Abstract
The past decade has witnessed a number of societal and political changes that have raised critical questions about the long-term impact of marijuana (Cannabis sativa) that are especially important given the prevalence of its abuse and that potential long-term effects still largely lack scientific data. Disturbances of the epigenome have generally been hypothesized as the molecular machinery underlying the persistent, often tissue-specific transcriptional and behavioral effects of cannabinoids that have been observed within one’s lifetime and even into the subsequent generation. Here, we provide an overview of the current published scientific literature that examined epigenetic effects of cannabinoids. Though mechanistic insights about the epigenome remain sparse, accumulating data in humans and animal models have begun to reveal aberrant epigenetic modifications in brain and the periphery linked to cannabis exposure. Expansion of such knowledge and causal molecular relationships could help provide novel targets for future therapeutic interventions.
Keywords: Cannabinoids, epigenetics, DNA methylation, addiction, CB1 receptor, neurodevelopment
Introduction
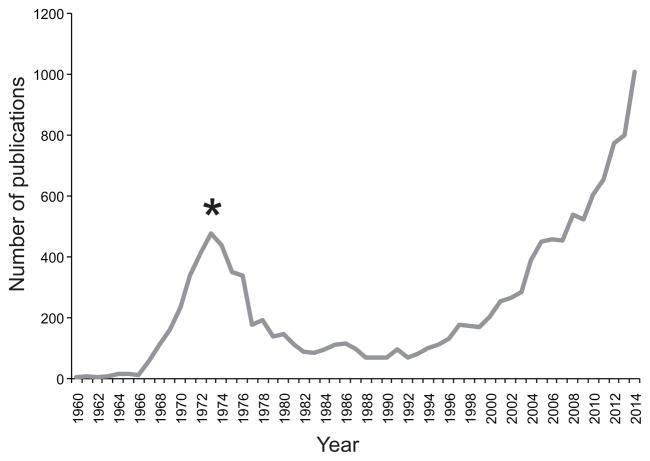
Extensive political and societal debates are currently being waged at state and federal levels regarding the legalization of marijuana (Cannabis sativa), which remains today the most commonly used illicit substance in the United States and in many countries worldwide. As evident in Figure 1, there has been a dramatic exponential increase of cannabis studies over the past two decades in response to the transformative implications resulting from the growing discussions and laws passed regarding legalization of recreational and medical marijuana use. Of the published studies to date, about 13% relate to the neurobiological effects of cannabis and approximately 27% is directed towards obtaining behavioral insights. Despite the perceived low health risk of cannabis use by the general public, there is growing clinical awareness about the spectrum of behavioral and neurobiological disturbances associated with cannabis exposure such as anxiety, depression, psychosis, cognitive deficits, social impairments, and addiction (1–7). The acute intoxication induced by cannabis consumption is strongly linked with concerns about its direct effects on cognition and motor function, but a central issue relates to its long-term impact especially when exposure occurs during critical periods of brain development. Key gaps of scientific knowledge pertain to the biological mechanisms that maintain persistent phenotypic and molecular alterations long after its acute use.
Figure 1.
Number of publications in PubMed between 1960 and 2014 related to ‘cannabis’ research. The data shows the exponential increase in research studies over recent decades that coincides with changes in the legalization status (starting ~1996) and debates of recreational and medical marijuana use. The drop in publications in the 1970s marks changes in state laws and local regulations banning possession or sale of cannabis and cannabis becoming a Schedule I drug (*).
The major psychoactive cannabinoid within cannabis, Δ9-tetrahydrocannabinol (THC), targets the endocannabinoid (eCB) system, which plays a key role in the development of the brain and several other organs. In recent years, various human and experimental animal studies have evaluated the long-term impact of cannabis and cannabinoids on neurodevelopment, behavior and several biological systems such as immunological mechanisms and reproductive processes (reviewed in (7–10)). Moreover, behavioral abnormalities and molecular impairments in the brain have also been demonstrated to extend even into subsequent generations of offspring whose parents were exposed to cannabinoids before mating (11–15).
The epigenome provides a cellular fingerprint of environmental experiences, including drug exposure history, and thus is a highly relevant biological candidate expected to maintain persistent abnormalities and aberrant neuronal processing over time. The role of epigenetics in psychiatric disorders has been a major scientific focus during the past few years. According to the classic definition, “an epigenetic trait is a stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence” (as proposed by Conrad Waddington in the 1950s); this view implies heritability resulting in a phenotype. In the molecular biological era of recent years, “epigenetic” typically has been used to refer to mechanisms that modulate gene expression without altering the genetic code. Our article provides an overview of research endeavors relevant to cannabis-related epigenetic mechanisms that could shed light about the biological processes that establish the molecular platform that maintains marijuana’s protracted effects on gene expression and ultimately behavior.
Epigenetic mechanisms
In a biological mechanistic context, knowledge of how gene expression is regulated by the cellular network of cis-acting elements and trans-acting factors has evolved substantially during the past decade. Generally, the interaction between genomic DNA elements (specific sequences with regulatory function), epigenetic modifiers and transcription factors determines the expression state of genes. This network of processes is tightly coordinated in space and time, in the specification of different cell, tissue and organ types, and throughout the lifespan of the individual (16–18).
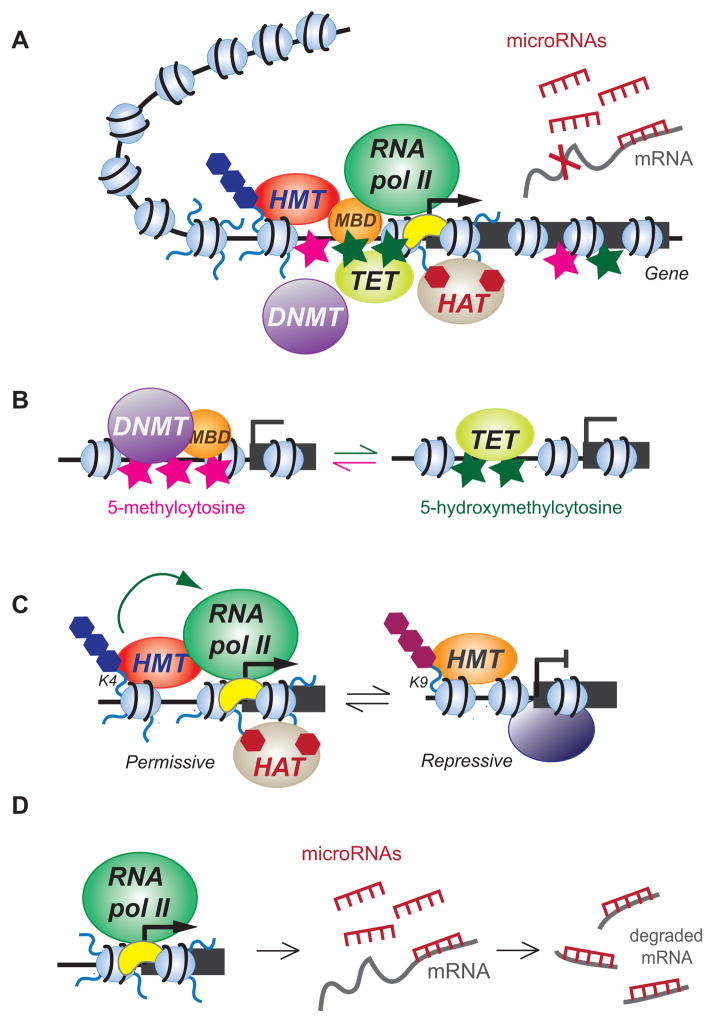
Some of the most important ontogenetic regulatory decisions take place in early development, and thus have critical implications for drug exposure during this period. Epigenetic modifications that can regulate gene expression levels include DNA methylation, nucleosomal structure and positioning, post-translational modifications of nucleosomal histones, histone replacement, and small RNA molecules that influence protein production (Figure 2A). Mechanistic implications of the specific epigenetic processes that have thus far been linked to the effects of cannabis are briefly summarized below.
Figure 2.
Several epigenetic mechanisms relevant to the effects of exogenous cannabinoids. (A) Gene expression is regulated by a network of DNA elements (e.g. promoters) and trans-acting factors (proteins that bind to the DNA) that interact physically and functionally to generate appropriate mRNA transcript levels from a gene. The resulting balance can be disrupted by drug exposure. Regulatory mechanisms include DNA methylation (Me), positioning and post-translational modifications of nucleosomes (small blue balls), recruitment of sequence-specific and basal transcription factors and RNA polymerase II, and non-coding RNAs. The DNA-protein structure forms three-dimensional structures (represented by the chromatin loop) that influence the expression of associated genes. (B) DNA methyltranserases (DNMT) generate 5-methylcytosine (pink stars) at CpG sites, facilitated by methyl-CpG binding domain (MBD)-containing proteins. Ten-eleven translocation (TET) proteins mediate the oxidation of 5-methylcytosine to 5-hydroxymethylcytosine (green stars), leading to demethylation of the DNA. (C) Modifications of nucleosomal histone tails such as methylation (Me) and acetylation (Ac) are mediated by histone methyltransferases (HMT) and histone acetyltransferases (HAT), respectively. Depending on modified amino acid residue, methylation can have either permissive (e.g. on lysine4, K4) or repressive (e.g. on lysine 9, K9) effects on transcription. Permissive modifications facilitate gene activation via the recruitment of the RNA polymerase II machinery. Acetylation is removed by histone deacetylases (HDAC) and can lead to transcriptional repression. (D) MicroRNAs are produced from specific genes and target protein-coding messenger RNAs (mRNA) for degradation, thereby prevention protein production.
DNA Methylation
The role of DNA methylation (Figure 2B) in the regulation of gene expression is still controversial and highly dependent on genomic location, developmental stage, cell type, or disease state. Historically, CpG methylation in promoter regions and transcriptional regulatory sequences has frequently been associated with gene silencing, whereas methylation within the gene body is less understood and may act as either positive or negative effectors (19, 20). Accumulating evidence now also indicates that DNA methylation in brain is reversible and its distribution changes throughout neuronal maturation and aging, in neurodevelopmental disorders, including addiction to drugs such as cocaine (21, 22). Mechanistically, DNA methylation (5-methylcytosine, 5mC) is generated by DNA methyltranserases (DNMTs). At promoter regions, 5mC is often associated with the binding of methyl-CpG binding domain (MBD)-containing proteins (e.g. Mecp2). The oxidation of 5mC to 5-hydroxymethylcytosine (5hmC) by ten-eleven translocation (TET) proteins can prevent access to DNMTs and thereby can maintain an unmethylated state of the promoter, leading to transcriptional activation (23). Interestingly, DNA methylation marks at specific gene loci have been shown to even persist during the maturation of germ cells (24, 25) and thus are interesting candidates for the propagation of the long-term effects of cannabis throughout multiple generations.
Histone modifications
On the protein level, the main epigenetic mechanism that has been implicated in neurobiological disturbances related to drug abuse is posttranslational modifications of nucleosomal histones (Figure 2C) which with the ~146bp of DNA that encircle them comprise the basic unit of chromatin. Histones are subject to a variety of modifications including but not limited to, lysine acetylation, lysine and arginine methylation, serine and threonine phosphorylation, and lysine ubiquitination and sumoylation (26). These modifications occur primarily within the histone amino-terminal tails protruding from the surface of the nucleosome as well as on the globular core region, and have been shown to influence both the accessibility of genomic regions and the binding of trans-acting factors to the DNA (27). Changes in acetylation and phosphorylation in response to drug exposure are often transient and appear to be associated with the quick activation of genes rather than the maintenance of an altered transcription state (28). However, histone lysine methylation is known to maintain stable gene expression alterations, and it is also the nucleosomal modification that has been associated with the long-term effects of marijuana and different cannabinoids in neurons and other cell types (29–32).
Non-coding RNAs (ncRNAs)
These functional RNA molecules are transcribed from DNA but are not translated into proteins. Many ncRNAs regulate gene expression at the transcriptional and post-transcriptional level. Those ncRNAs that are known to be involved in epigenetic processes can be divided into two main groups — short ncRNAs (<30 nucleotides) and long ncRNAs (>200 nucleotides). The three major classes of short ncRNAs are microRNAs (miRNAs), short interfering RNAs (siRNAs), and piwi-interacting RNAs (piRNAs) (33). Of these, alterations in miRNA profiles have been associated with cannabinoid exposure in the mammalian brain, peripheral blood cells, and the gut (Figure 2D) (34–37). While the exact genomic targets of specific cannabinoid-affected miRNAs remain to be characterized, these observations are mechanistically intriguing given the variety of tissue-specific cellular and developmental processes that are influenced by miRNAs.
The endocannabinoid (eCB) system
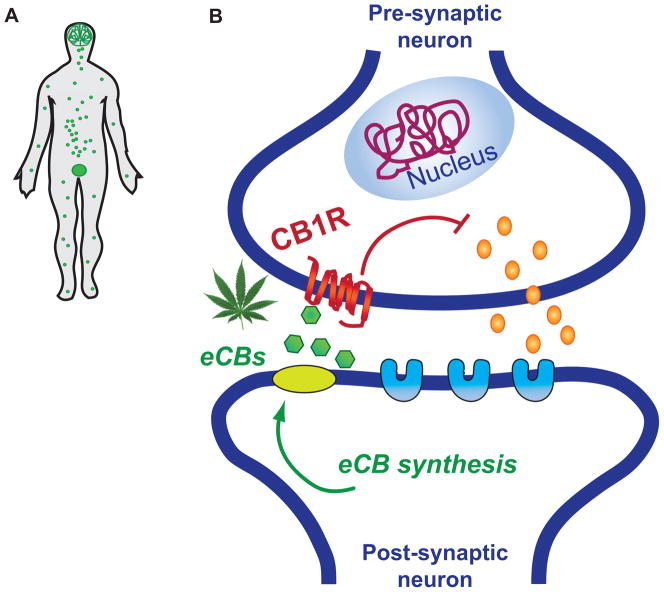
Cannabis targets the eCB system, which contributes to organogenesis as well as neurogenesis and gliogenesis of the CNS. It is well documented that the eCB system controls neuronal hardwiring during prenatal ontogeny, relevant to the development of neural pathways such as the corticostriatothalamic circuit, which are implicated in addiction and psychiatric disorders (38, 39) During postnatal developmental, the eCB system is known to be a critical regulator of synaptic plasticity. In mammals, two cannabinoid receptors have been identified (CB1R and CB2R), along with two major endocannabinoids as their ligands, N-arachidonoy-lethanolamine (anandamide) and 2-arachido-noylglycerol (2-AG) (40). During development these endogenous cannabinoid transmitters act as signaling molecules via a primarily autocrine activation of CB1Rs colocalized in the same developing neurons, whereas in the mature brain, eCBs are synthesized by postsynaptic neurons and travel retrogradely across the synapse to inhibit presynaptic neurotransmitter release via CBRs (41). CB1R is the most abundant G-protein-coupled receptor in the adult brain and mediates in large part the neurobehavioral effects of THC (Figure 3). Consistent with the known neurobiological and behavioral effects of the eCB system, CB1Rs are abundant in brain areas involved in learning and memory (e.g. hippocampus), motor function (e.g. basal ganglia, cerebellum), cognitive and emotional processes (e.g. striatum, amygdala, prefrontal cortex) (3), as well as the regulation of physiological and metabolic processes including feeding and stress response via the interaction of the Hypothalamic-Pituitary-Adrenal (HPA) and Gonadal (HPG) axes (42, 43). In neurons, CB1Rs are preferentially localized on the surface of presynaptic cells regulating both excitatory (glutamate) and inhibitory (GABA) transmission. Low expression of CB2Rs has recently been reported in the brain, frequently in association with inflammatory processes (44), and it has been detected in neurons within mesocorticolimbic brain regions relevant to cognition and motor function (45, 46). Despite its low abundance in brain, modulation of the CNS CB2R has been implicated in addiction-related behaviors (47, 48). Both CBRs are present in peripheral tissues, including the immune system, adipose tissue, liver, skeletal muscle and reproductive organs (49).
Figure 3.
Biological processes affected by cannabinoid exposure. (A) The active compounds of cannabis target cannabinoid receptors (CB1R and CB2R; expression pattern in the body is indicated by green dots in the human figure). (B) Cannabinoid receptors are trans-membrane receptors of the G protein-coupled family. The CB1R (shown in red), the primary target of THC, is expressed most abundantly in the brain, but also in the lungs, liver, kidneys, immune system, gut, and in germ cells such as the sperm. The CB2R is present mainly in the immune system and in hematopoietic cells with low expression in brain. Cannabinoid receptors can be activated by endocannabinoids (eCBs, green polygons; retrograde signaling), THC, or synthetic cannabinoids (see also Table 1). In the adult brain, activation of the CB1R on the surface of pre-synaptic neurons modulates the release of neurotransmitters (orange dots) that bind to their specific receptors (light blue shapes) in the post-synaptic cell, thereby changing the communication between neurons.
The normal epigenetic control of the eCB system has recently been reviewed (50). In the currect article, we focus on how cannabis, THC, and other exogenous cannabinoid receptor modulators alter epigenetic mechanisms and developmental regulation (Table 1). Briefly, however, various lines of evidence strongly suggest that the eCB anadamide and eCB signaling cascades mediated via CBRs regulate cellular functions in different tissues via epigenetic alterations in DNA methylation (e.g., cell differentiation in human keratinocytes, cells in the epidermis) (51), miRNA (regulating cells involved in interleukin production and inflammatory response) (36) and histone methylation (differentiation and inhibition of gliomagenesis) (29). These data highlight the role of the eCB system in regulating a repertoire of cellular functions in diverse tissues through multiple epigenetic modifications and suggest that exogenous modulation of these pathways with drugs may have long-lasting neurobiological impact.
Table 1. Epigenetic alterations related to the effects of cannabinoids in different organisms and biological systems.
Asterisks indicate the examples where cannabinoids have been shown to affect epigenetic regulation in brain or neurons.
| Cannabinoid | Epigenetic alteration | Biological target | Associated effect or consequence | References |
|---|---|---|---|---|
| Cannabis | Increased CpG DNA methylation at promoter | Human peripheral blood cells | Negative correlation between CB1R methylation and mRNA levels in schizophrenic cannabis users | (57) |
| Cannabis | Met/Met COMT gene genotype and promoter CpG DNA methylation | Human adolescent peripheral blood cells | Less likely cannabis dependence and decreased risk of psychosis | (61) |
| * THC | H3K4me3, H3K9me2; Promoter, gene body | Adult rat brain (NAc) | Decreased Drd2 gene mRNA levels in response to in utero THC exposure | (30) |
| * THC | H3K9me2, H3K9me3; Promoter, gene body | Adult rat brain (NAc shell) | Increased Penk gene mRNA levels in response to adolescent THC exposure | (31) |
| * THC | CpG DNA methylation at promoters, intergenic regions, especially in gene bodies | Adult rat NAc with parental THC exposure | Altered methylation enriched in genes implicated in synaptic plasticity | (15) |
| THC | H3K4me3, H3K9me3, H3K27me3, H3K36me3; Promoters, intergenic regions, gene bodies | Differentiating mouse lymph node cells | Genome-wide alterations in histone modifications associated with dysregulated genes and non-coding RNAs | (32) |
| THC | Increased HDAC3 expression | Human trophoblast cell line BeWo | Gene dysregulation during placental development | (70) |
| * THC | DNA methylation at CpG islands; miRNAs | Cerebellum and peripheral T cells of simian immunodeficiency virus-infected macaques | Altered DNA methylation, mRNA and miRNA expression profiles | (37) |
| THC | miRNAs | Mouse myeloid-derived suppressor cells | Altered mRNA, miRNA, and differentiation profile | (35) |
| THC | miRNAs | Intestine of simian immunodeficiency virus-infected macaques | Altered miRNA profile and intestinal epithelial cell composition | (34) |
| Exogenous anandamide | Increased global DNA methylation | Spontaneously immortalized human keratinocytes (HaCaT cell line) | Decreased expression of differentiation-related genes and altered cell differentiation | (51) |
| Exogenous anandamide | miRNAs | Mouse lymph node cells | Altered interleukin production and inflammatory response | (36) |
| HU-210, JWH-133 cannabinoid agonists | H3K9me3; Global levels | CB1R and CB2R-expressing human glioma stem-like cells (U87MG and U373MG lines) | Induction of differentiation, inhibition of gliomagenesis | (29) |
| * HU-210 cannabinoid agonist | miRNAs | Adolescent rat brain (entorhinal cortex) | Altered miRNA profile | (78) |
Epigenetic mechanisms relevant to the long-term effects of cannabis
The study of epigenetics in relation to drugs of abuse has been a rapidly emerging field during the past several years, yielding important mechanistic revelations about different addictions and related neuropsychiatric disorders (52, 53). However, experimental data about epigenetic effects associated with cannabis exposure are still sparse in spite of the relatively easy accessibility and frequent use and abuse of this drug. Of the few published studies, various epigenetic regulatory mechanisms that have been associated with cannabinoid exposure are summarized in Table 1. Epigenetic modifications have been shown to directly regulate the eCB system via targeting its individual components as well as downstream targets of eCB-associated pathways in a variety of cells types (Figures 2 and 3).
Human epigenetic studies
Of the different components of the eCB system, several investigations have focused on the epigenetic regulation of the CNR1 gene, which encodes the CB1R (Figure 3). Specific genomic elements of the CNR1 gene have been shown to interact with trans-acting factors, some of which are implicated in methylation of CpG sites in the DNA and histone posttranslational modifications (54–56). A few of these studies have revealed that CB1R expression is dysregulated in different pathological conditions and upon exposure to drugs of abuse. For example, CB1R expression is increased in peripheral blood lymphocytes of schizophrenic patients with cannabis abuse and is inversely correlated to methylation of the CNR1 promoter (Table 1) (57). However, that study had limitations in that most cannabis users also reported alcohol and cigarette use and were diagnosed with schizophrenia, making the direct delineation of any specific cannabis effect difficult. Nevertheless, CNR1 mRNA expression levels and promoter DNA methylation status detected in the blood was related to measures of cannabis craving, the severity of nicotine dependence and severity of cannabis (and alcohol) consumption that suggest a relationship to brain function. As such, lymphocyte CNR1 DNA methylation and CNR1 mRNA expression could potentially serve as peripheral biological marks. Clearly, a greater number of studies are needed to replicate these findings and to establish causal relationships in order to fully understand the functional relevance of peripheral epigenetic disturbances to neurobiological alterations induced by drug use. Moreover, whether such associations are evident in cannabis users without other comorbid neuropsychiatric conditions is also important to address.
One of the first gene x environment epigenetic associations described with cannabis use relevant to psychiatric vulnerability involved the COMT gene and schizophrenia risk. COMT (encodes catechol-O-methyltransferase that metabolizes catecholamine neurotransmitters such as dopamine) has also long been implicated in substance use. A well-known Val108/158Met COMT polymorphism increases COMT activity and thus levels of dopamine, which plays a critical role in reward, motivation, cognition and other behaviors linked to addiction. The Val allele has generally been associated with increased substance use disorder (58, 59) (but see meta-analysis in (60)). Recently, Val108/158Met genotype interaction with COMT DNA methylation status in blood was associated with non-daily cannabis use, which was not observed in either daily or non-users. Thus, adolescents with the Met/Met genotype in combination with high rates of COMT promoter methylation were less likely to be high-frequent cannabis users than adolescents with the Val/Val or Val/Met genotype (61). Given that the status of COMT DNA methylation depended on the frequency of cannabis use in active using adolescents, it remains unanswered whether such epigenetic alterations persist long after these individuals stop using the drug.
It is evident that a complex relationship exists between genetic and epigenetic interactions, and the relationship between peripheral epigenetic marks and methylation status in brain is still unknown. Despite the apparent associations of cannabis exposure with discrete molecular alterations in humans and the possibility to conduct studies on genetic associations, the specificity of the observed disturbances attributed to cannabis must be verified especially in the light of potential polysubstance exposure, which is common in humans. In addition, cannabis consists of over 60 cannabinoids, one of which is THC, and cannabis preparations can largely differ in amounts of these various cannabinoids, typically confounding clinical studies. Another important limitation is that given the low incidence of cannabis-related mortality that would allow postmortem brain molecular analyses, most human epigenetic studies can only be conducted in the periphery of live subjects and thus their relationships with brain changes remain unclear. Nevertheless, the accumulating data indicate epigenetic disturbances in human subjects relevant to cannabis use disorders that would predict the potential for long-term molecular alterations.
Cannabinoid animal models and epigenetic factors
Animal models provide more controllable experimental strategies in which the protracted molecular consequences of long-term cannabinoid exposure can be better explored with regard to epigenetic mechanisms that could potentially maintain abnormal gene regulation and related behavioral disturbances. Such preclinical animal studies also facilitate the direct causal investigation of protracted effects in the brain as a consequence of developmental exposure to cannabinoid drugs. A number of early seminal animal studies demonstrated prenatal THC exposure on offspring behaviors and some suggested changes in gene expression (62, 63), confirmed by subsequent investigations (64–66). More recent research efforts into the developmental effects of THC directly described epigenetic alterations germane to addiction disorders. These studies focused in large part on the NAc, a critical neuroanatomical substrate underlying the pathophysiology of addiction (67–69). The CB1R is abundantly expressed on medium spiny neurons that represent the most abundant striatal cell-type and constitute the differential output pathways (striatopallidal and striatonigral) that regulate specific behaviors. Interestingly, exposure to low-to-moderate THC dosing paradigms has generally induced significant alterations of the dopaminergic D2 receptor (D2R) and the opioid neuropeptide proenkephalin (PENK) genes (9, 30, 31, 66), which are preferentially expressed on the striatopallidal neurons and have been linked with epigenetic impairments. The sensitivity of D2R gene (DRD2) and PENK to cannabis/THC exposure in both the human fetus and animal models is intriguing given the role of these genes in drug addiction vulnerability. Both human and animal postmortem studies have revealed specific disturbances in the expression of the PENK and DRD2 genes in the NAc of subjects exposed to THC during either prenatal or adolescent developmental periods that persists into adulthood (30, 31). Of the multiple epigenetic mechanisms, the regulation of histone modification is unique because methylation of distinct residues can have antagonistic effects on transcription (Figure 2C). Indeed, our previous studies revealed disturbances in the histone modification profile in the NAc of adult rats with prenatal THC exposure. These studies identified decreased levels of the trimethylation of lysine 4 on histone H3 (H3K4me3), a transcriptionally permissive mark, increased levels of dimethylation of lysine 9 on histone H3 (H3K9me2), a repressive mark, as well as decreased RNA polymerase II association with the promoter and coding regions of the gene in the NAc (Table 1) (30). The combined epigenetic alterations were consistent with the observed reduction of the Drd2 gene expression and emphasize the enduring consequences of THC exposure following prenatal development. Similarly, persistent changes in repressive H3K9me2 and H3K9me3 were observed at the Penk locus in the NAc of adult rats following adolescent THC exposure in line with enduring upregulation of Penk mRNA levels (31). These findings emphasize an altered epigenetic landscape within the adult brain directly as a consequence of developmental cannabinoid exposure.
There is also evidence that THC exposure can affect the regulation of histone modification in other cell and tissue types during development. In differentiating mouse lymph node cells, alterations in H3K4me3, H3K9me3, H3K27me3, and H3K36me3 have been associated with dysregulated ncRNAs and mRNA genes (32). In addition, THC treatment dose-dependently increased the expression of HDAC3, a histone deacetylase, in a human trophoblast cell line indicating the possibility for cannabinoid exposure to affect placental development (70).
The studies discussed above highlight the long-term effects of cannabis exposure that influences the development of various cell and tissue types with functional and phenotypic consequences. Since these investigations so far have mainly been carried out at specific sets of candidate gene loci, rigorous future work will require comparisons between epigenomic and transcriptome alterations in order to address the mechanistic implications of these findings on the level of complex biological systems in different tissue types, and their dynamic regulation throughout development.
Multi-generational effects of cannabis
It has long been a subject of debates as to whether epigenetic disturbances that occurred during the lifespan of an individual are reprogrammed across most of the genome from parent to offspring, thereby establishing a new epigenetic “slate” for the next generation. Such concepts have been challenged in recent years by findings in various disease states where epigenetic aberrations that influence disease risk were shown to be inherited through the germline from parent to child (25, 71). More specifically, several cases of parent-child transmission regarding drugs of abuse have been published, describing both behavioral phenotypes and molecular disturbances in the offspring of parents that were exposed to drugs before mating (reviewed in (72)).
We have previously demonstrated that exposure of male and female adolescent rats before mating (“germline exposure”) leads to behavioral and molecular abnormalities in their unexposed offspring (11). Adult offspring of THC-exposed parents displayed increased work effort to self-administer heroin, with stereotyped behaviors during the period of acute heroin withdrawal. On the molecular level, parental THC exposure was associated with changes in the mRNA expression of cannabinoid, dopamine, and glutamatergic receptor genes in the striatum and altered synaptic plasticity in neurophysiological measures. In a more recent study and in line with the initial observations, DNA methylation disturbances were detected in the NAc of adult rats with parental germline THC exposure in an epigenome-scale investigation (15). The most significant finding was the identification of epigenetic alterations within an interaction network centered around the Dlg4 gene, encoding Psd-95, a membrane associated guanylate kinase scaffolding protein located in neural postsynaptic densities, involved in the regulation of dopamine-glutamate interactions. Psd-95 associates with the NMDA subtype of glutamate receptors and is required for synaptic plasticity associated with NMDA receptor function. A variety of genes involved in glutamatergic neurotransmission were also found to contain DNA methylation changes in the offspring of THC-exposed rats. Previously, epigenetic dysregulation of Dlg4 has been linked to abnormal glutamatergic transmission involved in morphine conditioning (73), consistent with the earlier observations of increased heroin self-administration in adult offspring with germline THC exposure (11). In other studies and in line with the above observations, adolescent female rats treated with the cannabinoid agonist WIN-55,212 before mating and pregnancy had progeny that exhibited increased morphine sensitivity (14, 74). These findings demonstrate that germline cannabinoid exposure can impact offspring phenotype, affect the molecular characteristics of the brain, and could possibly confer enhanced risk for addiction disorders.
Multi-generational epigenetic effects occur when an environmental trigger induces epigenetic changes that can be observed in at least one subsequent generation. The observations summarized above fit the classic concept of epigenetically inherited phenotypes. In-depth investigations are still needed to provide insights about epigenetic mechanisms underlying the transmission of cannabis effects through the germline. Moreover, important questions remain to be answered as to whether this represents a true transgenerational epigenetic transmission to subsequent generations (grandchildren and beyond) without direct germline exposure.
The eCB system plays important roles not only in the development of a variety of somatic cells and physiological systems, but also in reproduction. It is known that both male and female reproductive tissues express CBRs and eCBs and that in males, THC can disrupt gonadal functions (10, 75). Studies on the impact of cannabinoids on epigenetic changes in male fertility have been conducted in Cnr1 null mutant mice that displayed higher histone retention in germ cells compared to the wild type mice (76). In that study, CB1R expression was demonstrated to be necessary for spermiogenesis by controlling chromatin condensation in the developing sperm via the regulation of histone displacement during spermiogenesis, resulting in poor sperm quality. Adverse effects of cannabis use on the ovary of females have also been found to present a higher risk of primary infertility due to anovulation. Even when marijuana-using women undergo in vitro fertilization treatment, they produce poor quality oocytes and lower pregnancy rates (77). The effects of cannabis on the oocyte epigenome that could potentially lead to multi-generational transmission remain to be explored. Specifically, subsequent studies are required to assess how possible epigenetic processes (e.g. DNA methylation) are involved in the transmission of cannabinoid effects from parent to offspring.
Summary
Although still quite sparse in the number of studies and current mechanistic depth, there is solid scientific data that documents protracted effects of cannabinoids on brain as well as in other organs. Based on the current rapid growth in this scientific field, it is expected that significant developments in the near future will fill critical gaps of knowledge by focusing attention on long-term epigenetic processes and behavioral consequences of cannabis exposure.
The majority of addiction-related epigenetic neurobiological studies have targeted the adult brain. Even conceptually, very few studies have considered the potential lifelong or multi-generational epigenetic impact of cannabis. Although identifying mechanisms by which cannabis effects are maintained and transmitted is intriguing by itself, such explorations have potential far-reaching impact in the broader domain of developmental neurobiology since the identified epigenetic processes will no doubt be fundamental to transmission of other environmental insults across generations that bear on psychiatric vulnerability.
The mechanistic links between epigenetic modifications and gene expression impairments will require rigorous comparisons between epigenomic and transcriptome alterations. The overlay of results from approaches like RNA-sequencing, ChIP-sequencing and genome-scale DNA methylation studies in alignment to the genome will provide a unique potential to correlate epigenetic marks with the transcriptional regulation of neighboring genes. Moreover, the specific distribution and changes in 5-methylcytosine and 5-hydroxymethylcytosine (a demethylation intermediate, see Figure. 2B) has not yet been studied in the context of cannabis, and will likely be an interesting direction for in-depth mechanistic investigations. Importantly, direct causal relationships will be gained through the use of genomic editing tools to determine the impact of specific epigenetic disturbances in relation to gene expression. Providing causal links between gene expression impairments and specific behavioral phenotypes using in vivo gene manipulations offers important mechanistic value and the potential for developing targeted therapeutic solutions.
Overall, the integration of information garnered from clinical populations with data emerging from animal models will provide innovative insights to guide future translational studies and better inform clinical treatment and prevention strategies for the long-term impact of cannabis and even for the growing use of synthetic cannabinoids.
Acknowledgments
This work was supported by grants from NIH/NIDA DA030359 and DA033660.
Footnotes
Financial Disclosure. All authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alegria AA, Hasin DS, Nunes EV, Liu SM, Davies C, Grant BF, et al. Comorbidity of generalized anxiety disorder and substance use disorders: results from the National Epidemiologic Survey on Alcohol and Related Conditions. The Journal of clinical psychiatry. 2010;71:1187–1195. doi: 10.4088/JCP.09m05328gry. quiz 1252–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crean RD, Crane NA, Mason BJ. An evidence based review of acute and long-term effects of cannabis use on executive cognitive functions. Journal of addiction medicine. 2011;5:1–8. doi: 10.1097/ADM.0b013e31820c23fa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jutras-Aswad D, DiNieri JA, Harkany T, Hurd YL. Neurobiological consequences of maternal cannabis on human fetal development and its neuropsychiatric outcome. European archives of psychiatry and clinical neuroscience. 2009;259:395–412. doi: 10.1007/s00406-009-0027-z. [DOI] [PubMed] [Google Scholar]
- 4.Leweke FM, Koethe D. Cannabis and psychiatric disorders: it is not only addiction. Addiction biology. 2008;13:264–275. doi: 10.1111/j.1369-1600.2008.00106.x. [DOI] [PubMed] [Google Scholar]
- 5.Malone DT, Hill MN, Rubino T. Adolescent cannabis use and psychosis: epidemiology and neurodevelopmental models. British journal of pharmacology. 2010;160:511–522. doi: 10.1111/j.1476-5381.2010.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mechoulam R, Parker LA. The endocannabinoid system and the brain. Annual review of psychology. 2013;64:21–47. doi: 10.1146/annurev-psych-113011-143739. [DOI] [PubMed] [Google Scholar]
- 7.Morris CV, DiNieri JA, Szutorisz H, Hurd YL. Molecular mechanisms of maternal cannabis and cigarette use on human neurodevelopment. The European journal of neuroscience. 2011;34:1574–1583. doi: 10.1111/j.1460-9568.2011.07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Owen KP, Sutter ME, Albertson TE. Marijuana: respiratory tract effects. Clin Rev Allergy Immunol. 2014;46:65–81. doi: 10.1007/s12016-013-8374-y. [DOI] [PubMed] [Google Scholar]
- 9.Chadwick B, Miller ML, Hurd YL. Cannabis Use during Adolescent Development: Susceptibility to Psychiatric Illness. Front Psychiatry. 2013;4:129. doi: 10.3389/fpsyt.2013.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bari M, Battista N, Pirazzi V, Maccarrone M. The manifold actions of endocannabinoids on female and male reproductive events. Frontiers in bioscience. 2011;16:498–516. doi: 10.2741/3701. [DOI] [PubMed] [Google Scholar]
- 11.Szutorisz H, DiNieri JA, Sweet E, Egervari G, Michaelides M, Carter JM, et al. Parental THC exposure leads to compulsive heroin-seeking and altered striatal synaptic plasticity in the subsequent generation. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2014;39:1315–1323. doi: 10.1038/npp.2013.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byrnes JJ, Babb JA, Scanlan VF, Byrnes EM. Adolescent opioid exposure in female rats: transgenerational effects on morphine analgesia and anxiety-like behavior in adult offspring. Behavioural brain research. 2011;218:200–205. doi: 10.1016/j.bbr.2010.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byrnes JJ, Johnson NL, Carini LM, Byrnes EM. Multigenerational effects of adolescent morphine exposure on dopamine D2 receptor function. Psychopharmacology. 2013;227:263–272. doi: 10.1007/s00213-012-2960-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vassoler FM, Johnson NL, Byrnes EM. Female adolescent exposure to cannabinoids causes transgenerational effects on morphine sensitization in female offspring in the absence of in utero exposure. J Psychopharmacol. 2013;27:1015–1022. doi: 10.1177/0269881113503504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watson CT, Szutorisz H, Garg P, Martin Q, Landry JA, Sharp AJ, et al. Genome-Wide DNA Methylation Profiling Reveals Epigenetic Changes in the Rat Nucleus Accumbens Associated With Cross-Generational Effects of Adolescent THC Exposure. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2015 doi: 10.1038/npp.2015.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dambacher S, de Almeida GP, Schotta G. Dynamic changes of the epigenetic landscape during cellular differentiation. Epigenomics. 2013;5:701–713. doi: 10.2217/epi.13.67. [DOI] [PubMed] [Google Scholar]
- 17.Dillon N. Factor mediated gene priming in pluripotent stem cells sets the stage for lineage specification. Bioessays. 2012;34:194–204. doi: 10.1002/bies.201100137. [DOI] [PubMed] [Google Scholar]
- 18.Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nature reviews Genetics. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 19.Baubec T, Schubeler D. Genomic patterns and context specific interpretation of DNA methylation. Curr Opin Genet Dev. 2014;25:85–92. doi: 10.1016/j.gde.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 20.Kato T, Iwamoto K. Comprehensive DNA methylation and hydroxymethylation analysis in the human brain and its implication in mental disorders. Neuropharmacology. 2014;80:133–139. doi: 10.1016/j.neuropharm.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y, Bernstein A, Chen D, Jin P. 5-Hydroxymethylcytosine: A new player in brain disorders? Experimental neurology. 2015;268:3–9. doi: 10.1016/j.expneurol.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng J, Shao N, Szulwach KE, Vialou V, Huynh J, Zhong C, et al. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nature neuroscience. 2015;18:536–544. doi: 10.1038/nn.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nature reviews Genetics. 2012;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 24.Szyf M. The genome- and system-wide response of DNA methylation to early life adversity and its implication on mental health. Can J Psychiatry. 2013;58:697–704. doi: 10.1177/070674371305801208. [DOI] [PubMed] [Google Scholar]
- 25.Szyf M. Nongenetic inheritance and transgenerational epigenetics. Trends Mol Med. 2015;21:134–144. doi: 10.1016/j.molmed.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 27.Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol. 2004;11:1037–1043. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- 28.Ciccarelli A, Giustetto M. Role of ERK signaling in activity-dependent modifications of histone proteins. Neuropharmacology. 2014;80:34–44. doi: 10.1016/j.neuropharm.2014.01.039. [DOI] [PubMed] [Google Scholar]
- 29.Aguado T, Carracedo A, Julien B, Velasco G, Milman G, Mechoulam R, et al. Cannabinoids induce glioma stem-like cell differentiation and inhibit gliomagenesis. The Journal of biological chemistry. 2007;282:6854–6862. doi: 10.1074/jbc.M608900200. [DOI] [PubMed] [Google Scholar]
- 30.DiNieri JA, Wang X, Szutorisz H, Spano SM, Kaur J, Casaccia P, et al. Maternal cannabis use alters ventral striatal dopamine D2 gene regulation in the offspring. Biological psychiatry. 2011;70:763–769. doi: 10.1016/j.biopsych.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomasiewicz HC, Jacobs MM, Wilkinson MB, Wilson SP, Nestler EJ, Hurd YL. Proenkephalin mediates the enduring effects of adolescent cannabis exposure associated with adult opiate vulnerability. Biological psychiatry. 2012;72:803–810. doi: 10.1016/j.biopsych.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang X, Hegde VL, Rao R, Zhang J, Nagarkatti PS, Nagarkatti M. Histone modifications are associated with Delta9-tetrahydrocannabinol-mediated alterations in antigen-specific T cell responses. The Journal of biological chemistry. 2014;289:18707–18718. doi: 10.1074/jbc.M113.545210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iyengar BR, Choudhary A, Sarangdhar MA, Venkatesh KV, Gadgil CJ, Pillai B. Non-coding RNA interact to regulate neuronal development and function. Front Cell Neurosci. 2014;8:47. doi: 10.3389/fncel.2014.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandra LC, Kumar V, Torben W, Vande Stouwe C, Winsauer P, Amedee A, et al. Chronic administration of Delta9-tetrahydrocannabinol induces intestinal anti-inflammatory microRNA expression during acute simian immunodeficiency virus infection of rhesus macaques. J Virol. 2015;89:1168–1181. doi: 10.1128/JVI.01754-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hegde VL, Tomar S, Jackson A, Rao R, Yang X, Singh UP, et al. Distinct microRNA expression profile and targeted biological pathways in functional myeloid-derived suppressor cells induced by Delta9-tetrahydrocannabinol in vivo: regulation of CCAAT/enhancer-binding protein alpha by microRNA-690. The Journal of biological chemistry. 2013;288:36810–36826. doi: 10.1074/jbc.M113.503037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson AR, Nagarkatti P, Nagarkatti M. Anandamide attenuates Th-17 cell-mediated delayed-type hypersensitivity response by triggering IL-10 production and consequent microRNA induction. PLoS One. 2014;9:e93954. doi: 10.1371/journal.pone.0093954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molina PE, Amedee A, LeCapitaine NJ, Zabaleta J, Mohan M, Winsauer P, et al. Cannabinoid neuroimmune modulation of SIV disease. Journal of neuroimmune pharmacology : the official journal of the Society on NeuroImmune Pharmacology. 2011;6:516–527. doi: 10.1007/s11481-011-9301-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berghuis P, Rajnicek AM, Morozov YM, Ross RA, Mulder J, Urban GM, et al. Hardwiring the brain: endocannabinoids shape neuronal connectivity. Science. 2007;316:1212–1216. doi: 10.1126/science.1137406. [DOI] [PubMed] [Google Scholar]
- 39.Tortoriello G, Morris CV, Alpar A, Fuzik J, Shirran SL, Calvigioni D, et al. Miswiring the brain: Delta9-tetrahydrocannabinol disrupts cortical development by inducing an SCG10/stathmin-2 degradation pathway. The EMBO journal. 2014;33:668–685. doi: 10.1002/embj.201386035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luchicchi A, Pistis M. Anandamide and 2-arachidonoylglycerol: pharmacological properties, functional features, and emerging specificities of the two major endocannabinoids. Molecular neurobiology. 2012;46:374–392. doi: 10.1007/s12035-012-8299-0. [DOI] [PubMed] [Google Scholar]
- 41.Calvigioni D, Hurd YL, Harkany T, Keimpema E. Neuronal substrates and functional consequences of prenatal cannabis exposure. European child & adolescent psychiatry. 2014;23:931–941. doi: 10.1007/s00787-014-0550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirtcliff EA, Dismukes AR, Marceau K, Ruttle PL, Simmons JG, Han G. A dual-axis approach to understanding neuroendocrine development. Developmental psychobiology. 2015;57:643–653. doi: 10.1002/dev.21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koch M, Varela L, Kim JG, Kim JD, Hernandez-Nuno F, Simonds SE, et al. Hypothalamic POMC neurons promote cannabinoid-induced feeding. Nature. 2015;519:45–50. doi: 10.1038/nature14260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cabral GA, Raborn ES, Griffin L, Dennis J, Marciano-Cabral F. CB2 receptors in the brain: role in central immune function. British journal of pharmacology. 2008;153:240–251. doi: 10.1038/sj.bjp.0707584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Onaivi ES, Ishiguro H, Gong JP, Patel S, Perchuk A, Meozzi PA, et al. Discovery of the presence and functional expression of cannabinoid CB2 receptors in brain. Annals of the New York Academy of Sciences. 2006;1074:514–536. doi: 10.1196/annals.1369.052. [DOI] [PubMed] [Google Scholar]
- 46.Van Sickle MD, Duncan M, Kingsley PJ, Mouihate A, Urbani P, Mackie K, et al. Identification and functional characterization of brainstem cannabinoid CB2 receptors. Science. 2005;310:329–332. doi: 10.1126/science.1115740. [DOI] [PubMed] [Google Scholar]
- 47.Ignatowska-Jankowska BM, Muldoon PP, Lichtman AH, Damaj MI. The cannabinoid CB2 receptor is necessary for nicotine-conditioned place preference, but not other behavioral effects of nicotine in mice. Psychopharmacology. 2013;229:591–601. doi: 10.1007/s00213-013-3117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xi ZX, Peng XQ, Li X, Song R, Zhang HY, Liu QR, et al. Brain cannabinoid CB(2) receptors modulate cocaine’s actions in mice. Nature neuroscience. 2011;14:1160–1166. doi: 10.1038/nn.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Graham ES, Ashton JC, Glass M. Cannabinoid receptors: a brief history and “what’s hot”. Frontiers in bioscience. 2009;14:944–957. doi: 10.2741/3288. [DOI] [PubMed] [Google Scholar]
- 50.D’Addario C, Di Francesco A, Pucci M, Finazzi Agro A, Maccarrone M. Epigenetic mechanisms and endocannabinoid signalling. FEBS J. 2013;280:1905–1917. doi: 10.1111/febs.12125. [DOI] [PubMed] [Google Scholar]
- 51.Paradisi A, Pasquariello N, Barcaroli D, Maccarrone M. Anandamide regulates keratinocyte differentiation by inducing DNA methylation in a CB1 receptor-dependent manner. The Journal of biological chemistry. 2008;283:6005–6012. doi: 10.1074/jbc.M707964200. [DOI] [PubMed] [Google Scholar]
- 52.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sweatt JD. The emerging field of neuroepigenetics. Neuron. 2013;80:624–632. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee KS, Asgar J, Zhang Y, Chung MK, Ro JY. The role of androgen receptor in transcriptional modulation of cannabinoid receptor type 1 gene in rat trigeminal ganglia. Neuroscience. 2013;254:395–403. doi: 10.1016/j.neuroscience.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mukhopadhyay B, Liu J, Osei-Hyiaman D, Godlewski G, Mukhopadhyay P, Wang L, et al. Transcriptional regulation of cannabinoid receptor-1 expression in the liver by retinoic acid acting via retinoic acid receptor-gamma. The Journal of biological chemistry. 2010;285:19002–19011. doi: 10.1074/jbc.M109.068460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagre NN, Subbanna S, Shivakumar M, Psychoyos D, Basavarajappa BS. CB1-receptor knockout neonatal mice are protected against ethanol-induced impairments of DNMT1, DNMT3A, and DNA methylation. Journal of neurochemistry. 2015;132:429–442. doi: 10.1111/jnc.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, Chen J, Ehrlich S, Walton E, White T, Perrone-Bizzozero N, et al. Methylation patterns in whole blood correlate with symptoms in schizophrenia patients. Schizophr Bull. 2014;40:769–776. doi: 10.1093/schbul/sbt080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beuten J, Payne TJ, Ma JZ, Li MD. Significant association of catechol-O-methyltransferase (COMT) haplotypes with nicotine dependence in male and female smokers of two ethnic populations. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2006;31:675–684. doi: 10.1038/sj.npp.1300997. [DOI] [PubMed] [Google Scholar]
- 59.Vandenbergh DJ, Rodriguez LA, Miller IT, Uhl GR, Lachman HM. High-activity catechol-O-methyltransferase allele is more prevalent in polysubstance abusers. American journal of medical genetics. 1997;74:439–442. [PubMed] [Google Scholar]
- 60.Tammimaki AE, Mannisto PT. Are genetic variants of COMT associated with addiction? Pharmacogenetics and genomics. 2010;20:717–741. doi: 10.1097/FPC.0b013e328340bdf2. [DOI] [PubMed] [Google Scholar]
- 61.van der Knaap LJ, Schaefer JM, Franken IH, Verhulst FC, van Oort FV, Riese H. Catechol-O-methyltransferase gene methylation and substance use in adolescents: the TRAILS study. Genes, brain, and behavior. 2014;13:618–625. doi: 10.1111/gbb.12147. [DOI] [PubMed] [Google Scholar]
- 62.Rubio P, Rodriguez de Fonseca F, Martin-Calderon JL, Del Arco I, Bartolome S, Villanua MA, et al. Maternal exposure to low doses of delta9-tetrahydrocannabinol facilitates morphine-induced place conditioning in adult male offspring. Pharmacology, biochemistry, and behavior. 1998;61:229–238. doi: 10.1016/s0091-3057(98)00099-9. [DOI] [PubMed] [Google Scholar]
- 63.Vela G, Martin S, Garcia-Gil L, Crespo JA, Ruiz-Gayo M, Fernandez-Ruiz JJ, et al. Maternal exposure to delta9-tetrahydrocannabinol facilitates morphine self-administration behavior and changes regional binding to central mu opioid receptors in adult offspring female rats. Brain research. 1998;807:101–109. doi: 10.1016/s0006-8993(98)00766-5. [DOI] [PubMed] [Google Scholar]
- 64.Campolongo P, Trezza V, Cassano T, Gaetani S, Morgese MG, Ubaldi M, et al. Perinatal exposure to delta-9-tetrahydrocannabinol causes enduring cognitive deficits associated with alteration of cortical gene expression and neurotransmission in rats. Addiction biology. 2007;12:485–495. doi: 10.1111/j.1369-1600.2007.00074.x. [DOI] [PubMed] [Google Scholar]
- 65.Singh ME, McGregor IS, Mallet PE. Perinatal exposure to delta(9)-tetrahydrocannabinol alters heroin-induced place conditioning and fos-immunoreactivity. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2006;31:58–69. doi: 10.1038/sj.npp.1300770. [DOI] [PubMed] [Google Scholar]
- 66.Spano MS, Ellgren M, Wang X, Hurd YL. Prenatal cannabis exposure increases heroin seeking with allostatic changes in limbic enkephalin systems in adulthood. Biological psychiatry. 2007;61:554–563. doi: 10.1016/j.biopsych.2006.03.073. [DOI] [PubMed] [Google Scholar]
- 67.Everitt BJ, Robbins TW. From the ventral to the dorsal striatum: Devolving views of their roles in drug addiction. Neuroscience and biobehavioral reviews. 2013 doi: 10.1016/j.neubiorev.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 68.Girault JA. Integrating neurotransmission in striatal medium spiny neurons. Advances in experimental medicine and biology. 2012;970:407–429. doi: 10.1007/978-3-7091-0932-8_18. [DOI] [PubMed] [Google Scholar]
- 69.Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2010;35:217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khare M, Taylor AH, Konje JC, Bell SC. Delta9-tetrahydrocannabinol inhibits cytotrophoblast cell proliferation and modulates gene transcription. Mol Hum Reprod. 2006;12:321–333. doi: 10.1093/molehr/gal036. [DOI] [PubMed] [Google Scholar]
- 71.Bohacek J, Mansuy IM. Epigenetic inheritance of disease and disease risk. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2013;38:220–236. doi: 10.1038/npp.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vassoler FM, Sadri-Vakili G. Mechanisms of transgenerational inheritance of addictive-like behaviors. Neuroscience. 2014;264:198–206. doi: 10.1016/j.neuroscience.2013.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z, Yan P, Hui T, Zhang J. Epigenetic upregulation of PSD-95 contributes to the rewarding behavior by morphine conditioning. Eur J Pharmacol. 2014;732:123–129. doi: 10.1016/j.ejphar.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 74.Byrnes JJ, Johnson NL, Schenk ME, Byrnes EM. Cannabinoid exposure in adolescent female rats induces transgenerational effects on morphine conditioned place preference in male offspring. J Psychopharmacol. 2012;26:1348–1354. doi: 10.1177/0269881112443745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Banerjee A, Singh A, Srivastava P, Turner H, Krishna A. Effects of chronic bhang (cannabis) administration on the reproductive system of male mice. Birth defects research Part B, Developmental and reproductive toxicology. 2011;92:195–205. doi: 10.1002/bdrb.20295. [DOI] [PubMed] [Google Scholar]
- 76.Chioccarelli T, Cacciola G, Altucci L, Lewis SE, Simon L, Ricci G, et al. Cannabinoid receptor 1 influences chromatin remodeling in mouse spermatids by affecting content of transition protein 2 mRNA and histone displacement. Endocrinology. 2010;151:5017–5029. doi: 10.1210/en.2010-0133. [DOI] [PubMed] [Google Scholar]
- 77.Klonoff-Cohen HS, Natarajan L, Chen RV. A prospective study of the effects of female and male marijuana use on in vitro fertilization (IVF) and gamete intrafallopian transfer (GIFT) outcomes. American journal of obstetrics and gynecology. 2006;194:369–376. doi: 10.1016/j.ajog.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 78.Hollins SL, Zavitsanou K, Walker FR, Cairns MJ. Alteration of imprinted Dlk1-Dio3 miRNA cluster expression in the entorhinal cortex induced by maternal immune activation and adolescent cannabinoid exposure. Transl Psychiatry. 2014;4:e452. doi: 10.1038/tp.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]