Main Text
The reductionist movement of twentieth century biological science successfully used the tools of biochemistry, molecular biology, and structural biology to provide us with an increasingly detailed parts list of living systems. As the troves of molecular data grew, the advent of bioinformatics brought to bear information technologies that allowed biological scientists to annotate, query, search, and integrate these data with relative ease. This gave birth to systems biology, which seeks to reconstruct networks of the molecular interactions that give rise to the essential biochemical, biophysical, and regulatory functions of cells, and that give the different cell types the unique properties they need to build specialized organ systems such as the central nervous system, the musculoskeletal system, and the cardiovascular system. With these increasingly detailed, yet invariably still incomplete, molecular network reconstructions, the foundation has been laid for systems models of biological functions at all scales that simulate the dynamic physiology of living systems, especially cells, as large circuit diagrams of functional interactions. Great promise is held by these new quantitative, computer-driven approaches that can provide a new level of integrative scientific insight and identify promising new pharmacologic therapies.
Biological systems are exquisitely structured and depend critically on their dynamic three-dimensional organization to achieve their physiological functions. The challenge of building models that integrate structurally across physical scales of biological organization from molecule to cell to organ system and organism is a defining problem of modern biophysics. Like systems biology, this field of multiscale modeling is data-intensive. We depend on structural biology, microscopy, and medical imaging technologies to build high-quality, high-resolution data sets on molecular, cellular, tissue, and organ structures. But multiscale modeling also relies heavily on physics to define and constrain that ways that molecular and cellular processes can scale up to produce tissue and organ-scale physiology.
The need for multiscale modeling of organ systems is readily apparent when we put ourselves in the shoes of the physician. Patients present with symptoms and diseases that manifest at the tissue, organ, and whole body scales. But medical therapies target specific molecules. How can we diagnose and effectively treat illnesses without understanding the multiscale relationships between molecules and the whole body?
Take the case of the heart diseases. Cardiac arrhythmias are disorders of the heart’s electrical system. Many of them can be traced to alterations in specific ion channels in the heart cell membrane, yet all arrhythmias are organ-level phenomena. Their manifestation depends on electrical interactions between cells and are commonly associated with altered coupling between cells or structural changes in the extracellular matrix that organizes the cells into cardiac muscle tissue. Similarly, the engines for muscle contraction are molecular motors in the cardiac muscle cells. The efficiency with which the heart converts contractile forces into ventricular pumping is critically dependent on the arrangement of the motors within the cell, their regulation by electrical excitation, the architecture of muscle cells and matrix in the tissue, the size and shape of the ventricular walls, and the coupling via the heart valves of the ventricles of the atria and the vasculature. Any number of these properties can change in congenital or acquired heart diseases. Understanding how the many molecular, cellular, tissue, and organ scale alterations give rise to cardiac electrical and mechanical dysfunction has been a primary motivation for the development of multiscale biophysics models of the heart.
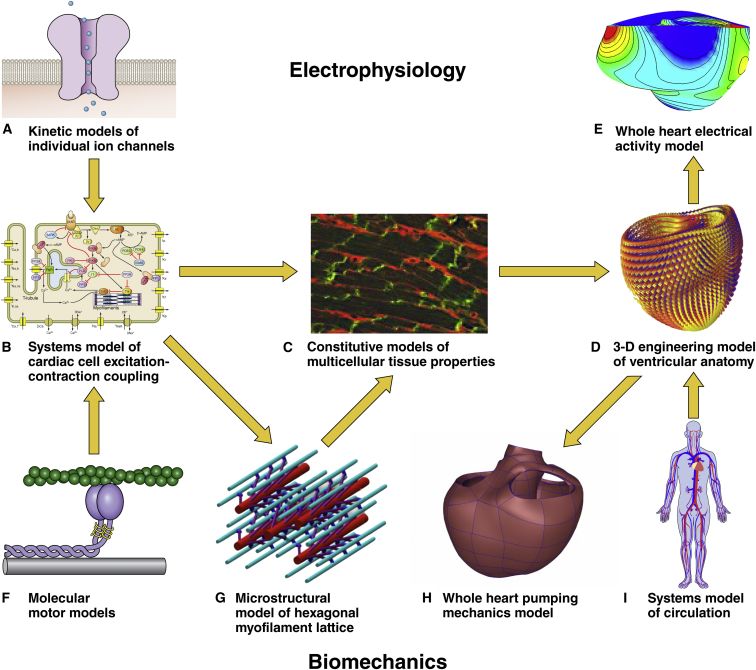
There is no single paradigm or recipe for multiscale modeling. Different organs are specialized for different functions involving different physics. At the same time, the same physical principles are exploited by many living systems for different purposes, so approaches developed for one system can often be applied to others. The heart has electrical, mechanical, and transport functions all linked together. Fig. 1 summarizes some of the popular paradigms that have been used to develop and integrate multiscale models of cardiac physiology. On the top, specialized proteins form channels in the cell membrane for specific ions to cross, carrying electrical charge with them, which in combination with each other can change the voltage across the whole cell membrane and allow electrical impulses to propagate through the heart tissue, giving rise to electric fields in the torso that are detected by electrodes on the body surface as electrocardiograms. Similarly, molecular motors are organized into contractile filaments in the muscle cells. Electrical depolarization of the muscle cell triggers brief releases of calcium, causing a rise in filament tension that is distributed in three dimensions through the hexagonal myofilament lattice and into the tissue. The chambers contract against the load of blood pressure in the circulation, pressure that was generated originally by the heart itself. This description shows that the top scale of our cardiac mechanical models is a model of pressures, flows, and resistances of the circulation. These electrical and mechanical systems are coupled through the intracellular calcium release system, and models of intracellular calcium fluxes provide a link between these systems, allowing us to build coupled models of cardiac electromechanics and hemodynamics. Because these systems all require energy, models of cell metabolism and oxygen delivery to the heart muscle also find a natural fit in this modeling framework. We explore each subsystem below with an emphasis on the governing physics and the driving scientific questions and clinical applications.
Figure 1.
(A–I) Multiscale models of cardiac electrophysiology and biomechanics can be combined to model cardiac electromechanical function in health and disease. To see this figure in color, go online.
The cardiac electrical system
It has long been known that the heart generates electrical current, a phenomenon that Dutch physiologist Willem Einthoven (1860–1927) successfully exploited in his invention of the electrocardiogram (ECG or EKG) in 1903, for which he received the Nobel Prize for Medicine in 1924. The origin of the electrical activity detected in the ECG is the flow of ions across the membranes of cardiac muscle cells. At rest, the muscle cells (myocytes) have negative electrical potential with respect to the outside. Each heartbeat is triggered by pacemaker cells that cause a wave of electrical excitation to propagate through the atria and then the ventricles. This excitation is a transient period of electrical depolarization carried by sodium ions rushing into cells, followed by a repolarization due mainly to the outward flux of potassium ions. The dynamics of this process is made possible by specialized ion channels in the cell membrane that can open and close as a function of the membrane potential itself.
Modern multiscale models of cardiac electrical activity take into account the voltage-dependent kinetics of dozens of different ion channels, pumps, and transporters that carry sodium, potassium, calcium, and chloride ions (Fig. 1 A). They can account for detailed knowledge of the numerous different states the channels can occupy, made possible by detailed single-channel recordings and even the specific effects of many drugs and gene mutations. They include the capacitance of the membranes in a whole cell model (Fig. 1 B) and the resistive electrical coupling between neighboring muscle cells at the tissue scale (Fig. 1 C) as well as the three-dimensional anatomy of the cardiac chambers and the complex spiral-wound laminar organization of the muscle fibers in the heart walls (Fig. 1 D). The most important underlying physics for these sophisticated integrated models of whole heart electrical activity (Fig. 1 E) is well established: Ohm’s law is used to relate the ion channel and intracellular resistances to the membrane voltage. Kirchhoff’s current law provides the other key physical principle that Alan Hodgkin and Andrew Huxley famously used in their 1952 mathematical model that explained the ionic mechanisms of the electrical impulse conduction along a nerve, work for which they received the Nobel Prize in 1963.
Today, sophisticated multiscale systems models of cardiac electrical activity are not only helping to elucidate basic scientific mechanisms, they are increasingly helping us to understand human cardiac arrhythmias, and they may soon become part of the cardiologist’s tool kit. Important ongoing questions being addressed include: How do cellular instabilities lead to arrhythmias and under what conditions? How important are the molecular alterations in the cell compared with the structural changes associated with heart disease at the tissue and organ scales? How can we design smarter and more reliable pacemakers and defibrillators? Will drugs be effective at terminating or preventing specific arrhythmias, and can we identify potentially dangerous proarrhythmic drugs before they reach the clinic? Finally, can we identify who is most at risk and most likely to benefit from therapies such as implantable cardioverter defibrillators?
The cardiac mechanical system
The basic function of the heart to pump blood through the body has been recognized since William Harvey’s publication in 1628 of Exercitatio Anatomica de Motu Cordis et Sanguinis in Animalibus in which he clearly established the concept of blood circulation and the central importance of the heart as a pump. The German physician and physiologist Otto Frank (1865–1944) and English physiologist Ernest Starling (1866–1927) separately performed the ground-breaking experiments on the pumping mechanics of the heart that established what is now known as the Frank-Starling law of the heart. This important law states that the more the heart fills and the longer the muscle fibers are stretched the more strongly the ventricular pumps contract. The most important applicable physics are again well established and originally due to Isaac Newton, namely the conservation of linear momentum. Modern multiscale models of cardiac mechanics solve Newton’s laws for the heart walls as continua subject to the additional constraints of mass and energy conservation.
The challenge is to link the pumping mechanics of the cardiac chambers (Fig. 1 H) both up in scale to explain the interactions between the filling and contraction of the cardiac chambers and the pressures and flows in the circulatory system (Fig. 1 I), and down in scale to the level of the molecular motors (Fig. 1 F) in the cardiac myocytes that convert biochemical energy to mechanical work. A critical intermediate mesoscale is the complex three-dimensional organization of the cells and matrix of the heart into a three-dimensional continuum capable of withstanding cycles of very large shape changes every second, uninterrupted, a billion times throughout a lifetime. Until recently, most computational models of cardiac tissue-scale mechanical properties were largely descriptive engineering models, but as quantitative three-dimensional microscopy techniques improve in resolution and molecular specificity (Fig. 1 C), we are starting to see new microstructural models of cardiac tissue mechanics that will replace these more traditional formulations. At the molecular level, Huxley’s famous 1957 model of muscle contraction (Fig. 1 F), which has been revised and extended many times, still forms the core of cardiac mechanical models. Recent work has focused on incorporating detailed models of the effects of the hexagonal myofilament lattice structure inside the myocytes (Fig. 1 G), the biochemistry of chemomechanical energy conversion, and the regulation of the strength of cardiac muscle, especially by calcium ions, which mediate the process known as excitation-contraction coupling (Fig. 1 B). Intracellular calcium transients triggered by the electrical action potential are the key link between cardiac electrical excitation and contraction. By including intracellular calcium dynamics in multiscale models of the heart, we now have fully coupled electromechanical models of the heart. As such, these models are both multiscale and multiphysics.
Some of the important scientific and clinical problems being addressed by modern multiscale multiphysics cardiac mechanical and electromechanical models include: How do specific drugs or defects in single genes lead to substantially altered whole organ pumping? When a patient has disease, how reversible is it and how much of the dysfunction is due to an initial insult or genetic defect versus subsequent alterations in the natural history of the disease? How do the three-dimensional microstructure of the heart walls and the changes associated with diseases such as myocardial infarction and ventricular hypertrophy affect the mechanical pumping performance of the heart in vivo? Can we use models based on clinical data to design clinical trials or predict outcomes of therapy?
Hemodynamics, oxygen delivery, and metabolism
Cardiac wall mechanics is a problem of solid mechanics, but blood flow through the cardiac chambers and coronary vessels is a fluid mechanics problem. Modern techniques in computational fluid dynamics (CFD) have made detailed analysis of blood flow through the coronary arteries, inside the atrial and ventricular chambers, across the heart valves, and in the great vessels highly practical when coupled with accurate anatomic reconstructions from cardiac computed tomography (CT) or magnetic resonance imaging (MRI). Again, the governing physics come from conservation of momentum, mass, and energy. The most important governing equations are the Navier-Stokes equations (named after Claude-Louis Navier, 1785–1836, and Sir George Stokes, 1819–1903), which express Newton’s second law for fluid flows. What makes blood flow, especially through the heart and valves? An interesting CFD problem is that the walls are moving, and in the case of the cardiac chambers, the motion of the walls is driving the flow itself. This is where the development recently of robust algorithms for modeling fluid-structure interactions (FSI) has had a great impact. It is now possible to make patient-specific models of blood flows through the heart, valves, and vessels and to use them to predict the effects of surgical procedures. Nowhere is the potential clinical impact of this computational modeling technology more promising than in the development of better surgical procedures for infants and toddlers born with congenital heart defects (the most common class of birth defect).
Models of blood flow in the coronary circulation must take into account the mechanical effects on the coronary blood vessels of the squeezing of the heart walls during each heartbeat. In every other circulation in the body, blood flow is highest during systole when the blood pressure is highest. This is the phase in the heart when the stresses in the wall are greatest, thereby squeezing the coronary blood vessels and restricting systolic flow. This defines another especially challenging problem that couples heart wall mechanics with regional coronary blood flow. The demand for blood is driven by the need for oxygenation of the cardiac myocytes, which is in turn driven by the regional mechanical work demand on the muscle cells. Current efforts are linking models of wall mechanics, contraction, and energy metabolism to models of coronary blood flow and oxygen transport. Many cases of heart disease are associated with ischemia and metabolic stress that the need for such new models is pressing.
Future prospects
More than 50 years of cardiac computational modeling starting with Denis Noble’s 1961 cardiac cell model, together with a great deal of experimental testing and validation, have laid the foundations for exciting progress in understanding the integrative mechanisms of human heart diseases, improving diagnosis and therapy planning, and discovering new therapeutics. Some of the developments that we expect to see in the near future include patient-specific computational cardiac modeling, augmented medical imaging technologies, new drug target identification and repurposing of existing drugs, the discovery of new combinatorial drug therapies, and the development of models that span the longer timescales of cardiac development, disease progression, and aging. We will also continue to see the growth of modeling closely connected with basic research as we develop more comprehensive models of animal cardiac cells and disease and new models of cardiac progenitor cells derived from human stem cells.
Among the most mature multiscale cardiac systems models are models of ventricular electrophysiology and mechanics. Excellent progress in this field shows promise of clinical impact in the not-too-distant future. Currently, to help protect patients at risk of sudden cardiac death, implantable cardioverter defibrillators (ICDs) are being used. ICDs are life-saving but also expensive and not without risk of complications. The consequences of shocks inappropriately delivered by ICDs can be harmful, and to avoid missing those patients who might need an ICD, many are implanted but never needed. The latest multiscale models of ventricular fibrillation can be customized to patient anatomy and myocardial infarct morphology and show exciting promise to better discriminate those patients at highest risk from those who may not need an ICD without the need for lengthy invasive clinical testing in the cardiac electrophysiology lab. Similarly, pacemaker implantation in patients with dyssynchronous heart failure can improve cardiac pumping performance by resynchronizing the electrical activation of the left and right ventricles. A significant fraction of patients receiving this cardiac resynchronization therapy (CRT) do not improve significantly. New patient-specific models customized with clinical measurements from cardiac imaging, electrocardiography, and cardiac catheterization are showing the potential to better predict CRT outcomes and optimize the performance of the therapy in those who receive it. These patient-specific models have been made possible by improved four-dimensional medical imaging technologies such as cardiac CT and MRI. As modeling based on these images becomes easier and more common, we can expect to see some of these model simulations built into the computers that process scanner images to allow more detailed functional properties of the heart to be visualized. Drugs target specific molecules, but their biological impacts are dependent on effects that propagate through large cellular networks of molecular interactions. The short- and long-term therapeutic and adverse effects of these molecular perturbations are dependent on responses at the level of the tissue, organ, and whole body. Multi-scale systems models will help to identify adverse drug effects earlier in the expensive drug development pipeline, identify promising drug candidates that might not have been found with more traditional approaches, and find potential new applications for existing drugs alone or in combination.
Acknowledgments
The author is grateful for the contributions of his trainees and collaborators to his work on cardiac systems modeling and to the NIH for grant support for his research.
The author is cofounder and equity holder in Insilicomed, Inc., a licensee of computational modeling software developed in his laboratory.