Abstract
Purpose
Define criteria for selection of optimal flip angle sets for T1 estimation and evaluate effects on T1 mapping.
Theory and Methods
Flip angle sets for SPGR-based T1 mapping were selected by minimizing T1 estimate variance weighted by the joint density of M0 and T1 in an initial acquisition. The effect of optimized flip angle selection on T1 estimate error was measured using simulations and experimental data in the human and rat brain.
Results
For 2-point acquisitions, optimized angle sets were similar to those proposed by other groups, and therefore performed similarly. For multi-point acquisitions, optimal angle sets for T1 mapping in the brain consisted of a repetition of two angles. Implementation of optimal angles reduced T1 estimate variance by 30–40% compared to a multi-point acquisition using a range of angles. Performance of the optimal angle set was equivalent to that of a repetition of the two-angle set selected using criteria proposed by other researchers.
Conclusion
Repetition of two carefully selected flip angles notably improves the precision of resulting T1 estimates compared with acquisitions using a range of flip angles. This work provides a flexible and widely applicable optimization method of particular use for those who repeatedly perform T1 estimation.
Keywords: T1 mapping, flip angle selection, error propagation, SPGR
Introduction
T1 estimation is a quantitative MRI tool with a variety of applications, including monitoring myelin-related disease, diagnosing Parkinson’s disease, and studying tissue perfusion (1–3). The gold standard for T1 estimation is an inversion recovery sequence with multiple inversion times (4). However, inversion recovery scans require repetition times (TRs) much longer than the longest T1 of interest, rendering them impractical for use in clinical scans or in vivo research settings. For this reason, a multiple flip angle spoiled gradient echo (SPGR) approach is commonly employed to reduce scan time for T1 mapping (5). More recently, several researchers have proposed supplemental methods to correct for common sources of error in resulting SPGR-based T1 estimates, such as incomplete spoiling and imperfect RF pulses (6–8).
For SPGR-based T1 mapping, a minimum number of flip angles must be carefully selected to maximize the accuracy and precision of the T1 estimate and minimize scan time. Several works addressing angle selection criteria settle on the same optimal flip angles in a 2-point acquisition despite differences in selection approach. These approaches include variance minimization (9), definition of an efficiency function related to both the T1 estimate variance and the scan time (10), and maximizing the product of the regression line dynamic range and the fractional signal with respect to the Ernst angle (11). Another approach uses criteria of T1-to-noise ratio maximization to select larger flip angle sets (12). However, these approaches address T1 estimation in the simplified setting of a single or average T1 value. Tissues and pathologies of interest contain a continuous distribution of T1 values, which can complicate the task of selecting optimal flip angles. Furthermore, most of these approaches address the optimized selection of just two flip angles and therefore do not develop approaches that can be generalized to the selection of larger angle sets.
The selection of flip angles for more realistic imaging tasks has also been addressed in several studies. Cheng and Wright propose selection of 3-point angle sets based on the minimum and maximum of the range of T1 values to be estimated (13). T1 variance minimization has also been used to select 2-point sets for several different ranges of T1 values to be estimated (14). In another study, researchers selected 3-point angle sets to maximize T1 mapping efficiency over a realistic range of T1 values (15). Although useful, these approaches lack flexibility to select any number of flip angles, and they do not take into account the full shape and range of the T1 and M0 distributions of the anatomy being imaged.
In this study, we aim to address these missing components of flip angle selection schemes with the goal of providing a robust flip angle selection method that can be applied in almost any setting. To do so, we develop a data-driven approach based on T1 variance minimization to determine the optimal set of any number of flip angles for T1 estimation. The T1 distribution in the imaging subject of interest is taken into account using an initial acquisition. We describe and apply a novel set of flip angle selection criteria, based on minimization of the variance in the T1 estimates weighted by the joint density of M0 and T1. Using experimental data and simulations, we evaluate the effects of this optimized acquisition design on the precision and accuracy of T1 estimates. The concepts behind the proposed method were previously described in abstract form (16).
Theory
In the general setting of nonlinear least squares-based (NLS) parameter estimation, as is used for T1 and M0 estimation in this work, the objective function can be represented by
| [1] |
where γ is the parameter vector to be estimated and ri(γ) are the error terms or the residual terms if ri are evaluated at the NLS estimate. Defining r = [r1 r2 … rn]T, the gradient vector and the Hessian matrix of fNLS(γ) can be expressed respectively as
| [2] |
and
| [3] |
where [Ti]kl = ∂2ri/∂γk∂γl is the second order derivative of the residual term, and the Jacobian matrix is defined as [Jγ(r)]ij ≡ ∂ri/∂γj. According to the framework of error propagation (17), the covariance matrix of the estimated parameter can be written as
| [4] |
where σ2 is the unknown noise variance. However, in a computational experimental design setting, the covariance matrix takes a much simpler form:
| [5] |
because the residual terms are assumed to be zero on the average. This assumption is based on that of Gaussian-distributed noise contributing to residuals that average to zero and are independent of the parameters of interest.
In the setting of T1 estimation, the appropriate term of this covariance matrix can be used to define an objective function for minimization of variance in the NLS-based T1 estimate. The NLS objective function for T1 estimation from variable flip angle SPGR acquisitions can be written as
| [6] |
where si are the observed signals, αi are the flip angles, M0 is the unknown equilibrium longitudinal magnetization, TR is the repetition time, and T1 is the unknown longitudinal relaxation time. Therefore, γ=[M0 T1]T. We note here that repetition time is held constant and held close to the minimum allowable by the MR system, echo time (TE) is similarly held close to the practical minimum, and therefore differences in T2* decay are assumed negligible for this specific setting.
Taking the derivative of the error with respect to both M0 and T1, we arrive at the expressions for the terms in the Jacobian matrix:
| [7] |
and
| [8] |
With some algebraic manipulation, as shown in Appendix A, the determinant of JγT(r)Jγ(r) and the components of the covariance matrix Σγ can be expressed as
| [9] |
| [10] |
| [11] |
| [12] |
where and E = exp(TR/T1). Note that [Σγ]11, [Σγ]22, and [Σγ]21 are the variance of M0, the variance of T1, and the covariance of M0 and T1, respectively. A similar derivation for the T1 estimate variance, along with the analytical result for the two-angle T1 estimate, has previously been published by Wood (18).
Methods
Study design
In order to test the described T1 variance minimization criteria for SPGR flip angle selection, we pursued following approach. First, an objective function for flip angle selection was defined based on minimizing T1 variance in the imaging volume of interest. Second, optimal flip angle sets and control flip angle sets were defined based on T1 and M0 maps calculated from an open-source IR data set in the human brain. Using these T1 and M0 maps as ground truth, simulations were performed to compare the relative performance of several flip angle sets for SPGR-based T1 estimation. Third, the proposed flip angle selection technique was tested in the ex vivo rat brain both with simulations and experimentally. Using an initial IR acquisition and resulting T1 and M0 maps, optimal flip angle sets and alternate flip angle sets were defined. To compare the performance of these flip angle sets, simulations were performed using IR-based T1 and M0 maps as ground truth. Last, experimental data was again acquired in the ex vivo rat brain using data-driven flip angle sets in order to compare the inter-scan variance when different angle sets were used.
Flip angle selection
In an NLS fitting problem, the covariance matrix of the parameters for estimation (in this case, M0 and T1) is given by the inverse of the Hessian matrix multiplied by the unknown noise variance, as described above. Therefore, assuming a constant noise variance σ2 in the voxels of interest (i.e., those representing the tissue in which T1 will be estimated), a single term from the inverse of the Hessian can be used to estimate the variance in the T1 estimate at each voxel, given by .
The selection of the two optimal flip angles for T1 estimation was guided by the goal to minimize the variance in the T1 estimate. Therefore, an objective function for flip angle selection, Ω, was defined according to the equation
| [13] |
where P(M0, T1) is the smoothed joint probability density estimate of M0 and T1 over the imaging volume or region of interest. To select optimal flip angles for any imaging setting, this objective function was minimized using the user inputs of TR, number of desired flip angles (Nα), and P(M0, T1). While TR and Nα can be selected based on the constraints of the imaging setting and time available, P(M0, T1) is more complicated to provide. In this work, we pursue a data-driven approach in which P(M0, T1) is estimated from an initial set of T1 and M0 maps based on a non-optimized SPGR acquisition. For all optimized flip angle sets in this work, P(M0, T1) was specified using the subset of image voxels in the brain tissue.
T1 estimation
In both simulations and analysis of experimental data, an estimate for T1 and M0 in each voxel was obtained using the Levenberg-Marquardt approach for non-linear least squares fitting (19) with the goal of minimizing the objective function in Eq. 6. It should be noted that the above nonlinear least squares fitting problem can also be reformulated as a simple iterative linear fitting problem (20). For experimental data, a calculated B1 map was taken into account in the parameter estimation procedure (6). This correction was used due to the variability of the relationship between nominal and actual flip angle and the potentially significant effect thereof on T1 estimation. Iterations were continued until the step size between successive estimates was extremely small (<10−15), the objective function was appropriately small (<10−6), or after 500 iterations. All image processing, simulations, and computational work was performed in MATLAB R2014b (The MathWorks, Natick, MA).
Simulations
Simulations were performed to evaluate the precision and accuracy of T1 mapping with the flip angles selected as described above. They were also used to compare these results with those obtained using previously proposed flip angle sets and selection techniques (11, 12). Using inversion recovery data from the Quantitative MRI Analysis Package (QMAP, http://www.medphysics.wisc.edu/~samsonov/qmap/), FSL’s Brain Extraction Tool (21), and FMRIB’s Linear Image Registration Tool (22, 23), T1 and M0 maps in the human brain were calculated and used as ground truth. From these maps, optimal flip angles were determined as described above. For comparison of the 2-point angle set, flip angle sets were selected using Deoni and colleagues’ 2-point selection criteria based on the mean T1 in the segmented human brain (11). For comparison of the 10-point angle set, two other flip angle sets were selected. One of these sets was based on repetition of the 2-point set described above (referred to as “10-point repeat”). The other set of angles, referred to as “10-point range,” is based on a previously proposed optimal 10-angle set for T1 mapping in the human brain that covers a range of angles, which is a common approach to T1 mapping with VFA SPGR (12, 24, 25). The “10-point range” set was selected in order to determine the impact of using a repetition of two carefully selected flip angles compared to a range of flip angles. The flip angle sets used in simulations are shown in Table 1.
Table 1.
Flip angle sets used in simulations and experiments of SPGR-based T1 estimation
| Optimal angle set | Human Simulation | Rat Simulation | Rat Experimental |
|---|---|---|---|
| Nα = 2 | 4.7°, 27.2° | 3.6°, 21.7° | Not tested |
| Nα = 10 | 5×4.7°, 5×27.2° | 5×3.6°, 5×21.7° | 5×3.2°, 5×18.6° |
| Control angle set | |||
| Nα = 2 | 5.1°, 30.0° | 4.8°, 27.1° | Not tested |
| Nα = 10 repeat | 5×5.1°, 5×30.0° | 5×4.8°, 5×27.1° | 5×3.5°, 5×19.8° |
| Nα = 10 range | 2°, 3°, 4°, 5°, 7°, 9°, 12°, 14°, 16°, 18° | 2°, 3°, 4°, 5°, 7°, 9°, 12°, 14°, 16°, 18° | 2°, 3°, 4°, 5°, 7°, 9°, 12°, 14°, 16°, 18° |
In simulations of SPGR acquisition, the signal in each voxel was calculated according to the SPGR signal equation included in Eq. 6. Noisy MR signals were modeled to follow a Rician distribution. Noise was added according to the model , where S is the signal magnitude and ε1 and ε2 are random values from a normal distribution of mean zero and standard deviation dependent upon S and user-defined signal-to-noise ratio (SNR). Noisy signals were calculated in 500 realizations, and T1 and M0 were estimated as described above. Results were used to calculate the bias and variance of the SPGR-based T1 estimates. The relative performance of optimal flip angles compared to other flip angle sets was measuring at 6 SNR values ranging from 5 to 40. Simulations were similarly performed using IR-based T1 and M0 maps in the ex vivo rat brain as the ground truth, using the flip angle sets shown in Table 1.
MR imaging
Imaging experiments were performed on a 4.7T small animal scanner (MRBR 4.7T/310, Agilent Technologies, Santa Clara, CA) with a quadrature volume RF coil for signal transmission and reception. An ex vivo rat brain was scanned because it has a continuous distribution of T1 and M0 values, as would be expected in a human subject. A 3D SPGR pulse sequence was performed using three individual flip angle sets, shown in Table 1. Each flip angle set was repeated three times to measure experimental variance in T1 estimates. To minimize scan time and maximize the MR signal collected, the TR and TE were minimized and had values of 9.08 ms and 3.81 ms, respectively. Other scan parameters were as follows: matrix size = 128 × 128 × 128, slab size = 30 mm × 20 mm × 20 mm, total scan time = 2 min 31 s per FA. (For clarity, in experiments with more than one acquisition at each FA, each acquisition is referred to as an individual FA rather than an additional average.) A flip angle map was acquired using an actual flip-angle imaging SPGR scan with scan parameters as follows: TR1/TR2 = 5.9/29.5 ms, TE = 2.22 ms, α = 55°, matrix size = 64 × 64 × 64 (6).
To calculate a T1 map in the ex vivo rat brain for simulations, an IR acquisition was used. A set of 2D spin-echo scans was acquired at a range of 8 inversion times between 8 and 4000 ms. Other scan parameters were held constant and were as follows: TR = 6 s, TE = 13.82 ms, NEX = 4, matrix size = 128 × 128, in-plane FOV = 20 × 20 mm, with 50 slices of 0.5 mm thickness.
Results
Selection of optimal flip angles
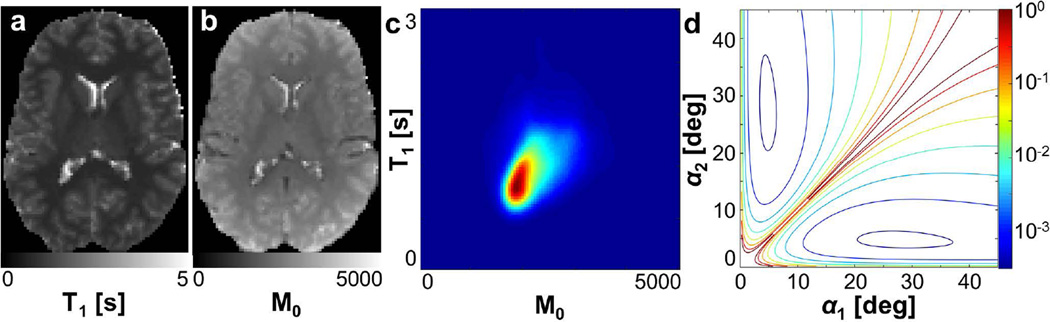
It was found that the optimal flip angles for minimizing T1 variance were, as expected, dependent upon the joint T1 and M0 distribution in the region of interest. Using the proposed flip angle selection approach, any number of optimal flip angles specific to reducing T1 variance in the ROI used to define P(M0, T1) could be determined. A representative set of parameter maps used for input to the flip angle selection algorithm is shown in Figure 1a–b. These maps were estimated from an open-source inversion recovery data set (QMAP). The resulting smoothed joint density function of T1 and M0 in the human brain is shown in Figure 1c. Finally, the objective function for selection of two flip angles is plotted in Figure 1d, and agrees well with the expected shape shown in previous works addressing selection of 2-angle sets (9–11, 14). In the limit of a single T1 value in the region used to define P(M0, T1), the proposed flip angle selection approach resulted in the same two recommended angles as the approach proposed by Deoni and colleagues (11).
FIG. 1.
Representative input to flip angle selection algorithm. Initial estimates of (a) T1 map and (b) M0 map in the human brain based on an inversion recovery data set. (c) Joint density distribution of T1 and M0 based on the maps in (a) and (b). (d) Flip angle minimization objective function, shown here for 2 flip angle selection for the purposes of visualization.
For even-numbered flip angle sets greater than two, it was found that the optimal flip angle sets were repetitions of the pair of two optimal flip angles. For odd-numbered flip angle sets greater than two, it was found that the optimal flip angle sets were repetitions of two angles. Interestingly, the specific angles varied based on the number of flip angles. Table 2 shows the specific flip angle sets selected for different set sizes using the human brain to define the smoothed joint density of T1 and M0 as algorithm input. It is important to note that, particularly for selection of an odd number of flip angles, local minima were present in the objective function landscape. In these cases, optimal angles were selected based on inputting several different initial points for the minimum search and selecting optimal angles that corresponded to the minimum of the objective function that was found. For odd-numbered angle sets, two local minima with the same objective function values were found (see Table 2). Generally, the angle selection algorithm selected erroneous local minima in the case of an unreasonable search input.
Table 2.
Representative optimal flip angle sets with varying flip angle set sizes. From this list, even-numbered sets were used in human simulations. Odd-numbered sets are included to illustrate typical results of the flip angle selection algorithm.
| Flip angle set size |
α1 | α2 | α3 | α4 | α5 | α6 | α7 | α8 | α9 | α10 |
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 4.7° | 27.2° | ||||||||
| 3 | 4.9° | 28.9° | 28.9° | |||||||
| 3 | 4.0° | 4.0° | 24.1° | |||||||
| 4 | 4.7° | 4.7° | 27.2° | 27.2° | ||||||
| 5 | 4.7° | 4.7° | 27.7° | 27.7° | 27.7° | |||||
| 5 | 4.2° | 4.2° | 4.2° | 24.9° | 24.9° | |||||
| 6 | 4.7° | 4.7° | 4.7° | 27.2° | 27.2° | 27.2° | ||||
| 8 | 4.7° | 4.7° | 4.7° | 4.7° | 27.2° | 27.2° | 27.2° | 27.2° | ||
| 10 | 4.7° | 4.7° | 4.7° | 4.7° | 4.7° | 27.2° | 27.2° | 27.2° | 27.2° | 27.2° |
Simulations
To test the effect of using the proposed angle sets on the variance and bias in T1 estimates, simulations were performed using IR-based T1 and M0 maps of a human brain and an ex vivo rat brain as digital phantoms. These parameter maps were also used as input to the angle selection algorithm for definition of P(M0, T1). Simulation results were used to calculate the bias, variance, and root mean squared error (RMSE) of the SPGR-based T1 estimate.
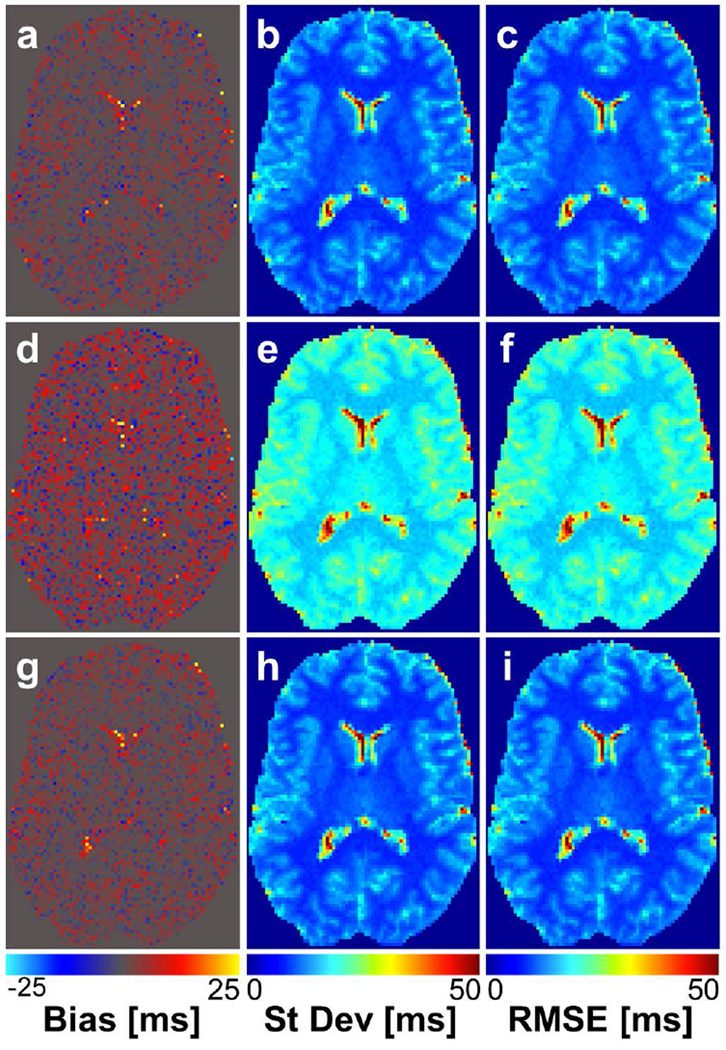
In human simulations using 2-angle estimates, it was found that optimized angles had very slightly improved performance compared to Deoni angles and greatly improved performance compared to arbitrarily selected angles. The improvement in performance at all SNR values, with the exception of SNR = 30, was measured by a RMSE reduction of 0.40 – 3.10%. The percent reduction in RMSE was greater at lower SNR values, indicating that the proposed method resulted in improved performance particularly for low SNR settings. In simulations using 10-angle acquisitions for T1 estimation, it was found that optimized angles had improved performance compared to the 10-point range set (Fig. 2). This improvement was measured by a reduction in T1 estimate RMSE of 42% at all SNR values. However, optimal angles had nearly identical performance to the 10-point repeat set, again with relative performance independent of SNR. The RMSE in human brain T1 estimates calculated for all SNR values and angle sets are shown in Table 3.
FIG. 2.
T1 bias (a, d, g), standard deviation (b, e, h), and RMSE (c, f, i) maps based on simulations of T1 mapping in the human brain using 10-angle acquisitions at SNR = 20. a–c correspond to simulation results using the 10 angles selected with our proposed method; d–f correspond to results using the 10-point range angle set; g–i correspond to results using a the 10-point repeat set. All measurements are in units of ms.
Table 3.
Results of simulations in the human brain. The RMSE in T1 estimates (units of s) are given for each angle set and SNR. RMSE % change is in comparison to optimal angle set for that same number of flip angles.
| Nα = 2 | Nα = 10 | |||||||
|---|---|---|---|---|---|---|---|---|
| SNR | Optimal angle set |
Control angle set | Optimal angle set |
10-point repeat | 10-point range | |||
| RMSE | RMSE | % change |
RMSE | RMSE | % change |
RMSE | % change |
|
| 5 | 5.94E-1 | 6.13E-1 | +3.20 | 2.15E-1 | 2.17E-1 | +0.93 | 3.74E-1 | +74.0 |
| 10 | 2.50E-1 | 2.51E-1 | +0.40 | 1.07E-1 | 1.07E-1 | -- | 1.87E-1 | +74.8 |
| 15 | 1.62E-1 | 1.63E-1 | +0.62 | 7.13E-2 | 7.13E-2 | -- | 1.24E-1 | +73.9 |
| 20 | 1.21E-1 | 1.21E-1 | -- | 5.36E-2 | 5.36E-2 | -- | 9.33E-2 | +74.1 |
| 30 | 8.02E-2 | 8.00E-2 | −0.25 | 3.57E-2 | 3.57E-2 | -- | 6.21E-2 | +73.9 |
| 40 | 6.00E-2 | 6.00E-2 | -- | 2.68E-2 | 2.68E-2 | -- | 4.66E-2 | +73.9 |
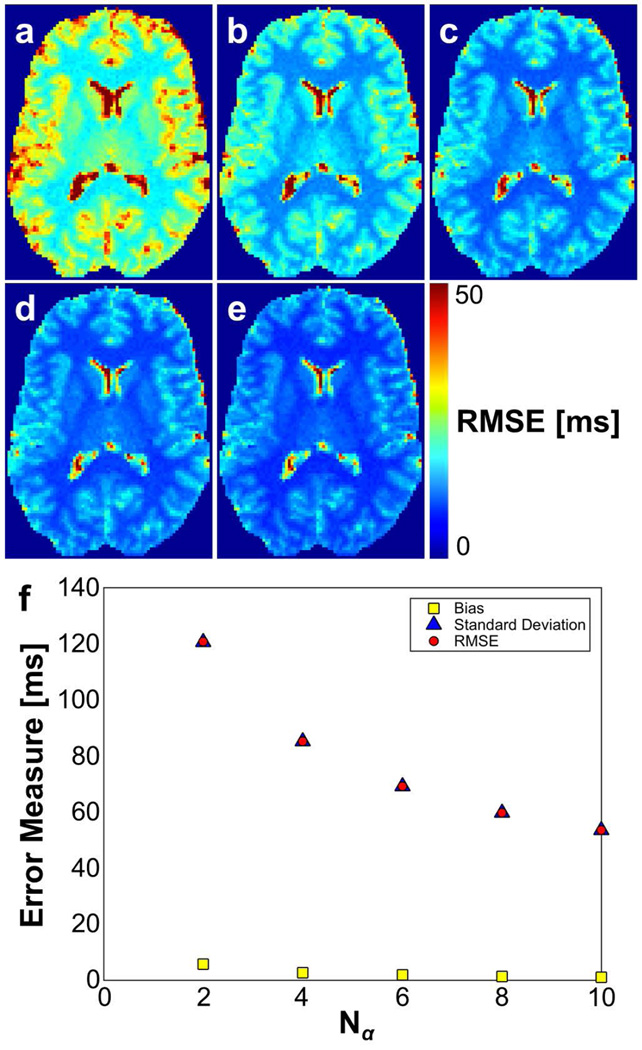
Simulations were also used to compare the relative performance of 2-, 4-, 6-, 8-, and 10-point optimal angle acquisitions for T1 estimation (angle sets listed in Table 2). As shown in Figure 3, increasing the number of flip angles used for acquisition reduces the RMSE in the T1 estimate primarily by reducing variance. It should be noted that this is equivalent to increasing the number of averages at 2 optimal flip angles. Variance was the primary contributor to RMSE, as evidenced by estimate standard deviation ranging from 20–50 times greater than estimate bias at the range of SNR used in these simulations (Table 4).
FIG. 3.
(a–e) RMSE in T1 estimates from simulations of 2, 4, 6, 8, and 10-angle SPGR acquisitions, respectively. (f) Bias, standard deviation, and RMSE in T1 estimates plotted versus number of acquisition flip angles. The estimate variance is the primary contributor to error in the T1 estimate. All measurements are in units of ms.
Table 4.
Effect of number of flip angles on bias, standard deviation, and RMSE of resulting T1 estimates (all measures in units of s). Results are based on simulations in the human brain with an SNR of 20 and using flip angle sets listed in Table 2. Percent change in RMSE is compared to that of the Nα = 2 estimate.
| Nα = 2 | Nα = 4 | Nα = 6 | Nα = 8 | Nα = 10 | |
|---|---|---|---|---|---|
| Bias | 5.77E-3 | 2.75E-3 | 1.92E-3 | 1.39E-3 | 1.13E-3 |
| Standard Deviation | 1.21E-1 | 8.52E-2 | 6.92E-2 | 5.98E-2 | 5.36E-2 |
| RMSE | 1.21E-1 | 8.52E-2 | 6.92E-2 | 5.98E-2 | 5.36E-2 |
| RMSE % change | −29.4 | −42.7 | −50.5 | −55.6 |
In 2-point ex vivo rat brain simulations, it was found that optimized angles only slightly improved performance (RMSE reduction of 2.37%) at the lowest SNR value. At higher SNR values, Deoni angles out-performed optimized angles, with an RMSE reduction of 2.22 – 3.05%. These results confirm the finding in human simulations that our angle selection method performs most strongly in low-SNR scenarios. For 10-angle estimates, it was found that optimized angles had improved performance compared to the 10-point range angle set, reflected in an RMSE reduction of approximately 38% at all SNR values. Compared to the 10-point repeat angle set, optimized angles had slightly worse performance, independent of SNR. Simulation results in the ex vivo rat brain are shown in Table 5.
Table 5.
Results of simulations in the rat brain. The RMSE in T1 estimates (units of s) are given for each angle set and SNR. RMSE % change is in comparison to optimal angle set for that same number of flip angles.
| Nα = 2 | Nα = 10 | |||||||
|---|---|---|---|---|---|---|---|---|
| SNR | Optimal angle set |
Control angle set | Optimal angle set |
10-point repeat | 10-point range | |||
| RMSE | RMSE | % change |
RMSE | RMSE | % change |
RMSE | % change |
|
| 5 | 1.93E-1 | 1.98E-1 | +2.59 | 7.31E-2 | 7.16E-2 | −2.05 | 1.20E-1 | +64.2 |
| 10 | 8.47E-2 | 8.29E-2 | −2.13 | 3.66E-2 | 3.56E-2 | −2.73 | 5.94E-2 | +62.3 |
| 15 | 5.54E-2 | 5.40E-2 | −2.53 | 2.44E-2 | 2.37E-2 | −2.87 | 3.96E-2 | +62.3 |
| 20 | 4.13E-2 | 4.01E-2 | −2.91 | 1.83E-2 | 1.77E-2 | −3.28 | 2.97E-2 | +62.3 |
| 30 | 2.74E-2 | 2.66E-2 | −2.92 | 1.22E-2 | 1.18E-2 | −3.28 | 1.99E-2 | +63.1 |
| 40 | 2.05E-2 | 1.99E-2 | −2.93 | 9.14E-3 | 8.87E-3 | −2.95 | 1.49E-2 | +63.0 |
Experimental T1 mapping
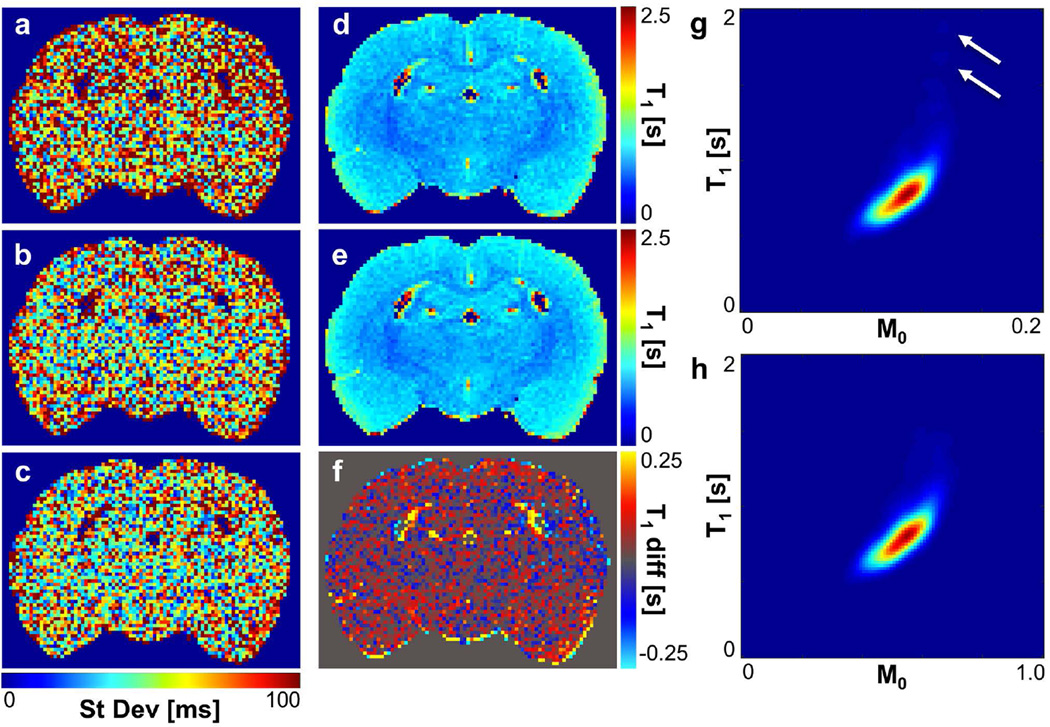
To test the impact of implementing optimal angles in an experimental scanning scenario, an ex vivo rat brain was scanned first using the 10-point range angle set. The resulting estimated T1 and M0 maps were used to define P(M0, T1) in the rat brain, and from this, optimal angle sets and control angle sets were selected (Table 1). The rat brain was then scanned again using these data-driven angle sets. To observe the effect of acquisition flip angles on the variance of T1 estimates, scan protocols for T1 mapping were repeated three times in order to calculate the experimental variance in the T1 estimate. T1 standard deviation maps for the three angle sets used 10-point range, optimal 10-point, and 10-point repeat) are shown in Fig. 4a–c.
FIG. 4.
Maps of inter-scan standard deviation in T1 show reduced variation when the 10-point optimal angle set (b) or 10-point repeat angle set (c) is used compared to the 10-point range angle set (a). See Table 1 for specific angle sets used for acquisition. T1 maps of the rat brain (c) prior to flip angle optimization and (d) after flip angle optimization 10-point optimal angle set) show an increase in the average T1 estimate in the rat brain by 15.8 ms, more easily observable in (e) the T1 difference map (difference = optimal – 10-point range). Smoothed joint density functions of T1 and M0 (f) prior to and (g) following flip angle optimization also show this slight shift, along with the elimination of a small population of voxels with high initial T1 estimates (T1 > 1.5 s, white arrows). (T1 map, difference map, and P(T1, M0) are not shown for the 10-point repeat angle set because results are nearly identical to those for the 10-point optimal angle set under visual inspection.)
The mean inter-scan standard deviation in the T1 estimate was measured to be 73.3 ms when data were acquired using the 10-point range angle set. This standard deviation decreased to 61.2 ms when using the optimal flip angle set (Nα = 10). Interestingly, a slight additional decrease in inter-scan standard deviation, to 59.0 ms, was observed when data were acquired with the 10-point repeat angle set based on Deoni and colleagues’ criteria for selection of two flip angles (11). Therefore, use of flip angle sets in which two carefully selected flip angles were each repeated 5 times reduced inter-scan T1 estimate standard deviation in the rat brain by 16–20% when compared with the 10-point range angle set (variance reduction of 30–35%). Notably, the angle sets composed of a repeat of two angles selected using Deoni and colleagues’ criteria out-performed those selected using our variance minimization objective function in terms of inter-scan variance reduction.
T1 mapping results showed that T1 estimates in the rat brain were, on average, 15.8 ms higher when estimated using the optimized flip angles rather than the range of flip angles (P < 0.0001). When estimated using the 10-angle repeat set, T1 estimates were, on average, 24.3 ms higher than the range of flip angles (P < 0.0001). The difference of 8.5 ms between the 10 optimal angles and 10 angle repeat set was also found to be significant (P < 0.0001). Estimation of the smoothed joint density function of T1 and M0 from pre- and post-optimization acquisitions (Fig. 4d–e) also showed a slight shift of the mean T1 estimate in the rat brain toward higher T1 values when optimized angles were used. Elimination of a small population of voxels with high, outlying T1 estimates (T1 > 1.5 s) was also observed.
Discussion and Conclusions
In this work, we have proposed and evaluated a new data-driven method for selection of SPGR flip angles for T1 estimation. In the proposed angle selection method, we aimed to provide increased flexibility compared to previously developed approaches. This flexibility is particularly designed (1) to select any number of acquisition flip angles based on available scan time and (2) to select flip angles when the T1 distribution of interest is non-Gaussian and is therefore not well-described by the mean of the distribution.
In selecting multi-point optimal flip angle sets in two settings, the human brain and the rat brain, we found that the optimal flip angle set consisted of the repetition of two optimal angles. This is in disagreement with the common practice of acquiring data at a range of flip angles to cover a wide range of the SPGR signal curve (12, 24, 25). Instead, it more strongly agrees with Deoni and colleagues’ 2-point angle selection method, which maximizes the product of the dynamic range and the fractional signal of the acquired data (11, 18). It is also in agreement with a previous study confirming improved T1 mapping performance when using a small set of carefully selected flip angles over a larger set covering a range of angles (13).
Simulations testing these optimized angle sets showed very similar RMSE performance when comparing two optimized angles to two angles selected with Deoni and colleagues’ criteria. For greater than two angles, simulations also show that a repetition of two carefully selected flip angles has significantly improved performance compared to using a range of angles. However, our simulations assumed perfect RF coil performance in which nominal and actual flip angle match exactly. In a true scanning scenario, the relationship between of nominal and actual flip angle can vary greatly, and system-specific RF performance should be considered when optimizing scan protocol (13). This could be achieved by scaling the optimal flip angles according to the expected RF performance or by adjusting flip angle sets to cover a small range close to the optimal angles within the expected range of flip angle variation.
Simulations further elucidated the relative performance of different flip angle sets with respect to both bias and variance. As shown in Figure 3f, variance contributed relatively much more (20–50×) than bias to the RMSE of the T1 estimate. This supports the work in our group and others in which flip angle selection is guided by an objective of T1 variance minimization. When studying the effect of using optimal flip angles experimentally, we indeed found that T1 variance was reduced by 30% when compared to T1 estimated from SPGR acquisitions at a range of angles. These simulations also agreed with previous work showing that T1 estimates from VFA SPGR acquisitions tend to have positive bias when compared with the gold standard IR-based estimation approach (4).
Experimental measurement of inter-scan variance using three different 10-point acquisition angle sets showed that the use of a repetition of two thoughtfully selected flip angles indeed reduced estimate variability when compared to an acquisition using a more broad range of angles. The out-performance of our proposed optimal flip angle set in comparison to the 10-point repeat angle set could be due to imperfect RF performance, which in our scans caused actual flip angles to be approximately 80% of the nominal prescribed flip angles. This reduction would cause the two angles in the 10-point repeat set to be closer in magnitude to the intended optimal angles. The effects of this imperfect RF performance on the variability in T1 estimates further emphasizes that it is an important consideration in the design of SPGR-based T1 mapping experiments.
Based on these simulation and experimental results, the application of this flip angle selection technique to T1 mapping in research and clinical settings could notably reduce variance in T1 estimates if used in place of a set consisting of a range of flip angles. However, in many situations, the implementation of such an algorithm may be excessively complicated to incorporate into experimental design. In the imaging settings similar to those presented in this paper, in which T1 distributions approach a Gaussian shape, we suggest that it would be as effective and much simpler to use Deoni and colleagues’ criteria for selection of two flip angles based on the mean T1 in the volume of interest. If time allows for additional data collection or increased SNR is desired, these two angles should be repeated (multiple averages collected at each flip angle) as necessary. It should be noted, however, that the use of just two repeated flip angles might not be appropriate in some scenarios. When more than one T1 component is present in a voxel, deviation from the SPGR signal equation cannot be identified unless a flip angle set containing more than two unique points is used. Researchers interested in detecting the presence of multiple T1 times in individual voxels may find it useful and necessary to incorporate additional acquisition angles in addition to the two optimal angles for T1 variance minimization.
Future extensions of this work will evaluate the performance of our proposed angle selection in situations with more distinctly non-Gaussian T1 distributions. In this way, we hope to determine whether there are situations in which this more simplified angle selection approach using Deoni and colleagues’ set of criteria is not appropriate. Furthermore, more specific definition of P(M0, T1) based on the tissue of interest may be appropriate in some research and clinical settings. For example, specific flip angles could be selected for T1 mapping in the white matter in the study of myelin-related disease by using only segmented white matter voxels to define P(M0, T1).
In conclusion, this work demonstrates that the precision of T1 estimates from SPGR acquisitions can be improved by using flip angles selected based on minimization of the T1 variance weighted by the smoothed joint density of T1 and M0. Specifically, our proposed approach significantly out-performed T1 estimation using a range of flip angles when the same total number of flip angles were used. Interestingly, when comparing the proposed angle selection method with previously proposed selection criteria, we found that Deoni and colleagues’ 2-angle selection criteria can also be effectively used to determine the two angles to be repeated for a multi-point acquisition with nearly identically improved performance (11). The proposed flip angle selection approach resulted in either equivalent or up to 42% reduced RMSE compared to other flip angle selection criteria in simulations. Experimentally, implementation of optimal angles reduced inter-scan variability in T1 estimates by 30%. The application of this data-driven technique for optimal flip angle selection has applications in clinical and research settings where scans are performed repeatedly in the same anatomical region (e.g., the human brain) or on the same equipment (consistent scanner and coil).
Acknowledgements
The authors thank Dr. Carlo Pierpaoli for his encouragement of the preliminary version of this work. We further thank Alexey Samsonov and the co-contributors of QMAP, data from which was used for simulations and is available at http://www.medphysics.wisc.edu/~samsonov/qmap/. We gratefully acknowledge our funding, including the NIH Biotechnology Training Grant (T32 GM008349 to CML).
Appendix A
From Eq. 7–8, the Jacobian matrix can be written as
where and . Therefore,
By defining E = exp(TR/T1), Eq. 7–8 can be rewritten as follows:
and
so that
and
The determinant of can then be written as a double summation:
Once the determinant of is obtained, the covariance matrix can be computed quite easily; therefore, the derivation will not be shown here.
References
- 1.Lutti A, Dick F, Sereno MI, Weiskopf N. Using high-resolution quantitative mapping of R1 as an index of cortical myelination. Neuroimage. 2014;93:176–188. doi: 10.1016/j.neuroimage.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Baudrexel S, Nurnberger L, Rub U, Seifried C, Klein JC, Deller T, Steinmetz H, Deichmann R, Hilker R. Quantitative mapping of T1 and T2* discloses nigral and brainstem pathology in early Parkinson's disease. Neuroimage. 2010;51:512–520. doi: 10.1016/j.neuroimage.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Salerno M, Kramer CM. Advances in parametric mapping with CMR imaging. JACC Cardiovasc Imaging. 2013;6:806–822. doi: 10.1016/j.jcmg.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stikov N, Boudreau M, Levesque IR, Tardif CL, Barral JK, Pike GB. On the accuracy of T1 mapping: searching for common ground. Magn Reson Med. 2015;73:514–522. doi: 10.1002/mrm.25135. [DOI] [PubMed] [Google Scholar]
- 5.Fram EK, Herfkens RJ, Johnson GA, Glover GH, Karis JP, Shimakawa A, Perkins TG, Pelc NJ. Rapid calculation of T1 using variable flip angle gradient refocused imaging. Magn Reson Imaging. 1987;5:201–208. doi: 10.1016/0730-725x(87)90021-x. [DOI] [PubMed] [Google Scholar]
- 6.Yarnykh VL. Actual flip-angle imaging in the pulsed steady state: a method for rapid three-dimensional mapping of the transmitted radiofrequency field. Magn Reson Med. 2007;57:192–200. doi: 10.1002/mrm.21120. [DOI] [PubMed] [Google Scholar]
- 7.Hurley SA, Yarnykh VL, Johnson KM, Field AS, Alexander AL, Samsonov AA. Simultaneous variable flip angle-actual flip angle imaging method for improved accuracy and precision of three-dimensional T1 and B1 measurements. Magn Reson Med. 2012;68:54–64. doi: 10.1002/mrm.23199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heule R, Ganter C, Bieri O. Variable flip angle T1 mapping in the human brain with reduced T2 sensitivity using fast radiofrequency-spoiled gradient echo imaging. Magn Reson Med. 2015 doi: 10.1002/mrm.25668. [DOI] [PubMed] [Google Scholar]
- 9.Wang HZ, Riederer SJ, Lee JN. Optimizing the precision in T1 relaxation estimation using limited flip angles. Magn Reson Med. 1987;5:399–416. doi: 10.1002/mrm.1910050502. [DOI] [PubMed] [Google Scholar]
- 10.Imran J, Langevin F, Saint-Jalmes H. Two-point method for T1 estimation with optimized gradient-echo sequence. Magn Reson Imaging. 1999;17:1347–1356. doi: 10.1016/s0730-725x(99)00092-2. [DOI] [PubMed] [Google Scholar]
- 11.Deoni SC, Rutt BK, Peters TM. Rapid combined T1 and T2 mapping using gradient recalled acquisition in the steady state. Magn Reson Med. 2003;49:515–526. doi: 10.1002/mrm.10407. [DOI] [PubMed] [Google Scholar]
- 12.Deoni SC, Peters TM, Rutt BK. Determination of optimal angles for variable nutation proton magnetic spin-lattice, T1, and spin-spin, T2, relaxation times measurement. Magn Reson Med. 2004;51:194–199. doi: 10.1002/mrm.10661. [DOI] [PubMed] [Google Scholar]
- 13.Cheng HL, Wright GA. Rapid high-resolution T(1) mapping by variable flip angles: accurate and precise measurements in the presence of radiofrequency field inhomogeneity. Magn Reson Med. 2006;55:566–574. doi: 10.1002/mrm.20791. [DOI] [PubMed] [Google Scholar]
- 14.Schabel MC, Morrell GR. Uncertainty in T(1) mapping using the variable flip angle method with two flip angles. Phys Med Biol. 2009;54:N1–N8. doi: 10.1088/0031-9155/54/1/N01. [DOI] [PubMed] [Google Scholar]
- 15.Samsonov A, Alexander AL, Jung Y, Field AS. Practical optimum experimental designs for fast T1 relaxometry with SPGR sequences. Proc Intl Soc Mag Reson Med. 2008;16 [Google Scholar]
- 16.Koay CG, Chang LC, Deoni S, Pierpaoli C. An optimal framework for T1 estimation in an SPGR acquisition. Proc Intl Soc Mag Reson Med. 2007;15 [Google Scholar]
- 17.Koay CG, Chang LC, Pierpaoli C, Basser PJ. Error propagation framework for diffusion tensor imaging via diffusion tensor representations. IEEE Trans Med Imaging. 2007;26:1017–1034. doi: 10.1109/TMI.2007.897415. [DOI] [PubMed] [Google Scholar]
- 18.Wood TC. Improved formulas for the two optimum VFA flip-angles. Magn Reson Med. 2015 doi: 10.1002/mrm.25592. [DOI] [PubMed] [Google Scholar]
- 19.Marquardt DW. An algorithm for least-squares estimation of nonlinear parameters. J Soc Ind Appl Math. 1963;11:431–441. [Google Scholar]
- 20.Chang LC, Koay CG, Basser PJ, Pierpaoli C. Linear least-squares method for unbiased estimation of T1 from SPGR signals. Magn Reson Med. 2008;60:496–501. doi: 10.1002/mrm.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001;5:143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- 23.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 24.Liu F, Chaudhary R, Hurley SA, Munoz Del Rio A, Alexander AL, Samsonov A, Block WF, Kijowski R. Rapid multicomponent T2 analysis of the articular cartilage of the human knee joint at 3.0T. J Magn Reson Imaging. 2014;39:1191–1197. doi: 10.1002/jmri.24290. [DOI] [PubMed] [Google Scholar]
- 25.Pineda FD, Medved M, Fan X, Karczmar GS. B and T mapping of the breast with a reference tissue method. Magn Reson Med. 2015 doi: 10.1002/mrm.25751. [DOI] [PMC free article] [PubMed] [Google Scholar]