Summary
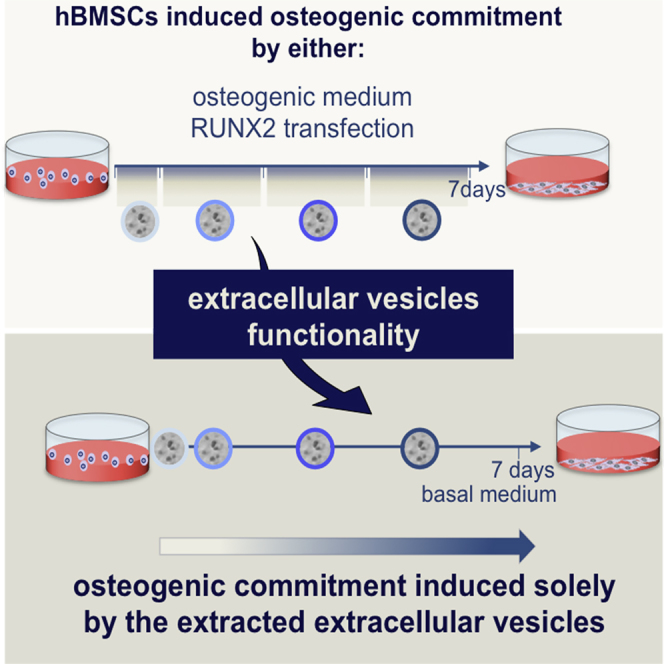
The effective osteogenic commitment of human bone marrow mesenchymal stem cells (hBMSCs) is critical for bone regenerative therapies. Extracellular vesicles (EVs) derived from hBMSCs have a regenerative potential that has been increasingly recognized. Herein, the osteoinductive potential of osteogenically induced hBMSC-EVs was examined. hBMSCs secreted negatively charged nanosized vesicles (∼35 nm) with EV-related surface markers. The yield of EVs over 7 days was dependent on an osteogenic stimulus (standard chemical cocktail or RUNX2 cationic-lipid transfection). These EVs were used to sequentially stimulate homotypic uncommitted cells during 7 days, matching the seeding density of EV parent cells, culture time, and stimuli. Osteogenically committed hBMSC-EVs induced an osteogenic phenotype characterized by marked early induction of BMP2, SP7, SPP1, BGLAP/IBSP, and alkaline phosphatase. Both EV groups outperformed the currently used osteoinductive strategies. These data show that naturally secreted EVs can guide the osteogenic commitment of hBMSCs in the absence of other chemical or genetic osteoinductors.
Graphical Abstract
Highlights
-
•
hBMSC-EV secretion during culture is osteogenic stimulus dependent
-
•
Osteogenically induced hBMSC-EVs are early osteoinductors
-
•
Osteogenically induced hBMSC-EVs outperform currently used osteoinductive strategies
In this article, Neves and colleagues show that hBMSCs secrete populations of nanosized EVs during the osteogenic commitment induced either by standard osteogenic medium or by Runx-2 transfection. Furthermore, the EVs obtained during both hBMSC osteogenic induction methods carry potent signals that are efficient in driving the osteogenic lineage progression of homotypic cells.
Introduction
Human bone marrow mesenchymal stem cells (hBMSCs) are attractive candidates for advanced cell therapies, including bone regeneration (Costa-Pinto et al., 2012). Satisfactory treatments are hampered by the difficulty in obtaining a well-defined population of terminally differentiated cells. This causes heterogeneity of hBMSCs, enhances the possibility of spontaneous differentiation into other lineages (Nombela-Arrieta et al., 2011), and may shorten the cells’ engraftment-activation time (Tsubota et al., 1999). Therefore, approaches beyond the standard chemical cocktails have been investigated (Heng et al., 2004) such as genetic modulation through the overexpression of genes (e.g., runt-related transcription factor 2, RUNX2) (Karsenty et al., 2009, Monteiro et al., 2014), and the use of synthetic/recombinant factors such as bone morphogenetic protein 4 or cell-derived conditioned medium (CM). Indeed, CM contains an array of growth factors, cytokines, proteins (Makridakis et al., 2013), and the recently highlighted extracellular vesicles (EVs) (Collino et al., 2010). EVs are nanosized particles (exosomes, 30–100 nm; microvesicles, 50–2000 nm) carrying lipids, proteins, and nucleic acids (Akers et al., 2013). It has been suggested that hBMSC-secreted EVs include differentiation cues (miRNA, Collino et al., 2010, Baglio et al., 2015; tRNA, Baglio et al., 2015; and proteins, Kim et al., 2012), even upon osteogenic induction (Xu et al., 2014). The regenerative potential of MSC-EVs is supported by preclinical studies showing the improvement of at least one clinical outcome associated with acute kidney/liver/lung injury, myocardial infarction, or hindlimb ischemia (Akyurekli et al., 2015). Furthermore, the literature shows that hBMSCs undergo osteogenic differentiation induced by EVs derived from monocytes (Ekstrom et al., 2013) or platelet lysate (Torreggiani et al., 2014). These data enable us to hypothesize that EVs may be vehicles of communication toward tissue regeneration. The knowledge on the bone regenerative potential of hBMSC-EVs is scarce. Therefore, the purpose of this study is to validate the functionality of hBMSC-EVs in the osteoinduction of hBMSCs. We hypothesized that if EVs mirror the content and fate of parent cells, then EVs derived from osteogenically committed hBMSCs will induce the osteogenic commitment of homotypic cells without further supplementation.
Results
hBMSCs Exposed to Osteogenic Stimuli Secrete EVs during Culture
hBMSCs were induced into the osteogenic lineage over 7 days by continuous chemical stimuli provided by standard osteogenic medium (OM) (Jaiswal et al., 1997) or by a single genetic stimulus in basal medium (RUNX-2). We next isolated EVs from the CM of chemically and genetically induced hBMSCs (OM-EVs and RUNX2-EVs, respectively) at specific time points using polymeric precipitation.
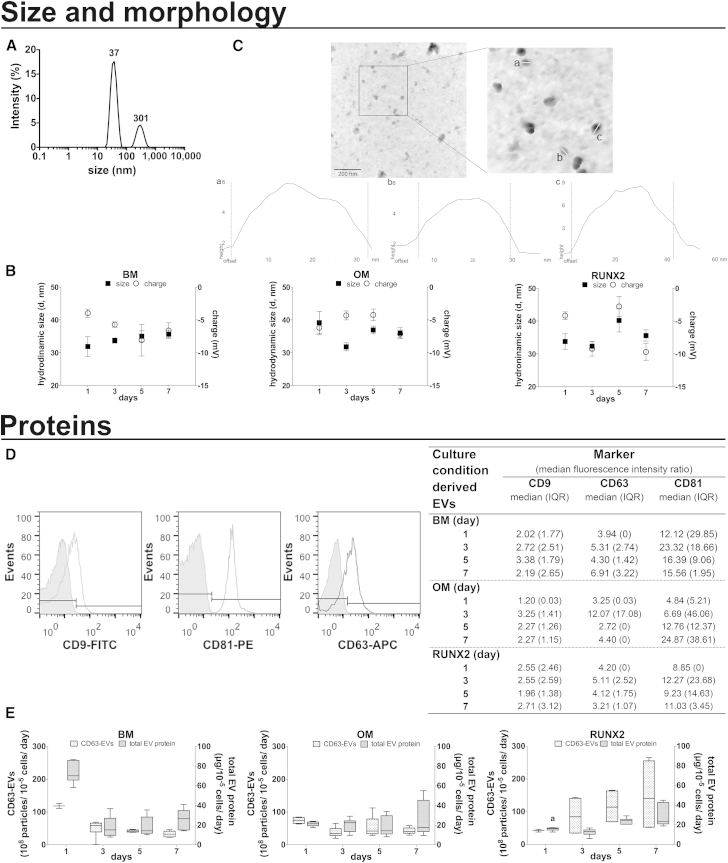
EVs showed a polydisperse size distribution by dynamic light scattering (Figure 1A), with polydispersity indexes between 0.15 and 0.6 (min and max), independently of the culture conditions and time. The main peak corresponded to ≥65% of the population (65.2%–88%, 68.7%–73.5%, 76.2%–84%, min-max for OM-, RUNX2-, basal medium [BM]-EVs, respectively). Within this population, the diameter of EVs was very homogeneous (31.9–40.2 nm), irrespective of culture time or stimulus (Figure 1B). In contrast, the size of EVs within the second peak (Figure 1A) ranged between 45 and 348.6 nm. These findings were corroborated by atomic force microscopy, which showed a population of intact rounded structures with estimated desiccated diameters of 32, 40, and 24 nm (Figure 1C, a–c). On the other hand, as assessed by laser Doppler micro-electrophoresis, the surface charge of OM-, RUNX2-, and BM-EVs averaged at −5.2, −6.4, and −5.9 mV, respectively, and was similar within the different groups (Figure 1B).
Figure 1.
Characterization of Osteogenically Induced hBMSC-EVs
EVs were isolated from the CM of hBMSCs under osteogenic medium (OM) or RUNX2 transcriptionally activated in basal medium (BM, RUNX2).
(A and B) EV size and charge. Representative size distribution profile highlighting the mean particle diameters (A); and EV size within the peak with main intensity (left axis) and charge (right axis), shown as the mean ± SEM (three donors, three technical replicates) (B).
(C) EV morphology. Representative atomic force micrographs (top) and graphical representation of individual EV size (a–c) (bottom).
(D) EV enrichment in CD9, CD63, and CD81 analyzed by flow cytometry. Representative histograms of labeled (open peak) and unlabeled (filled peak) EVs and the median fluorescence intensity ratio (median fluorescence intensity of the detected molecule divided by that of the non-labeled control), shown as the median (interquartile range [IQR]) (three donors, two technical replicates).
(E) EV yield in terms of normalized CD63-positive EVs (left axis) and normalized total EV protein (right axis), shown as the mean ± SEM (three donors, two and three technical replicates, respectively). a denotes significant differences compared with BM.
To shed light into the biochemical profile of EVs, we first used flow cytometry (Figure 1D) to detect the tetraspanins CD9/63/81, expected to be enriched in EVs fractions (Lötvall et al., 2014). These analyses showed that EVs labeled for each of the markers show a distinct positive shift of the fluorescence signal beyond the non-labeled controls. Furthermore, the data suggested abundant CD81-positive and weak CD9-positive EVs populations, despite the culture conditions (Figure 1D). The presence of CD63-positive EVs was further confirmed using ELISA (Figure 1E).
In addition, we aimed at estimating the release of EVs during culture, based on the normalized number of CD63 particles and total EV protein (Figure 1E). An osteogenic stimulus-dependent release was observed, differing from that observed in control conditions. Specifically, while for OM cultured cells the release of EVs remained at a steady level over time, for RUNX2 stimulated cells the secretion of EVs, in terms of total protein, increased over time peaking from the fifth day of culture onward (day 3 versus 5 or versus 7, p < 0.01; Figure 1E).
Collectively, these results show that hBMSCs secrete populations of nanosized particles with physical and biochemical features of EVs, and the yield is regulated by the osteogenic stimulus provided.
hBMSCs Osteogenically Derived EVs Outperform Current Strategies to Elicit hBMSCs Osteogenic Commitment
To test our hypothesis, OM- and RUNX2-EVs were isolated at set time points during culture (Figure 1E) and used to sequentially feed uncommitted homotypic hBMSCs, matching seeding density, culture time, and stimuli without further supplementation. Therefore, the concentration of EVs added to culture mimicked the specific release of parent cells into the CM.
We first assessed whether EVs could interact and deliver nucleic acids to homotypic recipient cells. RUNX2-EVs isolated from 1-day cultured cells expressing RUNX2-GFP tag, were added to uncommitted hBMSCs. After 1 day, the GFP expression was on average 233-fold greater versus BM in cells exposed to RUNX2-EVs (6.8–803, min-max), although at lower levels than that observed for parent cells transfected by lipofection (average 1.6 × 106 fold change, 4 × 104–7 × 106, min-max). This suggests that EVs are able to interact with the recipient cells and transfer functional plasmid DNA.
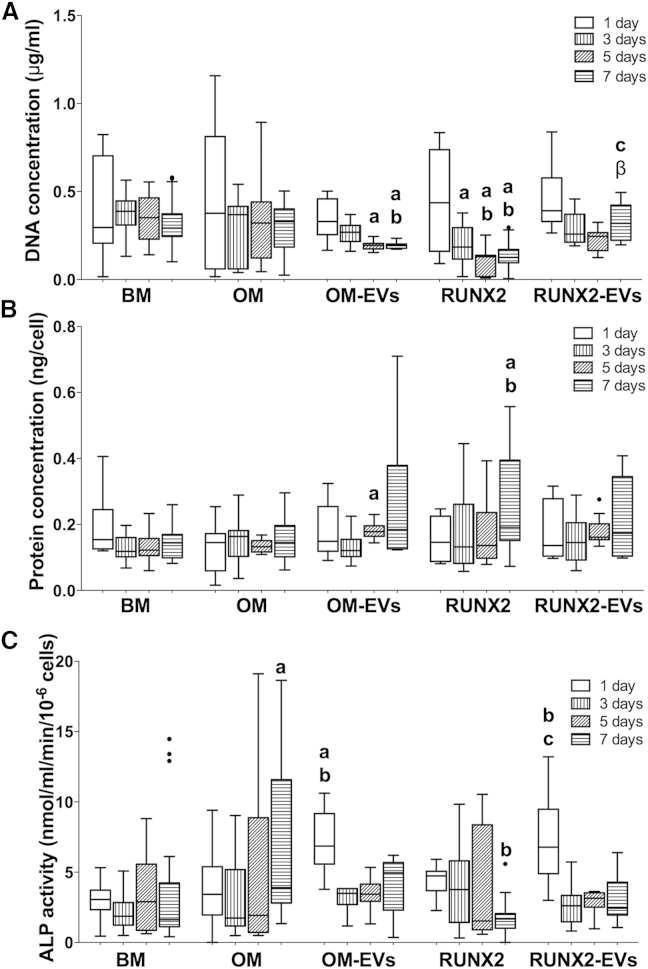
The ability of OM- and RUNX2-EVs to promote the onset of osteogenesis was examined and compared with currently available strategies. The differentiation process is known to impair cell proliferation, due to an increase in the length of the cell cycle (Roccio et al., 2013), and to induce changes in the protein synthesis rate (Kristensen et al., 2013). Therefore, we evaluated the profile of recipient hBMSC proliferation (Figure 2A) and total protein (Figure 2B). Notably, a high seeding density was used to ensure high transfection efficiency. Under these conditions, the proliferation capacity of OM-EV-treated cells decreased after day 3 versus BM and OM (Figure 2A), in parallel with a net protein increase (day 5, versus BM) (Figure 2B). Furthermore, RUNX2-EV treatment did not impair cell proliferation versus BM, in contrast to parent cells (RUNX2), attaining proliferation levels higher than those of parent and OM-EV-exposed cells at the seventh day (Figure 2A), without major changes in total protein (Figure 2B).
Figure 2.
Effect of Osteogenically Induced hBMSC-EVs on the Biochemical Parameters of Homotypic Cells
EVs derived from hBMSCs under osteogenic medium (OM), OM-EVs, or RUNX2 transcriptionally activated in basal medium (BM), RUNX2-EVs, were transferred to uncommitted hBMSCs without further supplementation. Cell lysates (A) DNA concentration, (B) normalized protein, (C) normalized ALP activity are shown as Tukey boxplots (three donors, three technical replicates). The black dots denote outliers; letters denote significant differences compared with: a, BM; b, OM; c, RUNX2; β, OM-EVs.
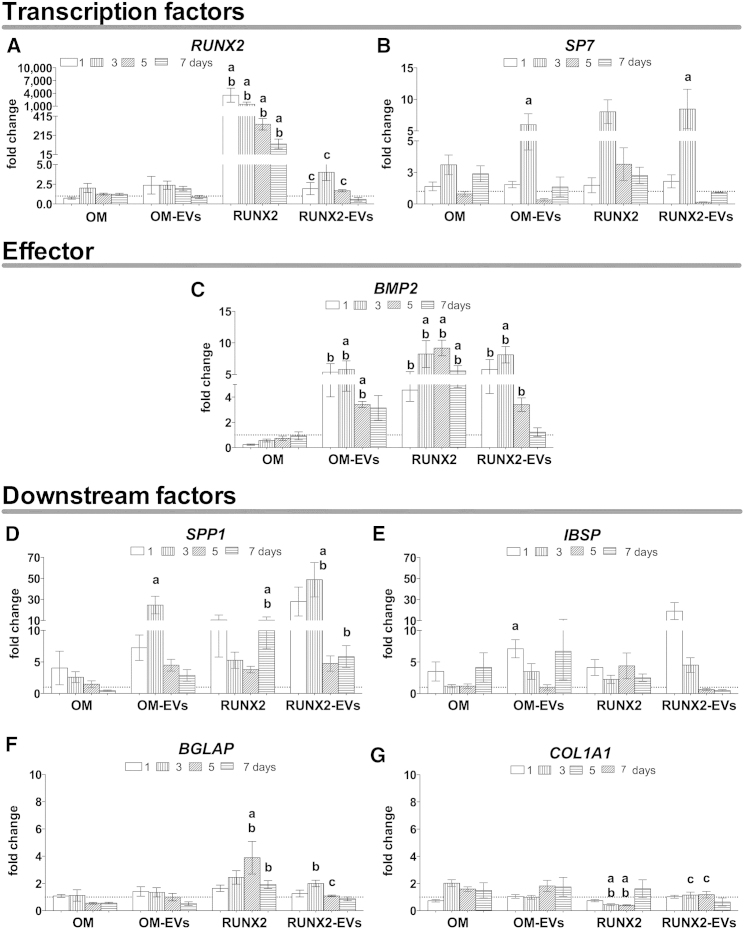
The osteoinductive potential of EVs was then evaluated in terms of the expression of the extracellular matrix marker alkaline phosphatase (ALP) (Figure 2C). OM- and RUNX-2 EVs induced early activation of ALP, in contrast to that exhibited by the parent cells. Notably, this was observed in the absence of the mineralization inductors dexamethasone and β-glycerophosphate (Langenbach and Handschel, 2013). To better define the osteogenic commitment of these cells, the temporal gene expression of osteogenic markers was investigated (Figure 3). OM-EV-cultured hBMSCs showed early overexpression of the activator bone morphogenetic protein 2 (BMP2) (Figure 3C) by ∼3- to 6-fold versus BM and OM, and a transient increase in the expression of the Sp7 transcription factor (osterix) (SP7) (Figure 3B) by 6-fold versus BM at day 3. Indeed, osteoblastic differentiation is determined by the overexpression of SP7 (Sinha and Zhou, 2013) through the BMP-2 signaling cascade (Ulsamer et al., 2008). Herein, the data suggest early activation of the cascade independent of the upstream factor RUNX2. In addition, at early culture times, the downstream factors secreted phosphoprotein 1 (osteopontin) (SPP1) (Figure 3D) and integrin-binding sialoprotein (bone sialoprotein) (IBSP) (Figure 3E) were strongly upregulated (approximately 25-fold and 7-fold, respectively) in comparison with BM. Furthermore, RUNX2-EV-cultured hBMSCs showed lower mRNA RUNX2 levels than those of parent cells stimulated to overexpress this gene, but similar to that observed in BM or OM (Figure 3A). As observed for OM-EVs, at early culture times, RUNX2-EVs induced the overexpression of BMP2 by 3- to 8-fold (versus BM and OM) (Figure 3C) and that of SP7 by 8-fold (versus BM) (Figure 3B), indispensable for progression onto the osteogenic lineage. Also consistent with osteogenic activation, the expression levels of SPP1 (Figure 3D), and importantly those of bone γ-carboxyglutamate (gla) protein (osteocalcin) (BGLAP) (Figure 3F) were increased by 48- and 2-fold versus OM and/or BM, respectively. However, IBSP expression (Figure 3E) was only slightly increased on the first day of culture (versus BM, p = 0.02), whereas collagen type I alpha 1 (COL1A1) expression (Figure 3G) remained at the BM level although at higher levels than those observed for parent cells.
Figure 3.
Effect of Osteogenically Induced hBMSC-EVs on Osteogenic Lineage-Related Markers of Homotypic Cells
EVs derived from hBMSCs under osteogenic medium (OM), OM-EVs, or RUNX2 transcriptionally activated in basal medium (BM), RUNX2-EVs, were transferred to uncommitted hBMSCs without further supplementation. BM was set as control. qPCR data for relative gene expression of (A and B) transcription factors, (C) effector, and (D–G) downstream factors, are shown as means ± SEM (three donors, three technical replicates). Letters denote significant differences compared with: a, BM; b, OM; c, RUNX2.
These results show that extracellular signaling derived from polymeric precipitated EVs obtained during hBMSCs osteogenic induction provides guidance for osteogenic lineage progression of homotypic cells.
Discussion
Bone regenerative therapies have been challenged by the limited understanding of the induction of hBMSC fate. This hinders the clinical translation of the most promising strategies. For example, genetically modified cells raise safety, efficacy, and fate issues (Kumar et al., 2008); synthetic/recombinant factors, used at supra-physiological doses, are associated with severe side effects and high costs (Jakob et al., 2012); the CM concentration of biotherapeutics is low, at least for some applications, and contains medium contaminants, such as phenol red (Tran and Damaser, 2015).
Our study demonstrates that hBMSCs secrete vesicles with features of EVs that have osteoinductive potential. We showed that hBMSCs secrete a population of EVs heterogeneous in size, as noted earlier (Sokolova et al., 2011, Lobb et al., 2015, Van Deun et al., 2014). Although this can be attributed to the method of EV isolation (Lobb et al., 2015, Van Deun et al., 2014) or to sample aggregation, the sizes obtained (Figures 1A–1C) roughly fall within the range reported for hBMSC-EVs, 47–180 nm (Bian et al., 2014, Bruno et al., 2013). Consistent with MSCs expressing CD9 (Chen et al., 2010), CD63 (Stewart et al., 2003), and CD81 (Lee et al., 2009), EVs also showed these surface markers (Figures 1D and 1E), suggesting that they originated at the parent cell plasma membrane lipid rafts (Tan et al., 2013). Therefore, it is reasonable to accept that the parent cell source affects the EVs’ surface proteins, their glycosylation or lipid composition, and consequently their charge. We found that the surface charge of hBMSC-EVs was relatively less negative (Figure 1B) than that reported for umbilical cord MSC-EVs, −52 mV (Sokolova et al., 2011). Furthermore, our data showed that the environmental stimuli affect the yield of EVs (Figure 1E) probably in parallel with cell activation (Figures 2A and 2B). This is in line with the oxygen-dependent placental secretion of MSC-EVs (Salomon et al., 2013). Due to their biophysical and biochemical properties, namely the size (Figures 1A–1C), EVs may interact differentially with cells. In general, the uptake of EVs is dose, time (Franzen et al., 2014), and recipient-cell (Feng et al., 2010) dependent, while the fate of the EV cargo varies with the particle subtype (Kanada et al., 2015). We indirectly showed that EVs added in concentrations comparable with those produced in vitro undergo uptake by hBMSCs, although the subtype and percentage of EVs being internalized was not determined.
The exogenous addition of osteogenically derived EVs outperformed the effect of current osteoinductive strategies in terms of the type and/or intensity of the signaling, with the early activation of the key osteogenic commitment genes SP7 (Figure 3B) and BMP2 (Figure 3C), considered necessary and sufficient to induce bone formation (Noel et al., 2004). In addition, EVs induced a transient upregulation of downstream matrix-associated genes and protein (Figures 3D, 3E, and 2C) that may be required to promote cell-cell and cell-matrix interactions and support long-term differentiation and mineralization (Langenbach and Handschel, 2013).
Notably, no major differences were noted between the exogenous addition of OM- or RUNX2-EVs, suggesting that the inherent cargo of hBMSC-EVs may also contribute to the observed effect. Indeed, the selective cargo of EVs derived from uncommitted hBMSCs suggests their involvement in the osteogenic differentiation. MSC-EVs were described to carry tRNA species putatively targeting the transcription factor RUNX2 and SOX11 (Baglio et al., 2015), miRNA, miR-22, that indirectly targets RUNX2 (Baglio et al., 2015) and several proteins involved in the BMP, mitogen-activated protein kinase, transforming growth factor β, and Wnt pathways (Kim et al., 2012). Furthermore, our experimental design, comprising the addition of a pool of OM-/RUNX2-EVs to hBMSCs, does not exclude the contribution of the EVs derived from recipient cells and a possible synergistic signaling.
Our results show the beneficial effects of cell-secreted factors, confirming that hBMSCs secrete EVs that are key players in the modulation of cell fate. We were able to identify and isolate unique factors that regulate the osteogenic commitment. Therefore, EVs are potential cell-free/secretome-based therapies for gene delivery applications that may circumvent the risks associated with current therapies and provide greater safety and efficacy benefits.
Experimental Procedures
hBMSC Isolation and Culture
hBMSCs were obtained from three patients (aged 55, 68, and 71 years) after informed consent, as approved by the Ethical Committee of the Hospital Center of Alto Ave, Guimarães, under the Cooperation Agreement established between the 3B's Research Group, University of Minho and this hospital. hBMSCs were characterized and cultured using our standard protocols (Monteiro et al., 2014).
Osteogenic Commitment of Chemically and Genetically Induced hBMSCs
The following experiments were carried out using minimum essential medium alpha medium with fetal bovine serum depleted from EVs (Thery et al., 2006). The osteogenic commitment of adhered hBMSCs (passage 4; 750 cells/coverslip mm2; 1 ml/well) was induced by chemical stimuli (BM with 10−8 M dexamethasone, 50 μg/ml ascorbic acid, and 0.01 M β-glycerophosphate; Sigma); or by genetic modulation (RUNX2 cationic-lipid transfection in BM). The transfection mixture comprised 500 ng of plasmid DNA and a lipid-DNA ratio of 2× (Lipofectamine LTX with Plus Reagents, Alfagene, Life Technologies). After 5 hr the transfection mixture was replaced by 1 ml of BM. The transfection efficiency was evaluated by GFP reporter gene expression, showing a transient overexpression of RUNX2 (Figure S1). See Supplemental Experimental Procedures for additional details. hBMSCs cultured in BM were set as control. For all the conditions, the CM was harvested at 1, 3, 5, and 7 days. The cells were rinsed, immersed in 1 ml of water or 0.8 ml of Tri-reagent (Sigma) and stored at −80°C.
Isolation of EVs
EVs were isolated from CM using a polymeric precipitation solution (ExoQuick-TC; System Biosciences, BioCat) according to the manufacturer's instructions. See Supplemental Experimental Procedures for additional details, including validation (Figure S2).
Characterization of Osteogenically Induced hBMSC-EVs
EV size was evaluated by DLS, charge by laser Doppler micro-electrophoresis (Malvern Nano ZS), and morphology by AFM (Dimension Icon, Bruker) at 15 μg of total protein/ml. EV total protein was evaluated by Micro BCA (Fisher Scientific) and CD63 by ELISA (EXOEL CD63, System Biosciences), according to the manufacturer's instructions; and CD9/63/81 by flow cytometry (Lasser et al., 2012). See Supplemental Experimental Procedures for additional details.
Transfer of Osteogenically Induced hBMSC-EVs into Uncommitted Homotypic Cells
Adhered hBMSCs (passage 4; 750 cells/coverslip mm2; 1 ml/well) were fed with BM containing EVs derived from osteogenically induced hBMSCs after 1 day of culture, matching the seeding density. At days 1, 3, and 5 the recipient cells were fed with BM containing EVs derived from osteogenically induced hBMSCs, matching the seeding density, culture time, and stimuli. Cells were retrieved at 1, 3, 5, and 7 days and processed as the EV parent cells.
Cellular Biochemistry Analyses
Cell proliferation was evaluated by DNA quantification (Quant-iT PicoGreen dsDNA assay, Invitrogen, Alfagene), cellular protein by Micro BCA assay, according to the manufacturer's instructions; and alkaline phosphatase specific activity by a colorimetric assay (Monteiro et al., 2014). The number of hBMSCs was estimated based on an experimental standard curve (DNA = 0.0017 × cell number + 95.25, R2 = 0.98).
Gene Expression Analyses
Total RNA was isolated and reverse transcribed into cDNA (qScript cDNA synthesis kit, Quanta BioSciences, VWR), followed by qPCR (PerfeCTa SYBR Green FastMix, Quanta BioSciences), according to the manufacturer's instructions. See Supplemental Experimental Procedures for additional details.
Statistical Analyses
Data normality was evaluated by the D'Agostino-Pearson test. A non-parametric Kruskal-Wallis test was applied followed by Dunn's test for multiple comparisons (GraphPad, version 6.0). Statistical significance was defined at p < 0.01.
Authors Contributions
Study Conception and Design, M.M., A.M., R.L.R., N.M.N.; Acquisition of Data, M.M., D.R., A.M.; Analysis and Interpretation of Data, M.M., D.R., A.M., N.M.N.; Drafting of manuscript, M.M., A.M., N.M.N.; Critical Revision, N.M.N., R.L.R.
Acknowledgments
The authors thank the financial support of QREN (RL1-ABMR-NORTE-01-0124-FEDER-000016 and RL3–TECT–NORTE-01-0124-FEDER-000020) co-financed by North Portugal Regional Operational Program (ON.2–O Novo Norte), under the NSRF, through the ERDF; the European Union's Seventh Framework Program (FP7/2007–2013), Grant No. REGPOT-CT2012-316331- POLARIS; and the laboratory assistance of Elsa Ribeiro, Magda Graça, and Sónia Zacarias.
Published: February 25, 2016
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, two figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.01.001.
Supplemental Information
References
- Akers J.C., Gonda D., Kim R., Carter B.S., Chen C.C. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol. 2013;113:1–11. doi: 10.1007/s11060-013-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akyurekli C., Le Y., Richardson R.B., Fergusson D., Tay J., Allan D.S. A systematic review of preclinical studies on the therapeutic potential of mesenchymal stromal cell-derived microvesicles. Stem Cell Rev. 2015;11:150–160. doi: 10.1007/s12015-014-9545-9. [DOI] [PubMed] [Google Scholar]
- Baglio S.R., Rooijers K., Koppers-Lalic D., Verweij F.J., Perez Lanzon M., Zini N., Naaijkens B., Perut F., Niessen H.W., Baldini N., Pegtel D.M. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 2015;6:127. doi: 10.1186/s13287-015-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian S., Zhang L., Duan L., Wang X., Min Y., Yu H. Extracellular vesicles derived from human bone marrow mesenchymal stem cells promote angiogenesis in a rat myocardial infarction model. J. Mol. Med. 2014;92:387–397. doi: 10.1007/s00109-013-1110-5. [DOI] [PubMed] [Google Scholar]
- Bruno S., Collino F., Deregibus M.C., Grange C., Tetta C., Camussi G. Microvesicles derived from human bone marrow mesenchymal stem cells inhibit tumor growth. Stem Cells Dev. 2013;22:758–771. doi: 10.1089/scd.2012.0304. [DOI] [PubMed] [Google Scholar]
- Chen T.S., Lai R.C., Lee M.M., Choo A.B., Lee C.N., Lim S.K. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res. 2010;38:215–224. doi: 10.1093/nar/gkp857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collino F., Deregibus M.C., Bruno S., Sterpone L., Aghemo G., Viltono L., Tetta C., Camussi G. Microvesicles derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PLoS One. 2010;5:e11803. doi: 10.1371/journal.pone.0011803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Pinto A.R., Correlo V.M., Sol P.C., Bhattacharya M., Srouji S., Livne E., Reis R.L., Neves N.M. Chitosan-poly(butylene succinate) scaffolds and human bone marrow stromal cells induce bone repair in a mouse calvaria model. J. Tissue Eng. Regen. Med. 2012;6:21–28. doi: 10.1002/term.391. [DOI] [PubMed] [Google Scholar]
- Ekstrom K., Omar O., Graneli C., Wang X., Vazirisani F., Thomsen P. Monocyte exosomes stimulate the osteogenic gene expression of mesenchymal stem cells. PLoS One. 2013;8:e75227. doi: 10.1371/journal.pone.0075227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D., Zhao W.L., Ye Y.Y., Bai X.C., Liu R.Q., Chang L.F., Zhou Q., Sui S.F. Cellular internalization of exosomes occurs through phagocytosis. Traffic. 2010;11:675–687. doi: 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- Franzen C.A., Simms P.E., Van Huis A.F., Foreman K.E., Kuo P.C., Gupta G.N. Characterization of uptake and internalization of exosomes by bladder cancer cells. Biomed. Res. Int. 2014;2014:619829. doi: 10.1155/2014/619829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng B.C., Cao T., Stanton L.W., Robson P., Olsen B. Strategies for directing the differentiation of stem cells into the osteogenic lineage in vitro. J. Bone Miner. Res. 2004;19:1379–1394. doi: 10.1359/JBMR.040714. [DOI] [PubMed] [Google Scholar]
- Jaiswal N., Haynesworth S.E., Caplan A.I., Bruder S.P. Osteogenic differentiation of purified, culture-expanded human mesenchymal stem cells in vitro. J. Cell. Biochem. 1997;64:295–312. [PubMed] [Google Scholar]
- Jakob M., Saxer F., Scotti C., Schreiner S., Studer P., Scherberich A., Heberer M., Martin I. Perspective on the evolution of cell-based bone tissue engineering strategies. Eur. Surg. Res. 2012;49:1–7. doi: 10.1159/000338362. [DOI] [PubMed] [Google Scholar]
- Kanada M., Bachmann M.H., Hardy J.W., Frimannson D.O., Bronsart L., Wang A., Sylvester M.D., Schmidt T.L., Kaspar R.L., Butte M.J. Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc. Natl. Acad. Sci. USA. 2015;112:1433–1442. doi: 10.1073/pnas.1418401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G., Kronenberg H.M., Settembre C. Genetic control of bone formation. Annu. Rev. Cell Dev. Biol. 2009;25:629–648. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- Kim H.S., Choi D.Y., Yun S.J., Choi S.M., Kang J.W., Jung J.W., Hwang D., Kim K.P., Kim D.W. Proteomic analysis of microvesicles derived from human mesenchymal stem cells. J. Proteome Res. 2012;11:839–849. doi: 10.1021/pr200682z. [DOI] [PubMed] [Google Scholar]
- Kristensen A.R., Gsponer J., Foster L.J. Protein synthesis rate is the predominant regulator of protein expression during differentiation. Mol. Syst. Biol. 2013;9:689. doi: 10.1038/msb.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Chanda D., Ponnazhagan S. Therapeutic potential of genetically modified mesenchymal stem cells. Gene Ther. 2008;15:711–715. doi: 10.1038/gt.2008.35. [DOI] [PubMed] [Google Scholar]
- Langenbach F., Handschel J. Effects of dexamethasone, ascorbic acid and beta-glycerophosphate on the osteogenic differentiation of stem cells in vitro. Stem Cell Res. Ther. 2013;4:117. doi: 10.1186/scrt328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasser C., Eldh M., Lotvall J. Isolation and characterization of RNA-containing exosomes. J. Vis. Exp. 2012;59:e3037. doi: 10.3791/3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.J., Choi B.H., Min B.H., Park S.R. Changes in surface markers of human mesenchymal stem cells during the chondrogenic differentiation and dedifferentiation processes in vitro. Arthritis Rheum. 2009;60:2325–2332. doi: 10.1002/art.24786. [DOI] [PubMed] [Google Scholar]
- Lobb R.J., Becker M., Wen S.W., Wong C.S., Wiegmans A.P., Leimgruber A., Moller A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles. 2015;4:27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lötvall J., Hill A.F., Hochberg F., Buzás E.I., Di Vizio D., Gardiner C., Gho Y.S., Kurochkin I.V., Mathivanan S., Quesenberry P.J. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makridakis M., Roubelakis M.G., Vlahou A. Stem cells: insights into the secretome. Biochim. Biophys. Acta. 2013;1834:2380–2384. doi: 10.1016/j.bbapap.2013.01.032. [DOI] [PubMed] [Google Scholar]
- Monteiro N., Ribeiro D., Martins A., Faria S., Fonseca N.A., Moreira J.N., Reis R.L., Neves N.M. Instructive nanofibrous scaffold comprising runt-related transcription factor 2 gene delivery for bone tissue engineering. ACS Nano. 2014;8:8082–8094. doi: 10.1021/nn5021049. [DOI] [PubMed] [Google Scholar]
- Noel D., Gazit D., Bouquet C., Apparailly F., Bony C., Plence P., Millet V., Turgeman G., Perricaudet M., Sany J., Jorgensen C. Short-term BMP-2 expression is sufficient for in vivo osteochondral differentiation of mesenchymal stem cells. Stem Cells. 2004;22:74–85. doi: 10.1634/stemcells.22-1-74. [DOI] [PubMed] [Google Scholar]
- Nombela-Arrieta C., Ritz J., Silberstein L.E. The elusive nature and function of mesenchymal stem cells. Nat. Rev. Mol. Cell Biol. 2011;12:126–131. doi: 10.1038/nrm3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccio M., Schmitter D., Knobloch M., Okawa Y., Sage D., Lutolf M.P. Predicting stem cell fate changes by differential cell cycle progression patterns. Development. 2013;140:459–470. doi: 10.1242/dev.086215. [DOI] [PubMed] [Google Scholar]
- Salomon C., Ryan J., Sobrevia L., Kobayashi M., Ashman K., Mitchell M., Rice G.E. Exosomal signaling during hypoxia mediates microvascular endothelial cell migration and vasculogenesis. PLoS One. 2013;8:e68451. doi: 10.1371/journal.pone.0068451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha K.M., Zhou X. Genetic and molecular control of osterix in skeletal formation. J. Cell. Biochem. 2013;114:975–984. doi: 10.1002/jcb.24439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova V., Ludwig A.K., Hornung S., Rotan O., Horn P.A., Epple M., Giebel B. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf. B Biointerfaces. 2011;87:146–150. doi: 10.1016/j.colsurfb.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Stewart K., Monk P., Walsh S., Jefferiss C.M., Letchford J., Beresford J.N. STRO-1, HOP-26 (CD63), CD49a and SB-10 (CD166) as markers of primitive human marrow stromal cells and their more differentiated progeny: a comparative investigation in vitro. Cell Tissue Res. 2003;313:281–290. doi: 10.1007/s00441-003-0762-9. [DOI] [PubMed] [Google Scholar]
- Tan S.S., Yin Y., Lee T., Lai R.C., Yeo R.W., Zhang B., Choo A., Lim S.K. Therapeutic MSC exosomes are derived from lipid raft microdomains in the plasma membrane. J. Extracell. Vesicles. 2013;2:22614. doi: 10.3402/jev.v2i0.22614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;Chapter 3 doi: 10.1002/0471143030.cb0322s30. Unit 3.22. [DOI] [PubMed] [Google Scholar]
- Torreggiani E., Perut F., Roncuzzi L., Zini N., Baglio S.R., Baldini N. Exosomes: novel effectors of human platelet lysate activity. Eur. Cell. Mater. 2014;28:137–151. doi: 10.22203/ecm.v028a11. [DOI] [PubMed] [Google Scholar]
- Tran C., Damaser M.S. Stem cells as drug delivery methods: application of stem cell secretome for regeneration. Adv. Drug Deliv. Rev. 2015;82-83:1–11. doi: 10.1016/j.addr.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota S., Tsuchiya H., Shinokawa Y., Tomita K., Minato H. Transplantation of osteoblast-like cells to the distracted callus in rabbits. J. Bone Joint Surg. Br. 1999;81:125–129. doi: 10.1302/0301-620x.81b1.9018. [DOI] [PubMed] [Google Scholar]
- Ulsamer A., Ortuno M.J., Ruiz S., Susperregui A.R., Osses N., Rosa J.L., Ventura F. BMP-2 induces Osterix expression through up-regulation of Dlx5 and its phosphorylation by p38. J. Biol. Chem. 2008;283:3816–3826. doi: 10.1074/jbc.M704724200. [DOI] [PubMed] [Google Scholar]
- Van Deun J., Mestdagh P., Sormunen R., Cocquyt V., Vermaelen K., Vandesompele J., Bracke M., De Wever O., Hendrix A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles. 2014;3:24858. doi: 10.3402/jev.v3.24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.F., Yang G.H., Pan X.H., Zhang S.J., Zhao C., Qiu B.S., Gu H.F., Hong J.F., Cao L., Chen Y. Altered MicroRNA expression profile in exosomes during osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. PLoS One. 2014;9:e114627. doi: 10.1371/journal.pone.0114627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.