Summary
During the reprogramming of mouse embryonic fibroblasts (MEFs) to induced pluripotent stem cells, the activation of pluripotency genes such as NANOG occurs after the mesenchymal to epithelial transition. Here we report that both adult stem cells (neural stem cells) and differentiated cells (astrocytes) of the neural lineage can activate NANOG in the absence of cadherin expression during reprogramming. Gene expression analysis revealed that only the NANOG+E-cadherin+ populations expressed stabilization markers, had upregulated several cell cycle genes; and were transgene independent. Inhibition of DOT1L activity enhanced both the numbers of NANOG+ and NANOG+E-cadherin+ colonies in neural stem cells. Expressing SOX2 in MEFs prior to reprogramming did not alter the ratio of NANOG colonies that express E-cadherin. Taken together these results provide a unique pathway for reprogramming taken by cells of the neural lineage.
Highlights
-
•
NANOG expression can be obtained without E-cadherin in cells of the neural lineage
-
•
Transgene independence and late markers only occur when colonies are E-cadherin+
-
•
NANOG+E-cadherin+ colonies also upregulate cell cycle-related genes
-
•
DOT1L inhibition increases NANOG+E-cadherin+ colony numbers
In this article, Sridharan and colleagues show that NANOG-expressing reprogramming colonies are obtained routinely in both multipotent and differentiated cells of the neural lineage that express SOX2 without the concomitant or prior expression of E-cadherin. Transgene independent colonies and late markers of reprogramming only occur when both NANOG and E-cadherin are expressed. Thus, the order of events of reprogramming observed from mouse embryonic fibroblasts is altered during reprogramming from the neural lineage.
Introduction
Overexpression of the four transcription factors, Oct4, Sox2, Klf4, and c-Myc (OSKM) is sufficient to reprogram somatic cells into induced pluripotent cells (iPSCs) (Jackson and Sridharan, 2013). The mechanism of reprogramming is incompletely elucidated due to the inefficiency of the process with about 5% of the cells reaching the iPSC state under standard serum or serum replacement culture conditions (Papp and Plath, 2013). While a variety of somatic cells have been used as a starting point for the reprogramming process (Hussein and Nagy, 2012), mechanistic studies have been largely limited to those using mouse embryonic fibroblasts (MEFs). Tracking reprogramming populations has delineated a series of events that take place in a timed manner such as the loss of somatic cell gene expression followed by mesenchymal to epithelial transition (MET) indicated primarily by the acquisition of the cell surface marker E-cadherin (Samavarchi-Tehrani et al., 2010, Li et al., 2010). This is followed by the gain of expression of pluripotency markers such as OCT4 and NANOG, by the appearance of stabilization markers such as DPPA4, and independence from exogenous reprogramming factor expression (Apostolou and Hochedlinger, 2013). Overlaid on these transitions, experiments on single cells have revealed an early stochastic phase of gene expression followed by a late hierarchical phase triggered by the activation of Sox2 (Buganim et al., 2012). Therefore, we were interested in determining if cells that expressed endogenous SOX2 followed the same pathway as MEFs and focused on reprogramming both adult stem cells (neural stem cells [NSCs]) and differentiated cells (astrocytes) from the neural lineage.
Both human and mouse NSCs can be reprogrammed with the omission of exogenous Sox2 in the reprogramming cocktail (Kim et al., 2008), and can even be reprogrammed with Oct4 alone (Kim et al., 2009). NSCs can also be more readily reprogrammed to intermediate stages, called partially reprogrammed cells, than MEFs (Silva et al., 2008). Remarkably, we found that upon induction of reprograming, in both NSCs and astrocytes, NANOG expression preceded or was concomitant with E-cadherin expression and the expression of SSEA1, an intermediate marker of pluripotency.
Abrogation of E-cadherin expression through shRNA-mediated knockdown reduces reprogramming efficiency from MEFs and compromises the quality of iPSCs obtained (Chen et al., 2010), while MEFs lacking E-cadherin cannot form Nanog+ colonies (Redmer et al., 2011). E-cadherin can also replace Oct4 in the reprogramming factor cocktail (Redmer et al., 2011). Truncations of E-cadherin in MEF reprogramming revealed the necessity of the extracellular domain (Chen et al., 2010). Interestingly, in the absence of E-cadherin in embryonic stem cells (ESCs), N-cadherin is able to functionally replace E-cadherin to maintain pluripotency (Hawkins et al., 2012).
We found that Nanog+ colonies from NSC reprogramming cultures can have N-cadherin, E-cadherin, or neither cadherin. However, colonies that expressed stabilization markers (Golipour et al., 2012), such as Dppa4, and that were transgene independent always co-expressed NANOG and E-cadherin. Gene expression analysis of populations sorted for expressing NANOG alone (N+) or NANOG and E-cadherin (N+E+) revealed that the N+E+ population expressed higher levels of cell cycle genes suggesting a greater propensity to expand. Finally, enhancing MET by inhibiting the histone methyltransferase, DOT1l (Onder et al., 2012) increased both N+ and N+E+ colony numbers.
Results
Nanog+ Colonies Emerge in Reprogramming in the Absence of E-Cadherin or SSEA1 Expression
We wanted to compare the routes to reprogramming taken by adult stem cells and differentiated cells from the same lineage that express endogenous Sox2. Therefore, we isolated postnatal day 4 (d4) NSCs and astrocytes that were 90% pure as determined by staining for PAX6 or GFAP, respectively (data not shown). These starting populations were derived from mice homozygous for the Yamanaka reprogramming factors Oct4, Klf4, Sox2, and c-Myc (OKSM) under the control of a doxycycline (dox) inducible promoter at a single locus and heterozygous for reverse tetracycline transactivator (rtta) ubiquitously expressed from the Rosa26 locus (Sridharan et al., 2013). All animal procedures were approved by the University of Wisconsin Medical School’s Animal Care and Use Committee.
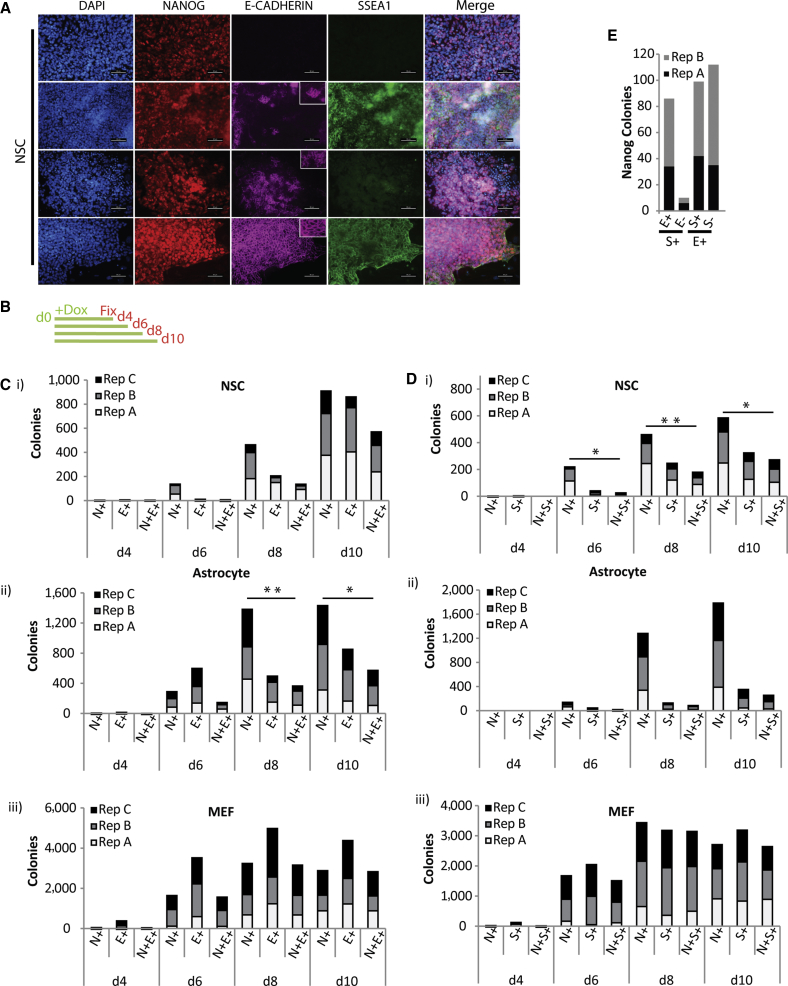
After induction with dox, reprogramming cultures were fixed at different time intervals and assessed for Nanog expression by immunofluorescence. Colonies were defined as closely clustered groups of at least four cells. Nanog (N+) colonies consistently emerged from NSCs on d6 (Figures 1A and 1B, 1Ci, 1Di) and accumulated until d10 of reprogramming (Figures 1Ci, Di); after which they became large and more difficult to define (Figure S1A). Astrocytes and MEFs display similar kinetics of N+ colony emergence (Figures 1Cii, 1Ciii and 1Dii, 1Diii). The total N+ colony number from MEF reprogramming was greater than that from both NSCs (3-fold lower) and astrocytes (2-fold lower) (Figures 1C and 1D). NSCs and astrocytes adhered to glass coverslips with a 3-fold lower frequency than MEFs (Figure S1B) and expressed slightly lower levels of exogenous Oct4 (Figure S1C), which may account for the lower numbers.
Figure 1.
Nanog+ Colonies from Neural Stem Cell and Astrocyte Reprogramming Can Emerge Independent of E-Cadherin or SSEA1
(A) Immunofluorescence (IF) images of NANOG colonies on day 10 of reprogramming NSC with E-cadherin and/or SSEA1. Scale bar, 50 μm. Insets, magnification of field.
(B) Scheme of experiment presented in (C) and (D). Dox was added to cells on day 0 (d0) and reprogramming cultures were fixed on indicated days.
(C) Counts of Nanog+ (N+), E-cadherin+ (E+) and Nanog+/E-cadherin+ (N+E+) colonies from (i) NSC, (ii) astrocyte, and (iii) MEF reprogramming cultures, fixed on days indicated on X axis. Counts from three independent experiments are stacked. Statistical significance of difference between N+ and N+E+ colonies by paired two-tailed t test; ∗p < 0.05, ∗∗p < 0.01. Non-significant differences are not marked.
(D) As in (C) above except for SSEA1+. SSEA1 = S+, Nanog+/SSEA1 = N+S+. t test; ∗p < 0.05, ∗∗p < 0.01.
(E) Combined counts of NANOG+/SSEA1+/E-cadherin. Counts from two independent experiments are stacked. Note that the number of S+E+ colonies and E+S+ colonies differ because of scoring between large fields of SSEA containing multiple E-cadherin+ colonies.
The typical pathway to pluripotency detailed from reprogramming MEFs indicates that MET, measured by acquisition of E-cadherin occurs prior to Nanog expression (Apostolou and Hochedlinger, 2013). We found that consistent with previous reports, both E-cadherin and SSEA1 colonies are visible by d4 of reprogramming from MEFs (Figures 1Ciii and Diii). The number of colonies that have E-cadherin or SSEA1 is equal to or exceeds the number of NANOG colonies; hence N+ colonies from MEF reprogramming cultures co-stain for E-cadherin and SSEA1. In striking contrast, several N+ colonies from both NSC and astrocyte reprogramming do not express E-cadherin (Figures 1A, 1Ci, 1Cii, and S1D). In addition, E-cadherin positive (E+) colonies emerge simultaneously with Nanog colonies during NSC and astrocyte reprogramming (Figures 1Ci, ii, and S1D). During MEF reprogramming, the expression of intermediate cell surface markers such as SSEA1 occurs after the MET and before Nanog expression. Analysis of SSEA1 colonies revealed remarkably that many N+ colonies were obtained without SSEA1 expression in both NSC and astrocyte reprogramming (Figures 1A, Di, ii, and S1D). Thus, Nanog expression can be obtained by skipping at least two early events described for MEF reprogramming. Further, analyzing the d10 N+ colonies from NSC reprogramming, almost all N+ SSEA1+ colonies also express E-cadherin, however the number of N+E+SSEA1+ colonies account for only half of the total N+E+ colonies (Figure 1E), suggesting that E-cadherin is activated before SSEA1. Note that while the absolute number of colonies obtained are different in each replicate experiment, as observed commonly in reprogramming experiments, the trends remain the same (Table S1). When E-cadherin levels are depleted, N+ colonies can emerge, but the total number of N+ colonies decreases suggesting that some colonies may require a threshold level of E-cadherin for Nanog expression (Figure S1E).
Reprogramming NSCs Gain Nanog without Loss of N-Cadherin
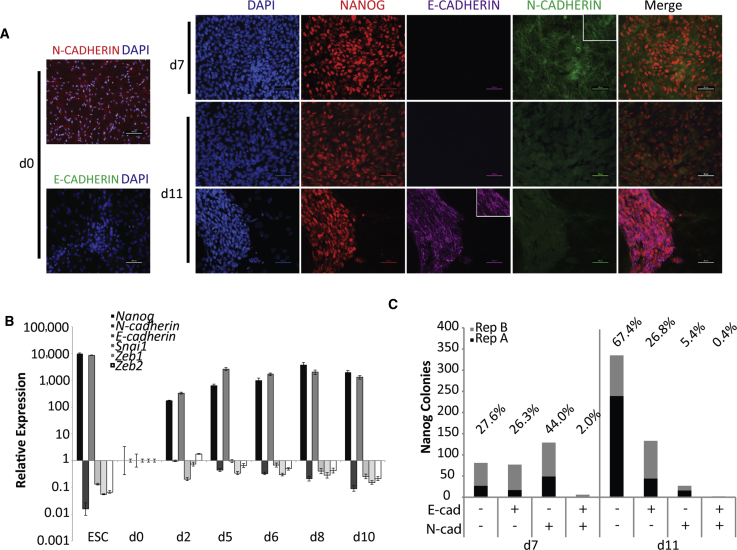
Neural-cadherin (N-cadherin) is expressed uniformly by NSC (Figure 2A) and can replace E-cadherin in promoting Nanog expression in ESC culture (Hawkins et al., 2012). Therefore, we interrogated whether early N+ cells were supported because of N-cadherin expression. Analysis of the reprogramming culture by RT-PCR indicated that, globally, N-cadherin transcript levels decreased by day 2 of reprogramming, concomitant with an increase in E-cadherin expression (Figure 2B). Mesenchymal genes commonly downregulated in MEF reprogramming (Li et al., 2010, Samavarchi-Tehrani et al., 2010), Snai1, Zeb1, and Zeb2 are also reduced during NSC reprogramming. Like E-cadherin, Nanog transcripts are detected after 2 days of reprogramming (Figure 2B). By d7 of reprogramming almost half (44%) of the N+ colonies maintain N-cadherin. The remaining N+ colonies are evenly divided between those that have E-cadherin or that express neither N-cadherin nor E-cadherin (Figures 2A and 2C). As reprogramming proceeds, the overall number of N+ colonies increases, while the percentage of N+N-cadherin+ colonies declines (Figures 2A and 2C). Thus, Nanog expression can be obtained independent of the expression of either cadherin assayed.
Figure 2.
Nanog Colonies Emerge Independent of N-Cadherin Expression
(A) Left: NSC on day 0 stained with N-cadherin or E-cadherin. Right: IF images of Nanog colonies from NSC reprogramming cultures at d7 and d11 co-stained with N-cadherin or E-cadherin. Scale bar, 50 μm; d0 N-cadherin = 100 μm. Insets, magnification of field.
(B) Time course of NSC reprogramming showing relative expression of Nanog and genes involved in MET, relative to d0 NSC = 1. Error bars indicate SD of three technical replicates from one representative experiment. ESC, embryonic stem cell.
(C) Quantification of d7 and d11 NANOG colonies co-stained with E-cadherin and N-cadherin. Counts from two independent experiments are stacked and average percentages presented.
Late Reprogramming Features Associate with E-Cadherin Expression
Since both the MET and expression of surface SSEA1 are early events in MEF reprogramming, we next wanted to determine whether the Nanog+ colonies that do not express E-cadherin support later hallmarks. We first tried to derive cell lines that were either N+E− or N+E+. For this purpose, we picked 20 colonies and maintained them over two passages either with or without dox. Within two passages, all colonies had E-cadherin expression on their surface (data not shown). This result suggests that the Nanog+E-cadherin− cells are in a transient stage that cannot be maintained.
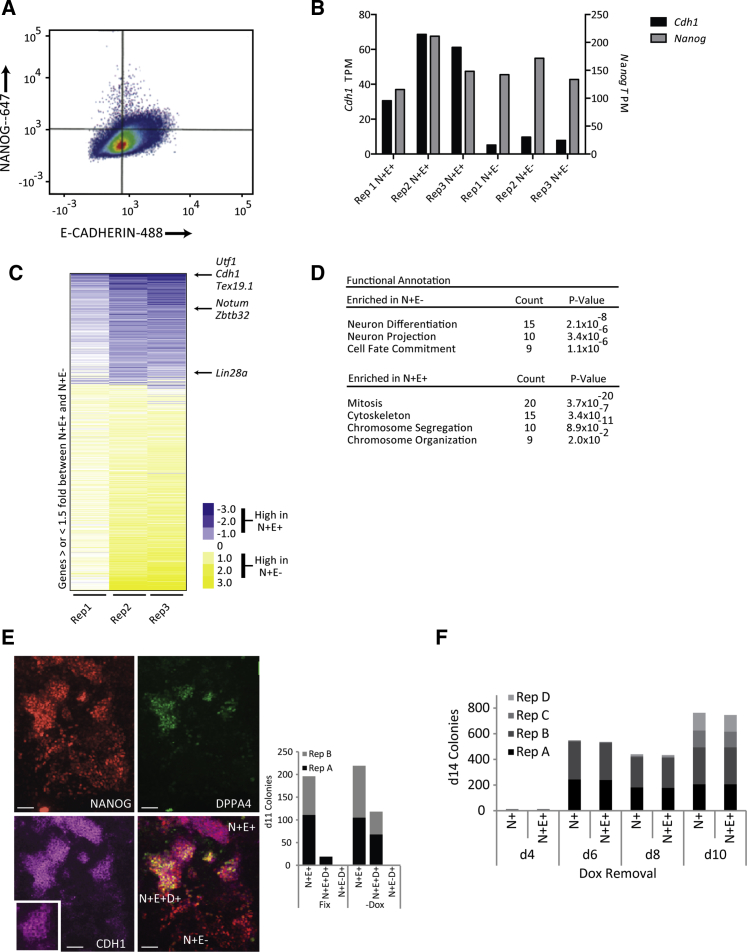
To obtain a molecular understanding of the characteristics of this transient population, we performed a flow cytometry-based sorting experiment on d7 (Figure 3A). Since the NSCs did not contain fluorescent reporter at either the Nanog or E-cadherin loci, we performed an intracellular staining for Nanog after fixing and staining for E-cadherin. Staining and sorting was performed under RNAse-free conditions and the cells decrosslinked before RNA sequencing was performed. In each of the three independent experiments, while Nanog levels remained the same between the N+E+ and N+E− populations (Figure 3B and Table S2), there was at least a 5-fold difference in E-cadherin expression suggesting that the sorted populations can be sufficiently distinguished. We next determined genes that were specifically upregulated at least 1.5-fold in the N+E+ population. Genes associated with the stabilization phase of reprogramming, including Tex19.1 and Notum, and pluripotency genes such as Utf1 were present in this group (Figure 3C). Functional annotation using gene ontology revealed that the N+E+ population was enriched for mitosis-enriched genes (Figure 3D), suggesting that the N+E+ cells may have a growth advantage over the N+E− cells at d7 of reprogramming. In contrast the N+E− cells were functionally enriched for neuronal differentiation-related genes (Figure 3D). This suggests that N+E− cells may not have completely lost their cellular identity and that E-cadherin expression increases after this change has occurred.
Figure 3.
Late Reprogramming Markers Are Enriched in the N+E+ Population
(A) Flow cytometry plot of reprogramming culture stained with Nanog-647 and E-cadherin-488.
(B) RNA-Seq read counts (transcripts per million [TPM]) for E-cadherin, labeled Cdh1 in figure, (left, Y axis) and Nanog (right, Y axis) for N+E+ and N+E− populations from three independent experiments.
(C) Heatmap of genes 1.5-fold differentially expressed between N+E+ and N+E− populations (set to 1) ranked by average TPM from three independent experiments.
(D) Functional annotation of gene expression in N+E+ and N+E− populations.
(E) Left: Immunofluorescence images of NANOG, E-cadherin, and DPPA4 on d11 of NSC reprogramming. Right: Counts from two independent experiments of Dppa4+ colonies in N+E+ and N+E− colonies are stacked. Scale bar, 50 μm. Insets, magnification of field.
(F) Comparison of d14 N+ and N+E+ colonies following withdrawal of dox on the day indicated on the X axis. Counts from four independent experiments are stacked. No significant difference between N+ and N+E+ colonies by paired two-tailed t test.
We further confirmed that the N+E+ population represented a more advanced stage of reprogramming by immunostaining for the stabilization protein DPPA4. Interestingly, DPPA4 positive colonies were only found within the N+E+ population (Figure 3E). Consistent with previous reports (Golipour et al., 2012), only sporadic DPPA4 colonies were observed when there was transgene expression and DPPA4 was upregulated upon dox withdrawal (Figure 3E).
A defining feature of the pluripotent state is the activation of the pluripotency regulatory network so that the reprogrammed colonies are no longer reliant on the exogenous transcription factors and become transgene independent. We analyzed a time course of NSC reprogramming, removing dox on consecutive days and allowing cells to remain in culture until d14 (Figure 3F and Scheme S2A). After dox withdrawal, exogenous reprogramming transcript and protein declined rapidly within 3 days (Figures S1C and S1F); almost all colonies that remained N+ now also co-stained for E-cadherin. Similar results were obtained during the reprogramming of astrocytes and MEFs (Figures S2A–S2C). Taken together, these data indicate that while N+E− colonies express early markers of MEF reprogramming, the late markers are only achieved in N+E+ cells.
Inhibition of Dot1L Alters the Ratio of N+E+ Colonies
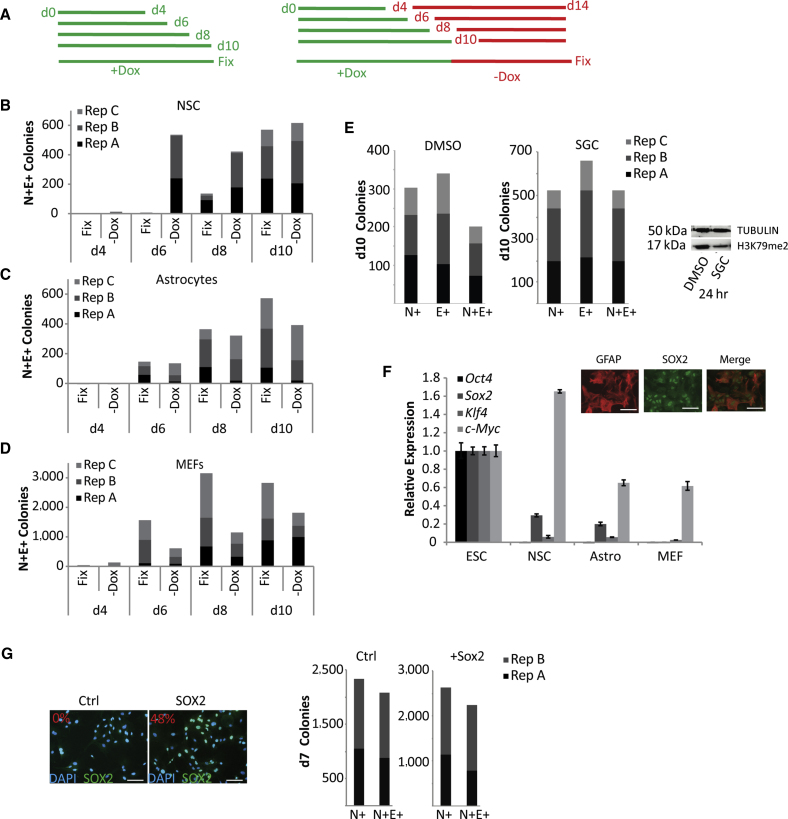
Since transgene independent colonies expressed both Nanog and E-cadherin, we wanted to determine whether early emerging N+E+ colonies were predictive of transgene independence. Comparing the number of N+E+ colonies obtained when reprogramming cultures were fixed on a particular day with how many remained after dox withdrawal on that day revealed an increase in N+E+ colonies following dox removal in reprogramming NSCs (Figure 4B). This phenomenon is reversed in MEF and astrocyte reprogramming, with more N+E+ colonies in the fix samples compared with samples with dox withdrawn on that day (Figures 4C and 4D). These data suggest that there may be differences in transgene dependence when starting with an adult stem cell compared with differentiated cells as a substrate for reprogramming. Therefore, we determined how enhanced acquisition of E-cadherin expression would affect NSC reprogramming.
Figure 4.
Effects of Reduced DOT1L Activity and Enhanced Sox2 Levels on the Ratio of Nanog+/E-Cadherin+ Colonies
(A) Left: Scheme of experiment for “Fix” samples in (B)–(D). Dox added on d0, fixed on days indicated. Right: Scheme of experiment for “-Dox” in (B)–(D). Dox added on d0, removed on days indicated, and fixed on d14. No significant difference between N+E+ colonies in (B), (C), (D).
(B) Comparison of N+E+ colonies in “Fix” and “-Dox” samples for NSCs. Counts from three independent experiments are stacked. Note the same data are presented in Figures 1C and 3C.
(C) As in (B) but for astrocytes. Note: same data are presented in Figures 1C and S2B.
(D) As in (B) but for MEFs. Note: same data are presented in Figures 1C and S2C.
(E) Counts of N+, E+ and N+E+ colonies from NSC reprogramming cultures on d10 in the presence of DMSO (left) or DOT1L inhibitor SGC0946 (middle). Counts from three independent experiments are stacked. Western blot analysis of H3K79me2 after SGC0946 treatment for 24 hr. No significant difference in the two conditions by t test.
(F) Relative expression of endogenous Oct4, Sox2, Klf4, and c-Myc in NSC, astrocytes, and MEFs. Error bars are SD of three technical replicates from one representative experiment. Inset, SOX2 expression in GFAP+ astrocytes. Scale bar, 50 μm.
(G) Left: Immunofluorescence images of MEFs following infection with pMX-Sox2 retrovirus. Percentages are SOX2+ cells from 100 nuclei (DAPI). Right: counts for N+ and N+E+ colonies following 7 days doxycycline treatment after pre-infection with pMX-Flag (Ctrl) or pMX-Sox2 (Sox2). Scale bar, 50 μm.
In a previous study, it had been demonstrated that inhibition of the histone H3K79 methyltransferase DOT1L in human fibroblast reprogramming leads to the activation of Nanog via increased MET (Onder et al., 2012). We utilized the small molecule SGC0946 to inhibit activity of DOT1L, resulting in reduced H3K79me2 after 24 hr of treatment (Figure 4E). Compared with the control DMSO condition, the number of N+ colonies obtained was higher on d10 in multiple reprogramming factor combinations (Figure 4E, S2D and S2E). In addition, in the presence of SGC0946, almost all N+ colonies also express E-cadherin (Figure 4E). The increase in numbers of both N+ and N+E+ colony numbers suggests that Dot1L may selectively be converting the N+E− colonies to N+E+ colonies and conferring the same growth advantage by activating cell cycle-related genes. Similar increases were also observed for N+SSEA1+ cells (data not shown).
Exogenous Sox2 Does Not Alter the Ratio of N+E+ Colonies from MEFs
NSCs and astrocytes (Zhang et al., 2014) express endogenous Sox2 while MEFs do not (Figures 4F, 4G, and S2F). SOX2 is bound to the promoter of Nanog in ESCs (Chen et al., 2008). Therefore, we determined if Sox2 expression could lead to the early emergence of Nanog-expressing cells. We infected MEFs with Sox2 retrovirus, which would lead to the overexpression of Sox2 (Figure 4G), prior to the addition of dox to express all four reprogramming factors. After 7 days of dox exposure, cultures were analyzed for NANOG and E-cadherin. The addition of Sox2 prior to reprogramming initiation had no effect on the ratio of N+E+ colonies (Figure 4G) indicating that the moderate levels of additional Sox2 expression obtained in these experiments are unlikely to be solely responsible for the differences we have observed between NSC and MEF reprogramming. The reciprocal experiment depleting the levels of Sox2 in NSC reprogramming yielded very high cell death and was inconclusive.
Discussion
Overall, this work has demonstrated that the classic sequential reprogramming pathway described for MEFs: loss of somatic markers, MET, gain of intermediate markers such as SSEA1, gain of pluripotency gene expression and transgene independence is not necessarily the path taken by all somatic cells. In some NSCs and astrocytes, these events can occur simultaneously or in reverse order such as the emergence of NANOG colonies that subsequently acquire E-cadherin. However, to achieve the final stages such as transgene independence, both Nanog and E-cadherin are expressed.
Different routes of reprogramming have been described even when starting with MEFs but these are restricted to late stages of reprogramming and have been demonstrated with different combinations of reprogramming factors (Parchem et al., 2014). Interestingly, recent reports also suggest that stalled clonal intermediates of MEF reprogramming can be obtained that express NANOG with lower levels of E-cadherin compared with fully reprogrammed iPSCs (Tonge et al., 2014). These clones also express very high levels of the reprogramming factor transgenes. An important distinction of our work is that half the reprogramming colonies, rather than a few clones that can be stabilized, transition through a cadherin-independent NANOG-expressing phase.
Interestingly, the inhibition of DOT1L in NSCs boosts the total number of N+ colonies and enhances the emergence of E-cadherin, resulting in all NANOG colonies also having E-cadherin. Since our assays used fixed reprogramming cultures, we could not estimate whether these increases were due to NANOG-positive colonies gaining E-cadherin or vice versa. E-cadherin plays an important signaling role in maintaining expression of Nanog in ESCs (Hawkins et al., 2012). However, stem cell lines can also be derived and maintained from the blastocysts that express Nanog, Oct4, and Sox2 under lower E-cadherin expression conditions (Chou et al., 2008). Taken together, these results and previously published work (Pieters and van Roy, 2014) suggest that there may be at least two independent but interlinked signaling pathways from E-cadherin to Nanog expression.
During MET in reprogramming MEFs, Sox2 suppresses the expression of Snai1, which is itself a suppressor of E-cadherin (Li et al., 2010). Therefore, increased SOX2 should lead to increased E-cadherin. However, NSCs, which express enough endogenous Sox2 that they can be reprogrammed without the addition of exogenous Sox2 (Kim et al., 2008), do not readily upregulate E-cadherin. By overexpressing Sox2 in MEF using retrovirus prior to induction of OSKM with dox, we demonstrated that additional Sox2 did not alter the ratio of Nanog colonies that had E-cadherin in this setting, although it is possible that the expression levels obtained may not have been sufficient. In initial experiments, we have found that SOX2 is not pre-bound to the Nanog promoter in NSCs (data not shown), although it remains possible that N+E− cells may acquire transient binding of SOX2 to facilitate Nanog expression. In conclusion, we demonstrate that different somatic cells types have a distinct order of getting to the pluripotent state, suggesting unique barriers to the cell fate change effected by reprogramming.
Experimental Procedures
NSC Astrocyte and MEF Isolation
Postnatal d4 mice were anesthetized with isofluorane, decapitated, and the brain removed from the skull. Tissue after removal of hindbrain, frontal lobes, and meninges was dissociated using a pipette. Cells were plated in DMEM with penicillin/streptomycin (Pen/Strep), L-glutamine, non-essential amino acids (NEAA), and 10% fetal bovine serum (FBS) for astrocyte outgrowth or plated in neurobasal medium with B27 (−Vit A), Pen/Strep, L-glutamine, NEAA, 20 ng/ml epidermal growth factor, and 20 ng/ml fibroblast growth factor 2 for NSC. After 4 days of culture, FBS was reduced to 1% for the astrocyte culture. MEFs were isolated as described (Tran et al., 2015). Mice were homozygous for the OKSM configuration and heterozygous for the reverse tetracycline transactivator (rtTA) allele (SSRW) except where indicated.
Cell Culture and Reprogramming
Reprogramming experiments were initiated by plating 1 × 105 cells per well onto gelatin-coated glass coverslips in a 12-well plate in ESC media (knockout DMEM, 15% knockout serum replacement, L-glutamine, Pen/Strep, NEAA, 2-mercaptoethanol, and leukemia inhibitory factor) with 2 μg/ml doxycycline. On d3, irradiated MEFs were seeded overnight in ESC media in FBS and switched on d4 back to ESC media with KSR. Where indicated, DMSO and 5 μM SGC0946 (Xcessbio) were added to the cultures.
Immunofluorescence
Immunofluorescence was performed as in Sridharan et al. (2009). Antibodies used Nanog− (1:100, Cosmobio, RCAB0002P-F), E-cadherin (1:100, Ebiosciences, 14-3249-80), and SSEA1− (1:100, Ebiosciences, 14-8813), N-cadherin (1:500, BD Biosciences, 610920), Dppa4 (1:100 Santa Cruz, sc-74616), and Sox 2 (1:100, R&D Systems, MAB2018) to stain with Dppa4 antibodies applied sequentially. Colony counting and imaging were performed on a Nikon Eclipse Ti using NIS Elements software.
Western Blot
Antibodies used α-tubulin (1:1000, Sigma T9026), H3K79me2 (1:1000, Abcam ab3594).
qRT-PCR
RNA was made using an ISOLATE II RNA Kit (Bioline) according to the manufacturer’s instructions, and converted to cDNA using Superscript II Reverse Transcriptase (Thermo). Relative gene expression was assayed with gene-specific primers using SYBR green master mix (Bio-Rad) on a Bio-Rad c1000 thermocycler.
Intracellular Fluorescence-Activated Cell Sorting
Cells were trypsinized and filtered to a single cell suspension. Cells were stained with Alexa 488-conjugated E-cadherin (Cell Signaling 3199) for 30 min, fixed with Cytofix/Cytoperm (BD Biosciences) for 15 min at room temperature followed by Permeabilization Buffer Plus (BD Biosciences) for 10 min at 4°C. From this step onward, cells received RNase out (Life Technologies) at each wash and staining step. Cells were stained with Alexa 647-conjugated Nanog (BD Biosciences 560279) for 30 min on ice. Sorting was performed on a BD FACS Aria III, and samples collected in tubes containing RNase out. Gates were set based on staining with negative controls for each isotype: Alexa-IgG-488 (Cell Signaling 4340) and Alexa-IgG-647 (BD Pharmingen 557732) and unstained samples.
RNA Isolation and Sequencing from Fixed Sorted Populations
Cross-linking was reversed by adding Buffer PKD from a Qiagen FFPE RNA Isolation Kit (Qiagen) and Proteinase K, incubation at 55°C for 15 min followed by 80°C for 15 min. RNA was isolated using Trizol (Thermo) and quantified by Qubit fluorimetric quantitation (Thermo). Quality was determined with an RNA 6000 Pico Kit (Agilent Technologies). An RNA-seq library was prepared from 5 to 300 ng of total RNA using a TruSeq RNA Sample Preparation Kit according to the manufacturer’s instructions. Reads were aligned to the mouse mm9 assembly and analyzed using RSEM to calculate transcripts per million. Expression in the N+E− population was set to 1. Functional annotation was analyzed using the DAVID algorithm.
Acknowledgments
We thank Prof. James Thomson and Mitch Probasco for use of the flow cytometer; Paul Ehrlich for technical assistance; and advice from Dr. Srikumar Sengupta on RNA isolation from fixed samples; Dr. Laura Moody and Prof. Albee Messing for astrocyte isolation; Dr. Weixiang Guo and Prof. Xinyu Zhao for NSC isolation. This work was supported by the UW-Madison, a Shaw Scientist award from the Greater Milwaukee Foundation, and a Basil O’Connor award from the March of Dimes Foundation to R.S. S.A.J. was partly supported by a postdoctoral fellowship from the Stem Cell and Regenerative Medicine Center of UW-Madison and K.A.T by the GRFP (NSF DGE-1256259).
Published: February 18, 2016
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes two figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.01.009.
Accession Numbers
The accession number for the gene expression data reported in this paper is GEO: GSE76130.
Primers used in this study
| Gene | Forward | Reverse |
|---|---|---|
| Actin | TGTTACCAACTGGGACGACA | TCTCAGCTGTGGTGGTGAAG |
| Nanog | CATCCCGAGAACTATTCTTGCT | GAGGCAGGTCTTCAGAGGAA |
| N-cadherin | GGGGATATTGGGGACTTCAT | CCGCTACTGGAGGAGTTGAG |
| E-cadherin | GCCACCAGATGATGATACCC | GGAGCCACATCATTTCGAGT |
| Snai1 | TGGAAAGGCCTTCTCTAGGC | CTTCACATCCGAGTGGGTTT |
| Zeb1 | AGACACCGCCGTCATTTATCC | GCGCTTGTAGCCTCTATCACAA |
| Zeb2 | TACCGCCACGAGAAGAATGAA | GCTCCTTGGGTTAGCATTTGG |
Supplemental Information
References
- Apostolou E., Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Faddah D.A., Cheng A.W., Itskovich E., Markoulaki S., Ganz K., Klemm S.L., van Oudenaarden A., Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chen T., Yuan D., Wei B., Jiang J., Kang J., Ling K., Gu Y., Li J., Xiao L., Pei G. E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells. 2010;28:1315–1325. doi: 10.1002/stem.456. [DOI] [PubMed] [Google Scholar]
- Chou Y., Chen H.-H., Eijpe M., Yabuuchi A., Chenoweth J.G., Tesar P., Lu J., McKay R.D.G., Geijsen N. The growth factor environment defines distinct pluripotent ground states in novel blastocyst-derived stem cells. Cell. 2008;135:449–461. doi: 10.1016/j.cell.2008.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golipour A., David L., Liu Y., Jayakumaran G., Hirsch C.L., Trcka D., Wrana J.L. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell. 2012;11:769–782. doi: 10.1016/j.stem.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Hawkins K., Mohamet L., Ritson S., Merry C.L.R., Ward C.M. E-cadherin and, in its absence, N-cadherin promotes nanog expression in mouse embryonic stem cells via STAT3 phosphorylation. Stem Cells. 2012;30:1842–1851. doi: 10.1002/stem.1148. [DOI] [PubMed] [Google Scholar]
- Hussein S.M., Nagy A.A. Progress made in the reprogramming field: new factors, new strategies and a new outlook. Curr. Opin. Genet. Dev. 2012;22:435–443. doi: 10.1016/j.gde.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Jackson S.A., Sridharan R. Peering into the black box of reprogramming to the pluripotent state. Curr. Pathobiol. Rep. 2013;1:129–136. [Google Scholar]
- Kim J.B., Zaehres H., Wu G., Gentile L., Ko K., Sebastiano V., AraUzo-Bravo M.J., Ruau D., Han D.W., Zenke M. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- Kim J.B., Greber B., AraUzo-Bravo M.J., Meyer J., Park K.I., Zaehres H., Schöler H.R. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–653. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Onder T.T., Kara N., Cherry A., Sinha A.U., Zhu N., Bernt K.M., Cahan P., Mancarci O.B., Unternaehrer J., Gupta P.B. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B., Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchem R.J., Ye J., Judson R.L., LaRussa M.F., Krishnakumar R., Blelloch A., Oldham M.C., Blelloch R. Two miRNA clusters reveal alternative paths in late-stage reprogramming. Cell Stem Cell. 2014;14:617–631. doi: 10.1016/j.stem.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters T., van Roy F. Role of cell-cell adhesion complexes in embryonic stem cell biology. J. Cell Sci. 2014;127:2603–2613. doi: 10.1242/jcs.146720. [DOI] [PubMed] [Google Scholar]
- Redmer T., Diecke S., Grigoryan T., Quiroga-Negreira A., Birchmeier W., Besser D. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 2011;12:720–726. doi: 10.1038/embor.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Golipour A., David L., Sung H.-K., Beyer T.A., Datti A., Woltjen K., Nagy A., Wrana J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T.W., Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R., Tchieu J., Mason M.J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R., Gonzales-Cope M., Chronis C., Bonora G., McKee R., Huang C., Patel S., Lopez D., Mishra N., Pellegrini M. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat. Cell Biol. 2013;15:872–882. doi: 10.1038/ncb2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonge P.D., Corso A.J., Monetti C., Hussein S.M.I., Puri M.C., Michael I.P., Li M., Lee D.-S., Mar J.C., Cloonan N. Divergent reprogramming routes lead to alternative stem-cell states. Nature. 2014;516:192–197. doi: 10.1038/nature14047. [DOI] [PubMed] [Google Scholar]
- Tran K.A., Jackson S.A., Olufs Z.P.G., Zaidan N.Z., Leng N., Kendziorski C., Roy S., Sridharan R. Collaborative rewiring of the pluripotency network by chromatin and signaling modulating pathways. Nat. Commun. 2015;6:6188. doi: 10.1038/ncomms7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O’Keeffe S., Phatnani H.P., Guranieri P., Caneda C., Ruderisch N. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.