Abstract
Insertions and deletions (indels) are important sequence variants that are considered as phylogenetic markers that reflect evolutionary adaptations in different species. In an effort to systematically study indels specific to the phylum Nematoda and their structural impact on the proteins bearing them, we examined over 340,000 polypeptides from 21 nematode species spanning the phylum, compared them to non-nematodes and identified indels unique to nematode proteins in more than 3,000 protein families. Examination of the amino acid composition revealed uneven usage of amino acids for insertions and deletions. The amino acid composition and cost, along with the secondary structure constitution of the indels, were analyzed in the context of their biological pathway associations. Species-specific indels could enable indel-based targeting for drug design in pathogens/parasites. Therefore, we screened the spatial locations of the indels in the parasite’s protein 3D structures, determined the location of the indel and identified potential unique drug targeting sites. These indels could be confirmed by RNA-Seq data. Examples are presented that illustrate the close proximity of the indel to established small-molecule binding pockets that can potentially facilitate selective targeting to the parasites and bypassing their host, thus reducing or eliminating the toxicity of the potential drugs. The study presents an approach for understanding the adaptation of pathogens/parasites at a molecular level, and outlines a strategy to identify such nematode-selective targets that remain essential to the organism. With further experimental characterization and validation, it opens a possible channel for the development of novel treatments with high target specificity, addressing both host toxicity and resistance concerns.
Keywords: insertions and deletions, adaptation, spatial structures of proteins, drug targets, selective targeting, parasites, nematodes
1. Introduction
The phylum Nematoda is one of the largest and most diverse phyla on the planet. At least 25,000 distinct nematodes species have been described and it is estimated that the actual species count may go well into the millions (Hugot et al., 2001). Members of this phylum are found in hot springs, polar ice, and almost everywhere in between, and the lifestyles of these organisms vary from free-living to parasitic organisms (which are found in plants, vertebrates, insects, and even other nematodes). Plant and animal parasitic nematodes are of special concern because of their detrimental effect on the economy and global health. It is estimated by the WHO that 2.9 billion people are infected with parasitic nematodes (Hotez et al., 2007). In addition parasitic nematodes cost the agricultural industry more than $80 billion per year in crop treatment and lost product (Nicol et al., 2011). Currently anthelminthic drugs utilized to treat and prevent nematode infections are becoming less effective as drug resistance increases among populations (e.g., (Wolstenholme et al., 2004; Wrigley et al., 2006)). As resistance increases, drugs with novel mechanisms of action and/or alternate therapeutic approaches for control are needed to combat these parasites.
In the past decade, fast-evolving DNA and RNA sequencing technology has greatly enriched our understanding of many organisms (including many nematodes) from a genomic perspective. The rapid growth of genome information for nematodes has led to many in-depth studies of their genetics, genomics and functional evolution (Brindley et al., 2009; Dieterich and Sommer, 2009; Mitreva et al., 2007; Sommer and Streit, 2011). This genomic data can also be exploited to better understand parasite adaptations at a molecular level, and to facilitate the pursuit of novel treatments for prevention and/or control. Parasite genes or proteins are often examined in terms of their potential to serve as targets of new treatments for parasite control. There are two main groups of proteins that can be exploited for these purposes: i) proteins that are specific to the parasite or ii) proteins that are highly homologous between the parasite and the host, but have diverged sufficiently to enable selective targeting in the parasite. These two groups of potential targets are non-overlapping and potentially provide promising targets for the development of drugs with low toxicity to the host.
Previous studies have examined drug targets unique to the target organism in order to minimize or eliminate toxic effects to the host (e.g. (Galperin and Koonin, 1999)), but if conserved (i.e. non-unique) essential proteins are eliminated from the target pool, then only a small fraction of the proteins are left for further exploration. For example, in a study of Bacillus subtilis, 96% of essential genes were found to be conserved in other bacteria and nearly 70% were found to be conserved in Archaea and Eukaryotes (Kobayashi et al., 2003). This indicates that if only the proteins that are unique to B. subtilis are examined for potential drugs, most of the proteome would need to be excluded. The majority of the nematode-specific proteins remain so distinct from host proteins that extrapolation of distant homology based on protein folds can only be used to infer putative functions for ~10% of the novel proteins (Yin et al., 2009). Therefore, selecting nematode-specific proteins as drug targets requires extensive experimental characterizations of their functions. This is also reflected in the fact that few of the current anthelmintics are targeted against species-specific or nematode-specific proteins.
On the other hand, proteins that are essential and conserved in multiple species are likely to be involved in core cellular processes (Kobayashi et al., 2003). A set of 458 core proteins shared among most eukaryotes has been previously defined (Parra et al., 2007), and these could prove to be more effective targets than species-unique proteins. However, unless differentiated regions are identified in order to facilitate specific targeting, there is a possibility of high toxicity to the host. The differential regions within these proteins can range from single amino acid changes to the insertion or deletion (indels) of multiple amino acids (Thorne, 2000). Indels have been shown to have a greater effect on protein structure and function than single amino acid changes that result from substitutions (Hormozdiari et al., 2009; Salari et al., 2008), and can also create a unique ligand binding site on the protein surface (Studer et al., 2013). It has been shown that indels rarely affect the structural scaffold of a protein, but much more often alter peripheral elements (Studer et al., 2013), which may lead to changes in binding sites that facilitate specific ligand binding.
A study (Wang et al., 2009) identified important roles of indels in nematode adaptation, but the focus was on the relevance of the indels for evolutionary adaptations, so many aspects related to the structural impact of the indels were not investigated. Comparisons of the homologous protein structures in proteins in the Protein Data Bank, has shown that the location of indels in a protein occur in a non-random manner; specifically, they tend to be located in loop regions more frequently than elsewhere (Fechteler et al., 1995). In one study up to 85% of indels were found in coiled regions of proteins (Pascarella and Argos, 1992). Indels have also been shown to vary in composition from other sections of proteins (Hsing and Cherkasov, 2008), and tend to be enriched in amino acids with small side chains and flanked by highly structured regions (Wrabl and Grishin, 2004). In a large scale study of bacterial/human homologs, sizeable indels were shown to exist in 5–10% of bacterial proteins with human homologs, and this number is even larger (~25%) for some protozoan pathogens (Cherkasov et al., 2006). Study including model species (Bacillus subtilis, Escherichia coli, and Saccharomyces cerevisiae), with available high-quality protein essentiality data, has shown that indels are more prevalent in essential than non-essential proteins (Chan et al., 2007).
By utilizing information about indels, it is possible to rationally design a ligand/drug that specifically targets a conserved protein in one organism without interfering with its homologue in another species by binding at the unique site (“indel targeting” (Cherkasov et al., 2005)). Indel targeting was successfully performed for the elongation factor 1-α (EF1-α) protein in the pathogen Leishmania donovani, a virulence factor that allows the intracellular pathogen to persist in the human macrophage (Nandan et al., 2007). EF-1α has greater than 80% sequence identity with its human homolog, but a 12 amino acid deletion in the L. donovani ligand binding site was exploited to design small molecules that selectively bind to L. donovani EF-1α. Other more recent reports have also explored sequence diversification among host and pathogen homologous proteins and studied their therapeutic potential (e.g. (Fox et al., 2009; Jansen et al., 2013; Kerr et al., 2010; Urbaniak et al., 2013; Wang et al., 2012)).
In this report, we present a systematic approach to identifying novel drug targets that leverages existing sequence information to expand and improve our knowledge of proteins bearing nematode-specific indels. The study is based on sequence data from 21 different nematode species (over 340,000 polypeptides derived from whole genome sequencing), and investigates nematode-specific indels in nematode proteins and their underlying biological function, amino acid composition, cost, druggability, and location in the spatial structure. We present a few cases that demonstrate whether specific inhibitions could be achieved by selectively targeting the indel regions on the protein structures. Our methodical evaluation and results provide key information useful for the selection of proteins to examine further as new drug targets, based on indel-selective targeting.
2. Methods
2.1. Protein datasets, building protein families and their classification and characterization
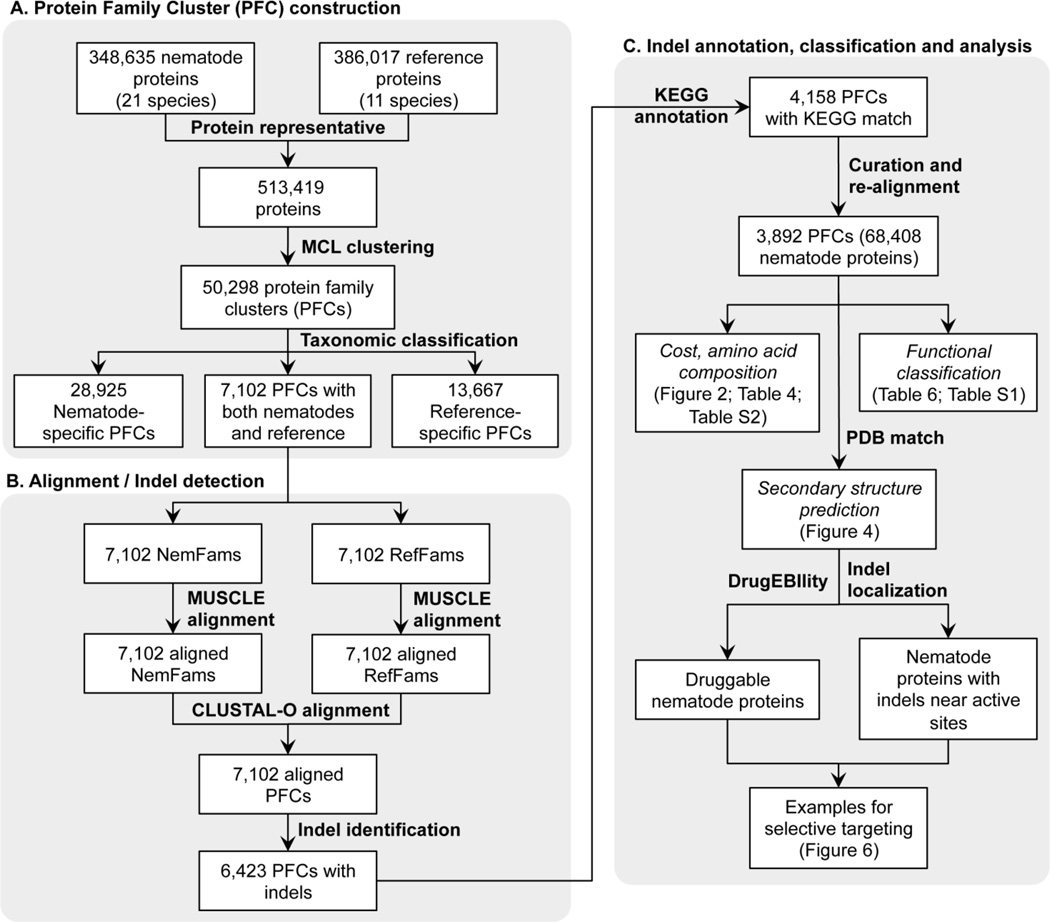
The workflow for the systematic analysis in this study is shown in Figure 1. Protein sequences from 21 nematodes (both parasitic and non-parasitic) were examined. The complete proteome datasets comprises 348,635 proteins generated in nematode genome-sequencing projects (http://nematode.net (Martin et al., 2015), and Wormbase-Parasite (Howe et al., 2015), Table 1). They were compared with 386,017 proteins from 11 outgroup or host species. For each species, isoforms of these protein sequences were examined against the coding genes, and only the longest were kept when applicable. Protein families (orthologous groups) were defined utilizing the Markov cluster algorithm (Enright et al., 2002) using the OrthoMCL package (Fischer et al., 2011; Li et al., 2003) with an inflation factor 1.5, based on the final proteome datasets. Each protein family consists of at least two proteins from one or more species. Among them, the final dataset for nematode specific indel analysis were those protein family clusters (PFC) containing both NemFams (having sequences from at least 1 nematode species) and RefFams (having at least 1 non-nematode homologs). The NemFams and RefFams within each PFC were then split for the sequence alignments as discussed below.
Fig. 1.
Systematic identification, analysis and evaluation of nematode specific indels.
Table 1.
The species and datasets used in this study.
| Reference Species | Common name/Host |
Source* | # Proteins |
Longest isoform |
|
|---|---|---|---|---|---|
| Outgroup | Candida dubliniensis | yeast | Genbank (NCBI v106) | 5,860 | - |
| Candida glabrata | yeast | Genbank (NCBI v106) | 5,213 | - | |
| Saccharomyces cerevisiae | yeast | Ensembl release 77 | 6,692 | - | |
| Drosophila melanogaster | fruit fly | Ensembl release 77 | 26,950 | 13,937 | |
| Hosts | Homo sapiens | human | Genbank (NCBI v106) | 71,338 | 20,190 |
| Canis familiaris | dog | Genbank (NCBI v103) | 42,467 | 19,877 | |
| Mus musculus | mouse | Genbank (NCBI v104) | 77,623 | 23,167 | |
| Bos taurus | cow | Genbank (NCBI v103) | 63,118 | 23,116 | |
| Ovis aries | sheep | Genbank (NCBI v100) | 22,692 | 19,108 | |
| Sus scrofa | pig | Genbank (NCBI v104) | 38,230 | 22,917 | |
| Mesocricetus auratus | hampster | Genbank (NCBI v100) | 25,834 | 19,305 | |
| Nematodes | Trichuris suis | human, pig | Nematode.net** | 9,832 | - |
| Trichuris muris | human, mouse | WormBase ParaSite | 11,004 | - | |
| Trichuris trichiura | human | WormBase ParaSite | 9,856 | - | |
| Trichinella spiralis | human | Nematode.net | 16,380 | - | |
| Ascaris suum | human, pig | Wormbase WS238 | 18,542 | - | |
| Ascaris lumbricoides | human | WormBase ParaSite** | 23,604 | - | |
| Brugia malayi | human | Wormbase WS238 | 17,959 | 14,216 | |
| Brugia pahangi | dog | WormBase ParaSite** | 14,747 | - | |
| Loa loa | human | Wormbase WS238 | 15,445 | - | |
| Wuchereria bancrofti | human | WormBase ParaSite** | 13,134 | - | |
| Onchocerca volvulus | human | Wormbase WS241 | 12,994 | - | |
| Onchocerca ochengi | human | WormBase ParaSite** | 13,990 | - | |
| Strongyloides ratti | human | Wormbase WS238 | 8,188 | - | |
| Strongyloides stercoralis | human | WormBase ParaSite** | 14,490 | 13,114 | |
| Caenorhabditis briggsae | non-parasite | Wormbase WS238 | 21,961 | - | |
| Caenorhabditis elegans | non-parasite | Wormbase WS241 | 26,983 | 20,493 | |
| Dictyocaulus viviparus | cow | Nematode.net** | 14,171 | - | |
| Teladorsagia circumcincta | sheep | Nematode.net** | 25,535 | - | |
| Haemonchus contortus | sheep | Wormbase WS241 | 24,775 | 21,897 | |
| Necator Americanus | human, hampster | Nematode.net | 19,153 | - | |
| Ancylostoma ceylanicum | human | Nematode.net** | 15,892 | - |
Genbank (Benson et al., 2015), Ensembl (Cunningham et al., 2015), WormBase ParaSite (http://parasite.wormbase.org), and Wormbase (Harris et al., 2014), Nematode.net (Martin et al, 2015; http://nematode.net/).
Unpublished and obtained from the referenced database
2.2. Multiple sequence alignment and indel detection
The whole alignment and indel detection process was done following published protocol (Wang et al., 2009). Briefly, aligning the NemFam sequences with the RefFam sequences was a multi-step process. Within each PFC, the NemFam and RefFam sequences were first each aligned using MUSCLE (Edgar, 2004). Before performing the alignment with the NemFam sequences, any RefFam sequence in a RefFam group that deviated from the mean length by more than 30% was removed. Once the alignments of the RefFam/NemFam sequences were complete, they were then combined again and aligned using the profile alignment function of CLUSTAL-Omega (Sievers et al., 2011). After the profile alignment, the NemFam and RefFam sequences were again split into separate files and the NemFam sequences were further curated to improve alignment and reduce redundancy. Exons in nematodes tend not to exceed 100 AA in length (average length), thus any gap larger than 2 times the average length (200 AA) was removed from consideration as it likely to be an artifact due to disparity in polypeptide and protein size. Automated sequence alignment programs sometimes return alignments containing stretches of gaps intervened by very short stretches of AA sequences. For easier data analysis, they were combined into a single, long stretch of gap sequence. The flanking regions (10 AA upstream and downstream) of each reported gap were then examined, and only gaps with flanking regions that were comprised of at least 10 total AA (i.e., 50%) were kept. Other gaps were combined with their peripheral gaps into a single gap for downstream analysis (Figure 1). Additionally sequences that exhibited a poor alignment were removed. Any sequence that had a maximum pairwise percent identity less than 10% of the average percent identity of the entire alignment or less than 0.34 fraction of length was removed. Alignments were rerun for the PFCs with erroneous sequences. The resulting improved alignments were used for insertion and deletion detection as previously described (Wang et al., 2009). For the purposes of this study, a gap absent from the RefFam sequences was recorded as being a ‘nematode specific deletion’, while gaps present only in the RefFam sequences were recorded as a ‘nematode specific insertion’. A gap was determined to be ‘shared’ in sequences within a multiple alignment if the gap overlapped by more than one third of their total length or more than half of any individual gap. The length of a shared gap is the average length of the member deletion. A ‘background’ sequence is defined as the areas of the protein alignments not containing gaps.
2.3. Associating insertions and deletions to cellular pathways
For each PFC, all the protein sequences were screened against the KEGG database v70.0 (Kanehisa and Goto, 2000) to associate them with functions and corresponding pathways using KEGGscan (Wylie et al., 2008) (Table S1). The associated KEGG Orthology pathways (KOs) for each PFC were assigned based on the KOs of all the protein sequences, in a step-wise approach similarly as previously reported (Wang et al., 2015). Each PFC was then assigned into one of the five major KEGG categories (Metabolism, Environmental information processing, Cellular processes, Genetic information processing and Organismal systems) and their subcategories based on the pathways it participates. If one PFC participates more than one pathways falling into multiple KEGG (sub)categories, the subcategory with the most KO association is assigned as the final subcategory the PFC belongs to. The total numbers of indels possessed by the proteins with associated KEGG categories and the mean number of families associated with each pathway and pathway category were then calculated (indel rate, total indel/total family members).
2.4. Analytical processing and mapping of the RNA-Seq reads
RNA-Seq reads of Brugia malayi across multiple life-cycle stages were obtained from previous published work (Choi et al., 2011) and downloaded from Array Express (http://www.ebi.ac.uk/arrayexpress/, accession number E-MTAB-811). Analytical processing of the Illumina short-reads was performed using in-house scripts to filter out regions of low compositional complexity and to convert them into Ns. Subsequently Ns were removed and reads were discarded without at least 25 bases of non-N sequence. Contamination screening was also carried out to filter out standard contaminants (bacteria, human and ribosomes). Gene expression for each sample was calculated by mapping the screened RNA-Seq reads to the whole genomic DNA sequences using Tophat2 (Kim et al., 2013) (version 2.0.8) and calculating depth and breadth of coverage per gene using Refcov (version 0.3, http://gmt.genome.wustl.edu/gmt-refcov/).
2.5. Composition and cost
The amino acid composition and cost, and the underlying structure of inserted, deleted, shared and background sequences were determined as follows. Sequences present in both NemFam and RemFam groups are defined as ‘background’. Regions with gaps in the sequence alignment for a subset of NemFam sequences as well as a subset of RefFam sequences were annotated as ‘shared’. Insertions and deletions for specific nematode sequences were compared to those of the rest of the NemFam group as well as the associated RemFam sequences.
Eight different methods to estimate amino acid biosynthetic cost (Barton et al., 2010; Craig and Weber, 1998; Heizer et al., 2006; Seligmann, 2003; Wagner, 2005) were used to estimate the difference in synthetic cost resulting from the indels in the study. The average cost was calculated by summing the cost of the individual amino acids in a position and dividing by the total number of amino acids. Amino acid composition was determined by counting the percentage of a specific amino acid appeared in a sequence.
2.6. Parasitic nematode specific indels and their structure
The nematode species included in this study represented parasitic and non-parasitic nematodes, including both animal and human parasitic nematodes. Parasitic nematode proteins containing indels were aligned with known tertiary structures from the PDB using BLAST (threshold 1e-05, 35% identity at over 50% fraction of length) to 300,191 sequences (including multiple chains for a single PDB) to identify homologs. Secondary structure annotations were downloaded from RCSB PDB (Joosten et al., 2011) as annotated by DSSP (Kabsch and Sander, 1983).The druggability of each PDB structure was assessed using the ChEMBL DrugEBllity portal (Bento et al., 2014; Gaulton et al., 2012) which predicts the suitability of the binding site for small molecules. If a PDB chain is reported by the database to have a positive score (including any of tractable, druggable or ensemble score), it is labeled as a druggable PDB structure. The druggability of the nematode proteins was then determined based upon the BLAST match with the PDB sequence.
2.7. Systematic evaluation of indels for selective drug targeting
In each PFC, proteins having a PDB hit were also evaluated for the possibility of specific targeting at the indel locations (relative to its top PDB hit structure) using SiteHound (Ghersi and Sanchez, 2009), to identify any potential ligand binding sites. The NemFam sequences were mapped to the matching PDB sequences. If an indel was detected within 3 AA of any binding site identified by SiteHound, the indel was classified as a target site of interest.
Modeling of protein structures was carried out for three selected candidates using the I-TASSER Suite 2.1 (Roy et al., 2010) using default parameters. Alignments were based on the indel identification process above. These models were refined using NAMD following a published protocol (Phillips et al., 2005). Molecular dynamics was run with 10 separate trajectories of 1ns, and the last 100 ps of each were averaged to create the refined models.
3. Results
In this study proteins from 21 nematode species were compared to 11 non-nematode reference species to identify indels that are specific to nematode proteins (Table 1). The overall workflow is presented in Figure 1 and details of the approach are presented in the Methods section.
3.1. Identification of insertions and deletions
Markov clustering of 513,419 nematode and reference proteins resulted in 50,298 homologous protein families, of which 35,922 had at least one nematode sequence (NemFams). Of these 7,102 NemFams had at least one homolog from the reference species (RefFam). Further alignment improvement resulted in the identification 6,423 protein family clusters PFCs (i.e. NemFam vs. RefFam sequence alignment) for further analysis. Out of these 6,423 PFCs, 4,158 were associated with biological pathways, with 3,892 PFCs matching the 5 main functional KEGG categories (see Methods). The sequences from 68,408 nematode proteins in these 3,892 PFCs were examined for indels specific to nematodes (Figure 1).
The number of deletions (70,704) observed was approximately 1.7 times higher than the number of observed insertions (41,062). On average, deletions were significantly longer than insertions (p-value < 2.2e-16, 19 vs 11 AA) and there was a higher frequency of long deletions than insertions (>10 AA; Table 2).
Table 2.
Size distributions of indels in PFCs.
|
Size bin (AA) |
3,892 PFCs |
|||
|---|---|---|---|---|
| Count | Percent | |||
| Deletions | Insertions | Deletions | Insertions | |
| 0~4 | 33447 | 19541 | 47.31 | 47.59 |
| 5~10 | 12005 | 8602 | 16.98 | 20.95 |
| 11~19 | 7626 | 6041 | 10.79 | 14.71 |
| 20~200 | 17626 | 6878 | 24.93 | 16.75 |
| Total | 70704 | 41062 | 100 | 100 |
| Average | 18.79 | 11.49 | - | - |
| SD | 32.31 | 17.67 | - | - |
| Sizeable indel/family | 9.57 | 5.53 | - | - |
| Short indels/family | 8.59 | 5.02 | - | - |
3.2. Sequence composition, cost and secondary structure of indels
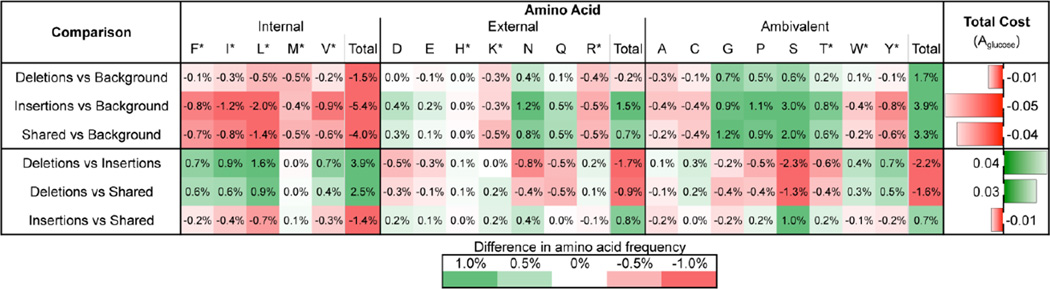
A detailed analysis of amino acid usage in insertions, deletions, shared and background sequences is summarized in Table 3 and Figure 2. Compared with insertions and shared sequences, deletions are highly enriched in seven amino acids (F, I, L, V, C, W, and Y), notably including the three amino acids with the highest synthetic cost (Aglucose; F, W, and Y). Most amino acids in insertions appear with lower frequency than background except a few: D, E, N, Q, G, P, S, and T. Among them, only T is auxotrophic in nematodes. Shared sequences almost always have an amino acid composition somewhere between deletions and insertions, as expected. The distribution of amino acids for shared gap regions were always between that of insertions and deletions.
Table 3.
Amino acid composition and cost analysis for the indels. Detailed analysis and statistics for amino acid composition and cost and P values are shown in Figure 2.
| Position | Amino Acid* |
Aglucose cost |
Deletions | Insertions | Shared | Background | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| mean | SD | mean | SD | mean | SD | mean | SD | |||
| Internal | F* | 1.84 | 4.24 | 11.31 | 3.53 | 11.89 | 3.69 | 11.68 | 4.38 | 2.07 |
| I* | 1.21 | 5.79 | 12.89 | 4.84 | 14.25 | 5.21 | 14.05 | 6.04 | 2.32 | |
| L* | 1.21 | 8.85 | 15.81 | 7.27 | 17.11 | 7.93 | 17.07 | 9.32 | 2.83 | |
| M* | 1.25 | 2.33 | 8.62 | 2.36 | 10.53 | 2.29 | 9.75 | 2.79 | 1.43 | |
| V* | 0.96 | 6.40 | 13.87 | 5.67 | 15.48 | 6.00 | 15.14 | 6.57 | 2.37 | |
| Total | 27.60 | 25.24 | 23.67 | 28.35 | 25.12 | 27.62 | 29.10 | 5.22 | ||
| External | D | 0.61 | 5.53 | 13.28 | 5.99 | 16.09 | 5.79 | 15.13 | 5.54 | 2.14 |
| E | 0.86 | 6.63 | 14.57 | 6.89 | 17.38 | 6.75 | 16.26 | 6.68 | 2.77 | |
| H* | 1.46 | 2.39 | 8.81 | 2.32 | 10.23 | 2.34 | 9.82 | 2.36 | 1.41 | |
| K* | 1.31 | 6.29 | 14.08 | 6.27 | 16.50 | 6.09 | 15.46 | 6.54 | 2.98 | |
| N | 0.79 | 4.83 | 12.74 | 5.60 | 15.59 | 5.23 | 14.50 | 4.44 | 1.92 | |
| Q | 0.92 | 3.88 | 11.28 | 4.33 | 14.02 | 4.35 | 13.52 | 3.81 | 1.97 | |
| R* | 1.39 | 5.53 | 12.99 | 5.37 | 15.08 | 5.44 | 14.34 | 5.91 | 2.51 | |
| Total | 35.07 | 28.02 | 36.79 | 32.99 | 36.00 | 31.37 | 35.29 | 6.37 | ||
| Ambivalent | A | 0.5 | 6.54 | 14.28 | 6.45 | 16.60 | 6.65 | 16.20 | 6.84 | 2.62 |
| C | 0.75 | 1.92 | 7.53 | 1.66 | 8.32 | 1.69 | 7.87 | 2.04 | 1.61 | |
| G | 0.31 | 6.33 | 14.84 | 6.53 | 17.68 | 6.74 | 17.25 | 5.59 | 2.54 | |
| P | 0.99 | 4.86 | 13.34 | 5.38 | 15.39 | 5.22 | 14.96 | 4.32 | 2.21 | |
| S | 0.49 | 7.90 | 16.09 | 10.23 | 20.41 | 9.21 | 18.96 | 7.25 | 2.72 | |
| T* | 0.69 | 5.36 | 13.16 | 5.98 | 16.16 | 5.76 | 15.31 | 5.15 | 1.96 | |
| W* | 2.39 | 1.23 | 6.33 | 0.80 | 6.07 | 0.91 | 6.04 | 1.15 | 1.02 | |
| Y* | 1.77 | 3.19 | 9.94 | 2.52 | 10.63 | 2.69 | 10.37 | 3.27 | 1.75 | |
| Total | 37.32 | 28.60 | 39.54 | 33.56 | 38.88 | 32.24 | 35.61 | 5.55 | ||
| Total Cost (Aglucose) | 0.981 | 0.251 | 0.940 | 0.284 | 0.951 | 0.274 | 0.994 | 0.048 | ||
Auxothrophs in nematodes (Barrett, 1991)
Fig. 2.
Differences in amino acid and usage and cost among insertions, deletions, shared gaps and background sequences.
The eight methods used to calculate biosynthesis cost (Barton et al., 2010; Craig and Weber, 1998; Heizer et al., 2006; Seligmann, 2003; Wagner, 2005) are all highly correlated with amino acid composition, so pairwise comparisons of the average costs for the four categories generated by these methods all show similar patterns (Table S2). We report one set of representative results in Table 3, which was obtained from a recently developed systems biology approach based on genome-scale metabolic models (Aglucose (Barton et al., 2010)). Insertions tended to incorporate amino acids with the lowest average biosynthetic cost (0.940) while deletions possess amino acids with the higher cost (0.981). Shared gap regions had an average cost between the insertions and deletions (0.951), and the cost in background sequences was the highest among the four categories examined, at 0.994. Overall the cost of the amino acids essential in nematodes (auxotrophs (Barrett, 1991)) was higher compared to the cost of the nonessential amino acids (Table 3 and Table S3).
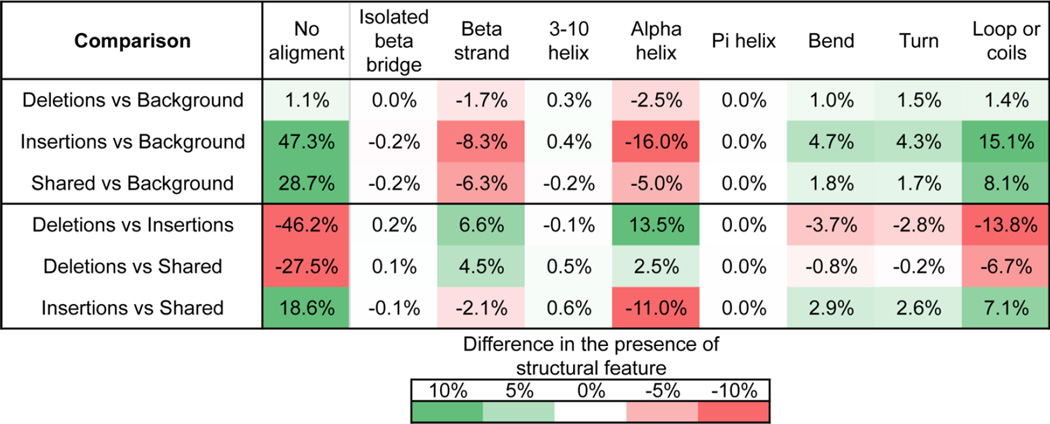
The secondary structure of the PFC proteins was determined by comparison to the PDB entries. In the aligned sequences, deletions have higher percentage of ordered structures than insertions (especially for the abundant structural categories α-helix and β-strands, and insertion regions have higher portions of bend, turn and loops (coils)(Table 4). Again, the compositions of the secondary structures for the shared sequences fall in between the values for insertions and deletions, while background sequences have a composition more similar to deletions.
Table 4.
Secondary structure constitution analysis for the indels. Detailed analysis and statistics for secondary structure constitution and P values are shown in Figure 4.
| Structure annotation |
label | Deletions | Insertions | Shared | Background | ||||
|---|---|---|---|---|---|---|---|---|---|
| mean | SD | mean | SD | mean | SD | mean | SD | ||
| didn’t align | - | 36.046 | 42.628 | 82.224 | 35.752 | 63.583 | 44.553 | 34.901 | 28.925 |
| Aligned region breakdown |
|||||||||
| isolated beta bridge | B | 0.966 | 4.999 | 0.762 | 6.805 | 0.827 | 5.438 | 0.981 | 1.587 |
| beta strand | E | 16.372 | 26.847 | 9.750 | 26.653 | 11.835 | 24.950 | 18.086 | 13.588 |
| 3–10 helix | G | 3.421 | 12.287 | 3.494 | 16.573 | 2.899 | 12.635 | 3.073 | 3.702 |
| alpha helix | H | 31.583 | 36.544 | 18.093 | 35.726 | 29.107 | 38.143 | 34.101 | 19.442 |
| pi helix | I | 0.017 | 0.853 | 0.005 | 0.682 | 0.025 | 1.069 | 0.018 | 0.277 |
| bend | S | 9.703 | 18.692 | 13.433 | 29.527 | 10.496 | 22.598 | 8.714 | 5.163 |
| turn | T | 12.002 | 20.988 | 14.773 | 31.111 | 12.175 | 24.204 | 10.483 | 5.333 |
| loop, or coils | C | 25.936 | 30.826 | 39.690 | 43.701 | 32.636 | 38.083 | 24.544 | 12.175 |
3.3. Associations of indel bearing proteins with cellular pathways and their druggability
Selective pressure can vary according to the pathway in which a protein functions, and this difference in selective pressure may result in a distribution of insertions and deletions that varies according to the biological pathway. The frequency of insertions and deletions was examined in five KEGG pathway categories (Table 5). PFCs that were identified as being involved in ‘Genetic Information Processing’ had the lowest frequency of both sizable (> 4 AA) and all insertions and deletions (Table 5 and Table S1), while ‘Environmental Information Processing’ had the highest frequency of deletions, and ‘Organismal Systems” had the highest frequency of insertions. The rates of sizable insertions/deletions show almost exactly the same trend in each category as indels of all sizes.
Table 5.
The distribution of insertions and deletions (gap regions) specific to nematodes among the different KEGG categories. Sizable indels are defined as gaps greater than 4 amino acid long. Indel rates are defined as indel/cluster numbers (the average frequency of indels in a cluster).
| KEGG category |
indel PFC |
del | ins | del rate |
ins rate |
sizable del |
small del |
sizable del rate |
small del rate |
sizable ins |
small ins |
sizable ins rate |
small ins rate |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolism | 1448 | 26306 | 15208 | 18.17 | 10.50 | 13887 | 12419 | 9.59 | 8.58 | 8012 | 7196 | 5.53 | 4.97 |
| Genetic Information Processing |
1638 | 29378 | 16508 | 17.94 | 10.08 | 15227 | 14151 | 9.30 | 8.64 | 8543 | 7965 | 5.22 | 4.86 |
| Environmental Information Processing |
414 | 7791 | 4479 | 18.82 | 10.82 | 4218 | 3573 | 10.19 | 8.63 | 2410 | 2069 | 5.82 | 5.00 |
| Cellular Processes |
305 | 5658 | 3643 | 18.55 | 11.94 | 3059 | 2599 | 10.03 | 8.52 | 1920 | 1723 | 6.30 | 5.65 |
| Organismal Systems |
87 | 1571 | 1224 | 18.06 | 14.07 | 866 | 705 | 9.95 | 8.10 | 636 | 588 | 7.31 | 6.76 |
| Total (excluding Human Diseases) |
3892 | 70704 | 41062 | 18.17 | 10.55 | 37257 | 33447 | 9.57 | 8.59 | 21521 | 19541 | 5.53 | 5.02 |
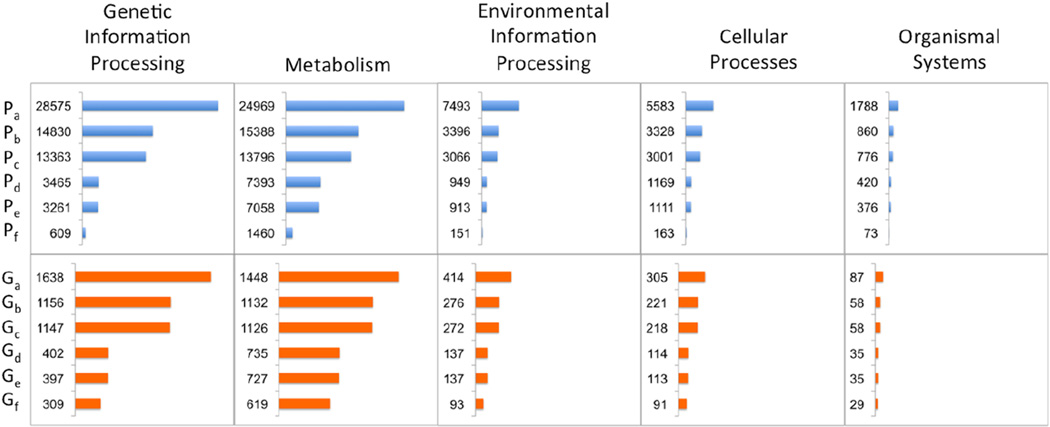
Among the previously identified 34,002 proteins from parasitic species (2,821 PFCs) with a match in the PDB, 13,396 proteins (1,423 PFCs) are identified as druggable. The vast majority of them (12,719 proteins in 1,409 PFCs) contained nematode-specific indels. The distribution of these druggable proteins in the KEGG categories follows the overall distribution of indel bearing proteins, with majority being involved in ‘Metabolism’ (Figure 3, Table 6).
Fig. 3.
Structure determination and indel target prioritization based on five KEGG functional categories. Label P for upper panel denotes protein count; label G for lower panel denotes PFC count. Subscripts denote different group definition in each prioritization step. Subscript A: number of proteins/PFCs within the KEGG category; subscript B: number of proteins/PFCs with a PDB match; subscript C: number of parasitic proteins/PFCs with a PDB match; 19ubscript D: number of parasitic proteins/PFCs that are druggable (based on EBI drugEBIlity analysis); subscript E: number of parasitic druggable proteins/PFCs s having an indel; subscript F: number of parasitic druggable proteins/PFCs having an indel close to potential ligand binding sites.
Table 6.
Druggable protein distributions in each KEGG category.
| KEGG category | All proteins | Proteins wi th indels | ||
|---|---|---|---|---|
| Proteins (#) | PFCs (#) | Proteins (#) | PFCs (#) | |
| Metabolism | 7393 | 735 | 7058 | 727 |
| Genetic Information Processing | 3465 | 402 | 3261 | 397 |
| Environmental Information Processing | 949 | 137 | 913 | 137 |
| Cellular Processes | 1169 | 114 | 1111 | 113 |
| Organismal Systems | 420 | 35 | 376 | 35 |
| Total (excluding Human Diseases) | 13396 | 1423 | 12719 | 1409 |
3.4. Localization of nematode-specific indels in spatial structures for selective targeting
Indels have been suggested to serve as candidates of pathogen-specific drug targeting to reduce the likelihood of host toxicity. In our approach, we compared all members of each PFC to the matching PDB structures, and determined proximity to predicted binding site residues. An indel at the immediate periphery of a ligand binding site could alter the size and residue locations of the site, sometimes creating a unique binding site compared to the homologous host proteins. Approximately 70% of the PFCs (2,843 out of 3,892) have at least one protein hitting a PDB structure. In about 30% of the PFCs (1,141), at least one protein had been identified with an indel close to a potential ligand binding site. Below we describe three examples to illustrate how these indel sites could be exploited to design specific ligand to achieve selectivity.
4. Discussion
Indel frequency has been shown to vary across different organisms. Studies of genetic variation in Drosophila melanogaster and Caenorhabditis elegans have shown that indels represent between 16% and 25% of all genetic polymorphisms in these species (Berger et al., 2001; Wicks et al., 2001). It is estimated that human populations typically harbor a minimum of 1.56 million indels (Mills et al., 2006). Not only does the frequency of indels vary, but in general, deletions are more prevalent than insertions. In a recent study examining 5,000 indel events in noncoding regions of 17 taxonomic groups across the three domains of life (Kuo and Ochman, 2009), deletion events outnumbered insertions in all groups. Deletions also outweighed insertions more in prokaryotes compared to eukaryotes. In the current study, we also found this to be the case for nematodes compared with the reference sets at the deduced proteome level. Deletions were ~1.7 times more abundant than insertions in proteins found to be associated with the five examined KEGG categories. We showed associations with different modes of existence and uneven functional evolution. However, given the draft nature of the available genomes when alternative splicing or exon skipping information becomes available for nematode species in the future (at present genome-wide alternative splicing isoform information is not available for any parasitic nematodes but only for the non-parasitic C. elegans) our findings could be refined.
In this present study, we focused on the structural categories the indels fall into, by comparing the nematode proteins with known protein structures to further understand their impact and the consequences in novel drug discovery. The highly specific indel content within nematodes are reflected in their overall amino acid composition. We observed that both deletions and insertions were comprised predominantly of amino acids with small side chains and high turn propensity, such as G, P, S, N and Q. In addition, insertions are also enriched in the two hydrophilic residues D and E. Notably, none of these amino acids are auxothropic in nematodes. Compared to background, both deletions and insertions tended to be depleted in hydrophobic amino acids (27.60% and 23.67% vs. 29.10%) but enriched in ambivalent amino acids (37.32% and 39.54% vs 35.61%). Deletions are also slightly depleted in hydrophilic amino acids than the background (35.07% vs 35.29%), while insertions are enriched for them (36.79% vs. 35.29%). These results are in line with what have been reported in previous work (Roth and Liberles, 2006) and other indel databases such as IndelFR (Zhang et al., 2012) and IndelPDB (Hsing and Cherkasov, 2008) (Table S5). In those databases, gaps were limited to short length (99.9% of gaps were < 100 AA, while 90% of gaps were < 10 AA long) in contrast to our longer allowed gap length. Also, the observations in those databases are limited to the protein structures in highly homologous species, so there may be some differences for a few individual amino acids.
Deletions have higher average biosynthetic cost than insertions due to the higher portion of residues with large side chains such as F, W, and Y. This suggests that setting aside the penalties of gap opening/closing, the cost for extending a gap region on a residual basis will be lower than the background sequences to compensate for the cost of opening it. The average cost ratio (using eight different methods) of essential (auxothrophic) to nonessential amino acids in nematodes is 2.27 (Table S3). Our analysis only considers the cost differences for indels in parasitic nematodes, and does not include differences with either the parasite hosts or free-living species. Such analysis may provide insight into the nature of parasitism, since differences in amino acid usage between parasites and their free-living relatives may be the result of parasitic adaptation.
Indels have been shown to vary not only in amino acid composition but also in structural constitutions. We found that for insertions, the majority of amino acids did not align with any portion of a PDB protein and thus it was impossible to directly determine the underlying structure. As loop regions of proteins are often not resolved in crystal structures and commonly have lower sequence identity, it is expected that many indels occur in loop regions. In contrast, deletions have significantly higher portions of sequences aligned with PDB structures. In the aligned deleted regions, about ¼ of the sequences adopt loop or random coil conformations, which is just slightly less than the portions of α-helical conformations in all secondary structure categories, while in the aligned inserted regions, loops take as much as 40% of the structures, further supporting the idea that loops play an important structural role for nematode specific indels.
There was a biased distribution of indel events within KEGG pathways, which is likely due to differences in selective pressure. Proteins that were associated with ‘Environmental information processing’ had the highest number of sizable insertions and deletions per PFC, while proteins that were involved in ‘Genetic information processing’ had the lowest average number of insertions and deletions per cluster, as previously reported (Wang et al., 2009). KEGG subdivides the category ‘Environmental information processing’ into three main subcategories (membrane transport, signal transduction, and signaling molecules and interaction), and it has been previously shown that mutations in proteins involved with signal transduction can result in an increased longevity and stress resistance (Longo, 1999). It is possible that the indels in these proteins occur as a result of positive selection. Proteins in ‘Genetic information processing’ have stringent selective constraints, and are under strong negative selection to preserve their functions (Bergmiller et al., 2012). Accordingly, these proteins have the least number of insertions and deletions per PFC.
Out of the 34,002 parasitic proteins with a match in PDB, 13,396 of them were identified as druggable and 12,719 were druggable parasite proteins with indels. Over half (7,058 out of 12,719, 55%) of these proteins were classified as being involved in metabolism. This suggests that it is possible that further optimization of the candidate compounds based on indel information may result in new approaches to control or prevent nematode infection.
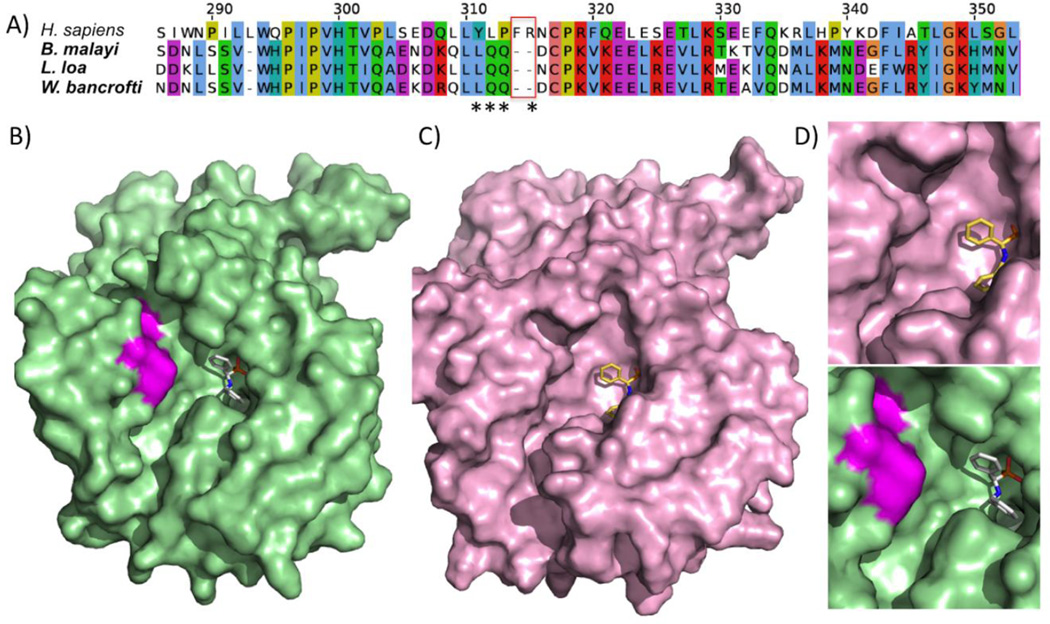
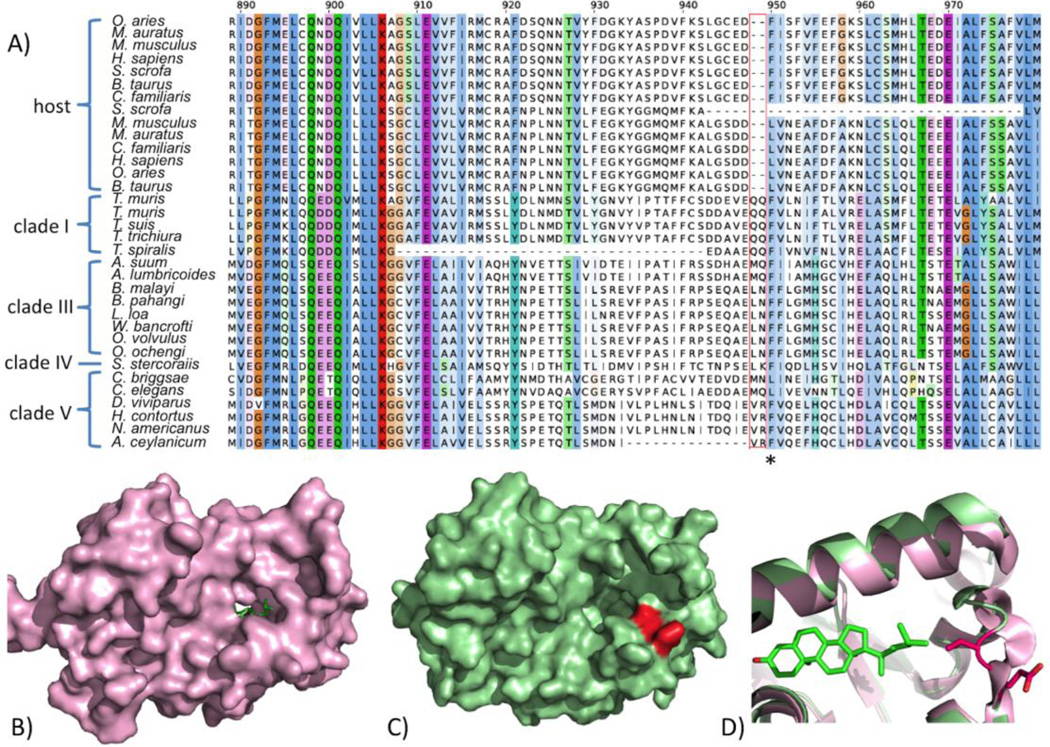
As an example from the ‘Metabolism’ category, prostatic acid phosphatase (PAP, EC: 3.1.3.2) is a ubiquitous lysosomal enzyme that hydrolyses organic phosphates at an acid pH (Muniyan et al., 2013), with 4 structures of the human protein available. Indel location analysis identified a 2 amino-acid deletion specific to the human filarial nematode Brugia malayi at the immediate periphery of its active site (Figure 5A). Using gene expression data we confirm the presence of this deletion and expression and relevant stages (i.e. parasitic stages). The mRNA sequences from multiple RNA-seq libraries from different developmental stages also validated the sequences flanking the gap regions, and showed that the expression of the protein is almost ubiquitous across multiple development stages with highest expression in adult stages (Table S4, Figure S1), hence validated the existence of the indel and providing expression profile of the indel bearing gene. Furthermore, to explore the consequence of the indel on the nematode protein structure, we built a homology model for PAP of B. malayi based on its alignment against the human structure (PDB code: 1ND5). The PAP protein structure from H. sapiens shares ~33% sequence identity with that of B. malayi. In comparison with the human crystal structure, the deletion clearly created a larger pocket (Figure 5B, C, and D), and a non-selective inhibitor could potentially be modified to be more specific at the active site of PAP for B. malayi.
Fig. 5.
The indel sequence and protein model of prostatic acid phosphatase (PAP), a protein suitable for indel-specific drug targeting. (A) Sequence alignment of the deleted region in PAP from a few filarial species compared to the host H. sapiens. The active site residues interacting with known ligands are marked with asterisks. (B) Human PDB structure in complex with a ligand (PDB code 1ND5), and the to be deleted region colored in magenta. (C) Refined model of PAP with the ligand. (D) Close-up view of the ligand binding site.
Another example is the NemFam-encoding, retinoic acid-related orphan nuclear hormone receptor (ROR) in the ‘Signaling molecules and interaction” category. In the host species, the RORs are involved in many physiological processes, including regulation of metabolism, development and immunity as well as the circadian rhythm (Kojetin and Burris, 2014). In C. elegans, ROR is required in all larval molts and the hypodermal expression of other proteins essential for larval development and adult morphogenesis (Kostrouchova et al., 2001). As shown in Figure 6A, our sequence alignment reveals a small, 2 aa insertion in the ligand binding domain (LBD) present in almost all of the parasitic nematode species within the NemFam. Using the human whipworm species Trichuris trichiura as an example, a homology model based on the human template (PDB code: 1N83) shows that the insertion results in the intrusion of the N-terminus of H7 into the tightly packed binding pocket (Figure 6B–D). The bound cholesterol in the crystal structure interacts with this region, so even this small change in AA composition could potentially be used to design parasite-specific ligands.
Fig 6.
The indel sequence and protein model of retinoic acid-related orphan nuclear hormone receptor (ROR), a protein suitable for indel-specific drug targeting in the ‘Environmental Information Processing’ category. (A) Sequence alignment of the inserted region in ROR LBD from all the species within the PFC. The active site residues interacting with known ligands are marked with asterisks. Species from the same nematode clade or host are marked by curly brackets. (B) Human PDB structure in complex with a ligand (PDB code: 1N83). (C) Model of T. trichiura ROR. The inserted residues are colored in red. (D) Close-up view of the ligand binding site with two structures overlaid together. The inserted regions in T. trichiura model are shown in red with side chains as stick.
5. Conclusions
With drug resistance and environmental concerns rising, there is an urgent need for new anthelmintic therapeutics In looking for new drug targets in parasites, two groups of protein candidates are of special interest, i) targets that are unique to the pathogen, avoiding proteins that were evolutionarily conserved between hosts and pathogen to reduce toxicity or ii) targets that share homology with the host proteins (essential proteins) that possess molecular features (such as indels) specific to the pathogen that enables selective targeting. In recent years, steady progress in genome sequencing projects has generated large amounts of genomic data for nematodes and provided an abundance of resources to study the evolution, adaptation, and unique features of nematode proteins, especially for parasites. Indel analysis (in combination with other approaches such as druggability analysis and structural and functional annotations) at a genome-wide scale provides a systematic method of identifying novel potential drug targets. Classification and understanding of indel location, structure, and composition is important, as it provides information on specific events that improve our understanding of protein evolution, and it allows researchers to take advantage of such an event in approaches such as selective targeting. By identifying and selectively targeting these structurally unique regions with small molecules, the method promises to open the door to a whole new standard for antihelmintic drug discovery.
We developed and applied a systematic approach for identifying, analyzing and evaluating specific indels present in the phylum Nematoda (in comparison with their host organisms) in order to understand the unique structural features of the indels. By scanning the indel locations for the parasitic druggable proteins in each cluster with its corresponding PDB structure, we were able to narrow down to about 20% of these proteins with interesting indel target sites. Because of their uniqueness resulting from various lengths of the gaps and 3D conformations of the cavities, not all sites may be feasible targets for small molecules. However, the results indicate that indels could indeed often be located at critical regions of proteins, hypothetically creating novel ligand binding sites through the alteration of the shapes and amino acid compositions of these sites. Among these, we presented three examples of indels in the binding sites of nematode proteins compared to those of the hosts. In each example, the indel creates a structural change in the binding site which may be a exploited to design small molecules capable of specific binding to the nematode target.
Future studies of the indel bearing proteins identified and characterized in this communication may improve our understanding of protein evolution in parasites (and nematodes in general), and may lead to new drug targets, anthelminthic drugs, and new strategies to control these parasites of global importance.
Supplementary Material
Figure S1. RNA-Seq mapping results of B malayi PAP gene. The gap region in Figure 5 is marked by the vertical line.
Fig. 4.
Relative over-representation and under-representation of secondary structures among insertions, deletions, shared gaps and background sequences. .
Highlights.
We systematically studied indels specific to the phylum Nematoda
Indels unique to Nematoda proteins were present in over 3,000 protein families.
The cost of indels is highly correlated with the amino acid composition.
There is a biased distribution of indel events within biological pathways.
Indel close to the binding pocket can facilitate selective drug targeting
Acknowledgments
The authors thank Zhengyuan Wang for assisting with technical issues relation with indel identification and John Martin for technical assistance with HMM model building. This work was supported by the National Institute of Health NIAID (R01 AI081803) and NIGMS (R01 GM097435) to M.M.. We thank the parasite genomics group at the Wellcome Trust Sanger Institute for making some of the unpublished reference genome used in this study available at WormBase-ParaSite.
Abbreviations
- PFC
Protein Family Cluster
- KO
KEGG Orthology pathway
- NemFam
PFC with at least 1 nematode species
- RefFam
PFC with at least 1 non-nematode species
- PDB
RCSB Protein Data Bank
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data access
The orthologous groups, the proteins, their annotation, indel location and related features have been made publicly available through an interactive web interface at http://nematode.net (http://nematode.net/indels.html).
Authors’ contributions
QW, EH and MM conceived and designed the experiments. QW, BAR and EH carried out experiments and analyses. QW, EH, BAR, SAW, JWJ and MM interpreted results and prepared the manuscript. All authors have read and approved the final manuscript.
References
- Barrett J. Amino acid metabolism in helminths. Advances in parasitology. 1991;30:39–105. doi: 10.1016/s0065-308x(08)60306-1. [DOI] [PubMed] [Google Scholar]
- Barton MD, Delneri D, Oliver SG, Rattray M, Bergman CM. Evolutionary systems biology of amino acid biosynthetic cost in yeast. PloS one. 2010;5:e11935. doi: 10.1371/journal.pone.0011935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Research. 2015;43:D30–D35. doi: 10.1093/nar/gku1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento AP, Gaulton A, Hersey A, Bellis LJ, Chambers J, Davies M, Kruger FA, Light Y, Mak L, McGlinchey S, Nowotka M, Papadatos G, Santos R, Overington JP. The ChEMBL bioactivity database: an update. Nucleic acids research. 2014;42:7. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J, Suzuki T, Senti KA, Stubbs J, Schaffner G, Dickson BJ. Genetic mapping with SNP markers in Drosophila. Nature genetics. 2001;29:475–481. doi: 10.1038/ng773. [DOI] [PubMed] [Google Scholar]
- Bergmiller T, Ackermann M, Silander OK. Patterns of evolutionary conservation of essential genes correlate with their compensability. PLoS genetics. 2012;8:e1002803. doi: 10.1371/journal.pgen.1002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindley PJ, Mitreva M, Ghedin E, Lustigman S. Helminth genomics: The implications for human health. PLoS Negl Trop Dis. 2009;3:e538. doi: 10.1371/journal.pntd.0000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SK, Hsing M, Hormozdiari F, Cherkasov A. Relationship between insertion/deletion (indel) frequency of proteins and essentiality. BMC bioinformatics. 2007:8. doi: 10.1186/1471-2105-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasov A, Lee SJ, Nandan D, Reiner NE. Large-scale survey for potentially targetable indels in bacterial and protozoan proteins. Proteins-Structure Function and Bioinformatics. 2006;62:371–380. doi: 10.1002/prot.20631. [DOI] [PubMed] [Google Scholar]
- Cherkasov A, Nandan D, Reiner NE. Selective targeting of indel-inferred differences in spatial structures of highly homologous proteins. Proteins. 2005;58:950–954. doi: 10.1002/prot.20391. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Ghedin E, Berriman M, McQuillan J, Holroyd N, Mayhew GF, Christensen BM, Michalski ML. A deep sequencing approach to comparatively analyze the transcriptome of lifecycle stages of the filarial worm, Brugia malayi. PLoS Negl Trop Dis. 2011;5:e1409. doi: 10.1371/journal.pntd.0001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig CL, Weber RS. Selection costs of amino acid substitutions in ColE1 and ColIa gene clusters harbored by Escherichia coli. Molecular biology and evolution. 1998;15:774–776. doi: 10.1093/oxfordjournals.molbev.a025981. [DOI] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Girón CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kähäri AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Aken BL, Birney E, Harrow J, Kinsella R, Muffato M, Ruffier M, Searle SMJ, Spudich G, Trevanion SJ, Yates A, Zerbino DR, Flicek P. Ensembl 2015. Nucleic Acids Research. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich C, Sommer RJ. How to become a parasite - lessons from the genomes of nematodes. Trends in genetics : TIG. 2009;25:203–209. doi: 10.1016/j.tig.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic acids research. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fechteler T, Dengler U, Schomburg D. Prediction of protein three-dimensional structures in insertion and deletion regions: a procedure for searching data bases of representative protein fragments using geometric scoring criteria. Journal of molecular biology. 1995;253:114–131. doi: 10.1006/jmbi.1995.0540. [DOI] [PubMed] [Google Scholar]
- Fischer S, Brunk BP, Chen F, Gao X, Harb OS, Iodice JB, Shanmugam D, Roos DS, Stoeckert CJ., Jr Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Current protocols in bioinformatics / editoral board, Andreas D. Baxevanis … [et al.] Chapter 6 Unit 6. 2011;12:11–19. doi: 10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox BA, Ristuccia JG, Bzik DJ. Genetic identification of essential indels and domains in carbamoyl phosphate synthetase II of Toxoplasma gondii. International journal for parasitology. 2009;39:533–539. doi: 10.1016/j.ijpara.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV. Searching for drug targets in microbial genomes. Current opinion in biotechnology. 1999;10:571–578. doi: 10.1016/s0958-1669(99)00035-x. [DOI] [PubMed] [Google Scholar]
- Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic acids research. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghersi D, Sanchez R. EasyMIFS and SiteHound: a toolkit for the identification of ligand-binding sites in protein structures. Bioinformatics. 2009;25:3185–3186. doi: 10.1093/bioinformatics/btp562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TW, Baran J, Bieri T, Cabunoc A, Chan J, Chen WJ, Davis P, Done J, Grove C, Howe K, Kishore R, Lee R, Li Y, Muller HM, Nakamura C, Ozersky P, Paulini M, Raciti D, Schindelman G, Tuli MA, Van Auken K, Wang D, Wang X, Williams G, Wong JD, Yook K, Schedl T, Hodgkin J, Berriman M, Kersey P, Spieth J, Stein L, Sternberg PW. WormBase 2014: new views of curated biology. Nucleic Acids Res. 2014;42:D789–D793. doi: 10.1093/nar/gkt1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heizer EM, Jr, Raiford DW, Raymer ML, Doom TE, Miller RV, Krane DE. Amino acid cost and codon-usage biases in 6 prokaryotic genomes: a whole-genome analysis. Molecular biology and evolution. 2006;23:1670–1680. doi: 10.1093/molbev/msl029. [DOI] [PubMed] [Google Scholar]
- Hormozdiari F, Salari R, Hsing M, Schonhuth A, Chan SK, Sahinalp SC, Cherkasov A. The effect of insertions and deletions on wirings in protein-protein interaction networks: a large-scale study. J Comput Biol. 2009;16:159–167. doi: 10.1089/cmb.2008.03TT. [DOI] [PubMed] [Google Scholar]
- Hotez PJ, Molyneux DH, Fenwick A, Kumaresan J, Sachs SE, Sachs JD, Savioli L. Control of neglected tropical diseases. N Engl J Med. 2007;357:1018–1027. doi: 10.1056/NEJMra064142. [DOI] [PubMed] [Google Scholar]
- Howe KL, Bolt BJ, Cain S, Chan J, Chen WJ, Davis P, Done J, Down T, Gao S, Grove C, Harris TW, Kishore R, Lee R, Lomax J, Li Y, Muller HM, Nakamura C, Nuin P, Paulini M, Raciti D, Schindelman G, Stanley E, Tuli MA, Van Auken K, Wang D, Wang X, Williams G, Wright A, Yook K, Berriman M, Kersey P, Schedl T, Stein L, Sternberg PW. WormBase 2016: expanding to enable helminth genomic research. Nucleic acids research. 2015 doi: 10.1093/nar/gkv1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing M, Cherkasov A. Indel PDB: a database of structural insertions and deletions derived from sequence alignments of closely related proteins. BMC bioinformatics. 2008;9:293. doi: 10.1186/1471-2105-9-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Baujard P, Morand S. Biodiversity in helminths and nematodes as a field of study: an overview. Nematology. 2001;3:199–208. [Google Scholar]
- Jansen C, Wang H, Kooistra AJ, de Graaf C, Orrling KM, Tenor H, Seebeck T, Bailey D, de Esch IJ, Ke H, Leurs R. Discovery of novel Trypanosoma brucei phosphodiesterase B1 inhibitors by virtual screening against the unliganded TbrPDEB1 crystal structure. Journal of medicinal chemistry. 2013;56:2087–2096. doi: 10.1021/jm3017877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten RP, te Beek TA, Krieger E, Hekkelman ML, Hooft RW, Schneider R, Sander C, Vriend G. A series of PDB related databases for everyday needs. Nucleic acids research. 2011;39:D411–D419. doi: 10.1093/nar/gkq1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr ID, Wu P, Marion-Tsukamaki R, Mackey ZB, Brinen LS. Crystal Structures of TbCatB and rhodesain, potential chemotherapeutic targets and major cysteine proteases of Trypanosoma brucei. PLoS Negl Trop Dis. 2010;4:e701. doi: 10.1371/journal.pntd.0000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O'Reilly M, O'Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JF, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N. Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojetin DJ, Burris TP. REV-ERB and ROR nuclear receptors as drug targets. Nature reviews. Drug discovery. 2014;13:197–216. doi: 10.1038/nrd4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrouchova M, Krause M, Kostrouch Z, Rall JE. Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2001;98:7360–7365. doi: 10.1073/pnas.131171898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CH, Ochman H. Deletional bias across the three domains of life. Genome biology and evolution. 2009;1:145–152. doi: 10.1093/gbe/evp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome research. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD. Mutations in signal transduction proteins increase stress resistance and longevity in yeast, nematodes, fruit flies, and mammalian neuronal cells. Neurobiol Aging. 1999;20:479–486. doi: 10.1016/s0197-4580(99)00089-5. [DOI] [PubMed] [Google Scholar]
- Martin J, Rosa BA, Ozersky P, Hallsworth-Pepin K, Zhang X, Bhonagiri-Palsikar V, Tyagi R, Wang Q, Choi YJ, Gao X, McNulty SN, Brindley PJ, Mitreva M. Helminth.net: expansions to Nematode.net and an introduction to Trematode.net. Nucleic acids research. 2015;43:D698–D706. doi: 10.1093/nar/gku1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, Devine SE. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome research. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitreva M, Zarlenga DS, McCarter JP, Jasmer DP. Parasitic nematodes - from genomes to control. Veterinary parasitology. 2007;148:31–42. doi: 10.1016/j.vetpar.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Muniyan S, Chaturvedi NK, Dwyer JG, Lagrange CA, Chaney WG, Lin MF. Human prostatic Acid phosphatase: structure, function and regulation. International journal of molecular sciences. 2013;14:10438–10464. doi: 10.3390/ijms140510438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandan D, Lopez M, Ban F, Huang M, Li Y, Reiner NE, Cherkasov A. Indel-based targeting of essential proteins in human pathogens that have close host orthologue(s): discovery of selective inhibitors for Leishmania donovani elongation factor-1alpha. Proteins. 2007;67:53–64. doi: 10.1002/prot.21278. [DOI] [PubMed] [Google Scholar]
- Nicol JM, Turner SJ, Coyne DL, Nijs Ld, Hockland S, Maafi ZT. Current Nematode Threats to World Agriculture. In: Jones J, Gheysen G, Fenoll C, editors. Genomics and Molecular Genetics of Plant-Nematode Interactions. Netherlands: Springer; 2011. pp. 21–43. [Google Scholar]
- Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Pascarella S, Argos P. Analysis of insertions/deletions in protein structures. Journal of molecular biology. 1992;224:461–471. doi: 10.1016/0022-2836(92)91008-d. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. Journal of computational chemistry. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C, Liberles DA. A systematic search for positive selection in higher plants (Embryophytes) BMC plant biology. 2006;6:12. doi: 10.1186/1471-2229-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salari R, Schönhuth A, Hormozdiari F, Cherkasov A, Sahinalp SC. The Relation between Indel Length and Functional Divergence: A Formal Study. In: Crandall K, Lagergren J, editors. Algorithms in Bioinformatics. Berlin Heidelberg: Springer; 2008. pp. 330–341. [Google Scholar]
- Seligmann H. Cost-minimization of amino acid usage. Journal of molecular evolution. 2003;56:151–161. doi: 10.1007/s00239-002-2388-z. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer RJ, Streit A. Comparative genetics and genomics of nematodes: genome structure, development, and lifestyle. Annu Rev Genet. 2011;45:1–20. doi: 10.1146/annurev-genet-110410-132417. [DOI] [PubMed] [Google Scholar]
- Studer RA, Dessailly BH, Orengo CA. Residue mutations and their impact on protein structure and function: detecting beneficial and pathogenic changes. The Biochemical journal. 2013;449:581–594. doi: 10.1042/BJ20121221. [DOI] [PubMed] [Google Scholar]
- Thorne JL. Models of protein sequence evolution and their applications. Curr Opin Genet Dev. 2000;10:602–605. doi: 10.1016/s0959-437x(00)00142-8. [DOI] [PubMed] [Google Scholar]
- Urbaniak MD, Collie IT, Fang W, Aristotelous T, Eskilsson S, Raimi OG, Harrison J, Navratilova IH, Frearson JA, van Aalten DM, Ferguson MA. A novel allosteric inhibitor of the uridine diphosphate N-acetylglucosamine pyrophosphorylase from Trypanosoma brucei. ACS chemical biology. 2013;8:1981–1987. doi: 10.1021/cb400411x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Energy constraints on the evolution of gene expression. Molecular biology and evolution. 2005;22:1365–1374. doi: 10.1093/molbev/msi126. [DOI] [PubMed] [Google Scholar]
- Wang H, Kunz S, Chen G, Seebeck T, Wan Y, Robinson H, Martinelli S, Ke H. Biological and structural characterization of Trypanosoma cruzi phosphodiesterase C and Implications for design of parasite selective inhibitors. The Journal of biological chemistry. 2012;287:11788–11797. doi: 10.1074/jbc.M111.326777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Rosa B, Jasmer DP, Mitreva M. Pan-Nematoda transcriptomic elucidation of essential intestinal functions and therapeutic targets with broad potential. EBioMedicine. 2015 doi: 10.1016/j.ebiom.2015.07.030. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Martin J, Abubucker S, Yin Y, Gasser RB, Mitreva M. Systematic analysis of insertions and deletions specific to nematode proteins and their proposed functional and evolutionary relevance. BMC evolutionary biology. 2009;9:23. doi: 10.1186/1471-2148-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nature genetics. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Wolstenholme AJ, Fairweather I, Prichard R, von Samson-Himmelstjerna G, Sangster NC. Drug resistance in veterinary helminths. Trends Parasitol. 2004;20:469–476. doi: 10.1016/j.pt.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Wrabl JO, Grishin NV. Gaps in structurally similar proteins: towards improvement of multiple sequence alignment. Proteins. 2004;54:71–87. doi: 10.1002/prot.10508. [DOI] [PubMed] [Google Scholar]
- Wrigley J, McArthur M, McKenna PB, Mariadass B. Resistance to a triple combination of broad-spectrum anthelmintics in naturally-acquired Ostertagia circumcincta infections in sheep. New Zealand veterinary journal. 2006;54:47–49. doi: 10.1080/00480169.2006.36604. [DOI] [PubMed] [Google Scholar]
- Wylie T, Martin J, Abubucker S, Yin Y, Messina D, Wang Z, McCarter JP, Mitreva M. NemaPath: online exploration of KEGG-based metabolic pathways for nematodes. BMC genomics. 2008;9:525. doi: 10.1186/1471-2164-9-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Martin J, Abubucker S, Wang Z, Wyrwicz L, Rychlewski L, McCarter JP, Wilson RK, Mitreva M. Molecular determinants archetypical to the phylum Nematoda. BMC genomics. 2009;10:114. doi: 10.1186/1471-2164-10-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xing C, Wang L, Gong B, Liu H. IndelFR: a database of indels in protein structures and their flanking regions. Nucleic acids research. 2012;40:D512–D518. doi: 10.1093/nar/gkr1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. RNA-Seq mapping results of B malayi PAP gene. The gap region in Figure 5 is marked by the vertical line.