Abstract
Prevention programs for β-thalassemia based on molecular diagnosis of heterozygous carriers and/or patients require the use of reliable mutation screening methods. The aim of this study was to compare between direct DNA sequencing, and reverse dot-blot PCR in detection of different β-globin gene mutations in Egyptian children with β-thalassemia. Forty children with β-thalassemia were subjected to mutation analysis, performed by both direct DNA sequencing and β-globin Strip Assay MED™ (based on reverse dot-blot PCR). The most frequent mutant alleles detected by reverse dot-blot PCR were; IVSI-110 G>A (31.25 %), IVS I-6 T > C (21.25 %), and IVS I-1 G>A (20 %). Relatively less frequent mutant alleles detected by reverse dot-blot PCR were “IVSII-1 G>A (5 %), IVSII-745 C>G (5 %), IVSII-848 C>A (2.5 %), IVSI-5 G>C (2.5 %), -87 C>G(2.5 %), and cd39 C>T (2.5 %)”, While the genotypes of three patients (6 alleles 7.5 %) were not detected by reverse dot-blot PCR. Mutant alleles detected by direct DNA sequencing were the same as reverse dot-blot PCR method except it revealed the genotypes of 3 undetected patients (one patient was homozygous IVSI-110 G>A, and two patients were homozygous IVS I-1 G>A. Sensitivity of the reverse dot-blot PCR was 92.5 % when compared to direct DNA sequencing for detecting β-thalassemia mutations. Our results therefore suggest that, direct DNA sequencing may be preferred over reverse dot-blot PCR in critical diagnostic situations like genetic counseling for prenatal diagnosis.
Keywords: β-Thalassemia, Genetic mutations, β-Globin Strip Assay MED™ (reverse dot-blot PCR), DNA sequencing
Introduction
β-Thalassemia is the most common genetically inherited hemoglobin disorder in Egypt with a carrier rate varying from 5.3 to ≥9 % [1]. It has been estimated that 1000 children out of the 1.5 million live births are born annually with β-thalassemia major [2].
The mainstay for treatment of severe forms of thalassemia is lifelong blood transfusion and iron chelation therapy. The complications of iron overload, together with the sequel of anemia, ineffective erythropoiesis and the chelation therapy itself, are the major causes of morbidity and mortality in the transfusion dependent β-thalassemia [3].
More than 200 β-thalassemia mutations have been characterized worldwide, however, a relatively small number of common β-thalassemia mutations can be found for each high risk population [4]. In Egypt, although more than 20 different mutations have been detected so far to cause the disease, the information available concerning the underlying molecular defects in β-thalassemia has not yet been completed [5].
The molecular defects resulting in β-thalassemia phenotype, in the Egyptian population, show a clear heterogenic pattern. Many studies have focused on the molecular detection and characterization of these mutations, using a wide array of the available techniques with successful detection of both known and unknown mutations. PCR based techniques, including direct DNA sequencing, are effective with some limitations considering the time, effort and high cost to reach a final diagnosis [6].
This study compared “direct DNA sequencing”, and “reverse dot-blot PCR” as screening tools for β-thalassemia.
Subjects and Methods
Forty children, 23 with β-thalassemia major and 17 with β-thalassemia intermediate, 20 males and 20 females (with mean age of 6.7 ± 3.43 years) were recruited from outpatient Clinic of Hematology Unit, Pediatric Department, Tanta University Hospital. Children with other types of hemolytic anemia and genetic diseases were excluded. An informed consent was obtained from all parents of children before the enrollment in the study. The study was approved by the Ethics Committee of Tanta Faculty of Medicine, Tanta University. They were subjected to meticulous history taking with reference to positive consanguinity, and clinical evaluation of all body systems. The following investigations were performed:
Routine hematological investigations e.g., complete blood count using ERMA PCE-210N cell counter, reticulocytes’ count, Hb electrophoresis using cellulose acetate in a tris EDTA borate buffer at PH 8.4 (Helena Laboratories, Beaumount, TX, USA), serum ferritin levels using MonobindInc ELISA Microwells kit (lake Forest, CA 92630, USA).
All affected patients were clinically classified into thalassemia major and thalassemia intermediate with consideration to: the age of disease onset, the age of first transfusion, frequency of blood transfusion, hemoglobin level, hepato-splenomegaly, facial and growth affection [7].
For direct fluorescent DNA sequencing of the β-globin gene, DNA was extracted from EDTA -whole blood using a QIA amp DNA blood mini kit (Qiagen, Hilden, Germany CA. No. 51104), and stored at −20 °C. DNA concentration and purity were determined spectrophotometrically as well as by QubitR ds DNA BR Assay kit (USA) with use QubitR 2.0 flurometer (Invitrogen™ by life tecnologies™). PCR amplification was performed using GMLRSeqFinder Sequencing System HBB (Applied Biosystem) and these products visualized under UV illumination (Biometra Germany) after gel electrophoresis. These products were then purified by QIA QuickRPCR Purification kit (Qiagen, Hilden, Germany cat. No. 28104) and subjected to cycle sequencing PCR using fluorescent dyes (Applied Biosystem, Foster City, CA, USA). They were then further purified using CENTRI-SEP columns (cat. No. CS-901) before analysis on automated sequencer (ABI PRISM™ 310 Genetic Analyzer). Finally, interpretations of the results were presented via SeqScape software version 2.7 Applied Biosystem.
Reverse dot-blot PCR was done using β-Globin StripAssay MED™, (ViennaLab Diagnostics GmbH). In this commercial assay, DNA isolated from anticoagulated blood using Lysis Solution and GEN×TRACT Resin was subjected to amplification and biotin-labeled β-globin sequences in a single (multiplex) PCR amplification reaction. The amplification products are then selectively hybridized to a test strip containing wild type and mutant oligonucleotide probes immobilized as parallel lines. Bound biotinylated sequences are detected using streptavidin–alkaline phosphatase and color substrates. The assay covers 22 mutations, characteristics for the Mediterranean area. www.viennalab.com.
Statistical Analysis
Data were analyzed using SPSS version 20. Data were expressed as mean ± SD for quantitative variables, number and percentage for qualitative ones. Chi square, ANOVA, and ROC curve were used when appropriate. P value < 0.05 was considered to be statistically significant.
Results
Demographic data, clinical features, and some Laboratory data (CBC, Hb electrophoresis, serum ferritin level) are presented in Tables 1 and 2. Different patients phenotypes and their mutations are presented in Tables 3 and Fig. 1, it was found that most frequent mutant alleles detected by reverse dot-blot PCR were IVSI-110 G>A (31.25 %), IVS I-6 T>C (21.25 %), and IVS I-1 G>A (20 %).Relatively less common set of mutations detected by reverse dot-blot PCR were IVSII-1 G>A (5 %), IVSII-745 C>G (5 %), IVSII-848 C>A (2.5 %), IVSI-5 G>C (2.5 %), -87 C>G (2.5 %), and cd39 C>T (2.5 %), but the genotypes of three patients (6 alleles 7.5 %)were not detected by reverse dot-blot PCR. Those 40 children classified phenotypically into, 23 patients (57.5 %) with thalassemia major (TM) and 17 patients (42.5 %) with thalassemia intermedia (TI).
Table 1.
Clinical data among studied cases
| Age/year | |
| Mean ± SD | 6.7 ± 3.43 |
| Range | 1–12 |
| Sex | Male (50 %)–female (50 %) |
| Consanguinity | Positive (50 %)–negative (50 %) |
| Family history | Positive (35 %)–negative (65 %) |
| Age at diagnosis/month | |
| Mean ± SD | 17.25 ± 11.1 |
| Range | 4–60 |
| Age of 1st transfusion/month | |
| Mean ± SD | 15.63 ± 12.1 |
| Range | 2–72 |
| Interval of transfusion/week | |
| Mean ± SD | 4.11 ± 2.41 |
| Range | 2–6 |
| Pallor (%) | 95 % |
| Jaundice (%) | 45 % |
| Hepatomegaly (%) | 70 % |
| Splenomegaly (%) | 45 % |
| Splenectomy (%) | 25 % |
| Time of splenectomy/year | |
| Mean ± SD | 6.96 ± 2.20 |
| Range | 4–10 |
SD standard deviation
Table 2.
Laboratory data of the studied patients
| CBC parameters | Mean ± SD | Range |
|---|---|---|
| Hb (g/dl) | 7.49 ± 1.52 | 4.9–9.4 |
| MCV (fl) | 65.11 ± 4.41 | 59.1–74 |
| MCH (pg/cell) | 21.95 ± 2.69 | 19–24 |
| Corrected Reticulocyte % | 3.06 ± 1.74 | 0.3–8.6 |
| WBCs (109/L) | 12.21 ± 2.61 | 6.2–29 |
| Platelet (109/L) | 491.10 ± 48.1 | 70–1217 |
| Hemoglobin electrophoresis | ||
| Hb F % | 59.51 ± 20.95 | 7.9–91 |
| Hb A % | 42.32 ± 21.62 | 2.7–74.4 |
| Hb A2 % | 2.90 ± 1.41 | 0–6.3 |
| Serum ferritin (ng/l) | 1772 ± 259.3 | 32–6833 |
Hb hemoglobin, MCV mean corpuscular volume, MCH mean corpuscular hemoglobin, MCHC mean corpuscular hemoglobin concentration, WBCs white blood cells
Table 3.
Different mutations with its phenotypes detected by reverse dot-blot PCR
| Mutation | Genotype | Phenotype | Patients no (%) |
|---|---|---|---|
| IVS I-110 (G>A)/IVS I-110 (G>A) | β+/β+ | 3TM and 3TI | 6 (15) |
| IVS I-110 (G>A)/IVS I-6 (T>C) | β+/β+ | 2TM and 3TI | 5 (12.5) |
| IVSI-I (G>A)/IVSI-6 (T>C) | β0/β+ | 2TM and 2TI | 4 (10) |
| IVSI-I (G>A)/IVSI-I (G>A) | β0/β0 | 3TM | 3 (7.5) |
| IVSI-I (G>A)/IVSI-110 (G>A) | β0/β+ | 3TM | 3 (7.5) |
| IVSII-I (G>A)/IVS I-110 (G>A) | β0/β+ | 2TM | 2 (5) |
| IVSI-I (G>A)/IVSII-848 (C>A) | β0/β+ | 2TM | 2 (5) |
| IVSII-I (G>A)/IVSI-6 (T>C) | β0/β+ | 1TM and 1TI | 2 (5) |
| IVSII-745 (C>G)/IVSII-745 (C>G) | β+/β+ | 2TI | 2 (5) |
| IVSI-6 (T>C)/-87 (C>G) | β+/β+ | 2TI | 2 (5) |
| Cd39 (C>T)/IVSI-110 (G>A) | β0/β+ | 2TM | 2 (5) |
| IVS I-6 (T>C)/IVS I-6 (T>C) | β+/β+ | 2TI | 2 (5) |
| IVS I-1 (G>A)/IVS I-5 (G>C) | β0/β+ | 1TI | 1 (2,5) |
| IVS I-110 (G>A)/IVS I-5 (G>C) | β+/β+ | 1TM | 1 (2,5) |
| Undetected | 2TM and 1TI | 3 (7.5) | |
| Total number of patients | 23 (57.5 %)TM and 17 (42.5 %)TI | 40 (100) |
Fig. 1.
Examples of reverse dot-blot PCR. a Compound heterozygous IVSI-1 G>A / IVSI-5 G>C, b homozygous IVSII-745 C>G
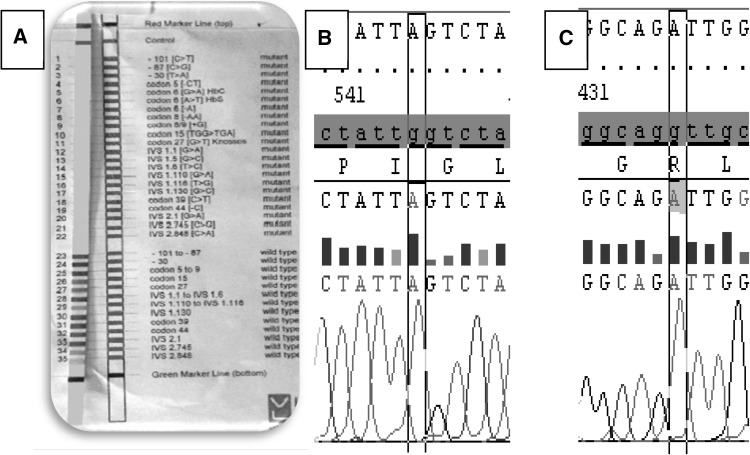
Mutant alleles detected by direct DNA sequencing were the same as reverse dot-blot PCR method except it revealed the genotypes of 3 undetected patients, (Fig. 2). Normal or undetected cases by reverse dot-blot PCR, their DNA sequencing result was “Homozygous IVSI-110 G>A” so their percentage became (33.75 %). Other two undetected cases were homozygous IVS I-1 G>A, so their percentage became (25 %). Sensitivity of reverse dot-blot PCR was 92.5 %, specificity was 100 %, positive predictive value was 100 % and negative predictive value was 99.63 %”.
Fig. 2.
Undetected cases by reverse dot-blot PCR (a) and its DNA sequencing results (Homozygous IVS 1-110 G > A (b) and homozygous IVS 1-1 G < A (c))
Discussion
β-Thalassemia is a group of hemoglobin diseases caused by a reduction (β+ or absence (β0) in the synthesis of β-globin chains. Affected individuals can be compound heterozygous, or homozygous for β-thalassemia [8, 9].
Despite efforts to develop gene therapy or bone marrow transplantation, prenatal diagnosis followed by termination of the affected fetus remains the best form of prevention [10]. In Egypt, combined effects of high carrier rates and high frequency of consanguineous marriages [11], make the incidence of β-thalassemia particularly high [12]. Prevention is the least expensive and the most effective means to deal with β-thalassemia [13]. So; it is a prerequisite to investigate the molecular basis and natural history of these disorders to establish the least costly effective methods for their control and management [14].
Considering demographic data, clinical features, and laboratory data of the present study, it was in agreement with the data published by Hussein et al. [2], Galanello and Origa [15], Kattamis [16], Thein and Rees [17], Soliman et al. [18], Rahim et al. [19], Mosca et al. [20], and El-Shanshory et al. [21].
The patient’s genotype distribution in our study was in agreement with Kaddah et al. [22], Waye et al. [23], Settin et al. [24], and Jiffri et al. 2010 [11] whose studies stated that the most common genetic mutations of the β-thalassemia evaluated in Egyptian studies are; IVSI-110 G>A, IVS I-1 G>A, IVSI-6 T>C, IVSII-1 G>A, IVSII-745 C>G, -87 C>G and cd39 C>T. IVS-I-5 G> C was previously found in Chinese, Asian Indian and Mediterranean populations [25].
The sensitivity of the reverse dot-blot PCR was in agreement with Othman et al. [26], who stated that the sensitivity of β-Globin StripAssay method was 90 %. In our study the undetected mutations of patients by the reverse dot-blot PCR were IVSI-110 G>A, and IVS I-1 G>A was included in the 22 mutations covered by the same method so they were not a rare one. The explanation of these undetected mutations may be strip instability “containing probes”, bad quality of DNA isolation of this technique which may be in accurate when compared to a QIA amp DNA blood mini kit, and also may be some reagent deterioration since it contains enzyme, color, and heat dependent reagents. Finally interpretation is personally, and visually subjective. DNA sequencing is more costly, time consuming, and labour- intensive than reverse dot-blot PCR but it depends on many accurate steps of extraction, purification, etc. Which are checked before entering the next one by sophisticated techniques, and finally software interpretation. So DNA sequencing method is more robust than β-Globin StripAssay.
Conclusion
The sensitivity of the reverse dot-blot PCR was 92.5 % when compared to direct DNA sequencing for detecting β-thalassemia mutations. Our results therefore suggest that, direct DNA sequencing may be preferred over reverse dot-blot PCR in critical diagnostic situations like genetic counseling for prenatal diagnosis.
Acknowledgments
Conflict of interest
None.
References
- 1.El-Beshlawy A, Youssry I. Prevention of hemoglobinopathies in Egypt. Hemoglobin. 2009;33(1):14–20. doi: 10.3109/03630260903346395. [DOI] [PubMed] [Google Scholar]
- 2.Hussein G, Fawzy M, El-Serafi T, et al. Rapid detection of β-thalassemia alleles in Egypt using naturally or amplified created restriction sites and direct sequencing: a step in disease control. Hemoglobin. 2007;31(1):49. doi: 10.1080/03630260601057088. [DOI] [PubMed] [Google Scholar]
- 3.Quek L, Thein S. Molecular therapies in β-thalassemia. Br J Hematol. 2006;136:353–365. doi: 10.1111/j.1365-2141.2006.06408.x. [DOI] [PubMed] [Google Scholar]
- 4.Li D, Liao C, Li J, et al. Prenatal diagnosis of β-thalassemia by reverse dot-blot hybridization in southern China. Hemoglobin. 2006;30(3):365. doi: 10.1080/03630260600755625. [DOI] [PubMed] [Google Scholar]
- 5.Omar A, Abdel Karim E, El Gendy W, Marzouk I, Wagdy M. Molecular basis of beta thalassemia in Egypt. Egypt J Immunol. 2005;12(1):15–24. [PubMed] [Google Scholar]
- 6.Christopoulos G, Ezzat GM, Kleanthous M. Use of denaturing gradient gel electrophoresis in screening unknown β-thalassemia mutations in Egyptian patients. Egypt J Med Hum Genet. 2012;13:343–349. doi: 10.1016/j.ejmhg.2012.06.008. [DOI] [Google Scholar]
- 7.Nadkarni A, Gorakshakar A, Colah R, Mohanty D, Ghosh K. Evaluation of the clinical severity of β thalassemia homozygous patients using a phenotypic scoring system. J Chin Clin Med. 2007;2(8):8. [Google Scholar]
- 8.Thein SL. β-thalassaemia. Baillieres Clin Haematol. 1998;11:91–126. doi: 10.1016/S0950-3536(98)80071-1. [DOI] [PubMed] [Google Scholar]
- 9.Weatherall DJ. Phenotype-genotype relationships in monogenic disease: lessons from the thalassaemias. Nat Rev Genet. 2001;2:245–255. doi: 10.1038/35066048. [DOI] [PubMed] [Google Scholar]
- 10.Jiffri EH, Bogari N, Zidan KH, et al. Molecular updating of β-thalassemia mutations in the upper Egyptian population. Hemoglobin. 2010;34(6):538–547. doi: 10.3109/03630269.2010.526440. [DOI] [PubMed] [Google Scholar]
- 11.Hafez M, El-Tahan H, Awadalla M, El-Khayat H, Gafar Abdul, Ghoneim M. Consanguineous matings in the Egyptian population. J Med Genet. 1983;20:58–60. doi: 10.1136/jmg.20.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Beshlawy A, Kaddah N, Moustafa A, Mouktar G, Youssry I. Screening for β-thalassemia carriers in Egypt: significance of the osmotic fragility test. East Mediterr Health J. 2007;13(4):780–786. [PubMed] [Google Scholar]
- 13.MahmoodBaig, S.M., A. Azhar, H. Hassan, J.M. Baig, A. Kiyani, U. Hameed, F. Rabbi, H. Bokhari, M. Aslam, M.A. Ud Din S.A. Baig, K.Hassan, J.A. Qureshi and T. Zaman (2006) Spectrum of beta-thalassemia mutations in various regions of Punjab and Islamabad, Pakistan: establishment of prenatal diagnosis. Haematologica 91(1):e13-e15 [PubMed]
- 14.Alan N Schechter. Hemoglobin research and the origins of molecular medicine. Blood. 2008;112(10):3927–3938. doi: 10.1182/blood-2008-04-078188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galanello R and Origa R (2010) Review of beta thalassemia. Orphanet J Rare Dis 5-11 [DOI] [PMC free article] [PubMed]
- 16.Kattamis CA. Management of thalassemias: growth and development, hormone substation, vitamin supplementation and vaccination. Semin Hematol. 1995;32:269–279. [PubMed] [Google Scholar]
- 17.Thein SL and Rees D (2011) Haemoglobin and the Inherited Disorders of Globin Synthesis. Postgraduate Haematology, Sixth edition. 83–108
- 18.Soliman HH, Kabbash IA, El-Shanshory MR, et al. Evaluation of immune status against hepatitis B in children with thalassemia major in Egypt. JMID. 2012;2:44–49. doi: 10.5799/ahinjs.02.2012.02.0041. [DOI] [Google Scholar]
- 19.Rahim F, Ski N, Jalai-Far MA. The role of gene mutations detection in defining the spectrum of beta-thalassemia in various ethnic regions. Hum Genetic Dis. 2011;6:109–120. [Google Scholar]
- 20.Mosca A, Paleari R, Leone D, et al. The relevance of hemoglobin F measurement in the diagnosis of thalassemia and related hemoglobinopathies. Clin Geochem. 2009;42:1797–1801. doi: 10.1016/j.clinbiochem.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 21.EL- MR, Kabbash IA, Soliman HH, et al. Prevalence of hepatitis C infection among children with β-thalassaemia major in Mid Delta, Egypt: a single centre study. Trans R Soc Trop Med Hyg. 2013;107(4):224–228. doi: 10.1093/trstmh/trs024. [DOI] [PubMed] [Google Scholar]
- 22.Kaddah N, Rizk S, Kaddah AM, Salama K, Lotfy H. Study of possible genetic factors determining the clinical picture of thalassemia Intermedia. J Med Sci. 2009;9:151–155. doi: 10.3923/jms.2009.151.155. [DOI] [Google Scholar]
- 23.Waye J, Bory S, Eng B, et al. Spectrum of β-thalassemia mutations in Egypt. Hemoglobin. 1999;23(3):255. doi: 10.3109/03630269909005706. [DOI] [PubMed] [Google Scholar]
- 24.Settin AA, Al-Haggar MM, Neamatallah M, Al-Said AM, Hafez MM. Detection of beta-thalassaemia mutations using primer-specific amplification compared to reversed dot blot hybridization technique in Egyptian cases. Haema. 2006;9(3):401–409. [Google Scholar]
- 25.Huisman THJ, Carver MFH, Baysal EA. Syllabus of thalassemia mutations. Augusta: The Sickle Cell Anemia Foundation; 1997. [Google Scholar]
- 26.Othman E, Sohier Y, Amani S, et al. Reverse hybridization StripAssay detection of beta-thalassemia mutations in northeast Egypt. Haematology. 2010;15(3):182–186. doi: 10.1179/102453310X12583347010214. [DOI] [PubMed] [Google Scholar]