Abstract
LC-MS using electrospray ionization is currently the method of choice in bio-organic analysis covering a wide range of applications in a broad spectrum of biological media. The technique is noted for its high sensitivity but one major limitation which hinders achievement of its optimal sensitivity is the signal suppression due to matrix inferences introduced by the presence of co-extracted compounds during the sample preparation procedure. The analysis of DNA adducts of common environmental carcinogens is particularly sensitive to such matrix effects as sample preparation is a multistep process which involves “contamination” of the sample due to the addition of enzymes and other reagents for digestion of the DNA in order to isolate the analyte(s). This problem is further exacerbated by the need to reach low levels of quantitation (LOQ in the ppb level) while also working with limited (2-5 μg) quantities of sample. We report here on the systematic investigation of ion signal suppression contributed by each individual step involved in the sample preparation associated with the analysis of DNA adducts of polycyclic aromatic hydrocarbon (PAH) using as model analyte dG-BaP, the deoxyguanosine adduct of benzo[a]pyrene (BaP). The individual matrix contribution of each one of these sources to analyte signal was systematically addressed as were any interactive effects. The information was used to develop a validated analytical protocol for the target biomarker at levels typically encountered in vivo using as little as 2 μg of DNA and applied to a dose response study using a metabolically competent cell line.
Keywords: Matrix effects, Benzo[a]pyrene, DNA adducts, Ion suppression, Polycyclic aromatic hydrocarbons
1. Introduction
Polycyclic aromatic hydrocarbons (PAHs) comprise a diverse class of largely anthropogenic compounds best represented by the human carcinogen benzo[a]pyrene, BaP [1] whose DNA adduct was used as a model in the present studies. PAHs have two or more aromatic rings, either substituted (e.g., nitroPAHs) or native (e.g. BaP) [2]. Several PAHs have been linked to various diseases including cancer and cardiovascular disease, and thus may serve as biomarkers to indicate exposure to harmful substances or risk of disease [3].
PAHs are ubiquitous, with the highest levels occurring in heavily industrialized areas. Exposure can be through inhalation (associated with particulate material or from smoking), ingestion (deposited on food from cooking fuels), or absorption through the skin (coke oven workers) [4-7]. In a cell, PAHs are processed as xenobiotics where CYP1A1, CYP1B1 and epoxide hydrolase activate the procarcinogen BaP to the ultimate reactive carcinogen, benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide, BPDE. This activated species most commonly reacts with the N2 position of deoxyguanosine [8, 9].
In detecting and quantifying DNA adducts of PAHs in biological samples, the analyst is faced with a number of major challenges as these biomarkers typically occur at very low (trace) levels in vivo (ppm-ppb). These analytical requirements are generally expressed in terms of the number of adducts that can be detected in a million or billion normal nucleobases. Improving detection limits and in particular reducing the amount of DNA needed for analysis has been a major goal in this field. Until relatively recently, much of the analytical work has relied on the use of microgram (20-100 μg) quantities of DNA and method sensitivity had been addressed through the use of microcapillary columns operated at flow rates in the range of 200-1000 nL/min. In order to decrease both the sample quantities and the limits of detection and quantitation (LOD/LOQ), nanoflow liquid chromatography (LC) has been successfully adopted in many labs including our own [10]. In an attempt to further improve on these latter advances and further reduce the amount of DNA required for the analysis, both we [11-14] and others [15-21] have now adopted a state-of-the-art microfluidic chip-based technology. While the approach also relies on the use of nanoflow LC, in addition, it incorporates on-line sample cleanup procedures that bypass much of the typical manual sample processing to reduce sample losses. This approach has been more recently employed in the analysis of 4-aminobiphenyl DNA adducts in bladder and liver tissues [22]
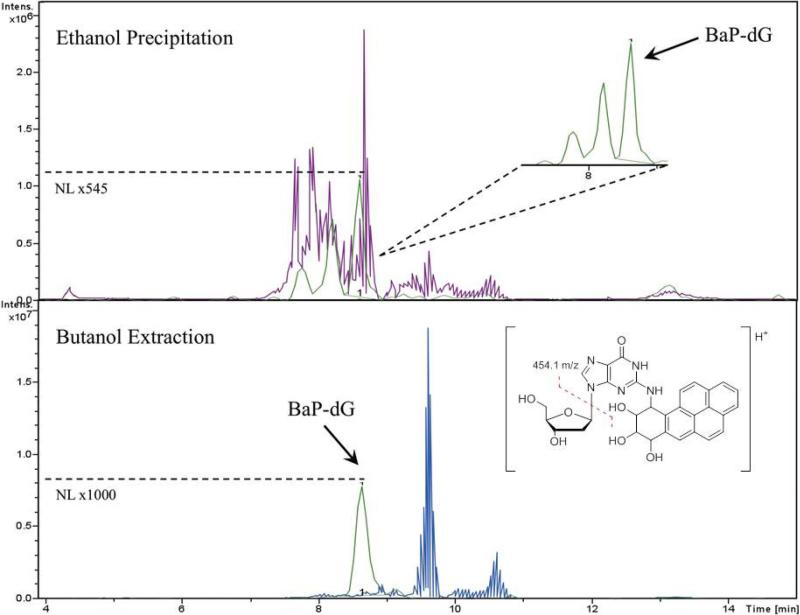
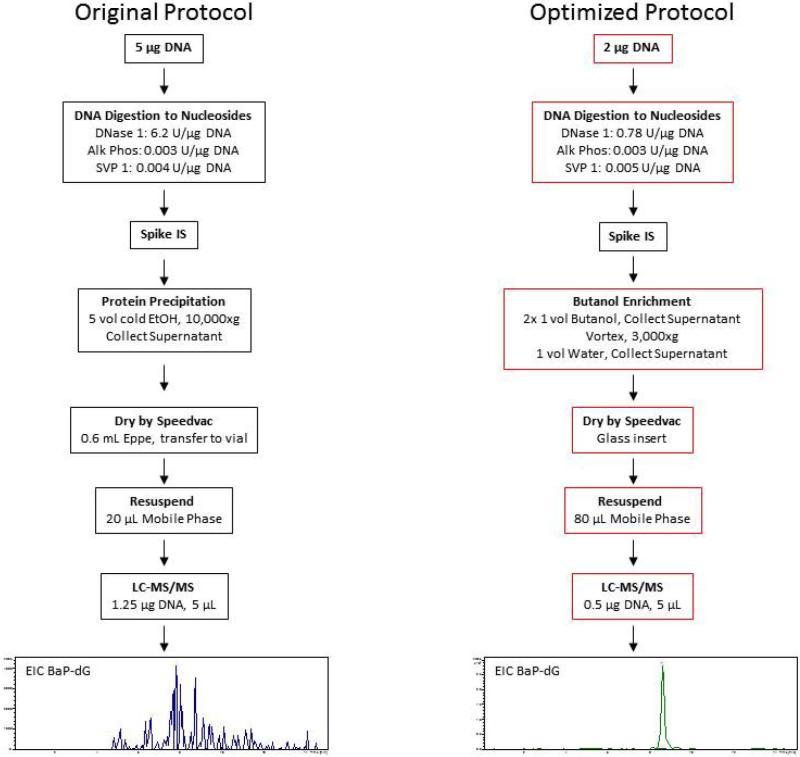
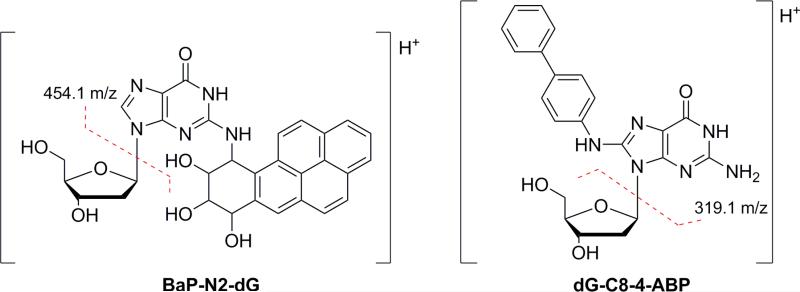
We present here a detailed account of the further development and optimization of the earlier protocols for use with the microfluidic chip-based technology with specific focus on the analysis of PAH adducts in relevant cell cultures. In this context, a major focus has been on addressing matrix effects, specifically analyte ion signal suppression, which is a key issue in biological mass spectrometric analysis at the trace level when using ESI. [23]. In fact, during the experiments it was quickly determined that the current protocol, which was adopted from dG-4-ABP analysis, did not provide adequate sensitivity for these purposes. While the compounds are not entirely different, both bulky, hydrophobic dG adducts with two or more aromatic rings, it is not surprising that the method did not transfer directly [24]. The structures of both adducts and predominant fragment ions that are formed by CID are shown in Figure 1. Figure 2 (left panel) shows the results obtained for the analysis of dG-BaP using the protocol originally adapted for targeted analysis of 4-aminobiphenyl-DNA adducts [12]. In the optimized protocol (Figure 2, right panel) every step has been modified to some degree from the original protocol, as indicated by red boxes. The rationale and specific steps taken for each modification in order to achieve the results illustrated in the final box of Figure 2 (right panel) are discussed below. After optimization of the sample preparation and LC-MS/MS conditions, the method was utilized in a dose response study in cultured human cells exposed to BaP with subsequent quantification of the N2-BPDE-deoxyguanosine adduct. It should be noted that, since the LC conditions did not provide for resolution of the different stereoisomers [25, 26], the quantities described refer to the sum of all related species.
Figure 1.
The structures of BaP-N2-dG (left) and dG-C8-4-ABP (right) are shown along with the primary fragment ions produced by CID.
Figure 2.
Comparison of the original and optimized protocols. The steps of DNA digestion to LC-MS/MS analysis are summarized with red boxes indicating steps that were modified. Sample chromatograms show the quality of BaP-dG analyte signal in a calf thymus (ctDNA) digest matrix.
2. Materials and methods
2.1. Reagents
Reagents including, sodium hydroxide pellets, phosphate buffered saline (PBS) tablets, chloroform, magnesium chloride hexahydrate, Trizma (1 M), deoxyribonucleic acid sodium salt from calf thymus (ctDNA), 2'-deoxyadenosine monohydrate (dA), 2'- deoxycytidine hydrate (dC), thymidine (dT), 2'-deoxyguanosine hydrate (dG), DNase I (bovine pancreas), phosphodiesterase I (Crotalus adamanteus venom), alkaline phosphatase (Escherichia coli), were purchased from Sigma Aldrich (St. Louis, MO). Water, methanol, acetonitrile, ethanol, isopropanol, acetic and formic acids were purchased from Fisher Scientific (Pittsburg, PA) in the highest purity or LC-MS grade when available. The adduct (±)-anti- benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide-N2- [15N5]-deoxyguanosine (15N-BaP-dG) was synthesized in our laboratory [27]. The activated carcinogen (±)-anti-benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide (BPDE) was purchased from MRI Global (Kansas City, MO). The Qiagen Blood and Cell Culture Midi® Kit and the Quant-IT™ Qubit® dsDNA BR assay kit were purchased from Invitrogen (Carlsbad, CA).
2.2. BEAS-2B cell culture
The BEAS-2B human bronchial epithelial cell line, developed from a human bronchial epithelial cell line transfected with a vector expressing the 3A4 isoform of cytochrome p450 (CYP-2B1) was obtained from Roger Coulombe [28, 29]. Tissue culture plates were plated by pre-incubated for 12-24 h with Plate Coat solution prepared by adding 5 mg bovine fibronectin (Sigma), 5 mL of collagen solution, and 50 mL of BSA stock solution (Biofluids, Rockville, MD) to LHC Basal medium (Biofluids) [28, 29]. Cells were cultured on coated plates with LHC-9 medium prepared by adding 1 mL of 3.3 mM retinoic acid and 1 mL of epinephrine to LHC-8 medium (all from Biofluids). Cell cultures were maintained at 37°C in a 5% CO2, water saturated atmosphere and passaged as previously described [28].
Cells were washed with ice cold PBS and harvested for simultaneous isolation of total RNA and DNA using All-Prep Kits™ (Qiagen, Valencia, CA) according to the manufacturer's protocol, and stored at −80°C until used for analysis.
2.3. Preparation of BaP-adducted DNA and purification of BaP-dG reference standard
The semi-synthetic preparation of BaP-dG has been described previously and was necessary for the thorough examination of matrix effects on this particular analyte [27]. Before digestion of the adducted polymeric DNA to monomers, a portion was reserved to prepare a standardized BaP-adducted DNA sample. This DNA was highly adducted, in the range of 1 adduct/103 nucleosides and was diluted using unadducted (untreated) ctDNA 1:100 and 1:1000 (w/w) to produce a sample of DNA with an adduction level on the order of 1 adduct/108 nucleosides for use in digestion efficiency studies.
2.4. DNA digestion
Five microgram aliquots of isolated DNA were prepared in digestion buffer (5 mM Tris, 10 mM MgCl2, pH 8.0) to 0.1 μg/μL in a final volume of 50 μL. DNase I (6.2 KU) was added per μg of DNA. Samples were incubated at 37 °C for 5 hours after which 0.003 U of alkaline phosphatase (AP) and 0.004 U of snake venom phosphodiesterase I (SVP I) were added per microgram of DNA. The mixture was incubated for another 18 hours at 37 °C. After incubation, offline sample cleanup was performed either by protein precipitation with organic solvent (such as cold ethanol) or butanol enrichment. Each procedure is described in detail below. After offline cleanup, the DNA digest was dried by speed-vac evaporation for 1 hour or until dry. Dried samples were reconstituted in 5:95 MeOH:Water (v/v) and analyzed by LC-MS/MS.
2.5. Butanol extraction
Butanol extraction was based on a published procedure but modified for these experiments [30]. To the digested DNA, 100 μL of water-saturated butanol (WSB) was added. Samples were vortexed for 30 s and centrifuged for 1 minute at 3,000 × g to facilitate phase separation. The top layer (butanol) was decanted into a clean 0.6 mL microcentrifuge tube. Another 100 μL of WSB was added to the digested DNA, samples were vortexed for 30 s and centrifuged for 1 minute at 3,000 × g. The top butanol layer was decanted and combined with the first. The bottom aqueous layer was discarded and the combined butanol layer was back-extracted with the addition of 200 μL of water to remove salts. The samples were again vortexed 30 s and centrifuged at 3,000 × g, after which the top layer (butanol) was collected and dried by speed-vac evaporation and the dried samples were reconstituted in 20 μL of mobile phase.
2.6. Protein precipitation
After DNA hydrolysis, samples were spiked with 8 μL of 15N-BaP-dG internal standard (10 fmol/μL in 5:95 MeOH:Water (v/v)), (0.2 pmol/μL stock) and 5-volumes of −80 °C EtOH (~100 μL sample vol., 500 μL EtOH). Samples were vortexed briefly and centrifuged at 10,000 × g for 10 min at 4 °C. After centrifugation, the supernatant was decanted into a clean microcentrifuge tube, or alternatively into glass silylated borosilicate glass inserts (Microliter, Wheaton). If decanted directly into glass inserts, 300 μL (volume of insert) of supernatant were added, dried in situ, then another 300 μL of supernatant was added and dried until complete sample transfer. Samples were reconstituted in 20 μL of 5:95 MeOH:Water (v/v), vortexed briefly and submitted for LC-MS/MS analysis. Samples dried in microcentrifuge tubes were dried by speed-vac evaporation (1 hour or until dry) and reconstituted in 20 μL of 5:95 MeOH:Water (v/v), vortexed briefly, transferred to 300 μL silylated borosilicate glass inserts and submitted for LC-MS/MS analysis.
2.7. Analysis by nanoLC-ESI-IT-MS/MS with HPLC chip
Analytical separation was done using an Agilent Technologies’ microfluidic HPLC chip that combined online enrichment, nanoLC, and ESI functionality. The platform was comprised of two Agilent Technologies’ 1100 Series HPLC pumps (one for sample loading and enrichment, and one for nanoLC) and a 1260 Series micro well-plate autosampler; a 1260 model chip-cube interface and small molecule chip with Zorbax SB-C18 80-Å, 5 μm particles packed in a 40 nL trapping column (online microSPE for analyte enrichment) and a 43 mm analytical column terminating in an emitter tip for nanoLC-nanoESI coupled to a model 6330 ion trap mass spectrometer. No special precautions were taken to ensure the durability of the chips other than as specified by the manufacturer.
The injection volume was held constant at 5 μL in all experiments (8 μL sample loop) which was loaded onto the trap column by the enrichment pump at 4 μL/min with the following solvent composition: 93% mobile phase A (water with 3% acetonitrile and 0.1% acetic acid), and 7% mobile phase B (methanol with 0.1% acetic acid). Samples were loaded onto the trapping column for 4 minutes (solvent diverted to waste), the enrichment pump was linked to the analytical column by the chip's switching valve, and the sample was back-flushed off the enrichment column by the nanoflow analytical pump (solvent to MS via ESI) at 300 nL/min with the following gradient: 10% B for 4 min to equilibrate (during enrichment) then to 90% B in 4 minutes, hold 2 minutes at 90% B, then back to 10% B in 2 min and hold 4 min to re-equilibrate.
The ion trap mass spectrometer, operated in positive ion mode, was calibrated by infusing Agilent calibration solution and tuning and optimization were done with a 10 μg/mL solution of BaP-dG reference standard in 70/30 MeOH/H2O with 0.1% acetic acid. Drying gas was 3.0 L/min N2 at 325 °C, and capillary voltage −1725 V. Ion optics were as follows: skimmer 1, 40 V; capillary exit, 94.3 V; octopole 1 DC, 12 V; octopole 2 DC, 1.7 V; octopole Rf, 170 V; trap drive, 64.5; lens 1, −5 V; and lens 2, −60 V. Spectral acquisition was done in ultra scan mode with 24,000 m/z per scan, accumulation was set to 50 msec or 500,000 ions and 3 averages per scan. Collision gas was ultrahigh purity He held at 3 mT, and dynamic ramping was used for optimal fragmentation, ranging from 0.45-3.0 V. ChemStation LC 3D Systems, Quant Analysis Version 1.8, and Data Analysis Version 3.4 software were used for operation and data analysis (Agilent Technologies, Wilmington, DE). In all cases, the analyte detection was based on the use of tandem MS, monitoring the transition 570.2 m/z → m/z 454.1 m/z for BaP-dG and the transition 575.2 m/z →m/z 459.1 m/z for the 15N-BaP-dG internal standard.
3. Results and discussion
3.1. Determination of cause(s) of decreased sensitivity
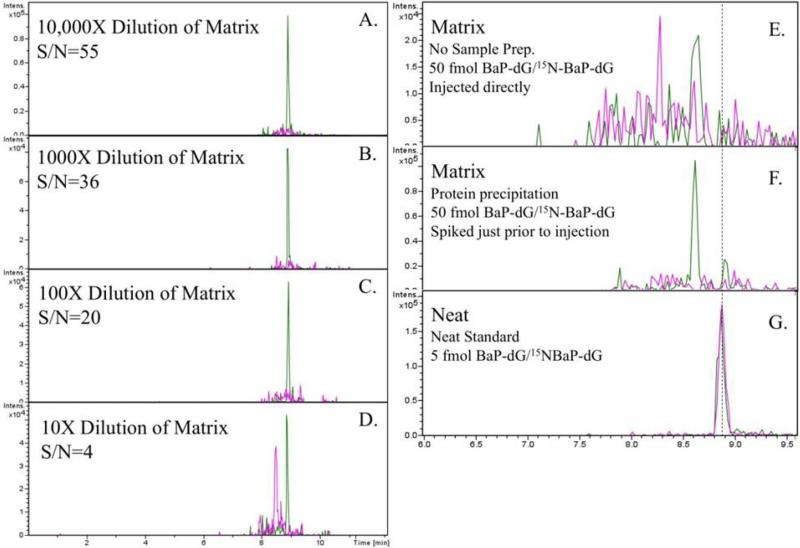
According to the IUPAC, matrix effects is defined as “...the combined effect of all components of the sample other than the analyte on the measurement of the ‘analyte ’quantity.” [31]. Most bioanalytical LC-MS methods involve some sample preparation (protein precipitation, solid phase extraction, etc.) and reconstitution of the analyte in some solvent compatible with MS detection. On that basis, the matrix effect may be quantified by determining the ratio between the analyte response measured in post–extraction spiked extracts and the response obtained from standards in a neat solvent. [32]. Invariably, sample components co-extracted with the analyte introduce ion suppression effects which result in analyte signal reduction and this is particularly observed in analyses conducted by electrospray ionization for reasons well documented in earlier publications [33, 34] and also reviewed by Trufelli, et al., [35]. In accordance with these considerations, in order to establish that matrix interference was the cause for the signal losses indicated in Figure 2, two straightforward experiments were carried out. In the first experiment a sample of ctDNA was digested, ethanol washed and reconstituted in 5 mM Tris, 10 mM MgCl2, pH 8.0 to 50 ng/μL. Then a 10X serial dilution was performed on the ctDNA digest matrix to produce a series of diluted matrix samples ranging from 10X to 10,000X. An equal volume (100 μL for 5 μg ctDNA digest) was removed from each diluted matrix, spiked with an equal concentration of BaP-dG analyte (10 fmol/μL), and analyzed by LC-MS/MS. Results are presented in Figure 3 (panels A-D) and show that as the matrix becomes more dilute, the analyte signal becomes stronger. In the second experiment, two aliquots of ctDNA digest (5 μg, 50 ng/μL) were spiked with BaP-dG analyte, one before and one after protein precipitation (ethanol wash). This simple test was intended to show that ion suppression by the matrix, rather than co-precipitation of BaP-dG with protein (during ethanol wash) was the principal cause for loss of signal. Results are also shown in Figure 3 (panels E and F). Since no BaP-dG analyte was observed when spiked into the matrix after all cleanup steps it was ascertained that ion suppression due to matrix interferences, rather than analyte recovery was the likely problem.
Figure 3.
Analyte spiking experiments that were used to demonstrate that ion suppression by matrix components, rather than analyte recovery was the primary contributor to loss of signal. Panels A-D show increasing analyte signal with increasing dilution of matrix. Panels E-F show that analyte was not detected even when spiked into matrix after cleanup, just prior to analysis. Panel G shows the LC-MS/MS analysis of 5 fmol of analyte and internal standard for comparison.
In view of these observations, a systematic study was undertaken in order to identify the degree or extent to which individual components associated with the processing of the sample may have contributed to signal suppression.
3.2. Determination of the specific causes of ion suppression: examination of sample components
To determine which component or components were responsible for loss of signal, the analyte was spiked into each component of the final sample including buffer, DNA, and enzymes. These components were washed with ethanol, dried and resuspended as per the standard protocol. In each experiment, analyte peak areas from LC-MS/MS analysis were compared to an appropriate control or to a neat, unprocessed dilution of BaP-dG analyte. Internal standard was in very short supply, and therefore was used sparingly in method development. However, these experiments were designed for relative (qualitative) comparison, with each component being tested for suppression of the BaP- dG analyte ion signal.
3.2.1. Buffer
The DNA digestion buffer (5 mM Tris, 10 mM MgCl2, pH 8) was developed for enzyme cocktails capable of hydrolyzing DNA to monomeric nucleo(sides/tides) [36, 37]. The buffer provides the essential divalent cations required for DNase I activity and buffers the solution to a pH at or near optimal for each enzyme. The impact of the buffer on the LC-MS/MS analysis was negligible when comparing standard BaP-dG in 5% MeOH vs. buffer and cleanup with ethanol precipitation.
3.2.2. DNA
DNA matrix effects were tested in two ways using both polymeric DNA and an equivalent mixture of deoxynucleosides, as would be produced by digestion of the DNA. Intact DNA was tested for two reasons: first, because the purified ctDNA purchased from Sigma has been reported to contain protein impurities (up to 21% protein by some measurements) [38], it was necessary to measure the effects of these impurities on the analyte signal, and second, to assess the impact of undigested DNA on the BaP-dG analyte (ion suppression or interactions such as base stacking) since there may be undigested DNA in the final sample.
To test matrix effects from deoxynucleosides, an equimolar (1:1:1:1) mixture of dA, dC, dT, dG equivalent to 5 μg of polymeric DNA (1.8 × 105 nucleosides/μg DNA) was prepared in 100 μL of digestion buffer. BaP-dG was spiked into the mixture to a final concentration of 10 fmol/μL followed by ethanol precipitation, evaporative drying in a speed-vac, and resuspension in 5% MeOH before LC-MS/MS analysis. To test matrix effects from polymeric DNA, 5 μL of ctDNA stock solution (1 mg/mL in 5 mM Tris, 10 mM MgCl2, pH 8) was diluted to 100 μL in digestion buffer and processed in the same way. The BaP-dG analyte signal in the presence of the mononucleoside mixture was reduced by 2-fold compared to neat analyte (10 fmol/μL in 5% MeOH), which is a reasonable depletion. A similar result was found with polymeric DNA, which showed a 4-fold depletion of BaP-dG analyte signal. Commonly, analytes spiked into sample matrix (followed by cleanup steps) will have signals decreased by at least 10-fold compared to a neat standard. It may be therefore concluded that the presence of unmodified nucleotides and undigested polymeric DNA is likely not a major contributor to ion suppression.
3.2.3. Enzymes
In comparison to the BaP-dG spiked DNA sample described above, when the 3- enzyme cocktail alone (no DNA or incubation) was spiked with BaP-dG analyte, a dramatic 380-fold decrease in analyte signal was observed relative to a neat sample, as well as an approximately three-fold decrease in S/N (Table 1). Therefore, it was determined that one or more of the enzymes was causing the overwhelming suppression of analyte signal. To determine which specific enzyme(s) were responsible, each enzyme was prepared separately in the digestion buffer and spiked with 10 fmol/μL BaP-dG analyte. Also, binary mixtures of each enzyme were prepared and spiked in the same way. Samples were ethanol washed, dried, reconstituted, and analyzed by LC-MS/MS and the results are summarized in Table 1. As the table shows, the strongest signal (largest peak area of BaP-dG) was obtained from alkaline phosphatase. However, when the DNase I or the SVP I enzymes were present in the sample, the analyte signal was only weak or moderate in comparison, ranging from 9- to 80-fold weaker, relative to the neat reference sample (10 fmol/μL BaP-dG).
Table 1.
Comparison of analyte signal when BaP-dG was spiked into preparations of each enzyme separately, and in combination with others. AP has the smallest impact on analyte signal whereas SVP I caused the largest decrease in analyte signal.
|
Enzyme |
||||
|---|---|---|---|---|
| DNase I | SVP I | AP | Peak Area | Analyte Signal |
| + | + | + | 1.18 × 105 | Weak |
| + | + | - | 4.53 × 104 | Weak |
| - | + | - | 1.09 × 105 | Weak |
| - | + | + | 1.15 × 105 | Weak |
| + | - | + | 4.06 × 105 | Moderate |
| + | - | - | 4.13 × 105 | Moderate |
| - | - | + | 3.63 × 106 | Strong |
Peak areas are average of duplicate analyses.
Analyte signals are listed as weak, moderate, and strong for facile comparison.
To address the issue of excessive ion suppression caused by DNase I and SVP I, we considered two options. The first option was to decrease the enzyme quantity, which should in turn decrease the ion suppression. This would also require an activity assay to ensure that a change in enzyme quantity did not adversely affect DNA digestion. The second option was to substitute with different enzymes, if available.
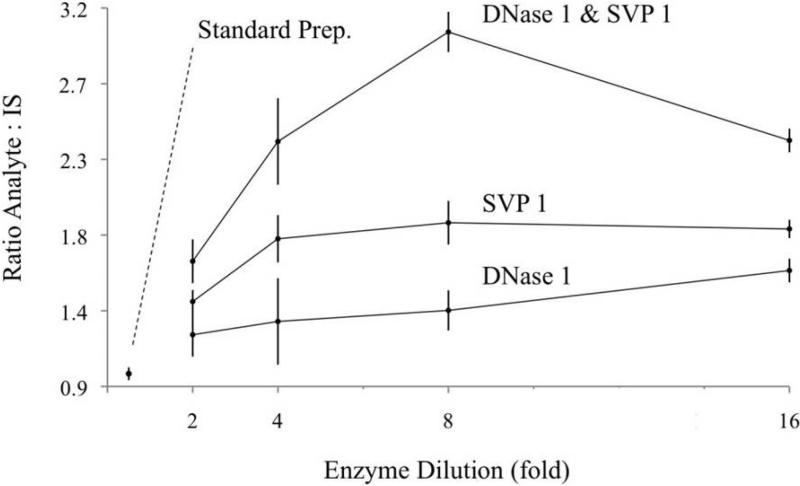
To determine the quantity of DNase I capable of providing optimal digestion efficiency and minimal ion suppression, enzyme cocktails (3 enzymes) were prepared with decreasing quantities of DNase I and were used to digest a 5 μg aliquot of standardized BaP-adducted ctDNA with a nominal adduction level (see Materials and Methods). In this way, DNA digestion efficiency was determined by LC-MS/MS quantitation of the liberated BaP-dG adducts. In the standard protocol, DNase I was prepared in the cocktail at 3.1 KU/μg DNA. Enzyme cocktails were also prepared with the following concentrations of DNase1: 1.6, 0.78, 0.39, 0.19 KU/μg DNA. These concentrations corresponded to 2, 4, 8, and 16-fold dilutions of the DNase I, while quantities of the other enzymes (SVP1 and alkaline phosphatase) remained constant. Results are shown in Figure 4, which includes data from the analogous experiment with SVP I (discussion follows). A small increase in signal (ratio of analyte:IS) was detected as the DNase I quantity was decreased. We interpret these results in the following way: DNA hydrolysis was complete in all preparations and the increasing ratio can be attributed to decreased suppression of the analyte ion.
Figure 4.
Correlation of enzyme quantity and analyte signal. Concentrations of DNase I (bottom) and SVP I (middle) were varied in the 3-enzyme cocktail to determine the optimal conditions for DNA digestion. In the top line, DNase I and SVP I were both diluted in the enzyme cocktail to determine the combined effects. Ion suppression is believed to cause the smaller ratios of analyte:IS at the lower dilutions (higher concentrations). For comparison, the standard 3-enzyme cocktail is included, labeled “Standard Prep.” and gave the weakest signal (closest to origin).
The experiment with SVP I was conducted in the same manner. Enzyme cocktails were prepared with decreasing quantities of SVP I and used to digest 5 μg aliquots of standardized BaP-adducted ctDNA. In the standard protocol, SVP I is added to the DNA at a concentration of 0.003 U/μg DNA, or 3 mU/μg DNA. Enzyme cocktails were also prepared at the following concentrations of SVP I: 1.5, 0.8, 0.4, and 0.2 mU/μg DNA equating to 2, 4, 8, and 16-fold dilutions, respectively. The digests were prepared in triplicate, spiked with IS and analyzed for liberated BaP-dG by LC-MS/MS. As Figure 4 (middle line) shows, decreasing the concentration of SVP I in the enzyme cocktail slightly increased the analyte signal up to an 8-fold dilution, but digestion efficiency may begin to decrease beyond this point.
A fourth experiment was also conducted in which quantities of DNase I and SVP I were both decreased in the enzyme cocktail while alkaline phosphatase remained constant. This ratio was then used to digest DNA in triplicate. The combined effects of decreasing both enzymes had a much more notable effect on ion suppression and digestion efficiency. A 3-fold increase in the analyte signal was observed with an 8-fold dilution (ratio 3.05) compared to the standard preparation (no dilution, ratio 0.98). As a result, these optimized enzyme concentrations were adopted into the final protocol.
As mentioned above, substitution of enzymes was also considered, but in the case of SVP I no alternative 5'-exonucleases were commercially available. Alternatives were available, however, for DNase I. The original digestion protocol developed in 1990 by Crain et al. utilized nuclease P1, rather than DNase I, as the endonuclease [37]. Unfortunately, nuclease P1 isn't ideal for this application because of its optimal pH (5-8), temperature (70 °C), and specificity (for ssDNA) which are generally incompatible with the other enzymes used in the cocktail. As a result, extra steps were required resulting in an overly complicated procedure that affects sample throughput. A more compatible and commercially available option was the recombinant endonuclease, benzonase. This enzyme is expressed in cell culture, in a controlled environment, and available in high (≥90%, SDS-PAGE) and ultra high (≥ 99%) purities. These higher purities would presumably impart less contamination than the DNase I purified from bovine pancreas (purity of ≥80%).
To test this latter hypothesis solutions of DNase I and benzonase were each prepared to 3.1 U/μL (so 1 μL enzyme equals 1 equivalent, or 3.1 U enzyme/μg DNA). Three concentrations were prepared, in duplicate, in 50 μL of digestion buffer: 6, 12, and 18 U/50 μL. BaP-dG (4.0 fmol/μL) was spiked into each aliquot and analyzed by LC- MS/MS. DNA, SVP I, and AP were excluded from the sample. The signal (peak area) for BaP-dG increased by 19%, 6 U; 33%, 12 U; and 97%, 18 U when ultrapure benzonase was used in place of DNase I. Next, the quantity of benzonase was optimized by preparing 3 concentrations of benzonase in the 3-enzyme cocktail according to the standard protocol. Aliquots of 5 μg of standard BaP-adducted ctDNA were treated with each of these cocktails in duplicate. No significant difference in liberated BaP-dG was detected by LC-MS/MS indicating that 6 U benzonase was sufficient to completely hydrolyze 5 μg of DNA and this condition was adopted into the final protocol.
3.3. Optimization of offline sample cleanup to address ion suppression
Offline cleanup of digested DNA for LC-MS analysis is typically a multi-step process. In the first step, after incubation with DNA, the hydrolytic enzymes are removed by ethanol precipitation. Other organic solvents such as acetonitrile are occasionally substituted for ethanol. In the second step, small molecules such as salts and unmodified nucleosides are removed by solid phase extraction (SPE) on a reversed-phase medium. For analysis of small samples containing trace analytes, such as PAH-DNA adducts present at ppm-ppb levels, it is advantageous to perform this step online with the LC separation and MS detection in order to minimize sample loss.
3.3.1. Protein precipitation solvents
Protein precipitation is a key step in sample cleanup of digested DNA to remove the enzymes that were added in the previous step. Siuzdak et al. conducted a thorough examination of various organic solvents and acids for protein precipitation in serum samples [39]. Their area of study, metabolomic profiling, was quite different but shared a common goal with our current application, namely protein depletion from a biological sample for the LC-MS/MS analysis of small molecule analytes. In a similar way, we tested the effectiveness of many of these solvents for the following criteria: first, removal of contaminants such as protein; and second, retention of the analyte (BaP-dG).
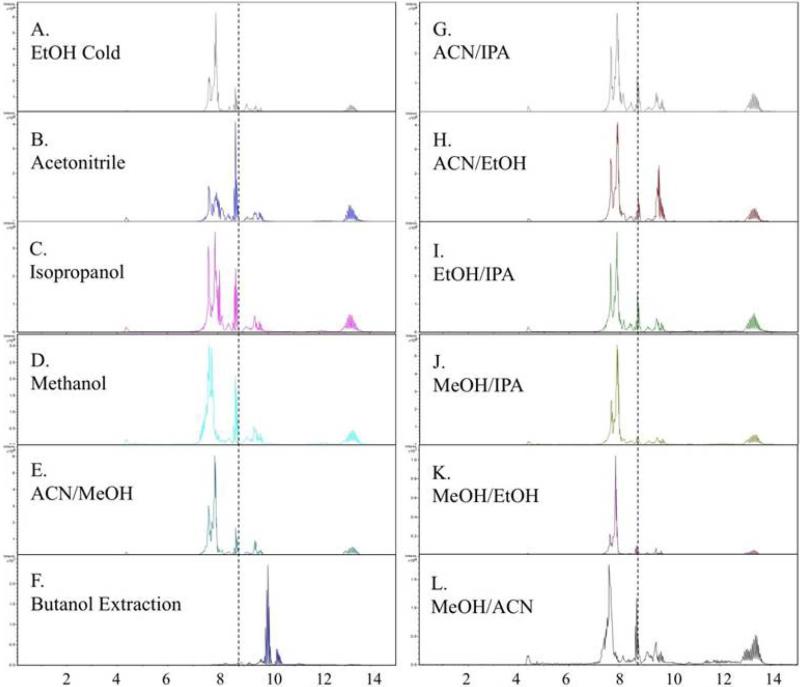
The first step was to test the effectiveness of the different solvents toward decreasing the background signal in a ctDNA digest. The solvents methanol, ethanol, isopropanol, and acetonitrile, as well as 50/50 (v/v) mixtures of each combination (e.g. MeOH/IPA, ACN/IPA, EtOH/IPA, and so on) were prepared at room temperature and cold (just above their freezing points). Aliquots of 5 μg ctDNA digest were prepared by digesting the DNA overnight with the standard 3-enzyme cocktail. BaP-dG was spiked into each sample before adding 5 volumes of the appropriate solvent. Samples were centrifuged, dried, resuspended in 5% MeOH and analyzed by LC-MS/MS according to the standard protocol. A comparison of the resulting chromatograms (some are excluded if TIC pattern is redundant) is shown in Figure 5. The background signal from contaminating ions can be compared by referring to the TICs that are aligned (x-axis, retention time) for visualization. The dashed vertical line indicates the retention time of the BaP-dG analyte. While subtle differences can be observed in the TICs of the different solvents, the BaP-dG signal did not exceed a S/N ratio of 5 in any of the precipitation experiments, which is likely due to the presence of high background signal spanning the range from 7-10 minutes (retention time of BaP-dG analyte is 8.6 min). By contrast, Panel F shows the resulting TIC after butanol extraction, which is much more effective at removing matrix components eluting between 7-9.5 minutes. A large background signal is still present, however it is shifted to 9.5-10.5 minutes, after elution of the BaP-dG analyte. This likely explains the much higher signals obtained when butanol extraction was used for offline cleanup rather than precipitation with organic solvents.
Figure 5.
Protein precipitation solvents. TIC chromatograms show how solvent choice used in offline cleanup affects the background ions that may contribute to analyte ion suppression. The dashed vertical line indicates the retention time of BaP-dG. Note that the BaP-dG EIC is not shown in these chromatograms and “peaks” are due to background matrix ions, not the analyte.
3.3.2. Butanol extraction
After observing that extraction in butanol introduced the lowest matrix effects, the second step was to verify that the BaP-dG analyte was also extracted efficiently from the matrix using the same solvent. Determination of analyte extraction efficiency independent from other factors such as ion suppression, adsorptive loss, etc. was difficult. The evaluation could be done by preparing numerous samples of matrix with analyte and spiking internal standard at each step along the sample preparation process. For quantitative results, this would have to be done as a single experiment and was determined to be excessively time-consuming for our purposes. Therefore, a simple comparison was conducted in which BaP-dG was prepared in digestion buffer and extracted with butanol; and conversely spiked into 5 μg of ctDNA digest and extracted with butanol. Both samples were analyzed by LC-MS/MS after drying and reconstitution. Peak areas were compared with a neat preparation of BaP-dG (no sample processing) to determine recovery. In a separate experiment (not shown), the relative signal of dG-C8-4-ABP as a model analyte was determined in the presence of different matrices. Equal quantities of analyte were spiked into equal volumes of 5% MeOH in water solvent (i.e. neat), 5 mM Tris / 10 mM MgCl2 digestion buffer, and 5 μg of ctDNA digest. Respectively, the ratio observed was 1:0.33:0.033. Because the neat BaP-dG control was not comparable to the matrix-containing samples and % recoveries would appear artificially low, a correction factor was also applied to account for signal loss due to the presence of matrix (normalize for matrix effects) as follows: buffer only: % recovery × 3; ctDNA digest: % recovery × 30. Butanol extraction recovered 56% of BaP-dG analyte prepared in buffer, and 45% of BaP-dG analyte in ctDNA digest. These values indicate that BaP-dG is being adequately extracted from the matrix. Butanol extraction also showed a significant 4.1-fold improvement over ethanol precipitation (Figure 6). Some fraction of this improvement is likely attributed to decreased matrix effects in butanol extraction because of the more efficient removal of co-eluting matrix ions, as discussed above.
Figure 6.
Comparison of cleanup strategies. TICs are overlaid with EICs of BaP-dG analyte and clearly show that butanol enrichment is more effective at removing co-eluting background ions than protein precipitation with ethanol. The single BaP-dG peak is indicated by an arrow. Multiple peaks in the top trace (ethanol precipitation) are from coeluting contaminants.
3.3.3. Sample drying
Before LC-MS/MS analysis, samples of digested DNA are typically dissolved in a solvent incompatible with MS, and excessively diluted from the preceding cleanup step (e.g. EtOH wash or butanol extraction). Evaporative drying using a speed-vac allows samples to be reconstituted in a desirable solvent of specific volume for LC-MS/MS analysis. However, this step generally reduces analyte recovery to some degree due to adsorption of the analyte to the walls of the tube in which the samples are dried. To minimize this loss, drying steps are another parameter that should also be optimized. Since DNA digests are prepared in polypropylene microcentrifuge tubes (1.5 or 0.6 mL, silylated or non-silylated), samples are also dried in these tubes, then resuspended in mobile phase and transferred to an autosampler vial. This extra sample-transfer step can result in some loss due to analyte adsorption to the tube walls, depending on their composition, or incomplete sample transfers due to pipetting errors. Therefore, in an effort to eliminate this step, we decided to dry the samples directly into the autosampler vials.
Three different glass inserts that were used in combination with the standard 2.0 mL, 12 mm × 32 mm screw-thread glass vials after evaporative speed-vac drying were compared for the recovery efficiency of Bap-dG. While these 2.0 mL vials are accommodated by most autosamplers, their large volume is not compatible with the low sample quantities required for our analyses and these inserts are used to reduce the volume of the vial to 300 μL. They are specially designed to be conical in shape with a drawn tip to ensure maximal sample recovery by the autosampler needle. Two inserts were obtained from Microliter (Wheaton, Millville, NJ), one standard glass and one silylated; and the third insert was also standard glass and was purchased from Supelco (Sigma, St. Louis, MO). All inserts were very similar: borosilicate glass, silylated or non-silylated, 300 μL volumes with conical point tips.
To test recovery in these inserts, both neat and 5 μg ctDNA digest preparations of BaP-dG were prepared in duplicate. Butanol extraction was done on neat and matrix samples and the extract (< 300 μL) was collected and dried directly in the glass inserts. Samples were reconstituted in 5% MeOH and analyzed by LC-MS/MS. As shown in Table 2, in the Microliter brand insert, silylation had very little effect on BaP-dG recovery in both neat and spiked matrix preparations. Interestingly, a considerable difference was observed between Microliter brand and Supelco brand inserts, the latter resulting in significant reduction of analyte recovery. Compared to non-silylated Microliter inserts, Supelco inserts recovered 3.8-times less analyte in neat and matrix preparations. Since both inserts have nearly identical specifications, it is difficult to identify the specific causes of this discrepancy but clearly the manufacture process varies in some way, and this experiment indicates the importance of keeping consistent supplies of consumables.
Table 2.
Effect of silylation on analyte recovery. BaP-dG recovery after speed-vac evaporative drying was only minimally affected by silylation but a significant difference was observed between brands. The presence of matrix (DNA digest) had no significant effect. Inserts from both manufacturers of similar shape: pulled-point conical, borosilicate glass. “Avg PA”, average peak area from duplicate analyses.
| Average PA BaP-dG | |||
|---|---|---|---|
| Brand | Silylation | Neat | Matrix |
| Microliter | Slz | 6.02 × 106 | 4.72 × 105 |
| Microliter | non-Slz | 4.54 × 106 | 4.51 × 105 |
| Supelco | non-Slz | 1.25 × 106 | 1.17 × 105 |
3.3.4. Reconstitution solvent
An important parameter that is easily overlooked is the solvent used for sample reconstitution after drying. For HPLC analysis, dry samples are typically reconstituted in mobile phase, with equal or lesser percentage of the organic component. In the present case, the sample containing the analyte standard and its isotopically labeled analog was loaded on the enrichment column with 10% B, which was 0.1% acetic acid in methanol and samples were reconstituted in 5% acid-free methanol. The analyte, BaP-dG, is fairly hydrophobic (logP ≈ 3.6, BP-7,8- dihydrodiol) thus it seemed plausible that a solvent with a higher percent organic was necessary for reconstitution. Acid is generally excluded from our sample preparation due to the labile ribose moiety, but acidic solvent could also help solubilize the analyte through protonation/ionization, and it was decided to test this as well.
Three reconstitution solvents were evaluated: Water containing 5% methanol with and without 0.1% formic acid and a 50/50 (v/v) mixture of DMSO/water (no acid), as DMSO is commonly used for solubilizing various nucleoside adducts [40]. In each case BaP-dG was spiked into 5 μg aliquots of DNA digest before protein precipitation and analyzed by LC-MS/MS and the results from duplicate analyses are summarized in Table 3. Peak areas of BaP-dG analyte in each sample were compared to a neat preparation of BaP-dG in order to calculate percent recoveries. In addition, because the neat BaP-dG control was not comparable to the matrix-containing samples and % recoveries would appear artificially low, a correction factor was also applied (% recovery × 30). As discussed above, this table includes a comparison of two different protein precipitation solvents, EtOH (−80 °C) and ACN (−20 °C) used in combination with the three different reconstitution solvents. The highest recoveries, 54- 55% were obtained when ACN was used in combination with 5% MeOH, either with or without 0.1% formic acid. To avoid potential complications from resolubilizing with acid (hydrolysis of ribose from nucleobase after extended incubation in autosampler), 5% MeOH was chosen as the reconstitution solvent.
Table 3.
Dependence of analyte recovery on resuspension solvent. Common solvents were tested for their ability to resolvate the analyte after evaporative drying by speed-vac. The combined effects with two protein precipitation solvents, ethanol and acetonitrile, are shown. Protein precipitation with acetonitrile followed by resuspension with 5% MeOH provided the best recovery in this comparison. FA, formic acid.
| Solvent | Resuspended In | % Recovery |
|---|---|---|
| EtOH | 5% MeOH | 9.6 |
| 50/50 DMSO / H2O | 13 | |
| 0.1% FA / 5% MeOH | 17 | |
| ACN | 5% MeOH | 55 |
| 50/50 DMSO / H2O | 17 | |
| 0.1% FA / 5% MeOH | 54 | |
% Recovery calculated from duplicate analyses
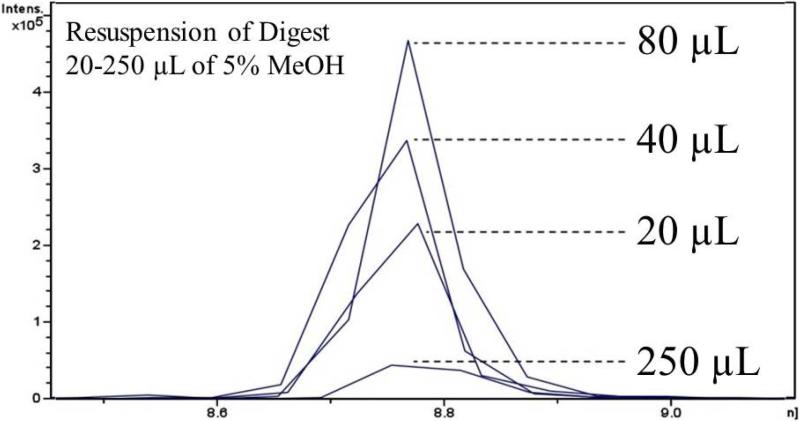
Sample dilution is often recommended as a means for reducing ion suppression. [41]. If the injection volume remains constant, sample dilution effectively results in less matrix entering the mass spectrometer. In turn, this also results in less analyte entering the mass spectrometer and so the dilution factor must be optimized to balance analyte signal with matrix effects. To examine the effect of dilution on the LC-MS/MS analysis of BaP-dG in DNA digest, five aliquots of 5 μg ctDNA digest were washed with ethanol, dried and spiked with standard BaP-dG (prepared in duplicate) in 20 μL of 5% MeOH. Subsequently, duplicate samples were diluted with 5% MeOH to final volumes of 40, 80, 160, and 250 μL. Five μL of each sample were injected and analyzed by LC-MS/MS. Figure 7 shows representative EICs overlaid for comparison. The peak areas increased linearly with increasing volume from 20 (S/N 23) to 80 μL (S/N 47). At a volume of 160 μL (S/N 35), the analyte signal decreased in intensity (not shown, approx. equal to 20 μL point, S/N 34 μL) and at 250 μL (S/N 4.1), the sample was excessively dilute resulting in virtually complete loss of signal.
Figure 7.
Matrix effect reduction via sample dilution. ctDNA digest was spiked with 4 fmol BaP-dG standard, dried and reconstituted in different volumes of 5% MeOH. As this figure demonstrates, injection of a more concentrated sample does not produce the strongest signal. Dilution of sample from 20-80 μL produced stronger analyte signals, likely due to decreasing ion suppression by the more dilute matrix. Above 80 μL, the signal deteriorated, likely due to over dilution, where too little analyte was being introduced into the mass spectrometer.
3.4. Optimization of online cleanup and chromatography to further address ion suppression
3.4.1. Online enrichment
When combined with optimized sample preparation, the microfluidic HPLC Chip from Agilent Technologies enables highly sensitive LC-MS/MS assays for DNA adducts using very small quantities of sample (2 μg DNA). The HPLC Chip combines online SPE with nanoLC and ESI via column switching, in which the sample is loaded on a C18 trap column and washed with mobile phase (10% B, methanol with 0.1% acetic acid) with effluent directed to waste, then after a defined time (4 min, original protocol) the trap column is linked to the analytical column via a switching valve, the analytical gradient begins and effluent is electrosprayed into the mass spectrometer. Online SPE improves throughput and, most significantly, increases recovery. For instance, offline SPE, which typically includes an evaporative speed-vac drying step, often has a recovery in the range of 50% [42]. For example, in previous studies dealing with the analysis of the deoxyguanosisne adduct of 4-aminobiphenyl (dG-ABP) using on-line SPE cleanup with the SPE with the HPLC Chip, analyte recoveries ranged from 71-95% [12].
To ensure maximal cleanup of the sample after injection onto the trap column, sample loading/washing time was varied from 4 to 8 minutes, with mobile phase compositions of 10% B and 30% B; the latter condition was used for offline SPE of dG- BaP by Feng et al. [43]. Despite the fact that two binary pumps are used, one for sample loading (online SPE), the other for analytical separation, the solvents used for sample loading/washing matched the solvent system used for the analytical pump.
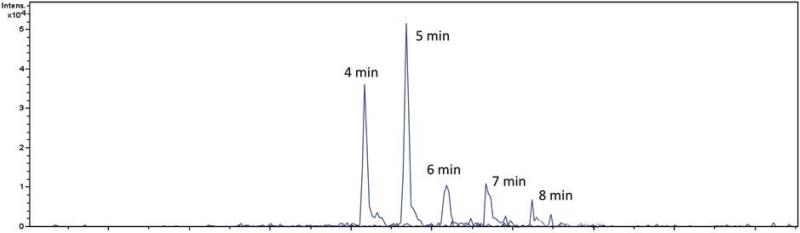
BaP-dG was spiked into 5 μg of digested ctDNA matrix and analyzed by LC- MS/MS under these variable conditions. With sample loading/washing at 30% B, some peak broadening was observed compared to loading/washing at 10% B at all enrichment times tested. Figure 8 shows the effect of varying the loading/washing time using 30% B on the analyte signal. After 6 minutes of isocratic washing with 30% B, the analyte signal is considerably reduced and at 8 minutes the signal is nearly undetectable (S/N < 3). This is likely due to elution of the BaP-dG from the trapping column, a presumption supported by the peak broadening observed at 30% B compared to 10% B. The signal intensity was not significantly affected by loading/washing time at 10% B, thus the original conditions were kept, namely 4 minutes of loading/washing with 10% B (methanol with 0.1% acetic acid).
Figure 8.
DNA digests were enriched on the HPLC chip by online SPE. Here the washing time (and volume) is varied to determine the effect on analyte signal. Flow rate is 4 μL/min with 30% methanol in water containing 0.1% acetic acid. Data show that the analyte begins to elute (to waste) after 6 minutes (24 μL) or more of enrichment. Note the enrichment column has a volume of 40 nL, therefore 6 minutes of washing equals 600 column volumes. Also note that the separation gradient was started after enrichment, therefore the increasing retention time observed is artificial (BaP-dG elutes at 45% B in each case).
3.4.2 Separation gradient and mobile phases
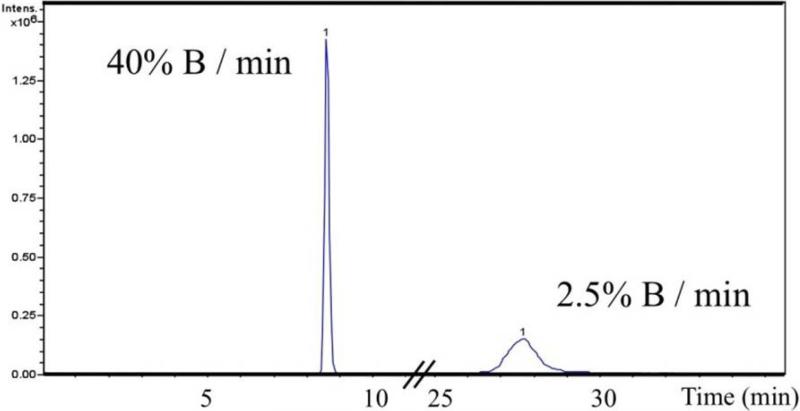
The gradient and solvent systems were selected for optimal sensitivity when the method was originally developed. This was verified by adjusting the gradient. Because a single analyte was being chromatographed, a very steep gradient was used (from 10-90% B in 2 minutes equaling 40% B/min), to generate a very sharp peak. This fast chromatography may cause many components to co-elute with the analyte resulting in ion suppression. To test this possibility, the gradient was slowed to 10-90% B in 4 min, 8 min, 16 min, and 32 min and BaP-dG spiked into ctDNA digest was resolved. The slower gradients resulted in lower S/N as the peak height decreased due to peak broadening. With a 4-minute gradient, broadening was slight, but this increased proportionally with time and at 16 and 32 minutes broadening became excessive. Figure 9 shows the dramatic peak broadening that occurred as the gradient was slowed from 40% B/min (2 minute, fast gradient) to 2.5% B/min (32 minute, gradual gradient). While the peak areas are nearly identical (PA=1.3 × 107, 40% B/min; PA=1.0 × 107, 2.5% B/min), the wider peak associated with the slow gradient has a S/N approximately 4-fold lower in these neat preparations of BaP-dG analyte, which is expected to have an even greater impact on the sensitivity in the presence of matrix, as the peak edges will become indistinguishable from the noise.
Figure 9.
Effect of separation gradient on peak shape. As the gradient is slowed from 40% B/min to 2.5% B/min, the analyte peak broadens reducing the S/N approximately 4-fold when analyzed in the absence of matrix (neat BaP-dG analyte).
Two solvent systems were compared with the solvent system in place (A: water/0.1% formic acid and B: methanol/0.1% formic acid). When 0.1% acetic acid was used in place of formic acid, no difference was observed. Solvent prepared with 0.1% ammonium formate was also tested and slightly lower sensitivity was observed. When acetonitrile was substituted for methanol as the organic solvent, a dramatic decrease in signal intensity was observed. This may be due simply to the fact that the ESI parameters required optimization after switching to ACN, but at the time this was not considered because the ESI parameters are generally optimized during tuning and calibration of the MS.
3.5. Applications to Cell Dosing
As described above, the sensitivity of the LC-MS/MS assay was improved primarily by decreasing ion suppression caused by contaminating matrix ions. The next step was to apply the optimized method to the analysis of cultured human cells.
3.5.1. Calibration curve
A calibration curve was prepared in calf-thymus DNA matrix as follows. A two- fold serial dilution of a 12.5 μM stock solution of synthetic BaP-dG (stored at −20 °C, protected from light, in dry DMSO) was prepared at the following concentrations: 0.030, 0.061, 0.12, 0.24, 0.49, 0.98, 2.0, and 3.9 fmol/μL. Ten (10) microliters (μL) of each dilution was spiked into 5 μg of ctDNA digest resulting in samples with the following adduction levels: 8.20, 16.3 32.7, 65.4, 131, 261, 523, and 1046 BaP-dG adducts per 108 nucleosides. Internal standard (0.2 pmol/μL stock) was spiked into each sample to maintain a constant concentration of 1 fmol/μL. Each sample was reconstituted into a final volume of 20 μL and triplicate analyses were carried out by injecting 5 μL aliquots (i.e., 1.5 μg of DNA) on-column. A calibration curve was generated by plotting the number of femtomoles of analyte injected on-column vs. the ratio of analyte to IS peak areas. The linear range was plotted to generate a best-fit line and the equation y=0.136× + 0.0776 (R2 = 0.995) was used to calculate the number of BaP-dG adducts. At the lowest point of the calibration curve this corresponded to the injection of 0.075 fmole (75 attomole per 1.5 μg of DNA) equivalent to an LOQ of 8.20 BaP-dG adducts per 108 nucleosides. These detection and quantitation limits are in line with previous reports on levels of dG-N2-BaP adducts observed in humans [44, 45] and in principle could be further improved by processing larger quantities of DNA.
3.5.2. BaP-dG adducts detected in cell culture
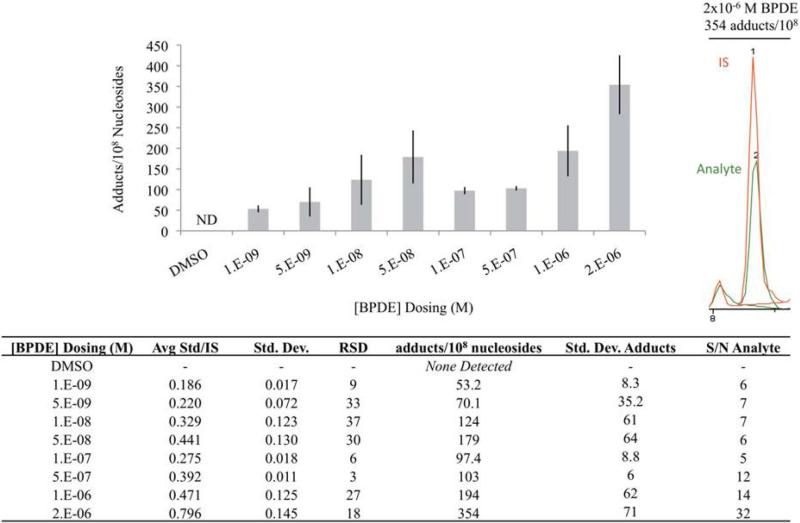
The general applicability of the optimized process was next tested in a study in which cultured human BEAS-2B bronchial epithelial cells were dosed in triplicate with BPDE at the following concentrations: 1.0 × 10−9, 5.0 × 10−9, 1.0 × 10−8, 5.0 × 10−8, 1.0 × 10−7, 5.0 × 10−7, 1.0 × 10−6, 2.0 × 10−6 M and a DMSO control. After 8 days of exposure (24 h for BPDE), cells were cultured in fresh medium for an additional 7 days (24 hours for BPDE) before the DNA was isolated from the cell cultures and dissolved in 5 mM Tris/10 mM MgCl2 buffer and quantified using Invitrogen's Qubit dsDNA BR assay kit according to the manufacturer's protocol. A volume corresponding to 2 μg of DNA was removed from each sample and diluted to 25 ng/uL in 5 mM Tris/10 mM MgCl2. A single aliquot of 2 μg DNA was digested from each of 3 biological replicates. The samples were digested and prepared for LC-MS/MS analysis using the optimized protocol.
Results of LC-MS/MS quantitation are shown in Figure 10 (table included in figure). DNA adducts were not detected in the DMSO negative control or the lowest concentration, 1.0−9 M. Adducts were detected from treatment with 5.0 × 10−9 M, but were below the LOQ (<15 fmol/uL) so could not be quantified. The range of BPDE treatments from 1.0 × 10-8 M to 2.0 × 10−6 M suggested the possibility of a bimodal distribution with maxima at 5.0 × 10−8 M (247 adducts/108) and 2.0 × 10−6M (603 adducts/108). However the limited sample size and the large amount of variability preclude any conclusion regarding the shape of the dose response curve. The dosing range from 1.0 × 10−7 M to 2.0 × 10−6 M showed a nearly linear increase in adduction levels (R2=0.980). It is not apparent whether the trend would continue for concentrations higher than 2.0 × 10−6 M BPDE because hydrolysis of the epoxide is a competing, deactivating reaction and expected to be rapid in the aqueous cell medium.
Figure 10.
Quantitation of dG-BaP IN BEAS-2B cells exposed to varying concentrations of BPDE. The bar graph shows BaP-dG adduct levels and data are tabulated in the table below. A sample chromatogram with integrated BaP-dG analyte and IS peaks is shown in the top right corner. Error bars represent biological variation from triplicate cell dosings of biological replicates, each analyzed once by LC-MS/MS.
4. Conclusion
Sample preparation is a critical component in the development of a bioanalytical protocol which frequently involves several steps each one of which needs to be optimized in order to achieve the best results possible in the analysis. The preparation of a sample for analysis by LC-MS employing electrospray ionization is particularly susceptible to matrix effects which may interfere – generally reduce – with the signal of the analyte(s) of interest. In the analysis of DNA adducts, in addition to endogenous species already present in the biological sample, further complexities are introduced due to the use of enzymes to digest the DNA. Moreover, this is on top of the solvents used for analyte extraction and reconstitution, the LC mobile phase and buffer selection. These effects may be especially detrimental when dealing with trace level analyses such as those associated with the detection and quantification of DNA adducts and introduce errors in quantitation and precision. Indeed, the results presented above demonstrate that in the analysis of DNA adducts matrix effects may originate from a wide variety of sources any one of which may significantly compromise the results of the analysis. In the course of the development of a protocol for analysis of the deoxyguanosine adduct of benzo[a]pyrene, the individual matrix contribution of each one of these sources to analyte signal was systematically addressed as were any interactive effects. Moreover, as recommended [46] the analyte concentrations used for validation of matrix effects were in line with those that may be encountered under “real world” conditions. This comprehensive evaluation of matrix effects resulted in the development of an optimized protocol which was tested and shown to be applicable to a dose response study using metabolically competent cell cultures and achieve detection limits compatible with those observed in vivo. While the optimized parameters described above are specific for reduction of matrix effects associated with the analysis of the N2-BPDE-dG, the strategy employed is directly applicable to the analysis of DNA adducts and, in a broader sense, to the field of bioanalysis in general. It is envisioned that, despite the empirical approach that needs to be taken, an evaluation of matrix effects as presented in this paper can provide a further means for improving sensitivity in the area of targeted DNA adduct analysis.
Highlights.
DNA adducts serve as important biomarkers of risk assessment from exposure to environmental carcinogens.
Sample preparation for their analysis is a multistep process in which each step can introduce matrix interferences that may have a detrimental effect on the sensitivity of the analytical method.
We show that systematic evaluation of matrix contribution associated with each step may be integrated toward the development of an optimized protocol for quantification of DNA adducts using low microgram quantities of DNA.
ACKNOWLEDGEMENTS
This work was supported by Public Health Services Grants: NIH RO1CA112231 (P. Vouros, P.I.), NIH RO1 069390 (P. Vouros, P.I.); NIEHS Center Grant P30ES005022 (H. Zarbl, P.I.) and Toxicology training grant T32ES007148 (H. Zarbl, P.I.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.A Review of Human Carcinogens: Chemical Agents and Related Occupations. International Agency for Research on Cancer; 2012. pp. 111–144. [Google Scholar]
- 2.D.o.N.T. Program, NTP Research Concept: Polycyclic Aromatic Hydrocarbons (PAHs) 2012:1–14. [Google Scholar]
- 3.Some non-heterocyclic polycyclic aromatic hydrocarbons and some related exposures. International Agency for Research on Cancer; 2010. pp. 1–853. [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips DH. Fifty years of benzo(a)pyrene. Nature. 1983;303:468–472. doi: 10.1038/303468a0. [DOI] [PubMed] [Google Scholar]
- 5.Tobacco smoke and involuntary smoking. International Agency for Research on Cancer; 2004. pp. 1–1438. [Google Scholar]
- 6.Chiang TA, Pei-Fen W, Ying LS, Wang LF, Ko YC. Mutagenicity and aromatic amine content of fumes from heated cooking oils produced in Taiwan. Food Chem Toxicol. 1999;37:125–134. doi: 10.1016/s0278-6915(98)00081-7. [DOI] [PubMed] [Google Scholar]
- 7.Bostrom CE, Gerde P, Hanberg A, Jernstrom B, Johansson C, Kyrklund T, Rannug A, Tornqvist M, Victorin K, Westerholm R. Cancer risk assessment, indicators, and guidelines for polycyclic aromatic hydrocarbons in the ambient air. Environ Health Perspect. 2002;110(Suppl 3):451–488. doi: 10.1289/ehp.110-1241197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimada T, Gillam EM, Oda Y, Tsumura F, Sutter TR, Guengerich FP, Inoue K. Metabolism of benzo[a]pyrene to trans-7,8-dihydroxy-7, 8-dihydrobenzo[a]pyrene by recombinant human cytochrome P450 1B1 and purified liver epoxide hydrolase. Chem Res Toxicol. 1999;12:623–629. doi: 10.1021/tx990028s. [DOI] [PubMed] [Google Scholar]
- 9.Shimada T, Oda Y, Gillam EM, Guengerich FP, Inoue K. Metabolic activation of polycyclic aromatic hydrocarbons and other procarcinogens by cytochromes P450 1A1 and P450 1B1 allelic variants and other human cytochromes P450 in Salmonella typhimurium NM2009. Drug Metab Dispos. 2001;29:1176–1182. [PubMed] [Google Scholar]
- 10.Klaene JJ, Sharma VK, Glick J, Vouros P. The analysis of DNA adducts: The transition from 32P-postlabeling to mass spectrometry. Cancer Lett. 2013;334:10–19. doi: 10.1016/j.canlet.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding Y, Paonessa JD, Randall KL, Argoti D, Chen L, Vouros P, Zhang Y. Sulforaphane inhibits 4-aminobiphenyl-induced DNA damage in bladder cells and tissues. Carcinogenesis. 2010;31:1999–2003. doi: 10.1093/carcin/bgq183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Randall KL, Argoti D, Paonessa JD, Ding Y, Oaks Z, Zhang Y, Vouros P. An improved liquid chromatography-tandem mass spectrometry method for the quantification of 4-aminobiphenyl DNA adducts in urinary bladder cells and tissues. J Chromatogr A. 2010;1217:4135–4143. doi: 10.1016/j.chroma.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paonessa JD, Ding Y, Randall KL, Munday R, Argoti D, Vouros P, Zhang Y. Identification of an unintended consequence of Nrf2-directed cytoprotection against a key tobacco carcinogen plus a counteracting chemopreventive intervention. Cancer Res. 2011;71:3904–3911. doi: 10.1158/0008-5472.CAN-11-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kafle A, Klaene J, Hall AB, Glick J, Coy SL, Vouros P. A differential mobility spectrometry/mass spectrometry platform for the rapid detection and quantitation of DNA adduct dG ABP. Rapid Commun Mass Spectrom. 2013;27:1473–1480. doi: 10.1002/rcm.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chitta KR, Landero-Figueroa JA, Kodali P, Caruso JA, Merino EJ. Identification of selenium- containing proteins in HEK 293 kidney cells using multiple chromatographies, LC-ICPMS and nano-LC ESIMS. Talanta. 2013;114:25–31. doi: 10.1016/j.talanta.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Grimley A, Bertram R, Roper MG. Microfluidic system for generation of sinusoidal glucose waveforms for entrainment of islets of Langerhans. Anal Chem. 2010;82:6704–6711. doi: 10.1021/ac101461x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bynum MA, Yin H, Felts K, Lee YM, Monell CR, Killeen K. Characterization of IgG N- glycans employing a microfluidic chip that integrates glycan cleavage, sample purification, LC separation, and MS detection. Anal Chem. 2009;81:8818–8825. doi: 10.1021/ac901326u. [DOI] [PubMed] [Google Scholar]
- 18.Vollmer M, van de Goor T. HPLC-Chip/MS technology in proteomic profiling. Methods Mol Biol. 2009;544:3–15. doi: 10.1007/978-1-59745-483-4_1. [DOI] [PubMed] [Google Scholar]
- 19.Brennen RA, Yin H, Killeen KP. Microfluidic gradient formation for nanoflow chip LC. Anal Chem. 2007;79:9302–9309. doi: 10.1021/ac0712805. [DOI] [PubMed] [Google Scholar]
- 20.Freire SL, Yang H, Wheeler AR. A practical interface for microfluidics and nanoelectrospray mass spectrometry. Electrophoresis. 2008;29:1836–1843. doi: 10.1002/elps.200700661. [DOI] [PubMed] [Google Scholar]
- 21.Dominical VM, Vital DM, O'Dowd F, Saad ST, Costa FF, Conran N. In vitro microfluidic model for the study of vaso-occlusive processes. Exp Hematol. 2015;43:223–228. doi: 10.1016/j.exphem.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Bhattacharya A, Klaene JJ, Li Y, Paonessa JD, Stablewski AB, Vouros P, Zhang Y. The inverse relationship between bladder and liver in 4-aminobiphenyl-induced DNA damage. Oncotarget. 2015;6:836–845. doi: 10.18632/oncotarget.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gosetti F, Mazzucco E, Zampieri D, Gennaro MC. Signal suppression/enhancement in high- performance liquid chromatography tandem mass spectrometry. J Chromatogr A. 2010;1217:3929–3937. doi: 10.1016/j.chroma.2009.11.060. [DOI] [PubMed] [Google Scholar]
- 24.Stephen S, Darryl R, Mary O, Joseph E. Pharmaceutical research and manufacturers association acceptable analytical practice for analytical method transfer. Pharmaceutical Technology. 2002;3:84–88. [Google Scholar]
- 25.Buening M, Wislocki P, Levin W, Yagi H, Thakker D, Akagi H, Koreeda M, Jerina DM, Conney A. Tumorigenicity of the optical enantiomers of the diastereomeric benzo [a] pyrene 7, 8-diol-9, 10- epoxides in newborn mice: exceptional activity of (+)-7beta, 8alpha-dihydroxy-9alpha, 10alpha-epoxy-7, 8, 9, 10-tetrahydrobenzo [a] pyrene. Proceedings of the National Academy of Sciences. 1978;75:5358–5361. doi: 10.1073/pnas.75.11.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh R, Gaskell M, Le Pla RC, Kaur B, Azim-Araghi A, Roach J, Koukouves G, Souliotis VL, Kyrtopoulos SA, Farmer PB. Detection and quantitation of benzo[a]pyrene-derived DNA adducts in mouse liver by liquid chromatography-tandem mass spectrometry: comparison with 32P- postlabeling. Chem Res Toxicol. 2006;19:868–878. doi: 10.1021/tx060011r. [DOI] [PubMed] [Google Scholar]
- 27.Flarakos CC. Applications of Mass Spectrometry Techniques to the Elucidation of Novel Metabolic Pathways of Vitamin D and the Quantification of DNA Adducts. ProQuest. 2008 [Google Scholar]
- 28.Van Vleet TR, Klein PJ, Coulombe RA., Jr. Metabolism and cytotoxicity of aflatoxin b1 in cytochrome p-450-expressing human lung cells. J Toxicol Environ Health A. 2002;65:853–867. doi: 10.1080/00984100290071216. [DOI] [PubMed] [Google Scholar]
- 29.Van Vleet TR, Mace K, Coulombe RA., Jr. Comparative aflatoxin B(1) activation and cytotoxicity in human bronchial cells expressing cytochromes P450 1A2 and 3A4. Cancer Res. 2002;62:105–112. [PubMed] [Google Scholar]
- 30.Savela K, Hemminki K. Analysis of cigarette-smoke-induced DNA adducts by butanol extraction and nuclease P1-enhanced 32P-postlabeling in human lymphocytes and granulocytes. Environmental health perspectives. 1993;101:145. doi: 10.1289/ehp.93101s3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNaught AD, McNaught AD. Compendium of chemical terminology. Blackwell Science Oxford. 1997 [Google Scholar]
- 32.Matuszewski B, Constanzer M, Chavez-Eng C. Strategies for the assessment of matrix effect in quantitative bioanalytical methods based on HPLC-MS/MS. Analytical chemistry. 2003;75:3019–3030. doi: 10.1021/ac020361s. [DOI] [PubMed] [Google Scholar]
- 33.Kebarle P, Tang L. From ions in solution to ions in the gas phase-the mechanism of electrospray mass spectrometry. Analytical Chemistry. 1993;65:972A–986A. [Google Scholar]
- 34.Enke CG. A predictive model for matrix and analyte effects in electrospray ionization of singly- charged ionic analytes. Analytical Chemistry. 1997;69:4885–4893. doi: 10.1021/ac970095w. [DOI] [PubMed] [Google Scholar]
- 35.Trufelli H, Palma P, Famiglini G, Cappiello A. An overview of matrix effects in liquid chromatography–mass spectrometry. Mass spectrometry reviews. 2011;30:491–509. doi: 10.1002/mas.20298. [DOI] [PubMed] [Google Scholar]
- 36.Heflich RH, Morris SM, Beranek DT, McGarrity LJ, Chen JJ, Beland FA. Relationships between the DNA adducts and the mutations and sister-chromatid exchanges produced in Chinese hamster ovary cells by N-hydroxy-2-aminofluorene, N-hydroxy-N′-acetylbenzidine and 1-nitrosopyrene. Mutagenesis. 1986;1:201–206. doi: 10.1093/mutage/1.3.201. [DOI] [PubMed] [Google Scholar]
- 37.Crain PF. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectometry. Methods in enzymology. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- 38.Welsh R. A method for the preparation of DNA from calf thymus nuclei in maximum purity and yield, analysis of this DNA compared with that obtained by other methods. Physiological chemistry and physics and medical NMR. 1992;25:125–135. [PubMed] [Google Scholar]
- 39.Want EJ, O'Maille G, Smith CA, Brandon TR, Uritboonthai W, Qin C, Trauger SA, Siuzdak G. Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry. Analytical Chemistry. 2006;78:743–752. doi: 10.1021/ac051312t. [DOI] [PubMed] [Google Scholar]
- 40.Rindgen D, Turesky RJ, Vouros P. Determination of in vitro formed DNA adducts of 2-amino-1- methyl-6-phenylimidazo [4, 5-b] pyridine using capillary liquid chromatography/electrospray ionization/tandem mass spectrometry. Chemical research in toxicology. 1995;8:1005–1013. doi: 10.1021/tx00050a003. [DOI] [PubMed] [Google Scholar]
- 41.Stahnke H, Kittlaus S, Kempe G.n., Alder L. Reduction of matrix effects in liquid chromatography–electrospray ionization–mass spectrometry by dilution of the sample extracts: how much dilution is needed? Analytical chemistry. 2012;84:1474–1482. doi: 10.1021/ac202661j. [DOI] [PubMed] [Google Scholar]
- 42.John H, Walden M, Schäfer S, Genz S, Forssmann W-G. Analytical procedures for quantification of peptides in pharmaceutical research by liquid chromatography–mass spectrometry. Analytical and bioanalytical chemistry. 2004;378:883–897. doi: 10.1007/s00216-003-2298-y. [DOI] [PubMed] [Google Scholar]
- 43.Feng F, Wang X, Yuan H, Wang H. Ultra-performance liquid chromatography-tandem mass spectrometry for rapid and highly sensitive analysis of stereoisomers of benzo[a]pyrene diol epoxide- DNA adducts. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:2104–2112. doi: 10.1016/j.jchromb.2009.05.054. [DOI] [PubMed] [Google Scholar]
- 44.Boysen G, Hecht SS. Analysis of DNA and protein adducts of benzo [a] pyrene in human tissues using structure-specific methods. Mutation Research/Reviews in Mutation Research. 2003;543:17–30. doi: 10.1016/s1383-5742(02)00068-6. [DOI] [PubMed] [Google Scholar]
- 45.Sangaraju D, Villalta P, Goggin M, Agunsoye MO, Campbell C, Tretyakova N. Capillary HPLC-accurate mass MS/MS quantitation of N7-(2, 3, 4-trihydroxybut-1-yl)-guanine adducts of 1, 3- butadiene in human leukocyte DNA. Chemical research in toxicology. 2013;26:1486–1497. doi: 10.1021/tx400213m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Eeckhaut A, Lanckmans K, Sarre S, Smolders I, Michotte Y. Validation of bioanalytical LC–MS/MS assays: evaluation of matrix effects. Journal of Chromatography B. 2009;877:2198–2207. doi: 10.1016/j.jchromb.2009.01.003. [DOI] [PubMed] [Google Scholar]