Abstract
K-Ras is a membrane-associated GTPase that cycles between active and inactive conformational states to regulate a variety of cell signaling pathways. Somatic mutations in K-Ras are linked to 15–20% of all human tumors. K-Ras attaches to the inner leaflet of the plasma membrane via a farnesylated polybasic domain; however, the structural details of the complex remain poorly understood. Based on extensive (7.5 μs total) atomistic molecular dynamics simulations here we show that oncogenic mutant K-Ras interacts with a negatively charged lipid bilayer membrane in multiple orientations. Of these, two highly populated orientations account for ∼54% of the conformers whose catalytic domain directly interacts with the bilayer. In one of these orientation states, membrane binding involves helices 3 and 4 of the catalytic domain in addition to the farnesyl and polybasic motifs. In the other orientation, β-strands 1–3 and helix 2 on the opposite face of the catalytic domain contribute to membrane binding. Flexibility of the linker region was found to be important for the reorientation. The biological significance of these observations was evaluated by initial experiments in cells overexpressing mutant K-Ras as well as by an analysis of Ras-effector complex structures. The results suggest that only one of the two major orientation states is capable of effector binding. We propose that the different modes of membrane binding may be exploited in structure-based drug design efforts for cancer therapy.
Introduction
H-, N-, and K-Ras proteins are membrane-associated molecular switches that regulate many signaling pathways involved in cell growth and division (1). Activating Ras mutations, typically at codons 12, 13, and 61, are associated with 15–25% of all human cancers (2); K-Ras mutations account for 85% of all oncogenic Ras mutations (3). Decades of sustained efforts produced a wealth of data that paints a detailed picture of Ras function at the molecular level (e.g., (1, 4, 5, 6, 7, 8, 9, 10, 11)). This includes insights into how conformational changes accompanying nucleotide exchange and GTP hydrolysis enable Ras to cycle between active and inactive conformational states. However, it remains unclear how Ras proteins that share a highly conserved catalytic domain (residues 1–166; >95% conserved) mediate several different signal outputs (12).
Differential membrane binding via the hypervariable region (HVR, residues 167–185/6) has been proposed as a possible mechanism by which Ras proteins achieve signaling specificity (11, 13). This is supported by the fact that K-Ras preferentially targets negatively charged membranes via a C-terminal farnesylated and polybasic domain, whereas in N- and H-Ras farnesylation is complemented by palmitoyl modification for attachment mostly to neutral membranes. Moreover, recent studies suggested that nucleotide-dependent membrane reorientation might lead to isoform-dependent regulation of function (14, 15, 16, 17, 18, 19). This suggests that the specific orientation of the catalytic domain (CD) can have an important role in Ras function due to the potential occlusion of functionally responsive surfaces by the membrane.
First proposed by molecular dynamics (MD) simulation studies (16), regulation of Ras function by membrane reorientation has been subsequently supported by experiments in cells (15), synthetic membranes (17, 18), and lipid nanodiscs (19). Additional studies have shown that Ras binding to downstream effectors such as Raf is sensitive to the specific membrane orientation of the CD and hence the relative accessibility of the canonical switches S1 (residues 30–40) and S2 (residues 60–75) to interaction partners (e.g., (14, 15, 19)). At the teleological level, the correlation between activity and membrane orientation may be understood from the structural differences between GDP- and GTP-bound Ras. However, understanding exactly what underpins this correlation at the atomic level is far from trivial.
A recent report proposed that GTP/GDP exchange gives rise to differential distribution of positively charged surface patches on the CD of K-Ras, leading to differential engagement with a negatively charged bilayer surface (19). Surprisingly, the effector-binding surface was found occluded by the membrane in the GTP-bound form. Moreover, K5N and D153V mutations, two Rasopathy mutations that do not affect the GTPase cycle, “relieve the occluded orientation by directly altering the electrostatics of two membrane interaction surfaces” (19). However, electrostatics alone may not explain how charge-neutral mutations, which represent the vast majority of oncogenic and Rasopathy mutations, would alter membrane orientation. Furthermore, as suggested previously (15, 16, 17, 18, 19), nucleotide- and mutation-dependent differences in membrane orientation likely involve changes in conformation and fluctuations that are communicated across long distances, leading to changes in the relative population of different orientation states. In other words, Ras membrane reorientation is a dynamic process underpinned by an intrinsic propensity to sample multiple conformations and hence orientation states; the effect of nucleotide and/or mutation would then be altering the population of states and/or the rate of interconversion between states. This hypothesis, namely, Ras has an intrinsic propensity to sample multiple orientation states, can be tested directly by showing that a single Ras protein spontaneously samples different orientation states. This is the goal of the current work.
As a model system, we chose the constitutively active oncogenic mutant K-Ras4B G12D (hereafter K-Ras) in its GTP-bound form (K-Ras.GTP). We chose this system both because of its intrinsic importance in cancer research and because oncogenic mutants often lead to enhanced dynamics (20). Building upon our previous successes in studying Ras dynamics at membranes (16), we applied a fully atomistic MD approach to generate extensive sets of trajectories (totaling >7.5 μs simulation time) of K-Ras attached to a negatively charged bilayer. We found that the catalytic domain directly interacts with the bilayer via multiple surfaces and in different orientations. Two of these orientations are highly populated within the simulations timescale. In one of these, the canonical switches S1 and S2 are available for interaction with other proteins, but they are partially or fully occluded in the other orientation. The bilayer-interacting surfaces are located at helices 3/4 (orientation state 1) and strands 1–3/helix 2 (orientation state 2), with a few residues at loop7 (and occasionally helix 2) providing a toehold as the CD rocks between these two states via the HVR tether. We also show that the linker region in the HVR plays a key role in the reorientation of the CD, and that enhanced dynamics of the whole protein or just the linker enables sampling of a range of membrane orientation states.
Materials and Methods
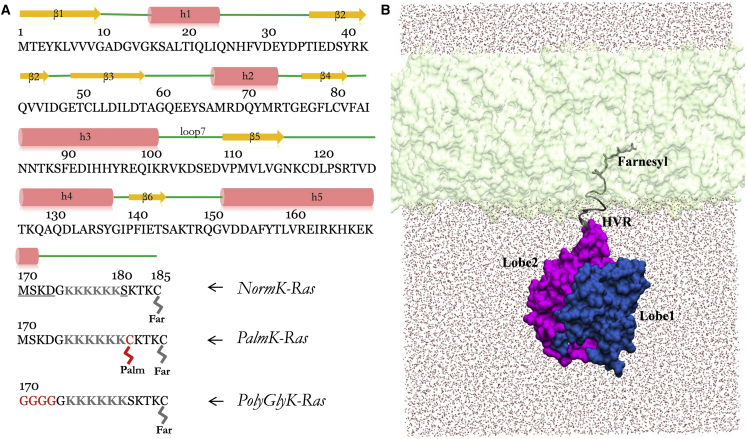
We performed 20 independent simulations on G12D K-Ras attached to a POPC/POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine) bilayer. The simulations were divided into four groups (Fig. 1). Unless stated otherwise, different initial conditions or sampling strategies were employed in simulations of different groups, whereas simulations within the same group differed only by the assignment of initial velocities. The first group (NormK-Ras) involved 10 runs of at least 400 ns each (minimum total length of 4 μs). The second group (NocmapK-Ras) of three 800 ns runs was the same as NormK-Ras except that no CHARMM CMAP correction was used (see Molecular Dynamics Simulation, below). The goal of the no-CMAP simulations was to allow increased flexibility at loop regions, as described in detail by Buck et al. (21); previously, this approach allowed us to sample open switch conformations of Ras in solution (22). The third group, referred to as PalmK-Ras, involved three runs of 100 ns each conducted on a system in which S181 was replaced by a palmitoylated Cys and no CMAP dihedral correction was applied. The fourth group consisted of four 200 ns simulations with CMAP on a K-Ras variant with residues 170–173 mutated to Gly (PolyGlyK-Ras). The aggregate simulation time was close to 8 μs.
Figure 1.
Sequence and structure of K-Ras and simulation setup. (A) Primary and secondary structure of G12D K-Ras, with structural elements shown in orange (β-strand), red (α-helix), and green (loop region). NormK-Ras refers to the unmodified G12D K-Ras in the GTP-bound form. Underlined residues were mutated and highlighted in red: S181 was mutated to a palmitoylated Cys in PalmK-Ras; residues 170–173 were mutated to Gly in PolyGlyK-Ras. Simulations in the NocmapK-Ras group are the same as in NormK-Ras with the only difference being application of CMAP dihedral correction in the latter but not the former. The hexa-lysine stretch (residues 175–180) and farnesyl are highlighted in gray. Lobe 1 encompasses residues 1–86 and lobe 2 residues 87–166. (B) Starting structure of the full-length farnesylated G12D K-Ras embedded in a POPC/POPS bilayer (light green), with lobe 1 in blue, lobe 2 in pink, HVR and farnesyl in gray and water shown as red dots. To see this figure in color, go online.
Model building
We simulated full-length (residues 1–185), farnesylated K-Ras in a negatively charged bilayer made up of 320 POPC and 96 POPS lipids (20%). Simulations were initiated from a model built by ligating the crystal structure of G12D K-Ras (23) (PDB: 4DSO, residues 1–173) to a K-Ras lipid anchor (tK, residues 174–185) bound to a POPC/POPG bilayer; the tK structure was the most stable conformation in a previous MD simulation of the peptide in a POPC/POPG (palmitoyloleoylphosphoglycerol) bilayer (24). Mg2+, GTP, and crystal waters were kept. The resulting K-Ras-POPC/POPG complex was solvated by adding TIP3P waters with the ionic strength set to 0.15 M by adding appropriate numbers of Na+ and Cl− ions, yielding a system size of ∼200,000 atoms (Fig. 1 B).
The system was then relaxed by running a 20 ns-long MD simulation without CMAP, the condition in which the previous tK-POPC/POPG complex was simulated (24). We used POPG in the previous work because it was one of the best-characterized anionic lipids at the time. The difference between the negatively charged lipids POPG and POPS is limited to the headgroup: −CHOHCH2(OH)2 versus −CHNH3CO2. However, K-Ras discriminates between anionic lipids and preferentially targets PS (Y.Z. and J.F.H., unpublished data; (25)), which is the most common anionic lipid in the plasma membrane of mammalian cells. We therefore swapped POPG with POPS. After replacing POPG by POPS, the last snapshot of this simulation was used as the starting point for NormK-Ras (except for four runs that were started directly from the manually built K-Ras-POPC/POPG system without a 20 ns relaxation) and NocmapK-Ras. The third group (PalmK-Ras) was also started from this configuration but after introducing palmitoylation at position 181 of the protein. In each of the three groups of simulations, the HVR was extended so that the CD was fully solvated and far away from the bilayer, with the helices roughly perpendicular to the bilayer surface (Fig. 1 B). To prepare the starting structure for the PolyGlyK-Ras simulations, we mutated residues 170–173 to glycine on a snapshot taken at 275 ns of one of the NormK-Ras runs in which the HVR was semiextended and the CD was in water.
Molecular dynamics simulation
After model building, each system was energy minimized for 2000 steps with lipid and protein heavy atoms fixed and equilibrated for 200 ps with the lipid phosphate and protein heavy atoms harmonically restrained with a force constant k = 4 kcal/mol/Å2, followed by four additional 100 ps equilibration steps with k scaled by 0.75, 0.50, 0.25, and 0; the time step was 1 fs. This was followed by production runs with a 2 fs time step using SHAKE (26) to restrain all bonds involving hydrogen atoms. Particle-mesh Ewald electrostatics (27) and 12 and 14 Å cutoffs for nonbonded interactions and pair-list update, were used with the switch function turned on at 10 Å. We used the NPT ensemble with constant pressure of 1 bar maintained by the Nosé-Hoover Langevin piston method and temperature of 310 K controlled by the Langevin thermostat method. The force field was CHARMM27 for proteins (28) and CHARMM36 for lipids (29), with the CMAP dihedral correction (30) applied in all simulations except NocmapK-Ras and PalmK-Ras. Parameters for the palmitoyl and farnesyl groups were from previous studies (16, 31). All simulations were run with the NAMD2.9 program (32).
Reaction coordinate and statistical analysis
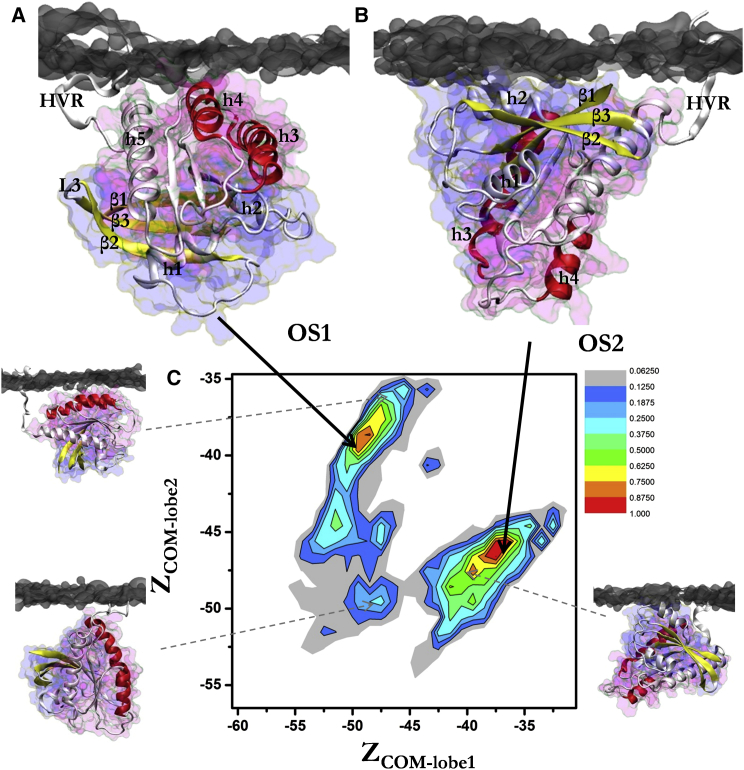
Displacement of the CD along the membrane normal (Z axis) was monitored by the Z coordinates of the centers-of-mass of lobe 1 (residues 1–86, ZCOM-lobe1) and lobe 2 (residues 87–166, ZCOM-lobe2) (Fig. S1 in the Supporting Material). ZCOM-lobe1 and ZCOM-lobe2 were measured after aligning the center of the bilayer to the origin. As schematically illustrated in Fig. S1, the centers-of-mass of lobes 1 and 2 are located at opposite sides of the CD so that once it is on the bilayer surface an increase in ZCOM-lobe1 would lead to a decrease in ZCOM-lobe2 and vice versa. The normalized two-dimensional probability distribution of these two variables, P(ZCOM-lobe1, ZCOM-lobe2), yielded two major populations (hot color in Fig. 3 C) centered at (−49, −39) and (−37.5, −46.5). Selecting only P(ZCOM-lobe1, ZCOM-lobe2) ≥ 0.2, we defined orientation state (OS1) as the population of conformers within dOrientation = 6 Å of (−49, −39) and orientation state 2 (OS2) as the population of conformers within dOrientation = 5.6 Å of (−37.5, −46.5). dOrientation was calculated as the root mean-square (RMS) displacement of (ZCOM-lobe1, ZCOM-lobe2) from the reference mean position (Zcen′, Zcen″) (Eq. 1):
| (1) |
With these strict criteria, 20 and 34% of the conformers were found to belong to OS1 and OS2, respectively, from a total dataset of 153,000 conformers derived from the merged NormK-Ras simulations. Note that in all of these snapshots the CD stably interacts with the membrane surface (Fig. 2 A). For reference, the starting structure was located at ZCOM-lobe1 = −69 and ZCOM-lobe2 = −67, highlighting the large displacement of the CD toward the bilayer surface.
Figure 3.
Two distinct membrane orientations of K-Ras. Two predominant orientation states are highlighted: OS1, where lobe 2 (h3/h4) contacts the bilayer (A), and OS2, where lobe 1 (β1–3, h2) is in direct contact with the negatively charged membrane (B). (C) Two-dimensional histogram of P(ZCOM-lobe1, ZCOM-lobe2) (see Materials and Methods) normalized by the maximum value, and examples of intermediate orientations (bottom, left and right) and a rare event in which lobe 2 is very close to the bilayer (top left) highlighted by snapshots at (−47.5, −49.5), (−39.5, −47.5), and (−47, −36), respectively. Helices 3 and 4 are in red, β1–3 in yellow, lobe 1 in blue, and lobe 2 in pink. Membrane is shown as gray surface. To see this figure in color, go online.
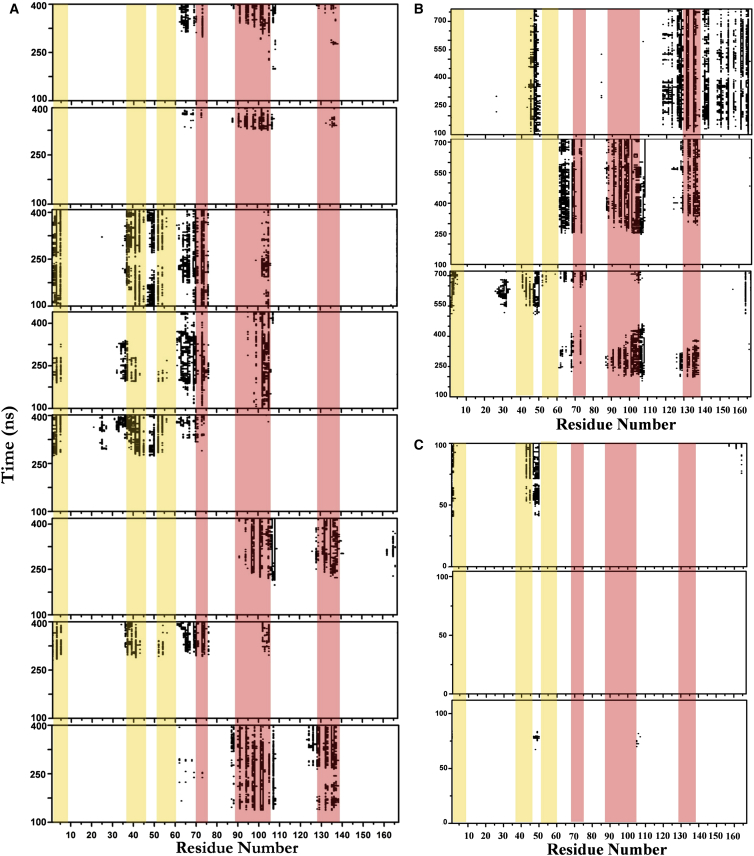
Figure 2.
Direct interaction of K-Ras catalytic domain with membrane. Shown are contacts of CD residues with bilayer lipids during NormK-Ras (A), NocmapK-Ras (B), and PalmK-Ras simulations (C). Based on the residues that are within 5 Å of the membrane, the data show that the CD made direct contact with the membrane within the simulations timescale in 8/10 of the NormK-Ras and 3/3 of the NocmapK-Ras runs. In (A) and (B), the initial 100 ns data was omitted as equilibration phase while (C) shows that even a K-Ras variant carrying a second lipid modification did not achieve significant CD-bilayer interaction within 100 ns. Strands β1–3 are highlighted in orange and helices 2, 3, and 4 in red. To see this figure in color, go online.
Structural analysis and surface electrostatic potential calculations
We used in-house scripts and VMD (33) to examine protein and bilayer structural properties using standard techniques such as calculation of RMS deviations, interatomic distances, and dihedral angles. Electrostatic potentials were calculated with the Adaptive Poisson Boltzmann Solver using default parameters, including an ionic strength of 150 mM and solute/solvent dielectric constants of 2/78.54. The Parse atomic charges and radii were used. Neutral pH was assumed with all Asp and Glu side chains negatively charged and all Lys and Arg residues positively charged; all other residues were neutral.
Electron microscopy and fluorescence lifetime imaging microscopy—fluorescence resonance energy transfer experiments
To examine the impact of K-Ras membrane reorientation on clustering and hence function, we used the complementary techniques of electron microscopy (EM) and fluorescence lifetime imaging microscopy-fluorescence resonance energy transfer (FLIM-FRET) that have proven useful in similar previous studies (14, 15). We refer the reader to those articles (14, 15) for the discussion of EM-spatial mapping and FLIM-FRET analyses. Only a very brief description is provided here.
For EM analysis, we used intact plasma membrane sheets of baby hamster kidney (BHK) cells expressing green fluorescent protein (GFP)-tagged K-Ras.G12V or its R73 mutants attached to EM grids, fixed with 4% PFA (paraformaldehyde) and 0.1% glutaraldehyde and labeled with 4.5 nm gold nanoparticles pre-coupled with anti-GFP antibody. Images were obtained using a transmission EM, and a 1 μm2 area of the plasma membrane sheet was selected and the x, y coordinates of each gold particle was assigned using ImageJ (National Institutes of Health, Bethesda, MD). The spatial distribution of the gold particles was calculated using Ripley’s K-function as described previously (34, 35, 36). At least 15 plasma membrane sheets were imaged and analyzed for each mutant. Bootstrap tests constructed as described before (35, 37) were used to measure statistical differences between replicated point patterns; statistical significance was evaluated against 1000 bootstrap samples.
For FLIM-FRET measurements, BHK cells transiently expressing GFP-tagged K-Ras variants with or without RFP-tagged cognate were washed with PBS and fixed in 4% PFA. Fluorescence lifetime imaging was conducted on a wide-field Eclipse microscope (Nikon, Melville, NY) with a FLIM module attachment (Lambert Instruments, Roden, the Netherlands). GFP fluorescence excitation was achieved using a sinusoidally simulated, modulating 3 W, 468-nm light-emitting diode at 40 MHz in combination with a 60 Plan-Apo/1.4-numerical-aperture oil immersion lens. At least 60 cells from three independent experiments were imaged for each mutant.
Results
Our simulations were started with the lipid anchor already embedded in the bilayer, and it remained embedded during each of the 20 simulations. In other words, no dissociation event was observed consistent with previous MD simulations of tK (24), full-length K-Ras (14), and other Ras proteins (16, 38, 39, 40). The previous simulations of full-length K-Ras were short (∼30 ns) and used a zwitterionic DMPC bilayer (14). The current simulations are significantly longer (100–800 ns) and are conducted under different conditions and in multiple copies using a POPC/POPS bilayer, which is a better model of mammalian cell membranes.
Membrane binding does not significantly affect the structure of K-Ras
The structure of membrane-bound full-length K-Ras has not been characterized in detail before. Given the conservation of the CD across the Ras family, we did not expect dramatic structural changes upon membrane binding. We wanted to check if this holds true in the simulations, especially in the no CMAP runs that could give rise to unphysical structures (21). Fig. S2 shows the time evolution of the CD backbone atoms RMS deviations using the equilibrated initial structure as a reference. One can see that all of the plots plateaued at generally small values of 0.8–1.5 Å (with maximum 2.0 Å; Fig. S2). Even in the case of one of the PolyGlyK-Ras runs where helix 3 elongated and straightened at ∼90 ns (inset of Fig. S2), the plot plateaued after ∼150 ns. These suggest that our systems are well equilibrated and the core structure of the CD is well preserved in both the CMAP (NormK-Ras/PolyGlyK-Ras) and no-CMAP (NocmapK-Ras/PalmK-Ras) simulations.
As expected, the protein was dynamic. This is reflected in the conformational transitions around 30 ns in some NormK-Ras and PalmK-Ras runs and at ∼150 ns in two NocmapK-Ras runs (Fig. S2). The Nocmap runs generally yielded higher fluctuations at switches 1 and 2 and some other loops similar to that shown in Fig. 6 of Prakash et al. (22). In fact, the switch regions and loop7 are flexible in almost all of the runs, consistent with previous observations (41, 42). Loop7 plays a critical role in bilayer interaction and will be discussed throughout the article. Overall, the core structure of the K-Ras CD was not significantly altered during the simulations but the use of multiple runs in different conditions led to diverse fluctuations. No major changes were observed in the bilayer structure either, aside from localized effects at the site of contact with the protein.
The catalytic domain of K-Ras interacts with the surface of a negatively charged bilayer
As expected the primary source of bilayer affinity is the interaction of the hexa-lysine and the farnesyl moieties with the bilayer and hydrophobic core, respectively. To check if membrane binding also involves other parts of the protein, we monitored the time evolution of contacts between lipids and protein residues at the CD. Fig. 2, A and B, shows that in 8 out of the 10 NormK-Ras runs and in all three of the NocmapK-Ras simulations the CD makes direct contact with the bilayer, which is in variance with a previous simulation of K-Ras in DMPC (14). One reason for the discrepancy is the extent of conformational sampling; the formation of stable CD-bilayer interaction took >100 ns (Fig. 2), three times longer than the previous simulations. Another obvious difference is the fact that the anionic POPC/POPS bilayer is a better host for the polybasic lipid anchor of K-Ras.
Once on the bilayer surface, the CD remains bound for the entire duration of the simulations. Furthermore, only a small subset of the CD residues interacts with lipids, primarily involving helix 2/β-strands 1–3 (β1–3/h2) or helices 3 and 4 (h3/4). This is remarkable for a number of reasons. First, despite the fact that the CD was completely solvated and away from the bilayer in the beginning, stable CD-bilayer contacts emerged within a relatively short period of time (several hundred nanoseconds). Second, CD-bilayer interaction occurred in 11/13 (85%) of the normal K-Ras simulations (NormK-Ras and NocmapK-Ras), irrespective of the initial condition or extent of conformational fluctuation. Third, the contacts are nonrandom and involve only a few surface patches (β1–3/h2 or h3/h4). Taken together, these observations indicate that the CD of K-Ras is attracted to and interacts with a negatively charged bilayer via specific sets of residues.
In previous studies, multiple copies of relatively short MD simulations on GTP-bound G12V H-Ras (16) and GDP-bound wild-type N-Ras (43) have led to direct interaction between the catalytic domain and a zwitterionic bilayer. In each case, the catalytic domain was almost nearly parallel to the membrane plane, with helix 4 sitting on the surface. All of the current simulations on G12D K-Ras, including those performed without CMAP as in the previous G12V H-Ras simulations (16), required a comparatively longer simulation for CD-bilayer contacts to occur (Fig. 2). We wondered if this difference could be due to the lack of palmitoylation on K-Ras, which exists in both H- and N-Ras. We tested this with three simulations on an artificial construct where S181 was mutated to palmitoylated Cys (PalmK-Ras); no CMAP was applied in these simulations as in the previous H-Ras simulations (16). Fig. 2 C shows that although one copy began to make a few contacts via turn β2-β3, none of the PalmK-Ras simulations led to a stable CD-bilayer interaction within 100 ns, which is about the same length as the N-Ras runs (43) and three times longer than those of H-Ras (16). We conclude that the apparently slow membrane binding of K-Ras CD is not due to the lack of palmitoylation.
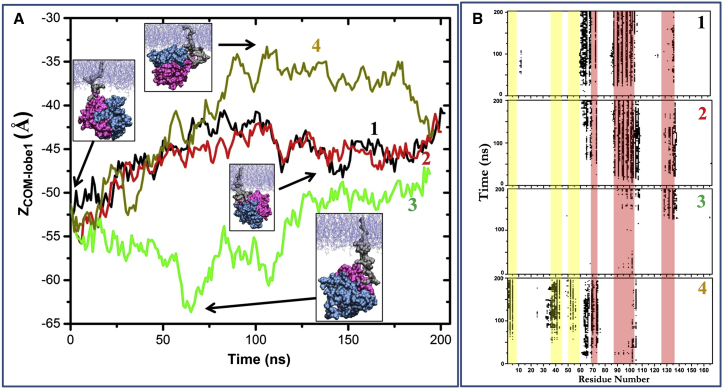
We then turned to the rest of the HVR excluding the hexa-lysine region, i.e., the linker residues 170–174. Inspection of this region yielded a curious observation: the backbone dihedral of G174, which is unique to K-Ras, adopts two distinct values (Fig. S3). Because this appeared to be the primary source of the HVR flexibility, we reasoned that increased dynamics of the linker region might lead to faster reorientation of the catalytic domain. We tested this by simulations in which the entire segment is converted to Gly (PolyGlyK-Ras, Fig. 1 A). The goal was to circumvent—to the extent possible—sampling limitations by enforcing faster dynamics of the HVR. Indeed, we found that the mutations led to a substantially faster reorientation; more orientation states were visited in <200 ns of the PolyGlyK-Ras runs than was achieved by 2–4 times longer simulations in the NormK-Ras and NocmapK-Ras series (Fig. 7). This shows that the HVR plays an important role in the membrane reorientation of K-Ras (discussed more in a later section). Given the sequence divergence of Ras isoforms at the HVR, we believe this observation has important implications for isoform-specific temporal regulation of Ras function via membrane orientation.
Figure 7.
Flexibility of the linker region enhances orientation dynamics. (A) Time evolution of ZCOM-lobe1 (A) and contacts of CD residues with the bilayer (B) for the PolyGlyK-Ras simulations numbered 1–4. (A, insets) Snapshots from different runs taken at the indicated ZCOM-Lobe1/time points illustrating the various orientations of K-Ras that were sampled, including cases where lobe 1 or lobe 2 are in close proximity to membrane. (Leftmost inset) Common starting structure of the simulations taken from one of the NormK-Ras simulations at 275 ns, where the linker was extended. Lobe 1 is in blue, lobe 2 in pink, bilayer is in light blue, and the HVR in gray. To see this figure in color, go online.
The K-Ras catalytic domain interacts with membrane primarily via two distinct surfaces
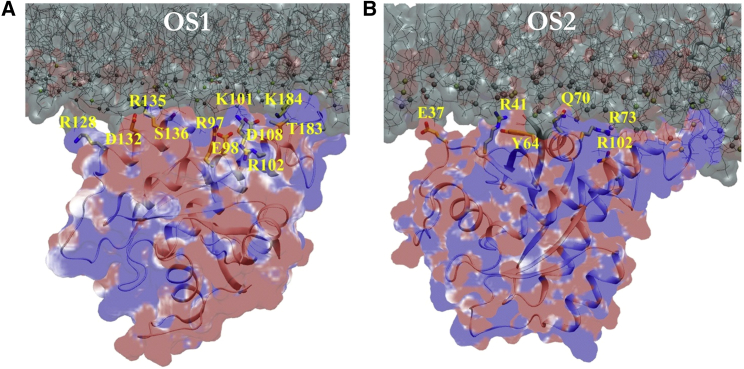
The contact maps in Fig. 2 suggest that CD-bilayer interaction involves either exclusively lobe 1 (residues 1–86) or exclusively lobe 2 (residues 87–166) but not both at the same time. This means that structures derived from the combined trajectory (separately for each simulation set) can be divided into conformations in which lobe 1 interacts with the bilayer and those in which the interaction is via lobe 2. This can be done by tracking the movement of the centers-of-mass of lobes 1 and 2 along the Z axis (see Materials and Methods; Fig. S1). A two-dimensional histogram along ZCOM-lobe1 and ZCOM-lobe2 yielded two dominant populations (Fig. 3 C). We refer to these two populations as membrane orientation state 1 (OS1) and membrane orientation state 2 (OS2). Note that in OS1, lobe 2 is in close proximity to the membrane, while, in OS2, lobe 1 interacts with the membrane (Fig. 3, A and B). Other intermediate states are also observed. Specifically, orientations that are sampled infrequently (colder color, Fig. 3 C) may represent a possible pathway for interconversion between the two predominant orientations.
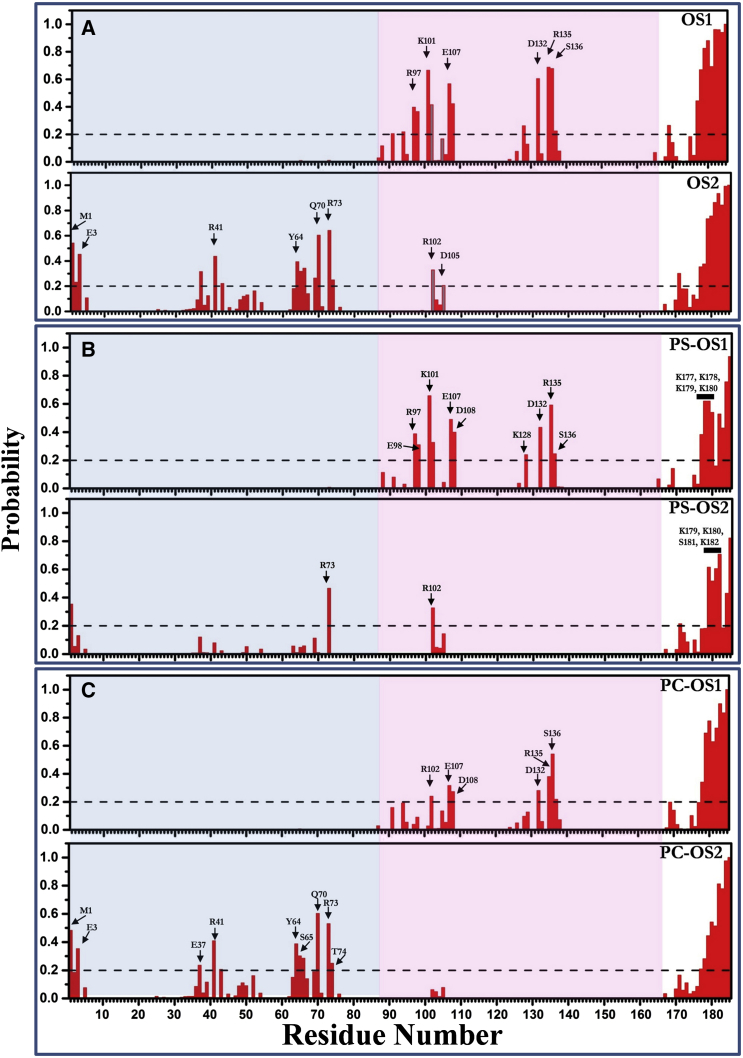
For further analysis, we separated conformers belonging to OS1 from those belonging to OS2 (see Materials and Methods) and calculated the frequency of bilayer contact made by individual residues separately for OS1 and OS2 (Fig. 4 A). Several important lessons can be drawn from this data. First, as expected, the farnesylated polybasic C-terminus of K-Ras contributes the most to membrane binding in both states. Second, in both OS1 and OS2, only a few (often positively charged) side chains are responsible for anchoring the CD to the surface of the bilayer; in each case these residues are spread out in sequence space but form only a single surface patch in the 3D structure. They involve residues from two amphipathic helices (h3/4, OS1) and one face of β1–3 and h2 (OS2) (Fig. 3, A and B). Third, a few residues from loop7, such as R102 (gray bars, Fig. 4 A), interact with the membrane roughly with equal probability in both orientation states. A closer inspection of the trajectories further shows that bilayer-interaction of loop7 (and in some cases h2) persist not only in the two states but largely also during the transition between them (see Movies S1 and S2 and Fig. S4). Lack of sufficient sampling (limited number of OS1/OS2 transitions) precludes a definitive conclusion about the mechanism of interstate transition. Nevertheless, this observation suggests that the CD rocks between the two orientation states using loop7 as a pliable toehold secondary to the tight anchor provided by the HVR. This sharply contrasts with a swinging motion that would occur if the HVR were the only anchor point. Conformations in which the CD is held to the bilayer only by the toehold can be considered intermediate (or transition) orientation states that are visited as the CD rocks between the two main states (Fig. S4 and Movies S1 and S2).
Figure 4.
Distinct sets of residues from lobe 1 (blue) and lobe 2 (pink), along with the lipid anchor (white), contribute to membrane binding of K-Ras in OS1 and OS2. Normalized probabilities of bilayer contact by K-Ras residues in OS1 and OS2 estimated based on a heavy atom distance cutoff of 4 Å: contacts with (A) all lipids; (B) POPS; (C) POPC. (Horizontal dashed lines) 20% arbitrary cutoff for significance. With this criterion, R102 is the only CD residue that is near the bilayer in both OS1 and OS2 (and in intermediate states; see Fig. S4). Note that OS1 and OS2 also differ in PS-interaction of specific lysines at the lipid anchor (labeled). To see this figure in color, go online.
Analyses of residue contacts separately for POPC and POPS (Fig. 4, B and C) provided additional insights into how OS1 and OS2 might be stabilized. K-Ras-PC interactions appear to involve predominantly polar (not necessarily basic) amino acids. These include E107/D108 of loop7 and D132/R135/S136/Y137 of h4 in OS1 and a number of polar/charged residues (plus M1) at the N-terminus, β2, and switch 2 in OS2. As might be expected, spatially proximal basic residues, such as K101 and R135 in OS1, dominate the interaction with PS (Fig. 4 B; see also Fig. 6). However, CD-PS charge complementarity is attenuated, at least in part, by anionic residues that are spatially close to the basic residues. Some of these effects are countered by sporadic interactions with selected basic residues at the HVR. Still, CD-bilayer interaction in either orientation does not appear to be solely electrostatic in origin (see Figs. 4 and 6).
Figure 6.
Surface potential. ±1 kT/e electrostatic surface potential calculated with Adaptive Poisson Boltzmann Solver on two snapshots representing OS1 (A) and OS2 (B). Key residues identified as bilayer-interacting in Fig. 4 are labeled. Here, k is Boltzmann’s constant and T is temperature in Kelvin. To see this figure in color, go online.
It is conceivable that in the cellular milieu interaction with other proteins or oligomerization modulates the type of motion as well as the population of orientation states. For instance, effector binding to OS1, where the effector-binding loop is not occluded by the bilayer (Fig. 3 A), might stabilize this orientation by increasing its population and decreasing the population of other states via a population shift mechanism (44, 45, 46, 47). Albeit indirect, our initial experiments support this conclusion (see Discussion). Our analysis also suggests the existence of intermediate conformations (Fig. 3 C), which are even more prominent in the PolyGlyK-Ras simulations (see Fig. 7) and will be discussed further later.
HVR dynamics is coupled with changes in orientation state
There is a negligible difference between OS1 and OS2 in terms of the interaction of the hexa-lysine residues 175–180 with the bilayer. Interestingly, however, the N-terminal K175 and K176 contact bilayer only in <20% of cases in both OS2 and OS1, respectively (Fig. 4 A). This is consistent with a previous study of tK (the isolated farnesylated polybasic domain) in a POPC/POPG bilayer (24), where we found that backbone structural constraints limit the number of Lys side chains that can directly interact with bilayer lipids. However, a detailed analysis of the contacts separately for POPC and POPS indicates small but significant differences. While contacts of the lipid anchor with PC are comparable (Fig. 4 C), K177 and K178 are better able to interact with PS in OS1 than in OS2 (Fig. 4 B). Also, S181 is proximal to PS in OS2 while T183 is solvated by PS in OS1 (Fig. 4 B). The linker residues 170–174 contact the bilayer 10–25% of the time in OS2 but not in OS1.
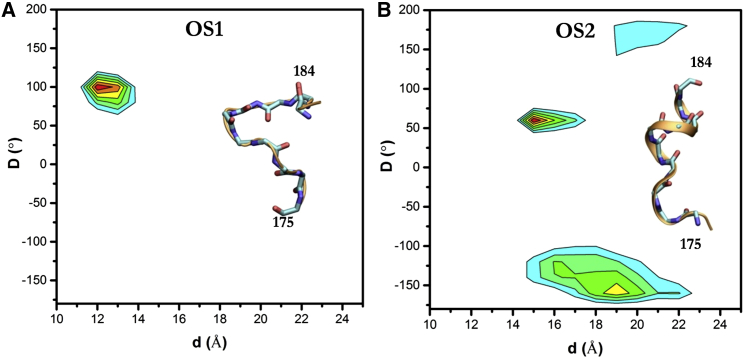
We wondered if this could be due to structural differences at the backbone level. To test this hypothesis, we followed a previously described procedure (24), where we calculate a dihedral angle defined by virtual bonds connecting the Cα atoms of consecutive residues 178–181 (D), and the distance between the Cα atoms of K175 and K184 (d). A plot of D versus d revealed that the HVR backbone is generally more extended in OS2 than in OS1 (Fig. 5). In both orientations, the HVR residues 175–184 approach (to within 5 Å) loop7 residues 102–108 and to a lesser extent the β1–3 surface in roughly similar proportions (Fig. S5). For example, K170 points toward loop7 in both states. However, there exist interesting variations as well: (1) the C-terminus and center of the lipid anchor approach loop7 in OS1 and OS2, respectively; (2) K175 and K177 make significant contacts with loop7 in OS2 but not in OS1; and (3) the sporadic contacts of the HVR with the β1–3 differ considerably between OS1 and OS2. These are related to the differences in the structure of the backbone, as illustrated in Fig. 5.
Figure 5.
HVR dynamics. The main plots and insets highlight the relatively compact structure of the HVR in OS1 (A) versus OS2 (B). Virtual bonds connecting the Cα atoms of residues 178–181 define the dihedral angle (D) and the distance (d) is between Cα atoms of residues 175 and 184. To see this figure in color, go online.
Fig. 6 shows the surface electrostatic potentials derived from the full-length protein in the two orientations. One can see that, in each state, multiple electropositive surface patches contact the bilayer but they are interspersed by electronegative or neutral surfaces, consistent with the presence of negatively charged and polar amino acids near the bilayer (Fig. 4). We realize that the protein is dynamic and no single snapshot can capture the behavior of the ensemble. However, visualization of numerous snapshots from different trajectories did not yield a contiguous, bilayer-facing electropositive surface such as that found in a Ras/Raf-RBD complex. In addition, the two membrane binding surfaces do not exhibit dramatic differences in electrostatic surface potential (Fig. 6, A and B) or charge distribution (Fig. 4). We propose that OS1/OS2 exchange is primarily a consequence of conformational change rather than a shift in charge distribution. This, however, does not exclude the possibility that long-range electrostatics facilitates the formation of an initial CD-bilayer encounter complex that is further modulated by conformational fluctuations.
The observations discussed above combined with previously observed communications between the canonical switches, loop7 and the C-terminus of the CD (41, 48, 49) prompted us to ask if the structure and dynamics of the HVR might dictate membrane reorientation of K-Ras. To examine this, we turned to our PolyGlyK-Ras simulations where the presence of five consecutive glycines (M170G, S171G, K172G, D173G, plus G174) made the HVR highly flexible. The enhanced flexibility allowed for sampling of multiple orientations, including OS1 and OS2 (Fig. 7, A and B) plus others, during comparatively short simulation times. Even orientations that were negligibly populated in NormK-Ras were sampled well in PolyGlyK-Ras (e.g., ZCOM-lobe1 ≈ −35). We conclude that the HVR and especially the linker play a critical role in the reorientation of the G-domain with respect to the bilayer plane, consistent with a previous suggestion that the orientation dynamics of K-Ras is encoded at HVR (14).
Discussion
A number of previous studies on Ras or Ras-related proteins have shown that differential membrane orientation impacts function (14, 15, 16, 17, 18, 19, 50, 51). For instance, nucleotide-dependent membrane reorientation, first predicted for G12V H-Ras by MD (16) and subsequently corroborated by multiple experimental studies (14, 15, 17, 18, 19), was found to affect effector binding. Similarly, a recent study of wild-type and different mutants of K-Ras in lipid nanodiscs found a nucleotide-dependent dynamics whereby an orientation with occluded switch loops was found to dominate the GTP analog GMPPNP-bound state (19). The focus of the current work, however, was on the potential diversity of orientations within a single nucleotide state of monomeric GTP-bound oncogenic mutant G12D K-Ras. We found that the catalytic domain of G12D K-Ras monomer directly interacts with our negatively charged bilayer model. The CD-bilayer interaction involved multiple orientations, including two predominant orientations referred to as membrane orientation states 1 and 2 (OS1 and OS2). In the former, helices 3 and 4 interact directly with the membrane while helix 2 and β1–3 are the main contributors in the latter. Loop7 serves as a hinge so that either lobe 1 or lobe 2, but not both simultaneously, is presented to the bilayer. Formation of dimers, as found in several recent studies (43, 52, 53, 54), will likely affect the distribution of the different orientation states. A simple thought experiment based on accessibility of surface patches to solvent would suggest that dimerization via the proposed helical interfaces (43, 53) might stabilize OS2 whereas dimerization via strand 2 (53) may stabilize OS1.
It is worth pointing out that the similar population sizes of OS1 and OS2 and the transition between them observed in some of our trajectories may suggest that the two states are iso-energetic and are separated by a relatively small barrier. However, we decided to limit our analysis to the structural aspects of the dynamics. This is because a reliable thermodynamic/kinetic analysis would require far more extensive sampling of phase space than achieved in this work.
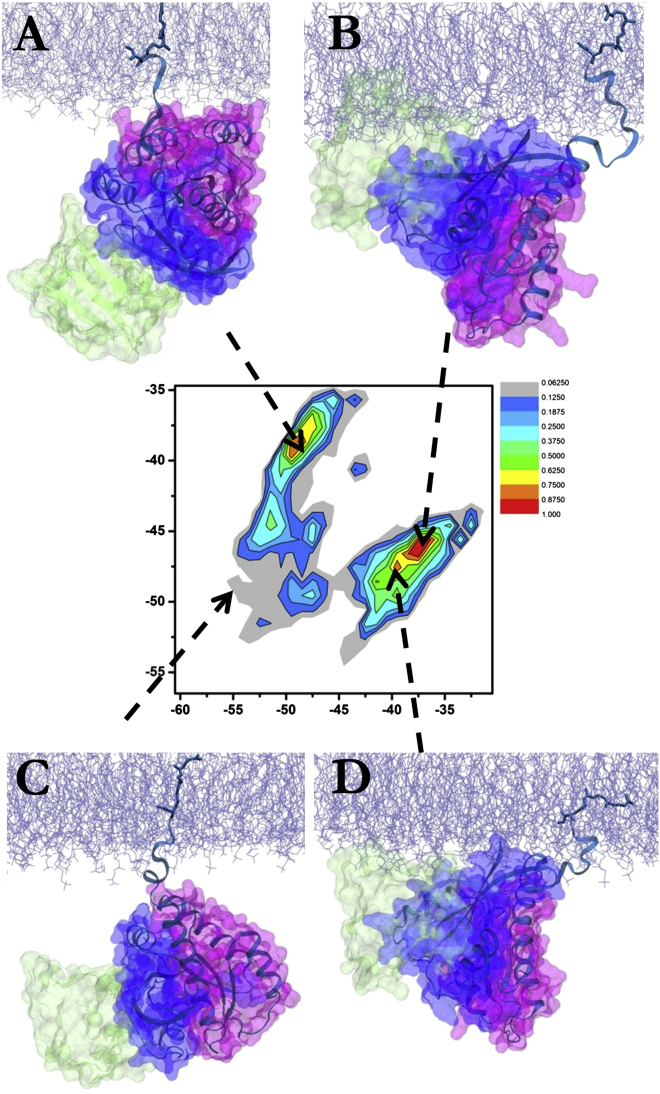
To assess the potential functional role of these diverse orientation states, we superposed representative structures for different orientations of simulated G12D K-Ras with the crystal structure of the Ras-Raf complex (PDB: 4G0N) (55). Fig. 8 A shows that OS1 would bind Raf without steric clashes whereas the effector protein extensively clashes with the bilayer when docked onto structures representing OS2 (Fig. 8 B). The effector-binding surface in OS2 is engaged with the membrane and not available for other interactions. In fact, OS2 is remarkably similar to the “occluded” conformation of K-Ras observed recently in lipid nanodiscs based on paramagnetic resonance enhancement measurements and HADDOCK calculations (19). However, OS1 is somewhat different from the “exposed” conformation found in the same study. The key difference is that helices 4/5 engage the bilayer in the nanodisc study instead of helices 3/4 we find here. Our simulations differ from the analysis in Mazhab-Jafari et al. (19) in several important ways. These include the presence of a spin-label (Gd+3) and a lipoprotein belt needed in the lipid nanodisc study. Moreover, all of the current simulations involved K-Ras.GTP whereas the h4/5 orientation was favored by the inactive K-Ras.GDP in the nanodisc study (19).
Figure 8.
Effector binding and orientation states. Superposition of representative structures from (A) OS1 and (B) OS2 indicates no steric clash and substantial overlap between Raf (green) and bilayer (ice blue), respectively. An orientation where neither lobe 1 nor lobe 2 is in close proximity with membrane is capable of interacting with effector (C), while an intermediate orientation that is similar to OS2 results in partial overlap of the effector with the membrane and therefore may not be favored (D). The crystal structure of Ras-Raf complex (PDB: 4G0N) was used for the superposition. To see this figure in color, go online.
The h4/5 orientation of K-Ras observed in lipid nanodiscs (19) was also somewhat different from G12V H-Ras’ parallel membrane orientation, where it interacts with lipids via only h4. In the current work, only a single run out of a total of 20 (see below) sampled an orientation in which h4/5 interacts with the bilayer (Fig. 2 B). None of the PolyGlyK-Ras simulations led to bilayer interaction via h4/5 (Fig. 7) despite the enhanced reorientation dynamics. Moreover, only one of the NocmapK-Ras runs sampled a h4/5 orientation (Fig. 2 B, top panel) even though these trajectories sampled open switch conformations that are similar to those found in the GDP-bound Ras and state 1 GTP Ras; this is interesting because GDP.K-Ras was shown to prefer the h4/h5 orientation (19). The second Nocmap run sampled a wide range of conformations, including intermediate orientations where either the tip of h3/loop7, h2, or h4 (but not h5) interacts with the membrane (Fig. 2 B, middle panel). We also saw interconversion between OS1 and OS2 in the third NocmapK-Ras run (Fig. 2 B, bottom panel) as well as one of the CMAP runs (fourth panel, Fig. 2 A), but none involved h5 (see also Movies S1 and S2). Therefore, we believe OS1 and OS2 are the dominant orientation states in G12D K-Ras.GTP, although many different orientations including h4/5 might still be accessible.
As an initial experimental test of the biological significance of our observations, we performed EM and FLIM-FRET experiments in cells expressing various mutants of R73 in the background of the constitutively active G12V K-Ras. G12V was chosen for consistency with previous studies (14, 15) and because we do not expect major differences in clustering between V and D at codon 12. We chose R73 because it is distal from the effector-binding loop and therefore unlikely to directly affect effector binding, and because it is one of the predominant residues involved in the stabilization of OS2 (Fig. 4 A). Our choice of this residue was also influenced by the observation that K-Ras-PS interaction, primarily via the hexa-lysine segment but potentially also through the CD, is critical for plasma membrane binding (Y.Z. and J.F.H., unpublished data; (25)). R73 is the only basic residue from lobe 1 that persistently interacts with PS (Fig. 4 B).
No microscopic/spectroscopic method is currently available to directly measure the interaction of a single side chain with cell membrane. Therefore, we adopted an indirect approach that measures changes in the overall organization of K-Ras on the plasma membrane. This entailed measurements of cluster formation with EM and protein-protein interaction with FLIM-FRET. Because the less conserved lobe 2 (41) and especially helices 3–5 tend to be more reactive (22) and possibly harbor a dimerization interface (43, 53), we hypothesized that K-Ras homo-oligomerization would be facilitated by the exposure of lobe 2 to solvent. This means that destabilization of OS2 by mutating R73 would shift the population to OS1 and thereby decrease the probability of cluster formation/oligomerization. We therefore mutated R73 to K and E to test the effect of the specific charge at position 73 in cellular K-Ras clustering and self-interaction.
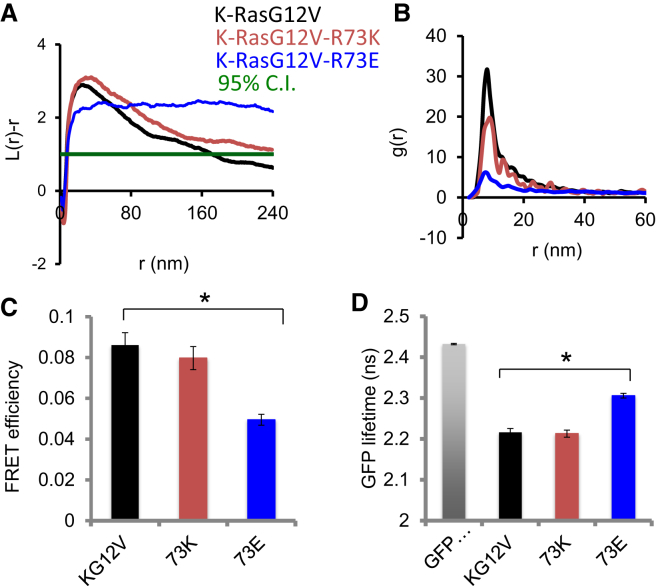
Fig. 9 shows that, relative to the reference construct G12V K-Ras, charge reversal via R73E significantly decreased nanoclustering (Fig. 9 A) and packing (Fig. 9 B), as well as self-association (Fig. 9, C and D). As expected, the charge neutral R73K mutation has no effect. For example, Fig. 9 B shows that packing densities measured by g(r) differ significantly between the two R73 mutants. Specifically, R73E had the lowest peak value indicating weaker packing ability, which is consistent with the reduced clustering shown in Fig. 9 A. Similarly, for R73E, the efficiency of FRET between the donor RFP and acceptor GFP is significantly reduced while the GFP lifetime is significantly increased. In comparison, there was no statistically significant difference between the reference and R73K. Together, these observations support our hypothesis that OS2/OS1 reorientation is coupled with clustering, and suggest that a positively charged residue at position 73 plays an important role in stabilizing OS2.
Figure 9.
Spatial mapping of K-Ras.G12V R73 mutants. (A) Nanoclustering of R73K and R73E mutants on the inner leaflet of the plasma membrane of cells, with statistical significance evaluated by bootstrap tests. L(r)−r is a measure of spatial clustering with the peak height corresponding to the extent of clustering (see Prior et al. (36) for more detail). (B) g(r) showing packing density within the nanoclusters where peak values summarize the extent of packing. (C) FRET efficiency. (D) Fluorescence lifetime. The data in (C) and (D) are shown as mean ± SE collected from at least 60 cells in three separate experiments. ∗p 0.05 using Student’s t-test. To see this figure in color, go online.
Clearly, more work is required to determine exactly how changes in membrane orientation would affect K-Ras function. When combined with previous studies of Ras (and related) proteins in lipid nanodiscs as well as in other synthetic and cell membranes (14, 15, 17, 18, 19, 50, 51), however, our results provide an intriguing picture of how intrinsic structural fluctuations and specific charge interactions work together to hold the catalytic domain on a membrane surface. This is remarkable because, given its water solubility, the catalytic domain might have been assumed to be staying away from membrane. More broadly, our findings reinforce the idea (16, 24, 56) that membrane binding of Ras GTPases involves much more than the catalytic domain passively hanging in water via the lipidated HVR that is glued to the membrane.
Conclusions
We have shown that G12D K-Ras.GTP interacts with a negatively charged POPC/POPS membrane with multiple distinct orientations. Among these, two orientations were found to be highly populated and are referred to as orientation states 1 and 2. Within the simulation lengths of hundreds of nanoseconds that we have employed, we found that surface residues from helices 3/4 (OS1) and helix 2/β1–3 (OS2) form direct contact with the membrane. A few residues at loop7 (and in some cases helix 2) provide a toehold as the catalytic domain rocks between the two major orientation states using the HVR as a tether. The switch region is completely solvent-exposed in OS1, which we propose is an effector-interacting orientation. The switches are occluded in OS2, which likely makes it a not effector interacting orientation. Flexibility of the linker region in the HVR affects the orientation of the G-domain with respect to the membrane plane and therefore may play a critical role in function as well. Because variations in the proximity of different surface patches of K-Ras to membrane can affect accessibility of ligand binding sites (57), we propose that the different modes of membrane interaction and dynamics should be taken into account in structure-based drug design efforts targeting membrane-bound K-Ras.
Author Contributions
A.A.G. and P.P. conceived and designed the project; P.P., H.L., Y.Z., J.F.H., and A.A.G. performed the experiments and analyzed the data; and P.P., Y.Z., and A.A.G. wrote the article.
Acknowledgments
We thank the Texas Advanced Computing Center for computational resources.
P.P. was supported by a postdoctoral training fellowship from the Keck Center Computational Cancer Biology Training Program of the Gulf Coast Consortia (CPRIT grant No. RP140113). This work was supported in part by grants from the Cancer Prevention and Research Institute of Texas (CPRIT grant No. DP150093) and the National Institutes of Health General Medical Sciences (grant No. R01GM100078).
Editor Michael Feig.
Footnotes
Five figures and two movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)00094-1.
Supporting Material
The movie was generated from one of the Nocmap-K-Ras simulations (Fig. 2 B), which is the only trajectory that underwent such a transition. The toehold region comprising the C-terminus of helix-3 and loop7 is shown in purple, helices 3 and 4 are green, and strands β1–3 and helix 2 are in orange. The residues in licorice are those within 4 Å of the bilayer (gray lines). The elements in licorice are colored as follows: nitrogen (blue), oxygen (red), carbon (cyan), and sulphur (yellow).
Color scheme is the same as in Movie S1.
References
- 1.Cox A.D., Der C.J. Ras history: the saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos J.L. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 3.Prior I.A., Lewis P.D., Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmick M., Kraemer A., Bastiaens P.I. Ras moves to stay in place. Trends Cell Biol. 2015;25:190–197. doi: 10.1016/j.tcb.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Parker J.A., Mattos C. The Ras-membrane interface: isoform-specific differences in the catalytic domain. Mol. Cancer Res. 2015;13:595–603. doi: 10.1158/1541-7786.MCR-14-0535. [DOI] [PubMed] [Google Scholar]
- 6.Ahearn I.M., Haigis K., Philips M.R. Regulating the regulator: post-translational modification of RAS. Nat. Rev. Mol. Cell Biol. 2012;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abankwa D., Gorfe A.A., Hancock J.F. Ras nanoclusters: molecular structure and assembly. Semin. Cell Dev. Biol. 2007;18:599–607. doi: 10.1016/j.semcdb.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock J.F. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y., Hancock J.F. Ras nanoclusters: versatile lipid-based signaling platforms. Biochim. Biophys. Acta. 2015;1853:841–849. doi: 10.1016/j.bbamcr.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Prakash P., Gorfe A.A. Overview of simulation studies on the enzymatic activity and conformational dynamics of the GTPase Ras. Mol. Simul. 2014;40:839–847. doi: 10.1080/08927022.2014.895000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prakash P., Gorfe A.A. Lessons from computer simulations of Ras proteins in solution and in membrane. Biochim. Biophys. Acta. 2013;1830:5211–5218. doi: 10.1016/j.bbagen.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrhardt A., Ehrhardt G.R., Schrader J.W. Ras and relatives—job sharing and networking keep an old family together. Exp. Hematol. 2002;30:1089–1106. doi: 10.1016/s0301-472x(02)00904-9. [DOI] [PubMed] [Google Scholar]
- 13.Prior I.A., Hancock J.F. Compartmentalization of Ras proteins. J. Cell Sci. 2001;114:1603–1608. doi: 10.1242/jcs.114.9.1603. [DOI] [PubMed] [Google Scholar]
- 14.Abankwa D., Gorfe A.A., Hancock J.F. Ras membrane orientation and nanodomain localization generate isoform diversity. Proc. Natl. Acad. Sci. USA. 2010;107:1130–1135. doi: 10.1073/pnas.0903907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abankwa D., Hanzal-Bayer M., Hancock J.F. A novel switch region regulates H-ras membrane orientation and signal output. EMBO J. 2008;27:727–735. doi: 10.1038/emboj.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorfe A.A., Hanzal-Bayer M., McCammon J.A. Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoylglycero-3-phosphocholine bilayer. J. Med. Chem. 2007;50:674–684. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
- 17.Kapoor S., Triola G., Winter R. Revealing conformational substates of lipidated N-Ras protein by pressure modulation. Proc. Natl. Acad. Sci. USA. 2012;109:460–465. doi: 10.1073/pnas.1110553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapoor S., Weise K., Winter R. The role of G-domain orientation and nucleotide state on the Ras isoform-specific membrane interaction. Eur. Biophys. J. 2012;41:801–813. doi: 10.1007/s00249-012-0841-5. [DOI] [PubMed] [Google Scholar]
- 19.Mazhab-Jafari M.T., Marshall C.B., Ikura M. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc. Natl. Acad. Sci. USA. 2015;112:6625–6630. doi: 10.1073/pnas.1419895112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukman S., Grant B.J., McCammon J.A. The distinct conformational dynamics of K-Ras and H-Ras A59G. PLOS Comput. Biol. 2010;6:e1000922. doi: 10.1371/journal.pcbi.1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buck M., Bouguet-Bonnet S., MacKerell A.D., Jr. Importance of the CMAP correction to the CHARMM22 protein force field: dynamics of hen lysozyme. Biophys. J. 2006;90:L36–L38. doi: 10.1529/biophysj.105.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prakash P., Hancock J.F., Gorfe A.A. Binding hotspots on K-ras: consensus ligand binding sites and other reactive regions from probe-based molecular dynamics analysis. Proteins. 2015;83:898–909. doi: 10.1002/prot.24786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurer T., Garrenton L.S., Fang G. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. USA. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janosi L., Gorfe A.A. Segregation of negatively charged phospholipids by the polycationic and farnesylated membrane anchor of Kras. Biophys. J. 2010;99:3666–3674. doi: 10.1016/j.bpj.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y., Liang H., Hancock J.F. Signal integration by lipid-mediated spatial cross talk between Ras nanoclusters. Mol. Cell. Biol. 2014;34:862–876. doi: 10.1128/MCB.01227-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryckaert J.P.C., Ciccotti G., Berendsen H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:15. [Google Scholar]
- 27.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 28.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 29.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackerell A.D., Jr., Feig M., Brooks C.L., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 31.Gorfe A.A., Pellarin R., Caflisch A. Membrane localization and flexibility of a lipidated ras peptide studied by molecular dynamics simulations. J. Am. Chem. Soc. 2004;126:15277–15286. doi: 10.1021/ja046607n. [DOI] [PubMed] [Google Scholar]
- 32.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 34.Kiskowski M.A., Hancock J.F., Kenworthy A.K. On the use of Ripley’s K-function and its derivatives to analyze domain size. Biophys. J. 2009;97:1095–1103. doi: 10.1016/j.bpj.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plowman S.J., Muncke C., Hancock J.F. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. USA. 2005;102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prior I.A., Muncke C., Hancock J.F. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diggle P.J., Mateu J., Clough H.E. A comparison between parametric and non-parametric approaches to the analysis of replicated spatial point patterns. Adv. Appl. Probab. 2000;32:13. [Google Scholar]
- 38.Janosi L., Li Z., Gorfe A.A. Organization, dynamics, and segregation of Ras nanoclusters in membrane domains. Proc. Natl. Acad. Sci. USA. 2012;109:8097–8102. doi: 10.1073/pnas.1200773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z., Janosi L., Gorfe A.A. Formation and domain partitioning of H-ras peptide nanoclusters: effects of peptide concentration and lipid composition. J. Am. Chem. Soc. 2012;134:17278–17285. doi: 10.1021/ja307716z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H., Gorfe A.A. Membrane remodeling by surface-bound protein aggregates: insights from coarse-grained molecular dynamics simulation. J. Phys. Chem. Lett. 2014;5:1457–1462. doi: 10.1021/jz500451a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorfe A.A., Grant B.J., McCammon J.A. Mapping the nucleotide and isoform-dependent structural and dynamical features of Ras proteins. Structure. 2008;16:885–896. doi: 10.1016/j.str.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grant B.J., Gorfe A.A., McCammon J.A. Ras conformational switching: simulating nucleotide-dependent conformational transitions with accelerated molecular dynamics. PLOS Comput. Biol. 2009;5:e1000325. doi: 10.1371/journal.pcbi.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Güldenhaupt J., Rudack T., Gerwert K. N-Ras forms dimers at POPC membranes. Biophys. J. 2012;103:1585–1593. doi: 10.1016/j.bpj.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grant B.J., Gorfe A.A., McCammon J.A. Large conformational changes in proteins: signaling and other functions. Curr. Opin. Struct. Biol. 2010;20:142–147. doi: 10.1016/j.sbi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grant B.J., McCammon J.A., Gorfe A.A. Conformational selection in G-proteins: lessons from Ras and Rho. Biophys. J. 2010;99:L87–L89. doi: 10.1016/j.bpj.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee G.M., Craik C.S. Trapping moving targets with small molecules. Science. 2009;324:213–215. doi: 10.1126/science.1169378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kar G., Keskin O., Nussinov R. Allostery and population shift in drug discovery. Curr. Opin. Pharmacol. 2010;10:715–722. doi: 10.1016/j.coph.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buhrman G., Holzapfel G., Mattos C. Allosteric modulation of Ras positions Q61 for a direct role in catalysis. Proc. Natl. Acad. Sci. USA. 2010;107:4931–4936. doi: 10.1073/pnas.0912226107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raimondi F., Orozco M., Fanelli F. Deciphering the deformation modes associated with function retention and specialization in members of the Ras superfamily. Structure. 2010;18:402–414. doi: 10.1016/j.str.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 50.Mazhab-Jafari M.T., Marshall C.B., Ikura M. Membrane-dependent modulation of the mTOR activator Rheb: NMR observations of a GTPase tethered to a lipid-bilayer nanodisc. J. Am. Chem. Soc. 2013;135:3367–3370. doi: 10.1021/ja312508w. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y., Kahn R.A., Prestegard J.H. Dynamic structure of membrane-anchored Arf∗GTP. Nat. Struct. Mol. Biol. 2010;17:876–881. doi: 10.1038/nsmb.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nan X., Tamgüney T.M., Chu S. Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc. Natl. Acad. Sci. USA. 2015;112:7996–8001. doi: 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muratcioglu S., Chavan T.S., Nussinov R. GTP-dependent K-Ras dimerization. Structure. 2015;23:1325–1335. doi: 10.1016/j.str.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin W.C., Iversen L., Groves J.T. H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc. Natl. Acad. Sci. USA. 2014;111:2996–3001. doi: 10.1073/pnas.1321155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fetics S.K., Guterres H., Mattos C. Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure. 2015;23:505–516. doi: 10.1016/j.str.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gorfe A.A., McCammon J.A. Similar membrane affinity of mono- and Di-S-acylated ras membrane anchors: a new twist in the role of protein lipidation. J. Am. Chem. Soc. 2008;130:12624–12625. doi: 10.1021/ja805110q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prakash P., Sayyed-Ahmad A., Gorfe A.A. pMD-membrane: a method for ligand binding site identifcation in membrane-bound proteins. PLOS Comput. Biol. 2015;11:e1004469. doi: 10.1371/journal.pcbi.1004469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The movie was generated from one of the Nocmap-K-Ras simulations (Fig. 2 B), which is the only trajectory that underwent such a transition. The toehold region comprising the C-terminus of helix-3 and loop7 is shown in purple, helices 3 and 4 are green, and strands β1–3 and helix 2 are in orange. The residues in licorice are those within 4 Å of the bilayer (gray lines). The elements in licorice are colored as follows: nitrogen (blue), oxygen (red), carbon (cyan), and sulphur (yellow).
Color scheme is the same as in Movie S1.