Abstract
The hallmarks of premalignant lesions were first described in the 1970s, a time when relatively little was known about the molecular underpinnings of cancer. Yet it was clear there must be opportunities to intervene early in carcinogenesis. A vast array of molecular information has since been uncovered, with much of this stemming from studies of existing cancer or cancer models. Here, examples of how an understanding of cancer biology has informed cancer prevention studies are highlighted and emerging areas that may have implications for the field of cancer prevention research are described. A note of caution accompanies these examples, in that while there are similarities, there are also fundamental differences between the biology of premalignant lesions or premalignant conditions and invasive cancer. These differences must be kept in mind, and indeed leveraged, when exploring potential cancer prevention measures.
Keywords: Preneoplastic lesions, Chemoprevention, Screening, Tumor subtypes, Risk
1. The hallmarks of premalignant conditions—an introduction
In 1976, Dr Michael Sporn outlined “...common, fundamental properties...shared by [premalignant] epithelial lesions...”[1]. These include: (1) being diffuse and multifocal; (2) having a statistical or probabilistic nature in progression to malignancy; (3) enhanced DNA synthesis; and (4) mechanisms of protection, as not all premalignant lesions or conditions progress to malignancy. Sporn also gave us a definition of chemoprevention, one that has since evolved to encompass using “...natural, synthetic, or biological agents to reverse, suppress, or prevent either the initial phases of carcinogenesis or the progression of premalignant cells to invasive disease” [1,2]. The supposition that progression of a premalignant condition to a malignant state can be prevented or the premalignant state reversed with the right intervention, assumes two key elements: that we can identify who has these lesions and know the appropriate way in which to intervene.
Coincidentally, the same year that Sporn described the properties of premalignant lesions, our understanding of the molecular origins of cancer profoundly changed. Careful study of the genetic etiology of an aggressive sarcoma in chickens implicated the SRC gene as the key genetic component that differed between the genome of a virus that transformed normal chicken cells into cancer cells and a similar virus that could not [3,4]. Intriguingly, the SRC gene was detected in normal, uninfected avian tissue, unequivocally demonstrating that the origins of cancer are within us and that, in most cases, it is our own cellular genome that accumulates sufficient alterations to eventually lead to cancer. This discovery initiated an intense interest in understanding the human cancer genome and was enabled by an exponential development in molecular technology (see Meerzaman et al, this issue).
At the time Sporn articulated his view of the characteristics or hallmarks of premalignant lesions, the diagnosis of cancer was largely based on pathologic characteristics. Today, pathological diagnoses are accompanied by a vast array of molecular detail [5,6]. Thus the question arises; can we leverage this molecular detail for the discrimination and “treatment” of premalignant conditions?
In this special issue of Seminars in Oncology, we focus on the molecular properties of cells early in the carcinogenic process and how this informs the design of cancer prevention studies and selection of chemopreventive agents. In this review, we first discuss the molecular basis for cancer prevention and provide a few key examples of how this knowledge has led to the development and implementation of cancer prevention strategies. Our goal is to (re-) acknowledge that although there is a continuum between premalignant and malignant lesions, they are two different entities [1] and with differing propensities for progression. We highlight a necessity to both study and recognize this distinction. Resolving the biological, molecular, and genetic hallmarks of cancer has entirely changed how we diagnose and treat cancer. Herein, we discuss the potential of leveraging this progress and paradigm for cancer prevention.
2. The molecular basis for cancer prevention
2.1. The hallmarks of cancer
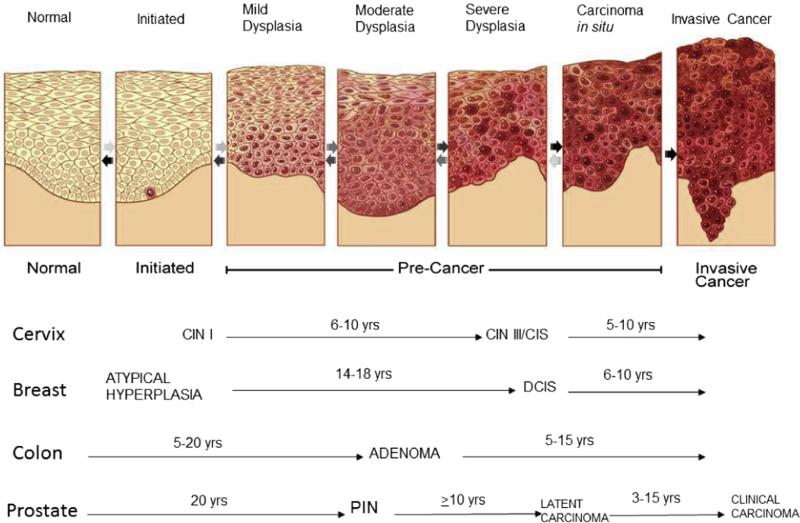
We know that whether from an underlying genetic predisposition, endogenous processes in the cell, bacterial or viral infection, and/or other exogenous factors such as sun, tobacco, certain chemical, or radiation exposure, it is our own cellular genome that collects enough genetic alterations to eventually transform a normal cell to a malignant one. Work by Vogelstein and others have shown us that many cancers have a natural history of progression, evolving from dysplasia to hyperplasia to in situ carcinoma and eventually to a malignant invasive tumor [7] (Fig. 1). To organize the growing volume of knowledge regarding the aberrant biology that characterizes cancer cells and to guide future studies of cancer biology, Hanahan and Weinberg first proposed the “Hallmarks of Cancer” framework in 2000 [8] (an updated version was published in 2011 [9]). The “Hallmarks of Cancer” proposal unifies the molecular pathways that contribute to the fundamental properties of cancer cells. The 2000 version of the “Hallmarks of Cancer” included: (1) self-sufficiency in growth signals; (2) insensitivity to anti-growth signals; (3) tissue invasion and metastasis; (4) limitless replication potential; (5) sustained angiogenesis; and (6) evading apoptosis. In 2011, two emerging hallmarks—deregulating cellular energetics and avoiding immune destruction—and two enabling characteristics—genome instability and mutation and tumor inflammation—were added to this framework.
Fig. 1.
Multi-step carcinogenesis [188].
However, the “Hallmarks of Cancer” characterize the last step of carcinogenesis (ie, cancer) and highlight a tension when comparing the biology of cancer to that of premalignant conditions. Some “Hallmarks of Cancer” are evident early in the course of carcinogenesis [10] and have bearing for chemoprevention efforts, while others may not wholly or accurately characterize premalignant conditions. For example, the hallmarks of ‘self-sufficiency in growth signals’ and ‘insensitivity to anti-growth signals’ are characteristics consistent with alterations evident in premalignant lesions, where increased DNA synthesis and a higher proliferation index have been documented [11,12]. Limitless replicative potential and evading apoptosis contribute to the sustained presence and proliferation of premalignant cells. Angiogenesis, one of the classical “Hallmarks of Cancer”, is also a characteristic of many premalignant conditions [10,13,14]. It provides a pathway for cells that have acquired the necessary alterations to metastasize. Of course, the ability to invade the basement membrane of a tissue is a clear demarcation between premalignant and malignant cells. Thus, it seems that many of the characteristics that we ascribe to cancer are also apparent in premalignant cells, in a way blurring the boundaries of distinction.
Beyond comparing the molecular or biological properties of a premalignant cell to a cancer cell, it is important to recognize that the microenvironment—including immune cells, stromal cells, other cell types and extracellular proteins—can influence the surrounding cells. In head and neck cancer for example, the premalignant microenvironment elicits pro-inflammatory cytokine production, in contrast to the tumor microenvironment, which is less immune stimulatory [15]. Alterations in the microenvironment can be present early before changes in the epithelial cells or these changes may be a reaction to excess proliferation of premalignant or malignant cells [16]. Indeed, as argued by the Tissue Organization Field Theory (TOFT), the progression of normal healthy tissue to a precancerous and eventually malignant state is primarily a problem of tissue organization. The theory proposes that deterioration of the microenvironment by carcinogenic agents or deleterious environmental exposures destroys the normal tissue architecture, disrupts cell-to-cell signaling, compromises genomic integrity and facilitates genetic mutation accumulation [17,18].
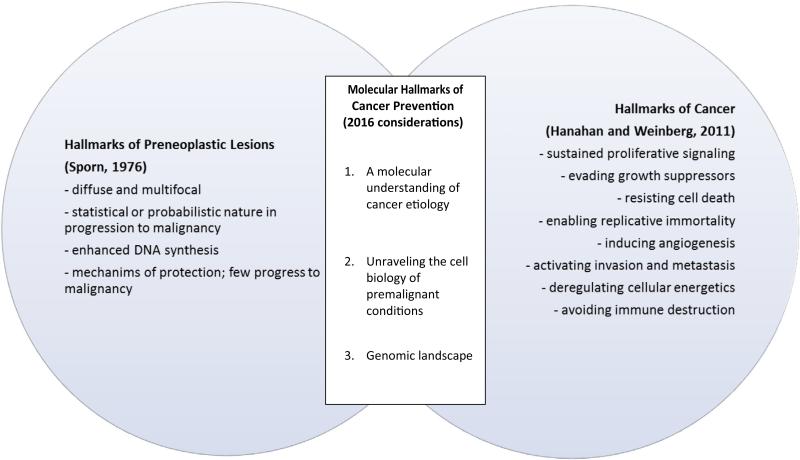
As shown in Fig. 2, the “Hallmarks of Cancer” do not necessarily equate to the hallmarks of premalignant conditions; yet, there are overlapping and emerging fields of research that bridge these areas. In truth, the field is not sufficiently advanced to collate a definitive molecular portrait of premalignant conditions but advances in molecular technology have enabled a greater understanding of cancer biology and have provided us with guideposts to begin to understand the trajectory of conditions that progress to a malignant state. Herein, we borrow from this framework and attempt to reorientate it towards elucidating the Molecular Hallmarks of Premalignant Conditions.
Fig. 2.
A paradigm for understanding and harnessing the molecular hallmarks of premalignant lesions—connection of 1976 hallmarks of preneoplastic lesions with 2011 hallmarks of cancer.
2.2. Translating a molecular understanding of cancer etiology into cancer prevention, including chemoprevention
Understanding the molecular origins of cancer provides opportunities for the design of precise chemoprevention strategies and other cancer prevention approaches (eg, nutritional, behavioral). For example, the viral etiology of certain cancers provided a direct path for demonstrating how a successful intervention can be developed once the molecular mechanisms are understood. Early studies of cervical cancer focused on lifestyle factors and describing the populations most likely to have an excess burden of disease, while later studies of the pathology of cervical lesions detected the presence of viral-like particles, subsequently identified as the human papilloma virus (HPV) [19]. The molecular studies that followed focused on how the virus infects cervical cells and disrupts normal cellular control of cell proliferation and genome stability. We now know that the protein products of the HPV viral genome inhibit the function of two important tumor suppressors; retinoblastoma protein (pRb) and p53. Although the development of vaccines to target these abnormalities in existing infections hold promise, this therapeutic approach does not yet exist.
Nevertheless, elucidating that HPV infection was the necessary, causative agent in cervical cancer etiology and understanding how HPV infection leads to the transformation to cervical cancer cells, unequivocally pointed to the potential of a prophylactic vaccine that would bolster the body's immune response to prevent HPV infection in the first place. Thus, in large part due to the seminal work of Lowy and Schiller, who discovered that a particular HPV protein called L1 could elicit such an immune response, several vaccines (Gardasil [Merck, Kenilworth, NJ], Cervarix [GlaxoSmith-Kline, Philadelphia, PA], and Gardasil-9 [Merck]) were developed that help the immune system make neutralizing antibodies that protect against HPV infection [20]. Importantly, population-based evidence shows that these vaccines are effective in reducing the incidence of the premalignant lesions, cervical intraepithelial neoplasia (CIN)2 and CIN3, as well as cervical cancer [21–23].
Liver cancer is associated with hepatitis B virus (HBV) and, as with HPV, prophylactic vaccines to thwart HBV infection have been developed. In 1984, Taiwan introduced the first national vaccination program targeting children and began vaccinating against HBV infection. A substantial decrease in liver cancer incidence and mortality [24] has mirrored this intervention, in itself a distinct success for cancer prevention efforts.
Conversely, the association of the bacterium Helictobacter pylori, a known carcinogen for non-cardia gastric cancer [25], demonstrates that understanding the etiological cause of cancer does not always lead to the development of an unequivocal preventative agent. While eradication of H plyori infection, especially cytotoxin-associated gene A (CagA) + H pylori, has been associated with a decreased incidence of gastric cancer [26], the same intervention is correlated with a rise in esophageal adenocarcinoma [27]. As such, H pylori eradication has been the subject of contentious debate [28,29]. One suggested mechanism for this undesirable association is the increase in acid production resulting from H pylori eradication, which leads in turn to an increase in esophageal adenocarcinoma [30]. H pylori could also skew immune and cytokine responses in ways that affect the adjacent esophagus [30]. From a public health perspective, the solution may lie in further study of focused eradication programs in areas such as South America and East Asia [28] where the prevalence of H pylori infection and incidence of stomach cancer are very high. This example highlights one of the challenges of cancer prevention strategies—the avoidance of unintended deleterious consequences.
Another example of how understanding disease etiology has led to successful chemoprevention is the elucidation of the role of estrogen in breast carcinogenesis, where efforts to disrupt estrogen pathways have led to Food and Drug Administration (FDA)-approved preventative interventions [31]. Although the hypothesis that estrogen blockade would be an effective breast cancer preventative strategy was first proposed by Lacassagne in 1936 [32], more contemporary experimental evidence that blockade of estrogen signaling could be chemopreventive came in the 1970s. In mice that were highly susceptible to mammary carcinomas, removal of the ovaries at an early age significantly reduced tumor incidence [31,33]. Later, estrogen was implicated as the circulating factor produced by the ovaries linked to the reduction in mammary tumor incidence, echoing the précis of both Schinzinger [34] and Beatson [35], who as far back as 1896 postulated an association between ovarian hormones and breast cancer. Further molecular elucidation of its mode of action demonstrated that estrogen binds to the estrogen receptor in the cellular cytoplasm. This receptor then translocates to the nucleus where it binds to DNA and recruits additional factors to turn on transcription of genes involved in cellular proliferation. Selective estrogen receptor modulators (SERMs) were developed to block estrogen binding to the estrogen receptor. The SERM tamoxifen was shown in adjuvant trials for early-stage breast cancer to be important not only for the treatment of existing cancers, ie, to prevent recurrence and metastases, but also for prevention of new primary cancers in the contralateral breast in the same individuals. These data led to implementation of several tamoxifen prevention trials in high-risk women. In the United States, a large multicenter randomized controlled clinical prevention trial provided the basis for the FDA's 1998 approval of tamoxifen for risk reduction of breast cancer in high-risk women [31,36].
Today, this seemingly old story continues to reinvent itself, as more is learned about the actions and signaling of estrogens [31]. For example, we now know that there are many bioactive estrogen metabolites in the circulation [37,38]. While some of these metabolites signal through the classical estrogen receptor pathway outlined above, others can promote carcinogenesis through receptor-independent mechanisms, such as direct binding to DNA and destabilization of chemical bonds, which leads to an abasic site. Such sites can lead to mutations if not repaired correctly. Additional proteins, so-called co-activators and co-repressors, can influence the action of estrogens, especially in the context of binding to the estrogen receptor and regulating gene transcription. Methods that target the function of estrogen will continue to evolve as the complexity of ligands, receptors, co-activators, and co-repressors and their expression, concentration, compartmentalization and action across tissues is unraveled.
Estrogen receptors are members of a larger nuclear, steroid-hormone receptor family. Recently, advances in estrogen receptor biology in breast cancer have been leveraged for chemoprevention strategies in other cancers where signaling from nuclear hormone receptors play an important role. In prostate cancer for example, Selective Androgen Receptor Modulators (SARMs) have been developed [39]. In mouse models, administration of SARMs leads to reduced tumor growth and reductions in PSA levels [39], suggesting they could be effective agents for prostate cancer treatment and perhaps prostate cancer prevention. However, it has yet to be tested whether or not these agents can be effective during the natural history of prostate cancer progression and whether they can prevent the progression of prostatic intraepithelial (PIN) lesions to malignancy.
Successful cancer preventative strategies and interventions that involve lifestyle and behavior changes have also evolved from understanding the molecular etiology of cancer. For example, in 1950, Richard Doll and Austin Hill published the first scientific evidence linking smoking to lung cancer [40]. Tobacco smoke contains thousands of chemicals and at least 60 known carcinogens. Many of these are known to form bulky DNA adducts, which lead to mutations and the loss of normal growth control mechanisms [41]. These epidemiological and extensive laboratory studies outlining how tobacco-specific constituents are carcinogenic has led to public health policies that restrict tobacco sales to minors, limit workplace exposure to secondhand smoke, and increase communication efforts that convey the risk of smoking to the population. Asbestos—which is a natural occurring fibrous hydrated magnesium silicate mineral—has also been linked with lung cancer incidence [42], in large part due to the seminal work of Blot and Fraumeni [43]. Asbestos, when inhaled into the lungs, causes scarring and inflammation and upregulation of the protein HMBG1, which over time contributes to the development of mesothelioma, a cancer that usually occurs in the pleural lining of the lungs and chest wall [44]. These research studies led to a series of Environmental Protection Agency (EPA) regulatory guidelines aimed at reducing exposure to asbestos within the population with the goal of reducing the future incidence of associated cancers.
2.3. Leveraging the molecular hallmarks of early and premalignant conditions for cancer prevention in the screening setting
As outlined above, there are several examples of how unraveling the complex etiology of cancer can lead to the development of successful primary cancer prevention strategies. The secondary prevention of cancer, which includes interventions designed to detect and remove early-stage cancer or premalignant lesions, is another path where understanding the molecular characteristics of cancer can be leveraged to prevent further progression. Cancer screening has had several successes, including use of the Papanicolaou test (also known as the Pap test) for the early detection and prevention of cervical cancer. Introduction of this test and colorectal cancer screening have led to significant decreases in incidence and mortality associated with the respective cancers. In both cases, the screening program detects premalignant lesions (CIN and adenomas), the identification and removal of which reduces the incidence of early cancers, thus reducing mortality. There is also now optimism that role-out of low-dose computed tomography (LDCT) screening will contribute to a reduction in lung cancer mortality among high-risk smokers [45].
The goal of having highly sensitive screening methods has led to some unintended consequences, primarily the detection of indolent lesions. Esserman and colleagues termed these lesions “indolent lesions of epithelial origin” (IDLES), which by and large pose little to no threat to the health of the individual [46] and which, if left alone, would not become clinically apparent or lead to death. Such overdiagnosis can place a significant psychological burden on the patient and places healthy people at risk of overtreatment. Thus, understanding the molecular hallmarks of premalignant conditions and early invasive lesions could facilitate their classification into aggressive versus indolent, which in the case of premalignancies could be considered “benign” conditions.
To address these limitations, many research groups are now working to marry the benefits of screening with that of biomarkers for more accurate and early diagnoses of cancer. By understanding and harnessing the characteristics of early-stage tumors, scientists are profiling coding and non-coding gene expression patterns [47,48], as well as assessing epigenetic [49], metabolomic [50], microbial [51], and inflammatory profiles [52]. Indeed, there is an emerging recognition that with recent advances in molecular biology, technology and bioinformatics, a precision medicine [53] approach should be developed. The concept of precision medicine or precision prevention [54] takes individual variability into account [53] and integrates all potential patient and “omic” data layers into a diagnostic algorithm that gives the most accurate (and early) diagnosis possible [55]. Successful examples of precision medicine for cancer treatment already exist [56]. However, the goal of a collective movement from traditional medicine to precision medicine has recently been placed at the center of the National Institute of Health's research mission [53]. Although much of the methodology necessary to reach this goal has yet to be developed [53], the initiative's success will undoubtedly lie in the synergy between technology development and many research fields.
2.4. Deciphering the genomic hallmarks of premalignant conditions
Whole genome and exome sequencing of cancer has demonstrated that most solid tumors have between 20–80 mutations [57]. In exploring these mutations, Vogelstein and colleagues have proposed a model for distinguishing the mutations “driving” the development of cancer from the mutations that are merely “passenger” or bystander mutations [57]. A driver mutation is defined as a “mutation that directly or indirectly confers a selective growth advantage to the cell in which it occurs.” Conversely, a passenger mutation has “no direct or indirect effect on the selective growth advantage of the cell in which it occur(s)” [57]. Through predetermined criteria for the frequency and type of mutation in a given gene, it was determined that, for cancer, there are currently around 140 genes that can fit the criteria for driver genes. Approximately 55% of these are categorized as tumor suppressors, while the remaining 45% are oncogenes. Whole genome and exome sequencing studies have illustrated the vast amount of genetic heterogeneity that exists among cancer cells in the same mass and between cancers in two different patients. Indeed, the ability to identify actionable mutations that inform novel treatment options has been transformative to the approach to cancer treatment.
Studies have focused predominantly on cancers and as such, the somatic landscape of premalignant conditions remains largely undefined. As premalignant conditions are the precursors to cancer, it is not surprising to find some overlap in genomic features. However, evidence suggests that there are differences in the genomic landscapes of these two distinct states. HER2, a key “driver” gene in breast cancer, is overexpressed in 40%–70% of ductal carcinoma in situ (DCIS) (a premalignant lesion of the breast) [58] and “just” 15%–30% of invasive breast cancers. In addition, KRAS mutations are detected in 14% of precursor adenoma lesions compared with 47% of colorectal cancers [59], making it highly likely that the actionable information we discern from these mutations in cancer will be different in premalignant conditions.
Is it possible that mutational profiles could help identify which lesions will progress to malignancy? One study found patients with DCIS that progressed to breast cancer had a significantly higher estrogen receptor-alpha level and Ki-67 labeling index (LI) than those that did not [12]. In addition, a low Ki-67 LI was associated with a 3% cumulative incidence of breast cancer at 10 years, compared with 14% in those with a high Ki-67 LI [11]. Increased mammographic density was also linked with DCIS progression to invasive cancers [60,61]. These data suggest that genomic and phenotypic profiles could be useful. However, findings across studies are not always consistent and many studies suffer from low power [62], something that again adds to the argument to include early and premalignant conditions in future large-scale population studies.
In addition to predicting whether or not premalignant conditions will progress to malignancy, could such conditions or lesions be targets for prevention? Again, evidence suggests that efforts to unravel the somatic landscape of premalignant conditions could be valuable. For example, short-term administration of lapatinib, which targets mutant epidermal growth factor receptor (EGFR), was found to decrease cell proliferation in ductal intraepithelial neoplasia [63] while, in mice, lapatinib prevented the development of estrogen receptor–positive mammary tumors [64]. Another tyrosine kinase inhibitor, gefitinib, might also be useful in the prevention setting. A patient-derived xenograft model of DCIS revealed a significant anti-proliferative effect of gefitinib [65] and may provide a rationale to eventually test this compound in a prevention trial. Emerging chemopreventative options also include administration of PARP inhibitors, which delay tumor formation in BRCA-deficient mice [66].
A concept related to that of “driver” and “passenger” mutations is that of oncogene addiction [67]. This concept posits that—for some cancers—activating oncogenic mutations occur during the course of the disease that confer a survival advantage to cancer cells. Cells then become dependent for survival on the signaling that results from the activating mutation. Identifying and targeting these mutations should, therefore, lead to death in cancer cells. A striking example of this is the BCR-ABL translocation in chronic myelogenous leukemia (CML). The BCR-ABL gene is formed via a fusion between chromosome 9 and chromosome 22. The resulting gene product is a fusion protein combining the breakpoint cluster region (BCR) with a constitutively active Abl kinase. It is an early event in the course of CML and the newly combined chromosome 9:22 (also known as the Philadelphia chromosome) is present in all transformed cells in CML [68]. There are now several drugs targeting the Bcr-Abl protein and its kinase activity. When individuals with CML develop resistance to these drugs, it is due to secondary mutations in BCR-ABL, not compensation by another pathway, suggesting that CML is addicted to the specific BCR-ABL oncogene and not evolving to become reliant on altered signaling from other molecules [69].
In CML, the BCR-ABL mutation is the initiating genetic change in the course of the disease, providing an early marker and target for intervention. This example brings up several questions spanning the areas of cancer treatment and cancer prevention. Would it be possible to identify the “driver” and “passenger” mutations in premalignant conditions? Are these concepts and that of oncogene addiction even valid in a premalignant context? Could this information be used to develop successful chemopreventive interventions?
Further detailed characterization is needed to understand the somatic landscape of premalignant conditions. It will not be sufficient to study isolated mutations in genes such as EGFR, KRAS and HER-2. As mentioned above, HER-2 is overexpressed in 40%–70% of DCIS lesions and is amplified in approximately 30% of breast cancers [70]. What this suggests is that the cellular context, timing, and order of genetic mutations are key components classifying a lesion as benign, premalignant or cancerous. Studies need to be conducted in a prospective manner so that we can learn which molecular features predict those that will progress to cancer, and moreover, which mutations might be targets for chemoprevention. We acknowledge, however, that these goals will not be easy to attain. Tissue biopsies will be needed to conduct these studies, and while powerful, whole genome sequencing technologies will need greater sample input than what is often available. There is the possibility of pushing the boundaries of technology even further to detect circulating tumor cells sloughed off from developing lesions, and to merge that with next generation sequencing to detect specific mutations that can portend a developing malignant tumor. Benign tumors from various lineages display a range of mutated genes, many of which may be considered as driver genes in a cancer context [71]; thus the timing of mutations and the collective repertoire of somatic changes may be the key to knowing whether a lesion is benign or whether it has a high probability of progression to malignancy. Will we be able to define a mutational spectrum that is associated with risk of colorectal cancer in patients with inflammatory bowel disease? And if so, can we detect such mutations in biopsies or stool samples and use them to predict which patients are at high risk of developing cancer? Can we study prostatic intraepithelial neoplasia and identify mutations that confidently predict disease progression? We think it is possible, but acknowledge that it will be difficult to fund cohorts large enough and with the necessary follow-up to accrue sufficient cancers for an analysis that is robust enough to answer these kinds of questions.
In examination of the molecular basis for cancer prevention, three key themes emerge. Firstly, understanding the etiology of cancer and resolving the molecular mechanisms that transform healthy cells into cancer cells has led to behavioral, policy, and medical interventions that have reduced the incidence of cancer. Additional efforts in this regard will undoubtedly yield additional successes for primary cancer prevention. Secondly, while not certain to be as effective, it is highly possible that leveraging the molecular biology of transformed cells can be used for secondary cancer prevention and the detection of premalignant lesions before they become invasive. Thirdly, sketching out the genomic landscape of premalignant conditions may well convey which lesions will develop to invasive cancers. Moreover, analagous to how these mutations are used to guide treatment for cancer, perhaps they can be used to “treat” premalignant conditions.
3. Emerging topics in the molecular hallmarks of cancer prevention
3.1. Linking specific exposures to tumor subtypes
Exposure–phenotype relationships are very heterogeneous. For years, we have failed to understand why only some smokers develop cancer, or why aspirin does not prevent cancer in all users. Such heterogeneity has been frustrating to understand. The advent of next generation sequencing and array technology means that we are now able to link specific exposures to molecular tumor subtypes, which are based on distinct expression and genomic signatures. These new technologies have given us a finer lens with which to resolve the relationship between specific exposures and tumor subtypes. For example, we have resolved that obesity is associated with triple-negative breast cancer [72], HPV is associated with a specific mutation pattern in head and neck cancer [73], benzene exposure leads to a distinct gene expression signature in acute myelogenous leukemia (AML) [74] and aflatoxin exposure is associated with R249S TP53 mutations in human hepatocellular carcinomas [75].
The field has also been called molecular pathological epidemiology (MPE), and is conceptually defined as the epidemiology of molecular pathology, pathogenesis and heterogeneity of disease [76]. The principle of MPE is to unravel the relationship between genetic and lifestyle exposures and the molecular processes of carcinogenesis. For example, regular use of aspirin after a diagnosis of colon cancer is associated with a better clinical outcome for some individuals. However, further study demonstrated that this advantage was restricted to those carrying PIK3CA mutations, suggesting that the PIK3CA mutation may serve as a predictive molecular biomarker for adjuvant aspirin therapy [77,78].
This approach to cancer research and cancer prevention lends an unparalleled opportunity to link specific exposures with tumor subtypes and disentangle some of the heterogeneity we find in exposure–cancer relationships. To fully do so, it will be necessary to catalog the entire human exposome, as defined by Wild in 2005 [79]. Such a map would only be possible with advances in methodological tools for environmental exposure assessment and would parallel the tools used for evaluation of the genome, which was investigated with The Cancer Genome Atlas (TCGA) and Genome Wide Association Studies (GWAS) [79,80]. Combined, these two instruments would constitute a powerful approach to precisely evaluate the consequences of environmental exposures for cancer and cancer prevention.
3.2. Cancer stem cells
In addition to the recent explosion in our understanding of the genetic complexity of cancer and a slow, but evolving understanding of how this relates to premalignant conditions, there is an increased understanding of the biology of cancer, and how this could also provide targets for cancer chemoprevention. One example of this involves recent revelations that much of cancer biology mirrors that of normal stem cell biology. The cancer stem cell (CSC) theory proposes that cancer is initiated and maintained by a sub-population of stem-like cells within a given tumor [81–83]. Mouse models support the hypothesis that CSCs arise from the malignant transformation of normal adult stem cells [84–86]. Indeed, studies of human lymphoid tumors also support this possibility [87,88].
Experimental evidence for CSCs first came from studies of human acute myeloid leukemia in the mid-1990s [89,90]. where normal haematopoietic stem cells were determined to be the cell of origin for AML. However, the theory itself was first proposed over a century ago by pathologists who, when reviewing slides from patients with cancer, noted that cancer cells had the appearance of embryonic cells. They even theorized that cancer was the result of a “reactivation” of remnant embryonic stem cells in the adult. The literature has not coalesced around a single theory for the derivation of “cancer stem cells”, and evidence variously suggests that they may arise from normal adult stem cells (an undifferentiated cell that is almost endlessly capable of giving rise to identical daughter [stem] cells and other cells that can differentiate into more specialized cell types), progenitor cells (intermediary cells derived from stem cells that have the ability to produce differentiated cells but cannot replicate indefinitely) and even adult differentiated cells. Indeed, this sub-population of cells does not have a unifying term to define them, with some resolving to call them cancer cells with stem-like properties.
The CSC hypothesis states that these particular cells are endowed with intrinsic chemo-resistant, tumor initiating, immune evading and metastasis-driving characteristics [82,83]; thus, targeting them for cancer treatment, is both attractive and quite likely to be successful. However, the field of cancer stem cells may also have relevance to cancer prevention.
Firstly, several models support the hypothesis that cancer stem cells derive from dysregulated normal stem cells [84–86,90,91]. Thus, given that cancer stem cells could be the cells of origin for several malignancies, strategies that prevent the conversion of a normal stem cell to a cancerous one could be a novel cancer prevention mechanism. In this regard, it is interesting to note that three of the main pathways that control the self-renewal of normal and cancer stem cells, ie, Notch, Sonic Hedgehog and Wnt/β-catenin, have been linked with chemopreventative agents. For example, the Wnt/β-catenin pathway is inhibited by curcum—a polyphenol found in the Indian spice turmeric [92–97]. Epigallocatechin-3-gallate (EGCG), the most abundant polyphenolic catechin in green tea, also inhibits the Wnt self-renewal pathway [98–101]. The link between high consumption of cruciferous vegetables (eg, broccoli and broccoli sprouts) and cancer prevention is supported by many epidemiological studies. Evidence suggests that sulforaphane, which is produced from a major glucosinolate in broccoli/broccoli sprouts [102,103] (see Yang et al in this issue), could exert its anti-cancer properties [104,105] by targeting the Wnt pathway [97,106–108]. Evidence also suggests that resveratrol, a polyphenol derived from a wide variety of plants such as grapes, berries, plums, and peanuts [109] and vitamin D3, may also target this pathway [97,101,110]. While the data presented above are promising and suggest that dietary constituents could have anti-tumor properties related to the targeting of cancer stem cells, most, if not all of the current studies use model systems that start with cancer. Thus, studying the ability of dietary compounds to prevent cancer via the suppression of cancer stem cell derivation will require a finer lens and an alternative experimental approach. For example, several well characterized mouse models with normal stem cells as the cells of origin for cancer have been developed (eg, LGR5+ cells in the intestinal crypt are the cells of origin for colon adenomas and carcinomas) [86,111,112]. These models can be used to interrogate whether dietary compounds can prevent the transformation of a normal stem cell into a malignant one.
Indeed, evidence is now emerging that chemopreventative agents are involved in cancer prevention via suppression of cancer stem cell evolution. Nonsteroidal anti-inflammatory drugs (NSAIDs) are well-known chemopreventive drugs in colorectal cancer (CRC) (see Umar et al in this issue), and recent studies have shown that NSAIDs may reduce the recurrence and mortality of CRC [78,113–116]. Work now suggests that this cancer prevention effect is mediated, at least in part, via control of dysregulated normal stem cells [117]. Leibowitz and colleagues have shown that NSAIDs prevent CRC by inducing the cell death of normal intestinal stem cells that have acquired mutations and lost functional APC [118]. These results suggest that effective chemoprevention of colon cancer by NSAIDs lies in the elimination of stem cells that are inappropriately activated by oncogenic events through induction of apoptosis [118].
Cancer stem-like cells have been detected in premalignant conditions [119]. Evidence supports the notion that the conversion of a normal stem cell to a cancer stem cell is an early event in carcinogenesis. It is important to note that not all cancers follow the cancer stem cell model [120]. However, as mentioned earlier, further understanding of the molecular basis for how chemopreventative agents might prevent the conversion of normal stem cells to cancer stem cells is critical and warrants further study.
3.3. The RNA world
Less than 2% of our genome accounts for our ~21,000 protein-coding genes. However, it is estimated that the human genome also encodes 9,000 small RNAs and 10–32,000 long non-coding RNAs and contains 11,000 pseudogenes [121]. The most studied class of small non-coding RNAs are the microRNAs, single-stranded mRNA sequences that are 19–25 nucleotides in length [122]. This RNA species functions mainly to repress protein translation. The first evidence showing a relationship between miRNAs and cancer came when Croce and Calin identified miR-15 and miR-16-1 as tumor suppressors in CML [123]. Since then, miRNAs have been associated with cancer etiology, progression and prognosis in multiple cancer types [47,124–134].
De-regulation of miRNA expression has been observed in premalignant conditions. For example, miR-21 is detected in DCIS [135] and adenomas [136], making it possible that such signatures could be used to detect cancer early and in premalignant stages. Indeed, a miRNA classifier has been identified that discriminates between benign and malignant lung lesions detected by low dose CT screening [137]. Moreover, as clinical trials are now underway targeting miRNAs, such as miR-21 [138], it is possible that such molecules could be targets of treatment in premalignant conditions that overexpress these “oncomiRs”.
As with the genome, the RNA world of early and premalignant conditions is largely unexplored; therefore, how the RNA landscape of these lesions compares with invasive cancers is not fully known. As mentioned above, the RNAome includes much more than miRNAs and RNA sequencing has led to a rapid rise in the discovery of new RNA species. Its potential translation to the detection of early cancers and premalignant conditions, however, lags behind.
There are many parallels between the genome and RNAome, yet it is worth remembering one key distinction. The genome has a relatively stable architecture. Profiles of RNAs—and indeed of microbes, epigenetics, metabolites and proteins—are variable with time [139]. This does not nullify their exploration or their potential value—temporal complexity is likely to increase the amount of information encoded in these profiles—but it does warrant additional caution when designing and interpreting studies of such entities.
3.4. Transgenerational inheritance and epigenetic modification
In this time of exponential growth in sequencing technologies, the field of epigenetics has also expanded and evolved. Recent reports that the establishment of epigenetic states can be altered by the environment, combined with the idea that epigenetic states can be inherited across generations, has generated interest in the hypothesis that epigenetic traits influenced by the environment and/or lifestyle may persist and subsequently be inherited across generations.
Unlike DNA, which from a heritability perspective is relatively stable, epigenetic marks are “erased” across the genome, initially following fertilization of the egg and secondly during the formation of germ cells. This is to remove imprints and epimutations and to ensure totipotency in early embryonic development. Thus, for transgenerational inheritance to occur at a specific locus, this reprogramming must somehow be bypassed.
Some of the clearest evidence for transgenerational epigenetic inheritance comes from studies in plants and mice [140–143]. In the dandelion, exogenous stress induces changes in DNA methylation that are transmitted to the next generation [141–143]. In the mouse, the Avy locus, a gene region that controls coat color, can be epigenetically modified by environmental exposures such as diet [142]. These epigenetic modifications can be transgenerationally inherited and influence phenotypes in the offspring [140,142], suggesting that there is a failure to erase the epigenetic marks that are established at the Avy locus in the germline.
The epigenetic marks that facilitate transgenerational epigenetic inheritance are poorly understood [144]. The simplest explanation has always been that DNA methylation at specific loci escapes the reprogramming events. But how? One possible mechanism might involve retrotransposons, as these DNA repeat elements seem to be resistant to the erasure of DNA methylation patterns during reprogramming [143–147]. In fact, the Avy locus is actually a retrotransposon inserted upstream of the transcription start site of the Agouti gene. Additionally, recent work in Caenorhabditis elegans suggests that epigenetic marks, such as histone H3 lysine 4 trimethylation could mediate transgenerational inheritance [148–150].
The recent discovery that germline cells contain large numbers of small RNA species suggests another novel way of transmitting epigenetic information through the germ line [143,144,151,152]. Research has shown that the maternal transmission of piwi-interacting RNAs (piRNAs) in Drosophila influences fertility of offspring via piRNA-directed silencing of transposable elements [143,153]. Recent work also demonstrated that early developmental trauma in mice leads to behavioral and metabolic changes that are inherited in offspring for several generations [143,154]. These phenotypes were conveyed to the offspring by miRNAs and piwiRNAs in the sperm [154]. The relevance of this observation to cancer is apparent when one considers that cigarette smoking induces differential miRNA expression in human spermatozoa [155], perhaps suggesting one mechanism by which tobacco exposure can influence not just the cancer risk of the exposed individual, but also that of the offspring.
One of the earliest examples of transgenerational inheritance of traits in humans was in 2001 when a Swedish study found that the nutritional and smoking habits of paternal grandparents could influence their descendants’ lifespan [156]. In addition, prenatal exposure—especially around the time of conception—to famine during the Dutch Hunger Winter (1944-1945) led to hypomethylation at the IGF2 gene among offspring conceived at that time [157]. A key future area to explore will be to determine what the phenotypic effects of such exposures and epigenetic changes are. Recent experimental evidence shed light on a potential mechanism for this observation and may also involve transmission of small RNAs. Starvation was found to induce expression of specific RNAs that were then inherited across multiple generations, targeting genes involved in nutrition [158].
So, can the food we eat, the air we breathe and our emotional experiences impact the health and happiness of our descendants? [144] While studies undoubtedly show evidence for an intergenerational effect of the environment on epigenetics and phenotypes, there is an important caveat to consider when conceptualizing the implications of transgenerational inheritance and generating experimental evidence for it: When an embryo is exposed to environmental stresses during pregnancy, not only the mother, but the F1 generation (embryo) and its developing germ line, which will give rise to the F2 generation are also exposed to these triggers [143,144]. Thus, to truly assess transgenerational epigenetic inheritance, one needs to look at further generations—something that is difficult for epidemiological and laboratory studies, but which is now being emphasized [143,144,155,159].
Understanding how the environment and our lifestyles impact epigenetic inheritance, and the extent to which it exists, could have significant implications for how we think about the molecular basis for cancer prevention. The oft-believed perception that the deleterious consequences of behavior and exposures are erased during the formation of the next generation may not be as steadfast as we originally thought, and as Luther Burbank put it in 1906, “heredity is only the sum of all past environments” [143,144,155,159]. In addition to genetic inheritance of phenotypes, it is possible that a system of epigenetic inheritance is superimposed [144,160], one that is both plastic and responsive to the external environment [144]. If this is the case, and the effects of chronic stress, carcinogens, or poor nutrition are epigenetically remembered and passed on through generations, the implications for cancer prevention, and how to conceptualize it, could be compelling.
4. Defining at-risk populations
As John Potter wrote, chemoprevention is predicated on the assumption that we can identify those at risk [161]. And, it is often acknowledged in prevention research that “... it is difficult to make healthy people healthier” [162]. It is likely there will be risks with any chemoprevention intervention and that individuals with an average likelihood of developing disease (ie, the general population) would not want to incur harm from an agent designed to prevent a disease the person does not yet have.
However, it may be possible to make healthy people healthier. The administration of cholesterol-lowering drugs, those that inhibit platelet aggregation and lower blood pressure, has led to a dramatic decline in deaths due to heart disease. In the mid-1970s the incidence of coronary heart disease (CHD) was twice that of cancer, but in 1999, the incidence rate fell below that of cancer for the first time, an unquestionable success for public health interventions. But within this success story, cautionary narratives emerge [163]. These drugs have many side effects that can limit use and uptake in the wider population. In addition, one of the main determinants of successful uptake of these drugs was the identification of reliable and measurable risk factors, such as cholesterolemia and hypertension. This has been a goal for many cancer biomarker researchers, but few robust biomarkers of risk have been identified and clinically translated. Chemoprevention strategies will therefore need to focus on individuals who have a greater likelihood of developing the disease and for whom the risk:benefit ratio from an intervention may be more favorable than that for the general population [163].
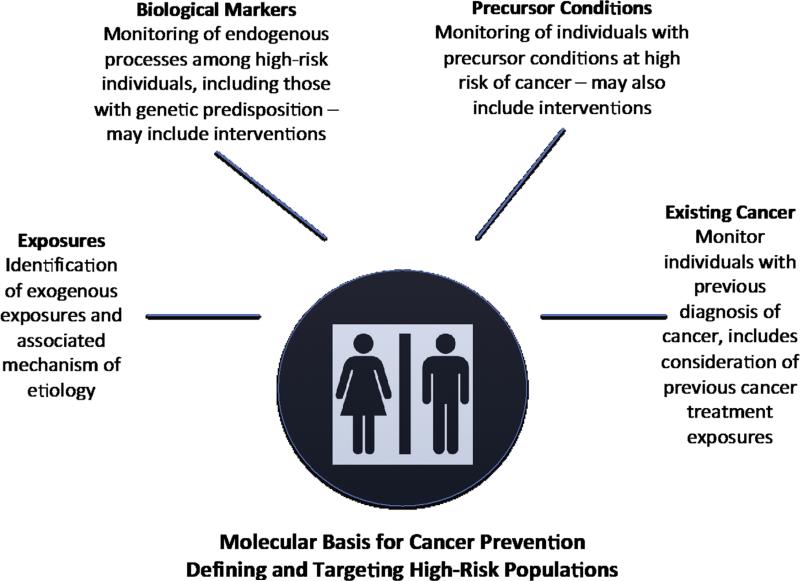
Thus conceptually, cancer prevention and the targeting of high-risk populations can be clustered in four main ways (Fig. 3). The first is identification of exposures, such as viruses, tobacco smoking, pollution, etc, that can increase cancer risk. Though validation of actual causality in exposure-disease relationships can be difficult, their identification is a powerful tool in prevention endeavors. Successful examples include smoking cessation, reduction in radiation exposure from the sun, and HBV/HPV eradication/prevention programs [22,24], to name a few. Some 16% of the 12.7 million new cases of cancer worldwide are attributable to infectious agents [164]. Thus, prophylactic measures against infections such as vaccination or antimicrobial treatments could have a substantial impact on reducing the cancer burden in areas endemic for these infections. In cases where avoidance of the primary exposure is not possible, another strategy is the use of agents that modify the carcinogenicity of an exposure. For example, aflatoxin B1 is a metabolite that is produced from the mold Aspergillus flavus commonly found in poorly stored foods and strongly associated with liver cancer. Studies are now being conducted to test dietary constituents [165], or chemical compounds [166] for their ability to neutralize the such toxins.
Fig. 3.
Defining high-risk groups within the context of the molecular basis for cancer prevention.
As mentioned, to fully leverage our ability to prevent cancer based on understanding exogenous environmental exposures, it will be necessary to catalog the entire human exposome [79]. Such a map would be challenging, but could be used in combination with genome-wide association studies, in so-called gene–environment-wide studies [167], to assess cancer risk due to environmental exposures. This would provide a finer lens and be akin to the MPE approach discussed earlier.
The second category of high-risk populations includes individuals who do not currently have cancer but have an underlying genetic susceptibility. Familial cases of cancer comprise, on average, 5%–10% of the total cancer burden in the population. Among the best-known genetic susceptibility mutations are those arising in the BRCA1 and BRCA2 genes. BRCA1 was first discovered in the early 1990s [168]. Presently, the United States Preventative Services Task Force has recommended genetic testing for those with a family history of breast, ovarian, fallopian tube, or peritoneal cancer [169]. However, it has been estimated that 50% of BRCA1/2 mutation carriers could be missed using these criteria [170,171]. Among those with no family history, the lifetime risk of developing cancer by age 80 years is 83% in BRCA1 carriers and 76% in BRCA2 carriers [171,172], in itself a potential argument for more widespread genetic testing for BRCA1 and BRCA2. For mutations that confer a high likelihood of breast or ovarian cancer development, there are prevention strategies an individual can take. These include prophylactic mastectomy and oophorectomy.
Similar to BRCA mutation carriers, individuals that carry germline mutations in TP53 carry a high lifetime risk of cancer development, including osteosarcoma, leukemia and soft tissue sarcoma. For these so-called Li-Fraumeni families (named after Fred Li and Joseph Fraumeni who discovered the disease) [172], a high level of monitoring aims to detect these cancers as early as possible. While there are many different forms of familial cancer where the underlying genetic predisposition is known, there are still others where the inherited molecular alteration is yet to be discovered.
TP53 and BRCA1/2 mutations are examples characterized by single high-penetrance genetic mutations, making cancer prevention interventions and recommendations possible. But most cancers are polygenic and, as such, they are much harder to predict. Many susceptibility mutations have been identified from GWAS studies, and unlike BRCA1 and BRCA2, most of the mutations identified are estimated to have small effect sizes on disease risk. In some ways, this limits the applicability and usefulness of these findings as it is difficult to suggest dramatic lifestyle/behavior changes to an individual based on a highly significant—yet small, increased absolute risk of cancer [173]. However, most GWAS findings are less than 5 years old and there are examples of GWAS loci that are in the process of clinical translation for several phenotypes, indcluding drug toxicity [174]. Therefore, although not without promise, it is not yet clear what degree of clinical utilty these highly significant susceptibility mutations will have for complex disease in low-risk populations.
Screening for a genetic pre-disposition to cancer in the genomics era brings additional challenges. While cancer screening with mammography or colonoscopy can lead to the early detection of a malignant or premalignant condition and genomic analysis of a cancer can lead to actionable mutations that guide treatment options, genetic testing of the population or high-risk individuals leads to the discovery of a predisposition to cancer [175]. In 2005, a non-federal working group was established to perform a systematic evaluation of genetic and genomic tests with a view towards establishing guidelines for their use. Their criteria extended beyond traditional outcomes, such as morbidity and mortality reduction, to consider societal implications as well [175]. Such broader benefits were included because mutation testing derives information that goes beyond the individual due to potential heritable implications for relatives and descendants. In addition, bearing such information can impact lifestyle and family planning decisions, among others. The “democratization of DNA” as described by Topol, proposes more effective ways to engage patients in their own healthcare management and provide treatments that will reduce the costs of healthcare, but it requires a balance between extensive sharing of molecular data and clear dissemination of the net benefits and harms such data can have for the patient's health [175,176]. While in the past, screening test evaluations considered clinical utility, many have recently advocated a shift towards appreciating “personal utility” [175]. The recent expansion of whole genome and exome sequencing, combined with increases in the marketing of such technologies to the public, is accentuating these challenges associated with genetic testing. Traditional BRCA1/2 testing has a narrow focus on a small region of the genome; however, current genetic sequencing services are affordable for many and are able to profile the entirety of the genome. This genomics revolution has also led to the identification of new susceptibility genes. PALB2 (partner and localizer of BRCA2) mutations, for example, are associated with a 35% cumulative risk of breast cancer by age 70 years [177]. However, in the case of BRCA1/2—while additional mutations have been discovered—some of them are exceedingly rare and the associations with risk of breast cancer are currently unknown. If applied to premalignant conditions in a systematic way, whole genome sequencing to detect somatic (as well as germline) mutations could lead to the identification of both known and unknown actionable mutations and, at present, there are scant guidelines for patients and doctors regarding how to deal with such information.
To consider the readiness of genomic applications for practice, a useful public health framework was proposed by Khoury and colleagues [178]. The framework considers a genomic test's utility through several useful lenses, including analytic validity, clinical validity, clinical utility, balance of benefits and harms, and the existence of an evidence-based recommendation. Indeed, a recent study suggested that genetic testing in high-risk populations does not adversely affect short-term psychological or quality-of-life outcomes [170]. However, screening for susceptibility to cancer by looking at mutations in TP53 and BRCA1/2 has been successful due to the high penetrance of these genes in terms of lifetime risk of cancer, and the net benefits of screening might be more difficult to translate to findings from GWAS or whole genome sequencing studies [178].
The third group of high-risk individuals includes those that have been diagnosed with a precursor condition, such as DCIS or adenomatous polyps. These individuals are a key group for whom interventions and monitoring are achievable. However, as discussed earlier, in the absence of a complete understanding of the natural history of the condition and knowledge of which premalignant lesions will progress to malignancy, it will be difficult to know for whom interventions are necessary and thus it will be important to conservatively manage such cases [60,179]. Furthermore, these individuals remain in a high-risk category, even following removal or regression of the premalignant condition. Screening tools such as mammography or LDCT can detect suspicious lesions that, while not indicative of cancer, could portend a developing cancer. Thus, these patients should also be grouped within a high-risk category and considered for cancer prevention and/or chemoprevention interventions.
The fourth category of high-risk populations includes those who have had a prior diagnosis of cancer. They constitute a key population for cancer and chemoprevention efforts and many are at high risk of developing a second cancer. Molecular information may be available from the initial cancer diagnosis that is informative when considering chemoprevention strategies. In the case of breast cancer, many chemoprevention agents beyond SERMs are being considered for women who have had a prior diagnosis of this disease. These strategies include other estrogen pathway targeting agents such as use of the drug metformin, which is currently approved for treatment of type 2 diabetes and has been shown to repress breast cancer cell proliferation [180], and cancer vaccines targeting known molecular alterations in specific breast cancer subtypes [181].
Treatment and therapy exposures among cancer patients are additional factors that should be considered in the context of patients with previous diagnoses of cancer. Studies have shown that risk of second cancers is higher among patients exposed to radiation and chemotherapy [182–184]. While data regarding the absolute risks of second cancers related to cancer treatment are still being unraveled [185,186], this remains an important factor to consider in defining and managing cancer prevention options in this group of patients.
5. Conclusions
The concept of cancer prevention is not new. As far back as 1727, Le Clerc recognized the potential to prevent colorectal cancer by removing polyps in the colon. Primary prevention strategies, such as vaccines, sanitation, and safer food and water, have extended life expectancy from 50 to 75 years over the space of a century, in itself proof that preventing disease can lead to gains in health outcomes. So why is it so difficult to prevent cancer? In a recent discussion by Fineberg, the “Paradox of Prevention” was considered [162]. He posed the question: If prevention is such a good idea, then why are we not getting more of it? Perhaps most endemic among these obstacles is the recognition that success in cancer prevention is, by and large, invisible, and the benefits of such preventative measures will not be seen for decades [162].
But there are other hurdles. As outlined in this review, the molecular hallmarks of premalignant conditions are largely undefined. Many of the successes in cancer research and treatment have come from advances in technology that unraveled the molecular complexity of cancer. In borrowing from the guideposts of advances in cancer research, we have conceptualized these hallmarks of premalignant conditions as falling within three main categories (Fig. 3). In doing so, we acknowledge that there is overlap in description and in purpose of each and that these are not mutually exclusive concepts. The first category includes epidemiology studies focused on cancer etiology, an understanding of which provides the basis for subsequent investigation of the molecular underpinnings for a particular cancer and a rich source of ideas for primary prevention strategies. The second category includes molecular (coding and non-coding genes, methylation, metabolism) characterization of premalignant conditions and early cancers to identify biomarkers for discrimination between indolent and high-risk lesions and for early detection in the primary and secondary prevention settings. The third category includes understanding the genetic alterations and mutations in premalignant lesions to identify those that have a high probability of progression to invasive cancer, and also to identify targets for novel chemoprevention agents.
To generate this atlas and to conduct studies on premalignant conditions, collaboration will be key, as assembly of sufficient, well-annotated biospecimens to provide meaningful data will be challenging. Can we envisage a premalignant condition atlas, cataloguing molecular, biological and genomic data, similar to the one that has been created for cancer? It would undoubtedly be a challenging endeavor. Such an atlas, however, could answer many additional questions: Are premalignant lesions multi-focal or heterogeneous? If we find a druggable target, how will we know which lesions will develop to cancer and thus merit therapeutic intervention? Building prospective cohorts that have multiple points at which exposures and biospecimens are collected will enable us to map out how disease progresses, understand the role that premalignant conditions play and how we may exploit them for preventative efforts. The Precision Medicine initiative may well represent one such resource which can be leveraged to gain insight into the natural history of cancer [53,55].
At the present time, cancer and chemopreventive efforts continue to evolve in a retrospective manner, working backward from what is known about invasive cancer. The ultimate goal is to have the molecular information, imaging capability, and model systems to reorient the field of cancer chemoprevention to begin with premalignant conditions and be able to predict future events in the carcinogenic pathway in order to intercede appropriately. Although there has been some frustration with the pace of cancer and chemoprevention research and the number of success stories, the complexity of this field is often vastly under-appreciated.
In a recent review, entitled, “Coming full circle—from endless complexity to simplicity and back again,” Weinberg outlined the trajectory of cancer research over the past 40 years [187]. The main theme of this article was the arc of cancer research: beginning with pathology, research revealed the complexity of various cancers at a cellular level. This was followed by reductionist approaches to molecular biology that elucidated the genes responsible for malignant transformation. The molecular alterations were categorized into simplified pathways to help organize the growing body of information. But new technologies have revealed an enormously complex network of genetic mutations, RNA and protein biology and with it, a challenge to understand the interplay between this biology and that of the cellular microenvironment—all of which exhibit considerable interindividual variation.
It seems cancer prevention is experiencing its own cycle of complexity to simplicity and back again and, in some ways, this complexity exceeds that of the treatment setting. As noted earlier, cancer treatment largely focuses on the end of the cancer progression spectrum while cancer prevention focuses on the earlier steps. This multistep pathway provides many opportunities for intervention long before an invasive cancer is detectable, but there is great difficulty in identifying these earlier lesions and in studying the key molecular alterations important for each step in the carcinogenesis process. Similar to what is seen in cancer, these early lesions are expected to be heterogeneous in cellular content and molecular alterations, both when comparing cells within a lesion in the same person and when comparing lesions of the same type across different people. The shifting cellular and molecular content as lesions progress to cancer make it likely that different interventions would be needed at different phases during the progression to cancer. The concept of precision medicine and precision prevention highlights this need to identify the right agent in the right dose for an individual depending on how far (ie, right time) their lesion has progressed. This continues to be the challenge and motivation for determining the underlying molecular alterations throughout the carcinogenesis process and also what may hinder the field of chemoprevention from having more notable success stories. Identifying the early lesions that are destined to progress to cancer, as opposed to the vast majority of non-progressing lesions poses challenges. The difficulties in identifying these lesions must be overcome, as molecular information cannot be obtained from a sample that cannot be collected.
In 1976, Sporn wrote that our understanding of premalignant lesions is “still in a very primitive state and that an immense amount of basic research remains to be done in this area” [1]. One could argue that this is still true today. And although the methodologies are difficult, and investment will need to be great, we would argue that there has never been such an opportunity to generate new molecular knowledge and to translate it into a foundation for cancer prevention.
Acknowledgments
The authors would like to thank Drs Curtis Harris and Michael Cook for the helpful comments and discussion during preparation of the manuscript.
Footnotes
Conflicts of interest
None.
References
- 1.Sporn MB. Approaches to prevention of epithelial cancer during the preneo-plastic period. Cancer Res. 1976;36:2699–702. [PubMed] [Google Scholar]
- 2.Steward WP, Brown K. Cancer chemoprevention: a rapidly evolving field. Br J Cancer. 2013;109:1–7. doi: 10.1038/bjc.2013.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stehelin D, Varmus HE, Bishop JM, Vogt PK. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature. 1976;260:170–3. doi: 10.1038/260170a0. [DOI] [PubMed] [Google Scholar]
- 4.Varmus HE, Weiss RA, Friis RR, Levinson W, Bishop JM. Detection of avian tumor virus-specific nucleotide sequences in avian cell DNAs (reassociation kinetics-RNA tumor viruses-gas antigen-Rous sarcoma virus, chick cells). Proc Natl Acad Sci U S A. 1972;69:20–4. doi: 10.1073/pnas.69.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le AT, Doebele RC. The democratization of the oncogene. Cancer Discov. 2014;4:870–2. doi: 10.1158/2159-8290.CD-14-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klauschen F, Andreeff M, Keilholz U, Dietel M, Stenzinger A. The combinatorial complexity of cancer precision medicine. Oncoscience. 2014;1:504–9. doi: 10.18632/oncoscience.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–41. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Lazebnik Y. What are the hallmarks of cancer? Nat Rev Cancer. 2010;10:232–3. doi: 10.1038/nrc2827. [DOI] [PubMed] [Google Scholar]
- 11.Hartmann LC, Reynolds C, Fritcher EB, et al. Ki67: A time-varying biomarker of risk of breast cancer in atypical hyperplasia. Cancer Res. 2009;69:533S–34SS. doi: 10.1007/s10549-009-0534-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaaban AM, Sloane JP, West CR, Foster CS. Breast cancer risk in usual ductal hyperplasia is defined by estrogen receptor-alpha and Ki-67 expression. Am J Pathol. 2002;160:597–604. doi: 10.1016/s0002-9440(10)64879-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raica M, Cimpean AM, Ribatti D. Angiogenesis in pre-malignant conditions. Eur J Cancer. 2009;45:1924–34. doi: 10.1016/j.ejca.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Sharma RA, Harris AL, Dalgleish AG, Steward WP, O'Byrne KJ. Angiogenesis as a biomarker and target in cancer chemoprevention. Lancet Oncol. 2001;2:726–32. doi: 10.1016/S1470-2045(01)00586-1. [DOI] [PubMed] [Google Scholar]
- 15.Johnson SD, De Costa AM, Young MR. Effect of the premalignant and tumor microenvironment on immune cell cytokine production in head and neck cancer. Cancers (Basel) 2014;6:756–70. doi: 10.3390/cancers6020756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casbas-Hernandez P, Sun X, Roman-Perez E, et al. Tumor intrinsic subtype is reflected in cancer-adjacent tissue. Cancer Epidemiol Biomarkers Prev. 2015;24:406–14. doi: 10.1158/1055-9965.EPI-14-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenfeld S. Are the somatic mutation and tissue organization field theories of carcinogenesis incompatible? Cancer Inform. 2013;12:221–9. doi: 10.4137/CIN.S13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soto AM, Sonnenschein C. The tissue organization field theory of cancer: a testable replacement for the somatic mutation theory. Bioessays. 2011;33:332–40. doi: 10.1002/bies.201100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–50. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 20.Kirnbauer R, Booy F, Cheng N, Lowy DR, Schiller JT. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci U S A. 1992;89:12180–4. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rana MM, Huhtala H, Apter D, et al. Understanding long-term protection of human papillomavirus vaccination against cervical carcinoma: cancer registry-based follow-up. Int J Cancer. 2013;132:2833–8. doi: 10.1002/ijc.27971. [DOI] [PubMed] [Google Scholar]
- 22.Muñoz N, Kjaer SK, Sigurdsson K, et al. Impact of human papillomavirus (HPV)-6/11/16/18 vaccine on all HPV-associated genital diseases in young women. J Natl Cancer Inst. 2010;102:325–39. doi: 10.1093/jnci/djp534. [DOI] [PubMed] [Google Scholar]
- 23.Lowy DR, Herrero R, Hildesheim A. Participants in the IARC/NCI workshop on Primary Endpoints for Prophylactic HPV Vaccine Trials. Primary endpoints for future prophylactic human papillomavirus vaccine trials: towards infection and immunobridging. Lancet Oncol. 2015;16:e226–33. doi: 10.1016/S1470-2045(15)70075-6. [DOI] [PubMed] [Google Scholar]
- 24.Chang MH, Chen CJ, Lai MS, et al. Universal hepatitis B vaccination in Taiwan and the incidence of hepatocellular carcinoma in children. Taiwan Childhood Hepatoma Study Group. N Engl J Med. 1997;336:1855–9. doi: 10.1056/NEJM199706263362602. [DOI] [PubMed] [Google Scholar]
- 25.Blaser MJ, Perez-Perez GI, Kleanthous H, et al. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111–5. [PubMed] [Google Scholar]
- 26.Ma JL, Zhang L, Brown LM, et al. Fifteen-year effects of Helicobacter pylori, garlic, and vitamin treatments on gastric cancer incidence and mortality. J Natl Cancer Inst. 2012;104:488–92. doi: 10.1093/jnci/djs003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Islami F, Kamangar F. Helicobacter pylori and esophageal cancer risk: a meta-analysis. Cancer Prev Res. 2008;1:329–38. doi: 10.1158/1940-6207.CAPR-08-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenks S. Renewed focus on preventing gastric cancer. J Natl Cancer Inst. 2015;107:501. doi: 10.1093/jnci/dju501. [DOI] [PubMed] [Google Scholar]
- 29.Herrero R, Parsonnet J, Greenberg ER. Prevention of gastric cancer. JAMA. 2014;312:1197–8. doi: 10.1001/jama.2014.10498. [DOI] [PubMed] [Google Scholar]
- 30.Blaser MJ. Disappearing microbiota: Helicobacter pylori protection against esophageal adenocarcinoma. Cancer Prev Res (Phila) 2008;1:308–11. doi: 10.1158/1940-6207.CAPR-08-0170. [DOI] [PubMed] [Google Scholar]
- 31.Jordan V. A century of deciphering the control mechanisms of sex steroid action in breast and prostate cancer: The origins of targeted therapy and chemoprevention. Cancer Res. 2009;69:1243–54. doi: 10.1158/0008-5472.CAN-09-0029. [DOI] [PubMed] [Google Scholar]
- 32.Lacassagne A. Hormonal pathogenesis of adenocarcinoma of the breast. Am J Cancer. 1936;27:217–25. [Google Scholar]
- 33.Jordan VC, Lababidi MK, Mirecki DM. Anti-oestrogenic and anti-tumour properties of prolonged tamoxifen therapy in C3H/OUJ mice. Eur J Cancer. 1990;26:718–21. doi: 10.1016/0277-5379(90)90125-d. [DOI] [PubMed] [Google Scholar]
- 34.Schinzinger A. Üeber carcinoma mammae. Verh Dtsch Ges Chir. 1889;18:28–9. [Google Scholar]
- 35.Beatson GT. On the treatement of inoperable cases of carcinoma of the mamma: Suggestion for a new method of treatment, will illustrative cases. Lancet. 1896;148:162–5. [PMC free article] [PubMed] [Google Scholar]
- 36.Fisher B, Costantino JP, Wickerham DL, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 study. J Natl Cancer Inst. 1998;90:1371–88. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 37.Fuhrman BJ, Xu X, Falk RT, et al. Assay reproducibility and interindividual variation for 15 serum estrogens and estrogen metabolites measured by liquid chromatography-tandem mass spectrometry. Cancer Epidemiol Bio-markers Prev. 2014;23:2649–57. doi: 10.1158/1055-9965.EPI-14-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziegler RG, Faupel-Badger JM, Sue LY, et al. A new approach to measuring estrogen exposure and metabolism in epidemiologic studies. J Steroid Biochem Mol Biol. 2010;121:538–45. doi: 10.1016/j.jsbmb.2010.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Sui Z. Deciphering the selective androgen receptor modulators paradigm. Expert Opin Drug Discov. 2013;8:191–218. doi: 10.1517/17460441.2013.741582. [DOI] [PubMed] [Google Scholar]
- 40.Doll R, Hill AB. Smoking and carcinoma of the lung; preliminary report. Br Med J. 1950;2:739–48. doi: 10.1136/bmj.2.4682.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.United States Public Health Service. Office of the Surgeon General, Office on Smoking and Health . The health consequences of smoking: a report of the Surgeon General. U.S. Dept. of Health and Human Services, Public Health Service, U.S. G.P.O; Washington, D.C.: 2004. [Google Scholar]
- 42.Sheers G, Templeton AR. Effects of asbestos in dockyard workers. Br Med J. 1968;3:574–9. doi: 10.1136/bmj.3.5618.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blot WJ, Harrington JM, Toledo A, Hoover R, Heath CW, Jr, Fraumeni JF., Jr Lung cancer after employment in shipyards during World War II. N Engl J Med. 1978;299:620–4. doi: 10.1056/NEJM197809212991202. [DOI] [PubMed] [Google Scholar]
- 44.Yang H, Rivera Z, Jube S, et al. Programmed necrosis induced by asbestos in human mesothelial cells causes high-mobility group box 1 protein release and resultant inflammation. Proc Natl Acad Sci U S A. 2010;107:12611–6. doi: 10.1073/pnas.1006542107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aberle DR, DeMello S, Berg CD, et al. Results of the two incidence screenings in the National Lung Screening Trial. N Engl J Med. 2013;369:920–31. doi: 10.1056/NEJMoa1208962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Esserman LJ, Thompson IM, Reid B, et al. Addressing overdiagnosis and overtreatment in cancer: a prescription for change. Lancet Oncol. 2014;15:e234–e242. doi: 10.1016/S1470-2045(13)70598-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okayama H, Schetter AJ, Ishigame T, et al. The expression of four genes as a prognostic classifier for stage I lung adenocarcinoma in 12 independent cohorts. Cancer Epidemiol Biomarkers Prev. 2014;23:2884–94. doi: 10.1158/1055-9965.EPI-14-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ooi AT, Gower AC, Zhang KX, et al. Molecular profiling of premalignant lesions in lung squamous cell carcinomas identifies mechanisms involved in stepwise carcinogenesis. Cancer Prev Res (Phila) 2014;7:487–95. doi: 10.1158/1940-6207.CAPR-13-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szyf M. DNA methylation biomarkers for colorectal carcinoma and adenomatous polyps. Epigenomics. 2012;4:14–5. [PubMed] [Google Scholar]
- 50.Mathe EA, Patterson AD, Haznadar M, et al. Noninvasive urinary metabolomic profiling identifies diagnostic and prognostic markers in lung cancer. Cancer Res. 2014;74:3259–70. doi: 10.1158/0008-5472.CAN-14-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu GQ, Gail MH, Shi JX, et al. Association between upper digestive tract microbiota and cancer-predisposing states in the esophagus and stomach. Cancer Epidemiol Biomarkers Prev. 2014;23:735–41. doi: 10.1158/1055-9965.EPI-13-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryan BM, Pine SR, Chaturvedi AK, Caporaso N, Harris CC. A combined prognostic serum interleukin-8 and interleukin-6 classifier 2014.for stage 1 lung cancer in the Prostate, Lung, Colorectal, and Ovarian cancer screening yrial. J Thorac Oncol. 2014;9:1494–503. doi: 10.1097/JTO.0000000000000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collins F, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–5. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rebbeck TR. Precision prevention of cancer. Cancer Epidemiol Biomarkers Prev. 2014;23:2713–5. doi: 10.1158/1055-9965.EPI-14-1058. [DOI] [PubMed] [Google Scholar]
- 55.Toward precision medicine: building a knowledge network for biomedical research and a new taxonomy of disease. Institute of Medicine; Washington (DC): 2011. [PubMed] [Google Scholar]
- 56.Richer AL, Friel JM, Carson VM, Inge LJ, Whitsett TG. Genomic profiling toward precision medicine in non-small cell lung cancer: getting beyond EGFR. Pharmgenomics Pers Med. 2015;8:63–79. doi: 10.2147/PGPM.S52845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van de Vijver MJ, Peterse JL, Mooi WJ, et al. Neu-protein overexpression in breast cancer. Association with comedo-type ductal carcinoma in situ and limited prognostic value in stage II breast cancer. N Engl J Med. 1988;319:1239–45. doi: 10.1056/NEJM198811103191902. [DOI] [PubMed] [Google Scholar]
- 59.Yamane LS, Scapulatempo-Neto C, Alvarenga L, et al. KRAS and BRAF mutations and MSI status in precursor lesions of colorectal cancer detected by colonoscopy. Oncol Rep. 2014;32:1419–26. doi: 10.3892/or.2014.3338. [DOI] [PubMed] [Google Scholar]
- 60.Habal MB, Castelano C, Hemkes N, Scheuerle J, Guilford AM. In search of causative factors of deformational plagiocephaly. J Craniofac Surg. 2004;15:835–41. doi: 10.1097/00001665-200409000-00025. [DOI] [PubMed] [Google Scholar]
- 61.Hwang ES, Miglioretti DL, Ballard-Barbash R, Weaver DL, Kerlikowske K, National Cancer Institute Breast Cancer Surveillance Consortium Association between breast density and subsequent breast cancer following treatment for ductal carcinoma in situ. Cancer Epidemiol Biomarkers Prev. 2007;16:2587–93. doi: 10.1158/1055-9965.EPI-07-0458. [DOI] [PubMed] [Google Scholar]
- 62.Lari SA, Kuerer HM. Biological markers in DCIS and risk of breast recurrence: a systematic review. J Cancer. 2011;2:232–61. doi: 10.7150/jca.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeCensi A, Puntoni M, Pruneri G, et al. Lapatinib activity in premalignant lesions and HER-2 positive cancer of the breast in a randomized, placebo-controlled presurgical trial. Cancer Prev Res. 2011;4:1181–9. doi: 10.1158/1940-6207.CAPR-10-0337. [DOI] [PubMed] [Google Scholar]
- 64.Strecker TE, Shen Q, Zhang Y, et al. Effect of lapatinib on the development of estrogen receptor-negative mammary tumors in mice. J Natl Cancer Inst. 2009;101:107–13. doi: 10.1093/jnci/djn436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chan KC, Knox WF, Gee JM, et al. Effect of epidermal growth factor receptor tyrosine kinase inhibition on epithelial proliferation in normal and premalignant breast. Cancer Res. 2002;62:122–8. [PubMed] [Google Scholar]
- 66.To C, Kim EH, Royce DB, et al. The PARP inhibitors, veliparib and olaparib, are effective chemopreventive agents for delaying mammary tumor development in BRCA1-deficient mice. Cancer Prev Res (Phila) 2014;7:698–707. doi: 10.1158/1940-6207.CAPR-14-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weinstein IB. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science. 2002;297:63–4. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 68.Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–17. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 69.Sawyers CL. Shifting paradigms: the seeds of oncogene addiction. Nat Med. 2009;15:1158–61. doi: 10.1038/nm1009-1158. [DOI] [PubMed] [Google Scholar]
- 70.Tandon AK, Clark GM, Chamness GC, Ullrich A, McGuire WL. HER-2/neu oncogene protein and prognosis in breast cancer. J Clin Oncol. 1989;7:1120–8. doi: 10.1200/JCO.1989.7.8.1120. [DOI] [PubMed] [Google Scholar]
- 71.Marino-Enriquez A, Fletcher CD. Shouldn't we care about the biology of benign tumours? Nat Rev Cancer. 2014;14:701–2. doi: 10.1038/nrc3845. [DOI] [PubMed] [Google Scholar]
- 72.Yang XR, Chang-Claude J, Goode EL, et al. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. J Natl Cancer Inst. 2011;103:250–63. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McHale CM, Zhang L, Lan Q, et al. Global gene expression profiling of a population exposed to a range of benzene levels. Environ Health Perspect. 2011;119:628–34. doi: 10.1289/ehp.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hsu IC, Metcalf RA, Sun T, Welsh JA, Wang NJ, Harris CC. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature. 1991;350:427–8. doi: 10.1038/350427a0. [DOI] [PubMed] [Google Scholar]
- 76.Kuller LH, Bracken MB, Ogino S, Prentice RL, Tracy RP. The role of epidemiology in the era of molecular epidemiology and genomics: summary of the 2013 AJE-sponsored Society of Epidemiologic Research Symposium. Am J Epidemiol. 2013;178:1350–4. doi: 10.1093/aje/kwt239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ogino S, Lochhead P, Giovannucci E, Meyerhardt JA, Fuchs CS, Chan AT. Discovery of colorectal cancer PIK3CA mutation as potential predictive biomarker: power and promise of molecular pathological epidemiology. Oncogene. 2014;33:2949–55. doi: 10.1038/onc.2013.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596–606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14:1847–50. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- 80.Wild CP, Bucher JR, de Jong BW, et al. Translational cancer research: balancing prevention and treatment to combat cancer globally. J Natl Cancer Inst. 2015;107:353. doi: 10.1093/jnci/dju353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Valent P, Bonnet D, De Maria R, et al. Cancer stem cell definitions and terminology: the devil is in the details. Nat Rev Cancer. 2012;12:767–75. doi: 10.1038/nrc3368. [DOI] [PubMed] [Google Scholar]
- 82.Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005;15:494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 83.O'Brien CA, Kreso A, Dick JE. Cancer stem cells in solid tumors: an overview. Semin Radiat Oncol. 2009;19:71–7. doi: 10.1016/j.semradonc.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 84.Snippert HJ, van Es JH, van den Born M, et al. Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology. 2009;136(2187-94):e2181. doi: 10.1053/j.gastro.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 85.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 86.Schepers AG, Snippert HJ, Stange DE, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–5. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 87.Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–35. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kabarowski JH, Witte ON. Consequences of BCR-ABL expression within the hematopoietic stem cell in chronic myeloid leukemia. Stem Cells. 2000;18:399–408. doi: 10.1002/stem.180399. [DOI] [PubMed] [Google Scholar]
- 89.Bhatia M, Wang JC, Kapp U, Bonnet D, Dick JE. Purification of primitive human hematopoietic cells capable of repopulating immune-deficient mice. Proc Natl Acad Sci U S A. 1997;94:5320–5. doi: 10.1073/pnas.94.10.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 91.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 92.Park CH, Hahm ER, Park S, Kim HK, Yang CH. The inhibitory mechanism of curcumin and its derivative against beta-catenin/Tcf signaling. FEBS Lett. 2005;579:2965–71. doi: 10.1016/j.febslet.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 93.Jaiswal AS, Marlow BP, Gupta N, Narayan S. Beta-catenin-mediated trans-activation and cell-cell adhesion pathways are important in curcumin (diferuylmethane)-induced growth arrest and apoptosis in colon cancer cells. Oncogene. 2002;21:8414–27. doi: 10.1038/sj.onc.1205947. [DOI] [PubMed] [Google Scholar]
- 94.Kakarala M, Brenner DE, Korkaya H, et al. Targeting breast stem cells with the cancer preventive compounds curcumin and piperine. Breast Cancer Res Treat. 2010;122:777–85. doi: 10.1007/s10549-009-0612-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yan C, Jamaluddin MS, Aggarwal B, Myers J, Boyd DD. Gene expression profiling identifies activating transcription factor 3 as a novel contributor to the proapoptotic effect of curcumin. Mol Cancer Ther. 2005;4:233–41. [PubMed] [Google Scholar]
- 96.Ryu MJ, Cho M, Song JY, et al. Natural derivatives of curcumin attenuate the Wnt/beta-catenin pathway through down-regulation of the transcriptional coactivator p300. Biochem Biophys Res Commun. 2008;377:1304–8. doi: 10.1016/j.bbrc.2008.10.171. [DOI] [PubMed] [Google Scholar]
- 97.Li Y, Wicha MS, Schwartz SJ, Sun D. Implications of cancer stem cell theory for cancer chemoprevention by natural dietary compounds. J Nutr Biochem. 2011;22:799–806. doi: 10.1016/j.jnutbio.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Landis-Piwowar KR, Huo C, Chen D, et al. A novel prodrug of the green tea polyphenol (-)-epigallocatechin-3-gallate as a potential anticancer agent. Cancer Res. 2007;67:4303–10. doi: 10.1158/0008-5472.CAN-06-4699. [DOI] [PubMed] [Google Scholar]
- 99.Kim J, Zhang X, Rieger-Christ KM, et al. Suppression of Wnt signaling by the green tea compound (-)-epigallocatechin 3-gallate (EGCG) in invasive breast cancer cells. Requirement of the transcriptional repressor HBP1. J Biol Chem. 2006;281:10865–75. doi: 10.1074/jbc.M513378200. [DOI] [PubMed] [Google Scholar]
- 100.Mishra L, Shetty K, Tang Y, Stuart A, Byers SW. The role of TGF-beta and Wnt signaling in gastrointestinal stem cells and cancer. Oncogene. 2005;24:5775–89. doi: 10.1038/sj.onc.1208924. [DOI] [PubMed] [Google Scholar]
- 101.Subramaniam D, Ramalingam S, Houchen CW, Anant S. Cancer stem cells: a novel paradigm for cancer prevention and treatment. Mini Rev Med Chem. 2010;10(5):359–71. doi: 10.2174/138955710791330954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fahey JW, Zhang Y, Talalay P. Broccoli sprouts: an exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc Natl Acad Sci U S A. 1997;94:10367–72. doi: 10.1073/pnas.94.19.10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Clarke JD, Dashwood RH, Ho E. Multi-targeted prevention of cancer by sulforaphane. Cancer Lett. 2008;269(2):291–304. doi: 10.1016/j.canlet.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chung FL, Conaway CC, Rao CV, Reddy BS. Chemoprevention of colonic aberrant crypt foci in Fischer rats by sulforaphane and phenethyl isothiocyanate. Carcinogenesis. 2000;21:2287–91. doi: 10.1093/carcin/21.12.2287. [DOI] [PubMed] [Google Scholar]
- 105.Jackson SJ, Singletary KW. Sulforaphane: a naturally occurring mammary carcinoma mitotic inhibitor, which disrupts tubulin polymerization. Carcino-genesis. 2004;25(2):219–27. doi: 10.1093/carcin/bgg192. [DOI] [PubMed] [Google Scholar]
- 106.Khor TO, Hu R, Shen G, et al. Phar macogenomics of cancer chemopreventive isothiocyanate compound sulforaphane in the intestinal polyps of ApcMin/+mice. Biopharm Drug Dispos. 2006 Dec;27(9):407–20. doi: 10.1002/bdd.522. [DOI] [PubMed] [Google Scholar]
- 107.Park SY, Kim GY, Bae SJ, Yoo YH, Choi YH. Induction of apoptosis by isothiocyanate sulforaphane in human cervical carcinoma HeLa and hepato-carcinoma HepG2 cells through activation of caspase-3. Oncol Rep. 2007 Jul;18(1):181–7. [PubMed] [Google Scholar]
- 108.Li Y, Zhang T, Korkaya H, et al. Sulforaphane, a dietary component of broccoli/ broccoli sprouts, inhibits breast cancer stem cells. Clin Cancer Res. 2010;16:2580–2590. doi: 10.1158/1078-0432.CCR-09-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Harikumar KB, Aggarwal BB. Resveratrol: a multitargeted agent for age-associated chronic diseases. Cell Cycle. 2008;7:1020–35. doi: 10.4161/cc.7.8.5740. [DOI] [PubMed] [Google Scholar]
- 110.Lee JE, Li H, Chan AT, et al. Circulating levels of vitamin D and colon and rectal cancer: the Physicians’ Health Study and a meta-analysis of prospective studies. Cancer Prev Res (Phila) 2011;4:735–43. doi: 10.1158/1940-6207.CAPR-10-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–30. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–6. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rothwell PM, Fowkes FG, Belch JF, Ogawa H, Warlow CP, Meade TW. Effect of daily aspirin on long-term risk of death due to cancer: analysis of individual patient data from randomised trials. Lancet. 2011;377:31–41. doi: 10.1016/S0140-6736(10)62110-1. [DOI] [PubMed] [Google Scholar]
- 114.Rothwell PM, Wilson M, Price JF, Belch JF, Meade TW, Mehta Z. Effect of daily aspirin on risk of cancer metastasis: a study of incident cancers during randomised controlled trials. Lancet. 2012;379:1591–601. doi: 10.1016/S0140-6736(12)60209-8. [DOI] [PubMed] [Google Scholar]
- 115.Rothwell PM, Wilson M, Elwin CE, et al. Long-term effect of aspirin on colorectal cancer incidence and mortality: 20-year follow-up of five randomised trials. Lancet. 2010;376:1741–50. doi: 10.1016/S0140-6736(10)61543-7. [DOI] [PubMed] [Google Scholar]
- 116.Rothwell PM, Price JF, Fowkes FG, et al. Short-term effects of daily aspirin on cancer incidence, mortality, and non-vascular death: analysis of the time course of risks and benefits in 51 randomised controlled trials. Lancet. 2012;379:1602–12. doi: 10.1016/S0140-6736(11)61720-0. [DOI] [PubMed] [Google Scholar]
- 117.Moon CM, Kwon JH, Kim JS, et al. Nonsteroidal anti-inflammatory drugs suppress cancer stem cells via inhibiting PTGS2 (cyclooxygenase 2) and NOTCH/HES1 and activating PPARG in colorectal cancer. Int J Cancer. 2014;134:519–29. doi: 10.1002/ijc.28381. [DOI] [PubMed] [Google Scholar]
- 118.Qiu W, Wang X, Leibowitz B, et al. Chemoprevention by nonsteroidal anti-inflammatory drugs eliminates oncogenic intestinal stem cells via SMAC-dependent apoptosis. Proc Natl Acad Sci U S A. 2010;107:20027–32. doi: 10.1073/pnas.1010430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Espina V, Mariani BD, Gallagher RI, et al. Malignant precursor cells pre-exist in human breast DCIS and require autophagy for survival. PLoS One. 2010;5:e10240. doi: 10.1371/journal.pone.0010240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–8. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 123.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zanetti KA, Haznadar M, Welsh JA, et al. 3’-UTR and functional secretor haplotypes in mannose-binding lectin 2 are associated with increased colon cancer risk in African Americans. Cancer Res. 2012;72(6):1467–77. doi: 10.1158/0008-5472.CAN-11-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ryan BM, Robles AI, McClary AC, et al. Identification of a Functional SNP in the 3’UTR of CXCR2 That Is Associated with Reduced Risk of Lung Cancer. Cancer Res. 2015;75(3):566–75. doi: 10.1158/0008-5472.CAN-14-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ryan BM, McClary AC, Valeri N, et al. rs4919510 in hsa-mir-608 is associated with outcome but not risk of colorectal cancer. PloS one. 2012;7:e36306. doi: 10.1371/journal.pone.0036306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ryan BM, Robles AI, Harris CC. KRAS-LCS6 genotype as a prognostic marker in early-stage CRC–letter. Clin Cancer Res. 2012;18:3487–8. doi: 10.1158/1078-0432.CCR-12-0250. author reply 3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Robles AI, Ryan BM. KRT81 miR-SNP rs3660 is associat ed with risk and survival of NSCLC. Ann Oncol. 2015 Nov; doi: 10.1093/annonc/mdv552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Robles AI, Yang P, Jen J, et al. A DRD1 polymorphism predisposes to lung cancer among those exposed to secondhand smoke during childhood. Cancer Prev Res (Phila) 2014;7(12):1210–8. doi: 10.1158/1940-6207.CAPR-14-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 133.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 135.Qi L, Bart J, Tan LP, et al. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer. 2009;9:163. doi: 10.1186/1471-2407-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yamamichi N, Shimomura R, Inada K, et al. Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin Cancer Res. 2009;15(12):4009–16. doi: 10.1158/1078-0432.CCR-08-3257. [DOI] [PubMed] [Google Scholar]
- 137.Sozzi G, Boeri M, Rossi M, et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol. 2014;32(8):768–73. doi: 10.1200/JCO.2013.50.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70(18):7027–30. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hoover RN, Chanock SJ. Opportunities-and hard work-ahead. J Natl Cancer Inst. 2015;107(1):398. doi: 10.1093/jnci/dju398. [DOI] [PubMed] [Google Scholar]
- 140.Dolinoy DC. The agouti mouse model: an epigenetic biosensor for nutritional and environmental alterations on the fetal epigenome. Nutr Rev. 2008;66(Suppl 1):S7–11. doi: 10.1111/j.1753-4887.2008.00056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rakyan VK, Chong S, Champ ME, et al. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci U S A. 2003;100(5):2538–43. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12(11):949–57. [PubMed] [Google Scholar]
- 143.Daxinger L, Whitelaw E. Transgenerational epigenetic inheritance: more questions than answers. Genome Res. 2010;20(12):1623–8. doi: 10.1101/gr.106138.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell Mar. 2014;157(1):95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hajkova P, Erhardt S, Lane N, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117(1-2):15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 146.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23(3):314–8. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 147.Lane N, Dean W, Erhardt S, et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35(2):88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- 148.Greer EL, Maures TJ, Ucar D, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479(7373):365–71. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460(7254):473–8. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hammoud S, Emery BR, Dunn D, Weiss RB, Carrell DT. Sequence alterations in the YBX2 gene are associated with male factor infertility. Fertil Steril. 2009;91(4):1090–5. doi: 10.1016/j.fertnstert.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 151.Aravin AA, Hannon GJ, Small RNA. silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:283–90. doi: 10.1101/sqb.2008.73.058. [DOI] [PubMed] [Google Scholar]
- 152.Mosher RA, Melnyk CW, Kelly KA, Dunn RM, Studholme DJ, Baulcombe DC. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature. 2009;460(7252):283–6. doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- 153.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008 Nov;322(5906):1387–92. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gapp K, Jawaid A, Sarkies P, et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;17(5):667–9. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Marczylo EL, Amoako AA, Konje JC, Gant TW, Marczylo TH. Smoking induces differential miRNA expression in human spermatozoa: a potential transgenerational epigenetic concern? Epigenetics. 2012;7(5):432–9. doi: 10.4161/epi.19794. [DOI] [PubMed] [Google Scholar]
- 156.Pembrey ME, Bygren LO, Kaati G, et al. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet. 2006;14(2):159–66. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- 157.Heijmans BT, Tobi EW, Stein AD, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105(44):17046–9. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rechavi O, Houri-Ze'evi L, Anava S, et al. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158(2):277–87. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Youngson NA, Whitelaw E. Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet. 2008;9:233–57. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- 160.Verhoeven KJ, Jansen JJ, van Dijk PJ, Biere A. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 2010;185(4):1108–18. doi: 10.1111/j.1469-8137.2009.03121.x. [DOI] [PubMed] [Google Scholar]
- 161.Potter JD. The failure of cancer chemoprevention. Carcinogenesis. 2014 May;35(5):974–82. doi: 10.1093/carcin/bgu063. [DOI] [PubMed] [Google Scholar]
- 162.Fineberg HV. The paradox of disease prevention: celebrated in principle, resisted in practice. JAMA. 2013;310(1):85–90. doi: 10.1001/jama.2013.7518. [DOI] [PubMed] [Google Scholar]
- 163.Howell A, Anderson AS, Clarke RB, et al. Risk determination and prevention of breast cancer. Breast Cancer Res. 2014;16(5):446. doi: 10.1186/s13058-014-0446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.de Martel C, Ferlay J, Franceschi S, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–15. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 165.Gross-Steinmeyer K, Eaton DL. Dietary modulation of the biotransformation and genotoxicity of aflatoxin B(1). Toxicology. 2012;299:69–79. doi: 10.1016/j.tox.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 166.Zhang Y, Munday R. Dithiolethiones for cancer chemoprevention: where do we stand? Mol Cancer Ther. 2008;7:3470–9. doi: 10.1158/1535-7163.MCT-08-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Thomas D. Gene–environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11:259–72. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hall JM, Friedman L, Guenther C, et al. Closing in on a breast cancer gene on chromosome 17q. American Journal of Human Genetics. 1992;50:1235–42. [PMC free article] [PubMed] [Google Scholar]
- 169.Moyer VA. Force USPST. Risk assessment, genetic counseling, and genetic testing for BRCA-related cancer in women: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2014;160:271–81. doi: 10.7326/M13-2747. [DOI] [PubMed] [Google Scholar]
- 170.Manchanda R, Legood R, Burnell M, et al. Cost-effectiveness of population screening for BRCA mutations in Ashkenazi jewish women compared with family history-based testing. J Natl Cancer Inst. 2015;107:380. doi: 10.1093/jnci/dju380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Manchanda R, Loggenberg K, Sanderson S, et al. Population testing for cancer predisposing BRCA1/BRCA2 mutations in the Ashkenazi-Jewish community: a randomized controlled trial. J Natl Cancer Inst. 2015;107:379. doi: 10.1093/jnci/dju379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li FP, Fraumeni JF., Jr Soft-tissue sarcomas, breast cancer, and other neoplasms. A familial syndrome? Annals of Internal Medicine. 1969;71:747–52. doi: 10.7326/0003-4819-71-4-747. [DOI] [PubMed] [Google Scholar]
- 173.Manolio TA. Bringing genome-wide association findings into clinical use. Nat Rev Genet. 2013;14:549–58. doi: 10.1038/nrg3523. [DOI] [PubMed] [Google Scholar]
- 174.Manolio TA, Weis BK, Cowie CC, et al. New models for large prospective studies: is there a better way? Am J. Epidemiol. 2012;175(9):859–66. doi: 10.1093/aje/kwr453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hamilton JG, Edwards HM, Khoury MJ, Taplin SH. Cancer screening and genetics: a tale of two paradigms. Cancer Epidemiol Biomarkers Prev. 2014;23:909–16. doi: 10.1158/1055-9965.EPI-13-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Topol EJ. Individualized medicine from prewomb to tomb. Cell. 2014;157:241–53. doi: 10.1016/j.cell.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Antoniou AC, Casadei S, Heikkinen T, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zimmern RL, Khoury MJ. The impact of genomics on public health practice: the case for change. Public Health Genomics. 2012;15:118–24. doi: 10.1159/000334840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sherman ME, Mies C, Gierach GL. Opportunities for molecular epidemiological research on ductal carcinoma in-situ and breast carcinogenesis: interdisciplinary approaches. Breast Dis. 2014;34:105–16. doi: 10.3233/BD-130359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zakikhani M, Dowling R, Fantus IG, Sonenberg N, Pollak M. Metformin is an AMP kinase-dependent growth inhibitor for breast cancer cells. Cancer Res. 2006;66:10269–73. doi: 10.1158/0008-5472.CAN-06-1500. [DOI] [PubMed] [Google Scholar]
- 181.Disis ML, Gad E, Herendeen DR, et al. A multiantigen vaccine targeting neu, IGFBP-2, and IGF-IR prevents tumor progression in mice with preinvasive breast disease. Cancer Prev Res (Phila) 2013;6:1273–82. doi: 10.1158/1940-6207.CAPR-13-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hauptmann M, Fossa SD, Stovall M, et al. Increased stomach cancer risk following radiotherapy for testicular cancer. Br J Cancer. 2015;112:44–51. doi: 10.1038/bjc.2014.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Berrington de Gonzalez A, Wong J, Kleinerman R, Kim C, Morton L, Bekelman JE. Risk of second cancers according to radiation therapy technique and modality in prostate cancer survivors. Int J Radiat Oncol Biol Phys. 2015;91:295–302. doi: 10.1016/j.ijrobp.2014.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Berrington de Gonzalez A, Gilbert E, Curtis R, et al. Second solid cancers after radiation therapy: a systematic review of the epidemiologic studies of the radiation dose-response relationship. Int J Radiat Oncol Biol Phys. 2013;86:224–33. doi: 10.1016/j.ijrobp.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Black A, Gibson TM, Shiels MS, et al. Pooling prospective studies to investigate the etiology of second cancers. Cancer Epidemiol Biomarkers Prev. 2014;23:1598–608. doi: 10.1158/1055-9965.EPI-14-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Elena JW, Travis LB, Simonds NI, et al. Leveraging epidemiology and clinical studies of cancer outcomes: recommendations and opportunities for translational research. J Natl Cancer Inst. 2013;105:85–94. doi: 10.1093/jnci/djs473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Weinberg RA. Coming full circle-from endless complexity to simplicity and back again. Cell. 2014;157:267–71. doi: 10.1016/j.cell.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 188.Dunn BK, Wagner PD, Anderson D, Greenwald P. Molecular markers for early detection. Semin Oncol. 2010;37:224–42. doi: 10.1053/j.seminoncol.2010.05.007. [DOI] [PubMed] [Google Scholar]