Abstract
Microorganisms produce a wide range of natural products (NPs) with clinically and agriculturally relevant biological activities. In bacteria and fungi, genes encoding successive steps in a biosynthetic pathway tend to be clustered on the chromosome as biosynthetic gene clusters (BGCs). Historically, “activity-guided” approaches to NP discovery have focused on bioactivity screening of NPs produced by culturable microbes. In contrast, recent “genome mining” approaches first identify candidate BGCs, express these biosynthetic genes using synthetic biology methods, and finally test for the production of NPs. Fungal genome mining efforts and the exploration of novel sequence and NP space are limited, however, by the lack of a comprehensive catalog of BGCs encoding experimentally-validated products. In this study, we generated a comprehensive reference set of fungal NPs whose biosynthetic gene clusters are described in the published literature. To generate this dataset, we first identified NCBI records that included both a peer-reviewed article and an associated nucleotide record. We filtered these records by text and homology criteria to identify putative NP-related articles and BGCs. Next, we manually curated the resulting articles, chemical structures, and protein sequences. The resulting catalog contains 197 unique NP compounds covering several major classes of fungal NPs, including polyketides, non-ribosomal peptides, terpenoids, and alkaloids. The distribution of articles published per compound shows a bias towards the study of certain popular compounds, such as the aflatoxins. Phylogenetic analysis of biosynthetic genes suggests that much chemical and enzymatic diversity remains to be discovered in fungi. Our catalog was incorporated into the recently launched Minimum Information about Biosynthetic Gene cluster (MIBiG) repository to create the largest known set of fungal BGCs and associated NPs, a resource that we anticipate will guide future genome mining and synthetic biology efforts toward discovering novel fungal enzymes and metabolites.
1. Introduction
Microorganisms produce natural products (NPs) with a remarkable range of structures and bioactivities. Fungi produce NPs with particularly diverse properties, including antibiotics (e.g. penicillin) and toxins (e.g. aflatoxins) (Amare and Keller, 2014; Hoffmeister and Keller, 2007; Newman and Cragg, 2007). NPs are often biosynthesized by multi-enzyme pathways, and the proteins comprising one pathway are often encoded within a clustered set of genes, termed a biosynthetic gene cluster (BGC) (Cimermancic et al., 2014; Keller et al., 2005; Medema et al., 2011). Many putative BGCs remain “cryptic” or “orphan”, meaning their encoded proteins and resulting chemicals are not expressed under standard conditions and are currently uncharacterized. The chemicals produced by these silent gene clusters represent untapped chemical diversity that could hold pharmacological and agricultural value (Walsh and Fischbach, 2010).
Researchers employ various approaches to determine the products encoded by cryptic fungal gene clusters (Wiemann and Keller, 2014). Synthetic biology and genetic engineering efforts include activation of transcriptionally silent clusters in natural hosts (Bouhired et al., 2007; Mao et al., 2015; Mattern et al., 2015) and expression of uncharacterized clusters in heterologous hosts (Chang et al., 2013; Heneghan et al., 2010; Kealey et al., 1998; Pel et al., 2007; Richter et al., 2014; Sakai et al., 2012, 2008; Yin et al., 2013). The synthetic biology toolset continues to expand with new methods, such as gene cluster refactoring (Shao et al., 2013) and the expression of multi-gene pathways (Unkles et al., 2014).
In the modern era of BGC research that benefits from the continuous flood of new sequencing data, computational approaches play a critical role in the discovery of NPs from uncharacterized BGCs. BGCs can be computationally identified by homology and other techniques, and their chemical product can sometimes be predicted based on the enzymes they encode (Donia et al., 2014; Medema and Fischbach, 2015; O'Brien et al., 2014; Pi et al., 2015; Throckmorton et al., 2015). These studies have revealed that fungal genomes encode several classes of BGCs capable of producing valuable, diverse NPs, including polyketides, non-ribosomal peptides, terpenoids, alkaloids, and others. Many of the basic biosynthetic mechanisms for producing the major classes of fungal NPs are well characterized, and new BGCs can therefore be detected in fungal genomic sequences via homology to core biosynthetic enzymes (Cimermancic et al., 2014; Inglis et al., 2013; Medema and Fischbach, 2015). The number of fungal BGCs yet to be discovered is vast, as a single fungal genome can house 40–80 gene clusters with homology to major BGC classes (Inglis et al., 2013). With rapid advancements in sequencing technologies, it is expected that within a decade, millions of bacterial and fungal sequences will be available to the public (Medema and Fischbach, 2015), which will be invaluable for NP genome mining efforts.
Given the wealth of genomic discovery opportunities at our disposal, the future of fungal NP research relies in part on our ability to make relevant information about BGCs accessible to the community (Medema and Fischbach, 2015; Medema et al., 2015). Large numbers of BGCs and NP structures have been characterized over decades of fungal NP research (Lim et al., 2012; Walsh and Fischbach, 2010), but these published results have not been curated and made available in a standardized format. A comprehensive catalog of fungal NPs with experimentally characterized biosynthetic genes would be of great value to the community. While many catalogs related to fungal genetics and metabolism exist, they either focus on primary metabolism (Cerqueira et al., 2014; Stajich et al., 2012), or do not directly link secondary metabolite structures to the encoding BGCs (Degtyarenko et al., 2008; Gaulton et al., 2012; Pence and Williams, 2010; Wang et al., 2009). DoBISCUIT (Ichikawa et al., 2013) and ClusterMine360 (Conway and Boddy, 2013) link molecular structure to DNA sequence, but focus on bacterial NPs.
The need for a fungal NP reference catalog inspired us to perform a comprehensive curation of published articles describing fungal NPs and associated biosynthetic genes. To identify all possible published articles related to fungal NP biosynthesis, we developed an automated search strategy to extract NP-related publications from the PubMed database, each of which was linked to a fungal nucleotide record in GenBank. We then employed the “many hands make light work” approach to annotate the fungal NPs described, by integrating this research challenge into the classroom. Under the close guidance of NP experts, trained undergraduate students curated those publications that experimentally verified biosynthesis of fungal NPs. The resulting catalog includes 197 unique fungal NPs from 217 peer-reviewed articles, and 779 nucleotide records. Here we present a systematic analysis of the resulting articles, chemical structures, and protein sequences. We contextualize our results within the framework of the recent MIBiG (Minimum Information about a Biosynthetic Gene cluster) effort, a parallel large-scale community annotation project (Medema et al., 2015). The information from our study is expected to guide the exploration and engineering of fungal systems for the biosynthesis of novel NPs. Furthermore, the significance of our findings extends into pedagogical space, as the student-led curation represents a unique educational process with direct benefits to the NP and synthetic biology research communities.
2. Materials and Methods
2.1 Identification of NP-related articles in PubMed linked to nucleotide records
To identify a preliminary set of NP-related peer-reviewed articles for manual curation, we employed an automated search of NCBI PubMed for articles with deposited nucleotide records. We refer to these as “article-nucleotide” pairs, since they have both a PubMed article (with a PubMed ID or PMID) and a NCBI nucleotide record ID.
As the first step in this process, we identified articles related to NP biosynthesis using three automated searches for: (a) nucleotide records with homology to known core enzymes, according to searches using antiSMASH 2.0 (Blin et al., 2013), (b) nucleotide records identified by text queries on full records or record titles in the NCBI nucleotide database, and (c) published articles identified by text queries of articles in the PubMed database. The text queries were composed of a list of compound classes, names of secondary metabolites, and keywords indicative of NP gene clusters. Full queries used in the searches are listed in Supplementary Materials section 1.1. From the candidate nucleotide records (a and b), we retained only those with linked PubMed abstracts or PubMed Central full text articles. From the candidate PubMed articles (c), only those with linked nucleotide records were retained.
We identified 2,449 articles linked to nucleotide records of fungal origin, 1,652 articles with nucleotide records of plant origin, and 5,904 articles linked to nucleotide records of bacterial origin. Manual examination of a subset of the fungal articles revealed that most were not relevant to this study: for example, some describe a fungal genome sequence with no mention of NPs, and some describe a non-biosynthetic gene. To remove such articles, we developed an automated “NP-filter”, the details of which are described in Supplementary Materials section 1.2.
2.2 Manual curation of NPs described in peer-reviewed articles
The “NP-filter” step reduced the number of fungal articles to 960, but only decreased the coverage of “gold standard” entries, i.e., the bacteria or fungal entries present in either DoBISCUIT or ClusterMine360, by 1% (Supplementary Materials, Section 1.3 and Table S1). Each of the 960 articles linked to fungal nucleotide records was assigned a score from 1–8.5 based on the types of supporting query strategies, with higher scores indicating stronger supporting evidence (see Supplementary Materials, Section 1.4). We observed a strong enrichment of ClusterMine360 entries in the highest-scoring articles. The 349 articles with scores > 2 covered 92% of the fungal “gold standard articles” (38 articles associated with the fungal entries in ClusterMine360).
The titles of the 960 articles were manually examined by two experts and assigned to three categories based on how likely the articles were to describe NP gene clusters: “Yes” (high likelihood, 251 articles), “Maybe” (99 articles), “No” (low likelihood, 610 articles). Among those marked “No”, 264 were noted as single enzyme papers, i.e. those that described a single, possibly NP-related, enzyme (such as a cytochrome P450). Other examples of articles marked “No” include whole-genome sequence articles that mentioned a NP in the article text but did not describe a BGC. We observed strong enrichment of ClusterMine360 articles in the “Yes” category (79.0%), while the “Maybe,” “Single Enzyme,” and “not Single Enzyme” categories covered 2.6%, 5.3%, and 13.2% of ClusterMine360, respectively. We selected 275 articles with score > 3 across all categories, or in the “Yes” category with a score > 2 for further full-text curation.
The first-pass curation of the 275 articles was conducted by 19 undergraduate students (junior and senior chemistry and biology majors at Haverford and Bryn Mawr colleges) with aid from the Pubtator (Wei et al., 2013) text-mining tool (http://www.ncbi.nlm.nih.gov/CBBresearch/Lu/Demo/PubTator/). Articles were assigned at random, with each article being assigned to two different students. Curators used a standardized workflow to ensure consistent methodologies. Curators first read the abstract and article to identify all relevant NPs linked to the nucleotide record(s) of interest. For each NP, curators catalogued the name, molecular structure (as a Simplified Molecular-Input Line-Entry System or SMILES string), chemical resource IDs (found on PubChem, ChEMBL, and ChemSpider), and NP classification (non-ribosomal peptide synthetase or NRPS, terpene, polyketide synthase or PKS, hybrid, or other). When a compound was not available in existing databases, curators drew the compound structure in ChemDraw software and extracted the SMILES string (Weininger, 1988). Curators also identified experimental evidence from the article confirming an association between the NP(s) and gene cluster(s). If the NP was a non-ribosomal peptide, curators identified adenylation domain substrate specificities. The link between nucleotide records and NPs was confirmed by searching for the associated nucleotide accession numbers in PubMed. An entry supported by two independent curators was viewed as confirmed. The remaining entries were examined by experts in a separate round of curation and were marked as either confirmed or rejected.
2.3. Confirmation of unique compound names and structures
To obtain the best-matched PubChem Compound Identifiers (CIDs), we converted each SMILES string to the International Chemical Identifier (InChI) key (Heller et al., 2013), and searched for the InChI key in PubChem. If an exact match of the isomeric SMILES (SMILES with isotopic and/or chiral specifications) was located in PubChem, we used the corresponding PubChem CID. Otherwise, we converted isomeric SMILES to non-isomeric canonical SMILES and used the corresponding PubChem CID. We crosschecked the compound names from the student curators with the synonyms from each PubChem entry and resolved conflict by manually re-examining the associated articles. For conflicting isomeric SMILES or CIDs of the same connectivity and compound name, we manually examined the chemical structures and retained only the SMILES with the most complete stereochemical information. Errors in structure or missing stereochemistry were corrected. Multiple conflicting isomeric SMILES bearing different compound names in a single research article were viewed as different compounds and all were retained; for example, aflaquinolone F and aflaquinolone G.
2.4. Chemical informatics analysis of compounds
For a given molecule, multiple valid SMILES strings can accurately represent the chemical structure. To ensure the uniqueness of each compound in our dataset, we converted all SMILES to the canonical SMILES string, which is a unique SMILES string among the many possible strings and is generated by the normalization (canonicalization) algorithm. We then used the canonical SMILES to generate compound fingerprints using the Chemistry Development Kit (Guha, 2007; Steinbeck et al., 2003). The fingerprint of a compound is represented as a bit vector indicating the presence of substructures in the compound. The similarity between two compounds is measured by the Tanimoto coefficient (Nikolova and Jaworska, 2003), defined as, T = a · b/[1 − (1 − a) · (1 − b)], where a · b is the dot product between two fingerprint bit vectors a and b for the two compounds. The Tanimoto coefficient is interpreted as the ratio of shared bits (substructures) to the total bits (substructures) in two compounds. We performed hierarchical clustering of the compounds using the neighbor-joining algorithm with 1 − T as the distance metric. Compound clusters were obtained with a cutoff 0.75 for Tanimoto coefficient. Based on our manual inspection, this provides a proper resolution that groups most compound analogs and intermediates (compounds from the same BGC) together (Table S2).
We compared the chemical similarity and performed hierarchical clustering of curated compounds together with other fungal NPs, which are represented by the ChEBI chemicals under ontology term ChEBI: 76946 (fungal metabolite) (Degtyarenko et al., 2008). An NP in ChEBI with a Tanimoto coefficient of similarity ≥0.75 to any compound in our catalog was considered covered by our catalog.
2.5. AntiSMASH annotation and phylogenetic analysis of biosynthetic genes
Certain types of core enzymes are known to biosynthesize the major classes of fungal NPs, and these enzymes can be readily identified by homology. We identified core enzymes in the nucleotide records from the curated catalog using the set defined by antiSMASH 2.0 (Blin et al., 2013). In an analogous fashion, we identified core enzymes in 581 annotated fungal genomes in GenBank. For phylogenetic analyses of these enzymes (or selected domains therein), we first performed multiple sequence alignment using MAFFT (Katoh, 2002), followed by approximate maximum-likelihood phylogenic inference using FastTree (Price et al., 2010). Phylogenetic trees were visualized in R using the APE package. The PKS tree was rooted at the common ancestor of the mitochondrial ketosynthase (KS) and type I fatty acid synthase (FAS) proteins.
3. Results
3.1. High-quality curation of articles that link fungal nucleotide sequences to natural products
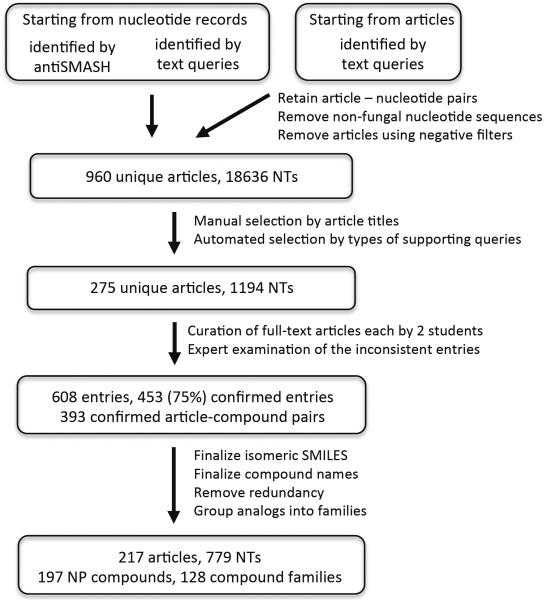
To identify publications that link fungal nucleotide sequences to NPs, we combined multiple query strategies on PubMed article and NCBI nucleotide databases (Figure 1). We identified 960 NP-related articles that were linked to one or more nucleotide records of fungal origin. Of these, we selected 275 articles with a high probability of linking gene sequences to NPs (see Methods for details). Undergraduate students manually curated these 275 full-text articles as part of a class assignment. Each student recorded the NP compound name(s) associated with each article, or “no compound” in cases where there was none. Structures of NPs were recorded as SMILES strings (Weininger, 1988).
Figure 1.
Workflow of article identification and curation for experimentally verified BGCs. NT refers to the nucleotide records in GenBank.
To ensure quality of curation, we randomly assigned each of the 275 articles to two independent student curators (see Methods). An “entry” was defined as an article-compound pair (or article-no-compound pair if there was no compound). The student curation resulted in 608 unique entries. An entry supported by two independent curators was marked as confirmed. The remaining entries were examined by experts in a separate round of curation, and were marked as either confirmed or rejected. 453 (75%) entries were confirmed, while 155 (25%) were rejected. Among the 453 confirmed entries, 393 had SMILES entered, corresponding to 217 articles (see Figure S1 for details). The 155 rejected entries spanned 83 articles, most of which were articles that also contained a confirmed entry. These tended to be compounds assigned by one curator because they had been mentioned in the article, but ultimately rejected because their biosynthesis was not definitively verified.
Stereochemistry is a vital consideration when looking at NP structure, with stereoisomers often possessing distinct biological activities. In some cases, multiple stereoisomers naturally co-occur and are assigned distinct names, e.g. solanapyrone A and solanapyrone D. To distinguish such compounds, we obtained isomeric SMILES when possible.
We compared collated gene clusters in various stages of our pipeline with those in the ClusterMine360 (Conway and Boddy, 2013), a crowd sourced BGC database that was the largest online resource for fungal BGCs available at the study outset. Our original list of 960 articles covers all 38 articles that are linked to 26 fungal nucleotides in ClusterMine360. Subsequent filtering dramatically reduced the total number of articles while retaining the majority of NP-related articles in this database. The set of 275 articles we selected for full text curation covered 33 of the 38 articles in ClusterMine360 (87%, Figure 2A) and 25 of their 26 fungal BGCs (96%, Figure 2C), while our final curation covers 29 (76%) of articles (Figure 2B), and 23 (88%) of the BGCs (Figure 2D).
Figure 2.
Overlaps between curated articles and nucleotide records in this study versus those in the ClusterMine360 database (CM360). The first row (A and B) represents the overlaps at the article level, while the second row (C and D) represents the overlaps at the nucleotide entry level. “Initial Set” (in A and C) includes the 275 articles and associated nucleotide records obtained by semi-automated selection. The “Final curation” set (in B and D) includes the confirmed article-nucleotide pairs from 217 articles, which are annotated with at least one compound.
Although we adopted a strict procedure to ensure curation quality, we still observed errors during manual post-processing, some resulting from errors in the literature itself. For example, curators included a gene cluster for viridiol and viridin from Trichoderma virens (Mukherjee et al., 2006) based on gene expression evidence, but later research suggested that the BGC is responsible for the synthesis of structurally distinct volatile terpene compounds (Crutcher et al., 2013). The former article (Mukherjee et al., 2006) was detected by our text-mining pipeline, and subsequently manually curated as describing viridiol/viridin. Such errors are a consequence of the evolving research.
We identified 10 NPs (5.1% of our catalog) that are currently not covered by PubChem. Among those NPs in PubChem, there are several cases where PubChem entries are incomplete or inaccurate, generally containing the compound structures but lacking the compound names. We also observed two cases where the structures provided in the original articles contained errors, and three cases where the PubChem entries provided the wrong structures. Our final catalog contains 217 articles for 197 unique compounds, which are linked to 779 nucleotide records from 174 species.
3.2. Literature bias towards certain compound families and fungal genera
To create a global view of existing literature on BGCs, we visualized the annotated article-compound relationships as a network between articles and compounds (Figure 3A). The nodes in the graph represent articles or compounds, and the edges represent confirmed article-compound relationships in our catalog.
Figure 3.
Global network view of confirmed article-compound relationships. A. An overview of the network between the articles and compounds. The network contains 393 article compound pairs between 217 articles (purple nodes) and 197 compounds (green nodes). B. The node degree distributions of the article nodes and compound nodes both follow the power-law distributions. C. The sub-network (a connected component) containing T-2 Toxin. D. The sub-network containing Aflatoxins.
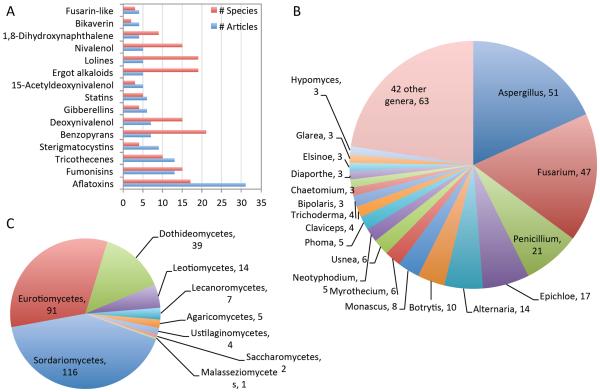
Plotting the node degrees in the log-scale revealed that both the number of compounds per article and the number of research articles per compound follow power-law distributions with exponents approximately equal to two (Figure 3B). According to theories in network biology, such a power-law distribution indicates a `rich-get-richer' circumstance (Barabási, 1999). The power-law distribution of articles per compound suggests a preferential attachment of research activities around popular compounds. For example, the top 15 (11.7%) compound families are described by 38.2% of the articles. Figure 4A shows the most frequently studied compounds. Aflatoxins alone are the focus of 31 BGC-associated research articles (14.3%).
Figure 4.
The 15 most frequently studied compounds (A), and the most frequently studied fungal genera (B) and classes (C) associated with compounds in the curated set of experimentally verified natural product gene clusters. For (A), Closely related compounds are manually merged into compound families. # Articles: number of unique articles covering one or more compounds in each family. # Species: number of unique species origins of the nucleotides associated with these articles.
Similarly, we found that certain fungal genera are disproportionally represented in the literature. For example, 51 of 197 compounds are reported from Aspergilli (26% of the compound set) (Figure 4B). At the class level, only 9 out of 46 fungal classes are covered, while the majority of compounds are produced by the Sordariomycetes, Eurotiomycetes, Dothideomycetes, and Leotiomycetes classes (Figure 4C). We observed a correlation between the number of articles per compound and the number of species producing the compound (Pearson correlation = 0.43, p-value 4 × 10−10 for individual compounds; Pearson correlation = 0.72, p-value 5 × 10−22 for compound families). Presence of a BGC in multiple species could explain why a compound would be studied more, although the reverse could also be true (those more extensively studied tend to be discovered in more species).
3.3. Chemical diversity of natural products in the curated catalog
We sought to understand the chemical diversity of compounds in our curated catalog, compared with a larger set of fungal metabolites in general. For comparison of chemical structures, we converted each compound to a chemical fingerprint representing the presence or absence of a string of substructures (O'Boyle et al., 2011). The Tanimoto coefficient of two such fingerprints reflects the chemical similarity between two compounds. Clustering compounds by these fingerprints revealed products and intermediate compounds from identical and related BGCs (Table S2). For example, one compound cluster includes aflatoxin B1, B2, G1, and G2, and a second compound cluster includes fumonisin C1-C3, B1-B4, fumonisin C1-OH, and isofumonisin C1.
To compare the compounds in our catalog to known fungal metabolites, we used the chemical database ChEBI, which contains a repository of 685 fungal metabolites (Degtyarenko et al., 2008). These include both primary metabolites (such as amino acids) and NPs. We clustered this set of 685 compounds together with compounds in our catalog according to their substructure fingerprints (Figure 5 and Supplemental Figure S5). The NPs in our catalog are spread fairly evenly across the larger set of fungal metabolites. Clades not represented in our catalog include many primary metabolites as well as fungal NPs for which the encoding biosynthetic pathways have not been characterized. For example, the genes encoding the strobilurin fungicides (Clough, 1993) do not appear in our literature-based catalog; these compounds originate from the understudied Basidiomycete phylum of fungi. Overall, 26% of the 685 fungal metabolites are similar to at least one compound in our catalog at a Tanimoto similarity cutoff of 0.75. We note that ChEBI focuses on high quality curation rather than completeness, so not all structurally characterized fungal NPs are present.
Figure 5.
Hierarchical clustering of 685 fungal metabolites in ChEBI and 197 compounds in our catalog (marked yellow).
To further examine the relationship between compound structures and biosynthetic genes, we compared the core enzyme annotation (from antiSMASH 2.0) with the chemical structure-based clustering of compounds (Figure S2). As expected, we observe a strong clustering of compounds according to the associated core-enzyme types. Specifically, compounds synthesized by single types of core enzymes (PKS, NRPS, or terpene cyclase) form separate clusters, while the compounds produced by multiple types of core enzymes (“hybrids”) tend to be more scattered.
3.4. Phyla of origin of nucleotide sequences
Nucleotides in our reference catalog originate primarily from Ascomycetes, especially from the Pezizomycotina subphylum (98%), which contains the filamentous fungi such as Aspergilli. This bias was expected, as most fungal NPs characterized to date have been identified in filamentous fungi. The bias highlights an opportunity for identifying novel NPs and enzyme activities in more diverse species, particularly those that are uncultivable.
Fungal genome sequencing projects are rapidly expanding our knowledge of fungal genetic diversity (Grigoriev et al., 2014). GenBank currently harbors 581 annotated fungal genomes (genomics set), comprising 338 unique species (accessed July 2015). We compared the phylogenetic diversity in our curated catalog with that in the genomics set, finding that the public genomes were more evenly distributed across phyla and subphyla than our catalog, though they were still biased towards Ascomycetes (Table 1).
Table 1.
Taxonomy origins of nucleotide records in curated set and in genomic set. Taxonomy is based on NCBI taxonomy levels 5 and 7 starting from level 1 Eukaryota
| Taxonomy | Fungal nucleotide records in curated set | Fungal genomes in GenBank | |
|---|---|---|---|
| Ascomycetes | Pezizomycotina (filamentous) | 765 (98%) | 253 (43.5%) |
| Schizosaccharomycetes | 0 | 4 (.7%) | |
| Saccharomycotina | 1 (.1%) | 182 (31.3%) | |
| Basidiomycetes | Agaricomycetes (mushrooms) | 9 (1.2%) | 55 (9.5%) |
| Ustilaginomycetes | 3 (.4%) | 10 (1.7%) | |
| Tremellomycetes | 0 | 24 (4.1%) | |
| Malasseziomycetes | 1 (.1%) | 2 (.3%) | |
| Other | 0 | 51 (8.8%) | |
| Total | 779 | 581 | |
3.5. Natural product biosynthetic core enzymes in curated catalog versus genomic sequences
Many known fungal NPs are biosynthesized in part by a core enzyme(s) that can be readily identified by homology, such as the polyketide synthase (PKS) for polyketide pathways and the non-ribosomal peptide synthetase (NRPS) for non-ribosomal peptide pathways. Alkaloid and terpenoid classes are broader, but their subclasses also share certain biosynthetic enzymes.
We searched for core enzymes in the curated set of nucleotide sequences using antiSMASH2.0, and identified five classes of common core enzymes: PKSs, NRPSs, dimethyl allyl tryptophan synthases (DMATSs, involved in the biosynthesis of certain ergot alkaloids), trichodiene synthases (involved in biosynthesis of a type of terpenoid), and geranylgeranyl pyrophosphate synthases (GGPPSs, also terpenoid). To evaluate the potential prevalence of these core enzymes across fungal species, we searched for proteins bearing homology to these five types of core enzymes in the 581 fungal genome sequences in GenBank. We identified 326 instances of these five enzymes in our curated set, and 8,879 instances in the genomics set (Table 2). The large number of enzyme homologs for which the NPs are not known suggests undiscovered chemical diversity.
Table 2.
Types of core enzymes identified in curated nucleotides in the catalog and in 581 fungal genomes in GenBank. Published Clusters: the experimentally verified clusters curated in this study. Predicted clusters: number of clusters predicted based on 581 fungal genomes in GenBank, majority of which are not experimentally verified.
| Core enzyme type | No. core enzymes in published clusters | No. core enzymes in predicted clusters |
|---|---|---|
| Ketosynthase-containing (PKS, FAS) | 127 | 4984 |
| Condensation-containing (NRPS) | 81 | 2983 |
| DMATS (alkaloid) | 44 | 550 |
| Trichodiene synthase (terpene) | 49* | 26 |
| GGPPS (terpene) | 25 | 336 |
| Total | 326 | 8879 |
The number is biased by one article that sequenced 39 homologous gene clusters
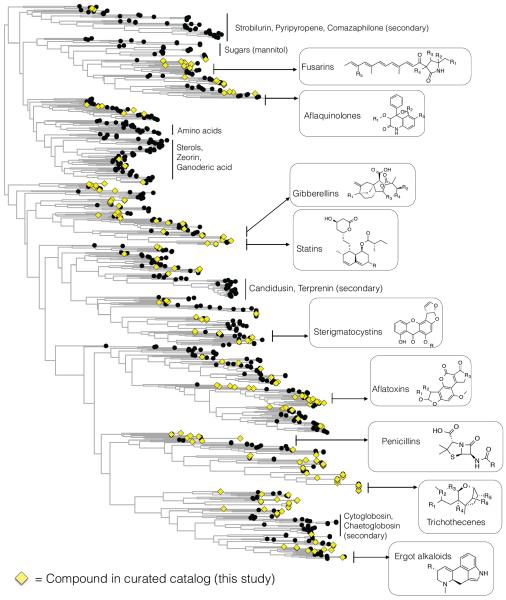
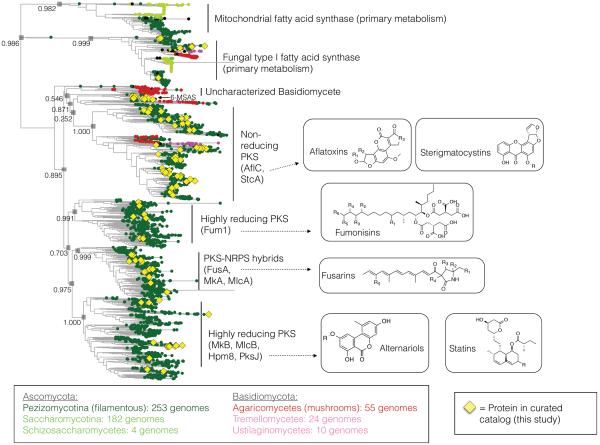
The large number of uncharacterized biosynthetic genes (Table 2) prompted us to investigate the phylogenetic distribution of characterized vs. uncharacterized core enzymes (Figure 6 and Figure S3). Alignment of the ketosynthase (KS) domains of all PKS proteins revealed that proteins generally group by phylum of origin (Figure 6 and Supplemental Figure S6). While many of the major Ascomycete clades harbor at least one characterized enzyme, there are numerous sub-clades that are “orphans,” or harbor no curated enzyme from our catalog. Similar trends were observed in the phylogenies of the other classes of core enzymes (Figure S3): homologs from the Basidiomycete phylum tend to represent uncharacterized clades, and enzymes from these clades may therefore encode novel compounds. Basidiomycetes are underrepresented in the genomics set (Table 1). As more genomes are sequenced, we anticipate that the Basidiomycete clades on the phylogenies (Figure 6) will become better resolved.
Figure 6.
Phylogenetic tree constructed using the KS domains of the PKS core enzymes identified by homology in sequenced fungal genomes from GenBank. Characterized PKSs from this study are highlighted with yellow diamonds. The numerical values associated with the internal nodes are estimated reliability values for the splits generated by the Shimodaira-Hasegawa test.
3.6. Integration with the MIBiG repository
The MIBiG repository, a community annotation of published, experimentally characterized BGCs, was a parallel effort to our catalog (Medema et al., 2015). Comparison of the two datasets indicates they are complementary, as different article selection criteria were used. The fungal entries in MIBiG correspond to 110 articles in total, 65 of which were included in the 275 articles of our final set, leaving 210 articles unique to our curation (Figure S4A). The remaining 45 MIBiG articles were not included in this study either because they did not have a deposited nucleotide record (required for our initial searches) or because they were linked with full genome sequences. Such clusters require software-based cluster predictions to locate in genome sequences, and hence are excluded from the curation pipeline by our filtering strategies. At the biosynthetic gene level (which corresponds to MIBiG entries), 76 BGCs (106 compounds) overlapped between our catalog and MIBiG, including two MIBiG entries that were updated based on our catalog (Figure S4B). 45 BGCs (69 compounds) were unique to our catalog. The remaining 22 entries in our catalog were mostly partial genes or non-core enzyme genes, not included in the MIBiG repository. There are 44 BGCs (46 compounds) in the MIBiG repository that are not in our catalog. 33 of these were linked to whole genome/chromosome sequence records, and hence did not meet our inclusion criteria for article-nucleotide pairs. Nine of the 44 BGCs were not linked with published articles (or were linked with articles not indexed by PubMed). One was linked to an article published after we finalized the article list. Only one BGC was truly missed by our standards (see Table S3 for details).
We converted our catalog into the minimal format of MIBiG and submitted it to the MIBiG repository (see Table S3 for the full list). The combination of this literature-based catalog and MIBiG resulted in 165 unique fungal BGCs with at least one core enzyme fully sequenced. To our knowledge, this represents the largest collection of experimentally validated fungal NP gene clusters to date. The complete resulting reference set of fungal BGCs is available from the MIBiG web page (http://mibig.secondarymetabolites.org) in GenBank, FASTA, and JSON formats.
4. Discussion
For natural product discovery and research to fully benefit from the genomic revolution, it is imperative that existing findings be straightforward to access and analyze in a systematic manner. Recent calls to the community to systematically catalog NP biosynthetic gene-associated metadata will facilitate the systematic deposition of newly-characterized NP gene clusters into databases (Medema et al., 2015). However, there remains a wealth of information spread out over the past few decades of scientific literature that will not be captured by these future efforts. Our work here addresses this challenge, by generating an extensive list of fungal biosynthetic genes published in peer-reviewed articles thus far. Given the scope of the cataloging process and the large number of potential peer-reviewed articles to be curated, we developed a unique approach that involved the computational identification of articles that both (1) discuss fungal metabolite biosynthesis and (2) contain a nucleotide record deposited in NCBI, linked to the article by the authors. We then leveraged the abilities and interests of motivated undergraduate students and experts to collate a comprehensive list of fungal NPs each with at least one associated gene sequence, and further integrated this list into a community repository. We note that our automated article prioritization pipeline was designed to retain only the most promising articles in order to minimize the workload for manual annotation. Although some true BGCs may be missed due to the stringent cutoff, our final curation covers over 88% of the fungal nucleotide entries in ClusterMine360 (see section 3.1) and virtually all of the fungal BGCs in MIBiG that are linked with non-full genome sequence records in GenBank and articles in indexed in PubMed (see section 3.6). In addition, our approach successfully retrieved a large number of BGCs otherwise missed by MIBiG.
The success of our approach is highlighted by the fact that we disclose 197 fungal NPs associated with sequenced genes and identify new areas ripe for investigation. We observed certain biases in experiments on fungal metabolite biosynthesis. First, we noted that certain molecules, such as aflatoxins, dominate the literature, with >10% of articles focused on this single class. Second, certain fungal genera are disproportionally studied, with compounds from Aspergilli alone comprising 25% of the compound set; by contrast, only 3.4% (20/581) of sequenced fungal genomes are from Aspergilli. Third, researchers have only sampled a small portion of the sequence diversity of important core enzymes such as PKSs. There are certain species and clades (e.g., Basidiomycete; Figures 6 and S3) that remain completely uncharacterized and may therefore represent untapped chemical diversity.
Our methods can be applied more generally to collate gene information for secondary metabolites produced by other microorganisms, such as plants and bacteria. Only the initial queries (text and homology) require adjustment; subsequent steps can be directly employed. By including undergraduate students in the curation process, we were able to make rapid progress while also providing students with a unique opportunity to contribute original research to the biosynthetic community. Notably, this challenge inspired several students to pursue postgraduate opportunities in natural product research. Our efforts to curate the history of research on fungal gene-molecule connections thus also helped to cultivate the future of the field. By sharing our positive experience with the community, we hope to inspire others to consider integrating original research challenges into the classroom.
Our collaborative effort yielded a reference catalog linking fungal nucleotide sequence to NP chemical structure, enabling researchers to more efficiently exploit past information to guide future research and discovery. By integrating new entries into the MIBiG data repository in several standardized formats, our data can be readily retrieved and analyzed by others (Medema et al., 2015). Modern genomic approaches to biosynthesis research require a systematic understanding of characterized molecules and biosynthetic genes, particularly in synthetic biology approaches involving comparative analyses of gene sequences. Such efforts include mining genomic DNA for new classes of molecules, testing putative NP gene clusters through heterologous expression, and rationally designing hybrid synthases to produce “unnatural natural products”. In the era of synthetic biology, that efforts like ours, combined with future MIBiG standard-compliant data, will provide fertile ground for future developments in fungal natural product research.
Supplementary Material
Highlights.
Through a close collaboration between a class of undergraduate students and experts in natural product research, we created a catalog of 197 characterized natural products along with associated biosynthetic genes. This catalog can be exploited for future fungal synthetic biology efforts.
We observed bias in the literature on natural product biosynthesis towards well-studied compounds such as aflatoxins, fumonisins, and trichothecenes. Meanwhile, many groups of known fungal natural products and clades of computationally identified biosynthetic genes are neglected.
We integrated our catalog with the MIBiG repository, resulting in a combined reference set of 158 fungal biosynthetic gene clusters with at least the core enzyme sequenced.
Acknowledgements
We thank Yi Tang and Mancheng Tang for helpful discussions. We gratefully acknowledge support from Haverford College, the Research Corporation for Science Advancement Cottrell College Scholars Award (L.K.C), and NIH grant U01 GM110706 (Y.F.L., M.E.H, C.J.B.H).
Abbreviations
- ACE
Abundance-based Coverage Estimators
- BGC
Biosynthetic Gene Cluster
- CI
Confidence Interval
- CID
Compound Identifiers
- FAS
fatty acid synthase
- InChI
International Chemical Identifier
- KS
ketosynthase
- MIBiG
Minimum Information about Biosynthetic Gene cluster
- NP
Natural Product
- NRPS
non-ribosomal peptide synthetase
- PKS
polyketide synthase
- SMILES
Simplified Molecular-Input Line-Entry System
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amare MG, Keller NP. Molecular mechanisms of Aspergillus flavus secondary metabolism and development. Fungal Genet. Biol. 2014;66:11–8. doi: 10.1016/j.fgb.2014.02.008. doi:10.1016/j.fgb.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Barabási A. Emergence of Scaling in Random Networks. Science (80-.) 1999;286:509–512. doi: 10.1126/science.286.5439.509. doi:10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Blackwell M. The fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 2011;98:426–38. doi: 10.3732/ajb.1000298. doi:10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- Blin K, Medema MH, Kazempour D, Fischbach M. a, Breitling R, Takano E, Weber T. antiSMASH 2.0--a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–12. doi: 10.1093/nar/gkt449. doi:10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhired S, Weber M, Kempf-Sontag A, Keller NP, Hoffmeister D. Accurate prediction of the Aspergillus nidulans terrequinone gene cluster boundaries using the transcriptional regulator LaeA. Fungal Genet. Biol. 2007;44:1134–1145. doi: 10.1016/j.fgb.2006.12.010. doi:10.1016/j.fgb.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Cerqueira GC, Arnaud MB, Inglis DO, Skrzypek MS, Binkley G, Simison M, Miyasato SR, Binkley J, Orvis J, Shah P, Wymore F, Sherlock G, Wortman JR. The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014;42:D705–10. doi: 10.1093/nar/gkt1029. doi:10.1093/nar/gkt1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S-L, Chiang Y-M, Yeh H-H, Wu T-K, Wang CCC. Reconstitution of the early steps of gliotoxin biosynthesis in Aspergillus nidulans reveals the role of the monooxygenase GliC. Bioorg. Med. Chem. Lett. 2013;23:2155–7. doi: 10.1016/j.bmcl.2013.01.099. doi:10.1016/j.bmcl.2013.01.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984:265–270. [Google Scholar]
- Chao A, Lee S-M. Estimating the Number of Classes via Sanple Coverage. J. Am. Stat. Assoc. 1992 [Google Scholar]
- Charlop-Powers Z, Owen JG, Reddy BVB, Ternei MA, Brady SF. Chemical-biogeographic survey of secondary metabolism in soil. Proc. Natl. Acad. Sci. U. S. A. 2014;111:3757–62. doi: 10.1073/pnas.1318021111. doi:10.1073/pnas.1318021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlop-Powers Z, Owen JG, Reddy BVB, Ternei MA, Guimarães DO, de Frias UA, Pupo MT, Seepe P, Feng Z, Brady SF. Global biogeographic sampling of bacterial secondary metabolism. Elife. 2015;4:e05048. doi: 10.7554/eLife.05048. doi:10.7554/eLife.05048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, Fischbach MA. Insights into Secondary Metabolism from a Global Analysis of Prokaryotic Biosynthetic Gene Clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. doi:10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough JM. The Strobilurins, Oudemansins, and Myxothiazols, Fungicidal Derivatives of B-Methoxyacrylic Acid. Nat. Prod. Rep. 1993;10:565–574. doi: 10.1039/np9931000565. [DOI] [PubMed] [Google Scholar]
- Conway KR, Boddy CN. ClusterMine360: A database of microbial PKS/NRPS biosynthesis. Nucleic Acids Res. 2013;41:402–407. doi: 10.1093/nar/gks993. doi:10.1093/nar/gks993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crutcher FK, Parich A, Schuhmacher R, Mukherjee PK, Zeilinger S, Kenerley CM. A putative terpene cyclase, vir4, is responsible for the biosynthesis of volatile terpene compounds in the biocontrol fungus Trichoderma virens. Fungal Genet. Biol. 2013;56:67–77. doi: 10.1016/j.fgb.2013.05.003. doi:10.1016/j.fgb.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcántara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–50. doi: 10.1093/nar/gkm791. doi:10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, Fischbach MA. A Systematic Analysis of Biosynthetic Gene Clusters in the Human Microbiome Reveals a Common Family of Antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. doi:10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroghazi JR, Albright JC, Goering AW, Ju K-S, Haines RR, Tchalukov KA, Labeda DP, Kelleher NL, Metcalf WW. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 2014;10:963–8. doi: 10.1038/nchembio.1659. doi:10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–7. doi: 10.1093/nar/gkr777. doi:10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42:D699–704. doi: 10.1093/nar/gkt1183. doi:10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha R. Chemical Informatics Functionality in R. J. Stat. Softw. 2007;18:1–16. doi:10.1109/LPT.2009.2020494. [Google Scholar]
- Heller S, McNaught A, Stein S, Tchekhovskoi D, Pletnev I. InChI - The worldwide chemical structure identifier standard. J. Cheminform. 2013;5:1. doi: 10.1186/1758-2946-5-7. doi:10.1186/1758-2946-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan MN, Yakasai AA, Halo LM, Song Z, Bailey AM, Simpson TJ, Cox RJ, Lazarus CM. First heterologous reconstruction of a complete functional fungal biosynthetic multigene cluster. Chembiochem. 2010;11:1508–12. doi: 10.1002/cbic.201000259. doi:10.1002/cbic.201000259. [DOI] [PubMed] [Google Scholar]
- Hoffmeister D, Keller NP. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat. Prod. Rep. 2007;24:393–416. doi: 10.1039/b603084j. doi:10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- Hong S-H, Bunge J, Jeon S-O, Epstein SS. Predicting microbial species richness. Proc. Natl. Acad. Sci. U. S. A. 2006;103:117–122. doi: 10.1073/pnas.0507245102. doi:10.1073/pnas.0507245102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa N, Sasagawa M, Yamamoto M, Komaki H, Yoshida Y, Yamazaki S, Fujita N. DoBISCUIT: a database of secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2013;41:D408–14. doi: 10.1093/nar/gks1177. doi:10.1093/nar/gks1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis DO, Binkley J, Skrzypek MS, Arnaud MB, Cerqueira GC, Shah P, Wymore F, Wortman JR, Sherlock G. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 2013;13:91. doi: 10.1186/1471-2180-13-91. doi:10.1186/1471-2180-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. doi:10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kealey JT, Liu L, Santi DV, Betlach MC, Barr PJ. Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc. Natl. Acad. Sci. U. S. A. 1998;95:505–9. doi: 10.1073/pnas.95.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller NP, Turner G, Bennett JW. Fungal secondary metabolism - from biochemistry to genomics. Nat. Rev. Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. doi:10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- Lim FY, Sanchez JF, Wang CCC, Keller NP. Methods in Enzymology. 1st ed. Elsevier Inc.; 2012. Toward awakening cryptic secondary metabolite gene clusters in filamentous fungi. doi:10.1016/B978-0-12-404634-4.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X-M, Xu W, Li D, Yin W-B, Chooi Y-H, Li Y-Q, Tang Y, Hu Y. Epigenetic Genome Mining of an Endophytic Fungus Leads to the Pleiotropic Biosynthesis of Natural Products. Angew. Chemie. 2015;127:7702–7706. doi: 10.1002/anie.201502452. doi:10.1002/ange.201502452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattern DJ, Valiante V, Unkles SE, Brakhage AA. Synthetic biology of fungal natural products. Front. Microbiol. 2015;6:775. doi: 10.3389/fmicb.2015.00775. doi:10.3389/fmicb.2015.00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach M. a, Weber T, Takano E, Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–46. doi: 10.1093/nar/gkr466. doi:10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Fischbach MA. Computational approaches to natural product discovery. Nat. Chem. Biol. 2015;11:639–648. doi: 10.1038/nchembio.1884. doi:10.1038/nchembio.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Chooi YH, Claesen J, Coates RC, Cruz-Morales P, Duddela S, Düsterhus S, Edwards DJ, Fewer DP, Garg N, Geiger C, Gomez-Escribano JP, Greule A, Hadjithomas M, Haines AS, Helfrich EJN, Hillwig ML, Ishida K, Jones AC, Jones CS, Jungmann K, Kegler C, Kim HU, Kötter P, Krug D, Masschelein J, Melnik AV, Mantovani SM, Monroe E. a, Moore M, Moss N, Nützmann H-W, Pan G, Pati A, Petras D, Reen FJ, Rosconi F, Rui Z, Tian Z, Tobias NJ, Tsunematsu Y, Wiemann P, Wyckoff E, Yan X, Yim G, Yu F, Xie Y, Aigle B, Apel AK, Balibar CJ, Balskus EP, Barona-Gómez F, Bechthold A, Bode HB, Borriss R, Brady SF, Brakhage A. a, Caffrey P, Cheng Y-Q, Clardy J, Cox RJ, De Mot R, Donadio S, Donia MS, van der Donk W. a, Dorrestein PC, Doyle S, Driessen AJM, Ehling-Schulz M, Entian K-D, Fischbach M. a, Gerwick L, Gerwick WH, Gross H, Gust B, Hertweck C, Höfte M, Jensen SE, Ju J, Katz L, Kaysser L, Klassen JL, Keller NP, Kormanec J, Kuipers OP, Kuzuyama T, Kyrpides NC, Kwon H-J, Lautru S, Lavigne R, Lee CY, Linquan B, Liu X, Liu W, Luzhetskyy A, Mahmud T, Mast Y, Méndez C, Metsä-Ketelä M, Micklefield J, Mitchell D. a, Moore BS, Moreira LM, Müller R, Neilan B. a, Nett M, Nielsen J, O'Gara F, Oikawa H, Osbourn A, Osburne MS, Ostash B, Payne SM, Pernodet J-L, Petricek M, Piel J, Ploux O, Raaijmakers JM, Salas J. a, Schmitt EK, Scott B, Seipke RF, Shen B, Sherman DH, Sivonen K, Smanski MJ, Sosio M, Stegmann E, Süssmuth RD, Tahlan K, Thomas CM, Tang Y, Truman AW, Viaud M, Walton JD, Walsh CT, Weber T, van Wezel GP, Wilkinson B, Willey JM, Wohlleben W, Wright GD, Ziemert N, Zhang C, Zotchev SB, Breitling R, Takano E, Glöckner FO. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. doi:10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee M, Horwitz BA, Sherkhane PD, Hadar R, Mukherjee PK. A secondary metabolite biosynthesis cluster in Trichoderma virens: evidence from analysis of genes underexpressed in a mutant defective in morphogenesis and antibiotic production. Curr. Genet. 2006;50:193–202. doi: 10.1007/s00294-006-0075-0. doi:10.1007/s00294-006-0075-0. [DOI] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 2007;70:461–77. doi: 10.1021/np068054v. doi:10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- Nikolova N, Jaworska J. Approaches to Measure Chemical Similarity– a Review. QSAR Comb. Sci. 2003;22:1006–1026. doi:10.1002/qsar.200330831. [Google Scholar]
- O'Boyle NM, Banck M, James C. a., Morley C, Vandermeersch T, Hutchison GR. Open Babel: An Open chemical toolbox. J. Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. doi:10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien RV, Davis RW, Khosla C, Hillenmeyer ME. Computational identification and analysis of orphan assembly-line polyketide synthases. J. Antibiot. (Tokyo) 2014;67:89–97. doi: 10.1038/ja.2013.125. doi:10.1038/ja.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H, et al. Package “vegan.”. 2015. [Google Scholar]
- Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JAE, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EGJ, Debets AJM, Dekker P, van Dijck PWM, van Dijk A, Dijkhuizen L, Driessen AJM, d'Enfert C, Geysens S, Goosen C, Groot GSP, de Groot PWJ, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JPTW, van den Hondel CAMJJ, van der Heijden RTJM, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJEC, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NNME, Ram AFJ, Rinas U, Roubos JA, Sagt CMJ, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, van de Vondervoort PJJ, Wedler H, Wösten HAB, Zeng A-P, van Ooyen AJJ, Visser J, Stam H. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 2007;25:221–31. doi: 10.1038/nbt1282. doi:10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- Pence HE, Williams A. ChemSpider: An Online Chemical Information Resource. J. Chem. Educ. 2010;87:1123–1124. doi:10.1021/ed100697w. [Google Scholar]
- Pi B, Yu D, Dai F, Song X, Zhu C, Li H, Yu Y. A genomics based discovery of secondary metabolite biosynthetic gene clusters in Aspergillus ustus. PLoS One. 2015;10:e0116089. doi: 10.1371/journal.pone.0116089. doi:10.1371/journal.pone.0116089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. doi:10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing. 2014. [Google Scholar]
- Richter L, Wanka F, Boecker S, Storm D, Kurt T, Vural Ö, Süßmuth R, Meyer V. Engineering of Aspergillus niger for the production of secondary metabolites. Fungal Biol. Biotechnol. 2014;1:1–13. doi: 10.1186/s40694-014-0004-9. doi:10.1186/s40694-014-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K, Kinoshita H, Nihira T. Heterologous expression system in Aspergillus oryzae for fungal biosynthetic gene clusters of secondary metabolites. Appl. Microbiol. Biotechnol. 2012;93:2011–22. doi: 10.1007/s00253-011-3657-9. doi:10.1007/s00253-011-3657-9. [DOI] [PubMed] [Google Scholar]
- Sakai K, Kinoshita H, Shimizu T, Nihira T. Construction of a citrinin gene cluster expression system in heterologous Aspergillus oryzae. J. Biosci. Bioeng. 2008;106:466–72. doi: 10.1263/jbb.106.466. doi:10.1263/jbb.106.466. [DOI] [PubMed] [Google Scholar]
- Smanski MJ, Schlatter DC, Kinkel LL. Leveraging ecological theory to guide natural product discovery. J. Ind. Microbiol. Biotechnol. 2015 doi: 10.1007/s10295-015-1683-9. doi:10.1007/s10295-015-1683-9. [DOI] [PubMed] [Google Scholar]
- Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS, Kissinger JC, Li W, Nayak V, Pinney DF, Stoeckert CJ, Roos DS. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res. 2012;40:D675–81. doi: 10.1093/nar/gkr918. doi:10.1093/nar/gkr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbeck C, Han Y, Kuhn S, Horlacher O, Luttmann E, Willighagen E. The Chemistry Development Kit (CDK): an open-source Java library for Chemo- and Bioinformatics. J. Chem. Inf. Comput. Sci. 2003;43:493–500. doi: 10.1021/ci025584y. doi:10.1021/ci025584y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throckmorton K, Wiemann P, Keller NP. Evolution of Chemical Diversity in a Group of Non-Reduced Polyketide Gene Clusters: Using Phylogenetics to Inform the Search for Novel Fungal Natural Products. Toxins (Basel) 2015;7:3572–607. doi: 10.3390/toxins7093572. doi:10.3390/toxins7093572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unkles SE, Valiante V, Mattern DJ, Brakhage AA. Synthetic biology tools for bioprospecting of natural products in eukaryotes. Chem. Biol. 2014;21:502–8. doi: 10.1016/j.chembiol.2014.02.010. doi:10.1016/j.chembiol.2014.02.010. [DOI] [PubMed] [Google Scholar]
- Walsh CT, Fischbach MA. Natural products version 2.0: connecting genes to molecules. J. Am. Chem. Soc. 2010;132:2469–93. doi: 10.1021/ja909118a. doi:10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–33. doi: 10.1093/nar/gkp456. doi:10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CH, Kao HY, Lu Z. PubTator: a web-based text mining tool for assisting biocuration. Nucleic Acids Res. 2013;41:518–522. doi: 10.1093/nar/gkt441. doi:10.1093/nar/gkt441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Model. 1988;28:31–36. doi:10.1021/ci00057a005. [Google Scholar]
- Wiemann P, Keller NP. Strategies for mining fungal natural products. J. Ind. Microbiol. Biotechnol. 2014;41:301–13. doi: 10.1007/s10295-013-1366-3. doi:10.1007/s10295-013-1366-3. [DOI] [PubMed] [Google Scholar]
- Yin W-B, Chooi YH, Smith AR, Cacho RA, Hu Y, White TC, Tang Y. Discovery of cryptic polyketide metabolites from dermatophytes using heterologous expression in Aspergillus nidulans. ACS Synth. Biol. 2013;2:629–34. doi: 10.1021/sb400048b. doi:10.1021/sb400048b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.