Main Text
Proteins form the molecular scaffolding of life and are essential to catalyzing the chemical reactions that sustain living systems. These characteristics have led us to think that proteins function only when folded into the right structure. The central dogma of molecular biology states that genetic information encoded in the DNA sequence is transcribed into messenger RNA and then translated into a sequence of amino acids, which folds into a protein. The mechanisms that govern how a linear sequence of amino acids folds into the correct three-dimensional structure are still not well understood. Biophysical techniques have been indispensable to unraveling how protein structures fold, and many of the major factors that determine how the amino-acid sequence codes for the folded protein structure are beginning to be understood.
The genomic era that began at the end of the 20th century gave scientists access to complete genome sequences. Scientists observed that some of the predicted protein sequences derived from genomes were not expected to fold into normal globular protein structures (1). At the same time, experimental studies began to uncover examples of important protein molecules and domains that were incompletely structured or completely disordered in solution yet remained perfectly functional (2, 3) (Fig. 1). In the following years, an explosion of experimental data and genome annotation studies mapped the extent of this intrinsic disorder phenomenon and explored the possible biological reasons for its widespread occurrence. Answers to the question of why a particular domain would need to be unstructured are as varied as the systems where such domains are found.
Figure 1.
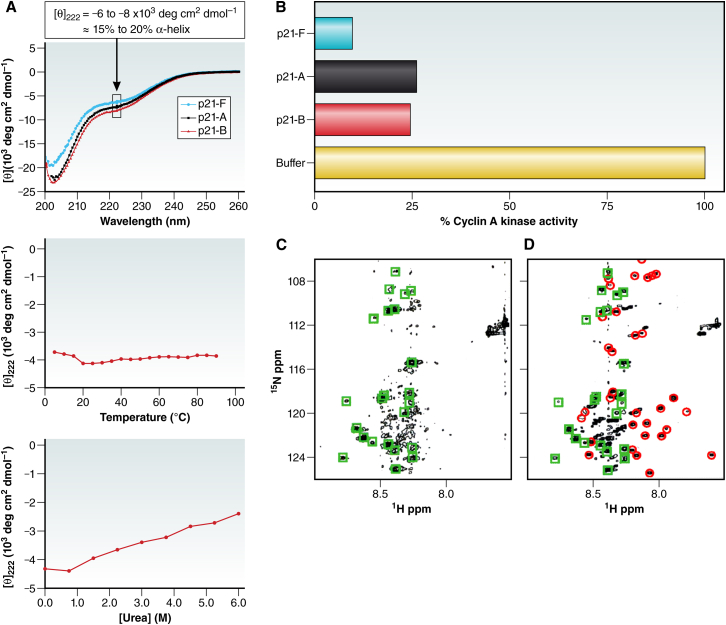
(A) The cyclin-dependent kinase inhibitor p21 does not form recognizable structure in solution, indicated by the far-UV circular dichroism spectrum of constructs of various lengths (top panel), by the absence of temperature-induced (middle panel), or urea-induced (bottom panel) unfolding transitions. (B) All of the p21 constructs are active in the inhibition of cyclin-A kinase activity. (C and D) 1H-15N HSQC NMR spectra of free p21 (C) and p21 bound to cyclin-dependent kinase-2. (D) Squares indicate cross peaks present in the same place in the two spectra, and circles denote new cross peaks at positions that indicate that folding has occurred (adapted from Kriwacki et al. (2) with permission, © 1996 National Academy of Sciences, USA). To see this figure in color, go online.
One of the hallmarks of intrinsically disordered proteins (IDPs) is a marked bias in the amino-acid composition, including a relatively low proportion of hydrophobic and aromatic residues, and a relatively high proportion of charged and polar residues (Fig. 2). The high frequency of small hydrophilic amino acids renders these sequences as unlikely candidates for membrane or scaffolding proteins. Yet many of the proteins identified in surveys, as well as in concurrent NMR experiments, showed that these proteins were involved in important cellular processes such as control of the cell cycle, transcriptional activation, and signaling (4, 5), and they frequently interacted with or functioned as central hubs in protein interaction networks (6). The amounts of various IDPs in the cell are tightly regulated to ensure fidelity in signaling. Altered abundance of IDPs is associated with disease (7).
Figure 2.
Illustration of the sequence bias found in disordered sequences. The amino-acid sequence of a portion of the multidomain transcriptional coactivator CBP is classified by amino-acid type: green, small hydrophilic amino acids (G, A, S, T, N, Q, P); yellow, hydrophobic amino acids (V, L, I, M, F, Y, W); red, acidic amino acids (D, E); blue, basic amino acids (K, R, H); pink, cysteine (C). The sequences of two folded domains, TAZ1 (blue) and KIX (yellow), show much greater sequence diversity than the disordered flanking and linker domains, which are predominantly green, indicating a heavy bias toward small hydrophilic amino acids (adapted from Dyson and Wright (4)).
Disordered sequences can also be found in proteins that contain ordered, structured domains, and these disordered sequences are termed intrinsically disordered regions (IDRs). Some IDRs function as linkers between interaction domains (Fig. 2), and in some cases, their properties as polymers contribute to their function (8). Many IDRs contain sequence elements that interact with partners and frequently fold upon binding. For example, the intrinsically disordered interaction domain of the transcription factor STAT2 folds upon binding to its partner, the TAZ1 domain of CREB-binding protein (CBP) (Fig. 3) (9). Backbone flexibility of an IDR in its free state enables it to bind to multiple targets, which increases its potential repertoire of responses, as exemplified in the binding of the hypoxia-inducible factor HIF-1α. The transactivation domain of HIF-1α binds to its partner TAZ1 as a helix (10), whereas the same HIF-1α sequence binds to the hydroxylating enzyme FIH as a β-strand (Fig. 4) (11).
Figure 3.
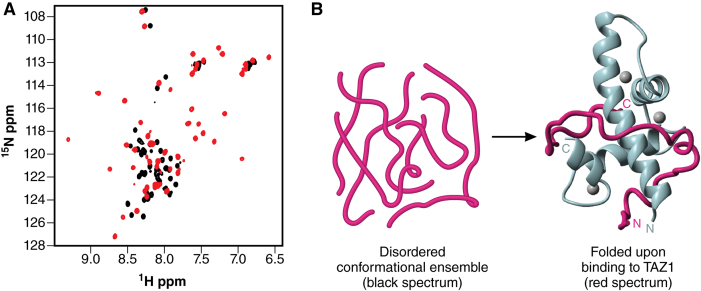
Coupled folding and binding of the transcription factor STAT2 on the TAZ1 domain of CBP. (A) Disorder in the free STAT2 is shown in the small resonance dispersion in the 1H dimension of the black 1H-15N HSQC spectrum. The structured nature of the bound STAT2 is shown by the increased 1H dispersion of the gray spectrum. (B) Schematic diagram illustrating the conversion of the disordered conformational ensemble of free STAT2 into a structured form on the TAZ1 (adapted from Wojciak et al. (9)). To see this figure in color, go online.
Figure 4.
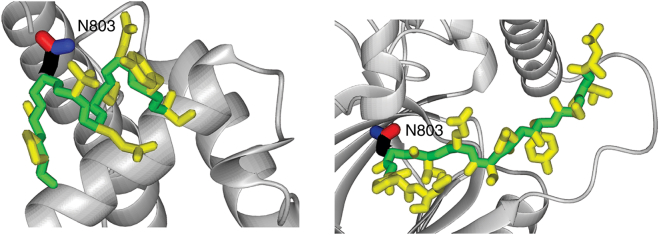
Structural differences between the transactivation domain of HIF-1α bound to the TAZ1 domain of CBP (10) (left panel), where the sequence containing the regulatory asparagines appears as an α-helix, and the same sequence bound to the hydroxylating enzyme FIH (11) (right panel), where it appears as a β-strand. To see this figure in color, go online.
Disorder makes IDR sequences accessible to posttranslational modification and IDRs are rich in modification sites. IDRs facilitate efficient protein-protein interactions using only a small number of residues. A folded protein would need to be much larger to provide an interaction surface area equivalent to that seen with IDRs, as illustrated in Fig. 4. This efficiency is important in signaling, as it translates into the ability to bind with high specificity but only modest affinity, enabling dissociation of the IDR after signaling is complete. Signaling can be turned off by competition between IDRs for a particular physiological partner, mediated by slightly different binding sites (Fig. 5). The reaction of cells to hypoxia (low oxygen) is a good example of this phenomenon (Fig. 6). Under normal conditions, the HIF protein is synthesized in the cell, but is degraded upon hydroxylation of two prolines. Interaction with the transcriptional coactivator CBP is further interdicted by the hydroxylation of an asparagine in the C-terminal activation domain (CTAD). Under hypoxic conditions, the hydroxylation reactions no longer occur, so the protein is stable to degradation and the CTAD can interact with the TAZ1 domain of CBP, leading to transcription of hypoxia-response genes such as VEGF, which promotes growth of blood vessels (12). Such a response is dangerous if not constrained, however, and the signal must be turned off before adverse physiological effects occur. One of the genes transcribed in response to the hypoxia signal encodes an inhibitor, CITED2, which removes the HIF CTAD from the CBP TAZ1 domain, turning off the hypoxia response. Several anti-cancer treatments include inhibition of the hypoxia response, which prevents vascularization of tumors, thus restricting their growth (12).
Figure 5.
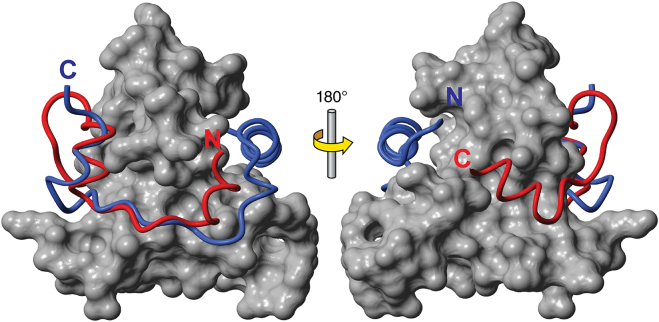
Comparison of the structures of ligands bound to the CBP TAZ1 domain. The surface of TAZ1 (almost identical in both complexes) is shown in gray, with the backbone of HIF-1α-C-terminal activation domain (10) in red (labeled N in the left image and C in the right image) and the CITED2-trans-activation domain (19) in blue (labeled C in the left image and N in the right image). (Left and right panels) 180° rotation around the vertical axis in the plane of the page (adapted from Wojciak et al. (9)). To see this figure in color, go online.
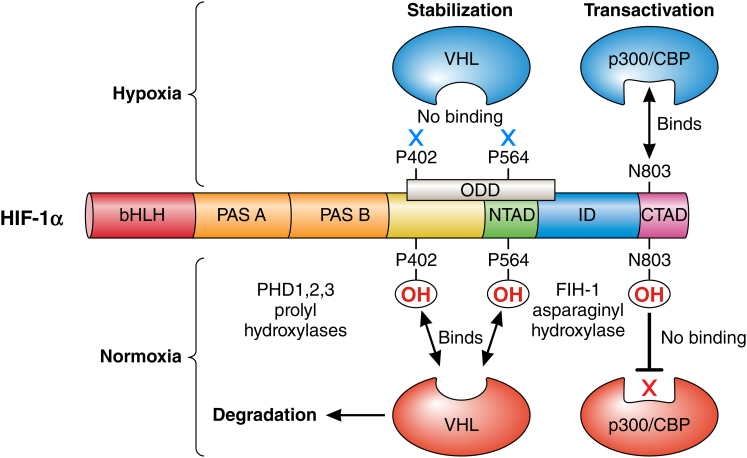
Figure 6.
Schematic diagram showing the regulation of the HIF-1α transcription factor under normal oxygenation conditions (bottom), where proline hydroxylation in the central oxygen-dependent degradation domain recruits the von Hippel-Lindau factor, leading to degradation, and asparagine hydroxylation in the C-terminal activation domain lowers the affinity for transcriptional activators. In hypoxic conditions (top), neither the prolines nor the asparagines are hydroxylated, with the result that HIF-1α is stabilized and binds to CBP/p300 to promote transcription of hypoxia-response genes. (Reproduced from Dyson (20) with permission.) To see this figure in color, go online.
IDPs offer novel advantages as therapeutic targets. Their central role in key cellular signaling pathways (5), their frequent association with disease (7), and the reversible nature of their intermolecular interactions, by which they bind with high specificity but modest affinity, makes them extremely attractive targets for small molecule drugs or stapled peptide mimetics (13, 14). Indeed, many viruses hijack the host cell by using their own viral IDPs, e.g., the adenovirus E1A or papillomavirus E7 oncoproteins (15, 16), to compete with cellular IDPs for binding to key regulatory proteins (17). IDPs commonly bind to concave grooves in the surface of their target proteins; the interactions are predominantly hydrophobic and the fit is more intimate than to their globular protein counterparts. Finally, it may prove possible to design drugs targeted against the IDP itself, rather than its globular target.
Disordered regions of proteins provide a uniquely versatile and useful toolbox for reactions in the cell. Interestingly, the majority of IDRs so far characterized are from eukaryotic systems, in which they are intimately involved with the signaling and physiological control required for multicellular organisms. IDRs are found in prokaryotes, but they tend to be associated with unusual functions in particular bacteria, for example the toxin-antitoxin systems of phage-infected Escherichia coli (18).
Protein molecules are rarely, if ever, completely rigid. Dynamic motions of backbone and side chains, independent of the tumbling of the whole molecule, can be estimated by various spectroscopic means, and are frequently associated with the function of enzymes. Disordered regions and fully disordered proteins can be thought of as a continuation of this characteristic, by which functional disorder, an extreme form of local protein dynamics, is functional through the particular advantages bestowed by the disordered state. The continuum between rigidity and complete disorder provides an expanded proteome, allowing proteins to perform multiple tasks through interactions with different partners or under different conditions. Disorder occupies an important biological niche that promises to yield important new insights into how biological systems operate.
Acknowledgments
This work was supported by grant Nos. GM071862 and GM113251 from the National Institutes of Health.
References
- 1.Romero P., Obradovic Z., Dunker A.K. Identifying disordered regions in proteins from amino acid sequences. Proc. IEEE. 1997;1:90–95. [Google Scholar]
- 2.Kriwacki R.W., Hengst L., Wright P.E. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc. Natl. Acad. Sci. USA. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daughdrill G.W., Chadsey M.S., Dahlquist F.W. The C-terminal half of the anti-σ factor, FlgM, becomes structured when bound to its target, σ28. Nat. Struct. Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- 4.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 5.Wright P.E., Dyson H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunker A.K., Cortese M.S., Uversky V.N. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 7.Babu M.M., van der Lee R., Gsponer J. Intrinsically disordered proteins: regulation and disease. Curr. Opin. Struct. Biol. 2011;21:432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 8.Tompa P. Chapman & Hall; Boca Raton, FL: 2010. Structure and Function of Intrinsically Disordered Proteins. [Google Scholar]
- 9.Wojciak J.M., Martinez-Yamout M.A., Wright P.E. Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains. EMBO J. 2009;28:948–958. doi: 10.1038/emboj.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dames S.A., Martinez-Yamout M., Wright P.E. Structural basis for Hif-1 α/CBP recognition in the cellular hypoxic response. Proc. Natl. Acad. Sci. USA. 2002;99:5271–5276. doi: 10.1073/pnas.082121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elkins J.M., Hewitson K.S., Schofield C.J. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1 α. J. Biol. Chem. 2003;278:1802–1806. doi: 10.1074/jbc.C200644200. [DOI] [PubMed] [Google Scholar]
- 12.Krock B.L., Skuli N., Simon M.C. Hypoxia-induced angiogenesis: good and evil. Genes Cancer. 2011;2:1117–1133. doi: 10.1177/1947601911423654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vassilev L.T., Vu B.T., Liu E.A. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 14.Chang Y.S., Graves B., Sawyer T.K. Stapled α-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl. Acad. Sci. USA. 2013;110:E3445–E3454. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreon J.C., Martinez-Yamout M.A., Wright P.E. Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein. Proc. Natl. Acad. Sci. USA. 2009;106:13260–13265. doi: 10.1073/pnas.0906770106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansma A.L., Martinez-Yamout M.A., Wright P.E. The high-risk HPV16 E7 oncoprotein mediates interaction between the transcriptional coactivator CBP and the retinoblastoma protein pRb. J. Mol. Biol. 2014;426:4030–4048. doi: 10.1016/j.jmb.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davey N.E., Travé G., Gibson T.J. How viruses hijack cell regulation. Trends Biochem. Sci. 2011;36:159–169. doi: 10.1016/j.tibs.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Pino A., Balasubramanian S., Loris R. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell. 2010;142:101–111. doi: 10.1016/j.cell.2010.05.039. [DOI] [PubMed] [Google Scholar]
- 19.De Guzman R.N., Martinez-Yamout M.A., Wright P.E. Interaction of the TAZ1 domain of the CREB-binding protein with the activation domain of CITED2: regulation by competition between intrinsically unstructured ligands for non-identical binding sites. J. Biol. Chem. 2004;279:3042–3049. doi: 10.1074/jbc.M310348200. [DOI] [PubMed] [Google Scholar]
- 20.Dyson H.J. Expanding the proteome: disordered and alternatively folded proteins. Q. Rev. Biophys. 2011;44:467–518. doi: 10.1017/S0033583511000060. [DOI] [PMC free article] [PubMed] [Google Scholar]