Abstract
Proper expression of genes requires communication with their regulatory elements that can be located elsewhere along the chromosome. The physics of chromatin fibers imposes a range of constraints on such communication. The molecular and biophysical mechanisms by which chromosomal communication is established, or prevented, have become a topic of intense study, and important roles for the spatial organization of chromosomes are being discovered. Here we present a view of the interphase 3D genome characterized by extensive physical compartmentalization and insulation on the one hand and facilitated long-range interactions on the other. We propose the existence of topological machines dedicated to set up and to exploit a 3D genome organization to both promote and censor communication along and between chromosomes.
Keywords: domains, polymers, enhancer, promoter, gene expression, gene regulation, CTCF, cohesin, condensin, Hi-C, simulations, biophysics
Chromosomal communication
Communication involves transfer of information from one party to another. This can be achieved in at least two mechanistically distinct ways: first, the parties directly interact, e.g. two or more people directly speaking to each other. Second, information can be transmitted from one location to another via media or intermediates and it is then received by the appropriate partner(s) at their respective locations. For the first mechanism, the two parties need to be physically close, for the second, there needs to be a means to send, transport and receive information from one place to another. Do similar mechanisms operate inside the cell nucleus where genes are regulated by communicating with regulatory elements that can be located elsewhere in the genome? Here we explore the idea that the spatial organization of a genome, and its physical properties, could constitute an effective mechanical communication device.
Genes do not work as single, isolated units. Their expression is modulated by regulatory elements that can be located from as little as a kb up to as much as several Mb away, although the precise distance distribution between genes and their regulatory elements is still poorly known (Bickmore, 2013; Bulger and Groudine, 1999; Carter et al., 2002; Gibcus and Dekker, 2013; Kleinjan and van Heyningen, 2005; Li et al., 2012; Sanyal et al., 2012; Schwarzer and Spitz, 2014; Tolhuis et al., 2002; West and Fraser, 2005). Since in a given cell thousands of genes are expressed throughout the genome, there is a corresponding abundance of long-range communication between genes and regulatory elements occurring at any moment in each cell nucleus. Over the last decade much has been learned about how this is achieved, revealing critical roles for the spatial organization of chromosomes.
Microscopy-based technologies, such as Fluorescence In Situ Hybridization (FISH) and live cell imaging, and increasingly high resolution chromosome conformation capture (3C) - based methods (Bickmore, 2013; Dekker et al., 2013; Dekker et al., 2002; Fraser et al., 2015; Hsieh et al., 2015; Kalhor et al., 2011; Lieberman-Aiden et al., 2009; Rao et al., 2014; Shachar et al., 2015; Tang et al., 2015) have been instrumental in determining how chromosomes are folded at different length scales (kb up to Gb), and this in turn is starting to provide answers to some long-standing questions related to gene regulation and other chromatin - templated processes. One mechanism by which distal regulatory elements can control genes located far away in the genome is through long-range physical interactions (Figure 1). For instance, enhancer and insulator elements often engage in physical contacts with their target promoters (Carter et al., 2002; Li et al., 2012; Sanyal et al., 2012; Tolhuis et al., 2002), pointing to direct molecular association as a means for long-range communication.
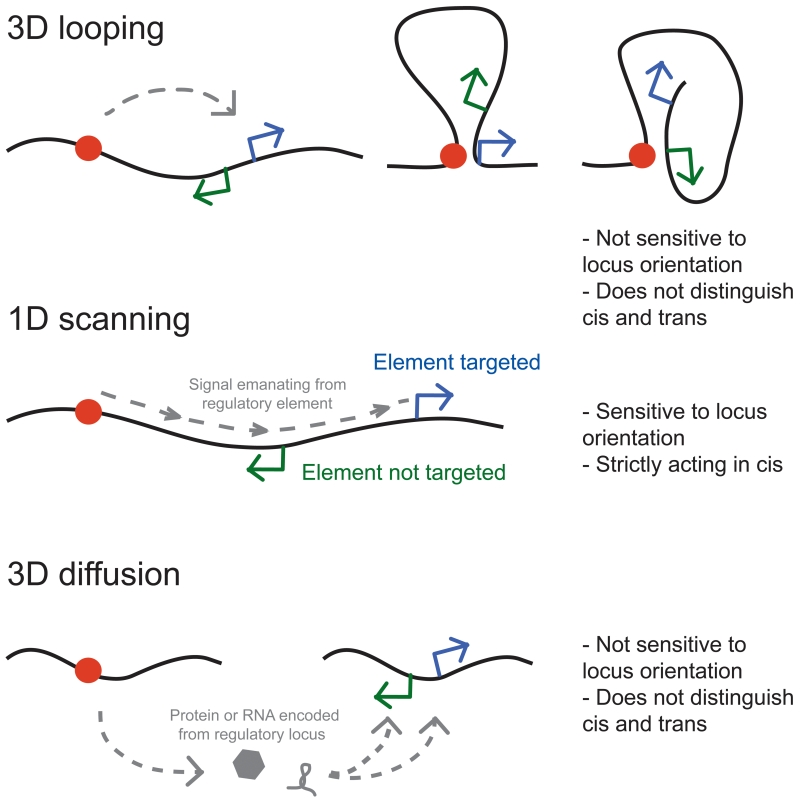
Figure 1. Chromosomal communication.
Top: communication between genomic loci by 3D looping interactions. For large loops, e.g. tens to hundreds of Kb, such interactions are not sensitive to locus orientation. Also, 3D interactions do not readily distinguish interactions with loci located on the same chromosome (in cis) or on different chromosomes (in trans). Middle: Singles emanating from one locus (e.g. RNA transcribed at that locus, a protein complexes recruited at that site) can spread in cis along the chromatin fiber till a target locus is reached. This mode of communications can be sensitive to relative orientation of the target locus, and is strictly in cis. Bottom: Communication by 3D diffusion of factors such as RNA or proteins released from one locus till they reach target loci. This mode of communication is not sensitive to target orientation and cannot distinguish cis from trans.
Although such physical associations appear to account for a significant fraction of long-range gene regulatory events, not all chromosomal communications involve direct contacts between the corresponding loci (Figure 1). An example is the case of X chromosome inactivation in female mammals. In this case, the Xist RNA is expressed from one X chromosome only and this RNA spreads along the length of the entire chromosome resulting in gene repression through the Xist-dependent recruitment of a set of silencing complexes (Chu et al., 2015; Galupa and Heard, 2015; Gendrel and Heard, 2014; Jeon et al., 2012; Wutz et al., 2002). Here communication along the inactivated X chromosome occurs not by direct physical interactions, but by cis-spreading of a signal, a non-coding RNA, that delivers silencing proteins to most of the genes linked in cis to the Xist gene. X-chromosome inactivation also requires initial inter-chromosomal communication to ensure that only one X chromosome expresses Xist. Though Xist loci of the two X chromosomes do transiently interact (Augui et al., 2007; Masui et al., 2011; Xu et al., 2006), implying physical communication, critical information is transmitted by diffusible proteins such as Rnf12 (Barakat et al., 2014; Galupa and Heard, 2015). The latter mode of communication includes the general, and widespread action of transcription factors encoded at one locus but acting throughout the genome. Thus, communication involves direct physical associations, cis-spreading of information, as well as diffusional signals including proteins and RNAs, that can move between chromosomes (Figure 1). In this perspective we do not discuss diffusion-based communication through transcription factors, and instead focus on communication through long-range chromatin interactions and spreading of signals in cis along chromosomes. In addition, we mostly discuss chromosome organization and long-range communication in mammalian genomes, even though other organisms including bacteria may employ similar mechanisms.
Not all chromosomal communication is for regulating gene expression. An interesting example is intra-chromosomal communication to control somatic recombination in the immunoglobulin loci, such as V(D)J recombination and antibody class switching. During these processes specific pairs of double-stranded breaks located up to 200 kb apart need to interact to be joined for successful recombination events. Recent studies (Dong et al., 2015; Gostissa et al., 2015) have revealed a surprising orientation bias in the IGH class switch recombination process where genomic orientation is preserved in the vast majority of recombination events prior to any further selection. During the process recombination occurs between recombination sequences that undergo AID-dependent DNA break formation. Interestingly, re-joining of ends is orientation-specific implying long-range communication between the break sites in a manner that maintains the relative orientation of the sites even when they are separated by hundreds of Kb. Thus, communication between two sites where double-stranded breaks are initiated requires not only direct proximity between them, but also preservation of their genomic orientation, pointing to specific processes to facilitate and or mediate their association in a directional manner (discussed below in more detail).
With the phenomenon of long-range communication well established, and the roles of chromosome structure and dynamics becoming increasingly clear, many new questions arise: First, how are long-range interactions established, i.e. how do distal elements find one another inside the crowded nucleus? Further, what determines specificity of such interactions and what prevents any of the thousands of active regulatory elements in the genome from inappropriately engaging in contacts with any of the thousands of genes? How do signals spread along chromosomes and how can such spreading be contained to a single chromosome (e.g. X-chromosome inactivation)? How is robustness and precision achieved so that important communication is efficiently and rapidly established in most or all cells? Answers to these questions start to emerge now that deeper knowledge is obtained about nuclear organization, the structural compartmentalization of chromosomes, the physical and mechanical properties of chromosomes and their dynamics, and the identification of molecular machines that can actively fold chromosomes to orchestrate and guide long-range communication.
Physics of chromosomal communication
Chromosomes are long polymers and many of their structural properties, dynamics and cell-to-cell variability in folding can be understood from their polymer nature. In fact, the polymer state of chromosomes has critical consequences for which pairs of loci have an opportunity to interact, the kinetics of their search for each other, and the number of cells in the population in which interactions occur (Figure 2). To illustrate this we will first explore the scenario in which the 3D genome is determined solely by the physical polymer state of chromosomes.
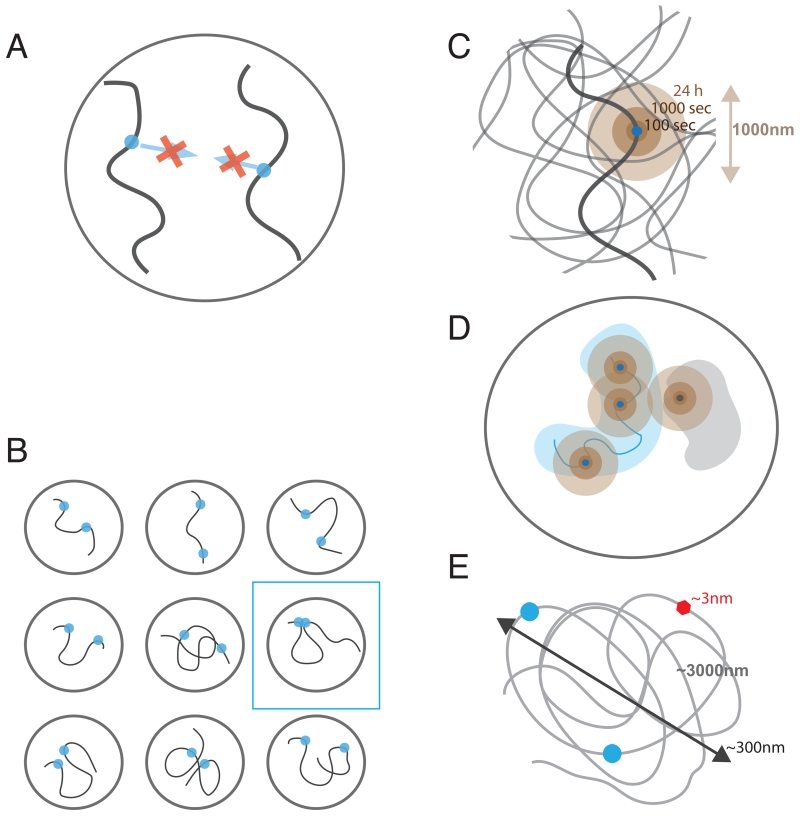
Figure 2. Physical aspects of chromosomal communication.
A. Short-range character of molecular interactions: molecular affinity is limited to 1-10nm range, and long-range attraction between chromosomal loci are impossible. B. Polymeric nature of chromatin made interactions even between relatively close loci infrequent, leading to large cell-to-cell variation. C. Localized dynamics of chromatin leads to mostly local exploration limited to 1-2um swept during a cell cycle. D. Chromosomal territories make cis interactions are much more likely and showing less cell-to-cell variation, while trans being highly variable. E. Individual proteins are much small (~3nm) than sizes of even modest (100-150Kb) chromatin region that has ~300nm in diameter and ~1-3um in length, making it difficult to insulate 3D interactions between chromosomal loci.
Three physical phenomena are central to our understanding of spatial genomic communication: the short-range character of molecular interactions, the polymeric nature of chromosomes, and the localized dynamics of chromosomal loci (Figure 2). Below we discuss implications of these aspects to genomic communications.
Short-range character of molecular interactions
Interactions between genomic loci rely on affinity of protein-protein and protein-DNA interactions. Hydrophobic, electrostatic, hydrogen-bonded and van der Waals in nature, these interactions are either short-range or screened by the high ionic strength of the nucleoplasm. As a result, protein and DNA interphases of a pair of genomic loci can attract each other only if located closer than ~1-5 nm. The affinity of two DNA-bound proteins will not attract these loci to each other unless they are already very close to each other in space (Figure 2A). Thus formation of most genomic interactions will rely on initially stochastic contacts between genomic loci.
Polymeric nature of chromatin
Since genomic communications rely on contacts that are already formed, it is the frequency of these contacts that determines possible genomic communications. The polymeric nature of chromosomes makes loci close along the linear genome interact much more frequently than more distant loci or loci located on different chromosomes (trans interactions). Despite of this most of interactions are extremely infrequent due to a large volume that is explored by any one locus (Figure 2B). If chromosomes were a melt of polymers in a spherical nucleus of volume V=300 um3 (Rnucleus≈4um (Milo et al., 2010)) two loci from different chromosomes would be in a Hi-C contact (Rc≈100-150nm) with the probability Ptrans≈(R/cRnucleus)~10−5, i.e. in only a few out of 100,000 cells. In an otherwise unconstrained polymer melt, two loci separated by 10Mb would be on average R(10Mb)~4um apart and interact as infrequently as trans loci. Correspondingly, loci separated by 1Mb or 100Kb (R(1Mb) ≈ 1.4um and R(100Kb) ≈ 0.4um) would interact with the probability P(1Mb)≈(Rc/R)3~10−3−10−4, P(100Kb)~10−2 and thus in a few out of 10,000 and 100 cells respectively. Two factors would lead to higher contact frequencies: chromosome dynamics and local compaction of chromatin at all levels.
Localized dynamics of chromatin
The polymeric nature of chromosomes significantly limits mobility of individual loci: moving one locus would require moving its neighbors and their neighbors etc., which is slow and may be further limited by steric and topological interactions with other nearby chains. As a result, polymers show highly localized mobility with the displacements increasing as time to the power of approximately ¼ (either due to Rouse diffusion or reptation, compared to normal diffusion where the power is ½). Such diffusion has been observed for chromosomal loci in yeast (Hajjoul et al., 2013) and mammalian cells (Bronstein et al., 2009; Lucas et al., 2014). There are two important consequences of this localized diffusion (Figure 2C): (i) in a given cell, a locus extensively explores its spatial neighborhood (~150-300nm in 100 sec, 0.5-0.8um in 1 hour and ~1-1.5um in 24h, i.e. the length of a typical cell cycle), thus allowing communications between spatially proximal loci; (ii) communications at a distance, however, are strongly suppressed since only small spatial distances are explored by a locus during a single cell cycle. Thus loci that happen to be sufficiently close in space upon exit from mitosis can interact, while those that are further apart would not have sufficient time to find each other and will have to wait for the next cell cycle to get a chance of interacting (Strickfaden et al., 2010). Even when interacting loci are within a distance that can be spanned within a cell cycle (e.g. ~1um), communication between them would require them first to find each other by this localized diffusive process, which makes time of the response highly variable.
Difficulty to insulate interactions
Difference in scales between sizes of chromosomal loops and sizes of individual proteins makes it challenging for a single protein to insulate long-range interactions. Even 100Kb of genomic separation between an enhancer and a promoter implies about ~2000-4000nm of 10-20nm fiber that is folded into an area of about ~300-500nm in radius. It’s mysterious how 3-5nm size protein bound somewhere along this chain can significantly influence frequencies of interactions between its monomers (Figure 2D). Recent simulation studies have shown that although formation of a 30Kb chromatin loop can facilitate intra-loop interactions, insulation of the loop interior from the exterior is very modest with about 30% reduction in the contact frequency Benedetti et al., 2014; Doyle et al., 2014). Polymer simulations (Fudenberg et al., 2015) have also shown that even a bulky protein assembly on a chromatin fiber cannot serve as a reliable insulator providing no insulation beyond the size of the bulky assembly. Similarly, local changes in the flexibility of the chromatin fiber that can be induced by an insulator cannot provide robust insulation between regions distant from the insulator along the genome (Fudenberg et al., 2015).
Taken together, these physical considerations demonstrate that the polymeric nature of chromosomes leads to spatial insulation of distal genomic regions and high cell-to-cell variation of their contacts, while at the same time allowing frequent contacts between genomically proximal regions. For small genomes such as yeast and C. elegans, this may be sufficient to ensure appropriate gene regulation. However for larger genomes this will become a highly stochastic process leading to tremendous cell-to-cell variation in gene expression.
Hence, in order to achieve robust, precise and reproducible cell type-specific gene expression patterns across the genome additional layers of chromosome organization are required so that communications between more distal regions can be actively facilitated while interactions between more proximal loci can be moderated both ways: they can be facilitated in some cases, but may need to be actively prevented (insulated) in other cases to prevent inappropriate gene – regulatory element interactions. It is now becoming clear that cells have evolved mechanisms to compartmentalize chromosomes at all scales, which allow more precise control of interactions between some sets of loci, while preventing others in the majority of cells.
3D Genome Compartmentalization
Years of microscopic observations and 3C-based studies have revealed that the spatial organization of the genome is not just a melt of otherwise uniform polymers: chromosomes are characterized by structural compartmentalization at many levels (Bickmore, 2013; Bickmore and van Steensel, 2013; Bouwman and de Laat, 2015; Gibcus and Dekker, 2013; Sexton et al., 2007). At the level of the whole nucleus, individual chromosomes occupy 1-2um territories that change their nuclear position upon every cell division, thus randomizing pairs of neighboring chromosomes (Branco and Pombo, 2006; Cremer and Cremer, 2001; Kind et al., 2013). Since inter-chromosomal interactions are mostly restricted to the zones where adjacent territories touch (Branco and Pombo, 2006), and a limited spatial range of interactions that can be formed within a cell cycle (~1um, see above), interactions between chromosomes should be highly cell-to-cell variable. Communication within chromosomes, however, can be more robust as a single locus explores a good fraction of its chromosomal territory during interphase. Within territories chromosomes are compartmentalized in different types of sub-chromosomal domains.
At the scale of several Mb, animal chromosomes show characteristics of space-filling polymers, i.e. continuous genomic regions occupy continuous chromosomal volumes (Shopland et al., 2006). This space-filling character of 0.1-10Mb of chromosomes is evident from microscopy data that show linear (or sub-linear) scaling of occupied volume with the length s of a stained genomic region (V(s)~sα α≤1), and from the scaling R(s)~s1/3 of the spatial distance between pairs with their genomic separation s (Rosa and Everaers, 2008; Halverson et al., 2014), As a result of this space-filling organization, spatial distances between chromosomal regions become much smaller than they would be in a polymer melt with R(10Mb)≈1.5-2um, R(1Mb)≈0.5um (for inactive chromosomal region and 1.5um for active ones) and R(100Kb)≈0.2um (Jhunjhunwala et al., 2008; Tark-Dame et al., 2014). Combined with the ranges that can be explored within 1h or 24h (see above) we estimate that loci separated by 10Mb are still unlikely to find each other within a cell cycle, while loci separated by less than 1Mb can find each other within a cell cycle and likely within a couple of hours. Faster (minute scale) temporal response would require ~100Kb separation between loci that based on these estimates are expected to interact very frequently.
Chromosome organization at the megabase level is also characterized by interactions between functionally distinct compartments: large blocks of active chromatin (on average 3-5 Mb in size) associate with other active chromatin domains, while inactive chromatin associates with other inactive regions (Lieberman-Aiden et al., 2009; Zhang et al., 2012). At the scale of several hundreds kb smaller domains, often referred to as topologically associating domains (TADs) can be detected (Crane et al., 2015; Dixon et al., 2012; Nora et al., 2012; Sexton et al., 2012). These domains are defined by the preferential interaction of loci located within them and the relative (about two-fold) depletion of interactions between loci located in different TADs. Though TADs can be distinct in structure, function and cell-to-cell variability in different organisms, in mammals (Dekker and Heard 2015), TADs are to a large extent tissue invariant, whereas the larger compartments are related to the cell type and the set of genes and regulatory elements that are active. It has been proposed that TADs are the invariant building blocks of chromosomes and that in a given cell type TADs of similar chromatin status form the larger cell type-specific compartments through a process of self-assembly (Dekker, 2014; Gibcus and Dekker, 2013).
The 3D genome as censor
Compartmentalization of chromosomes into structural domains has significant consequences for chromosomal communication. Loci located within TADs are relatively insulated from loci outside the domain, while they can readily interact with other loci within the domain. Several recent studies have now shown that the formation of insulated topological domains indeed prevents, or censors, physical and functional communication between genes and distal regulatory elements. In one elegant series of experiments Lupianez and co-workers engineered CRISPR/Cas – mediated genomic rearrangements and found that relocating TAD boundaries and regulatory elements can have major impact on gene expression by allowing otherwise inappropriate, or preventing normal long-range communication with distal regulatory elements (Lupiáñez et al., 2015). For instance, an inversion around a TAD boundary that results in repositioning of a set of limb enhancers within the same TAD as the wnt6 gene, leads to inappropriate interactions between them and the gene and up-regulation of wnt6 in limb tissues where the enhancers are active.
These studies suggest that there is only limited specificity to enhancer-promoter interactions and that a critical factor in determining which enhancers regulate any gene is the co-location within the same insulated chromosomal domain (de Laat and Duboule, 2013; Gibcus and Dekker, 2013; Schwarzer and Spitz, 2014). This in turn would predict that enhancers act on the entire domain. Several lines of evidence suggest that this is indeed the case. First, analysis of gene expression during differentiation of ES cells into neural progenitor cells showed that genes located in the same TAD tend to be more correlated in their expression pattern than genes located in adjacent TADs (Nora et al., 2012). Second, Symmons and co-workers used a functional genomic approach in which they used transposable elements to insert reporter genes in a large number of positions along the chromosomes (Symmons et al., 2014). They then analyzed where in the mouse these reporter genes were expressed and found that sets of reporter genes integrated in contiguous domains displayed highly similar tissue specific expression. Strikingly, these domains displayed strong correlations with TADs. Third, more recently analysis of enhancer-promoter interactions around the CFTR locus showed that the CFTR promoter engages with distinct cell-type specific distal enhancers and CTCF-bound loci in different tissues. Intriguingly, all these are contained within one tissue-invariant TAD (Smith et al., 2016; Yang et al., 2015). ChIA-PET analyses of CTCF-anchored loops between TAD boundaries and RNA polymerase-anchored chromatin loops also showed that gene regulatory interactions between gene promoters and their distal regulatory elements occur mostly within TADs (Tang et al. 2015). Thus, TADs appear to represent functional domains and TAD boundaries act as censors of communication by not allowing enhancers to reach genes located in adjacent TADs.
How do TAD boundaries prevent long-range physical and functional communication? This remains a poorly understood process, but recent genome editing experiments and chromatin folding simulations have led to some intriguing insights. First, TAD boundaries often contain CTCF binding sites (Dixon et al., 2012; Rao et al., 2014) or related architectural proteins in flies (Hou et al., 2012). Recently several studies independently found that the two boundaries of many TADs contain CTCF sites that are positioned in opposite orientation and that these sites engage in long-range interactions with each other (de Wit et al., 2015; Gómez-Marín et al., 2015; Guo et al., 2015; Rao et al., 2014; Vietri Rudan et al., 2015). Thus, one view of a TAD is that it involves formation of a chromosomal loop between two boundaries and that CTCF site orientation is of critical importance in setting up these loops. There is now direct experimental evidence for this model. Guo and co-workers (Guo et al., 2015) and de Wit and co-workers (de Wit et al., 2015) showed that changing the orientation of CTCF sites disrupts chromatin loops with distal CTCF sites that were in the opposite orientation. Further Sanborn and co-workers showed that genetic perturbation of CTCF sites re-organizes loops configurations as predicted by the orientation of the sites (Sanborn et al., 2015).
Physical and molecular mechanisms underlying structural and functional insulation of genomic communications by TAD boundaries remain to be understood (Fig 2E). TADs are characterized by about two-fold increase in the frequency of chromatin contacts inside a TAD as compared to contacts between TADs. Such modest increase of intra-TAD contacts cannot significantly insulate loci that belong to neighboring TADs. Formation of loops between TAD boundaries is expected to have similarly modest effect on contacts between elements that are not located right at the contacting boundaries. Recent simulations (Doyle et al., 2014; Benedetti et al., 2014) have shown that formation of a chromatin loop can facilitate interactions located within a loop or suppress interactions between the loop and the rest of the chromosomes, but the effect is still limited to about two-fold change in contact frequencies. It is possible, however, that the two-fold change in the frequency of interactions can be amplified by highly cooperative mechanism of gene activation taking place at promoters (Mirny, 2010; Ptashne, 2014). Such amplification of response still cannot address cell-to-cell variation as some cells may simply not get a contact between an enhancer and its target gene within a TAD, while others can get contacts outside of a TAD, making it hard to attribute critical regulatory roles to mere compaction or a looping of the TAD. It is likely that other intra-TAD mechanisms are involved.
A few models of TAD formation have been recently proposed. Two studies (Giorgetti et al., 2014; Hofmann and Heermann, 2015) suggested that TADs should be formed by dynamic intra-TAD interactions that lead to a highly diverse ensemble of TAD conformations. They however, did not propose a specific molecular mechanism for such preferential intra-TAD dynamic interactions. Two other studies suggested that TADs could result from preferential interactions between two or more types of interacting loci, with a TAD corresponding to a continuous segment of loci of a single type (Barbieri et al., 2012; Jost et al., 2014). Such mechanism ultimately produces alternating patterns at the genome-wide scale that are characteristic of compartments, rather than TADs, but nevertheless may be an adequate model for Drosophilla Hi-C data where TADs and compartments may be hard to delineate or distinguish from each other (Ulianov et al., 2015). A major limitation of such models for mammalian TADs is that a boundary deletion would not produce a merger of neighboring TADs as have been recently experimentally demonstrated. A different model of TAD formation in Drosophilla was proposed in (Ulianov et al., 2015) where each monomer can interact is at most one more (saturating bonds), and inter-TAD regions that are enriched in highly expressed genes are non-interacting. Although rather artificial, the assumption of saturating bonds turned out to be critical for success of the model. Another intriguing mechanism giving good agreement to the observed TAD organization and relying on the special role of boundaries relies on supercoiling (Benedetti et al., 2013), though the role of supercoiling in eukaryotes remains to be understood. Observed domain organization in Caulobacter (Le at el., 2013) has been attributed to transcription that can lead to local unwinding of supercoiled DNA, thus creating a linker between neighboring domains. It remains to be seen whether transcription can play a role in formation of TADs in mammalian chromosomes.
3D genome as an active moderator
Several considerations presented above suggest that spontaneous 3D interactions among genomic elements in interphase chromosomes would not be able to provide several important aspects of genomic communications such as (a) robust and timely interactions among elements separated by up to ~1Mb; (b) reliable insulation between elements that are sufficiently close (~100Kb, Fig 2E) on the chromosomes (e.g. insulation across TAD boundaries), and spreading of such interactions in cis when insulators are altered (de Wit et al., 2015; Guo et al., 2015; Nora et al., 2012; Sanborn et al., 2015); (c) preferential interactions among genomic elements that preserve their genomic orientations (e.g. CTCF sites (above) or elements of the IgH locus involved in VD(J) recombination and class switching (Dong et al., 2015; Gostissa et al., 2015; Hu et al., 2015). We argue that a recently proposed loop-extrusion model of chromosome organization during interphase (Fudenberg et al., 2015), (Bouwman and de Laat, 2015; Nichols and Corces, 2015; Sanborn et al., 2015), can demonstrate many of these characteristics.
Central to this model is the active (ATP consuming) process of loop extrusion, where a loop-extruding factor, possibly cohesin during interphase and condensin during mitosis, associates with the chromatin fiber and starts creating a progressively larger loop (Figure 3A). A loop-extrusion mechanism (under similar names of “processive loop enlargement” (Nasmyth, 2001), “loop enlargement” (Kimura et al., 1999)) has been suggested as a mechanism of chromosome compaction and chromatid segregation (Alipour and Marko, 2012; Nasmyth, 2001) in mammalian cells, and a mechanism of chromosome segregation [see (Gruber, 2014; Reyes-Lamothe et al., 2012), and most recently (Wang et al., 2015)] and repair (Allen et al., 1997) in bacteria. Despite of these earlier proposals, the loop-extrusion mechanism remains largely hypothetical.
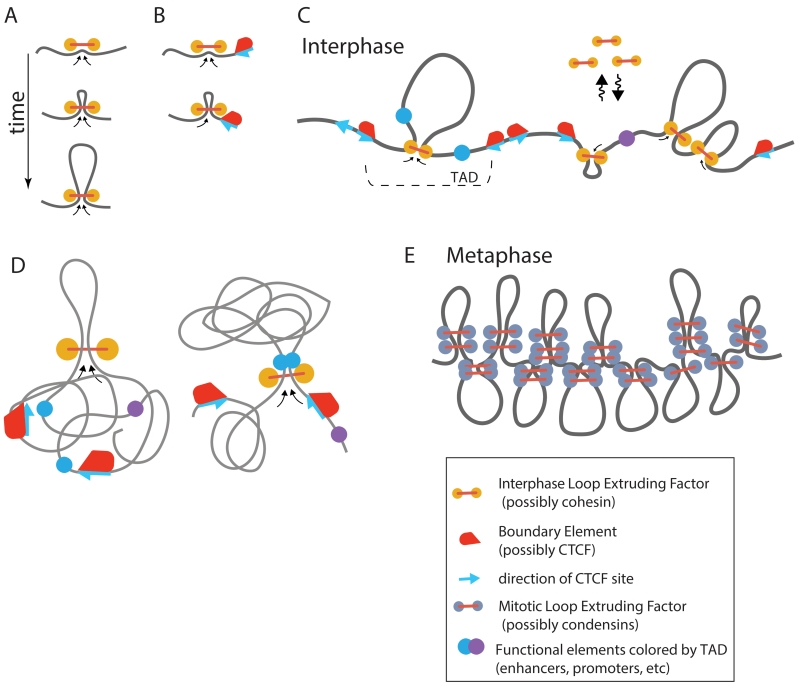
Figure 3. Loop extrusion as a moderator of chromosomal communications.
A. The mechanism of loop extrusion. B. Blocking of loop extrusion by boundary element (e.g. CTCF) that can halt cohesin only if properly oriented, as determined by orientation of its binding site. Fudenberg et al. assumed that interactions with CTCF would halt loop-extruding activity of only one of two cohesin motors, while the unobstructed motor can continue extruding a loop. C. In this picture, loop extrusion is a universal mechanism that provides both formation of TADs and cis communication during interphase and chromosome compaction during mitosis, both by about 100Kb loops but with different spacing between the loops. During interphase loop extrusion is performed by cohesin and blocking at TAD boundaries can be performed by several factors, including binding by CTCF. Blocking of loop extrusion allows small insulating factors to insulate interactions between distal elements (Fig 2E). D. We propose that loop extrusion can also facilitate close-range contacts between functional elements, by bringing them in direct molecular contact. Such interactions are possibly only within a TAD and are completely insulated by extruding-blocking boundary elements. E. During mitosis, loop extrusion is performed by condensins that should be more abundant on DNA making each loop reinforced by multiple condensins (Goloborodko et al., 2015, 2016).
Alipour and Marko have introduced a 1D model of multiple loop-extruding factors binding to DNA, which demonstrated that exchanging loop-extruding factors can form stacked configurations where a single loop is stabilized by multiple factors (Alipour and Marko, 2012). More recently Goloborodko et al. (Goloborodko et al., 2015; Goloborodko el al., 2016) have demonstrated by simulations that loop-extruding factors that exchange between the nucleoplasm and chromatin fiber self-organize chromatin into a dynamic array of consecutive loops. Depending on processivity and density of loop-extruding factors, the system self-organizes into one of the two steady state regimes: a dense (mitotic) regime where loop-extruding factors drastically compact a long chromatin fiber forming an array of consecutive loops to generate mitotic chromosomes (Fig 2E), and a sparse (interphase) regime where loops are separated by gaps and provide moderate compaction (Fig 2C). Our Hi-C study of human mitotic chromosome (Naumova et al., 2013) have provided a strong support to these theoretical predictions, demonstrating that an array of consecutive stochastically positioned ~100Kb loops that can be formed by loop-extruding condensins, and further longitudinally compacted, has a 3D structure that quantitatively agrees with mitotic Hi-C data. These studies have not considered its role in TAD formation in interphase.
Fudenberg et al. (Fudenberg et al., 2015) and Sandborn et al. (Sanborn et al., 2015) have proposed that TADs can be formed by loop extrusion activity of multiple exchanging cohesins that are stalled at TAD boundaries. When bound, a cohesin forms a progressively larger loop until cohesin encounters an obstacle, either another cohesin or due to interactions with boundary proteins, including CTCF (Fig 3A-C). This minimal model of (Fudenberg et al., 2015) doesn’t require loading or unloading of cohesins at specific sites or additional stabilization of cohesin upon binding to CTCF .. This mechanism was tested by polymer simulations, which showed formation of TADs that recapitulate qualitative and quantitative characteristics of TADs observed in Hi-C data. Such characteristics include the decay of the contact probability curve Pwithin(s) with genomic separation s, when both loci are located within the same TAD, and the same curve Pbetween(s) for interactions between neighboring TADs. Fudenberg et al., (2015) showed that the best quantitative agreement was achieved when each cohesin can extrude a loop of ~100-200Kb each, and cohesin density of DNA is about one per 100-200Kb, corresponding to about 30,000-60,000 cohesin molecules per diploid genome.
Interestingly, the model of human mitotic chromosome (Naumova et al., 2013) suggested mitotic loops to be of about the same ~100Kb size, while requiring much higher (×10-20) number of loop-extruders to achieve significant linear compactions of the chromosome (Goloborodko et al., 2015). In fact, Naumova et al (Naumova et al., 2013) were the first to demonstrate that an array of consecutive loops has a shallow scaling of contact probability P(s)~s−0.5, which is similar to that within TADs Pwithin(s)~s−0.6..−0.7 as was more recently demonstrated (Fudenberg et al., 2015; Sanborn et al., 2015). In summary, this loop-extruding model suggests that a TAD is composed by several dynamic loops that are constantly extruded by cohesins and dispersed when cohesins dissociate.
A very similar model has been put forward most recently in Sanburn et al. (Sanborn et al., 2015). This model additionally requires that upon formation of the border-to-border loop, interactions with CTCF stabilize a cohesin complex preventing its dissociation, making such loop practically irreversible. This model also shows excellent agreement with the Hi-C data, with Pwithin(s)~s−0.6..−0.7 scaling, and was able to reproduce qualitatively maps obtained by deletions and inversions of specific CTCF sites. The study claims to be able to compute Hi-C maps from CTCF occupancy data. According to this model, a TAD is a stable large border-to-border loop, while other intra-TAD loops are transient.
These models can explain observed preferential inward orientation of CTCF sites flanking a TAD (de Wit et al., 2015; Gómez-Marín et al., 2015; Guo et al., 2015; Rao et al., 2014; Vietri Rudan et al., 2015). If cohesin-halting function requires a proper orientation of CTCF relative to cohesin, then only those CTCF sites that provide such (inward) orientation will serve as TAD boundaries, thus explaining enrichment of such sites at TAD boundaries. Interestingly, TADs formed by loop extrusion can also reproduce not only enrichment of interactions within a TAD, but also a striking feature observed in about 50% of TADs: an enrichment of contacts between two TAD borders, i.e. a border-to-border loop (Rao et al., 2014).
Nichols and Corces (Nichols and Corces, 2015) and Bouwman and de Laat (Bouwman and de Laat, 2015) have also put forward hypotheses that loop extrusion, possibly mediated by cohesin, is a mechanism underlying formation of loops between TAD boundaries. In their models, cohesin binds to CTCF residing at one of the borders and processively extrude a single loop until CTCF residing at another border is reached. Similar to Fudenberg et al. (Fudenberg et al., 2015), the model would explain preferential orientation of CTCF sites at the borders. These models have not yet been tested by simulations and it remains to be seen whether they can reproduce other characteristics of TADs. Based on simulations of Fudenberg et al. (2015), stable loops between domain boundaries are inconsistent with TADs as a single loop cannot reproduce relatively uniform contact enrichment with a TAD.
What features allow a loop extrusion mechanism to produce TADs with their characteristic ~two-fold enrichment of contact probabilities? In essence, loop extrusion facilitates formation of 3D contacts by an effectively one-dimensaional process that can be controlled by proteins bound at TAD boundaries. Insulating action of these boundary elements ensures that extruded loops bring together only elements located within a single TAD and not pairs located in different domains, leading to enrichment of interactions within TADs. Note that effective linear insulation between TADs does not prevent formation of 3D contacts between them, but makes them less likely than intra-TAD contacts. Most importantly, extrusion-based models provide a molecular mechanism of how DNA-bound proteins, e.g. at TAD boundaries, and that are much smaller in size than the formed loops, can reduce the frequency of interactions between TADs (Fig 2E, Fig 3C).
We also anticipate that loop extrusion can facilitate functional communications by bringing loci (genes and regulatory elements) together in a specific close molecular arrangement (Fig 3D, e.g. small distance and orientation specific). Such close range contacts may be quite different, and functionally distinct, from a stochastic 3D collision between the two loci. Although 3D collisions are abundant within as well as between TADs, close molecular arrangement provided by loop extrusion would be limited only to elements located within the same TAD (Fig 3C,D). This can be a mechanism by which insulating elements at TAD boundaries can prevent formation of functional contacts between different TADs and hence provide functional insulation that is much more significant than the modest 2-fold difference in observed 3D contact frequencies. Importantly, the active process of loop extrusion could also make such functional interactions less stochastic, with smaller cell-to-cell and temporal variation, as they are moderated by an active 1D- rather than a diffusive 3D process.
Taken together, these arguments suggest that if functional interactions require specific molecular arrangement that can be created by loop extruding factors, rather than 3D contacts, then effectively 1-dimensional active loop extrusion can provide features that are hard to achieve by 3D contacts. Such features include: (a) smaller cell-to-cell and temporal variation of functional interactions between elements located <1Mb apart; (b) reliable insulation of functional interactions by CTCF and other boundary occupying proteins, despite much small sizes of such proteins than the formed loops; (c) possibility of linear spreading of such interactions when boundary elements are removed; (d) preservation of genomic orientation between elements when interactions are created by loop extrusion.
Other communication in cis
One critical feature of active moderation of contacts within TADs through tracking and loop extrusion is that it ensures that loci communicate only in cis. Would such mechanism also be involved in other communications? For instance, any tracking mechanism may transport and deliver signals, e.g. ncRNAs from one location to another, either within a TAD, or across larger sections of chromosomes. One particularly well-studied example is the process of X-inactivation. In female mammals one X chromosome expresses Xist RNA from one copy of the X, and this RNA will ultimately cover the entire X chromosome. The mechanism by which Xist spreads is not well understood. One model is that Xist is simply diffusing in 3D from its source, the Xist locus (Engreitz et al., 2013). This model is based on the observation that upon Xist induction from a still active X, the Xist RNA is found to be associated initially with gene dense loci that are also close in 3D to the Xist locus when the X chromosome is active. Although this can indeed point to 3D spreading, it may also be related to the fact that Xist generally binds more with gene dense regions (Sarma et al., 2014; Simon et al., 2014). Also, a 3D spreading mechanism cannot prevent spreading of Xist to other chromosomes. Therefore a cis-spreading mechanism remains very likely, and may involve similar processes as loop extrusion and tracking.
Another exciting example of spreading and tracking through TADs was recently described by a set of publications from the Alt lab (Dong et al., 2015; Hu et al., 2015). Alt and co-workers studied RAG-dependent V(D)J recombination. They found that RAG-mediated recombination is constrained within a TAD defined by pairs of convergent CTCF-bound elements. Like enhancer-promoter interactions, RAG-dependent recombination involves interactions between pairs of sites (recombination signal sequences, RSS). Intriguingly, these authors found an RSS orientation dependence of RAG off-target activity within CTCF loops spanning up to 2Mb and proposed that RAG complexes initially bind one RSS and then track linearly along the chromatin fiber till it encounters a convergent RSS.
Topological machines
The studies highlighted above provide intriguing evidence for directional and cis-guided long-range interactions. While DNA extrusion by FtsK and SpoIIIE proteins in bacterial has been well established (Gruber, 2014), there is as of yet no direct evidence that eukaryotic complexes are tracking in cis, or extruding loops in eukaryotes. We propose that the cell has evolved multiple machineries that we refer to as topological machines that can perform this action. We predict that such machines need to have the following characteristics: 1) Bind DNA, possibly in a directional manner. 2) Translocate along the chromatin fiber, possibly in a directional manner. 3) For looping, the machinery needs two motors moving in opposite directions. 4) Their migration can be blocked by other complexes, sometimes in a directional manner. Such blocking complexes may be located at TAD boundaries (e.g. CTCF), but can be any other complex such as those associated with enhancers, promoters and other regulatory elements that will lead to those elements becoming juxtaposed.
Are there any known protein complexes that have such properties? Several proteins are known to have translocase activity, several helicase can translocate along dsDNA without unwinding the duplex (see (Singleton et al., 2007) for review). These helicases and translocases share similarity to mammalian SMC proteins (including cohesin and condensin) in the P-loop containing ATPase domains. As was suggested in 1990s (Guacci et al., 1993; Hirano et al., 1995; Peterson, 1994) domain architecture of SMC proteins (ATPase, coiled coil arm, hinge, coiled coil arm, ATPase) resembles those of cytoplasmic motors such as kinesins and myosins, leading to a proposal that SMC proteins constitute chromatin mechanochemical proteins that actively drive mitotic condensation. Since then, however, the main focus has been on ability of SMC proteins to form a ring that can encircle that DNA. SMC complexes have been implicated in a variety of chromosome architectural processes such chromatin looping (Crane et al., 2015; Hirano, 2012; Kagey et al., 2010; Nasmyth, 2001), TAD formation (Crane et al., 2015; Seitan et al., 2013; Zuin et al., 2014), sister chromatid cohesion (Guacci et al., 1997; Michaelis et al., 1997; Nasmyth and Haering, 2009), and chromosome condensation and dosage compensation (Crane et al., 2015; Hirano, 2012).
Direct evidence that SMC - containing complexes can track in cis, and make orientation-specific loops is still scant, though some recent in vitro observations suggest SMC complexes can slide along DNA (Kim and Loparo, 2016). Interestingly, early work showed that condensin could induce compaction of an isolated stretched DNA by dynamically introducing loops along the DNA (Strick, 2004). Similarly, at about the same time the cohesin complex was shown to be loaded at one chromosomal position and then to be moved along the chromatin fiber to other positions (Lengronne et al., 2004).
There are several aspects of this proposed mechanism that are currently difficult to explain in molecular terms. First, how can such a machine travel along a highly complex chromatin fiber containing nucleosomes and a large set of additional non-histone factors? How fast can such chromosome motors move along DNA extruding loops, and how is motion of motors synchronized and regulated? Does this process introduce topological stress that needs to be relieved by topoisomerases, as was suggested earlier (Kimura et al., 1999)? Interestingly, bacterial DNA-extruding translocase FtsK is able to translocate at an astonishing speed of ~5Kb/s and to displace DNA-bound roadblocks (Crozat et al., 2010), supporting feasibility of translocation along chromatinized DNA and possibility of extruding ~200Kb loops during ~10-20min turnover time of cohesion (Gerlich et al., 2006).
Finally, we consider it likely that there are multiple machines that can perform this mechanism, besides SMC complexes. For instance, RNAP polymerase II is obviously tracking along the chromatin while transcribing genes. In an interesting recent study Blobel and co-workers found evidence that the moving polymerase may remain associated also with the promoter leading to dynamic loop formation between the promoters and body of gene (Lee et al., 2015). Tracking and extruding genes by active polymerases has also been reported by the Cook laboratory (Larkin et al., 2013). Could such polymerase-based loop extrusion also lead to enhancer-promoter interactions, perhaps related to previously proposed linking models (Bulger and Groudine, 1999)?. Further, as mentioned above Alt and co-worked have proposed that an RAG-containing complex scans chromatin within TADs during V(D)J recombination.
3D genome and noisy and stochastic gene expression
Although compartmentalization and potential cis-tracking canalizes interactions in cis and towards pairs of loci located within insulated domains, the spatial separation of such domains is by no means absolute. This is apparent from genome-wide chromatin interaction maps where one can detect interactions between loci located in different TADs, compartment or even on different chromosomes. These interactions are of low frequency. Thus chromatin interaction data, e.g. obtained with Hi-C or TCC, reflect the sum of many different 3D folding states of the genome in the population (Kalhor et al., 2011). Possibly, TAD boundaries are not implemented in all cells in the population and may be stochastic due to dynamic dissociation-re-association of CTCF, leading to fusion of adjacent domains in a subset of cells. It is tempting to propose that these low-frequency inter-domain, and inter-chromosomal interactions may contribute to gene expression noise and cell-to-cell variability in expression patterns and levels (Krijger and de Laat, 2013). In one careful study it was found that ectopic insertion of a strong enhancer, the beta-globin locus control locus, on chromosome 8 could affect expression of the endogenous beta-globin genes located on chromosome 7, but this activation only occurred in the very small subpopulation of cells where the ectopic locus control regions physically interacted with the globin genes (Noordermeer et al., 2011). These authors coined the term “Spatial Effect Variegation” to describe this phenomenon.
In cases where cells aim to express a gene in a highly stochastic manner, this may be one mechanism to achieve this. Regulation of olfactory receptor genes in neurons represents an interesting example of this, as proposed and carefully documented by the Lomvardas laboratory (Lomvardas et al., 2006; Markenscoff-Papadimitriou et al., 2014; Monahan and Lomvardas, 2015). Olfactory neurons contain several thousand olfactory receptor genes but each neuron expresses only one of these, and different neurons express different receptors. How can cells stochastically pick and express only one olfactory gene? The full answer to that question is not known yet, and many different processes appear to play a role including signaling feedback loops to repress expression of any additional receptor genes once a receptor is active (Dalton et al., 2013). But a role for stochastic inter-chromosomal interactions in initial picking a single receptor gene seems likely. Lomvardas and co-workers found that although there are thousands of receptor genes spread all over the genome, they are regulated by only a small number of enhancers (Markenscoff-Papadimitriou et al., 2014). To regulate most of the receptor genes, the enhancers will often have to act in trans, and as outlined above, such inter-chromosomal interactions occur in a highly stochastic manner. Thus, in any neuron the enhancers could interact with and activate only a very small subset of all receptor genes. This would be an example where the cell takes advantage of the incomplete spatial insulation of sub-chromosomal domains, and the stochastic sub-nuclear positioning of chromosomes.
Other roles for cis-communications: mitotic chromosome condensation
Chromosome condensation during prophase involves establishment of long-range interactions along entire chromosomes, which also involves cis-communication in order to prevent such interactions from occurring between different chromosomes. Extensive imaging experiments, mostly led by Laemmli and co-workers, and more recent 5C and Hi-C analyses combined with polymer simulations have led to a model where mitotic chromosomes fold as linearly organized longitudinally compressed arrays of randomly positioned consecutive chromatin loops (Dekker, 2014; Earnshaw and Laemmli, 1983; Marsden and Laemmli, 1979; Naumova et al., 2013). We had proposed that this structure is formed through loop extrusion along the chromosome followed by longitudinal compression (Naumova et al., 2013). One important feature of such a model is that looping interactions are ensured to occur in cis only. Furthermore, simulations show that loop extrusion mediated mitotic condensation leads to segregation and individualization of chromatids (Goloborodko et al., 2016). Transition from G2 to mitosis (Figure 3) can then be manifested by replacement of one class of loop-extruding enzymes (cohesins) with another (condensins), which should be present about 10-fold higher abundance, and loss of boundary elements (including CTCF) to provide uniform condensation and segregation of sister chromatids. If correct, loop extrusion may be a general mechanism for chromosome folding throughout the cell cycle.
Outlook
General principles guiding the spatial conformation of the chromosomes, such as compartmentalization, are now becoming increasingly understood and this is leading to a better understanding of long-range chromosomal communication. However, important gaps in our understanding remain especially regarding the dynamics of loci with respect to each other, cell-to-cell variability in chromosome folding and the identity and activity of machineries that drive these processes. A deeper understanding into the dynamics of chromatin within domains and along chromosomes, the mechanisms by which loci move (e.g. through loop extrusion and tracking), and how complexes track along chromatin, will lead to more quantitative models for gene regulation through long-range interactions over time and to mechanistic insights into cellular variability in transcription. Importantly, though loop extrusion is an appealing model that explains many experimental observations, this model remains to be tested experimentally. Further characterization of the mechanism of action of candidate topological machines such as cohesin and condensin in facilitating and preventing long-range interactions, and identification of other such machineries that no doubt exist, promise to reveal how the genome communicates.
Acknowledgements
We thank members of the Dekker and Mirny labs for discussion. Work in our labs is supported by the National Human Genome Research Institute (R01 HG003143, U54 HG007010, U01 HG007910), the National Cancer Institute (U54 CA193419), the NIH Common Fund (U54 DK107980, U01 DA 040588), the National Institute of General Medical Sciences (R01 GM 112720), and the National Institute of Allergy and Infectious Diseases (U01 R01 AI 117839). J.D. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that they have no competing interests.
References
- Alipour E, Marko JF. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012;40:11202–11212. doi: 10.1093/nar/gks925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen DJ, Makhov A, Grilley M, Modrich P, Griffith JD. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augui S, Filion GJ, Huart S, Nora E, Guggiari M, Maresca M, Stewart AF, Heard E. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318:1632–1636. doi: 10.1126/science.1149420. [DOI] [PubMed] [Google Scholar]
- Barakat TS, Loos F, van Staveren S, Myronova E, Ghazvini M, Grootegoed JA, Gribnau J. The trans-activator RNF12 and cis-acting elements effectuate X chromosome inactivation independent of X-pairing. Mol Cell. 2014;53:965–978. doi: 10.1016/j.molcel.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Bickmore WA. The spatial organization of the human genome. Annu Rev Genomics Hum Genet. 2013;14 doi: 10.1146/annurev-genom-091212-153515. [DOI] [PubMed] [Google Scholar]
- Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Bouwman BA, de Laat W. Getting the genome in shape: the formation of loops, domains and compartments. Genome Biol. 2015;16:154. doi: 10.1186/s13059-015-0730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Pombo A. Intermingling of Chromosome Territories in Interphase Suggests Role in Translocations and Transcription-Dependent Associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronstein I, Israel Y, Kepten E, Mai S, Shav-Tal Y, Barkai E, Garini Y. Transient anomalous diffusion of telomeres in the nucleus of mammalian cells. Phys Rev Lett. 2009;103:018102. doi: 10.1103/PhysRevLett.103.018102. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai Y.-f., Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. Condensin-driven remodeling of X-chromosome topology during dosage compensation. Nature. 2015 doi: 10.1038/nature14450. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Crozat E, Meglio A, Allemand JF, Chivers CE, Howarth M, Vénien-Bryan C, Grainge I, Sherratt DJ. Separating speed and ability to displace roadblocks during DNA translocation by FtsK. EMBO J. 2010;29:1423–1433. doi: 10.1038/emboj.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton RP, Lyons DB, S. L. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 2013;155:321–332. doi: 10.1016/j.cell.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- de Wit E, Vos ES, Holwerda SJ, Valdes-Quezada C, Verstegen MJ, Teunissen H, Splinter E, Wijchers PJ, Krijger PH, de Laat W. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell. 2015;60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- Dekker J. Two ways to fold the genome during the cell cycle: insights obtained with chromosome conformation capture. Epigenetics Chromatin. 2014;7:25. doi: 10.1186/1756-8935-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing Chromosome Conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Panchakshari RA, Zhang T, Zhang Y, Hu J, Volpi SA, Meyers RM, Ho YJ, Du Z, Robbiani DF, et al. Orientation-specific joining of AID-initiated DNA breaks promotes antibody class switching. Nature. 2015;525:134–139. doi: 10.1038/nature14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle B, Fudenberg G, Imakaev M, Mirny LA. Chromatin loops as allosteric modulators of enhancer-promoter interactions. PLoS Comput Biol. 2014;10:e1003867. doi: 10.1371/journal.pcbi.1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Laemmli UK. Architecture of metaphase chromosomes and chromosome scaffolds. J Cell Biol. 1983;96:84–93. doi: 10.1083/jcb.96.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J, Williamson I, Bickmore WA, Dostie J. An Overview of Genome Organization and How We Got There: from FISH to Hi-C. Microbiol Mol Biol Rev. 2015;79:347–372. doi: 10.1128/MMBR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Adbennur N, Mirny LA. Formation of chromosomal domains by loop extrusion. 2015 doi: 10.1016/j.celrep.2016.04.085. bioRxiv doi: http://dx.doi.org/10.1101/024620. [DOI] [PMC free article] [PubMed]
- Galupa R, Heard E. X-chromosome inactivation: new insights into cis and trans regulation. Curr Opin Genet Dev. 2015;31:57–66. doi: 10.1016/j.gde.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Heard E. Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annu Rev Cell Dev Biol. 2014;30:561–580. doi: 10.1146/annurev-cellbio-101512-122415. [DOI] [PubMed] [Google Scholar]
- Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol. 2006;16:1571–1578. doi: 10.1016/j.cub.2006.06.068. [DOI] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloborodko A, Marko JF, Mirny LA. Mitotic chromosome compaction via active loop etrusion. 2015 doi: 10.1016/j.bpj.2016.02.041. bioRxiv doi: http://dx.doi.org/10.1101/021642. [DOI] [PMC free article] [PubMed]
- Goloborodko A, Marko JF, Mirny LA. Compaction and segregation of sister chromatids via active loop extrusion. 2015 doi: 10.7554/eLife.14864. bioRxiv doi: http://dx.doi.org/10.1101/038281. [DOI] [PMC free article] [PubMed]
- Gómez-Marín C, Tena JJ, Acemel RD, López-Mayorga M, Naranjo S, de la Calle-Mustienes E, Maeso I, Beccari L, Aneas I, Vielmas E, et al. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc Natl Acad Sci U S A 2015. 2015;112:7542–7547. doi: 10.1073/pnas.1505463112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostissa M, Schwer B, Chang A, Dong J, Meyers RM, Marecki GT, Choi VW, Chiarle R, Zarrin AA, Alt FW. IgH class switching exploits a general property of two DNA breaks to be joined in cis over long chromosomal distances. Proc Natl Acad Sci U S A. 2015;111:2644–2649. doi: 10.1073/pnas.1324176111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S. Multilayer chromosome organization through DNA bending, bridging and extrusion. Curr Opin Microbiol. 2014;22:102–110. doi: 10.1016/j.mib.2014.09.018. [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Yamamoto A, Strunnikov A, Kingsbury J, JHogan E, Meluh P, Koshland D. Structure and function of chromosomes in mitosis in budding yest. Cold Spring Harb Symp Quant Biol. 1993;58:677–685. doi: 10.1101/sqb.1993.058.01.075. [DOI] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halverson JD, Smrek J, Kremer K, Grosberg AY. From a melt of rings to chromosome territories: the role of topological constraints in genome folding. Rep Prog Phys. 2014;77(2):022601. doi: 10.1088/0034-4885/77/2/022601. [DOI] [PubMed] [Google Scholar]
- Hajjoul H, Mathon J, Ranchon H, Goiffon I, Mozziconacci J, Albert B, Carrivain P, Victor JM, Gadal O, Bystricky K, et al. High-throughput chromatin motion tracking in living yeast reveals the flexibility of the fiber throughout the genome. Genome Res. 2013;23:1829–1838. doi: 10.1101/gr.157008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 2012;26:1659–1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ, Swedlow JR. The SMC family: from chromosome condensation to dosage compensation. Curr Opin Cell Biol. 1995;7:329–336. doi: 10.1016/0955-0674(95)80087-5. [DOI] [PubMed] [Google Scholar]
- Hou C, Li L, Qin ZS, Corces VG. Gene Density, Transcription, and Insulators Contribute to the Partition of the Drosophila Genome into Physical Domains. Mol Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TS, Weiner A, Lajoie BR, Dekker J, Friedman N, Rando OJ. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell. 2015;162:108–119. doi: 10.1016/j.cell.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Zhang Y, Zhao L, Frock RL, Du Z, Meyers RM, Meng FL, Schatz DG, Alt FW. Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes. Cell. 2015;163:947–959. doi: 10.1016/j.cell.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Sarma K, Lee JT. New and Xisting regulatory mechanisms of X chromosome inactivation. Curr Opin Genet Dev. 2012;22:62–71. doi: 10.1016/j.gde.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2011;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Loparo JJ. Multistep assembly of DNA condensation clusters by SMC. Nat Commun. 2016;7:10200. doi: 10.1038/ncomms10200. doi: 10210.11038/ncomms10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Rybenkov VV, Crisona NJ, Hirano T, Cozzarelli NR. 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijger PH, de Laat W. Identical cells with different 3D genomes; cause and consequences? Curr Opin Genet Dev. 2013;23:191–196. doi: 10.1016/j.gde.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Larkin JD, Papantonis A, Cook PR, Marenduzzo D. Space exploration by the promoter of a long human gene during one transcription cycle. Nucleic Acids Res. 2013;41:2216–2227. doi: 10.1093/nar/gks1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Hsiung CC, Huang P, Raj A, Blobel GA. Dynamic enhancer-gene body contacts during transcription elongation. Genes Dev. 2015;29:1992–1997. doi: 10.1101/gad.255265.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Lucas JS, Zhang Y, Dudko OK, Murre C. 3D trajectories adopted by coding and regulatory DNA elements: first-passage times for genomic interactions. Cell. 2014;158:339–352. doi: 10.1016/j.cell.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markenscoff-Papadimitriou E, Allen WE, Colquitt BM, Goh T, Murphy KK, Monahan K, Mosley CP, Ahituv N, S. L. Enhancer interaction networks as a means for singular olfactory receptor expression. Cell. 2014;159:543–557. doi: 10.1016/j.cell.2014.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden MP, Laemmli UK. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979;17:849–858. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- Masui O, Bonnet I, Le Baccon P, Brito I, Pollex T, Murphy N, Hupé P, Barillot E, Belmont AS, Heard E. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell. 2011;145:447–458. doi: 10.1016/j.cell.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Milo R, Jorgensen P, Moran U, Weber G, Springer M. BioNumber - the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010;38(Suppl 1):D750–D753. doi: 10.1093/nar/gkp889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA. Nucleosome-mediated cooperativity between transcription factors. Proc Natl Acad Sci U S A. 2010;107:22534–22539. doi: 10.1073/pnas.0913805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan K, Lomvardas S. Monoallelic Expression of Olfactory Receptors. Annu Rev Cell Dev Biol. 2015;31:721–740. doi: 10.1146/annurev-cellbio-100814-125308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43 doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols MH, Corces VG. a CTCF code for 3D genome architecture. Cell. 2015;162:703–705. doi: 10.1016/j.cell.2015.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, de Wit E, Klous P, van de Werken H, Simonis M, Lopez-Jones M, Eussen B, de Klein A, Singer RH, de Laat W. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat Cell Biol. 2011;13:944–951. doi: 10.1038/ncb2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL. The SMC family: novel motor proteins for chromosome condensation. Cell. 1994;79:389–392. doi: 10.1016/0092-8674(94)90247-x. [DOI] [PubMed] [Google Scholar]
- Ptashne M. The chemistry of regulation of genes and other things. J biol Chem. 2014;289:5417–5435. doi: 10.1074/jbc.X114.547323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Nicolas E, Sherratt DJ. Chromosome replication and segregation in bacteria. Annu Rev Genet. 2012;46:121–143. doi: 10.1146/annurev-genet-110711-155421. [DOI] [PubMed] [Google Scholar]
- Rosa A, Everaers R. Structure and dynamics of interphase chromosomes. PLoS Comput Biol. 2008;4:e1000153. doi: 10.1371/journal.pcbi.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Cifuentes-Rojas C, Ergun A, Del Rosario A, Jeon Y, White F, Sadreyev R, Lee JT. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell. 2014;159:869–883. doi: 10.1016/j.cell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Spitz F. The architecture of gene expression: integrating dispersed cis-regulatory modules into coherent regulatory domains. Curr Opin Genet Dev. 2014;27:74–82. doi: 10.1016/j.gde.2014.03.014. [DOI] [PubMed] [Google Scholar]
- Seitan V, Faure A, Zhan Y, McCord R, Lajoie B, Ing-Simmons E, Lenhard B, Giorgetti L, Heard E, Fisher A, et al. Cohesin-based chromatin interactions enable regulated gene expression within pre-existing architectural compartments. Genome Res. 2013;23:2066–2077. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Schober H, Fraser P, Gasser SM. Gene regulation through nuclear organization. Nat Struct Mol Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Shachar S, Voss TC, Pegoraro G, Sciascia N, Misteli T. Identification of Gene Positioning Factors Using High-Throughput Imaging Mapping. Cell. 2015;162:911–923. doi: 10.1016/j.cell.2015.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Lynch CR, Peterson KA, Thornton K, Kepper N, Hase J.v., Stein S, Vincent S, Molloy KR, Kreth G, et al. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol. 2006;174:27–38. doi: 10.1083/jcb.200603083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2014;504:465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, D.B. W. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Smith EM, Lajoie BR, Jain G, Dekker J. Invariant TAD boundaries constrain cell type-specific looping interactions between promoters and distal elements around the CFTR locus. Am J Hum Genet. 2016;98:185–201. doi: 10.1016/j.ajhg.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick TR. Real-time detection of single-molecule DNA compaction by condensin I. Curr Biol. 2004;14:874–880. doi: 10.1016/j.cub.2004.04.038. [DOI] [PubMed] [Google Scholar]
- Strickfaden H, Zunhammer A, van Koningsbruggen S, Köhler D, Cremer T. 4D chromatin dynamics in cycling cells: Theodor Boveri’s hypotheses revisited. Nucleus. 2010;1:284–297. doi: 10.4161/nucl.1.3.11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, Ettwiller L, Spitz F. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24:390–400. doi: 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tark-Dame M, Jerabek H, Manders EM, Heermann DW, van Driel R. Depletion of the chromatin looping proteins CTCF and cohesin causes chromatin compaction: insight into chromatin folding by polymer modelling. PLoS Comput Biol. 2014;10:e1003877. doi: 10.1371/journal.pcbi.1003877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P, Duong T, Dujon B, Zimmer C, Fabre E. Chromosome arm length and nuclear constraints determine the dynamic relationship of yeast subtelomeres. Proc Natl Acad Sci U S A. 2010;107:2025–2030. doi: 10.1073/pnas.0914187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjong H, Gong K, Chen L, Alber F. Physical tethering and volume exclusion determine higher-order genome organization in budding yeast. Genome Res. 2012;22:1295–1305. doi: 10.1101/gr.129437.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and Interaction between Hypersensitive Sites in the Active beta-globin Locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Ulianov SV, Khrameeva E,E, Gavrilov AA, Flyamer IM, Kos P, Mikhaleva EA, Penin AA, Logacheva MD, Imakaev MV, Chertovich A, Gelfand MS, Shevelyov YY, Razin SV. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 2015;26(1):70–84. doi: 10.1101/gr.196006.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Le TBK, Lajoie BR, Dekker J, Laub MT, Rudner DZ. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 2015;29:1661–1675. doi: 10.1101/gad.265876.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AG, Fraser P. Remote control of gene transcription. Hum Mol Genet. 2005;14:R101–111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- Wong H, Marie-Nelly H, Herbert S, Carrivain P, Blanc H, Koszul R, Fabre E, Zimmer C. A predictive computational model of the dynamic 3D interphase yeast nucleus. Curr Biol. 2012;22:1881–1890. doi: 10.1016/j.cub.2012.07.069. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- Yang R, Kerschner JL, Gosalia N, Neems D, Gorsic LK, Safi A, Crawford GE, Kosak ST, Leir SH, Harris A. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1358. pii: gkv1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IJcken WF, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]