Summary
Over the past years, microRNAs (miRNAs) have emerged as crucial factors that regulate self-renewal and differentiation of embryonic stem cells (ESCs). Although much is known about their role in maintaining ESC pluripotency, the mechanisms by which they affect cell fate decisions remain poorly understood. By performing deep sequencing to profile miRNA expression in mouse ESCs (mESCs) and differentiated embryoid bodies (EBs), we identified four differentially expressed miRNAs. Among them, miR-191 and miR-16-1 are highly expressed in ESCs and repress Smad2, the most essential mediator of Activin-Nodal signaling, resulting in the inhibition of mesendoderm formation. miR-23a, which is also down-regulated in the differentiated state, suppresses differentiation toward the endoderm and ectoderm lineages. We further identified miR-421 as a differentiation-associated regulator through the direct repression of the core pluripotency transcription factor Oct4 and the bone morphogenetic protein (BMP)-signaling components, Smad5 and Id2. Collectively, our findings uncover a regulatory network between the studied miRNAs and both branches of TGF-β/BMP-signaling pathways, revealing their importance for ESC lineage decisions.
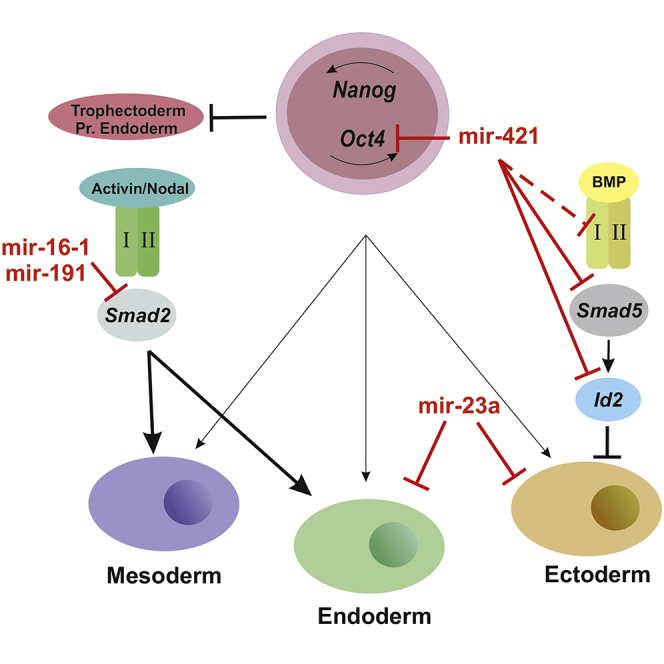
Graphical Abstract
Highlights
-
•
miR-16-1 and miR-191 suppress mesendoderm differentiation by Activin/Smad2 targeting
-
•
miR-23a represses endoderm and ectoderm differentiation
-
•
miR-421 promotes ectoderm and endoderm differentiation by TGF-β and Oct4 inhibition
In this article, Kretsovali and colleagues identify four miRNAs (miR-16-1, miR-191, miR-23a, and miR-421) regulating mESC differentiation toward the three germ layers. Their pivotal function in mouse ESC lineage commitment is revealed by interplay between the aforementioned miRNAs and the two branches of the TGF-β signaling pathway (Activin/Nodal and BMP).
Introduction
Embryonic stem cells (ESCs), derived from pre-implantation embryos, share two unique properties: the ability to grow indefinitely in culture and to differentiate into all cell types (Evans and Kaufman, 1981). ESC self-renewal is regulated by a complex network of transcription factors and signaling pathways (Ng and Surani, 2011). The transforming growth factor β (TGF-β) pathway plays a pivotal role in cell fate determination during mouse embryonic development, such as primitive streak formation (Oshimori and Fuchs, 2012). Both Smad1/5/8 and Smad2/3 branches are involved in pluripotency and differentiation of ESCs. Activin/Nodal/Smad2/3 signaling is important for proper differentiation toward the mesendoderm lineage (Fei et al., 2010), whereas bone morphogenetic protein (BMP)/Smad1/5/8 signaling promotes self-renewal in mouse ESCs (mESCs) (Ying et al., 2003).
Accumulating evidence reveals that microRNAs (miRNAs) are crucial in controlling the pluripotent stem cell state. Their important regulatory role in mouse and human ESCs has been identified using Dicer and DGCR8 knockout mice. Dicer and DGR8 deletion resulted in embryonic lethality (Bernstein et al., 2003), while DGCR8-deficient mESCs were viable but defective in proliferation and differentiation (Wang et al., 2007). Several studies reported on miRNAs maintaining the ESC state, whereas others reported miRNAs as promoting differentiation. miR-290–295 and miR-302–367 clusters include the most abundant miRNAs in mouse and human ESCs and are characterized as ES cell-specific cell cycle miRNAs (Gangaraju and Lin, 2009, Melton et al., 2010). In contrast, miR-134, miR-296, and miR-470 are related to ESC differentiation and self-renewal silencing (Tay et al., 2008). Although there is no doubt that miRNAs regulate ESC self-renewal and lineage commitment, their role in relevant signaling pathways that determine ESC function remains unclear.
In this study, we report the identification of four miRNAs as critical regulators of ESC fate. miR-16-1 (miR-16-1/15a cluster) and miR-191 (miR-191/425 cluster), which are highly expressed in mESCs, directly target Smad2, an Activin/Nodal signaling important mediator, leading to the inhibition of mesendoderm lineage. Another miRNA expressed in the undifferentiated state, miR-23a (miR-27/24a/23a cluster), inhibits the endodermal and ectodermal differentiation. On the contrary, miR-421 (miR-421/374b/c cluster) was identified as a differentiation regulator, by suppressing BMP signaling and the critical pluripotency factor, Oct4. Altogether, the mechanisms incorporating the two branches of TGF-β signaling pathway and miRNAs are highlighted, unraveling their importance to ESC lineage commitment.
Results and Discussion
Global miRNA Analysis of mESCs and Day-8 Embryoid Bodies
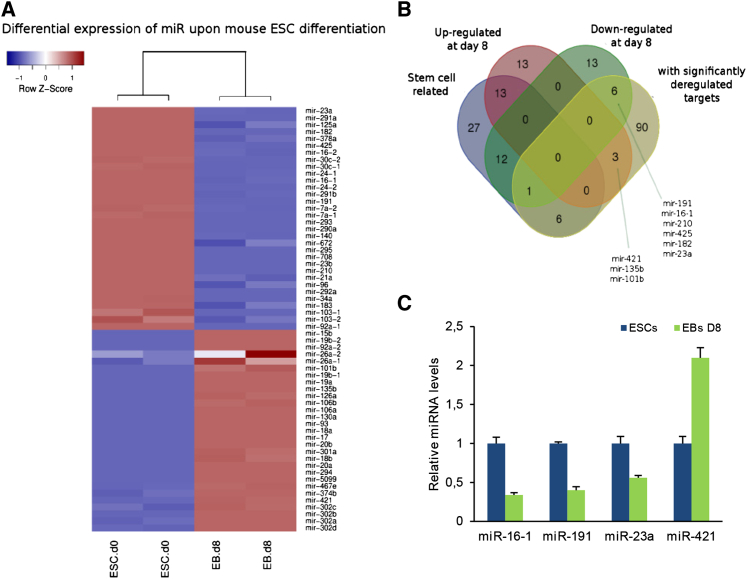
To identify miRNAs pivotal for ESC function and biology, we performed a global miRNA analysis from mESCs and day-8 embryoid bodies (EBs D8). Although the analysis revealed a large number (442) of differentially expressed miRNAs (Table S1), we restricted it by narrowing it down to highly abundant miRNAs. Thus, a total of 61 miRNAs with high abundance at either time point (D0 or D8) was further analyzed in terms of relative expression, relationship to the developmental process, and expression of their target genes. Of the 61 differentially expressed miRNAs, 32 were down- and 29 were up-regulated at D8, with this behavior being fairly consistent between replicates (Figure 1A). Among them, well-studied miRNAs crucial for pluripotent state were identified, such as miR-290–295 and miR-302 clusters (Gangaraju and Lin, 2009, Melton et al., 2010). In addition, those 61 miRNAs overlapped with previous published data for ESCs and EBs D5 or D7 (Table S2) (Lewis et al., 2005, Lee et al., 2011).
Figure 1.
miRNA Profiling of mESCs and Differentiated Cells
(A) Heatmap of log2-transformed miRNA abundances in replicate samples of undifferentiated ESCs and differentiated cells (EBs D8).
(B) Venn diagram showing common miRNAs between 59 stem cell-related miRNAs based on literature compiled from miRbase, our 32 down- and 29 up-regulated miRNAs, and 107 miRNAs whose mRNA targets were found to be significantly deregulated between days 0 and 9 in an independent study. Short lists of primary miRNA candidates for down- and up-regulated species are included.
(C) The RT-PCR verification of the four selected miRNA levels. Data are shown as mean ± SD of three independent experiments.
Screening the literature for mouse miRNAs (miRBase, Rel. 21), a list of 59 miRNAs reported to be implicated in the ESC differentiation process (Kozomara and Griffiths-Jones, 2014) was obtained (Table S3). Comparing the above list with our deregulated miRNAs, 43% of them were identical (26 of 61). However, we focused on the remaining miRNAs (57%), which have not been previously involved in ESC identity (Table S3).
Following a different approach, we performed a combination of in silico target analysis coupled with gene-expression data. Predicted miRanda (Betel et al., 2008) and TargetScan (Lewis et al., 2005) miRNA targets were gathered to form a concise table of genes targeted by our differentially expressed miRNAs. We obtained expression values of mRNA genes from a genome-wide expression profiling of mESC differentiation (Hailesellasse Sene et al., 2007). Scanning the list of all miRNA measured in our study, we obtained the mean log (fold change) of mRNA expression between D9 and D0 in the aforementioned study. By comparing this value for each of the miRNA targets with the overall mean of expression change, we pinned down 106 miRNAs whose targets were significantly deregulated during differentiation. The intersection of these 106 miRNAs with our deregulated ones, not reported to be related to stem cell differentiation, led to two short lists containing three up-regulated and six down-regulated miRNAs (Figure 1B). After further searching the literature for the predicted targets of selected miRNAs and following validation of the expression level changes, we ended up with four miRNAs. miR-16-1, miR-191, and miR-23a are down-regulated upon differentiation, whereas miR-421 is up-regulated in EBs D8 (Figure 1C).
miR-16-1 and miR-191 Inhibit Mesendoderm Differentiation by Targeting Activin/Smad2 Signaling Pathway
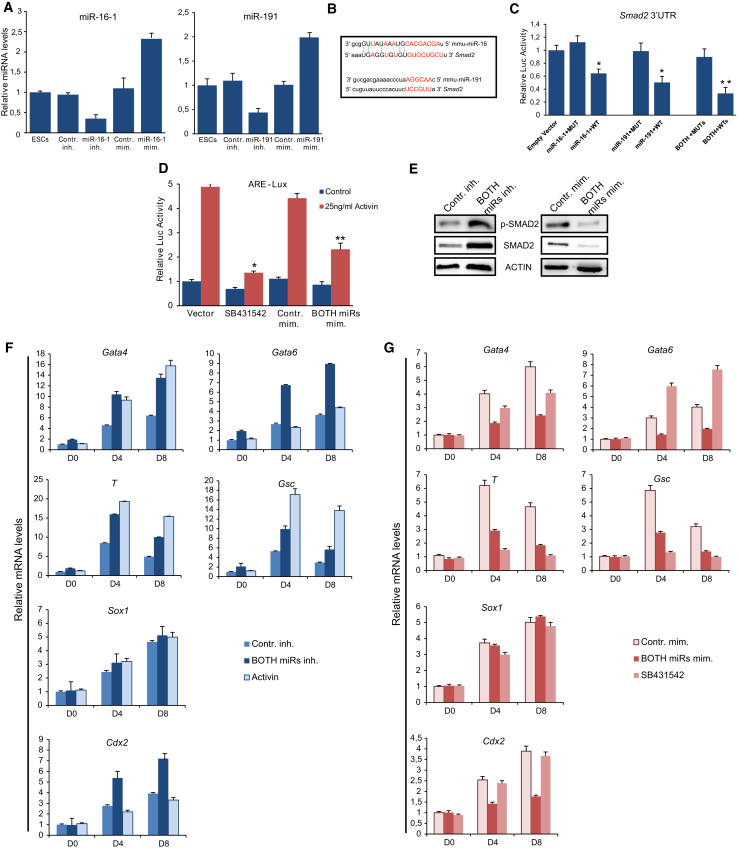
To evaluate the functional role of miR-16-1 and miR-191 in mESCs we used miR-16-1, miR-191 inhibitors, or miR-16-1, miR-191 mimics (Figure 2A), and examined their impact on self-renewal and pluripotency. Neither the inhibition nor the overexpression of these miRNAs caused any changes at the expression levels of Oct4 and Nanog (Figure S1A). In addition, no effect on mESC morphology (data not shown) and cell cycle (Figure S1B) was observed. These data suggest that miR-191 and miR-16-1 do not play a crucial role in mESC self-renewal.
Figure 2.
miR-16-1 and miR-191 Antagonize Activin/Smad2 Signaling in mESCs and Repress Mesendoderm Differentiation
(A) Measurement of miR-16-1 and miR-191 levels by RT-PCR after transient transfection with miR mimics or inhibitors. Error bars indicate SD of three independent experiments.
(B) miR-16-1 and miR-191 target sites in the 3′ UTR of Smad2. Red indicates complementarity between miRNA and the target gene. Error bars indicate SD of three independent experiments.
(C) miR-16-1 and miR-191 specifically repress their target in the luciferase assay. Data are shown as mean ± SD of four independent experiments. ∗p < 0.05 ∗∗p < 0.01.
(D) Ectopic expression of miR-16-1/miR-191 inhibits ARE-luc activity. Data are shown as mean ± SD of four independent experiments. ∗p < 0.05 ∗∗p < 0.01.
(E) Total SMAD2 and p-SMAD2 protein levels detected.
(F and G) Relative mRNA levels of genes associated with the three germ layers at EBs D0, D4, and D8 in response to miR-16-1/miR-191 repression (F) or overexpression (G). Error bars indicate SD of three independent experiments.
We next examined the potential effect of miR-191 and miR-16-1 on the induction of differentiation markers in the undifferentiated state. We found that after 72 hr of their inhibition, characteristic endodermal (Gata4, Gata6) and mesodermal (T, Gsc, Lhx1, Bmp4) markers were slightly up-regulated, whereas ectodermal (Pax6, Sox1) markers did not seem to be affected (Figure S1C). Conversely, miR-16-1 and miR-191 overexpression did not exert changes on lineage markers compared with negative control mimic (data not shown).
To study the mechanism by which these miRNAs regulate mESC differentiation, we focused on their targets. Smad2 mRNA is predicted to have binding sites for miR-16-1 and miR-191 (Figure 2B). Since it is known that Activin/Smad2 signaling is crucial for mesoderm and endoderm development in vivo (Moustakas and Heldin, 2009) and mESC differentiation in vitro (Fei et al., 2010), we hypothesized that miR-16-1 and miR-191 may compete with Activin/Smad2 signaling. To analyze whether Smad2 is a direct target of these miRNAs, we performed luciferase reporter assays using constructs that harbor wild-type (WT) or mutant (MUT) 3′ UTR of Smad2. We found that either miR-191 or miR-16-1 suppressed the WT but not MUT 3′ UTR reporter activity, and a combination of both miRNAs led to higher levels of suppression (Figure 2C).
To examine whether miR-16-1 and miR-191 interfere with Activin/Smad2 signaling, we employed the Activin Response Element reporter (pARE-Lux) in mESCs and analyzed the effect of a mixture of miR-16-1/miR-191 mimics on the activity upon stimulation with 25 ng/ml activin A. Whereas activin A enhanced the reporter activity, simultaneous addition of 10 μM SB431542 (an inhibitor of activin receptors) abolished the effect. Interestingly, the combined miR-16-1/miR-191 mimics inhibited the activation of the reporter by 47% (Figure 2D). To further confirm that miR-16-1 and miR-191 influenced Activin/Smad2 signaling, we examined the effect on SMAD2 and p-SMAD2 protein levels. miR-16-1/miR-191 knockdown mESCs had higher levels of SMAD2 and p-SMAD2, while mESCs transfected with miR-16-1/miR-191 mimics exhibited lower levels compared with controls (Figure 2E). These data reinforced the hypothesis that miR-16-1 and miR-191 diminish the activity of Activin/Smad2 signaling through Smad2 downregulation.
To examine whether the aforementioned miRNAs affect the mESC differentiation program, we transfected mESCs with a mixture of miR inhibitors or mimics and induced them to differentiate. As a control, mESCs treated with activin A or SB431542 was used. The efficiency of miR-16-1, miR-191 knockdown or overexpression (Figure S1D), as well as the expression of several lineage markers, was measured at EBs D0, D4, and D8. The induction of mesodermal (T, Gsc) and endodermal (Gata4, Gata6) markers were up-regulated upon inhibition of miR-16-1 and miR-191 (Figure 2F). Activin A caused an increase of mesodermal markers (T, Gsc) and the endodermal marker Gata4 while Gata6 was not affected, in line with previously published data (Lee et al., 2011). The significant increase of Gata6 induction by the addition of miR inhibitors may be attributed to Smad2 up-regulation (Fei et al., 2010). In contrast, the Sox1 ectodermal marker showed no significant changes (Figure 2F). Conversely, miR mimics reduced endoderm and mesoderm induction, similarly to the activity of SB431542 (Figure 2G). Contrary to miR mimics, SB431542 increases Gata6 induction (Lee et al., 2011). Interestingly, due to the alteration of Smad2 expression levels, the induction of trophectoderm marker (Cdx2) was significantly elevated by the miR inhibitors and lowered by the miR mimics (Figures 2F and 2G), while it remained unaffected by activin A and SB431542 (Fei et al., 2010, Lee et al., 2011).
Based on the above data, we conclude that miR-191 and miR-16-1 repress mesendoderm differentiation of mESCs through direct targeting of Smad2 and subsequent post-transcriptional control of Activin/Nodal signaling. In different settings, miR-16-1 and miR-191 are reported to regulate cell proliferation and/or cell cycle. In detail, miR-191 acts mainly as an oncomiR, but can also serve as a tumor suppressor (Nagpal and Kulshreshtha, 2014). miR-16-1 has a well-defined tumor-suppressor and cell cycle-arresting role in leukemia (Pekarsky and Croce, 2015). Our data revealed that these miRNAs did not affect the ESC cell cycle, and this difference may be attributed to the peculiar ESC cell cycle profile. It would be interesting to investigate whether Activin signaling is also involved in the tumor-regulatory functions of these miRNAs.
miR-23a Represses Ectoderm and Endoderm Differentiation of mESCs
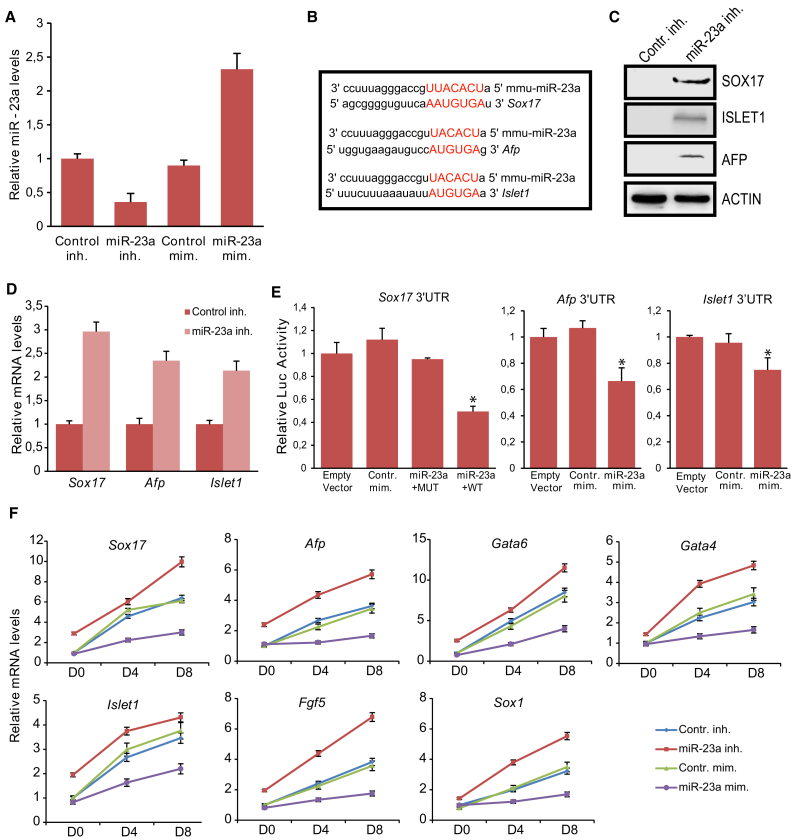
To gain insights into the potential role of miR-23a in mESCs, we used an miR-23a inhibitor and an miR-23a mimic (Figure 3A). To assess the influence of miR-23a on mESC self-renewal, we analyzed the expression levels of stemness markers (Oct4, Nanog, Nr0b1) in mESCs transfected with miR-23a inhibitor or mimic, but no difference compared with the controls was observed (Figures S2A and S2B). Furthermore, miR-23a inhibition or overexpression did not cause any changes in mESC morphology (data not shown) or cell cycle (Figure S2C).
Figure 3.
miR-23a Represses Endoderm and Ectoderm Differentiation
(A) miRNA expression levels in miR-23a mimic or inhibitor transfected mESCs. Error bars indicate SD of three independent experiments.
(B) Prediction of the binding sites of miR-23a on the 3′ UTR of the indicated differentiation-associated genes. Red indicates complementarity between miRNA and the target gene.
(C and D) miR-23a inhibition led to induction of SOX17, AFP, and ISLET1 protein (C) and mRNA (D) levels in mESCs.
(E) Relative luciferase activity of the WT 3′ UTR reporter co-transfected with mir-23a mimic. Data are shown as mean ± SD of four independent experiments. ∗p < 0.05.
(F) Relative mRNA levels of differentiation genes after EB formation in response to miR-23a inhibition or overexpression. Error bars indicate SD of three independent experiments.
Following in silico research, we identified three differentiation markers, Afp, Sox17, and Islet1, that were predicted to be targets of miR-23a (Figure 3B). Indeed, compared with controls, their protein and mRNA expression levels were induced in mESCs transfected with miR-23a inhibitor (Figures 3C and 3D), while remained constant in overexpressing miR-23a mESCs (Figure S2D). Next, through a luciferase reporter assay, we verified the direct link between miR-23a and the three differentiation markers. To verify the specificity of miR-23a binding to Sox17, we used a mutated 3′ UTR (Figure 3E). In addition, two endodermal (Gata6, Gata4) and three ectodermal (Pax6, Sox1, Fgf5) markers were up-regulated 72 hr after miR-23a inhibition (Figure S2E), whereas no effect was detected on their levels in miR-23a mimic-transfected mESCs (Figure S2F).
Sox17 has been previously reported to drive the up-regulation of the primitive endoderm-associated program, giving rise to endodermal progenitors (Niakan et al., 2010). The suppression of Sox17 and Afp, another endoderm marker gene, by miR-23a reinforces the hypothesis that miR-23a inhibits differentiation toward this lineage.
To test this assumption, we allowed mESCs transfected by miR-23a inhibitor or mimic to differentiate as EBs. miR-23a inhibition or overexpression was verified on EBs D0, D4, and D8 (Figure S2G). A significant increase in the induction of endodermal (Afp, Sox17, Gata6, Gata4) and ectodermal (Islet1, Fgf5, Sox1) genes was observed (Figure 3F) upon miR-23a inhibition, whereas trophectoderm and mesoderm lineage markers were not affected (Figure S2H). Interestingly, in miR-23a overexpressing mESCs the differentiation toward these lineages is suppressed, suggesting that the expression level of miR-23a is critical for pluripotency maintenance (Figure 3F).
The above results clearly show that miR-23a is an additional regulator of ESC differentiation. Recently, the miR-23a/24-2/27a cluster has been reported to be regulated by BMP4 and target Smad5 to protect mESCs from apoptosis during the transition to epiblast stem cells (Musto et al., 2014). In addition, miR-23a inhibits the osteoblast differentiation by targeting Runx2 (Hassan et al., 2010). In line with these observations, our results strongly support that miR-23a is a pivotal regulator of differentiation and controls ESC-specific germ-layer commitment and subsequent lineage decisions.
With respect to cancer, miR-23a has been considered either as an oncomiR (Chhabra et al., 2010) or a tumor suppressor (He et al., 2014). Apoptosis, migration, and invasion are some of its effects in cancer through regulation of molecular targets (PTEN, DAPP), while TGF-β/BMP has been implicated in the control of miR-23a expression in human cancers (Chandran et al., 2014).
In conclusion, miR-23a has a role in both tumor progression and mESC function, and the cross-regulatory relationship with TGF-β/BMP signaling awaits further investigation.
miR-421 Regulates Distinct Fate Choices of ESCs through Oct4 Repression and Competition with BMP Signaling
In contrast to the above miRNAs, miR-421 was identified as a differentiation-associated regulator, and its expression level was up-regulated during EB formation.
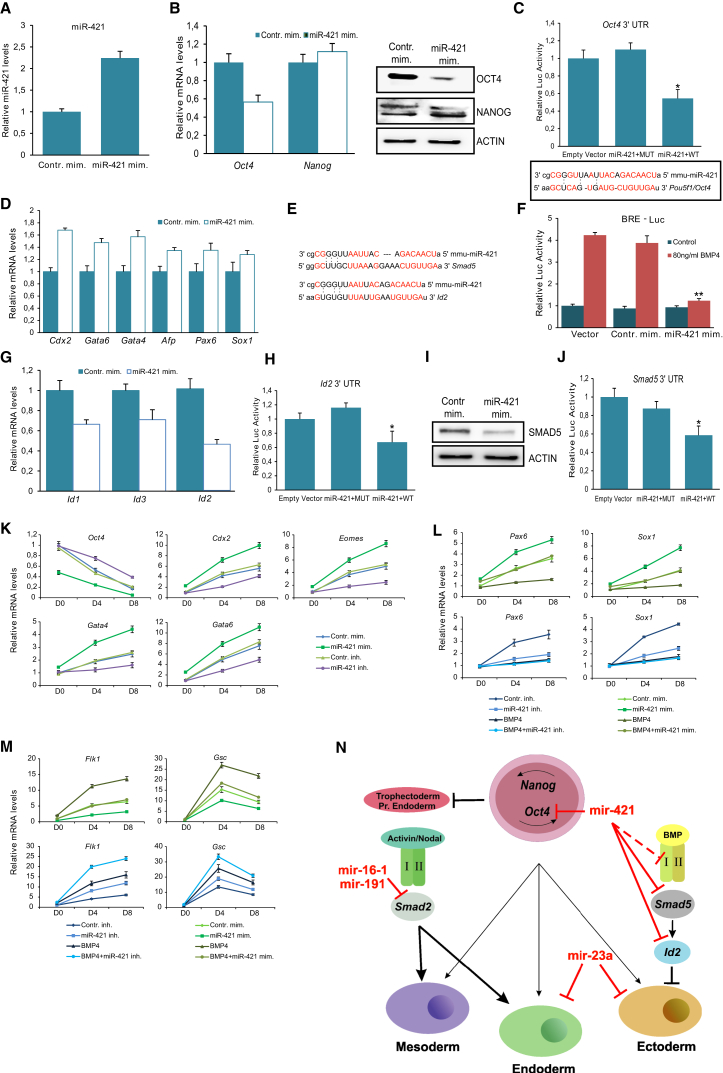
To study whether miR-421 is a crucial player in controlling differentiation, we ectopically expressed miR-421 in mESCs by using its mimic (Figure 4A). Compared with the control, miR-421 mimic had no effect on cell morphology (data not shown) and cell cycle progression (Figure S3A), but its addition significantly reduced the Oct4 expression levels (Figure 4B) while other pluripotency genes remained constant. Using bioinformatics tools (Miranda, TargetScan), miR-421 was predicted to bind the Oct4 3′ UTR, and the direct link between the two was further confirmed by luciferase reporter assay (Figure 4C).
Figure 4.
miR-421 Induces Differentiation by Suppressing Oct4 and Regulating BMP-Signaling Pathway
(A) Measurement of miRNA levels by RT-PCR after transient transfection with miR-421 mimic. Error bars indicate SD of three independent experiments.
(B) mRNA and protein levels of stemness factors (Oct4 and Nanog) after miR-421 overexpression. Error bars indicate SD of three independent experiments.
(C) mir-421 target sites in the 3′ UTR of Oct4. Luciferase activity of Oct4 3′ UTR upon miR-421 mimic supplementation. Red indicates complementarity between miRNA and the target gene. Data are shown as mean ± SD of four independent experiments. ∗p < 0.05.
(D) Relative mRNA levels of differentiation markers in miR-421-induced mESCs. Error bars indicate SD of three independent experiments.
(E) miR-421 binding sites in the 3′ UTR of Smad5 and Id2. Red indicates complementarity between miRNA and the target gene.
(F) miR-421 overexpression inhibits BRE-Luc activity. Data are shown as mean ± SD of four independent experiments. ∗∗p < 0.01.
(G) RT-PCR analysis of BMP4 target gene (Ids) expression levels in miR-421 overexpressed mESCs. Error bars indicate SD of three independent experiments.
(H) Overexpression of miR-421 decreased the luciferase activity of Id2. Data are shown as mean ± SD of four independent experiments. ∗p < 0.05.
(I and J) SMAD5 protein levels (I) and Smad5 3′ UTR luciferase activity (J) were reduced by miR-421 mimic. Data are shown as mean ± SD of four independent experiments. ∗p < 0.05.
(K) Relative mRNA levels of differentiation genes at EBs D0, D4, and D8 upon miR-421 overexpression or inhibition. Error bars indicate SD of three independent experiments.
(L and M) Relative mRNA levels of ectodermal (L) and mesodermal (M) differentiation genes at EBs D0, D4, and D8 upon miR-421 overexpression or inhibition in the presence of BMP4. Error bars indicate SD of three independent experiments.
(N) Proposed mechanism for the regulation of mESC differentiation by the aforementioned miRNAs.
To test the effect of miR-421 overexpression on differentiation, we analyzed the expression levels of several lineage markers. Interestingly, the trophectoderm marker Cdx2 was up-regulated (Figure 4D), in agreement with previous studies showing the repression of trophectoderm by Oct4 (Strumpf et al., 2005). Moreover, miR-421 overexpression was accompanied by a slight induction of primitive endoderm markers (Gata4, Gata6, Afp), which is consistent with previously published data analyzing the changes of gene expression upon inhibition of Oct4 (Hay et al., 2004, Strumpf et al., 2005). Interestingly, ectoderm-associated markers (Pax6, Sox1) were also up-regulated (Figure 4D), indicating that miR-421 might exert its action through an additional mechanism.
Due to the fact that several components of BMP signaling were predicted as candidate targets of miR-421 (Bmpr1, Smad5, Id2) (Figure 4E), we hypothesized that miR-421 may regulate this signaling and, thereby, lineage specification. Since the BMP pathway plays an important role in maintaining mESCs in the pluripotent state (Ying et al., 2003), through the activation of Id proteins acting as neuronal differentiation inhibitors (Ying et al., 2003, Zhang et al., 2010), the effect of miR-421 on BMP activity was investigated. Firstly, we confirmed that miR-421 overexpression significantly repressed the luciferase activity of the BRE-Luc reporter gene in response to BMP4 treatment compared with control (Figure 4F). Overexpression of miR-421 also reduced the mRNA expression levels of the endogenous targets of BMP signaling Id1, Id2, and Id3 (Figure 4G). Moreover, a luciferase reporter assay confirmed that miR-421 targeted directly the Id2 3′ UTR (Figure 4H). Interestingly, SMAD5 protein levels were decreased in mESCs expressing miR-421 mimic (Figure 4I), while Smad5 3′ UTR reporter assays verified the direct regulation of Smad5 by miR-421 (Figure 4J). To further analyze the function of miR-421 in differentiation, we differentiated mESCs transfected with miR-421 mimic or miR-421 inhibitor (Figure S3B). The overexpression of miR-421 favored the suppression of Oct4 and at the same time enhanced the induction of trophectoderm (Cdx2, Eomes) and endoderm (Gata4, Gata6) differentiation (Figure 4K). Concerning the induction of ectodermal markers (Pax6, Sox1), miR-421 elevation caused a significant increase, whereas the addition of BMP4 did not allow differentiation toward this lineage. miR-421 inhibitor up-regulated Oct4 expression and down-regulated the expression of trophectoderm, endoderm, and ectoderm differentiation markers (Figures 4K and 4L). Moreover, mesodermal markers (Flk1, Gsc) were not induced upon miR-421 overexpression, in contrast to miR-421 inhibition or BMP4 treatment whereby their induction was significantly raised (Figures 4M and S3C). In agreement with these data, the concurrent addition of miR-421 mimic and BMP4 did not affect the differentiation induction. It is noteworthy that ectodermal genes appeared to be decreased, while mesodermal markers were significantly increased in BMP4/miR-421 inhibitor-treated cells (Figures 4L, 4M, and S3C).
The above experimental results suggest that miR-421 is a positive regulator of mESC differentiation through two mechanisms, suppression of Oct4 and competition with BMP signaling.
Contrary to its function in mESCs, miR-421 has been previously characterized as an oncomiR in several cancers. In neuroblastoma, miR-421 suppresses ataxia-telangiectasia mutated uncoupling DNA damage from cell cycle check points (Hu et al., 2010). In pancreatic tumor cells, miR-421 represses Smad4, which is critical for BMP signal transduction, and represses its target gene Id3, promoting cell proliferation and colony formation (Hao et al., 2011). Therefore, miR-421 regulates Smad4-mediated signaling pathways in cancer cells. In addition, miR-421 is regulated by the TGF-β and BMP4 pathway in pulmonary artery smooth muscle cells, via a conserved Smad binding element (Marchand et al., 2012).
To conclude, this study unveils an miRNA-mediated mechanism for miRNAs that regulate ESC fate decisions (Figure 4N). Regarding miR-16-1, miR-191, and miR-421, this effect is due to competition with TGF-β family signaling. Inhibition of Activin/Nodal pathway by miR-16-1 and miR-191 promotes mESC maintenance, whereas competition of miR-421 with the BMP pathway results in exit of mESCs from pluripotency and their commitment to ectodermal fate. Conversely, miR-23a is itself regulated by TGF-β/BMP. Taken together, our work reveals a reciprocal antagonism between the investigated miRNAs and TGF-β signaling pathways in regulating ESC differentiation (Figure 4M). Our findings link these miRNAs with TGF-β/BMP signaling and may have implications in cancer biology, as the TGF-β pathway is a critical regulator of tumor growth, invasion, and metastasis (Drabsch and ten Dijke, 2012). miRNAs that have a parallel function in cancer and stem cells may be useful candidate molecules to advance the basic knowledge and design combinatorial strategies for cancer and cell replacement therapies.
Experimental Procedures
Cell Culture
The murine feeder-independent ESC line CGR8 was cultured in gelatin-coated flasks in Glasgow minimal essential medium (Gibco) supplemented with 500 U/ml leukaemia inhibitory factor (LIF; ESGRO-Millipore), 2 mM L-glutamine (Gibco), 100 μM β-mercaptoethanol (Gibco), and 15% heat-inactivated HyClone fetal bovine serum (FBS; GE Healthcare Life Sciences). For EB formation, cells were trypsinized and diluted in Iscove's modified Dulbecco's medium (Gibco) supplemented with the above components, to a final concentration of 1,000 cells/20 μl. EBs were cultured without LIF as hanging drops for 2 days, then collected and cultured in suspension for 6 more days.
Acknowledgments
We would like to thank V. Makatounakis and G. Vretzos for technical assistance, as well as I. Stratidaki for her contribution in miRNA sequencing profiling by the Ion Torrent platform (IMBB). We also thank Prof. D. Kardasis and Prof. C. Stournara for providing antibodies and reagents used in these experiments. This work was funded by Thalis-MIS380247-MIREG (NSRF 2007–2013), Umbistem 11SYN_10_668 and FP7-REGPOT-2012-CT2012-316223-InnovCrete.
Published: February 11, 2016
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, three figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.01.004.
Accession Numbers
The accession number for small RNA-sequencing data stated in this report is GEO: GSE76375.
Supplemental Information
References
- Bernstein E., Kim S.Y. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Betel D., Wilson M., Gabow A., Marks D.S., Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran P.A., Keller A., Weinmann L., Seida A.A., Braun M., Andreev K., Fischer B., Horn E., Schwinn S., Junker M. The TGF-beta-inducible miR-23a cluster attenuates IFN-gamma levels and antigen-specific cytotoxicity in human CD8(+) T cells. J. Leukoc. Biol. 2014;96:633–645. doi: 10.1189/jlb.3A0114-025R. [DOI] [PubMed] [Google Scholar]
- Chhabra R., Dubey R., Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23a∼27a∼24-2 cluster and its implication in human diseases. Mol. Cancer. 2010;9:232. doi: 10.1186/1476-4598-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabsch Y., ten Dijke P. TGF-beta signalling and its role in cancer progression and metastasis. Cancer Metastasis Rev. 2012;31:553–568. doi: 10.1007/s10555-012-9375-7. [DOI] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Fei T., Zhu S., Xia K., Zhang J., Li Z., Han J.D., Chen Y.G. Smad2 mediates Activin/Nodal signaling in mesendoderm differentiation of mouse embryonic stem cells. Cell Res. 2010;20:1306–1318. doi: 10.1038/cr.2010.158. [DOI] [PubMed] [Google Scholar]
- Gangaraju V.K., Lin H. MicroRNAs: key regulators of stem cells. Nat. Rev. Mol. Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailesellasse Sene K., Porter C.J., Palidwor G., Perez-Iratxeta C., Muro E.M., Campbell P.A., Rudnicki M.A., Andrade-Navarro M.A. Gene function in early mouse embryonic stem cell differentiation. BMC Genomics. 2007;8:85. doi: 10.1186/1471-2164-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J., Zhang S., Zhou Y., Liu C., Hu X., Shao C. MicroRNA 421 suppresses DPC4/Smad4 in pancreatic cancer. Biochem. Biophys. Res. Commun. 2011;406:552–557. doi: 10.1016/j.bbrc.2011.02.086. [DOI] [PubMed] [Google Scholar]
- Hassan M.Q., Gordon J.A., Beloti M.M., Croce C.M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. A network connecting Runx2, SATB2, and the miR-23a∼27a∼24-2 cluster regulates the osteoblast differentiation program. Proc. Natl. Acad. Sci. USA. 2010;107:19879–19884. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay D.C., Sutherland L., Clark J., Burdon T. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells. 2004;22:225–235. doi: 10.1634/stemcells.22-2-225. [DOI] [PubMed] [Google Scholar]
- He Y., Meng C., Shao Z., Wang H., Yang S. MiR-23a functions as a tumor suppressor in osteosarcoma. Cell Physiol. Biochem. 2014;34:1485–1496. doi: 10.1159/000366353. [DOI] [PubMed] [Google Scholar]
- Hu H., Du L., Nagabayashi G., Seeger R.C., Gatti R.A. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc. Natl. Acad. Sci. USA. 2010;107:1506–1511. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.L., Lim S.K., Orlov Y.L., Yitle Y., Yang H., Ang L.T., Poellinger L., Lim B. Graded Nodal/Activin signaling titrates conversion of quantitative phospho-Smad2 levels into qualitative embryonic stem cell fate decisions. PLoS Genet. 2011;7:e1002130. doi: 10.1371/journal.pgen.1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Marchand A., Proust C., Morange P.E., Lompré A.M., Trégouët D.A. miR-421 and miR-30c inhibit SERPINE 1 gene expression in human endothelial cells. PLoS One. 2012;7:e44532. doi: 10.1371/journal.pone.0044532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C., Judson R.L., Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A., Heldin C.H. The regulation of TGFbeta signal transduction. Development. 2009;136:3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- Musto A., Navarra A., Vocca A., Gargiulo A., Minopoli G., Romano S., Romano M.F., Russo T., Parisi S. miR-23a, miR-24 and miR-27a protect differentiating ESCs from BMP4-induced apoptosis. Cell Death Differ. 2014;22:1047–1057. doi: 10.1038/cdd.2014.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal N., Kulshreshtha R. miR-191: an emerging player in disease biology. Front. Genet. 2014;5:99. doi: 10.3389/fgene.2014.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H.H., Surani M.A. The transcriptional and signalling networks of pluripotency. Nat. Cell Biol. 2011;13:490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- Niakan K.K., Ji H., Maehr R., Vokes S.A., Rodolfa K.T., Sherwood R.I., Yamaki M., Dimos J.T., Chen A.E., Melton D.A. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 2010;24:312–326. doi: 10.1101/gad.1833510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N., Fuchs E. The harmonies played by TGF-beta in stem cell biology. Cell Stem Cell. 2012;11:751–764. doi: 10.1016/j.stem.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarsky Y., Croce C.M. Role of miR-15/16 in CLL. Cell Death Differ. 2015;22:6–11. doi: 10.1038/cdd.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D., Mao C.A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F., Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Tay Y., Zhang J., Thomson A.M., Lim B., Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Nichols J., Chambers I., Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Zhang K., Li L., Huang C., Shen C., Tan F., Xia C., Liu P., Rossant J., Jing N. Distinct functions of BMP4 during different stages of mouse ES cell neural commitment. Development. 2010;137:2095–2105. doi: 10.1242/dev.049494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.