Turjanski A.G., Pastore A. Ab initio study of NMR 15N chemical shift differences induced by Ca2+ binding to EF-hand proteins. Biochemistry. 2011;43:6554–6564. doi: 10.1021/bi0497852. [DOI] [PubMed] [Google Scholar]
- 62.Cieplak P., Dupradeau F.-Y., Wang J. Polarization effects in molecular mechanical force fields. J. Phys. Condens. Matter. 2009;21:333102. doi: 10.1088/0953-8984/21/33/333102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ye Y., Lee H.W., Shealy S. Probing site-specific calmodulin calcium and lanthanide affinity by grafting. J. Am. Chem. Soc. 2005;127:3743–3750. doi: 10.1021/ja042786x. [DOI] [PubMed] [Google Scholar]
- 64.Lipari G., Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 65.Maniccia A.W., Yang W., Yang J.J. Inverse tuning of metal binding affinity and protein stability by altering charged coordination residues in designed calcium binding proteins. PMC Biophys. 2009;2:11. doi: 10.1186/1757-5036-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ben-Amotz D., Underwood R. Unraveling water’s entropic mysteries: a unified view of nonpolar, polar, and ionic hydration. Acc. Chem. Res. 2008;41:957–967. doi: 10.1021/ar7001478. [DOI] [PubMed] [Google Scholar]
- 67.Nonner W., Gillespie D., Eisenberg B. Ion accumulation in a biological calcium channel: effects of solvent and confining pressure. J. Phys. Chem. B. 2001;105:6427–6436. [Google Scholar]
- 68.Nonner W., Catacuzzeno L., Eisenberg B. Binding and selectivity in L-type calcium channels: a mean spherical approximation. Biophys. J. 2000;79:1976–1992. doi: 10.1016/S0006-3495(00)76446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beard H., Cholleti A., Loving K.A. Applying physics-based scoring to calculate free energies of binding for single amino acid mutations in protein-protein complexes. PLoS One. 2013;8:1–11. doi: 10.1371/journal.pone.0082849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lindert S., Cheng Y.H., McCammon J.A. Effects of HCM cTnI mutation R145G on troponin structure and modulation by PKA phosphorylation elucidated by molecular dynamics simulations. Biophys. J. 2015;108:395–407. doi: 10.1016/j.bpj.2014.11.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zou X., Sun Y., Kunz I.D. Inclusion of solvation in ligand binding free energy calculations using the generalized-Born model. J. Am. Chem. Soc. 1999;121:8033–8043. [Google Scholar]
- 72.Zhukov I., Ejchart A., Bierzynski A. Structural and motional changes induced in apo-S100A1 protein by the disulfide formation between its Cys 85 residue and β-mercaptoethanol. Biochemistry. 2008;47:640–650. doi: 10.1021/bi701762v. [DOI] [PubMed] [Google Scholar]
- 73.Long D., Bruschweiler R. Atomistic kinetic model for population shift and allostery in biomolecules. J. Am. Chem. Soc. 2011;133:18999–19005. doi: 10.1021/ja208813t. [DOI] [PubMed] [Google Scholar]
- 74.McClendon C.L., Friedland G., Jacobson M.P. Quantifying correlations between allosteric sites in thermodynamic ensembles. J. Chem. Theory Comput. 2009;5:2486–2502. doi: 10.1021/ct9001812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Wart A.T., Durrant J., Amaro R.E. Weighted implementation of suboptimal paths (WISP): an optimized algorithm and tool for dynamical network analysis. J. Chem. Theory Comput. 2014;10:511–517. doi: 10.1021/ct4008603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miyamoto S., Kollman P.A. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 77.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of cartesian equations of motion of a systerm with constraints: molecular dynamics of N-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 78.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 79.Darden T., York D., Pedersen L. Particle mesh Ewald: an N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 80.Case D., Babin V., Kollman P.A. University of California; San Francisco, CA: 2014. AMBER 14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.