Abstract
FtsX is an integral membrane protein from Streptococcus pneumoniae (pneumococcus) that harbors an extracellular loop 1 domain (FtsXECL1Spn) that interacts with PcsB, an peptidoglycan hydrolase that is essential for cell growth and division. Here, we report nearly complete backbone and side chain resonance assignments and a secondary structural analysis of FtsXECL1Spn (residues 47–168 of FtsX) as first steps toward structure determination of FtsXECL1Spn.
Keywords: bacterial cell wall, divisome, peptidoglycan hydrolysis, ABC transporter, allostery, extracellular signaling
Biological context
Proper spatiotemporal regulation of the hydrolysis of the peptidoglycan (PG) layer of the bacterial cell wall is essential to cell growth and cell division (Sham et al. 2013). In Streptococcus pneumoniae D39, PcsB is an essential PG hydrolase whose activity is subject to regulation by protein-protein interactions (Sham et al. 2011), the details of which remain poorly defined (Bartual et al. 2014). The extended coiled-coil domain of PcsB (Bartual et al. 2014) has been shown to physically interact with FtsXECL1Spn and it is known that FtsX is essential for pneumococcal cell growth and division (Sham et al. 2011). A current model is that FtsXECL1Spn is an allosteric modulator of PcsB-catalyzed cell wall hydrolysis, in a manner that is linked to ATP hydrolysis by the ATPase FtsE, which itself interacts with the FtsZ divisome (Sham et al. 2013). FtsEX structurally resembles an ATP-binding cassette (ABC) transporter, but is not thought to transport ligands across the membrane. The mechanism by which ATP hydrolysis by FtsE is linked to a conformational change in FtsX transduced across the plasma membrane and thus unlocking the auto-inhibited state of PcsB is unknown, but of high interest as a potential target for the development of novel antibiotics that target this essential process.
FtsXSpn is a 303 residue homodimer, and a topology model suggests that residues 42–182 connecting transmembrane helices 1 and 2 comprise the large extracellular loop 1 (ECL1) (Sham et al. 2013). A second smaller loop 2 domain (ECL2; 28 residues) connects transmembrane helices 3 and 4 on the same, extracellular side of the membrane, and both loop domains appear to be involved in some way in PcsB allosteric activation. An initial construct encompassing the entire ECL1 domain yielded protein characterized by excellent chemical shift dispersion, but uneven cross peak intensities, and complete chemical exchange broadening of C-terminal residues 171–182, proximal to transmembrane helix 2. Interestingly, this region harbors three of the six ftsX mutants that suppress a temperature-sensitive mutation in the coiled-coil domain of PcsB (Sham et al. 2013). These data, coupled with a subsequent sequence and structure-based alignment with the extracellular domain (ECD) of Mycobacterium tuberculosis FtsX (Mavrici et al. 2014) (29% similarity; 17% identity) suggested that FtsXECL1Spn spanning residues 47–168 (122 residues) would be characterized by more favorable relaxation properties. Here, we report nearly complete 1H, 15N, 13C backbone and side chain resonance assignments of FtsXECL1Spn as a basis for further structural characterization of this key pneumococcal protein-protein interaction domain.
Methods and experiments
Protein expression and characterization
The region of the gene encoding FtsXECL1Spn (residues 47–168 from FtsXSpn) was PCR-amplified from genomic DNA of Streptococcus pneumoniae D39 strain and subcloned into a pHis-parallel expression vector (Sheffield et al. 1999). The protein was expressed as a His6-tag fusion protein in E. coli BL21 (DE3) cells in LB medium, with expression induced by addition of 0.4 mM IPTG and growth continued at 16 °C for 20 h. Cells were harvested by centrifugation at 6,000 rpm for 10 min and were resuspended in lysis buffer (25 mM Tris, 200 mM NaCl, pH 8.0) and lysed by sonication on ice. The lysate was clarified centrifugation for 30 min at 12,000 rpm at 4 °C and the supernatant was filtered through a 0.45 μm filter membrane. The filtered supernatant was loaded onto a His-Trap FF column (GE Healthcare) equilibrated with low imidazole buffer A (25 mM Tris, 200 mM NaCl, 25 mM imidazole, pH 8.0). The bound His6 tagged fusion protein was eluted with high imidazole buffer B (25 mM Tris, 200 mM NaCl, 300 mM imidazole, pH 8.0). The His6 tag was cleaved with TEV protease and the uncleaved protein and liberated His6 tag removed by re-loading onto the His-Trap FF column equilibrated with low imidazole buffer A (25 mM Tris, 200 mM NaCl, 25 mM imidazole, pH 8.0). The flow-through, containing cleaved FtsXECL1Spn, was further purified by size-exclusion chromatography on a HiLoad 16/600 Superdex 75 column (GE Healthcare). FtsXECL1Spn purified in this way encompasses residues 47–168 with a non-native N-terminal GAMA tetrapeptide sequence (denoted residues 43–46 here) as a consequence of the subcloning and expression strategy used.
Uniformly 15N, 13C double-labeled FtsXECL1Spn protein was expressed in bacteria growing in M9 minimal medium containing 1.0 g/L of 15NH4Cl and 3.0 g/L [13C6]-glucose, as the sole nitrogen and carbon sources, respectively. The double-labeled protein was purified following the protocol described above. Protein purity was assessed by 18% SDS-PAGE, and protein concentration was estimated by UV-Vis spectroscopy. Uniformly 15N, 13C labeled FtsXECL1Spn ranging from 0.5 to 0.7 mM were used for NMR data acquisition.
NMR spectroscopy
All spectra were acquired at 298 K on a Varian (Agilent) DDR 600 MHz spectrometer equipped with cryogenic probe in the METACyt Biomolecular NMR Laboratory at Indiana University Bloomington. Typical NMR samples contained 50 mM potassium phosphate, pH 7.0, 50 mM NaCl and 10 % v/v D2O. For experiments used to obtain side chain assignments, the NMR sample was buffer exchanged into 50 mM KPi, pH 7.0, 50 mM NaCl and 99.8 % v/v D2O. Backbone sequential resonance assignments were carried out using a standard suite of double and triple resonance NMR experiments including the 15N-HSQC, 13C-HSQC, HNCACB, CBCA(CO)NH, HNCO, HN(CA)CO, HNCA and HN(CO)CA. Side chain resonance assignments were obtained using C(CO)NH-TOCSY (Grzesiek et al. 1993), H(CCO)NH-TOCSY (Montelione et al. 1992), HCCH-TOCSY, HCCH-COSY (Bax et al. 1990), 15N-edited NOESY and 13C-edited NOESY spectra. nmrPipe (Delaglio et al. 1995) and ccpNMR ver. 2.3.0 were used for data processing and analysis, respectively. Secondary structure predictions were derived from backbone resonance assignments using TALOS+ (Shen et al. 2009) and backbone dynamics were assessed through measurement of 15N heteronuclear 15N–{1H} (hNOE) values.
Assignment extension and data deposition
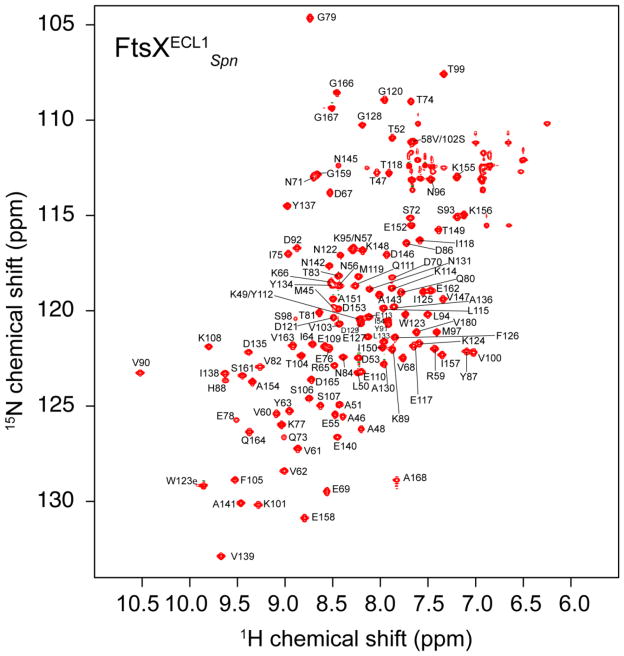
The 1H,15N HSQC spectrum of FtsXECL1Spn is shown in Fig. 1. Backbone amide resonances were assigned for all non-proline residues, with the exception of two non-native N-terminal residues (G43, A44) and N85. Resonance assignments for the 15Nε1–1Hε2 pair of W123 were also obtained. 92% of 13Cα, 13CO, side chain aliphatic and aromatic 13C and 1H resonances were assigned. The resonance assignments of FtsXECL1Spn have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu/) under the accession number 26534.
Figure 1.
1H, 15N HSQC spectrum of the uniformly 15N, 13C FtsXECL1Spn recorded at 600 MHz 1H frequency at 298 K. Solution conditions: 50 mM KPi, pH 7.0, 50 mM NaCl, 10 % D2O (v/v). The resonance assignments for backbone amides are shown using the one-letter amino acid code..
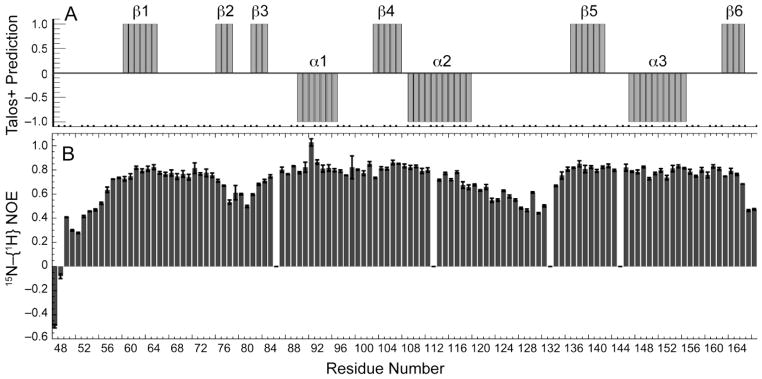
Secondary structural elements of FtsXECL1Spn were predicted using TALOS+ (Fig. 2A) with residues 59–64, 75–77, 81–83, 102–106, 136–141 and 162–165 forming short β-stranded regions (β1-β6), while residues 89–95, 108–118 and 146–155 form helical segments (α1-α3). The predicted secondary structure elements are supported by heteronuclear 15N–{1H} values that are generally ≥ 0.6 (Fig. 2B). Despite the low overall sequence similarity to M. tuberculosis FtsX ECD, this distribution of secondary structural units in FtsXECL1Spn is generally similar to that defined by the x-ray structure of FtsXECDMtb (Mavrici et al. 2014) with the major exception being what appears to be a β2-β3 β-hairpin insertion in FtsXECL1Spn in place of a short helical element (denoted α1) in FtsXECDMtb. FtsXECDMtb adopts a two-lobed structure in which a lower lobe is thought to be connected to the mixed α-β upper lobe via a “hinge-like’ linkage (Mavrici et al. 2014). If the global structures of FtsXECDMtb, and FtsXECL1Spn are similar, the smaller lower lobe in FtsXECL1Spn would correspond to residues ≈108–135, flanked by the β4 and β5 strands (Fig. 2). It will be of interest to determine the degree to which these two domains are structurally and dynamically oriented relative to one another in FtsXECL1Spn in solution since the cleft between the lobes is projected to define a major region of interaction with the cognate regulated PG hydrolase (Mavrici et al. 2014).
Figure 2.
Secondary structural analysis of FtsXECL1Spn. (A) TALOS+ predictions with probable β-strands and α-helices represented by +1 and − 1, respectively. Probabilities are based on TALOS+ probability values ≥ 0.6, with the exception of the β3 strand, in which only the middle residue (V82) meets this criterion. (B) Heteronuclear 15N–{1H} values for 15N, 13C-labeled FtsXECL1Spn. The error bar on each hNOE value was estimated from the noise in the spectra. Gaps correspond to the N85 (unassigned), Y112 (overlapped), P132 and P144.
Acknowledgments
This work was supported by NIH grants GM042569 (to D. P. G.), GM114315 (to M. E. W.) and a predoctoral Quantitative and Chemical Biology (QCB) institutional training grant fellowship supported by T32 GM109825 (to B.R.)
References
- Bartual SG, Straume D, Stamsas GA, Munoz IG, Alfonso C, Martinez-Ripoll M, Havarstein LS, Hermoso JA. Structural basis of PcsB-mediated cell separation in Streptococcus pneumoniae. Nat Commun. 2014:53842. doi: 10.1038/ncomms4842. [DOI] [PubMed] [Google Scholar]
- Bax A, Clore GM, Gronenborn AM. 1H–1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J Magn Reson. 1990;88 (2):425–431. [Google Scholar]
- Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6 (3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Grzesiek S, Anglister J, Bax A. Correlation of Backbone Amide and Aliphatic Side-Chain Resonances in 13C/15N-Enriched Proteins by Isotropic Mixing of 13C Magnetization. J Magn Reson. 1993;101 (1):114–119. [Google Scholar]
- Mavrici D, Marakalala MJ, Holton JM, Prigozhin DM, Gee CL, Zhang YJJ, Rubin EJ, Alber T. Mycobacterium tuberculosis FtsX extracellular domain activates the peptidoglycan hydrolase, RipC. Proc Natl Acad Sci U S A. 2014;111 (22):8037–8042. doi: 10.1073/pnas.1321812111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelione GT, Lyons BA, Emerson SD, Tashiro M. An efficient triple resonance experiment using carbon-13 isotropic mixing for determining sequence-specific resonance assignments of isotopically-enriched proteins. J Am Chem Soc. 1992;114 (27):10974–10975. [Google Scholar]
- Sham LT, Barendt SM, Kopecky KE, Winkler ME. Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsX(Spn) cell division protein in Streptococcus pneumoniae D39. Proc Natl Acad Sci U S A. 2011;108 (45):E1061–E1069. doi: 10.1073/pnas.1108323108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham LT, Jensen KR, Bruce KE, Winkler ME. Involvement of FtsE ATPase and FtsX extracellular loops 1 and 2 in FtsEX-PcsB complex function in cell division of Streptococcus pneumoniae. MBio. 2013;4(4):D39. doi: 10.1128/mBio.00431-13. pii: e00431–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purific. 1999;15 (1):34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS plus : a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44 (4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]