Main Text
Every so often news about a viral outbreak goes viral and catches widespread public attention in the media. Human immunodeficiency virus (HIV), West Nile virus, avian influenza (bird flu), Ebola, Middle East respiratory virus, and Zika virus have each become, in a flurry of headlines and broadcasts and interviews, the focus of the media’s spotlight. And then, like other crises, they fade from view leaving the public with a new health concern to worry about but little knowledge of the actual factors involved in the problem. Behind the scenes, however, scientists are continually at work trying to understand and defend against these insidious infectious agents.
Viruses are perfect parasites. It has been known for decades that once a virus gets inside a cell, it hijacks the cellular processes to produce virally encoded protein that will replicate the virus’s genetic material. Viral mechanisms are capable of translocating proteins and genetic material from the cell and assembling them into new virus particles. Contemporary research has revealed specific mechanisms viruses use to get inside cells and infect them.
An individual viral particle, called a virion, is a far simpler structure than a bacterium. It has often been questioned whether a virus is alive. It is certainly not living in the everyday sense of the word. Virions consist of genetic material—DNA or RNA enclosed in a protein coating. Many viruses, called enveloped viruses, have an additional outer membrane that encloses the protein coat. This membrane envelope is material co-opted from the cell’s own membrane. As the new virion buds out from an infected host cell, it is wrapped by the cell’s bilayer membrane and carries with it any protein that happens to be embedded in the membrane at the budding site. Enveloped viruses are then free to begin a new cycle of infection by fusing their cell-derived envelope with the cellular membrane of an uninfected cell.
Some types of enveloped virus fuse directly to the cell’s outer (plasma) membrane, whereas others are engulfed whole by endocytosis or similar processes and then fuse their envelope with the membrane of the engulfing internal organelle (e.g., an endosome) to gain access to the interior of the cell. In either case, the genetic material of the virus has invaded the cell through the barrier of its membrane, and infection will inevitably follow (Fig. 1). Infection can be prevented if fusion of the viral envelope with the cell or endosomal membrane can be blocked. Similarly, if a vaccine can be directed against the viral fusion protein, infection can be prevented. Vaccines against the influenza virus, for example, target the fusion proteins of the virus.
Figure 1.
Viral entry pathways. Virus can fuse either directly to the plasma membrane (receptor-mediated fusion) or after being swallowed into an endosome. Which of these routes is followed depends on the type of virus. In fusion with the plasma membrane, the virus binds to a protein in the cell membrane. The function of this cellular protein (a receptor for the virus, shown in green) is perverted to induce a conformational change in the viral fusion protein, leading to fusion. For virus that is triggered within an endosome, the endosome’s acidic conditions induce fusion. In either case, the viral genome passes through a fusion pore into cytosol, and infection is initiated. To see this figure in color, go online.
Viral genetic material is relatively small, encoding only a few proteins. All enveloped viruses contain fusion proteins, which are the molecules responsible for fusing the envelope to a cellular membrane. These proteins are derived from the virion’s genetic sequence. The precise genetic material, the amino acid sequence, and details in structure of a fusion protein are unique for each type of virus. Consequently, broad-spectrum antiviral drugs do not exist, and specific vaccines and drugs typically need to be developed for each virus type. The viral surface of an individual virion contains multiple copies of its fusion protein. Influenza virus, for example, typically contains 500–1000 copies, whereas HIV contains only about a dozen copies (1, 2). A virion’s machinery is so efficient that each cell infected by even a single virion can produce about a million new virions. Because enveloped viruses use similar mechanisms for delivery of genetic material into cells, there may be ways to prevent infection before viral entry that would be effective for large numbers of different viruses.
The membrane that is the skin of a cell and an enveloped virion, and is the gateway of viral entry, consists of lipids and proteins. Lipids are roughly linear molecules of fat that are attached at one end to a water-soluble headgroup. Lipids provide the cohesion that keeps biological membranes intact. They spontaneously arrange themselves into a lipid bilayer because oily fat does not mix with water. The headgroups of one monolayer face an external aqueous solution, whereas the headgroups of the other monolayer face the interior of the cell. Integral membrane proteins, such as viral fusion proteins, are inserted into the bilayer and project out from the lipid surface into the external solution-like icebergs. Membranes are generally 50% lipids and 50% proteins by weight, but proteins are much heavier than lipids, and so there are about a hundred times more lipids than proteins in a membrane. Membranes are able to fuse to each other because they are fluid (3), and the lipids provide fluidity to the membrane.
Viruses initially stick to cell membranes through interactions unrelated to fusion proteins. The virus surfs along the fluid surface of the cell and eventually the viral fusion proteins bind to receptor molecules on the cell membrane (4). If only binding occurred, the two membranes would remain distinct. Fusion does not happen spontaneously because bilayers are stable. Fusion proteins do the work of prodding lipids from their initial bilayer configuration. These proteins cause discontinuities in the bilayers that induce the lipids of one membrane (e.g., the viral envelope) to connect with lipids of another (e.g., a cellular membrane), converting two bilayers into one.
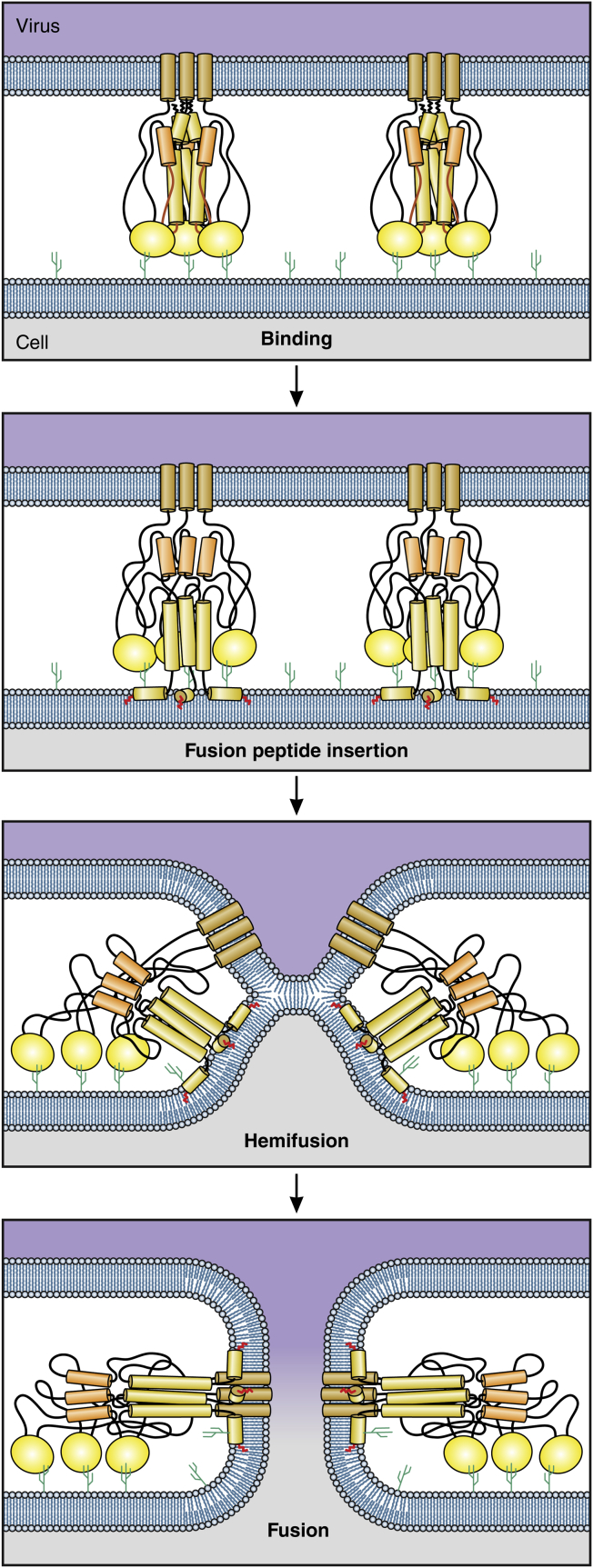
Fusion proceeds in two major steps (Fig. 2). First, the two monolayers from opposite membranes that touch each other merge, a process known as “hemifusion.” The two unmerged monolayers collapse onto each other to create a single bilayer, known as a hemifusion diaphragm, which continues to prevent the viral genome from entering cytosol. In the second step, the fusion proteins disrupt this single bilayer to create a pore that provides an aqueous pathway between the virus and the cell interior. It is through this fusion pore that the viral genome gains entry into a cell and begins infection.
Figure 2.
The steps of fusion. Virus binds to specific receptors (each illustrated as a small cactus) on a cell membrane. Initially, four monolayers (in blue) separate the two interior aqueous compartments. After fusion peptides insert into the target membrane, monolayers that face each other merge and clear from the merged region. The noncontacting monolayers bend into the cleared region and come into contact with each other, forming a new bilayer membrane known as a hemifusion diaphragm. At this point (hemifusion), only two monolayers separate the compartments. The fusion protein acts as a nutcracker to force the formation of a pore within the hemifusion diaphragm. This establishes continuity between the two aqueous compartments and fusion is complete. To see this figure in color, go online.
Hemifusion and pore formation appear to require comparable amounts of work, but the exact amount of energy needed for each step is not yet known (5). These energetic details may be important because the more work required to achieve a step, the easier it may be to pharmacologically block that step. These energies are supplied by the viral fusion proteins, which are essentially molecular machines. Some of their parts move long distances during the steps of fusion. Fusion proteins can be thought of as a complex assembly of wrenches, pliers, drills, and other mechanical tools.
Because fusion is not spontaneous, discontinuities must be transiently created within the bilayer that allows water to reach the fatty, oily interior of the membrane. Even a short-lived exposure of a small patch of the fatty interior to water is energetically costly. Similarly, creating a pore in a hemifusion diaphragm requires exposure of the bilayer interior to water (6). In contrast, pore enlargement needs no such exposure. Nevertheless, pore enlargement requires the most amount of work in the fusion process.
Energy is also needed because of another fundamental property of bilayer membranes. Though bilayers are fluid, they don’t entirely behave like water or oil, in that they do not assume the shape of their container. Biological membranes have shapes that are determined by their precise lipids and the proteins associated with them (7). Work is required to force membranes out of their spontaneous shape, which is the shape of lowest energy. The fusion pore that connects the virus and cell is roughly an hourglass shape (8). The wall of a fusion pore is a membrane with components that are a mixture of the two original membranes. An hourglass shape deviates significantly from the spontaneous shape of the initial membranes that constitute the pore. The greater the diameter of the pore, the greater is the area of the lining membrane, and so pore expansion is a highly energy consuming process. Viral genetic material, the genome, is rather large, on the order of ∼100 nm. The initial fusion pore is only ∼1 nm, so considerably more membrane must line a pore as it enlarges to a size sufficient to allow passage of a viral genome from a virus to a cell interior. In fact, it appears that more energy is required for pore expansion than for hemifusion or pore formation.
All viral fusion proteins contain a greasy segment of amino acids, referred to as a fusion peptide or fusion loop. Soon after activation of the fusion protein, the fusion peptide inserts into the target membrane (either plasma or endosomal). At this point, two extended segments of amino acids are anchored to the membranes: the fusion peptides in the target membrane and the membrane-spanning domains of the fusion proteins in the viral envelope (Fig. 2). The fusion proteins continue to reconfigure, causing the two membrane-anchored domains to come toward each other. This pulls the viral envelope and cellular membrane closely together (9). The fusion proteins exert additional forces, but exactly what these forces are and how they promote fusion remains unknown.
An initial step in a cell’s digestive system is to internalize extracellular materials through engulfment by endosomes. A virion engulfed into an endosome is like a Trojan horse, because the cell perceives the virus particle as food. Endosomes become increasingly acidified as they move from the cell surface further into the cell’s interior. Fusion of viruses within endosomes depends critically on the acidic environment. By breaking molecular bonds, acid triggers the conformational changes in the fusion protein that lead to the sequential steps of membrane fusion.
The hemifusion diaphragm is a bilayer membrane that is unusual in that each of its lipid monolayers is derived from different membranes, and it does not contain any membrane-spanning proteins (10). Several copies of the fusion protein within a virus are required to induce both hemifusion and pore formation. During hemifusion, the proteins form a ring just outside the diaphragm and act cooperatively to create stresses that lead to a local rupture in the diaphragm, thereby creating the initial fusion pore. The universality of this mechanism is remarkable when one considers that the primary amino acid sequences and structures of fusion proteins are quite diverse.
Influenza, HIV, and Ebola are enveloped viruses of significant public health concern. Each virus encodes a unique fusion protein: hemagglutinin (HA) for influenza, envelope glycoproteins for HIV (Env), and glycoprotein for Ebola (GP).
The earliest descriptions of an illness that was likely influenza were written in the 1500s and were called “catarrhal fever” (11). The flu pandemic of 1918 resulted in the deaths of some 20 million people and arguably accelerated the end of World War I (12). Flu pandemics have continued to occur periodically, as they did in 1947, 1957, 1968, and 2009, but were far less deadly.
Influenza virus is not free to infect other cells upon budding because HA binding to specific sugars, sialic acids, that protrude from cell surfaces prevents a virus from freeing itself from the cell. Another envelope protein, neuraminidase (NA), cleaves sialic acids off the cell, setting the influenza free. Drugs that are NA inhibitors, such as the well-known Tamiflu (oseltamivir), stop further infection within an individual by eliminating the cleavage of sialic acids (13).
Research efforts for influenza, HIV, and Ebola virus have focused on targeting their fusion proteins. But particular properties of the viruses and their proteins have hindered the successful development of vaccines that protect against infection.
Standard vaccines against envelope viruses prime the immune system to generate antibodies (Abs) against the envelope proteins. In the case of influenza, Ab binding is mainly to HA, and secondarily to NA. Abs bind to exposed outer portions of envelope proteins and are large, thereby hindering close engagement of the virus with a cell membrane. Some antigenic sites surround an indented pocket within the surface of HA that is responsible for binding sialic acids on cell surfaces. Abs thus block the binding of HA to plasma membranes, eliminating the membrane fusion that leads to infection. HA readily mutates, and although the accumulated individual mutations lead to only small changes in the conformation of HA, these mutations greatly reduce binding of Abs to HA. Hence, a new vaccine must be developed each year (14).
Influenza presents another problem: its genome is not one continuous strand of RNA, like most viruses, but is segmented into multiple strands. Segmentation allows the genes for HA and NA to reassort: the RNA strands of different flu viruses—such as genes from an avian flu virus and a mammalian flu virus—combine to make what is essentially a new virus. Reassorted viruses are described in terms of HA and NA types and are termed H1N1, H3N2, H5N1, and so forth. Some reassortments cause periodic influenza pandemics that are characterized by an unusually large number of severe, and sometimes fatal, infections (15).
HIV-1 is clinically, to date, the most important retrovirus. Retroviruses transcribe RNA into DNA in a process called reverse transcription, and the viral DNA is incorporated into the genome of the host cell. HIV is a relatively recent emerging virus, appearing in the last 70 years or so. It has independently jumped to humans at least four times, probably due to the bush meat trade of gorillas and chimpanzees, and from chimps kept as pets (16). Currently, ∼35 million people are infected, with about two-thirds of them living in Sub-Saharan Africa. Viruses not only cause diseases, but have also been important in evolution. Retroviruses can move large gene segments from one organism to another, and some 100,000 pieces of retroviral DNA make up ∼8% of the human genome (17).
The traditional approach of using attenuated or inactivated virus, and by extension, envelope proteins, as vaccines has been ineffective against HIV-1 for a number of reasons. The fidelity of the reverse transcriptase of HIV-1 is low and therefore mutations in the viral protein occur frequently. As a result, HIV-1 Env mutates so rapidly that it quickly evades a static vaccine. Furthermore, Env is highly glycosylated, effectively sugarcoating the exposed portion of the protein, and Abs do not bind well to sugars. There is a small unglycosylated region on the surface of Env, and efforts were directed against this bald spot but did not lead to clinically effective approaches. Many nontraditional vaccine approaches have been developed and tested and these efforts continue, but none have yet been sufficiently successful. Modern biology and public health measures have combined to develop positive methods to prevent and treat the acquired immunodeficiency syndrome.
Antiretroviral therapies have largely eliminated the progression of viral infection to AIDS in individuals for whom these therapies have been available. Relatively soon after HIV-1 was identified, blood supplies were able to be accurately screened for HIV contamination. This was achieved only because prior advancements in the biological sciences allowed the development of new diagnostic methods that were sensitive enough to detect HIV. More recently it has been shown that HIV infection can be eliminated from the body: the Berlin Patient infected with HIV (and suffering from leukemia) received a stem cell transplant and was thereafter free of the virus (18).
The recent Ebola outbreak was caused by the deadliest of the three types of Ebola virus strains known to infect humans, with a fatality rate exceeding 50%. It typically takes ∼4–10 days from the time of infection to the appearance of symptoms, but symptoms can manifest in as little as 2 days or as long as 3 weeks. It appears that with Ebola, unlike influenza, infected individuals do not become contagious until they exhibit symptoms. Trial vaccines using virus inactivated by traditional methods have proven unsuccessful, but viruses using recombinant technologies are showing considerable promise. Several other approaches may also be effective, including a cocktail of humanized murine monoclonal Abs, which have been shown to be statistically effective in protecting nonhuman primates.
Acidification of endosomes causes Ebola fusion in an unusual manner. Influenza HA, HIV-1 Env, and Ebola GP are cleaved into two subunits before viral-cell binding. This cleavage confers to HA and Env the full ability to induce fusion. In contrast, Ebola GP must be cleaved at an additional site to cause fusion. This cleavage occurs within endosomes by a protease (cathepsin) that is effective at low pH (19). A conformational change ensues, allowing Ebola GP to bind to an endosomal receptor, Niemann-Pick type C1. Binding activates GP, and a merger between the viral and endosomal membranes then proceeds. The identification of Niemann-Pick type C1 as a receptor opens up a new potential target for a small molecule drug to block binding and prevent infection (20).
The most reliable way to prevent infection caused by any virus is to eliminate entry in the first place. Intellectual and technological progress has been great, but recurrent viral outbreaks highlight the need for more innovative approaches. In addition to the proteins responsible for viral entry, many other targets are being explored, including genetic variations that increase susceptibility to infection, proteins that bind to viral proteins, and host immunity proteins. Genomic and proteomic analysis of cellular factors and their interactions, manipulation of experimental animals, live cell and molecular imaging, and analysis and integration of protein and gene data sets will identify host factors that viruses exploit in their life cycle. Because viruses make use of cellular machinery—and invariably do so in a streamlined and robust manner—future viral studies will provide new understandings that will apply not only to virally induced diseases but to other diseases as well. Biophysics has been an integral part of understanding viral entry mechanisms, which have brought new insights and discoveries that just a few years ago could not have been imagined.
Acknowledgments
I thank Patrick Lane, ScEYEnce Studios, for producing the figures.
This work was supported by National Institutes of Health R01 GM101539.
References
- 1.Harris A., Cardone G., Steven A.C. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. USA. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu P., Liu J., Roux K.H. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 3.Chernomordik L.V., Zimmerberg J., Kozlov M.M. Membranes of the world unite! J. Cell Biol. 2006;175:201–207. doi: 10.1083/jcb.200607083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehmann M.J., Sherer N.M., Mothes W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J. Cell Biol. 2005;170:317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen F.S., Melikyan G.B. The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J. Membr. Biol. 2004;199:1–14. doi: 10.1007/s00232-004-0669-8. [DOI] [PubMed] [Google Scholar]
- 6.Kuzmin P.I., Zimmerberg J., Cohen F.S. A quantitative model for membrane fusion based on low-energy intermediates. Proc. Natl. Acad. Sci. USA. 2001;98:7235–7240. doi: 10.1073/pnas.121191898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozlov M.M., Campelo F., McMahon H.T. Mechanisms shaping cell membranes. Curr. Opin. Cell Biol. 2014;29:53–60. doi: 10.1016/j.ceb.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryham R.J., Ward M.A., Cohen F.S. Teardrop shapes minimize bending energy of fusion pores connecting planar bilayers. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2013;88:062701. doi: 10.1103/PhysRevE.88.062701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White J.M., Delos S.E., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melikyan G.B., White J.M., Cohen F.S. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J. Cell Biol. 1995;131:679–691. doi: 10.1083/jcb.131.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson T. Sydenham Society; London, UK: 1852. Annuals of Influenza or Epidemic Catarrhal Fever in Great Britain from 1510 to 1837. [Google Scholar]
- 12.Barry J.M. Penguin Books; London, UK: 2004. The Great Influenza. [Google Scholar]
- 13.Kim J.H., Resende R., Withers S.G. Mechanism-based covalent neuraminidase inhibitors with broad-spectrum influenza antiviral activity. Science. 2013;340:71–75. doi: 10.1126/science.1232552. [DOI] [PubMed] [Google Scholar]
- 14.Nelson M.I., Holmes E.C. The evolution of epidemic influenza. Nat. Rev. Genet. 2007;8:196–205. doi: 10.1038/nrg2053. [DOI] [PubMed] [Google Scholar]
- 15.Fuller T.L., Gilbert M., Smith T.B. Predicting hotspots for influenza virus reassortment. Emerg. Infect. Dis. 2013;19:581–588. doi: 10.3201/eid1904.120903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharp P.M., Hahn B.H. The evolution of HIV-1 and the origin of AIDS. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010;365:2487–2494. doi: 10.1098/rstb.2010.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zahn J., Kaplan M.H., Contreras-Galindo R. Expansion of a novel endogenous retrovirus throughout the pericentromeres of modern humans. Genome Biol. 2015;16:74–98. doi: 10.1186/s13059-015-0641-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rennie S., Siedner M., Moodley K. The ethics of talking about ‘HIV cure’. BMC Med. Ethics. 2015;16:18–25. doi: 10.1186/s12910-015-0013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandran K., Sullivan N.J., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Côté M., Misasi J., Cunningham J. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]