Summary
Primordial germ cell (PGC) specification occurs early in development. PGC specifiers have been identified in Drosophila, mouse, and human but remained elusive in most animals. Here we identify the RNA-binding protein Dnd as a critical PGC specifier in the medaka fish (Oryzias latipes). Dnd depletion specifically abolished PGCs, and its overexpression boosted PGCs. We established a single-cell culture procedure enabling lineage tracing in vitro. We show that individual blastomeres from cleavage embryos at the 32- and 64-cell stages are capable of PGC production in culture. Importantly, Dnd overexpression increases PGCs via increasing PGC precursors. Strikingly, dnd RNA forms prominent particles that segregate asymmetrically. Dnd concentrates in germ plasm and stabilizes germ plasm RNA. Therefore, Dnd is a critical specifier of fish PGCs and utilizes particle partition as a previously unidentified mechanism for asymmetric segregation. These findings offer insights into PGC specification and manipulation in medaka as a lower vertebrate model.
Keywords: asymmetric segregation: dnd, lineage tracing, primordial germ cell, RNA particle, single-cell culture
Highlights
-
•
The medaka RNA-binding protein Dnd specifies primordial germ cells
-
•
Cells from medaka cleavage embryos can be singly cultured for lineage tracing
-
•
The dnd RNA forms particles as a new mechanism for asymmetric segregation
-
•
These findings offer new insights into PGC specification and manipulation
Primordial germ cells are precursors for eggs and sperm. Hong, Song, and colleagues identify an RNA-binding protein called Dnd that specifies primordial germ cells in the medaka fish and a new mechanism that controls asymmetric segregation by particle partition. These findings offer new insights into PGC specification and manipulation in fish.
Introduction
Primordial germ cells (PGCs) are germ stem cells capable of generating precursors of eggs and sperm and thus offer the basis for reproduction and fertility (Ko et al., 2010, Lin, 2007, Lin, 2012). PGC specification demarcates the soma-germline separation and thus sets a balance between individual life and species continuity. PGCs form early in development and migrate into the developing gonad (Wylie, 1999). In adult animals, gonadal germ cells undergo meiosis and produce eggs and sperm. PGC development has been extensively studied in several model organisms including Drosophila, Caenorhabditis elegans, and Xenopus (Houston and King, 2000), zebrafish (Lin et al., 1992, Raz, 2003), and mouse (Hayashi et al., 2011, Ohinata et al., 2005, Saitou et al., 2002, Tam and Zhou, 1996). In these model invertebrates and lower vertebrates, PGCs are cell autonomously preformed by maternally supplied germ plasm, a membrane-less organelle composed mainly of RNA-binding proteins and their mRNAs (Houston and King, 2000, Raz, 2003). In mouse, PGCs are epigenetically induced by cell-cell interactions in the absence of germ plasm (Tam and Zhou, 1996), where signaling molecules such as bone morphogenetic factor 4 play a critical role (Ying et al., 2001).
Dozens of genes essential for PGC development are known in the model invertebrates and lower vertebrates (Houston and King, 2000); clearly defined specifiers of the PGC fate has, however, been limited to Drosophila. In this organism, oskar acts as the PGC specifier, as it is necessary for PGC formation and more importantly, sufficient for ectopic PGC induction in a dose-dependent manner (Ephrussi and Lehmann, 1992). However, osk is restricted to certain insects. An evolutionarily conserved gene, piwi, is also necessary for PGC formation and sufficient for tripling the PGC number but incapable of ectopic PGC induction (Megosh et al., 2006). In vertebrates, piwi is dispensable for PGC specification but essential for subsequent PGC development, such as spermatogenesis in mouse (Deng and Lin, 2002), germ cell maintenance in zebrafish (Houwing et al., 2007), and PGC migration in medaka (Li et al., 2012). In mice, blimp1 (encoded by prdm1) and prdm14 are transcriptionally induced by BMP4 in the epiblast at E6.25, which together with tfap2c constitute a tripartite genetic network to induce the PGC fate in vivo (Magnusdottir et al., 2013, Ohinata et al., 2005) and in vitro from embryonic stem (ES) cells (Magnusdottir et al., 2013, Nakaki et al., 2013). In human, SOX17 has most recently been identified as a critical specifier of PGCs in ES cells (Irie et al., 2015). Accumulated data from Drosophila, mouse, and human suggest that PGC specifiers show remarkable diversity and do not follow the evolutionary history, although many genes involved in subsequent germ cell development are highly conserved across animal phyla.
A vertebrate-specific germ gene, dead end (dnd), was first identified in zebrafish as a germ plasm component encoding an RNA-binding protein crucial for PGC migration and survival (Tzung et al., 2015, Weidinger et al., 2003). Mouse dnd mutations do not prevent PGC formation (Youngren et al., 2005). The medaka fish (Oryzias latipes) is an excellent model for studying vertebrate development (Wittbrodt et al., 2002), stem cells (Centanin et al., 2011, Hong et al., 1996, Hong et al., 1998b), germ cells, and reproductive technologies. This fish has haploid ES cells capable of whole-animal production by semicloning (Yi et al., 2009), male germ stem cells capable of test-tube production (Hong et al., 2004a), and transgenic lines for PGC visualization (Li et al., 2009, Tanaka et al., 2001). Unusually, maternal germ plasm components distribute widely in medaka (Shinomiya et al., 2000, Xu et al., 2009), rather than locally as in zebrafish (Weidinger et al., 2003, Yoon et al., 1997). This suggests the presence of an unknown key factor that determines the PGC fate. Here we identify Dnd as such a critical PGC specifier in medaka. Interestingly, dnd RNA uses particle formation and partition as a mechanism for asymmetric segregation and cell fate decision in early developing embryos. These results provide insights into our understanding of PGC formation and manipulation in medaka as a lower vertebrate model.
Results
Dnd Dosage Determines the PGC Number In Vivo
Targeted dnd disruption in medaka embryos led to normal survival and development to adulthood (Wang and Hong, 2014). The dnd mutant medaka adults are sterile and not suitable for studying its role in PGC development during embryogenesis. Conditional knockout of genes essential for early development such as PGC formation is not yet available in fish. We thus adopted direct embryo microinjection of dnd-targeting morpholino oligos for gene depletion and dnd mRNAs for gene overexpression. Two morpholino oligos were used for dnd depletion: MOdnd targets the medaka dnd mRNA and inhibits its translation, and MOddm is a mutant derivative of MOdnd by introducing four mismatches (Figure S1A). For overexpression, mRNAs dnd:ch and dndΔ1:ch were synthesized from pCSdnd:chDD and pCSdndΔ1:chDD (Figure S1B); the former encodes a cherry fluorescent protein-tagged wild-type Dnd and the latter a tagged deletion mutant Dnd. The morpholino oligos and mRNAs were microinjected alone or in combination into one-cell embryos of transgenic medaka NgVg expressing GFP specifically in PGCs (Hong et al., 2010, Li et al., 2009).
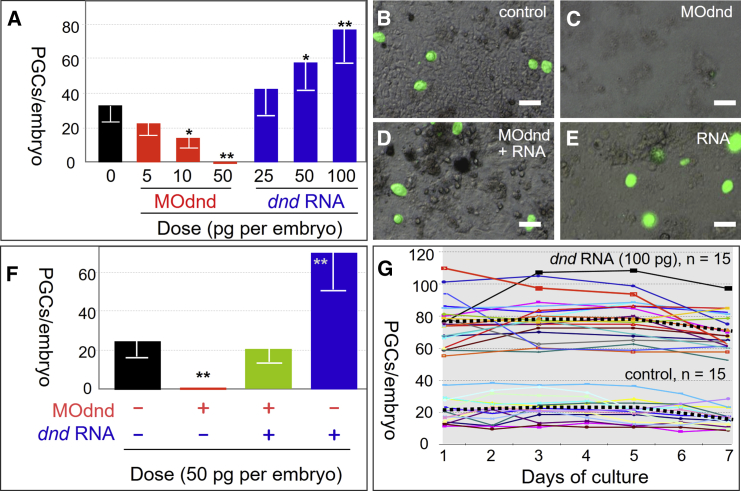
A medaka embryo at stages 18–22 has ∼32 PGCs that are recognizable by GFP expression (Figure 1A). Injection of mismatch-containing MOddm had no effect on the PGC number (Figure S2A). Remarkably, dnd depletion by injection with 50–100 pg of MOdnd caused the complete absence of PGCs in all (n = 333) manipulated embryos (Figures 1A and S2B). When MOdnd was coinjected with dnd:ch RNA, the PGC number was rescued (Figure S2C), whereas dndΔ1:ch mRNA did not rescue (Figure S2D). Moreover, injection with 50 pg of dnd:ch RNA alone was sufficient to increase the PGC number (Figure S2E). Injection of dnd:ch RNA at 100 pg considerably boosted the PGC number, so that a significant number of PGCs were located in ectopic sites as well as numerous PGCs in the gonad (Figure S2F). Clearly, altering dnd expression by injection of either MOdnd or dnd:ch RNA altered the PGC number, ranging from the complete absence to an increase by ∼2-fold (Figure 1A). The effect of MOdnd is specific, as it does not affect somatic development by morphological criteria, and dnd RNA but not its deletion mutant is capable of phenotypic rescue. Taken together, dnd is essential for PGC development and its dosage determines the PGC number in vivo.
Figure 1.
Dnd Dosage Determines the PGC Number
(A) Number of PGCs in embryos. Data are means ± SD (bars) of 12–21 embryos at stage 20.
(B–E) PGCs from midblastula cells at 2 days of culture. Scale bars, 20 μm.
(F) Number of PGCs at day 2 post culture from ≥12 embryos. ∗Significant difference (p ≤ 0.05); ∗∗very significant difference (p ≤ 0.01) compared with non-injected control.
(G) Time course of PGC numbers from individual blastula embryos in culture.
Dnd Depletion Does Not Cause PGC Death
To determine whether the absence of PGCs observed from stage 21 onward was due to the absence of PGC formation or loss of established PGCs, we examined NgVg embryos earlier at stage 15 when PGCs are unambiguously visible by the GFP signal. Cell counting in more than 12 embryos revealed that the average PGC number was 22.3, 9.7, 0., and 49.8 in the MOddm-injected control embryos, embryos injected with 20 pg and 50 pg of MOdd, and embryos injected with 50 pg of dnd:ch RNA, respectively (Figures S3A–S3E). These results indicate that a PGC decrease and loss in MOdd-injected embryos occurred as early as at stage 15, arguing against PGC loss between stage 15 and subsequent stages. To rule out the possible loss of PGCs at even earlier stages, we examined cell death by acridine orange staining (Li et al., 2009). No difference was observed at the blastula stage between control (Figure S3F) and MOdd-injected embryos (Figures S3G and S3H), indicating that MOdd depletion does not cause cell death. Collectively, the absence of PGC caused by dnd knockdown is not due to PGC loss by death prior to observation.
Dnd Dosage Determines the PGC Number In Vitro
In medaka, PGCs can be formed in culture from dissociated single cells of gastrulae (Li et al., 2009) and midblastula embryos (Li et al., 2014). We made use of this cell culture system to analyze the effect of dnd dosage on the PGC number in vitro. To this end, NgVg embryos were injected at the one-cell stage with MOdnd and/or dnd:ch mRNA and dissociated into single cells at the midblastula stage. The dissociated cells were seeded into 96-well plates, one embryo per well, and the cell fate decision was observed at day 2 post culture (dpc). PGC formation was seen in all embryos except for those dnd-depleted embryos injected with MOdnd (Figures 1B–1E). Cell counting revealed that the PGC number was dependent on Dnd dosage, as Dnd overexpression via injecting 50 pg of dnd:ch RNA increased the number of PGCs from 24 to 69, whereas dnd knockdown via injecting 50 pg of MOdnd led to the absolute absence of PGCs (Figure 1F). We observed a time course change in the PGC number per embryo in culture. In 12 control embryos, the PGC number per embryo ranged from 10 to 38 at 1 dpc and changed during subsequent days of culture (Figure 1G), revealing the individual variability in the PGC number per embryo and dynamics of PGC proliferation and survival. Injection of dnd RNA significantly increased the PGC number ranging from 55 to 109 (n = 15) at 1 dpc, without altering the variability and dynamics (Figure 1G). Thus, Dnd dosage controls the PGC number in vitro without influencing PCG proliferation and/or survival.
Dnd Dosage Determines the pPGC Number In Vitro
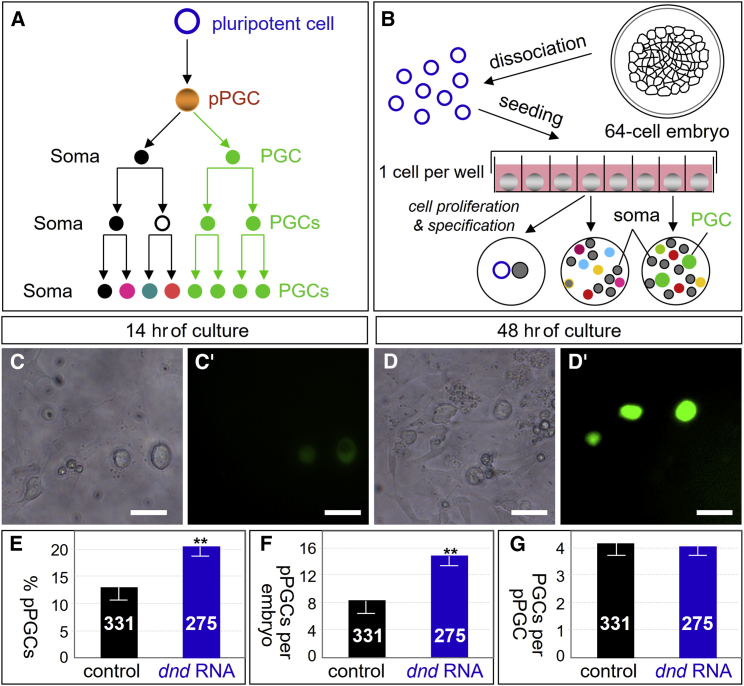
The first embryonic cells that will produce exclusively germ cells by clonal mitotic divisions are PGCs, and the PGC precursors, which are often morphologically indistinguishable from the surrounding somatic cells, are called presumptive PGCs (pPGCs) (Nieuwkoop and Sutasurya, 1979). At the final stage of fate decision, a pPGC divides to produce one PGC and one somatic cell. Hence, the pPGC number is equal to the initial PGC number (Figure 2A). We developed a procedure for lineage tracing of PGC development in single-cell culture (Figure 2B). Medaka PGCs can be formed in culture from gastrulae (Li et al., 2009) and midblastulae (Li et al., 2014). Cleavage blastomeres from 32-cell embryos are capable of cultivation (Li et al., 2011). We asked whether cleavage blastomeres were capable of PGC formation in vitro. To this end, blastomeres were dissociated from NgVg embryos at the 32- and 64-cell stage and individually seeded into 96-well plates, and PGCs were identified by phenotype and GFP expression. PGCs were found in culture from blastomeres isolated from embryos at the 32-cell stage (n = 24; Figures S4A and S4B) and 64-cell stage (n = 33; Figures S4C and S4D). Thus, dissociated cleavage blastomeres can form PGC in culture.
Figure 2.
Lineage Tracing of PGC Formation
(A) Conceptual PGC formation.
(B) Strategy of single blastomere culture.
(C–D′) 64-cell embryo blastomeres in single cell culture, showing PGCs (green) at 14 and 48 hr post culture. Scale bars, 50 μm.
(E) Percentage of pPGCs in culture.
(F) Number of pPGCs per embryo in culture.
(G) Number of PGCs per pPGC in culture. Numbers of blastomeres analyzed are shown in the columns. Data are means ± SD (error bars) from ≥7 independent experiments. ∗∗Very significant difference (p ≤ 0.01) compared with control.
We then tested the ability of cleavage blastomeres to form PGCs in single-cell culture. A proportion of blastomeres individually cultured in 96-well plates produced PGCs. These PGCs were first visible at 14 hr of culture by a larger size and faint GFP expression (Figures 2C and C′), which became more distinct with a brighter GFP signal at 48 hr (Figures 2D and D′). We made use of this single-cell culture system to lineage trace pPGCs in vitro. A total of 378 blastomeres from 64-cell NgVg embryos were seeded for single-cell culture; 331 survived PGC observations at day 2 of culture (Table S1), 42 of which were pPGCs (Figure 2E). This suggests that 12.7% of 64-cell blastomeres are pPGC, equivalent to 8.1 pPGCs per embryo at this stage (Figure 2F). These 42 pPGCs produced a total of 181 PGCs, corresponding to 4.3 PGCs per pPGC (Figure 2G). By extrapolation from these values, a medaka is able to produce 37.3 PGCs at 3 dpf (Table S1), comparable with 35.3 for developing embryos (Figure 1A). Hence, ∼8 pPGCs are committed in a medaka embryo already at the 64-cell cleavage stage, which demonstrates PGC preformation in this organism.
We analyzed dnd-dependent PGC development in single-cell culture. Since dnd knockdown causes the complete absence of PGCs in vivo and in vitro (Figures 1A, 1F, and S1), we analyzed how dnd overexpression enhanced the PGC number. To this end, 64-cell embryos injected at the one-cell stage with 50 pg of dnd:ch RNA were used for single-cell culture. A total of 308 blastomeres from dnd:ch RNA-injected 64-cell embryos were seeded, 275 survived observation at day 2 of culture (Table S1), 20.7% (n = 57) of which were pPGCs (Figure 2E). This corresponds to 13.3 pPGCs per embryo (Figure 2F). Therefore, dnd RNA injection at 50 pg increases the pPGCs number by up to 64%. On average, a pPGC from dnd:ch RNA-injected embryos produced 4.2 PGCs (Figure 2G), which is fully comparable with that of pPGCs from control embryos. Consequently, the PGC number per embryo increased by 53% to 57.1 upon dnd:ch RNA injection. Taken together, Dnd overexpression increases PGCs through increasing pPGCs without increasing PGC survival and/or proliferation.
dnd RNA Forms Particles for Asymmetric Segregation
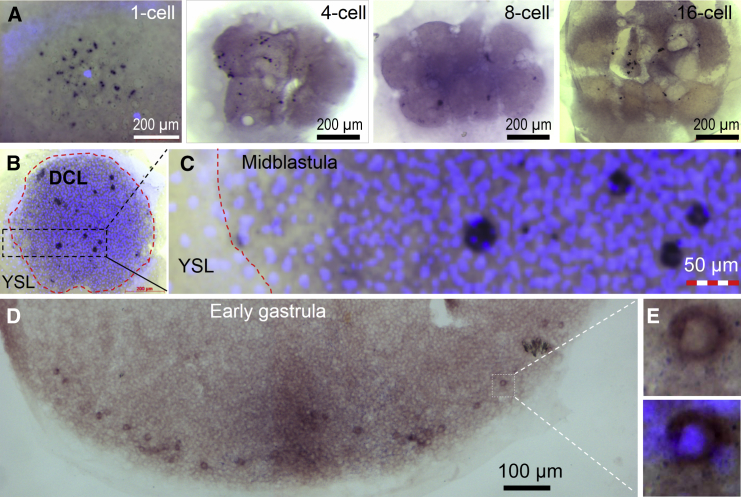
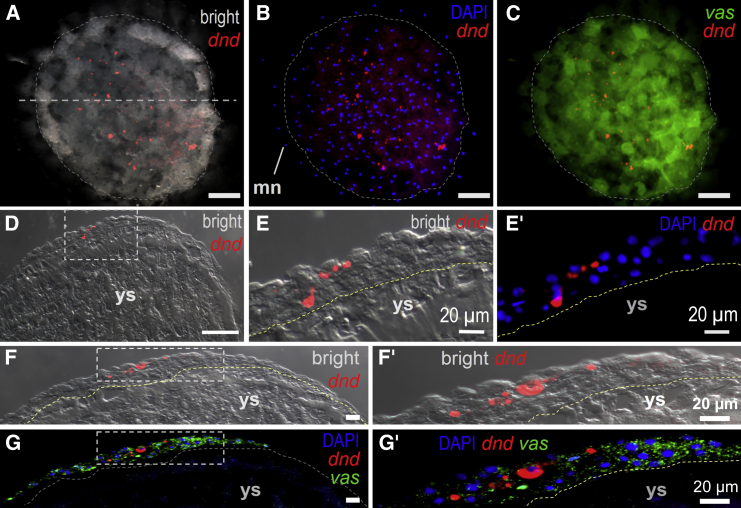
The medaka dnd RNA is specific to germ cells (Liu et al., 2009). We analyzed its temporospatial expression during early embryogenesis. In a majority (91%; n = 145) of cleavage embryos, dnd RNA existed predominantly in prominent cytoplasmic particles (Figures 3A–3C), which has never been seen for mRNAs of other germ genes such as boule, dazl, and vasa (Xu et al., 2009). As early as in the one-cell embryo, there were ∼40 particles of varying shapes and sizes ranging from 1 μm to 30 μm, which were not localized to a particular region of the embryo (Figure 3A). In 2- to 16-cell embryos, cells were different in the absence and presence of particles, suggesting irregular asymmetry of particle distribution between daughter cells (Figure 3A). In midblastula embryos comprising ∼2,000 cells, dnd RNA was seen in 6–11 prominent particles (Figure 3B). A closer inspection at larger magnification revealed that each prominent particle was frequently associated with 3–7 nuclei (Figure 3C). Until stage 13, dnd RNA eventually became dispersed and concentrated in the perinuclear region of PGCs (Figures 3D and 3E). We furthered our observation at better sensitivity and resolution by fluorescence in situ hybridization (FISH). This revealed 6–11 big particles 10–20 μm in size, ∼5 medium-sized (6–9 μm) particles, and ∼30 small particles ≤5 μm in medaka embryos at the early blastula stage (Figures 4A–4C). At this stage, dnd RNA particles were seen in all three to four cell layers on cryosections (Figures 4D–4G′). The vasa RNA did not form particles and displayed wide distribution among deep cells (Figures 4G and 4G′). Taken together, maternal dnd mRNA exists predominantly in cytoplasmic particles and segregates irregularly into a randomly positioned subset of blastomeres before its perinuclear localization in PGCs.
Figure 3.
dnd RNA Particles
(A) Cleavage embryos at 1- to 16-cell stages showing dnd RNA particles and their intercellular distribution.
(B) Midblastula embryo showing prominent dnd RNA particles in the deep cell layer (DCL) but absent in the extraembryonic yolk syncytial layer (YSL).
(C) Large magnification of the area framed in (B), highlighting association of prominent particles with multiple nuclei (blue).
(D) Early gastrula embryo squash showing PGCs scattered at the submarginal region.
(E) Large magnification of the boxed area in (D), highlighting perinuclear localization of dnd RNA within a PGC.
Figure 4.
Asymmetric Segregation of dnd RNA Particles
(A–C) Early blastula with one layer of marginal cell nuclei (mn).
(D) Cryosection through the broken line in (A).
(E and E′) Large magnification of the boxed area in (D), showing dnd RNA in all cell layers but absent in the yolk sac (ys).
(F and G) Early blastula cryosection.
(F′ and G′) Large magnification of the boxed areas in (F) and (G), highlighting fewer dnd-positive cells than vas-positive cells.
Scale bars, 100 μm unless indicated.
Dnd Protein Is a Component of Germ Plasm
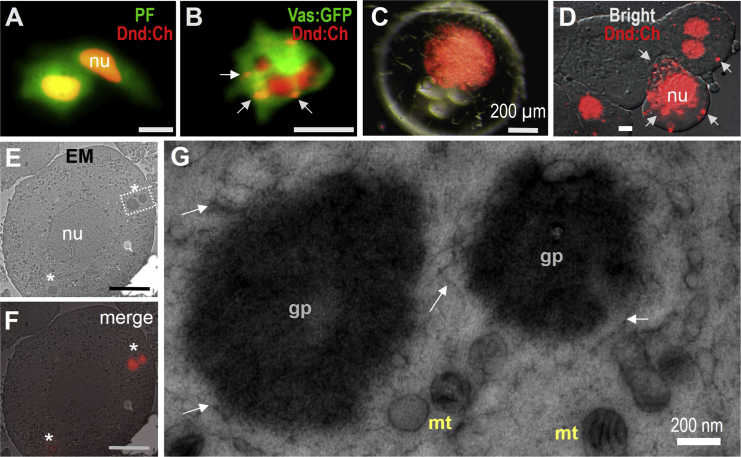
Many germ cell markers are localized in germ plasm, a membrane-less organelle that is closely associated with mitochondria (Houston et al., 1998, Kloc et al., 1998, Knaut et al., 2000). Our finding that Dnd determines the PGC fate provoked us to examine its intracellular localization. Fusion proteins PF (puromycin acetyltransferase:GFP) and Vas:GFP were exclusively cytoplasmic (Figures 5A and 5B), whereas Dnd:Ch was both nuclear and cytoplasmic in ES cells (Figures 5A and 5B) and early embryos (Figures 5C and 5D). We performed correlative light and electron microscopy to precisely determine the intracellular localization of Dnd protein. Embryos were microinjected with the dnd:ch RNA, single blastomeres were dissociated at the blastula stage and fixed by high-pressure freezing to preserve cellular architecture. After freezer substitution, ultrathin sections were transferred onto a copper grid for light microscopy (LM) and electron microscopy (EM). Dnd:Ch was seen mostly in cytoplasmic particles of 0.5–5 μm in size under LM, which appeared as dense bodies under EM (Figures 5E and 5F). At a larger magnification, these dense bodies lacked a membrane, possessed granulofibrillar structures, and were surrounded by many mitochondria (Figure 5G). These features are diagnostic of germ plasm that has been described in embryos of diverse organisms including Xenopus (Houston et al., 1998) and zebrafish (Knaut et al., 2000). Thus, Dnd protein is a component of germ plasm present in certain blastomeres prior to PGC formation.
Figure 5.
Intracellular Dnd Localization
(A and B) ES cells.
(C) Midblastula injected with dnd:ch RNA showing Dnd:Ch in many cells.
(D) Cells dissociated from the midblastula shown in (C), highlighting Dnd:Ch in the nucleus (nu) and cytoplasmic particles (arrows).
(E–G) Correlative microscopy, (E) EM micrograph showing cellular structure and dense bodies (asterisks).
(F) Correlative micrographs between fluorescent LM and EM.
(G) Large magnification of the area framed in (E), highlighting the fine structure of germ plasm (gp) as dense bodies in association with mitochondria (mt). Germ plasm lacks a membrane and has many granulofibrillar structures (arrows).
Scale bars, 10 μm unless indicated.
Dnd Stabilizes Germ Plasm RNA
In diverse animals, early development is controlled by maternal gene products in the absence of zygotic transcription. miRNAs play an essential role in maternal RNA clearance (Schier, 2007). Specifically, zebrafish MiR-430 promotes degradation of maternal mRNAs (Giraldez et al., 2006), and this degradation is suppressed by Dnd in cultured human germ cells and zebrafish PGCs through inhibiting miRNAs to target mRNAs (Kedde et al., 2007). Dnd interacts with its target RNAs via the DND-binding site. This binding protects the target RNAs from degradation by the MiR430 pathway (Giraldez et al., 2006). The binding sites for Dnd and MiR430 are present in the medaka dnd (Figure 6A; Figure S5A) and the zebrafish nos3 (initially nos1) (Kedde et al., 2007; Figure S5B). To determine the effect of Dnd binding on target RNAs in medaka, we exploited reporter assays by microinjection of gfpMiR430 and gfpNOS reporter RNAs (Figures 6A–6C). The gfpMiR430 RNA contains three copies of the MiR-430 target site (Giraldez et al., 2006). Upon coinjection into medaka embryos at the one-cell stage, GFP expression from injected gfpMiR430 RNA was monitored at stages 16 and 30. A preferential loss of GFP expression in many somatic cells was observed upon coinjection with dnd:ch but not dndΔ1:ch RNA, whereas coinjection of MOdnd reduced GFP expression to a barely detectable level (Figure 6B). A similar and more evident result was obtained by injection of gfpNOS reporter RNA (Figure 6C), which encodes GFP and contains the zebrafish nos3 3′-UTR capable of localization in PGCs (Koprunner et al., 2001). Thus, medaka Dnd regulates the level of germ plasm mRNA translation.
Figure 6.
Dnd Protects Germ Plasm RNAs from Degradation
(A) Strategy of a reporter assay. Embryos at the one-cell stage were injected with RNAs from the reporter and effector constructs illustrated and monitored for GFP expression at stages indicated.
(B) Dnd stabilizes gfpMir430 RNA.
(C) Dnd stabilizes gfpNOS RNA. Scale bars, 200 μm.
Discussion
In this study we provide several lines of evidence that Dnd serves as a critical PGC specifier in medaka. First, dnd knockdown is capable of specifically and completely abolishing PGC formation and its overexpression is sufficiently capable of inducing ectopic PGCs in embryos and significantly increasing the PGC number. Second, we reveal that dnd overexpression boosts PGCs via increasing the number of pPGCs without increasing PGC proliferation and survival in vitro. Third, we show that dnd RNA forms prominent particles that irregularly segregate, and thus limits the number of cells for PGC fate decision. Fourth, correlative microscopy reveals that Dnd protein is a component of germ plasm. Finally, we present evidence that Dnd stabilizes germ plasm RNAs essential for PGC development. Our findings pinpoint the mechanism by which Dnd exerts its functions: Dnd specifies the PGC fate through specifying pPGCs and thus determines the initial PGC number. dnd is a vertebrate-specific gene that is conserved from fish to mammals in germ cell-specific expression and requirement (Weidinger et al., 2003). In zebrafish and mouse, dnd knockdown or mutation does not prevent PGC formation. Knockdown of zebrafish dnd and several other germ genes leads to defects in PGC migration and/or survival but does not entirely prevent PGC formation, and their overexpression by RNA injection is unable to markedly induce additional PGCs (Knaut et al., 2000, Koprunner et al., 2001, Weidinger et al., 2003). In medaka, vasa knockdown severely affects PGC migration and vasa overexpression does not increase the PGC number (Li et al., 2009). Together with the fact that zebrafish null mutant for nos3 normally specifies PGCs and is defective in maintaining oocyte production (Draper et al., 2007), all the germ genes functionally analyzed so far in fish embryos act primarily on subsequent steps of PGC development, but not the first step, namely PGC fate decision. An exception to this is the zebrafish buc (Bontems et al., 2009). buc overexpression is able to increase the PGC number by up to ∼50%. buc cannot be a PGC specifier, as it is highly expressed in early embryos and its RNA distributes widely in all blastomeres rather than co-segregating with pPGCs. Our finding that dnd acts as a PGC specifier in medaka reveals a previously unidentified role for dnd in the initial step of PGC development, namely PGC fate decision rather than subsequent proliferation and survival, because the number of PGCs produced by each pPGC remains unchanged between normal and dnd RNA-injected embryos. It remains to be elucidated whether dnd acts as a PGC specifier also in other organisms.
Zebrafish makes use of the preformation mode for PGC specification (Raz, 2003). Two previous studies suggest that PGC formation occurs cell autonomously also in medaka (Herpin et al., 2007, Li et al., 2014). Our work here corroborates and extends these studies by providing convincing evidence that, as early as at cleavage stages, the medaka embryo contains pPGCs capable of PGC formation in single-cell culture. Therefore, PGC preformation operates in medaka as in zebrafish and Drosophila.
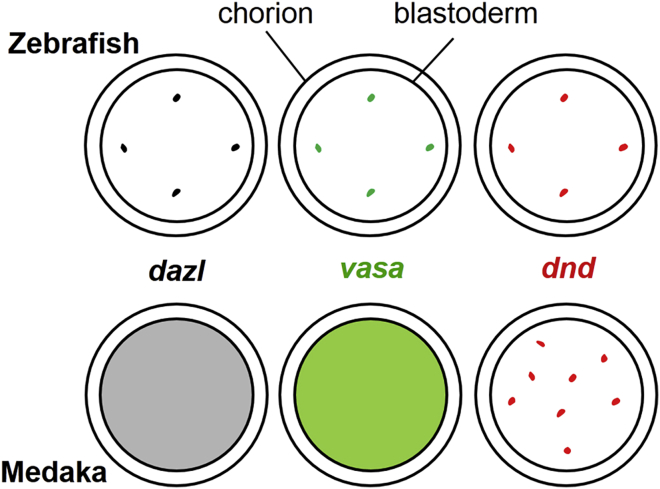
A striking finding from this study is how medaka segregates dnd as a cell fate specifier in the absence of a cellular mechanism for asymmetric segregation. In all organisms with PGC preformation, germ plasm components are either prelocalized to the posterior pole of the oocyte as in Drosophila, or localized and asymmetrically segregated in early embryos as in C. elegans, Xenopus, and zebrafish (Raz, 2003, Wylie, 1999). Our results in this study together with accumulated data reveal the presence of two distinct modes for asymmetric segregation of germ plasm RNAs in lower vertebrates as exemplified by zebrafish and medaka (Figure 7). In the zebrafish mode, RNAs of dnd, nanos, and vasa are co-localized in four cells that are regularly positioned and committed to the PGC fate (Raz, 2003, Weidinger et al., 2003). In the medaka mode, RNAs of all germ genes studied so far, including boule, dazl, and vasa (Li et al., 2009, Shinomiya et al., 2000, Xu et al., 2009) as well as piwi (Li et al., 2012), lack localization but exhibit wide distribution, which apparently suggests the absence of a cellular mechanism for prelocalization or localized segregation of maternal germ plasm components in medaka. This study has established that dnd is a unique exception to wide distribution. In the absence of such a cellular mechanism, the dnd RNA makes use of particle formation and partition as a mechanism for asymmetric segregation into pPGCs to specify the PGC fate.
Figure 7.
Modes of Germ Plasm RNA Segregation in Fish
Shown here are two distinct modes that regulate the intercellular distribution of germ plasm RNAs such as dazl (black), vasa (green), and dnd (red) RNAs in the embryo until the blastula stage. In the zebrafish mode, germ plasm RNAs are co-localized and asymmetrically segregated into four dots. In the medaka mode, dazl and vasa are widely distributed, whereas dnd is randomly segregated as particles into a subset of cells for the PGC fate decision.
A major challenge in analyzing cell fate decision is the absence of markers to trace steps before bona fide fate decision. An ideal situation is to put a cell under defined conditions and monitor its changes into a state diagnostic of a particular cell. In this study, we have established a single-cell culture system and a procedure to lineage trace PGC formation. This enables us to continuously monitor PGC development by precisely counting the number of potential pPGCs per 64-cell embryo as early as at 4 hr post fertilization, long before 13 hpf when PGCs become visible. We anticipate that this in vitro lineage-tracing system will also be applicable to other cell lineages and other organisms.
An increase in the PGC number caused by dnd RNA injection does not exceed 3-fold, even if a high level of Dnd expression occurs in many cells. It is likely that an additional factor essential for PGC specification exists in medaka, which shows expression and PGC-specifying activity similar to dnd but dnd-independent asymmetric segregation. Alternatively, PGC-competent cells are limited in the medaka embryo for the ultimate fate decision. It is also possible that only a limited number of PGCs are permissive for embryonic development and survival until observation for PGCs. This possibility is small, because dnd RNA-injected embryos are not different from controls in survival rate and cell death as well as survival rate of their blastomeres in single-cell culture. A similar situation has also been recorded for the Drosophila PGC specifier oskar in vivo (Ephrussi and Lehmann, 1992) and the human PGC specifier Sox17 in vitro (Irie et al., 2015).
During early development of diverse animals, miRNAs play an essential role in maternal-zygotic transcription and PGC development (Giraldez et al., 2006, Schier, 2007). Through inhibiting the accessibility of miRNAs including MiR-430 to target mRNAs, Dnd protects maternal RNAs from clearance in cultured human germ cells and PGCs of zebrafish (Kedde et al., 2007) and medaka (Tani et al., 2009). In this study, by reporter assays we reveal that Dnd counteracts with miRNA-430 to regulate maternal RNA turnover and translation for medaka PGC formation. This suggests that Dnd protects germ plasm RNAs from miRNA-mediated degradation to ensure PGC specification.
Our identification of Dnd as a critical PGC specifier in medaka provides insights into cell fate decisions during germline-soma separation in medaka as a lower vertebrate model. Altering Dnd activity could be a strategy for analyzing and manipulating germ cell formation. Our finding that dnd RNA asymmetrically segregates via particle partition reveals the presence of a previously unidentified mechanism that controls asymmetric segregation of macromolecules in early development.
Experimental Procedures
Fish
Work with fish was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Advisory Committee for Laboratory Animal Research in Singapore and approved by this committee (permit number 27/09). Medaka was maintained under an artificial photoperiod of 14-hr light and 10-hr dark at 26°C (Hong et al., 1996, Hong et al., 2010, Hong et al., 2012). Medaka strains (HB32C and af) and transgenic lines (Ng and Vg) were described previously (Hong et al., 2010). Ng and Vg express GFP specifically in germ cells from the medaka nanos and vasa promoters, respectively (Li et al., 2009). Vg embryos were produced by crossing homozygous Vg males to non-transgenic females. NgVg were produced by crossing homozygous Ng males to homozygous Vg females. Embryos were reared and staged as described (Li et al., 2009).
Morpholino Oligos
Two antisense morpholino oligos (Figure S1A), MOdnd (CAC CAC CTT GCT CTG ATT GTC CAT C) and MOddm containing four mismatches (CAC CAC CTT GCT tTG gTT aTC CAT c, mismatches underlined), were synthesized (Gene Tool) and dissolved in water at 10 mg/ml as stock for microinjection as described (Li et al., 2009).
Plasmids
Plasmids pGEMdnd and pCSdnd:chNS (Liu et al., 2009), pGEMvas and pCVvas:gfpNS (Li et al., 2009), and pCVpf expressing a fusion between puromycin acetyltransferase and GFP (Zhao et al., 2012) have been described. pCSdnd:chDD was made by replacing the 3′ UTR of the medaka nanos3 gene (NS) with the medaka dnd 3′ UTR (DD). pCSdndΔ1:chDD was constructed by replacing the full-length dnd open reading frame (ORF) with a partial dnd ORF for the 127 N-terminal amino acids in pCSdnd:chDD. In pCSdnd:chNS, pCSdnd:chDD, and pCSdndΔ1:chDD, four mismatch nucleotides were introduced to the target sequence of MOdnd around the ATG codon without changing the amino acid sequence (Figure S1A). Consequently, the synthetic mRNAs from these constructs were targeted by MOddm but not MOdnd. Plasmid pH2Bgfp contains the fusion between the cDNA encoding the bovine histone H2B and EGFP. The fusion gene was PCR-cloned using primers (CGG AGA TCT ATG CCA GAG CCA GCG A and CCG CTC GAG TTA CTT GTA CAG CTC GTC CAT GC; the restriction sites are underlined) from pBOS-H2Bgfp (BD Biosciences) and inserted between BamHI and XhoI to replace the cherry cDNA in pCScherry. The 3′ UTRs of the medaka dnd, dazl, and nanos3 were used to localize synthetic RNAs in germ cells. Structures of these plasmids are illustrated in Figure S1B.
RNA Synthesis and In Situ Hybridization
Sense and antisense dnd and vasa riboprobes were synthesized from linearized plasmids pGEMdnd (Liu et al., 2009) and pGEMvas (Li et al., 2009). Capped dnd and vasa mRNA were synthesized by using the mMessage Machine kit (Ambion) from linearized pCSdnd:chDD (Liu et al., 2009) and pCSvas:gfp and stored at −80°C until use. Chromogenic and two-color FISH and nuclear staining with DAPI were performed as described (Liu et al., 2009, Xu et al., 2009).
Microinjection
Medaka embryos were injected at the one-cell stage (Hong et al., 2004b, Li et al., 2009). MOddm and MOdnd (10–5,000 ng/μl), synthetic mRNAs (10–100 ng/μl), and plasmid DNA (50 ng/μl) were injected alone or in combination.
Lineage Tracing in Single-Cell Culture
Embryo preparation, cell isolation, and culture in gelatin-coated plastic ware were done essentially as described (Hong et al., 1996, Hong et al., 1998a, Hong et al., 1998b, Hong et al., 2010, Yi et al., 2009). For single-embryo culture, disperse cells were collected from a single embryo and seeded into a single well of a 96-well plate containing 150 μl of ES cell culture medium ESM2 (Hong et al., 1996), one embryo per well. For single-cell culture, blastomeres were dissociated from embryos at the 32- and 64-cell stages and seeded by pipetting into 96-well plates, one cell per well. PGCs were monitored by GFP expression from 12 hr post culture onward.
Cell Death Detection
Cell death in developing embryos were detected by vital dye acrinding orange (AO, Sigma) as decribed in (Mei et al., 2008). Briefly, embryos were dechorionated by proteinase K (Sigma) and hatching ezyme treatment as described (Yi et al., 2009), then embryos were incubated in 5 µg/ml AO for 15-20 min in BSS-1%PEG. After three times washes for 5 min each in BSS-1%PEG, the embryos were then observed under a fluorescent stereo microscopy.
Microscopy
Standard LM for observation and photography was done on a Leica MZFIII stereo microscope, and Zeiss Axiovert invert and Axiovert upright microscopes with a Zeiss AxioCam M5Rc digital camera (Zeiss Corp) as described previously (Hong et al., 2010, Xu et al., 2009, Yi et al., 2009). Preparation of ultrathin cryosections from blastomeres of medaka blastula embryos and correlative light EM were done as described (Yuan et al., 2014).
Statistics
Statistical analyses were calculated by using GraphPad Prism v4.0. Consolidated data were presented as means ± SD and p values were calculated using the non-parametric Student t test.
Author Contributions
N.H., H.Z., J.S., and Y.H. designed the research. N.H., M.L., Y.Y., T.W., M.Y., and H.X. performed the experiments. N.H., J.S., and Y.H. wrote the paper.
Acknowledgments
We thank Dr. A.F. Schier for pGFPmiR430, Jiaorong Deng for fish breeding, Veronica Wong and Choy Mei Foong for laboratory management. This work was supported by grants to Y.H. from the National Research Foundation of Singapore (NRF-CRP7-2010-03), to J.S. from the Ministry of Education of Singapore (R-154-000-678-112), and to M.L. from the National Natural Science Foundation of China (31372520) and the Shanghai Universities First-Class Disciplines Project of Fisheries.
Published: February 4, 2016
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes five figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.01.002.
Contributor Information
Jianxing Song, Email: dbssjx@nus.edu.sg.
Yunhan Hong, Email: dbshyh@nus.edu.sg.
Supplemental Information
References
- Bontems F., Stein A., Marlow F., Lyautey J., Gupta T., Mullins M.C., Dosch R. Bucky ball organizes germ plasm assembly in zebrafish. Curr. Biol. 2009;19:414–422. doi: 10.1016/j.cub.2009.01.038. [DOI] [PubMed] [Google Scholar]
- Centanin L., Hoeckendorf B., Wittbrodt J. Fate restriction and multipotency in retinal stem cells. Cell Stem Cell. 2011;9:553–562. doi: 10.1016/j.stem.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Deng W., Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- Draper B.W., McCallum C.M., Moens C.B. nanos1 is required to maintain oocyte production in adult zebrafish. Dev. Biol. 2007;305:589–598. doi: 10.1016/j.ydbio.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A., Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358:387–392. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- Herpin A., Rohr S., Riedel D., Kluever N., Raz E., Schartl M. Specification of primordial germ cells in medaka (Oryzias latipes) BMC Dev. Biol. 2007;7:3. doi: 10.1186/1471-213X-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Winkler C., Schartl M. Pluripotency and differentiation of embryonic stem cell lines from the medakafish (Oryzias latipes) Mech. Dev. 1996;60:33–44. doi: 10.1016/s0925-4773(96)00596-5. [DOI] [PubMed] [Google Scholar]
- Hong Y., Winkler C., Schartl M. Efficiency of cell culture derivation from blastula embryos and of chimera formation in the medaka (Oryzias latipes) depends on donor genotype and passage number. Dev. Genes Evol. 1998;208:595–602. doi: 10.1007/s004270050220. [DOI] [PubMed] [Google Scholar]
- Hong Y., Winkler C., Schartl M. Production of medakafish chimeras from a stable embryonic stem cell line. Proc. Natl. Acad. Sci. USA. 1998;95:3679–3684. doi: 10.1073/pnas.95.7.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Liu T., Zhao H., Xu H., Wang W., Liu R., Chen T., Deng J., Gui J. Establishment of a normal medakafish spermatogonial cell line capable of sperm production in vitro. Proc. Natl. Acad. Sci. USA. 2004;101:8011–8016. doi: 10.1073/pnas.0308668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Winkler C., Liu T., Chai G., Schartl M. Activation of the mouse Oct4 promoter in medaka embryonic stem cells and its use for ablation of spontaneous differentiation. Mech. Dev. 2004;121:933–943. doi: 10.1016/j.mod.2004.03.028. [DOI] [PubMed] [Google Scholar]
- Hong N., Li M., Zeng Z., Yi M., Deng J., Gui J., Winkler C., Schartl M., Hong Y. Accessibility of host cell lineages to medaka stem cells depends on genetic background and irradiation of recipient embryos. Cell. Mol. Life Sci. 2010;67:1189–1202. doi: 10.1007/s00018-009-0247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong N., Chen S., Ge R., Song J., Yi M., Hong Y. Interordinal chimera formation between medaka and zebrafish for analyzing stem cell differentiation. Stem Cells Dev. 2012;21:2333–2341. doi: 10.1089/scd.2011.0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston D.W., King M.L. Germ plasm and molecular determinants of germ cell fate. Curr. Top Dev. Biol. 2000;50:155–181. doi: 10.1016/s0070-2153(00)50008-8. [DOI] [PubMed] [Google Scholar]
- Houston D.W., Zhang J., Maines J.Z., Wasserman S.A., King M.L. A Xenopus DAZ-like gene encodes an RNA component of germ plasm and is a functional homologue of Drosophila boule. Development. 1998;125:171–180. doi: 10.1242/dev.125.2.171. [DOI] [PubMed] [Google Scholar]
- Houwing S., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Irie N., Weinberger L., Tang W.W., Kobayashi T., Viukov S., Manor Y.S., Dietmann S., Hanna J.H., Surani M.A. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M., Strasser M.J., Boldajipour B., Oude Vrielink J.A., Slanchev K., le Sage C., Nagel R., Voorhoeve P.M., van Duijse J., Orom U.A. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Kloc M., Larabell C., Chan A.P., Etkin L.D. Contribution of METRO pathway localized molecules to the organization of the germ cell lineage. Mech. Dev. 1998;75:81–93. doi: 10.1016/s0925-4773(98)00086-0. [DOI] [PubMed] [Google Scholar]
- Knaut H., Pelegri F., Bohmann K., Schwarz H., Nusslein-Volhard C. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J. Cell Biol. 2000;149:875–888. doi: 10.1083/jcb.149.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko K., Arauzo-Bravo M.J., Tapia N., Kim J., Lin Q., Bernemann C., Han D.W., Gentile L., Reinhardt P., Greber B. Human adult germline stem cells in question. Nature. 2010;465:E1. doi: 10.1038/nature09089. [DOI] [PubMed] [Google Scholar]
- Koprunner M., Thisse C., Thisse B., Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877–2885. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Hong N., Xu H., Yi M., Li C., Gui J., Hong Y. Medaka vasa is required for migration but not survival of primordial germ cells. Mech. Dev. 2009;126:366–381. doi: 10.1016/j.mod.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Li Z., Bhat N., Manali D., Wang D., Hong N., Yi M., Ge R., Hong Y. Medaka cleavage embryos are capable of generating ES-like cell cultures. Int. J. Biol. Sci. 2011;7:418–425. doi: 10.7150/ijbs.7.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Hong N., Gui J., Hong Y. Medaka piwi is essential for primordial germ cell migration. Curr. Mol. Med. 2012;12:1040–1049. doi: 10.2174/156652412802480853. [DOI] [PubMed] [Google Scholar]
- Li Z., Li M., Hong N., Yi M., Hong Y. Formation and cultivation of medaka primordial germ cells. Cell Tissue Res. 2014;357:71–81. doi: 10.1007/s00441-014-1867-z. [DOI] [PubMed] [Google Scholar]
- Lin H. piRNAs in the germ line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- Lin H. Capturing the cloud: UAP56 in nuage assembly and function. Cell. 2012;151:699–701. doi: 10.1016/j.cell.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Long W., Chen J., Hopkins N. Production of germ-line chimeras in zebrafish by cell transplants from genetically pigmented to albino embryos. Proc. Natl. Acad. Sci. USA. 1992;89:4519–4523. doi: 10.1073/pnas.89.10.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Hong N., Xu H., Li M., Yan Y., Purwanti Y., Yi M., Li Z., Wang L., Hong Y. Medaka dead end encodes a cytoplasmic protein and identifies embryonic and adult germ cells. Gene Expr. Patterns. 2009;9:541–548. doi: 10.1016/j.gep.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Magnusdottir E., Dietmann S., Murakami K., Gunesdogan U., Tang F., Bao S., Diamanti E., Lao K., Gottgens B., Azim Surani M. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat. Cell Biol. 2013;15:905–915. doi: 10.1038/ncb2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megosh H.B., Cox D.N., Campbell C., Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr. Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Mei J., Zhang Q., Li Z., Lin S., Gui J. C1q-like inhibits p53-mediated apoptosis and controls normal hematopoiesis during zebrafish embryogenesis. Dev Bio. 2008;319:273–284. doi: 10.1016/j.ydbio.2008.04.022. [DOI] [PubMed] [Google Scholar]
- Nakaki F., Hayashi K., Ohta H., Kurimoto K., Yabuta Y., Saitou M. Induction of mouse germ-cell fate by transcription factors in vitro. Nature. 2013;501:222–226. doi: 10.1038/nature12417. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P.D., Sutasurya L.A. Cambridge University Press; 1979. Primordium Germ Cells in the Chordates. [Google Scholar]
- Ohinata Y., Payer B., O'Carroll D., Ancelin K., Ono Y., Sano M., Barton S.C., Obukhanych T., Nussenzweig M., Tarakhovsky A. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Raz E. Primordial germ-cell development: the zebrafish perspective. Nat. Rev. Genet. 2003;4:690–700. doi: 10.1038/nrg1154. [DOI] [PubMed] [Google Scholar]
- Saitou M., Barton S.C., Surani M.A. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Schier A.F. The maternal-zygotic transition: death and birth of RNAs. Science. 2007;316:406–407. doi: 10.1126/science.1140693. [DOI] [PubMed] [Google Scholar]
- Shinomiya A., Tanaka M., Kobayashi T., Nagahama Y., Hamaguchi S. The vasa-like gene, olvas, identifies the migration path of primordial germ cells during embryonic body formation stage in the medaka, Oryzias latipes. Dev. Growth Differ. 2000;42:317–326. doi: 10.1046/j.1440-169x.2000.00521.x. [DOI] [PubMed] [Google Scholar]
- Tam P.P., Zhou S.X. The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev. Biol. 1996;178:124–132. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Kinoshita M., Kobayashi D., Nagahama Y. Establishment of medaka (Oryzias latipes) transgenic lines with the expression of green fluorescent protein fluorescence exclusively in germ cells: a useful model to monitor germ cells in a live vertebrate. Proc. Natl. Acad. Sci. USA. 2001;98:2544–2549. doi: 10.1073/pnas.041315498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani S., Kusakabe R., Naruse K., Sakamoto H., Inoue K. Genomic organization and embryonic expression of miR-430 in medaka (Oryzias latipes): insights into the post-transcriptional gene regulation in early development. Gene. 2009;449:41–49. doi: 10.1016/j.gene.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Tzung K.W., Goto R., Saju J.M., Sreenivasan R., Saito T., Arai K., Yamaha E., Hossain M.S., Calvert M.E., Orban L. Early depletion of primordial germ cells in zebrafish promotes testis formation. Stem Cell Rep. 2015;4:61–73. doi: 10.1016/j.stemcr.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Hong Y. Direct gene disruption by TALENs in medaka embryos. Gene. 2014;543:28–33. doi: 10.1016/j.gene.2014.04.013. [DOI] [PubMed] [Google Scholar]
- Weidinger G., Stebler J., Slanchev K., Dumstrei K., Wise C., Lovell-Badge R., Thisse C., Thisse B., Raz E. dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr. Biol. 2003;13:1429–1434. doi: 10.1016/s0960-9822(03)00537-2. [DOI] [PubMed] [Google Scholar]
- Wittbrodt J., Shima A., Schartl M. Medaka–a model organism from the far East. Nat. Rev. Genet. 2002;3:53–64. doi: 10.1038/nrg704. [DOI] [PubMed] [Google Scholar]
- Wylie C. Germ cells. Cell. 1999;96:165–174. doi: 10.1016/s0092-8674(00)80557-7. [DOI] [PubMed] [Google Scholar]
- Xu H., Li Z., Li M., Wang L., Hong Y. Boule is present in fish and bisexually expressed in adult and embryonic germ cells of medaka. PLoS One. 2009;4:e6097. doi: 10.1371/journal.pone.0006097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M., Hong N., Hong Y. Generation of medaka fish haploid embryonic stem cells. Science. 2009;326:430–433. doi: 10.1126/science.1175151. [DOI] [PubMed] [Google Scholar]
- Ying Y., Qi X., Zhao G.Q. Induction of primordial germ cells from murine epiblasts by synergistic action of BMP4 and BMP8B signaling pathways. Proc. Natl. Acad. Sci. USA. 2001;98:7858–7862. doi: 10.1073/pnas.151242798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon C., Kawakami K., Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157–3165. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
- Youngren K.K., Coveney D., Peng X., Bhattacharya C., Schmidt L.S., Nickerson M.L., Lamb B.T., Deng J.M., Behringer R.R., Capel B. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–364. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Li M., Hong N., Hong Y. Correlative light and electron microscopic analyses of mitochondrial distribution in blastomeres of early fish embryos. FASEB J. 2014;28:577–585. doi: 10.1096/fj.13-233635. [DOI] [PubMed] [Google Scholar]
- Zhao H., Hong N., Lu W., Zeng H., Song J., Hong Y. Fusion gene vectors allowing for simultaneous drug selection, cell labeling, and reporter assay in vitro and in vivo. Anal. Chem. 2012;84:987–993. doi: 10.1021/ac202541t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.