Abstract
Backgrounds
Despite reported discordance between the mutational status of primary lung cancers and their metastases, metastatic sites are rarely biopsied and targeted therapy is guided by genetic biomarkers detected in the primary tumor. This situation is mostly explained by the apparent stability of EGFR-activating mutations. Given the dramatic increase in the range of candidate drugs and high rates of drug resistance, rebiopsy or liquid biopsy may become widespread. The purpose of this study was to test genetic biomarkers used in clinical practice (EGFR, ALK) and candidate biomarkers identified by the French National Cancer Institute (KRAS, BRAF, PIK3CA, HER2) in patients with metastatic non-small-cell lung cancer for whom two tumor samples were available.
Methods
A retrospective study identified 88 tumor samples collected synchronously or metachronously, from the same or two different sites, in 44 patients. Mutation analysis used SNaPshot (EGFR, KRAS, BRAF missense mutations), pyrosequencing (EGFR and PIK3CA missense mutations), sizing assays (EGFR and HER2 indels) and IHC and/or FISH (ALK rearrangements).
Results
About half the patients (52 %) harbored at least one mutation. Five patients had an activating mutation of EGFR in both the primary tumor and the metastasis. The T790M resistance mutation was detected in metastases in 3 patients with acquired resistance to EGFR tyrosine kinase inhibitors. FISH showed discordance in ALK status between a small biopsy sample and the surgical specimen. KRAS mutations were observed in 36 % of samples, six patients (14 %) having discordant genotypes; all discordances concerned sampling from different sites. Two patients (5 %) showed PI3KCA mutations. One metastasis harbored both PI3KCA and KRAS mutations, while the synchronously sampled primary tumor was mutation free. No mutations were detected in BRAF and HER2.
Conclusions
This study highlighted noteworthy intra-individual discordance in KRAS mutational status, whereas EGFR status was stable. Intratumoral heterogeneity for ALK rearrangement suggests a limitation of single-biopsy analysis for therapeutic strategy with crizotinib.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-016-2249-6) contains supplementary material, which is available to authorized users.
Keywords: Non-small-cell lung cancer, Metastatic lesion, Rebiopsy, Genetic biomarkers, Targeted therapy
Background
Chemotherapy is still the standard treatment for metastatic non-small-cell lung cancer (NSCLC), which represents 60 % of cases at diagnosis. Overall and progression-free survival rates have gradually improved, notably with the use of different drugs or combinations and more precise histological classification [1,2]. However, median survival in advanced disease is still less than 12 months, even among patients receiving platinum-based doublet chemotherapy and bevacizumab [1,3]. The recent introduction of tyrosine kinase receptor inhibitors for the EGFR and ALK genes [4–8], which are mutated in respectively 10 and 4 % of non-small-cell lung tumors in Caucasian patients, has had a major impact, despite occasional resistance mutations such as T790M in the EGFR gene, which is found in more than 50 % of patients treated by tyrosine kinase inhibitor (TKI) [9,10]. Several clinical trials are underway, based on genetic biomarkers and activation pathway inhibitors. The intracellular oncogene KRAS is a particularly attractive target because of its high mutation rate (>25 % of patients), especially in current and former heavy smokers [11].
A major issue raised by targeted therapies is potential discordance between the mutational status of the primary tumor and its metastases, or between two regions of the same tumor. This is particularly important in lung cancer: repeat biopsy is rarely performed [12], even though various studies have shown discrepancies in EGFR, ALK and KRAS mutational status [13–18].
The present study examined discordance between repeat samples from the same tumor site or samples from two different sites, collected synchronously or metachronously. The principal mutations of EGFR, ALK, KRAS, BRAF, PIK3CA and HER2 were analyzed in 44 patients with non-small-cell lung cancer. The KRAS, BRAF, PIK3CA and HER2 oncogenes were selected because they represented potential drug targets [19]. They were identified as potentially predictive biomarkers in NSCLC by the French National Cancer Institute (INCa) and were introduced in the French nationwide initiative for tumor molecular profiling during the 2010–2014 period [20].
Methods
Patients
This retrospective cohort study included patients with non-small-cell lung cancer (adenocarcinoma or squamous cell carcinoma) for whom two tumor samples were available, collected synchronously or metachronously either from the same site or from two different sites during disease course between 2005 and 2012. Patients were identified by cross-matching information from surgical files (surgical biopsy of metastasis, analysis of lobectomy or pneumonectomy specimen, or bronchial biopsy) with the medical codes of the institution. The corresponding tissue blocks were identified in each case. Samples were obtained by simple biopsy (n = 47; 53.4 %), surgical excision or biopsy (n = 39; 44.3 %), or fine-needle aspiration cytology (n = 2; 2.3 %).
Ethics statement
The study was conducted in accordance to the Declaration of Helsinki principles. It was approved by the Human Research Ethics Committee of Brest University Hospital (“Comité de Protection des Personnes - Ouest VI”; January 18, 2012). Written informed consent for the use of tissues and clinical data for research was taken from patients at the time of procurement of tumor specimens.
DNA extraction
All tumor samples were formalin-fixed and embedded in paraffin (FFPE). In each case, the percentage of tumor cells was determined by an experienced pathologist on a representative histological cross-section. Samples from at least three serial 10-μm sections were macrodissected and pooled for DNA extraction. DNA was extracted using the Maxwell® 16 FFPE Plus LEV DNA purification kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Mutational analyses
EGFR, KRAS, BRAF and PI3KCA status
Fragment-length analysis was used to screen for deletions and insertions in EGFR exons 19 and 20 and in HER2 exon 20. Genomic tumor DNA was amplified using the Qiagen™ Multiplex PCR kit (Qiagen, Hilden, Germany) with the following primers: 5′-N-CTG-GAT-CCC-AGA-AGG-TGA-GA-3′ and 5′-GAT-TTC-CTT-GTT-GGC-TTT-CG-3′ (EGFR exon 19), 5′-N-CTC-CAG-GAA-GCC-T AC-GTG-AT-3′and 5′-CTG-CGT-GAT-GAG-CTG-CAC-3′ (EGFR exon 20), and 5′-N-CCT-CTC-AGC-GTA-CCC-TTG-TC-3′ and 5′-AGG-GCA-TAA-GCT-GTG-TCA-CC-3′ (HER2 exon 20). For universal labeling, the forward primers were tailed with a short nucleotide sequence (N) that matched a universal FAM-labeled probe [21]. The labeled PCR products were subjected to capillary electrophoresis on an ABI PRISM 3100 XL genetic analyzer (Applied Biosystems, Courtabœuf, France) and compared with the wild-type PCR product to determine whether differences in length were present and represented deletion or insertion. Positive samples were re-amplified and sequenced using the BigDye Terminator v.1 cycle sequencing kit (Applied Biosystems), according to the manufacturer’s protocol. Sequence electrophoregrams were interpreted using SeqPatient analysis software version 3.5.2 (JSI Medical Systems, Ettenheim, Germany).
The EGFR, KRAS and BRAF genes were analyzed for presence of missense mutations using the ABI PRISM SNaPshot Multiplex kit (Applied Biosystems). Briefly, three multiplex PCRs were designed, the first for KRAS exon 2 (codons 12 and 13) and BRAF exon 15 (codon 600), the second for KRAS exon 3 (codon 61) and 4 (codon 146) and the third for EGFR exons 18 (codon 719) and 20 (codon 790). Multiplex PCR used the Qiagen™ Multiplex PCR kit with a total volume of 20 μL. PCR products were treated with Exonuclease I (ExoI) and shrimp alkaline phosphatase (SAP) (USB, Cleveland, Ohio, USA). Each extension primer (SNaPshot primer) was designed to anneal to the reverse strand of its targeted PCR product adjacent to the mutation site of interest. SNaPshot primers contained an additional tail at their 5’ end for simultaneous detection. Mutation detection reactions were performed in a total volume of 5 μL, comprising 1.5 μL SAP/ExoI-treated PCR product, 2 μL SNaPshot Multiplex Ready Reaction mix and 1.5 μL SNaPshot primer mix (each primer at a final concentration of 0.5 to 1.5 μM). Products were treated with SAP before automated sequencing (ABI PRISM 3500 Dx Genetic Analyzer, Applied Biosystems). Data were analyzed with GeneMapper Analysis software version 4.0 (Applied Biosystems).
The EGFR L848R mutation and PIK3CA exons 10 and 21 (codons 542, 545, 546, 1043, 1044, 1047 and 1049) were analyzed by pyrosequencing. For each target, PCR amplification was performed using the Qiagen™ Multiplex PCR kit with one of the primers biotinylated. Biotinylated products were immobilized on streptavidin-coated beads. After washing steps, DNA samples were released by denaturation in NaOH and annealed into single strands to a sequencing primer. Pyrosequencing was performed on a PyroMark Q24 system (Qiagen) following the manufacturer’s instructions. PCR primers, sequencing primers and dispensing orders are available upon request.
ALK status
ALK rearrangement was investigated in FFPE samples (3 μm thick) by Fluorescent In Situ Hybridization (FISH) and/or ImmunoHistoChemistry (IHC). FISH was performed on Superfrost ® Plus slides (Thermo Scientific, Saint-Herblain, France) with the Vysis LSI ALK Dual Color break-apart rearrangement probe (Abbott Molecular, Abbott Park, IL, USA). The slides were read using a Carl Zeiss epifluorescence microscope and the ISIS digital image analysis system (Isis in situ imaging system v.5.3, Metasystems, Altlussheim, Germany). FISH-positive cases were defined as those presenting >15 % split signals or an isolated orange signal in tumor cells. At least 100 nuclei were assessed for each tumor sample. Immunostaining with ALK monoclonal antibody (1:25, clone 5A4, Clinisciences, Nanterre, France) was performed using the Ventana Benchmark XT® automated slide preparation system and the OptiView DAB IHC Detection Kit according to the manufacturer’s instructions (Roche Diagnostics, Mannheim, Germany). ALK IHC scores, based on staining intensity and the percentage of tumor cells with positive cytoplasmic staining, were assigned as follows: 0, no stained cells; 1, faint or weak staining in >5 % cells or any staining intensity in ≤5 % of tumor cells; 2, moderate staining intensity in >5 % of tumor cells; or 3, strong granular staining intensity in >5 % of tumor cells.
Availability of data and materials
A dataset (mutations detection) supporting the conclusions of this article is available in Additional file 1: Figure S1.
Statistical analyses
Fisher’s exact test was used to identify significant factors for discordance in mutational status between two samples from a given patient.
Results
Study population
Table 1 summarizes demographic characteristics. There were 44 patients (27 male, 17 female) for whom two samples could be analyzed by molecular biology (primary tumor and metastasis or two samples from the same site). Current or former heavy smoking (>10 pack years) was reported in the majority of cases (32/44). Mean age at diagnosis was 60.5 years, slightly below the French national average [22]. More than half the patients had stage IV disease at diagnosis. All patients had non-small-cell lung cancer; the predominant histological type was adenocarcinoma (41/44). Synchronous samples were defined as being obtained within a 3-month period during which no cancer treatment was administered.
Table 1.
Patient characteristics
| Number | Total (%) | |
|---|---|---|
| Age (years) | 60.5 [39–76] | |
| gender | ||
| men | 27 | 61.3 |
| women | 17 | 38.7 |
| Never smokers | 7 | 16 |
| Current and former smokers | 32 | 72.7 |
| unknown | 5 | |
| Histology | ||
| Adenocarcinoma | 41 | 93 |
| Squamous cell | 3 | 7 |
| Stage at diagnosis | ||
| I | 6 | 13.7 |
| II | 4 | 9.1 |
| III | 9 | 20.4 |
| IV | 25 | 56.8 |
| Average time between samples (months) | ||
| Synchronous | 1.2 (n = 13) | |
| métachronous | 18 (n = 31) | |
The population was divided into patients sampled twice at the same pulmonary site (18 patients: 12 synchronous and six metachronous samples), and patients sampled at two different sites: lung and another organ, two different pulmonary sites, or two organs other than the lung (26 patients: six synchronous and 20 metachronous samples) (Fig. 1). Remote metastases were situated in the brain (n = 8), lung (n = 5), pleura (n = 4), bone (n = 3), lymph node (n = 4), liver (n = 3) or skin (n = 2).
Fig. 1.
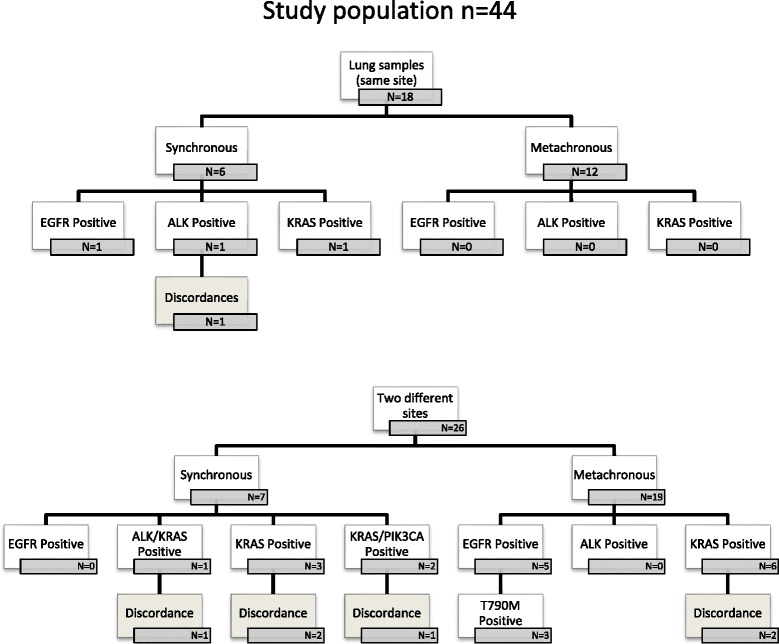
Distribution of multigene mutations and discordances (EGFR, KRAS, BRAF, HER2 and PIK3CA) in 44 French patients with metastatic non-small-cell lung cancer. The study population was composed of 26 patients sampled at different sites and 18 patients sampled twice at the same primary lung cancer location. Synchronous or metachronous status was taken into account in each group
Discordance in mutational status
About half the patients (52 %) harbored at least one mutation. The distribution of the mutations detected in the 44 patients and 88 samples is shown in Fig. 1.
EGFR, KRAS, BRAF and PI3KCA mutations
Five patients had an activating mutation of EGFR (L858R, or exon 19 deletion) in both the primary tumor and the metastasis. No discordances were found in these cases. None of these patients had additional mutations in KRAS, BRAF or PI3KCA. One discordance was not taken into account for analysis, as the patient, a non-smoking female, appeared to have two synchronous tumors; both these primary tumors were tested for EGFR activating mutations. As shown in Fig. 2, the tumor in the right upper lobe harbored an apparent 12 bp deletion corresponding to the p.Glu746_Thr751delinsValAla mutation (COSMIC mutation ID: COSM53205), while the tumor in the left upper lobe harbored a 15 bp deletion resulting in the p.Glu746_Ala750del mutation (COSM6225). These two distinct EGFR mutations were confirmed on repeat biopsy several months later. It is noteworthy that both tumors progressed synchronously under TKI treatment, and were subsequently found to be p.Thr790Met (T790M)-positive (COSM6240).
Fig. 2.
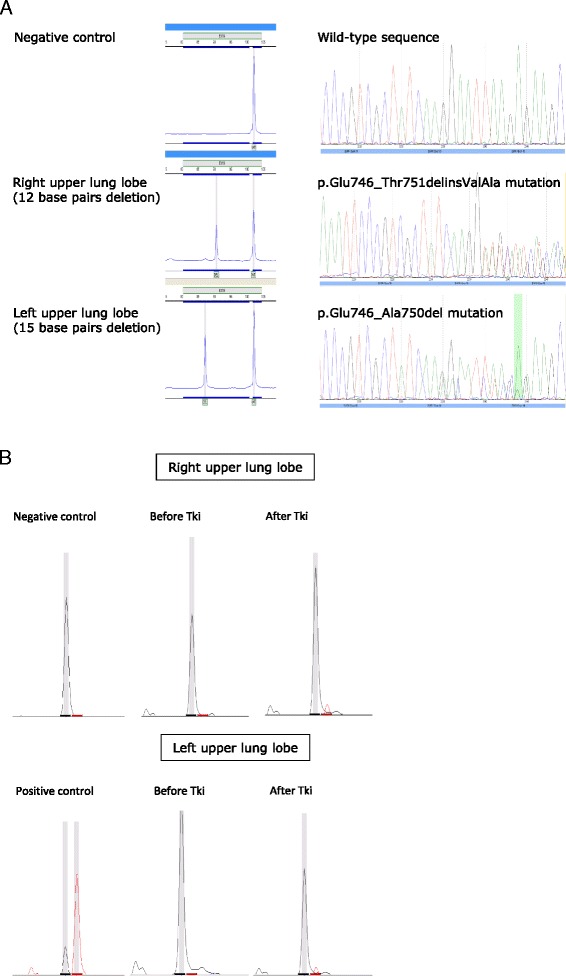
Screening for EGFR exon 19 deletions and the T790M resistance mutation in two independent tumors (right versus left upper lung lobe) diagnosed synchronously. a Fragment-length analysis and Sanger sequencing demonstrating two different EGFR deletions in the two independent tumors. b SNaPshot analysis showing simultaneous emergence of the T790M resistance mutation in the two tumors after completion of EGFR TKI therapy
In all, the p.Thr790Met (T790M) resistance mutation was found in metastases in three patients, but this was not considered as discordance because the initial activating mutation was still present in the metastasis.
Six cases of discordance in KRAS status were found: 3 between synchronous samples and 3 between metachronous samples. Three of the discordances involved KRAS mutation in the metastasis, while mutations were detected only in the primary tumor in another patient. The remaining two patients harbored different mutations in the primary tumor and in the metastasis. Interestingly, the G12C mutation (COSM516) was detected in eight of the 12 patients with KRAS mutation in the primary tumor and/or metastasis. All these patients were current heavy smokers.
Two patients showed PI3KCA mutations: p.His1047Arg (H1047R; COSM775) and p.Glu542Lys (E542K; COSM760). It is noteworthy that PI3KCA E542K mutation was concomitant with KRAS G12C mutation in the metastasis of a man in whom the synchronously sampled primary tumor was mutation-free.
ALK gene rearrangements
ALK status was introduced in routine practice in 2011 after it was recognized as a molecular target of crizotinib in non-small-cell lung cancer [23]. Due to lack of material, we were not able to retrospectively study all the samples obtained between 2005 and 2011 included in the study. In total, ALK status was evaluated in 25 patients using immunochemistry (n = 19), fluorescence in situ hybridization (n = 7) or both (n = 2). Discordance was observed in two former smokers. A female showed ALK rearrangement in the primary tumor (17 % break-apart signals) on FISH, but was negative in the metastatic brain tumor (FISH: 1 %); interestingly, KRAS testing led to the opposite result: detection of the G12C mutation in the metastatic but not in the primary lesion. The second case was a male, positive for ALK FISH in a small biopsy specimen (FISH: 25 %), whereas the ALK alteration was not detected in the resection specimen (FISH: 2 %): i.e., discrepancy between biopsy sample and surgical specimen of a regionally localized stage II lung cancer (Fig. 3).
Fig. 3.
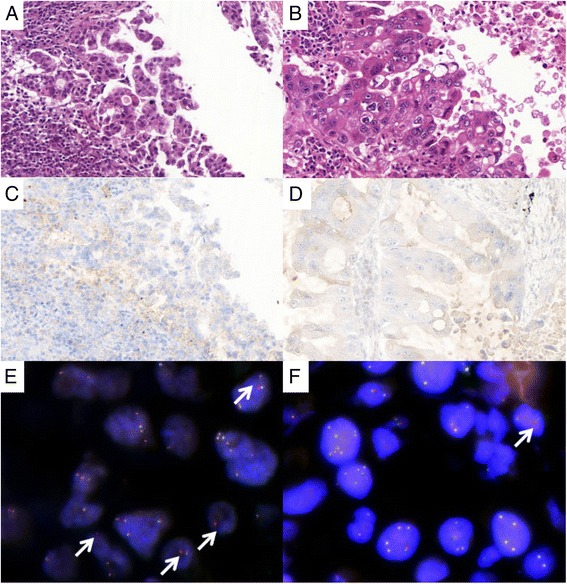
ALK rearrangement status and ALK expression determined by fluorescence in-situ hybridization (FISH) and immunohistochemistry (IHC) in two regions of the same primary lung tumor. a, b examination after hematoxylin-eosin-safran (HES) staining (×400): histological features of adenocarcinoma from the bronchoscopy (a) and the surgical resection (b) specimens. c, d ALK expression: very faint positive immunoreactivity (score, 0/1) in both the small biopsy and the corresponding surgical resection specimen. e, f Typical break-apart pattern observed by FISH (arrow): 25 % of rearranged tumor nuclei detected in the biopsy sample, versus 2 % in the excision specimen
Discordance analysis
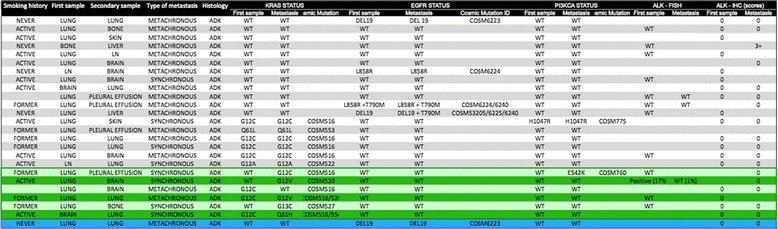
The seven cases of discordance (eight mutational discordances in a total seven patients, one patient showing discordances in both ALK and KRAS mutation status) were analyzed with respect to gender, smoking status, cancer treatment, time between the two samples from a given patient, and type of second sample (repeat primary biopsy versus biopsy of metastasis). Six discordances were found in the 26 patients sampled at two distinct sites Table 2; however, the study lacked power to demonstrate any significant difference in risk compared to the 18 patients sampled twice at the same site (6/26 versus 1/18 cases of discordance; p = 0.24, Fisher’s exact test). The metastases involved in these cases of discordance were located in the lung (1/6), brain (3/6), bone (1/6) or pleura (1/6). There were no other trends.
Table 2.
Distribution of mutations in patients with samples from different sites. Ln: lymph node; WT: wild type,  patients with discordant status
patients with discordant status  one patient with two distinct synchronous lung cancers
one patient with two distinct synchronous lung cancers
Discussion
Despite many recent studies of discordance in mutation status between primary lung tumors and their metastases and the feasibility of tumor rebiopsy [12], metastatic sites are rarely biopsied, as the results would currently have few if any direct therapeutic implications. Discordant responses to chemotherapy or tyrosine kinase inhibitors have also been described between primary tumors and their metastases, suggesting the existence of biological differences [24]. However, targeted therapies are usually administered on the basis of the mutational status of the primary tumor, which is most amenable to biopsy (bronchoscopy).
Major KRAS mutations were frequent in the present cohort (36.4 %), as generally reported in northern and western France [25]. EGFR activating mutations were also frequent (13.6 %): this may be explained by the fact that patients with EGFR activating mutations are more likely to be rebiopsied, as they progress under TKI therapy.
There was only one discordance in EGFR mutation, in a patient with different activating mutations at two distinct pulmonary sites considered as two synchronous primary tumors (Fig. 2). Other activating mutations were present at both sites. The p.Thr790Met (T790M) resistance mutation was found in metastases in three patients, but this was not considered as discordance because the initial activating mutation was still present in the metastasis. Our findings are slightly at odds with those of several other series which reported discordances in EGFR status [26–28]. Recent series also showed a very low rate of discordance in EGFR status, consistent with the present results [29]. The lack of discordance with respect to EGFR activating mutations is consistent with their “driver” status.
The present rate of discordance in KRAS mutational status (13.6 %) is consistent with that observed in several other studies [14,15,24]. We found three types of discordance: i) mutation at the primary but not the metastatic site, ii) mutation at the metastatic site, and iii) one mutation at the primary site and a different mutation in the metastasis. This variability of KRAS mutational status may reflect the fact that it behaves as a “passenger” mutation that, by definition, barely influences tumor outcome. We found a high rate of mutations in codons 12 and 13 of the KRAS gene (37.2 %), particularly the p.Gly12Cys (G12C) mutation. This mutation is usually associated with tobacco smoking [30–32]. Consistently, in the present study, at least 32 of the 44 patients had history of heavy smoking.
In one patient, the sampled metastasis harbored two different mutations: KRAS p.Gly12Cys (G12C) and PI3KCA p.Glu542Lys (E542K). This is in agreement with recent studies [29], confirming the possible coexistence of mutations, a concept that was long contested.
None of the study parameters (gender, smoking, age, anticancer treatment between samples) was significantly associated with overall risk of discordance. Although the difference could not be shown to be statistically significant, it is noteworthy that all six discordances involved patients sampled at two different sites, synchronously or metachronously, while only one involved patients sampled twice at the same site.
The discordances observed in patients sampled at two different sites might result from divergent evolution over time, with a possible influence of microenvironment and/or treatment effects. The advent of NGS (next generation sequencing) will probably extend such studies to larger populations, with dependence on the quality of the tumor tissue sampled. These new techniques are also expected to determine the molecular profile of each tumor site and to determine affiliation between two tumor sites with certainty.
Discordances between ALK status in primary lesions and their corresponding metastases and in multiple primary lesions have been reported in a subset of patients with NSCLC [16,17]. The present study found different types and levels of discordance in two patients unresponsive to crizotinib.
The first patient was a female who had at least a 30 pack-year history of smoking. She underwent surgical resection of brain metastasis (ALK negative) and, thereafter, received crizotinib for 6 months. The best response to ALK TKI administration was stable disease. ALK TKI was switched to radio-chemotherapy, which was well tolerated and led to reduction of the primary lung cancer. Although FISH is widely used as a gold standard method to diagnose ALK-rearranged NSCLC, it is important to remember that testing by FISH does not have 100 % sensitivity and specificity and shows cellular false-negatives and false-positives [33,34]. Here, the patient’s primary tumor tissue sample was borderline positive on FISH; the percentage of ALK rearrangements fell in the 15 to 20 % range. Giving the clinical history of the patient, it may be assumed that FISH led to a false-positive result.
The second patient was a male lifelong heavy smoker (35 packs-years) who developed lung cancer at 68 years of age. He was diagnosed after bronchial biopsy of a pT2N0M0 non-small-cell lung carcinoma. He underwent surgical resection of the lung cancer with standard lymphadenectomy. The disease relapsed 10 months after surgery, with lymph nodes and cerebral metastasis. At the first attempt, the patient received cisplatin-pemetrexed chemotherapy. CT scan showed remarkable lung tumor shrinkage (>50 %) and a stable cerebral lesion. After a few weeks, however, the patient asked to stop maintenance therapy, and the disease progressed rapidly. At this stage, the patient was treated with crizotinib. Despite dose reductions, the patient experienced severe renal failure that forced us to prematurely stop the targeted therapy. After 2 months of daily ALK TKI administration, the tumor manifested no response. This case is different in that the patient displayed discordance in ALK status between two regions of the same tumor: a small biopsy specimen showed an ALK rearrangement that was not detected in the surgical specimen of the corresponding regionally localized lung cancer. This illustrates the clonal evolution of lung tumors and the fact that ALK-positive clones sampled by biopsy may not necessarily be representative of the entire tumor. A situation exactly opposite was very recently reported by Abe and collaborators [18]. While our manuscript was under review, a paper appeared showing an intratumor heterogeneity of ALK rearrangement in a total of 7 NSCLC tumor samples (seven out of ten ALK positive tumors detected in a series of 105 mixed adenocarcinomas and 17 adenosquamous carcinomas). The authors attributed the observed differences in ALK status to the existence of different cell populations within the tumor. In contrast to the apparent stability of EGFR-activating mutations, they have evidenced a relationship between ALK status and certain histologic subtypes. As stated by the authors, this could suggest that rearrangement of the ALK gene is a late event of tumorigenesis [35]. Together, these observations suggest a limitation of single biopsy-based analyses for predictive biomarker tracking and personalized medicine.
Conclusions
In conclusion, this study confirms the substantial rate of discordance in mutational status between primary tumors and their metastases in patients with non-small-cell lung cancer. Discordance mainly concerned KRAS, an oncogene frequently mutated in lung cancer, particularly in smokers. KRAS-positive lung cancer patients are among the most refractory to available treatments, but efforts to develop new therapies for these patients, including anti-MEK drugs, are particularly intensive. The present findings indicate that, with the development of successful targeted therapies, KRAS-positive patients would benefit from genetic testing of different samples. Currently, and by contrast, the stability of EGFR status between primary sites and metastases confirms that there is no reason to systematically rebiopsy all patients, as the results would have no direct therapeutic implications other than in clinical trials. The discordance in ALK rearrangement found between a small biopsy and the corresponding surgical specimen shows that it is important to accumulate information about the biological behavior of infrequent genetic alterations with predictive value. Intra-tumor heterogeneity is another major source of concern in therapeutic resistance.
Acknowledgements
This study was supported by funds from La Ligue Contre le Cancer.
The authors thank Pr. Cédric Le Marechal for critical reading.
Abbreviations
- ALK
anaplastic lymphoma kinase
- BRAF
V-RAF murine sarcoma viral oncogene homolog
- CT scan
computed tomography scan
- EGFR
epidermal growth factor receptor
- FFPE
formaldehyde fixed and paraffin embedded
- FISH
fluorescence in situ hybridization
- HER2
V-ERB-B2 avian erythroblastic leukemia viral oncogene homolog 2
- KRAS
V-KI-RAS2 kirsten rat sarcoma viral oncogene homolog
- NSCLC
non-small-cell lung cancer
- PIK3CA
phosphatidylinositol 3-kinase, catalytic alpha
- TKI
tyrosine kinase inhibitor
Additional file
SNaPshot Multiplex analysis of KRAS codons 12 and 13 (HUGO Gene Nomenclature Committee #6407). (ODP 128 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GQ and GLG conceived the study, and participated in its design and coordination. RD, GR, CF and IDQ participated in the design of the study and interpretation of data. SA, OR and BF carried out the molecular genetics studies. PA and AU performed the FISH analyses. GLG and GQ wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Gilles Quéré, Email: gilles.quere@chu-brest.fr.
Renaud Descourt, Email: renaud.descourt@chu-brest.fr.
Gilles Robinet, Email: gilles.robinet@chu-brest.fr.
Sandrine Autret, Email: sandrine.autret@chu-brest.fr.
Odile Raguenes, Email: odile.raguenes@chu-brest.fr.
Brigitte Fercot, Email: brigitte.fercot@chu-brest.fr.
Pierre Alemany, Email: pierre.alemany@chu-brest.fr.
Arnaud Uguen, Email: arnaud.uguen@chu-brest.fr.
Claude Férec, Email: claude.ferec@univ-brest.fr.
Isabelle Quintin-Roué, Email: isabelle.quintin-roue@chu-brest.fr.
Gérald Le Gac, Phone: (+33) 2 98 44 50 64, Email: gerald.legac@univ-brest.fr.
References
- 1.Scagliotti GV, Parikh P, von Pawel J, Biesma B, Vansteenkiste J, Manegold C, Serwatowski P, Gatzemeier U, Digumarti R, Zukin M, Lee JS, Mellemgaard A, Park K, Patil S, Rolski J, Goksel T, de Marinis F, Simms L, Sugarman KP, Gandara D. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2008;26:3543–51. [DOI] [PubMed]
- 2.Toschi L, Cappuzzo F. Impact of biomarkers on non-small cell lung cancer treatment. Target Oncol. 2010;5:5–17. doi: 10.1007/s11523-010-0132-y. [DOI] [PubMed] [Google Scholar]
- 3.Soria J-C, Mauguen A, Reck M, Sandler AB, Saijo N, Johnson DH, Burcoveanu D, Fukuoka M, Besse B, Pignon J-P. Systematic review and meta-analysis of randomised, phase II/III trials adding bevacizumab to platinum-based chemotherapy as first-line treatment in patients with advanced non-small-cell lung cancer. Ann Oncol Off J Eur Soc Med Oncol ESMO. 2013;24:20–30. [DOI] [PubMed]
- 4.Inoue A, Kobayashi K, Usui K, Maemondo M, Okinaga S, Mikami I, Ando M, Yamazaki K, Saijo Y, Gemma A, Miyazawa H, Tanaka T, Ikebuchi K, Nukiwa T, Morita S, Hagiwara K. First-line gefitinib for patients with advanced non-small-cell lung cancer harboring epidermal growth factor receptor mutations without indication for chemotherapy. J Clin Oncol Off J Am Soc Clin Oncol. 2009;27:1394–400. [DOI] [PubMed]
- 5.Jackman DM, Miller VA, Cioffredi L-A, Yeap BY, Jänne PA, Riely GJ, Ruiz MG, Giaccone G, Sequist LV, Johnson BE. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res Off J Am Assoc Cancer Res. 2009;15:5267–73. [DOI] [PMC free article] [PubMed]
- 6.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM, Porta R, Cobo M, Garrido P, Longo F, Moran T, Insa A, De Marinis F, Corre R, Bover I, Illiano A, Dansin E, de Castro J, Milella M, Reguart N, Altavilla G, Jimenez U, Provencio M, Moreno MA, Terrasa J, Muñoz-Langa J, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–46. [DOI] [PubMed]
- 7.Mok TS, Wu Y-L, Thongprasert S, Yang C-H, Chu D-T, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang J-J, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. [DOI] [PubMed]
- 8.Shaw AT, Kim D-W, Nakagawa K, Seto T, Crinó L, Ahn M-J, De Pas T, Besse B, Solomon BJ, Blackhall F, Wu Y-L, Thomas M, O’Byrne KJ, Moro-Sibilot D, Camidge DR, Mok T, Hirsh V, Riely GJ, Iyer S, Tassell V, Polli A, Wilner KD, Jänne PA. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–94. [DOI] [PubMed]
- 9.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 10.Hammerman PS, Jänne PA, Johnson BE. Resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2009;15:7502–9. doi: 10.1158/1078-0432.CCR-09-0189. [DOI] [PubMed] [Google Scholar]
- 11.Thunnissen FB, Prinsen C, Hol B, Van der Drift M, Vesin A, Brambilla C, Montuenga L, Field JK. Smoking history and lung carcinoma: KRAS mutation is an early hit in lung adenocarcinoma development. Lung Cancer Amst Neth. 2012;75:156–60. [DOI] [PubMed]
- 12.Chouaid C, Dujon C, Do P, Monnet I, Madroszyk A, Le Caer H, Auliac JB, Berard H, Thomas P, Lena H, Robinet G, Baize N, Bizieux-Thaminy A, Fraboulet G, Locher C, Le Treut J, Hominal S, Vergnenegre A. Feasibility and clinical impact of re-biopsy in advanced non small-cell lung cancer: A prospective multicenter study in a real-world setting (GFPC study 12-01). Lung Cancer Amst Neth. 2014;86:170. [DOI] [PubMed]
- 13.Gow C-H, Chang Y-L, Hsu Y-C, Tsai M-F, Wu C-T, Yu C-J, Yang C-H, Lee Y-C, Yang P-C, Shih J-Y. Comparison of epidermal growth factor receptor mutations between primary and corresponding metastatic tumors in tyrosine kinase inhibitor-naive non-small-cell lung cancer. Ann Oncol Off J Eur Soc Med Oncol ESMO. 2009;20:696–702. [DOI] [PubMed]
- 14.Kalikaki A, Koutsopoulos A, Trypaki M, Souglakos J, Stathopoulos E, Georgoulias V, Mavroudis D, Voutsina A. Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Br J Cancer. 2008;99:923–9. [DOI] [PMC free article] [PubMed]
- 15.Monaco SE, Nikiforova MN, Cieply K, Teot LA, Khalbuss WE, Dacic S. A comparison of EGFR and KRAS status in primary lung carcinoma and matched metastases. Hum Pathol. 2010;41:94–102. doi: 10.1016/j.humpath.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 16.Kim H, Xu X, Yoo S-B, Sun P-L, Jin Y, Paik JH, Choe G, Jheon S, Lee C-T, Chung J-H. Discordance between anaplastic lymphoma kinase status in primary non-small-cell lung cancers and their corresponding metastases. Histopathology. 2013;62:305–14. [DOI] [PubMed]
- 17.Wu C, Zhao C, Yang Y, He Y, Hou L, Li X, Gao G, Shi J, Ren S, Chu H, Zhou C, Zhang J. High discrepancy of driver mutations in patients with NSCLC and synchronous multiple lung ground-glass nodules. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2015;10:778–83. [DOI] [PubMed]
- 18.Abe H, Kawahara A, Azuma K, Taira T, Takase Y, Fukumitsu C, Murata K, Yamaguchi T, Akiba J, Ishii H, Okamoto I, Hoshino T, Takamori S, Kage M. Heterogeneity of anaplastic lymphoma kinase gene rearrangement in non-small-cell lung carcinomas: a comparative study between small biopsy and excision samples. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2015;10:800–5. [DOI] [PubMed]
- 19.Coate LE, John T, Tsao M-S, Shepherd FA. Molecular predictive and prognostic markers in non-small-cell lung cancer. Lancet Oncol. 2009;10:1001–10. doi: 10.1016/S1470-2045(09)70155-X. [DOI] [PubMed] [Google Scholar]
- 20.Nowak F, Soria J-C, Calvo F. Tumour molecular profiling for deciding therapy-the French initiative. Nat Rev Clin Oncol. 2012;9:479–86. doi: 10.1038/nrclinonc.2012.42. [DOI] [PubMed] [Google Scholar]
- 21.Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol. 2000;18:233–4. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- 22.Locher C, Debieuvre D, Coëtmeur D, Goupil F, Molinier O, Collon T, Dayen C, Le Treut J, Asselain B, Martin F, Blanchon F, Grivaux M. Major changes in lung cancer over the last ten years in France: the KBP-CPHG studies. Lung Cancer Amst Neth. 2013;81:32–8. [DOI] [PubMed]
- 23.Kwak EL, Bang Y-J, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou S-HI, Dezube BJ, Jänne PA, Costa DB, Varella-Garcia M, Kim W-H, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vignot S, Soria J-C. Discrepancies between primary tumor and metastasis: impact on personalized medicine. Bull Cancer (Paris) 2013;100:561–8. doi: 10.1684/bdc.2013.1767. [DOI] [PubMed] [Google Scholar]
- 25.Barlesi F, Blons H, Beau-Faller M, Rouquette I, Ouafik L, Mosser J, Merlio JP, Bringuier PP, Jonveaux P, Le Marechal C, Denis M, Penault-Llorca F, Debieuvre D, Soria JC, Cadranel J, Mazieres J, Missy P, Morin F, Nowak F, Zalcman G. Biomarqueurs France : résultats de l’analyse en routine de EGFR, HER2, KRAS, BRAF, PI3K, et ALK sur 10 000 patients (pts) atteints de cancer bronchique non à petites cellules (CBNPC). Rev Mal Respir. 2014;31:A13–4.
- 26.Italiano A, Vandenbos FB, Otto J, Mouroux J, Fontaine D, Marcy P-Y, Cardot N, Thyss A, Pedeutour F. Comparison of the epidermal growth factor receptor gene and protein in primary non-small-cell-lung cancer and metastatic sites: implications for treatment with EGFR-inhibitors. Ann Oncol Off J Eur Soc Med Oncol ESMO. 2006;17:981–5. [DOI] [PubMed]
- 27.Bozzetti C, Tiseo M, Lagrasta C, Nizzoli R, Guazzi A, Leonardi F, Gasparro D, Spiritelli E, Rusca M, Carbognani P, Majori M, Franciosi V, Rindi G, Ardizzoni A. Comparison between epidermal growth factor receptor (EGFR) gene expression in primary non-small cell lung cancer (NSCLC) and in fine-needle aspirates from distant metastatic sites. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2008;3:18–22. [DOI] [PubMed]
- 28.Han C, Ma J, Zhao J, Zhou Y, Jing W, Zou H. EGFR mutations, gene amplification, and protein expression and KRAS mutations in primary and metastatic tumors of nonsmall cell lung cancers and their clinical implications: a meta-analysis. Cancer Invest. 2011;29:626–34. doi: 10.3109/07357907.2011.621914. [DOI] [PubMed] [Google Scholar]
- 29.Vignot S, Frampton GM, Soria J-C, Yelensky R, Commo F, Brambilla C, Palmer G, Moro-Sibilot D, Ross JS, Cronin MT, André F, Stephens PJ, Lazar V, Miller VA, Brambilla El. Next-generation sequencing reveals high concordance of recurrent somatic alterations between primary tumor and metastases from patients with non-small-cell lung cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2013;31:2167–72. [DOI] [PubMed]
- 30.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–50. [DOI] [PMC free article] [PubMed]
- 31.Garassino MC, Marabese M, Rusconi P, Rulli E, Martelli O, Farina G, Scanni A, Broggini M. Different types of K-Ras mutations could affect drug sensitivity and tumour behaviour in non-small-cell lung cancer. Ann Oncol Off J Eur Soc Med Oncol ESMO. 2011;22:235–7. doi: 10.1093/annonc/mdq680. [DOI] [PubMed] [Google Scholar]
- 32.Fritz JM, Dwyer-Nield LD, Russell BM, Malkinson AM. The Kras mutational spectra of chemically induced lung tumors in different inbred mice mimics the spectra of KRAS mutations in adenocarcinomas in smokers versus nonsmokers. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2010;5:254–7. [DOI] [PMC free article] [PubMed]
- 33.Camidge DR, Skokan M, Kiatsimkul P, Helfrich B, Lu X, Barón AE, Schulte N, Maxson D, Aisner DL, Franklin WA, Doebele RC, Varella-Garcia M. Native and rearranged ALK copy number and rearranged cell count in non-small cell lung cancer: implications for ALK inhibitor therapy. Cancer. 2013;119:3968–75. [DOI] [PMC free article] [PubMed]
- 34.Hutarew G, Hauser-Kronberger C, Strasser F, Llenos IC, Dietze O. Immunohistochemistry as a screening tool for ALK rearrangement in NSCLC: evaluation of five different ALK antibody clones and ALK FISH. Histopathology. 2014;65:398–407. doi: 10.1111/his.12399. [DOI] [PubMed] [Google Scholar]
- 35.Zito Marino F, Liguori G, Aquino G, La Mantia E, Bosari S, Ferrero S, Rosso L, Gaudioso G, De Rosa N, Scrima M, Martucci N, La Rocca A, Normanno N, Morabito A, Rocco G, Botti G, Franco R. Intratumor heterogeneity of ALK-rearrangements and homogeneity of EGFR-mutations in mixed lung adenocarcinoma. Plos One. 2015;10:e014152. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A dataset (mutations detection) supporting the conclusions of this article is available in Additional file 1: Figure S1.


