Abstract
The first exon of Huntingtin—a protein with multiple biological functions whose misfolding is related to Huntington’s disease—modulates its localization, aggregation, and function within the cell. It is composed of a 17-amino-acid amphipathic segment (Htt17), an amyloidogenic segment of consecutive glutamines (QN), and a proline-rich segment. Htt17 is of fundamental importance: it serves as a membrane anchor to control the localization of huntingtin, it modulates huntingtin’s function through posttranslational modifications, and it controls the self-assembly of the amyloidogenic QN segment into oligomers and fibrils. Experimentally, the conformational ensemble of the Htt17 monomer, as well as the impact of the polyglutamine and proline-rich segments, remains, however, mostly uncharacterized at the atomic level due to its intrinsic flexibility. Here, we unveil the free-energy landscape of Htt17, Htt17Q17, and Htt17Q17P11 using Hamiltonian replica exchange combined with well-tempered metadynamics. We characterize the free-energy landscape of these three fragments in terms of a few selected collective variables. Extensive simulations reveal that the free energy of Htt17 is dominated by a broad ensemble of configurations that agree with solution NMR chemical shifts. Addition of Q17 at its carboxy-terminus reduces the extent of the main basin to more extended configurations of Htt17 with lower helix propensity. Also, the aliphatic carbons of Q17 partially sequester the nonpolar amino acids of Htt17. For its part, addition of Q17P11 shifts the overall landscape to a more extended and helical Htt17 stabilized by interactions with Q17 and P11, which almost exclusively form a PPII-helix, as well as by intramolecular H-bonds and salt bridges. Our characterization of Huntingtin’s amino-terminus provides insights into the structural origin of its ability to oligomerize and interact with phospholipid bilayers, processes closely linked to the biological functions of this protein.
Introduction
Huntingtin is a large, ubiquitous protein of >3000 amino acids (1, 2). It is essential to embryonic development (3), interacts with many proteins through its 36 HEAT repeats (4, 5), and is involved in intracellular organelles and vesicular trafficking, (6) as well as transcription and axonal transport (7). The exon 1 of huntingtin—consisting of an amphipathic sequence of 17 amino acids (Htt17), an amyloidogenic polyglutamine region (QN), and a segment of 36 amino acids rich in prolines—is closely linked to Huntingtin’s functions. This segment contains a nuclear export sequence that controls the localization of huntingtin between the cytoplasm and the nucleus (8, 9), it can undergo posttranslational modifications affecting the localization and function of huntingtin (10, 11, 12, 13, 14), and it is responsible for the localization of huntingtin to the mitochondria and the Golgi (15, 16).
Over the past years, Huntingtin has attracted considerable attention, as it is an amyloid protein associated with Huntington’s disease, an autosomal dominant genetic disorder (17). Its assembly into amyloid fibrils is triggered in vivo by the expansion of the consecutive segment of glutamines at its first exon above a specific threshold. This characteristic behavior, which is shared by at least 10 other proteins, is termed the polyglutamine/CAG repeat disorder and is associated with several disorders (18, 19, 20). More specifically to Huntington’s disease, the huntingtin protein misfolds, self-assembles, and mislocalizes in the cell when the QN region has >36 repeats, causing deleterious effects by gain- and loss-of-function through various nuclear and extranuclear pathways (21, 22, 23). Huntingtin amino-terminus fragments, which can be generated in vivo by proteolytic cleavage, are found in postmortem brain tissue (24) and are involved in the pathogenesis of Huntington’s disease (25, 26). The first exon, more precisely, is closely linked to the cytotoxicity, as it is sufficient to cause Huntington’s phenotype both in vivo (27, 28) and in vitro (29, 30, 31). This segment also controls the toxicity, localization, and clearance of mutant huntingtin through posttranslational modifications (10, 11, 12, 13, 14). It furthermore interacts with the TRiC chaperonin mainly through its Htt17 segment suppressing the misfolding and aggregation of huntingtin (32).
The neighboring regions of QN in the first exon are crucial to control of its misfolding and amyloidogenesis (33). In fact, the aggregation of full-length huntingtin exon 1 is very similar to that of Htt17QNP10, showing the importance of the amino acids right next to QN (34). For instance, the presence of the Htt17 segment accelerates the fibrillation kinetics of QN (35, 36), whereas the PN segment decelerates it (37, 38). The nucleus size, as well as the overall aggregation pathways of QN, is also strongly affected by the presence of Htt17 (32, 39, 40, 41). Some experimental results indicate that it causes the aggregation to split into two main pathways in direct kinetic competition: it proceeds either 1) through the formation of α-helical tetrameric bundles of Htt17 that increase the local concentration of QN, favoring the nucleation of β-sheeted structures in it, or 2) through an unfavorable nucleation in the monomeric QN (40). The Htt17 segment could also facilitate the formation and stability of β-sheeted structures in QN by interacting directly with it (32, 42). A less direct role for Htt17 has also been suggested, where it could destabilize nonfibrillar aggregates by reducing the entanglement of QN (43), thus accelerating the formation of fibrils (41).
Given the importance of Htt17, a characterization of its conformational ensemble at the monomer level could shed light on the atomistic features responsible for its aggregation. Experiments suggest that Htt17 samples transient helical configurations in aqueous solution, as circular dichroism (CD) data indicate the presence of helical structures (15, 35, 44, 45), and as solution NMR suggests no stable secondary-structure motif (35). Due to its intrinsic flexibility and the absence of a stable secondary-structure motif, the Htt17 monomer yields too few NMR constraints in aqueous solution to extract any viable three-dimensional structural model (35). An x-ray model of the chimeric maltose-binding protein-huntingtin exon 1 (MBP-Htt17Q17-Ex1) protein also suggests that Htt17 can adopt helical structures, whereas the QN region is mostly disordered and the P11 is a polyproline type-II helix (46). A few notable simulations of Htt17 complemented these experimental results by describing, to some extent, its conformational ensemble using 1) all-atom, explicit-solvent simulated tempering molecular dynamics (47), 2) all-atom, implicit-solvent Monte Carlo (44), or 3) all-atom, explicit-solvent bias-exchanged metadynamics (48) simulations. All agree that Htt17 samples a broad ensemble of helix/coil structures. Other simulations were aimed at characterizing the overall effect of increasing the QN length in the context of huntingtin exon 1 (44, 49, 50).
The Htt17 is also crucial for the localization of huntingtin in the cell, in part through direct membrane interactions (15, 16). More recently, the structure of Htt17 in the presence of dodecylphosphocholine (DPC) micelles has been resolved using solution NMR: it is an α-helix from residues 6 to 17, whereas the rest of the sequence is disordered and highly flexible (45, 51). Results from solid-state NMR (51) and Hamiltonian replica-exchange (HREX) all-atom simulations (52) indicate that Htt17 is also an α-helix in the context of a membrane bilayer. As the formation of α-helical structures in Htt17 before its binding seems to favor its membrane partitioning (53), understanding the conformational ensemble of Htt17 in aqueous solution could then unveil motifs beneficial to membrane binding. In the context of exon 1, the effect of QN and P11 on the occurrence of such motifs could explain their modulation of Htt17 binding affinity, as observed experimentally (54).
Focusing on the identification of Htt17’s structural features at the origin of its oligomerization and membrane partitioning, we investigate the free-energy landscape of the monomeric Htt17 using all-atom, explicit-solvent HREX metadynamics (MetaD). Such a simulation protocol favors correct sampling of the entire conformational space physically available to the protein. We also quantify the effect on Htt17’s global free-energy landscape of adding the amyloidogenic QN region as well as the P11 segment. Overall, such detailed information is necessary to rationalize the importance of Htt17 in addition to paving the way for the investigation of the oligomerization and membrane-binding processes per se using such a similarly stringent simulation protocol.
Materials and Methods
In this study, we use HREX and parallel tempering (PT) combined with well-tempered MetaD (55, 56, 57, 58, 59) simulations to investigate the free-energy landscape of the 17-amino-acid amino-terminus segment (Htt17) of the Huntingtin protein in aqueous solution. We also quantify the impact on Htt17’s free-energy landscape of adding the amyloidogenic polyglutamine (Q17) and polyproline (P11) segments. The amino acid sequence of Htt17 is MATLEKLMKAFESLKSF and an amidated carboxy-terminus is used for all peptide constructs. All simulations are summarized in Table 1. We focus on the HREXMetaD simulations in the main text, whereas the PTMetaD simulations are presented in the Supporting Material.
Table 1.
Summary of the Performed Simulations
| Simulations | Type | Initial Conformation | Time per Replica (μs) | Time (μs) |
|---|---|---|---|---|
| Htt17_nmr | HREXMetaD | NMR | 0.9 × 16 | 14.4 |
| Htt17_coil | HREXMetaD | coil | 0.9 × 16 | 14.4 |
| Htt17Q17 | HREXMetaD | NMR/coil | 0.9 × 24 | 21.6 |
| Htt17Q17P11 | HREXMetaD | NMR/coil/coil | 0.9 × 24 | 21.6 |
| Htt17_nmr_remd | PTMetaD | NMR | 0.9 × 64 | 57.6 |
| Htt17_coil_remd | PTMetaD | coil | 0.9 × 64 | 57.6 |
| Htt17_grf | HREXMetaD | NMR | 0.25 × 16 | 4 |
| Htt17Q17P11_pro | HREXMetaD | NMR/coil/coil | 0.2 × 24 | 4.8 |
All simulations are done in the NVT ensemble in a rhombic dodecahedron periodic cell (; ; ; nm and 3500 water molecules for Htt17, nm and 7000 water molecules for Htt17Q17, and nm 10000 water molecules for Htt17Q17P11). This set-up is sufficient to ensure that our system, which is monomeric, interacts very little with its periodic image. Despite a formal dilution of 3.5 mM, it is therefore largely equivalent to a highly diluted system, allowing comparison to NMR studies done at 40 μM concentrations. We combined well-tempered MetaD to two other sampling enhancing simulation types, HREX and PT. The simulations on Htt17 are started from two different initial configurations, a fully random-coil structure and its NMR model in the presence of DPC detergent micelles (PDB: 2LD2). The latter configuration is disordered from residues 1 to 5 and an α-helix for the rest of the sequence (45, 51). In the initial state of the simulations on Htt17Q17 and Htt17Q17P11, Htt17 is taken as the NMR model, whereas Q17 and P11 are completely disordered. We focus on the HREX simulations in the main text, and we present the PT simulations in the Supporting Material. The last two simulations, Htt17_grf and Htt17Q17P11 _pro, are tests on the validity of the potentials and are presented and discussed in the Supporting Material.
Simulations protocols
Our molecular dynamics simulations are done with the Gromacs package version 4.6.5 (60, 61, 62, 63) combined with the PLUMED plug-in version 2.0.2 (64) to perform the well-tempered MetaD (56) and HREX (59) parts of our simulations, as described below. We use the all-atom forcefield AMBER99sb∗-ILDN (65, 66, 67), as it offers helix/coil-balanced sampling for the conformational ensemble of small and mostly disordered peptides with transient α-helical structures (66, 68), which is similar to Htt17 in aqueous solution (15, 35, 44, 45). It is also recognized as one of the best force fields for studying protein folding (69, 70, 71, 72). Our simulations are performed in the NVT ensemble and the temperature is maintained by the Bussi-Donadio-Parrinello thermostat with a coupling constant of 0.1 ps (73). The cutoff for van der Waals and short-range electrostatic interactions is 1.0 nm. Long-range electrostatics are computed using smooth particle-mesh Ewald (74, 75). Bond lengths and TIP3P water geometry are constrained using LINCS (76) and SETTLE (77), respectively, allowing an integration time step of 2 fs. The center-of-mass motion is removed every 20 fs. Configurations are saved every 4 ps for analysis.
We use HREXMetaD to efficiently sample the conformational ensemble and unveil the free-energy landscape of Htt17 in aqueous solution. This method combines two sampling enhancing techniques, HREX (78, 79) and MetaD (55, 56). MetaD introduces a history-dependent bias constructed by adding gaussians in the energy space to previously visited states along a set of specified collective variables (CVs). This increases the overall sampling at the same time as it reconstructs the free-energy landscape along those CVs as the introduced history-dependent biased potential converges to
| (1) |
where is the biased potential, F is the free energy, T is the temperature, C is an irrelevant constant, and ΔT is a parameter that controls the extent of barrier heights sampled in the well-tempered flavor of MetaD (56). It is also possible, as we do in this study, to reconstruct the free-energy landscape along any omitted CV given sufficient sampling (57, 80). The free-energy landscape is normalized so that all free energies are measured with respect to the most stable structure for each simulation, which is set at 0 kJ/mol.
For correct free-energy landscape reconstruction, all slow CVs need to be considered, but the maximum number of CVs computationally accessible for MetaD is ∼2–3, which is clearly not enough to model protein folding (81, 82, 83). One of the most efficient ways to avoid this limitation is to couple MetaD with a replica-exchange scheme such as HREX, as this technique, which is widely used to simulate protein folding on its own, increases the probability of escaping free-energy minima by allowing exchanges between simultaneous MD simulations at different Hamiltonians (59, 78). Using replica-exchange schemes such as HREX and PT together with MetaD allows correct sampling of other CVs not explicitly taken into account by the time-dependent biased potential, as demonstrated for proteins with conformational ensembles similar to that of Htt17 (84, 85, 86).
For the MetaD part of our hybrid simulations, we use two CVs to bias the α-helical character (Sα) and the radius of gyration (Sgyr) of the peptide:
| (2) |
| (3) |
where the sum in Sα is over the 13 possible α-helix hydrogen-bond distances, di, between main-chain HN-O couples separated by four residues, d0 is 0.3 nm, the sum in Sgyr is over all Cα atoms, ri and mi are the current Cα coordinate and mass, respectively, and rCOM is the center-of-mass coordinate. Note that by construction = 13, but the single α-helix has 12.0, as di is ∼0.20–0.25 nm for a hydrogen bond. This choice of CVs is motivated by the fact that the Htt17 peptide in aqueous solution has an average α-helix probability of 10–55% according to circular dichroism (CD) spectroscopy measurements (15, 35, 44, 45), but no stable α-helix, as determined by solution NMR experiments (35). The free-energy landscapes of peptides with conformational ensembles similar to that of Htt17 were also characterized using this set of CVs (84, 85, 86). During our simulations, a new gaussian is added to the biased potential every 4 ps, with standard deviations of 0.1 and 0.01 nm along Sα and Sgyr, respectively, and the initial height of these gaussians is 0.5 kJ/mol. The bias factor of the well-tempered scheme is set to 15.
The HREXMetaD simulations are performed at 303 K using 16 scales for Htt17 and 24 scales for Htt17Q17 and Htt17Q17P11 spanning 1.0 to 0.3 with intermediate scales specified by a geometric distribution. as previously done (59). Exchanges between neighboring scales are attempted every 4 ps, resulting in an exchange rate of ∼20–40%.
Simulated systems
All performed simulations are summarized in Table 1. The three investigated fragments of the huntingtin amino-terminus—Htt17, Htt17Q17, and Htt17Q17P11—are simulated using HREXMetaD at 303 K. The P11 segment corresponds to the first complete proline repeat sequence of the 36-amino-acid proline-rich segment connected to QN in huntingtin. Addition of both Htt17 and P11 are sufficient to reproduce the main characteristics of the aggregation of Huntingtin’s first exon (34). The two initial states for Htt17 are a random-coil structure and the solution NMR model (PDB: 2LD2) determined in the presence of DPC micelles (45, 51). In this latter state, Htt17 is an α-helix from residues 6 to 17 and disordered for the first five residues. For Htt17Q17 and Htt17Q17P11, Htt17 is the NMR model and Q17 and P11 are disordered. Random-coil configurations are generated with 100 ns high-temperature (600 K) simulations starting from initially totally extended structures.
Peptides are solvated in a rhombic dodecahedron periodic cell and neutralized by the addition of two chloride ions. All systems are energy minimized using the conjugate gradient algorithm and are equilibrated in the NPT ensemble at 303 K for 5 ns, restraining the protein backbone atoms to their initial positions using a harmonic potential. All replicas are further independently equilibrated at their respective Hamiltonian scale in the NVT ensemble for 10 ns.
Our analysis is performed using in-house GROMACS and PLUMED utilities. The secondary structure is computed using STRIDE (87) and chemical shifts using SPARTA+ (88) and Camshift (89). All computed quantities are reweighted to remove the bias introduced during the MetaD simulations, as previously described (80), using a python implementation by Ludovico Sutto that is available to the PLUMED community. The free energies are reweighted using a recently developed time-independent free-energy estimator (80). GROMACS utilities are used to compute the structural clusters using the gromos algorithm with a root mean-square deviation (RMSD) cutoff of 0.15 nm on the backbone atoms (g_cluster) (90), the solvent-accessible surface area (SASA) of the nonpolar residues (g_sas) (91), and the occurrence of H-bonds using a cutoff of 0.35 nm on the donor-acceptor distance and of 30° on the hydrogen-donor-acceptor angle (g_hbond). Salt bridges are considered when the distance between two oppositely charged moieties is <0.4 nm (92).
Errors correspond to one statistical standard deviation computed on the converged interval with 20 ns subsets.
Convergence
We assess the convergence of our simulations using three quantitative criteria, as shown in Fig. S1 for Htt17, Fig. S2 for Htt17Q17, and Fig. S3 for Htt17Q17P11. First, we track the evolution of the global uncertainty on the free energy to identify the time at which it becomes small enough. Second, we monitor the total free-energy bias added each 10 ns as a function of time to confirm that the added biases become small enough at some time. Third, we compute the two-dimensional free-energy uncertainty landscape as a function of Sα and Sgyr on the time interval of convergence determined from the two previous criteria to confirm that the errors are located in unimportant regions of the landscape.
We confirm that the choice of initial state does not impact our results on Htt17, as described in detail in the Supporting Material. To do so, we confirm that the convergence analysis and free-energy landscape generated starting from the NMR model obtained in the presence of DPC micelles and a random-coil state are very similar (compare Figs. S1 and S4). Moreover, as described in detail in the Supporting Material, we ensure that our results on Htt17 are mostly independent of the sampling method by comparing HREXMetaD to PTMetaD, since the latter is the one most often used. Additional simulations probing the effect of a generalized-reaction field on Htt17 (Htt17_grf) and the effect of the AMBER99sb∗-ILDNP force field, with revised proline parameters, on Htt17Q17P11 (Htt17Q17P11_pro) were done using slightly modified parameters to accelerate the simulation; a new gaussian is added to the potential every 1 ps (instead of 4 ps) and the standard deviation of Srg was increased to 0.03 nm (instead of 0.01 nm). An in-depth discussion on these simulations can be found in the Supporting Material.
In complement, we probe the quality of the sampling of our HREX simulations by monitoring the replicas visiting the first scale that is used in our analysis, as well as the secondary-structure content as a function of the scaling (Fig. S5). We find great diffusion in the replica space and a more disordered peptide at larger scales.
Overall, our convergence evaluation indicates that the following analysis time intervals are suitable: 400–900 ns for Htt17_nmr (Fig. S1); 500–900 ns for Htt17Q17 (Fig. S2); and 500–900 ns for Htt17Q17P11 (Fig. S3). Thus, all analyses presented are performed using these intervals.
Results
Htt17
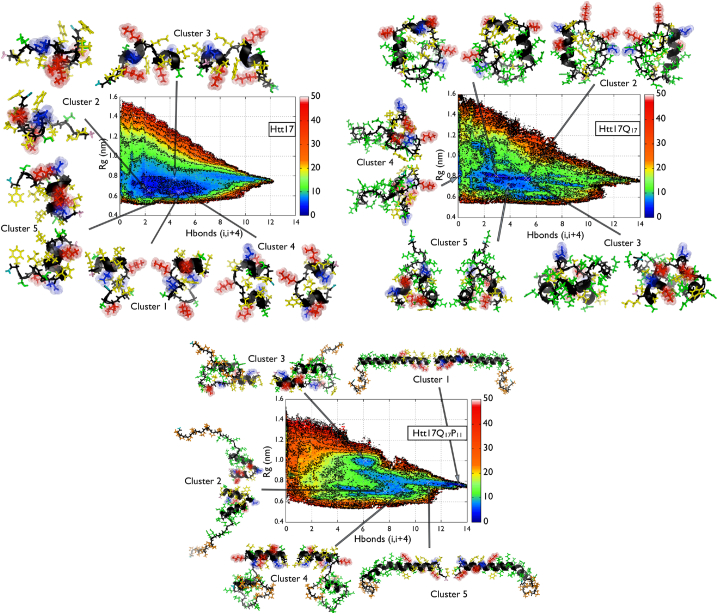
The two-dimensional free-energy surface (FES) of the Htt17 sequence in terms of the two biased CVs (Sα, number of helical H-bonds; Sgyr, gyration radius) for the Htt17_nmr simulation is shown in Fig. 1. It is characterized by a single large basin with the configurations below the 5 kJ/mol isoline being bound by two to six helical H-bonds and a gyration radius between 0.6 and 0.8 nm. In this region, the free-energy average is 4.8 kJ/mol and gradually rises to an average of 9.1 kJ/mol as the number of helical H-bonds increases. These latter conformations are less collapsed than those below the 5 kJ/mol isoline, as indicated by their larger gyration radius.
Figure 1.
The free-energy landscapes of Htt17 (Htt17_nmr) (top left), Htt17Q17 (top right) and Htt17Q17P11 (bottom). The horizontal and vertical axes represent the number of helical H-bonds (Sα) and the gyration radius (Sgyr), respectively. The number of helical H-bonds is computed on the first 13 residues for Htt17 and the first 17 residues for Htt17Q17 and Htt17Q17P11 to include possible H-bond formation with the Q17 domain. The energy isolines are drawn every 5 kJ/mol. The uncertainty on the free-energy landscapes of Htt17, Htt17Q17, and Htt17Q17P11 are shown in Figs. S1, S2, and S3, respectively. The uncertainty is always <1 kJ/mol on the relevant parts of the landscapes. Also shown is the cluster analysis of the most representative conformations populating the FES below 5 kJ/mol and below 8 kJ/mol for Htt17Q17P11. The negatively charged, positively charged, nonpolar, and polar residues of Htt17 are shown in blue, red, yellow, and green. The Q17 and the P11 segments are colored in green and orange, respectively. The backbone is colored in black, the amino-terminus in pink, and the carboxy-terminus in teal. To see this figure in color, go online.
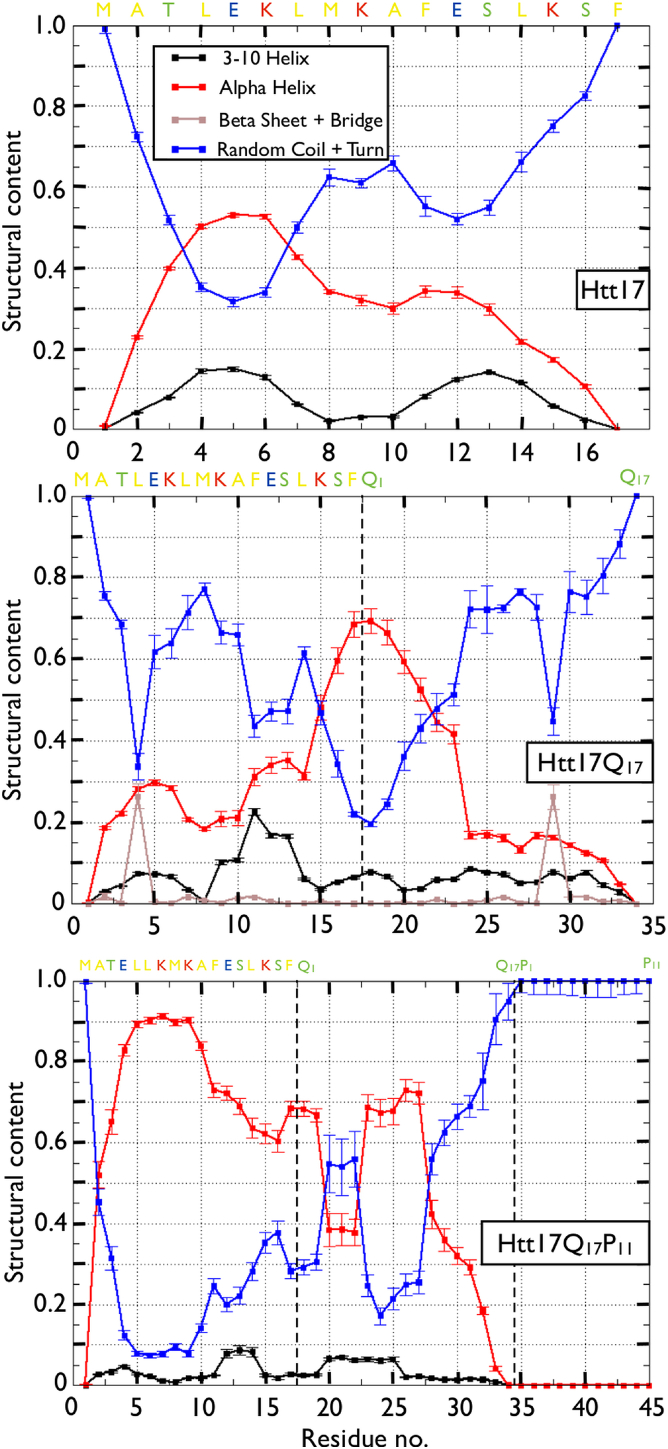
The per-residue secondary structure of Htt17 is shown in Fig. 2. We find that the first half of the peptide forms an α-helix (residues 3–7, ∼40–55%) more often than the second half (<35%). We also observe that residues 10–13 are very likely to form a turn, indicating a population of two-helix-bundle conformations. Overall, the peptide is mostly unstructured, with only 29.3 ± 0.7% α-helix probability, in agreement with measurements from several CD experiments (15, 35, 44, 45), and a negligible amount of β-sheet and β-bridge totalizing 1.2 ± 0.2%.
Figure 2.
From top to bottom are the per-residue secondary structures of Htt17 (Htt17_nmr), Htt17Q17, and Htt17Q17P11. The probability of α-helix, 3-10 helix, β-bridge and β-strand, and turn/coil are shown in red, black, brown, and blue, respectively. The vertical black dotted lines for Htt17Q17 and Htt17Q17P11 indicate the end of the Htt17 segment (center) and the ends of the Htt17 and Q17 segments (bottom). To see this figure in color, go online.
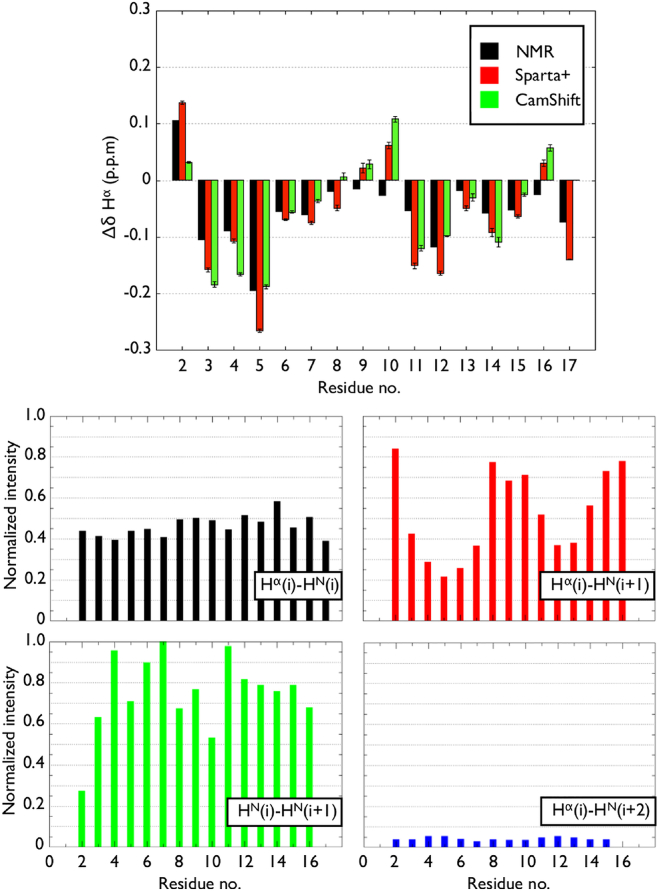
We also evaluate the presence of structural elements using the secondary Hα chemical shift and nuclear Overhauser effect (NOE) signal sampled in our simulations (Fig. 3) to compare with the solution NMR experiment of Wetzel and co-workers (35). The secondary chemical shifts are in very good agreement with the NMR measurements; except for residues Lys9, Ala10, and Ser16, which are more extended in our simulations. For the most part, the secondary chemical shifts are small and negative, indicating weak helical features. The high HN(i)-HN(i + 1) and low Hα(i)-HN(i + 2) NOE signals combined with small Hα(i)-HN(i + 3) NOE intensities indicate a global α-helix average of ∼30%, without any individual residue showing >55% probability, which slightly overestimates the α-helical propensity but remains compatible with the NMR data (see the Supporting Material for an in-depth discussion).
Figure 3.
(Top) The Hα secondary chemical shift per residue computed with SPARTA+ (red) and CamShift (green) are compared to the NMR measurements (black) on Htt17 in aqueous solution (35). The secondary shifts are obtained by subtracting the corrected coil value specific to each amino acid type from its Hα chemical shift (98). (Bottom) The computed intensities of the interproton NOEs for all residues between the Hα of residue i and the HN of residues i, i + 1, and i + 2, as well as between the HN of residues i and i + 1, are compared to the equivalent NMR measurements on Htt17 in aqueous solution (35). To see this figure in color, go online.
The main configurations sampled by Htt17 in basins below the 5 kJ/mol isoline are depicted by their cluster’s center in Fig. 1. In line with our previous analysis, the first residues of the Htt17 peptide have a greater tendency to be helical, whereas the last residues are mostly unstructured, and the central part of Htt17 (residues 10–13) forms a turn bringing the amino- and the carboxy-termini close to each other (see clusters 1, 3, and 4). The nonpolar residues are mainly accessible to the solvent This observation is confirmed by the analysis of the two-dimensional FES as a function of the SASA of Htt17’s nonpolar residues and the number of helical H-bonds (Fig. S6).
The map of contacts between residues is indicative of the mostly flexible and disordered tertiary structure of Htt17, as most contacts are between neighboring residues (Fig. S7). We observe, nevertheless, the presence of three electrostatic contacts: Glu5–Lys9, Glu12–Lys9, and Glu12–Lys15, with a probability of 50.3%, 42.3%, and 64.0%, respectively. The formation of a stable salt bridge occurs less often, however, with a probability of 4.2 ± 0.1%, 11.0 ± 1.0% and 12.3 ± 0.3%, respectively. We also note a long range nonpolar contact between Met8 and Phe17 in 24.1% of the sampled conformations that seems to be involved in the formation of the turn between residues 10 and 13, as well as in the destabilization of the secondary structure in the second half of the peptide (see clusters 1 and 4 of Fig. 1).
Htt17Q17
We now investigate the changes induced on the FES of Htt17 due to the addition of the Q17 segment at its carboxy-terminus. The two-dimensional FES of Htt17Q17 in terms of Sα and Sgyr—only on the Htt17 region—is shown in Fig. 1. In the largest basin, we observe three distinct minima characterized by a similar gyration radius around 0.8 nm, but with a different number of helical H-bonds (either 0.0 or between 2.0 and 6.0). Addition of the Q17 region modifies key features of the FES of Htt17 as it becomes more extended (more configurations having a larger gyration radius) and less structured (fewer configurations having a large number of helical H-bonds), as shown in Fig. 1. More precisely, the free energy of the conformations with a large number of helical H-bonds significantly increases from an average of 9.1 kJ/mol for Htt17 alone to 14.6 kJ/mol for Htt17Q17.
In terms of secondary structure, we observe a significant loss of helical propensity for residues 2–10, whereas that of the remaining residues in Htt17 greatly increases upon addition of the Q17 segment (Fig. 2). Even though the Htt17 segment in Htt17Q17 has an overall α-helix probability that is unchanged with respect to Htt17 alone (30.4 ± 1.4% vs. 29.3 ± 0.7%, respectively), the per-residue probability is very different: there is a significant amino- to carboxy-terminus shift of the helical probability that is directly due to the presence of the Q17 as the α-helix in Htt17 continues up to the first six glutamines. The remaining part of the Q17 is, however, mostly disordered. We also note the presence of a turn between Glu5 and Leu7 (∼40–45%). The amount of β-structure is still negligible (except for Leu4).
A cluster analysis of the structures characterized by a free energy <kJ/mol deepens the atomistic insights (Fig. 1). The first five clusters can be classified in three main categories: 1) no helical H-bond in Htt17 and a fully disordered Q17 (cluster 2); 2) two small α-helix fragments at both ends of Htt17 and a disordered Q17 (clusters 4 and 5); and 3) an α-helix spanning the last residues of Htt17 and the first glutamines of Q17 (clusters 1 and 3). In these clusters, the Htt17 segment adopts a u-shaped topology with a turn around Leu7 bringing its nonpolar residues close together, as shown by Htt17Q17’s contact map (Fig. S7). More precisely, we identify long-range nonpolar contacts between Leu4 and Phe17 (20.8%), Met8 and Phe17 (20.4%), and Phe11 and Phe17 (29.1%) that form a nonpolar cluster, which is further isolated from the solvent by the aliphatic carbons of the glutamines (for example, see clusters 1 and 3 of Fig. 1). As in Htt17 alone, we observe electrostatic contacts/salt-bridges between Glu5 and Lys9 (44.5%/6.9 ± 0.2%), Glu12 and Lys9 (51.1%/23 ± 2%), and Glu12 and Lys15 (48.9%/12.9 ± 0.2%). The charged residues Glu5, Lys6, Lys9, Glu12, and Lys15 of the Htt17 domain also interact with the Q17 domain with a probability of 54.6%, 50.8%, 48.2%, 40.7%, 83.9%, respectively. For its part, the glutamines aliphatic carbons of the Q17 segment interact a lot with the nonpolar residues of the Htt17 segment. As a consequence, the resulting nonpolar SASA decreases in contrast to Htt17 alone (Fig. S6). We also note the presence of several main-chain/side-chain and main-chain/main-chain interactions between Htt17 and Q17 (∼30%, Fig. S7).
The FES of the Q17 region shows that it is mostly disordered and collapsed (Fig. S8). A cluster analysis of the configurations below 4 kJ/mol shows that the first glutamines are α-helical, whereas the remaining region is disordered independently of Htt17’s structure, which is an α-helix either from residues Ser13 to Phe17 (clusters 1, 4, and 5) or from residues Ala2 to Met8 (clusters 2). At high helical H-bonds, we note the presence of a very narrow minimum characterized by an almost fully α-helical Q17, with Htt17 adopting the same structure up to residue Glu12.
Htt17Q17P11
We finally probe the effects of the addition of the Q17 and P11 segments on the FES of Htt17. The two-dimensional FES of Htt17Q17P11 in terms of Sα and Sgyr—only on the Htt17 region—is shown in Fig. 1. The resulting FES unveils a striking shift toward the α-helix, as the FES is characterized by a single minimum with a number of helical H-bonds between 12 and 14 and a gyration radius between 0.75 and 0.85 nm. Most of the structures sampled by Htt17 alone or Htt17Q17 are therefore less stable upon the addition of P11.
Analysis of the secondary structure indicates that the Htt17 region adopts an α-helical conformation 70.9 ± 1.6% of the time, a drastic increase compared to both Htt17 (29.3 ± 0.7%) and Htt17Q17 (30.4 ± 1.4%) (Fig. 2). The probability is notably high for residues 5–9 at >90%. The Q17 domain has 44.8 ± 2.5% of α-helical content, with the residues near Htt17 (glutamines 1, 2, and 6–10) having the largest probability, whereas glutamines 3 to 5 and those near P11 mostly form turn/coil structures. Finally, the P11 domain almost exclusively forms a PPII-helix characterized by Φ and Ψ dihedral angles near −75° and 150°, respectively, according to an analysis of the Ramachandran plot for every proline (data not shown).
The addition of P11 causes drastic changes in structure, as it doubles the helical content of both Htt17 and the Q17 domain. We further quantify its effect from a cluster analysis on the structures found below 8 kJ/mol. In contrast to the important structural diversity of Htt17 and Htt17Q17 in solution, Htt17 within Htt17Q17P11 has a strong tendency to form an α-helix, as the first cluster is composed of >25% of the sampled structures. The depicted conformation for the first cluster shows the Q17 as a fully formed α-helix and the P11 region extends away from Htt17 and Q17. We note a clear separation between the polar and nonpolar residues of Htt17. The former are interacting mostly with Q17, as shown on the Htt17Q17P11 contact map (Fig. S7). More specifically, there are contacts between Glu5, Lys6, Lys9, Glu12, and Lys15 of the Htt17 domain and the glutamines with a probability of 25.0%, 21.0%, 61.1%, 57.6%, and 85.7%, respectively. Salt bridges are also present between Lys9 and Glu12 (45 ± 4%) and Lys15 and Glu12 (14 ± 2%) stabilizing the Htt17 conformation. For its part, the P11 domain interacts mostly with residues surrounding Lys9 (33.5%), Ser13 (50.0%), Leu14 (24.0%), and Phe17 (48.8%), mostly via their side chains. As for the nonpolar residues in Htt17, they are all located on the same side of the peptide and fully accessible to the solvent, as shown by a drastic increase of the nonpolar SASA when compared to the Htt17 and Htt17Q17 peptides (Fig. S6).
The FES of Q17 unveils three minima with distinct numbers of H-bonds (0, 2.5, and 7.5), as shown in Fig. S8. A cluster analysis of the structures found inside those regions (below 4 kJ/mol) shows that the most important cluster of the Q17 domain has an important helical propensity up to the 10th glutamine and that the remaining glutamines are mostly unstructured. The other clusters depict the Q17 domain as fully unstructured independent of Htt17’s structure, which is either a fully formed α-helix (clusters 2, 4, and 5) or mostly unstructured (cluster 3).
Discussion
Numerous experiments indicate that the Huntingtin amino-terminus is crucial for its biological functions. More specifically, the first 17-amino-acid segment (Htt17), which is right before the amyloidogenic polyglutamine segment (QN), is directly involved in the membrane interactions and aggregation of Huntingtin. In this study, we quantify the conformational ensemble of three fragments of Huntingtin amino-terminus—Htt17, Htt17Q17, and Htt17Q17P11—in terms of FES, secondary structure, contact maps, and clusters. Our results demonstrate the effects of Q17 and P11 on the conformational ensemble of Htt17, and taken together with those of other studies, they provide insights on motifs at the origin of Htt17’s membrane binding and oligomerization.
Htt17 samples a wide variety of coil/helix structures
Experiments using CD indicate that Htt17 has a helical population of ∼10–55% in aqueous solution (15, 35, 44, 45), but solution NMR measurements show no stable structural motif (35). Recently, results from ion-mobility-spectrometry-mass-spectrometry coupled to molecular dynamics simulations suggest that Htt17 populates two kinds of helical monomer with an α-helix from the amino-terminus to residues Lys9–Ala10: 1) a compact structure characterized by an unstructured region between residues Phe11 and Phe17 that turns back on itself and brings the amino- and carboxy-termini closer to each other; and 2) an extended structure characterized by a 3-10 helix spanning residues Ala10 to Glu12 and where the carboxy-terminus region is extended away from Htt17 (93). Taken together, these observations suggest that the structural ensemble of Htt17 consists of a wide variety of flexible helix/coil conformations.
Previous simulations of Htt17 suggest such a conformational ensemble (44, 47, 48). More precisely, simulated tempering simulations with the AMBER03 force field and explicit-solvent (TIP3P) show that the conformational ensemble of Htt17 contains ∼70% of diverse two-helix bundles with a loop around Ala10, whereas the rest of the ensemble populates a single straight helix or disordered configurations (47). In this work, residues 3–6 have the highest α-helix propensity and the sampled configurations are mostly stabilized either by charged interactions or sequestration of the nonpolar residues. Other simulations performed using Monte Carlo with the ABSINTH implicit-solvent force field show that Htt17 has an α-helix probability of 34% and that it is mostly collapsed upon itself to sequester its nonpolar residues (44). Bias-exchange MetaD, for its part, unveiled the free-energy landscape of Htt17 using explicit-solvent, all-atom simulations (AMBER99/TIP3P) (48). In this work, Carloni et al. observe that the free-energy landscape using six CVs is mainly made of four basins and that the transitions from one basin to the others occur on the microsecond timescale. The resulting conformational ensemble of Htt17 is largely disordered (75%) and helical (25%) with a global α-helix probability of 29%, notably for residues 1–7. They also note that the disordered configurations of the largest basin have their nonpolar residues largely accessible to the solvent.
This is in line with the trend depicted by our simulations in terms of the free-energy landscape (Fig. 1) and the secondary-structure propensity (Fig. 2). Our predicted secondary structure is characterized by a global α-helix probability of 29.3 ± 0.7%, which is similar to the values obtained in the aforementioned simulations—43% (47), 34% (44), and 29% (48)—and CD experiments—10% (45), 34% (44), 45% (15), and 55% (35)—on Htt17 in aqueous solution. Our results also indicate that residues 3–5 have the highest helix propensity (Fig. 2, ∼40–50%), in agreement with other simulation protocols (47, 48). Finally, we find that Htt17 forms various two-helix, single helix, helix/coil, and coil conformations, as previously observed (47, 48, 93). The probability of structured conformations is, however, lower in our simulations than in (47), which might be due to AMBER03 slightly overstabilizing helical structures in helix/coil peptides when compared to the AMBER99sb∗-ILDN force field, as indicated by other studies (66, 68).
In terms of tertiary structure, our simulations indicate that Htt17’s nonpolar residues are mostly accessible to the solvent (Fig. S6), in agreement with previous bias-exchange MetaD simulations (48). In addition, mainly short-range contacts between neighboring residues are populated in Htt17. Still, a nonpolar contact between Met8 and Phe17 occurs in 24.1% of the sampled structures. It could be crucial in the formation of the turn between residues Ala10 and Ser13, therefore leading to the destabilization of the second half of Htt17.
Finally, we provide a detailed analysis indicating that the structural ensemble sampled in our simulations is consistent with the only solution NMR experiment done on Htt17 in an aqueous environment (35) in terms of secondary Hα chemical shifts and interproton NOE distances (Fig. 3 and Supporting Material). We are thus confident that our results yield relevant insights into the structural ensemble of Htt17.
Addition of Q17 reduces solvent accessibility for Htt17’s nonpolar residues
Fluorescence-based resonance energy transfer experiments indicate that Htt17 is in a collapsed state and that it becomes more extended upon addition of the polyglutamine segment (35). CD spectra suggest an increase of helix propensity with the polyglutamine length, but it is unknown whether this is due to the QN or Htt17 segments (39). Data from x-ray crystallography on a chimeric protein containing Huntingtin exon 1 supplement this by indicating that an α-helix in Htt17 can extend to the QN region (46). In both studies, absence of β-sheet is observed. For their part, Monte Carlo simulations using the implicit-solvent ABSINTH potential show that addition of the QN domain disorders Htt17 in a length-dependent manner, whereas the QN segment itself remains disordered (44). Pappu et al. also find that the nonpolar residues of Htt17 lie in the interdomain interface between Htt17 and QN.
Our results complement these experiments by showing that the QN region in Htt17Q17 is mostly disordered, but that it can sample α-helices with a probability of 27.8 ± 1.3%, particularly for the first glutamines (Fig. 2). The QN region also induces an amino-to-carboxy-terminus shift of the helical probability in the Htt17 region but leaves unchanged its global α-helical probability from 29.3 ± 0.7% to 30.4 ± 1.4%. In line with experimental results, we find a negligible amount of β structure for both the Htt17 and QN regions at the monomer level. We also find that the aliphatic carbons of the QN domain interact directly with the nonpolar residues of Htt17, dramatically reducing their solvent accessibility (Figs. S6 and S7), in agreement with previous simulations (44).
Overall, we observe that the QN region modifies the conformational ensemble of Htt17 already at the monomer level, which could have a direct impact on its aggregation and membrane-binding affinities, as discussed next. This indicates that not only does Htt17 influence QN, as previously determined experimentally, but that the opposite also occurs and that the interplay between Htt17 and QN might be more complex than previously thought. As demonstrated in the Supporting Material, these results are replicated qualitatively using a different electrostatic scheme as well as a newly proposed set of proline parameters; they are not, therefore, an artifact of the simulation conditions.
Htt17 is more structured upon addition of Q17P11
CD measurements show that the addition of a P10 domain to Htt17Q37 reduces the α-helix probability from >50% to ∼30% (39). However, CD is unable to tell the localization of these structural changes. Using a high-performance liquid chromatography sedimentation assay, Wetzel et al. also reported that the aggregation of Htt17Q35 is quicker than that for Htt17Q37P10, although the latter is still much faster than a QN domain of similar length alone. A ThT fluorescence kinetic profile monitoring the growth of the fibril showed mostly no difference between Htt17Q30 and Htt17Q30C38, where C38 is the full-length proline-rich region starting with P11, indicating that the aggregation mechanism is dominated by Htt17 (44). It is also found that C38 acts as a solubilizing module that weakens the driving force toward the formation of insoluble aggregates. X-ray crystallography on a chimeric protein containing Huntingtin exon 1 suggests that the Htt17 can populate α-helix configurations, whereas the QN region is mostly unstructured except for the first glutamines, which can populate an α-helix (46). The first prolines in the proline-rich region, for their part, are characterized by a PPII-helix.
Only one set of simulations has been performed on Htt17QNP11 to our knowledge (50). These all-atom replica-exchange molecular dynamics simulations with the FF03 force field and implicit solvent suggested that both Htt17 and QN adopt mostly α-helical conformations, whereas the P11 forms a PPII-helix. In these simulations, the α-helical content is especially large between residues 4 and 17 of Htt17, and the P11 region lies antiparallel to the Htt17 region when there are 17 glutamines in QN, but not when there are 55 glutamines (above the pathological threshold of 36 repeats).
In our simulation, the P11 domain stabilizes Htt17 as an almost fully formed α-helix with >70% helical content. The Q17 domain adopts an α-helix conformation 44.8 ± 2.5% of the time. Our results differ from the secondary-structure signal from CD (39) and are surprising overall. Indeed, the increase of the nonpolar SASA and the decreased number of contacts between Htt17 and Q17 are two strong destabilizing factors present in our simulations. The difference with experiment is perhaps due to the length of the QN domain used; longer QN (as in the CD experiment) might mitigate the stabilizing effects of P11. Another simulation protocol shows a similar α-helical population in Htt17 in the context of Htt17Q17P11, but significantly more α-helix in Q17 (50). Ultimately, other simulation and experimental protocol will be needed to unveil the origin of this dissimilarity.
Motifs relevant to membrane binding and oligomerization
Htt17 is crucial to the localization of Huntingtin in the cell (8, 9, 10, 11, 12, 13, 14, 15, 16) and adopts an α-helical conformation in the presence of micelles, vesicles, and phospholipid membranes, as shown by CD spectroscopy (15, 45), solution NMR (45), solid-state NMR (51) and HREX simulations (52). Solid-state NMR and HREX simulations also indicate that the amphipathic plane of Htt17 is aligned parallel to the membrane surface with its nonpolar residues facing the hydrophobic core of the membrane. The presence of α-helical structures in Htt17 before membrane binding has been shown to ease its insertion into the membrane (53).
Htt17 also drastically modifies the aggregation of the amyloidogenic QN segment. Three main models have been proposed to describe Htt17’s role in the aggregation mechanism of Huntingtin: 1) the formation of tetrameric α-helical bundles of Htt17, which increases the local concentration of QN, favoring the nucleation of β-sheeted structures in the latter region (39); 2) the reduction in the entanglement of QN destabilizing the nonfibrillar aggregates (41); and 3) the direct interaction between Htt17 and QN favoring formation of extended structures in the latter region (32). Solid-state NMR indicates, for its part, that the core of a Htt17Q30P10K2 amyloid fibril is formed by QN, whereas Htt17 and P10 form an α-helix and a PPII-helix, respectively (94, 95).
The first aggregation model, more specifically, is based on sedimentation velocity experiments that indicate that Htt17 and Htt17Q10K2 are mostly monomeric in solution with a low level of compact oligomers that correspond to, in decreasing order of population, tetramer, octomer, dodecamer, and so on (39). The aggregation-enhancing property of Htt17 with respect to QN alone can be then explained by the formation of reversible α-helical tetrameric bundles via Htt17. Namely, these tetramers assemble into higher-order oligomers that increase the local concentration of the amyloidogenic QN segment, easing the nucleation of the β-sheeted structures necessary to the formation of amyloid fibrils.
These previous investigations indicate common motifs in Htt17—the formation of helical structures and the sequestration of its nonpolar residues—that are fundamental to both its aggregation and membrane-binding enhancing properties. We now discuss how our observations on the Htt17, Htt17Q17, and Htt17Q17P11 monomers are related to these models.
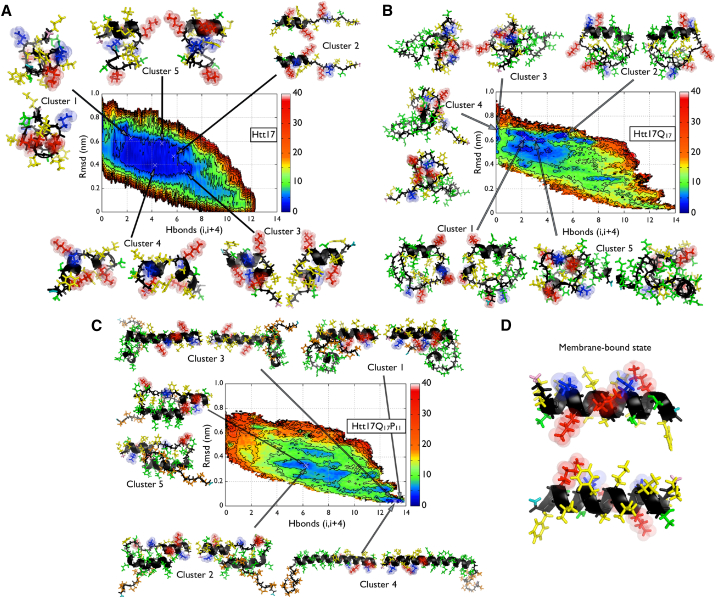
We observe in our simulations the presence of α-helical conformations (29.3 ± 0.7%) in Htt17. We also quantify, more specifically, the presence of highly α-helical structures in Htt17 by comparing the sampled ensemble in aqueous solution to its membrane-bound state. We compute the RMSD with respect to the α-helical structure sampled in a POPC bilayer (52) and reconstruct the two-dimensional FES of Htt17 in terms of this RMSD and the number of helical H-bonds (Fig. 4). We observe a broad basin between 1.0 and 7.5 helical H-bonds and 0.3 and 0.7 nm RMSD, corresponding to structures that are different from their membrane counterpart. The free energy gradually rises as the structural similarity to the membrane state increases, indicating that a highly α-helical structure similar to the membrane-bound state is not stable in aqueous solution.
Figure 4.
Comparison of the conformational ensemble of Htt17 to the membrane-bound state (52). (A–C) The FES of the Htt17 segment as a function of the backbone RMSD measured between Htt17 in solution and the membrane-bound state (vertical axis) and the number of helical H-bonds (Sα, horizontal axis) are shown along with the representative principal conformation clusters. The membrane-bound state is depicted in (D). The negatively charged, positively charged, nonpolar, and polar residues of Htt17 are shown in blue, red, yellow, and green. The Q17 and P11 segments are colored in green and orange, respectively. The backbone is colored in black, the amino-terminus in pink, and the carboxy-terminus in teal. Energy isolines are drawn every 4 kJ/mol. To see this figure in color, go online.
We note, nonetheless, that some configurations in the basin below the 4 kJ/mol isoline possess a motif that could initiate the membrane binding and the formation of the tetrameric α-helical bundle: the first residues of Htt17, particularly between residues Thr3 and Lys6, can form an α-helix (∼50%; Fig. 2). The presence of such a motif has also been observed in other simulations (48). Moreover, our results show that the nonpolar residues of Htt17, particularly Met8, Phe11, and Phe17, are mostly accessible to the solvent (Fig. S6), which could promote the association of Htt17 with other Htt17 segments and its anchoring to a phospholipid membrane.
Upon addition of Q17, the population of a highly helical Htt17 segment is significantly reduced (Fig. 2), moving away from the membrane-bound state (Fig. 4). This is explained by the α-helix motif between residues 3 and 7 being less stable than for Htt17 alone, even though a new α-helical motif is formed starting at residue 15 in Htt17 and extending to the first glutamines of Q17. This shift of the position of the α-helical motif could result in a shift of Htt17QN’s primary interaction site with phospholipid membranes and its nucleation site for the tetrameric bundle formation during oligomerization.
We also observe that the nonpolar residues of Htt17 are globally less exposed due to interactions with the aliphatic carbons of the glutamines (Figs. S6 and S7). Together with the α-helical shift, this could affect the aggregation and membrane-binding pathways, as the QN region needs to move away from the Htt17 region to free the nonpolar residues for these events to proceed. This might be one of the rate-limiting steps for the tetrameric bundle formation, as intrapeptide Htt17-QN interactions need to be dominated by interpeptide interactions between Htt17 and QN or QN and QN during dimerization. Previous simulations indeed indicate that QN-QN interactions dominate in the dimer (44).
Our results also suggest that the stability of Htt17 in a fully formed α-helix state drastically increases with the addition of the Q17P11 domain (Figs. 1 and 2). This results in an important population of membrane-bound-like states characterized by an RMSD <0.1 nm (Fig. 4) and a high solvent accessibility for the nonpolar residues of Htt17 (Fig. S6). Both the α-helical character and the nonpolar residue accessibility of Htt17 due to the combined addition of QN and P11 could promote membrane binding, as observed experimentally (54) and numerically (96), as well as the formation of an Htt17 tetrameric bundle.
A priori, this observation from our simulation on the P11 role seems to contradict previous experiments. In some studies, the addition of a P10 domain decreases the rate of formation and the stability of amyloid-like aggregates, leaving the nucleation mechanism unchanged compared to the QN domain alone (37). P10 would therefore stabilize conformations incompatible with aggregation. Other studies show, however, that the proline-rich segment C38—starting with P11 and located at the carboxy-terminus of QN—increases the overall solubility of Htt17QNC38, weakening the driving force toward the formation of insoluble aggregates but preserving a rate of fibril formation similar to that of Htt17QN (41). Consequently, the formation of structural features that could favor the aggregation of Htt17 in our simulations might not be sufficient to enhance the overall oligomerization. The slowing effect of P11 could then occur later in the aggregation process or be caused by another phenomenon, such as induced structural changes in the QN domain. In that regard, our simulations on the monomer indicate that the structural flexibility of QN is reduced by the incorporation of P11 (Fig. S8).
Other models have been proposed for huntingtin aggregation. In the first one, both Htt17 and C38—the proline-rich segment starting with P11 and located at the carboxy-terminus of QN—modulate the aggregation of QN by controlling the intrinsic structural heterogeneity of this amyloidogenic segment (41). Such a model is corroborated by dynamic light scattering experiments done on a polyglutamine domain fused to a heterotetrameric coiled-coil system that show that even a disordered N-terminal can facilite the structuring of the polyQ domain (97). Fibrillation is promoted by Htt17 destabilizing the intermediate nonfibrillar structures and P11 destabilizing the intermediate insoluble aggregates. The role for Htt17 was unveiled using Monte Carlo (44) and mesoscopic (43) simulations that investigated the dimerization and large-scale aggregation, respectively, of Htt17QN. The Htt17 segment would then reduce the entanglement within the QN segment and introduces a barrier to intermolecular associations that brings the formation of small spherical structures (soluble oligomers) and large linear aggregates (insoluble fibrils) on a similar timescale. In the second model, the amyloidogenic QN segment would interact directly with Htt17 to promote fibrillation through the formation of extended motifs in QN (32).
The relations between our simulations and these models remain, nevertheless, more limited, as we have focused our investigation on the Htt17 segment and do not have a simulation on the QN segment alone. We observe, nonetheless, that Q17 in the presence of Htt17 adopts a variety of structures that are mostly disordered (Fig. S8). It also interacts directly with Htt17 (Fig. S7), as previously suggested experimentally (32). Addition of P11 leads to a more compact Q17 region that interacts much more with itself, and this could reduce entanglement during oligomerization.
Conclusions
We studied three fragments of the amino-terminus of Huntingtin—Htt17, Htt17Q17, and Htt17Q17P11—with special consideration of the first 17-amino-acid segment (Htt17) that is crucial for its oligomerization and membrane binding. We applied, to our knowledge, a novel combination of two sampling enhancing techniques—HREX and well-tempered MetaD (HREXMetaD)—to have a thorough understanding of the modifications on Htt17’s structural ensemble due to the addition of the amyloidogenic QN segment and the polyproline segment (P11).
We find that the structural ensemble of Htt17 is characterized by a wide variety of helix/coil conformations. The addition of the Q17 domain results in an amino-to-carboxy-terminus shift of the helical content and it decreases the solvent accessibility of Htt17’s nonpolar residues by interacting directly with it. The addition of both Q17 and P11 drastically changes the structural ensemble of Htt17 toward more structured conformations with more exposed nonpolar surfaces.
Careful comparison with experimental aggregation and membrane-binding models reveals that Htt17 possesses crucial features essential to these processes whether alone or combined with Q17 or Q17P11. We find that the position and the type of motifs are very different depending on the adjacent sequences to Htt17, showing that all these neighboring regions strongly impact each other already at the monomer level.
Our results also provide a strong basis for further study of more complex situations, such as Htt17QNP11 oligomerization and membrane binding using a similar simulation protocol (HREXMetaD). We find that this, to our knowledge, novel protocol offers good sampling at a moderate computational cost and scales very well with the number of particles, as it is essentially size independent.
Author Contributions
Designed research, V.B., S.C., and N.M.; Performed research, V.B. and S.C.; Contributed analytic tools, V.B. and S.C.; Analyzed data, V.B., S.C. and N.M. Wrote the article, V.B., S.C., and N.M.
Acknowledgments
The authors thank the PLUMED community—particularly Giovanni Bussi, Pratyush Tiwary, and Ludovico Sutto—for helpful discussions and advices. They also thank Ronald Wetzel and In-Ja Byeon for kindly providing their NMR data and for their insight on our results.
Computations were made on the supercomputers Briarée from Université de Montréal and Mammouth from Université de Sherbrooke, managed by Calcul Québec and Compute Canada. The operation of these supercomputers is funded by the Canada Foundation for Innovation, Nano Québec, Réseau de Médecine Génétique Appliquée, and the Fonds de Recherche Québécois sur la Nature et les Technologies. This work was funded by the Canada Research Chairs program, the Natural Sciences and Engineering Research Council of Canada, the Fonds de Recherche Québécois sur la Nature et les Technologies, and the Fonds de Recherche en Santé du Québec.
Editor: Michael Feig.
Footnotes
Vincent Binette and Sébastien Côté contributed equally to this work.
Supporting Materials and Methods and thirteen figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)00090-4.
Supporting Material
References
- 1.Sharp A.H., Loev S.J., Ross C.A. Widespread expression of Huntington’s disease gene (IT15) protein product. Neuron. 1995;14:1065–1074. doi: 10.1016/0896-6273(95)90345-3. [DOI] [PubMed] [Google Scholar]
- 2.DiFiglia M., Sapp E., Aronin N. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14:1075–1081. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- 3.Duyao M.P., Auerbach A.B., Ge P. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- 4.Andrade M.A., Bork P. HEAT repeats in the Huntington’s disease protein. Nat. Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- 5.MacDonald M.E. Huntingtin: alive and well and working in middle management. Sci. STKE. 2003;2003:pe48. doi: 10.1126/stke.2003.207.pe48. [DOI] [PubMed] [Google Scholar]
- 6.Caviston J.P., Holzbaur E.L.F. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009;19:147–155. doi: 10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng Z., Diamond M.I. Huntington disease and the huntingtin protein. Prog. Mol. Biol. Transl. Sci. 2012;107:189–214. doi: 10.1016/B978-0-12-385883-2.00010-2. [DOI] [PubMed] [Google Scholar]
- 8.Zheng Z., Li A., Diamond M.I. An N-terminal nuclear export signal regulates trafficking and aggregation of Huntingtin (Htt) protein exon 1. J. Biol. Chem. 2013;288:6063–6071. doi: 10.1074/jbc.M112.413575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maiuri T., Woloshansky T., Truant R. The huntingtin N17 domain is a multifunctional CRM1 and Ran-dependent nuclear and cilial export signal. Hum. Mol. Genet. 2013;22:1383–1394. doi: 10.1093/hmg/dds554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steffan J.S., Agrawal N., Marsh J.L. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 11.Gu X., Greiner E.R., Yang X.W. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aiken C.T., Steffan J.S., Marsh J.L. Phosphorylation of threonine 3: implications for Huntingtin aggregation and neurotoxicity. J. Biol. Chem. 2009;284:29427–29436. doi: 10.1074/jbc.M109.013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson L.M., Aiken C.T., Steffan J.S. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J. Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atwal R.S., Desmond C.R., Truant R. Kinase inhibitors modulate huntingtin cell localization and toxicity. Nat. Chem. Biol. 2011;7:453–460. doi: 10.1038/nchembio.582. [DOI] [PubMed] [Google Scholar]
- 15.Atwal R.S., Xia J., Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 16.Rockabrand E., Slepko N., Thompson L.M. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum. Mol. Genet. 2007;16:61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- 17.Bates G.P. History of genetic disease: the molecular genetics of Huntington disease—a history. Nat. Rev. Genet. 2005;6:766–773. doi: 10.1038/nrg1686. [DOI] [PubMed] [Google Scholar]
- 18.Zoghbi H.Y., Orr H.T. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 19.Gatchel J.R., Zoghbi H.Y. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 20.Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 21.Benn C.L., Landles C., Bates G.P. Contribution of nuclear and extranuclear polyQ to neurological phenotypes in mouse models of Huntington’s disease. Hum. Mol. Genet. 2005;14:3065–3078. doi: 10.1093/hmg/ddi340. [DOI] [PubMed] [Google Scholar]
- 22.Ritch J.J., Valencia A., Kegel K.B. Multiple phenotypes in Huntington disease mouse neural stem cells. Mol. Cell. Neurosci. 2012;50:70–81. doi: 10.1016/j.mcn.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuccato C., Valenza M., Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol. Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 24.DiFiglia M., Sapp E., Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 25.Kim Y.J., Yi Y., DiFiglia M. Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington’s disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc. Natl. Acad. Sci. USA. 2001;98:12784–12789. doi: 10.1073/pnas.221451398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratovitski T., Nakamura M., Ross C.A. N-terminal proteolysis of full-length mutant huntingtin in an inducible PC12 cell model of Huntington’s disease. Cell Cycle. 2007;6:2970–2981. doi: 10.4161/cc.6.23.4992. [DOI] [PubMed] [Google Scholar]
- 27.Mangiarini L., Sathasivam K., Bates G.P. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 28.Davies S.W., Turmaine M., Bates G.P. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 29.Cisbani G., Cicchetti F. An in vitro perspective on the molecular mechanisms underlying mutant huntingtin protein toxicity. Cell Death Dis. 2012;3:e382. doi: 10.1038/cddis.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q.C., Yeh T.-L., Poirier M.A. A compact β model of huntingtin toxicity. J. Biol. Chem. 2011;286:8188–8196. doi: 10.1074/jbc.M110.192013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nucifora L.G., Burke K.A., Poirier M.A. Identification of novel potentially toxic oligomers formed in vitro from mammalian-derived expanded huntingtin exon-1 protein. J. Biol. Chem. 2012;287:16017–16028. doi: 10.1074/jbc.M111.252577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tam S., Spiess C., Frydman J. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat. Struct. Mol. Biol. 2009;16:1279–1285. doi: 10.1038/nsmb.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wetzel R. Physical chemistry of polyglutamine: intriguing tales of a monotonous sequence. J. Mol. Biol. 2012;421:466–490. doi: 10.1016/j.jmb.2012.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahoo B., Singer D., Wetzel R. Aggregation behavior of chemically synthesized, full-length huntingtin exon1. Biochemistry. 2014;53:3897–3907. doi: 10.1021/bi500300c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thakur A.K., Jayaraman M., Wetzel R. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat. Struct. Mol. Biol. 2009;16:380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liebman S.W., Meredith S.C. Protein folding: sticky N17 speeds huntingtin pile-up. Nat. Chem. Biol. 2010;6:7–8. doi: 10.1038/nchembio.279. [DOI] [PubMed] [Google Scholar]
- 37.Bhattacharyya A., Thakur A.K., Wetzel R. Oligoproline effects on polyglutamine conformation and aggregation. J. Mol. Biol. 2006;355:524–535. doi: 10.1016/j.jmb.2005.10.053. [DOI] [PubMed] [Google Scholar]
- 38.Darnell G., Orgel J.P.R.O., Meredith S.C. Flanking polyproline sequences inhibit β-sheet structure in polyglutamine segments by inducing PPII-like helix structure. J. Mol. Biol. 2007;374:688–704. doi: 10.1016/j.jmb.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 39.Jayaraman M., Kodali R., Wetzel R. Slow amyloid nucleation via α-helix-rich oligomeric intermediates in short polyglutamine-containing huntingtin fragments. J. Mol. Biol. 2012;415:881–899. doi: 10.1016/j.jmb.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jayaraman M., Mishra R., Wetzel R. Kinetically competing huntingtin aggregation pathways control amyloid polymorphism and properties. Biochemistry. 2012;51:2706–2716. doi: 10.1021/bi3000929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crick S.L., Ruff K.M., Pappu R.V. Unmasking the roles of N- and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. USA. 2013;110:20075–20080. doi: 10.1073/pnas.1320626110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Côté S., Wei G., Mousseau N. All-atom stability and oligomerization simulations of polyglutamine nanotubes with and without the 17-amino-acid N-terminal fragment of the Huntingtin protein. J. Phys. Chem. B. 2012;116:12168–12179. doi: 10.1021/jp306661c. [DOI] [PubMed] [Google Scholar]
- 43.Ruff K.M., Khan S.J., Pappu R.V. A coarse-grained model for polyglutamine aggregation modulated by amphipathic flanking sequences. Biophys. J. 2014;107:1226–1235. doi: 10.1016/j.bpj.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williamson T.E., Vitalis A., Pappu R.V. Modulation of polyglutamine conformations and dimer formation by the N-terminus of huntingtin. J. Mol. Biol. 2010;396:1295–1309. doi: 10.1016/j.jmb.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michalek M., Salnikov E.S., Bechinger B. Membrane interactions of the amphipathic amino terminus of huntingtin. Biochemistry. 2013;52:847–858. doi: 10.1021/bi301325q. [DOI] [PubMed] [Google Scholar]
- 46.Kim M.W., Chelliah Y., Bezprozvanny I. Secondary structure of Huntingtin amino-terminal region. Structure. 2009;17:1205–1212. doi: 10.1016/j.str.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelley N.W., Huang X., Pande V.S. The predicted structure of the headpiece of the Huntingtin protein and its implications on Huntingtin aggregation. J. Mol. Biol. 2009;388:919–927. doi: 10.1016/j.jmb.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossetti G., Cossio P., Carloni P. Conformations of the Huntingtin N-term in aqueous solution from atomistic simulations. FEBS Lett. 2011;585:3086–3089. doi: 10.1016/j.febslet.2011.08.036. [DOI] [PubMed] [Google Scholar]
- 49.Lakhani V.V., Ding F., Dokholyan N.V. Polyglutamine induced misfolding of huntingtin exon1 is modulated by the flanking sequences. PLOS Comput. Biol. 2010;6:e1000772. doi: 10.1371/journal.pcbi.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Długosz M., Trylska J. Secondary structures of native and pathogenic huntingtin N-terminal fragments. J. Phys. Chem. B. 2011;115:11597–11608. doi: 10.1021/jp206373g. [DOI] [PubMed] [Google Scholar]
- 51.Michalek M., Salnikov E.S., Bechinger B. Structure and topology of the huntingtin 1-17 membrane anchor by a combined solution and solid-state NMR approach. Biophys. J. 2013;105:699–710. doi: 10.1016/j.bpj.2013.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Côté S., Binette V., Mousseau N. Probing the Huntingtin 1-17 membrane anchor on a phospholipid bilayer by using all-atom simulations. Biophys. J. 2015;108:1187–1198. doi: 10.1016/j.bpj.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Côté S., Wei G., Mousseau N. Atomistic mechanisms of huntingtin N-terminal fragment insertion on a phospholipid bilayer revealed by molecular dynamics simulations. Proteins. 2014;82:1409–1427. doi: 10.1002/prot.24509. [DOI] [PubMed] [Google Scholar]
- 54.Burke K.A., Kauffman K.J., Legleiter J. The interaction of polyglutamine peptides with lipid membranes is regulated by flanking sequences associated with huntingtin. J. Biol. Chem. 2013;288:14993–15005. doi: 10.1074/jbc.M112.446237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laio A., Parrinello M. Escaping free-energy minima. Proc. Natl. Acad. Sci. USA. 2002;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barducci A., Bussi G., Parrinello M. Well-tempered metadynamics: a smoothly converging and tunable free-energy method. Phys. Rev. Lett. 2008;100:020603. doi: 10.1103/PhysRevLett.100.020603. [DOI] [PubMed] [Google Scholar]
- 57.Bonomi M., Barducci A., Parrinello M. Reconstructing the equilibrium Boltzmann distribution from well-tempered metadynamics. J. Comput. Chem. 2009;30:1615–1621. doi: 10.1002/jcc.21305. [DOI] [PubMed] [Google Scholar]
- 58.Crespo Y., Marinelli F., Laio A. Metadynamics convergence law in a multidimensional system. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2010;81:055701. doi: 10.1103/PhysRevE.81.055701. [DOI] [PubMed] [Google Scholar]
- 59.Bussi G. Hamiltonian replica exchange in GROMACS: a flexible implementation. Mol. Phys. 2014;112:379–384. [Google Scholar]
- 60.Pronk S., Páll S., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 62.Van Der Spoel D., Lindahl E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 63.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 64.Tribello G.A., Bonomi M., Bussi G. PLUMED 2: new feathers for an old bird. Comput. Phys. Commun. 2014;185:604–613. [Google Scholar]
- 65.Hornak V., Abel R., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Best R.B., Hummer G. Optimized molecular dynamics force fields applied to the helix-coil transition of polypeptides. J. Phys. Chem. B. 2009;113:9004–9015. doi: 10.1021/jp901540t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Piana S., Lindorff-Larsen K., Shaw D.E. How robust are protein folding simulations with respect to force field parameterization? Biophys. J. 2011;100:L47–L49. doi: 10.1016/j.bpj.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Piana S., Klepeis J.L., Shaw D.E. Assessing the accuracy of physical models used in protein-folding simulations: quantitative evidence from long molecular dynamics simulations. Curr. Opin. Struct. Biol. 2014;24:98–105. doi: 10.1016/j.sbi.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 69.Lindorff-Larsen K., Maragakis P., Shaw D.E. Systematic validation of protein force fields against experimental data. PLoS One. 2012;7:e32131. doi: 10.1371/journal.pone.0032131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Piana S., Lindorff-Larsen K., Shaw D.E. Protein folding kinetics and thermodynamics from atomistic simulation. Proc. Natl. Acad. Sci. USA. 2012;109:17845–17850. doi: 10.1073/pnas.1201811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beauchamp K.A., Lin Y.-S., Pande V.S. Are protein force fields getting better? A systematic benchmark on 524 diverse NMR measurements. J. Chem. Theory Comput. 2012;8:1409–1414. doi: 10.1021/ct2007814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cino E.A., Choy W.-Y., Karttunen M. Comparison of secondary structure formation using 10 different force fields in microsecond molecular dynamics simulations. J. Chem. Theory Comput. 2012;8:2725–2740. doi: 10.1021/ct300323g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 74.Darden T., York D., Pedersen L. Particle mesh Ewald: an N log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 75.Essmann U., Perera L., Pedersen L. A smooth particle mesh Ewald potential. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 76.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 77.Miyamoto S., Kollman P.A. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 78.Wang L., Friesner R.A., Berne B.J. Replica exchange with solute scaling: a more efficient version of replica exchange with solute tempering (REST2) J. Phys. Chem. B. 2011;115:9431–9438. doi: 10.1021/jp204407d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Laghaei R., Mousseau N., Wei G. Effect of the disulfide bond on the monomeric structure of human amylin studied by combined Hamiltonian and temperature replica exchange molecular dynamics simulations. J. Phys. Chem. B. 2010;114:7071–7077. doi: 10.1021/jp100205w. [DOI] [PubMed] [Google Scholar]
- 80.Tiwary P., Parrinello M. A time-independent free energy estimator for metadynamics. J. Phys. Chem. B. 2015;119:736–742. doi: 10.1021/jp504920s. [DOI] [PubMed] [Google Scholar]
- 81.Laio A., Gervasio F.L. Metadynamics: a method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Rep. Prog. Phys. 2008;71:126601. [Google Scholar]
- 82.Barducci A., Bonomi M., Parrinello M. Metadynamics. WIREs Comput. Mol. Sci. 2011;1:826–843. [Google Scholar]
- 83.Sutto L., Marsili S., Gervasio F.L. New advances in metadynamics. WIREs Comput. Mol. Sci. 2012;2:771–779. [Google Scholar]
- 84.Barducci A., Bonomi M., Derreumaux P. Assessing the quality of the OPEP coarse-grained force field. J. Chem. Theory Comput. 2011;7:1928–1934. doi: 10.1021/ct100646f. [DOI] [PubMed] [Google Scholar]
- 85.Camilloni C., Schaal D., De Simone A. Energy landscape of the prion protein helix 1 probed by metadynamics and NMR. Biophys. J. 2012;102:158–167. doi: 10.1016/j.bpj.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Camilloni C., Provasi D., Broglia R.A. Exploring the protein G helix free-energy surface by solute tempering metadynamics. Proteins. 2008;71:1647–1654. doi: 10.1002/prot.21852. [DOI] [PubMed] [Google Scholar]
- 87.Frishman D., Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 88.Shen Y., Bax A. SPARTA+: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J. Biomol. NMR. 2010;48:13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kohlhoff K.J., Robustelli P., Vendruscolo M. Fast and accurate predictions of protein NMR chemical shifts from interatomic distances. J. Am. Chem. Soc. 2009;131:13894–13895. doi: 10.1021/ja903772t. [DOI] [PubMed] [Google Scholar]
- 90.Daura X., Suter R., van Gunsteren W.F. Validation of molecular simulation by comparison with experiment: rotational reorientation of tryptophan in water. J. Chem. Phys. 1999;110:3049–3055. [Google Scholar]
- 91.Eisenhaber F., Lijnzaad P., Scharf M. The double cube lattice method: efficient approaches to numerical integration of surface area and volume and to dot surface contouring of molecular assemblies. J. Comput. Chem. 1995;16:273–284. [Google Scholar]
- 92.Barlow D.J., Thornton J.M. Ion-pairs in proteins. J. Mol. Biol. 1983;168:867–885. doi: 10.1016/s0022-2836(83)80079-5. [DOI] [PubMed] [Google Scholar]
- 93.Arndt J.R., Kondalaji S.G., Valentine S.J. Huntingtin N-terminal monomeric and multimeric structures destabilized by covalent modification of heteroatomic residues. Biochemistry. 2015;54:4285–4296. doi: 10.1021/acs.biochem.5b00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sivanandam V.N., Jayaraman M., van der Wel P.C.A. The aggregation-enhancing huntingtin N-terminus is helical in amyloid fibrils. J. Am. Chem. Soc. 2011;133:4558–4566. doi: 10.1021/ja110715f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hoop C.L., Lin H.-K., van der Wel P.C.A. Polyglutamine amyloid core boundaries and flanking domain dynamics in huntingtin fragment fibrils determined by solid-state nuclear magnetic resonance. Biochemistry. 2014;53:6653–6666. doi: 10.1021/bi501010q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nagarajan A., Jawahery S., Matysiak S. The effects of flanking sequences in the interaction of polyglutamine peptides with a membrane bilayer. J. Phys. Chem. B. 2014;118:6368–6379. doi: 10.1021/jp407900c. [DOI] [PubMed] [Google Scholar]
- 97.Kokona B., Rosenthal Z.P., Fairman R. Role of the coiled-coil structural motif in polyglutamine aggregation. Biochemistry. 2014;53:6738–6746. doi: 10.1021/bi500449a. [DOI] [PubMed] [Google Scholar]
- 98.Schwarzinger S., Kroon G.J., Dyson H.J. Sequence-dependent correction of random coil NMR chemical shifts. J. Am. Chem. Soc. 2001;123:2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.