Abstract
Small molecule secondary metabolites produced by organisms such as plants, bacteria, and fungi form a fascinating and important group of natural products, many of which have shown promise as medicines. Fungi in particular have been important sources of natural product polyketide pharmaceuticals. While the structural complexity of these polyketides makes them interesting and useful bioactive compounds, these same features also make them difficult and expensive to prepare and scale-up using synthetic methods. Currently, nearly all commercial polyketides are prepared through fermentation or semi-synthesis. However, elucidation and engineering of polyketide pathways in the native filamentous fungi hosts are often hampered due to a lack of established genetic tools and of understanding of the regulation of fungal secondary metabolisms. Saccharomyces cerevisiae has many advantages beneficial to the study and development of polyketide pathways from filamentous fungi due to its extensive genetic toolbox and well-studied metabolism. This review highlights the benefits S. cerevisiae provides as a tool for mining, studying, and engineering fungal polyketide synthases (PKSs), as well as notable insights this versatile tool has given us into the mechanisms and products of fungal PKSs.
1. Introduction
Small molecule secondary metabolites produced by organisms such as plants, bacteria, and fungi form a fascinating and important group of natural products. Many of these natural products with diverse bioactivities have been important sources of medicines. Approximately 50% of all new chemical entity small molecules approved by the U.S. Food and Drug Administration (FDA) from 2000–2010 and nearly half of the drugs approved from 1994–2008 were derived from natural products (Newman and Cragg, 2012; Harvey, 2008).
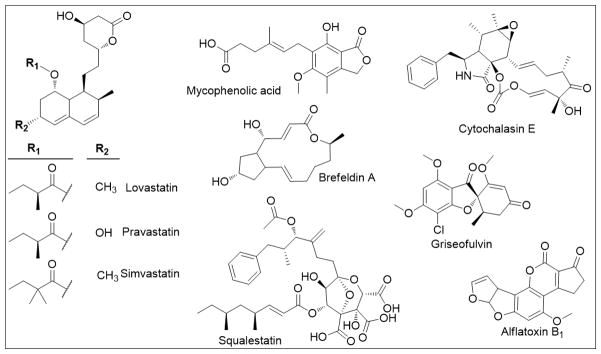
Polyketides are one of the major classes of natural products with diverse chemical structures and biological activities. Fungi in particular have been important sources of polyketide pharmaceuticals or virulence factors (Hertweck, 2009), and several examples of well-known fungal polyketides are included in Figure 1. Many of these compounds display clinically relevant activities against human diseases. For example, simvastatin, lovastatin, and mevastatin are all structurally related, natural product-derived cholesterol-lowering medications currently on the market (Barrios-González and Miranda, 2010). Squalestatin is an inhibitor of squalene synthases and also shows potential as a cholesterol-lowering medication (Baxter et al., 1992). In addition to the compounds displaying anti-hypercholesterolemia properties, several polyketides have been studied for their potential anticancer activities. Specifically, cytochalasin E has been shown to inhibit angiogenesis and tumor growth, and has been considered for use in cancer treatment and age-related macular degeneration (Udagawa et al., 2000). Brefeldin A has also shown potential as an anticancer agent due to its induction of apoptosis in several cancer cell lines, shown to be via activation of the mitochondria-mediated cell death pathway in ovarian cancer cells (He et al., 2013; Lee et al., 2013). Mycophenolic acid is a known immunosuppressant for organ-transplant patients but has shown potential as a method of inducing apoptosis in tumor cells through inosine monophosphate dehydrogenase (IMPDH) inhibition (Bentley, 2000). Griseofulvin was launched as antifungal agent in the 1950s and has now attracted renewed attention for its anticancer and antiviral activities (Petersen et al., 2014). However, there are fungal polyketides hazardous to human health, such as aflatoxins, which are the primary mycotoxins produced by some Aspergillus sp. and considered to be the most potent naturally occurring carcinogens (Roze et al., 2013).
Figure 1.
Examples of important fungal polyketides
While the structural complexity of these polyketides makes them interesting and useful bioactive compounds, these same features also make them difficult and expensive to prepare and scale-up using synthetic methods. Currently, nearly all commercial polyketides are prepared through fermentation or semi-synthesis (Dechert-Schmitt et al., 2014; Wong and Khosla, 2012). Thus, elucidating the biosynthetic pathways of polyketides and characterizing the pathway enzymes is necessary for the production and diversification of natural products for pharmaceutical applications (Tibrewal and Tang, 2014). However, efforts to elucidate and engineer polyketide pathways in the native filamentous fungi hosts are often hampered for the following reasons: 1) many fungal hosts lack established genetic tools; 2) the native hosts of desired natural products may have low biomass accumulation and produce low concentrations of the desired product; 3) fungal hosts often produce many secondary metabolites that can lead to significant background, allowing the possibility of cross-reactivity between pathways, and complicating the analysis of specific pathways; 4) transcriptional regulation of fungal natural product clusters is complex and not fully understood; and 5) there is no universal expression system with specified culture conditions that can be applied uniformly to fungal natural product pathways and their native hosts (Anyaogu and Mortensen, 2015; Brakhage, 2013; Schümann and Hertweck, 2006; Siddiqui et al., 2012). For these reasons, a versatile heterologous host for the study and engineering of fungal polyketide synthases (PKSs) is desired.
As one of the most intensively studied single-celled eukaryotes in fundamental and applied molecular biology research, Saccharomyces cerevisiae has proven to be a useful and prominent industrial host for recombinant protein production (Mattanovich et al., 2012). S. cerevisiae is widely used not only in the food and beverage industry, but also in the production of bioethanol and fine chemicals (Nevoigt, 2008). Specifically, S. cerevisiae has many advantages beneficial to the elucidation and engineering of heterologous biosynthetic pathways from filamentous fungi: 1) A number of genetic tools for protein expression and pathway construction in yeast have been developed; 2) it is a unicellular organism well-suited for large scale fermentation; 3) S. cerevisiae has a limited native secondary metabolism, which minimizes the background and potential interference with heterologous pathways; 4) S. cerevisiae grows more rapidly than most filamentous fungi and it is considered a GRAS (generally regarded as safe) organism by the FDA (Nevoigt, 2008). In addition, yeast naturally produces common polyketide building blocks such as acetyl-CoA and malonyl-CoA; as well cofactors such as NADPH and S-adnesylmethioine, which facilitate the production of fungal polyketides with minimal integration of heterologous genes (Kealey et al., 1998). Like filamentous fungi, yeast is also classified as a fungus and is expected to be a capable expression host for fungal proteins that are important for polyketide pathways. For example, S. cerevisiae can functionally express eukaryotic cytochrome P450s because these enzymes often anchor in the endoplasmic reticulum, which is absent in prokaryotes (Pompon et al., 1996).
Using yeast as a heterologous host presents some challenges, such as required genes for PKS activation, an inability to splice most fungal introns, low production of necessary precursors, a lack of compartmentalization, and potential toxicity. However, the advantages of S. cerevisiae have allowed increased understanding of fungal polyketide pathways and biosynthesis (Tsunematsu et al., 2013). For example, of the polyketides in Figure 1, the lovastatin and brefeldin A biosynthetic pathways were identified using heterologous yeast expression (Barriuso et al., 2011; Ma et al., 2009; Zabala et al., 2014), and the effects of statins, brefeldin A, mycophenolic acid, and griseofulvin have all been studied in yeast (Athlin et al., 1987; Desmoucelles et al., 2002; Kuranda et al., 2010; Leszczynska et al., 2009; Maciejak et al., 2013; Shah and Klausner, 1993). Aflatoxin pathway genes have been expressed heterologously in yeast in order to study aflatoxin biosynthesis, and its mode of action has also been studied using yeast (Fasullo et al., 2008; Kelly et al., 2002; Yabe et al., 2012). This review will focus on the use of S. cerevisiae as a tool for the discovery, study, and production of fungal PKSs and their natural products.
2. Introduction to polyketide synthases
The diverse structures of polyketides are biosynthesized from short-chain carboxylic acid units by PKSs (Hertweck, 2009). PKSs have been classified into type I, type II and type III based on their product profiles and catalytic domain architecture (Shen, 2003) Type I PKSs are large multidomain enzymes in which catalytic sites are juxtaposed in an assembly line fashion. The three essential domains for the elongation of the polyketide chain are β-ketoacyl synthase (KS), acyltransferase (AT) and acyl carrier protein (ACP). Other domains that control the degree of reduction of β-keto groups may be present. These are ketoreductase (KR), dehydratase (DH) and enoyl reductase (ER) domains. Other frequently found tailoring domains include the methyltransferase (MT) domain, which introduces an α-methyl group immediately after a round of chain elongation; the thioesterase (TE) domain, which releases the polyketide product from the enzyme by hydrolysis, esterification or macrocyclization (Keatinge-Clay, 2012); the reductase (R) domain which releases the polyketide product from the enzyme by two or four electron reduction or Dieckmann condensation (Du and Lou, 2010); a special type of TE domain called TE/CLC (Claisen-like cyclase) which catalyses Claisen-type condensations to release the products (Chooi and Tang, 2012). Type II PKSs are a set of multienzyme complexes that act iteratively and are frequently responsible for the biosynthesis of aromatic polyketides in bacteria. Type III PKSs are homodimers of KSs which catalyze the priming, extension, and cyclization of small polyketides, such as chalcone, in an ACP-independent fashion. (Shen, 2003; Yu et al., 2012).
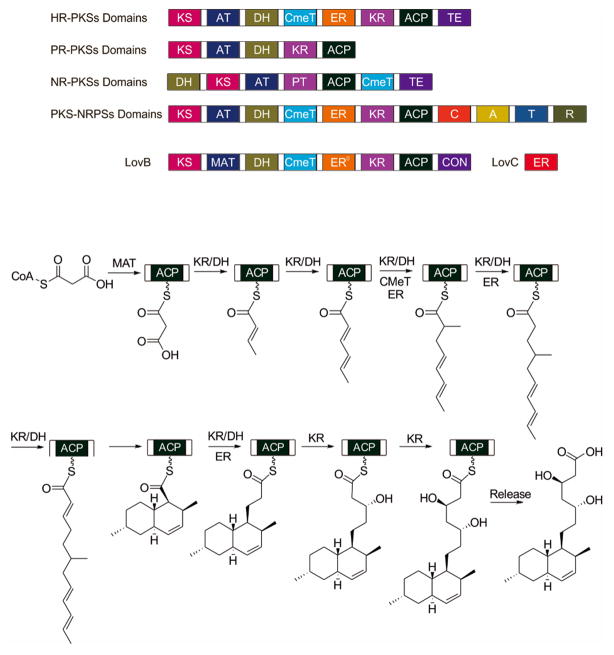
The majority of PKSs from filamentous fungi are type I PKSs. Unlike the multimodular bacterial type I PKSs that operate in a collinear fashion in which each set of domains (a module) are used once in the construction of the polyketide, fungal type I PKSs contain only one set of catalytic domains which are used iteratively in a well-programmed fashion to biosynthesize the final products. Hence, fungal PKSs are also known as iterative type I PKSs (iPKSs) (Figure 2). Furthermore, based on the extent of β-keto reduction catalyzed by the iPKSs, the fungal iPKSs can be classified into three subgroups: nonreducing PKSs (NR-PKSs) that produce aromatic compounds such as norsolorinic acid, the precursor to aflatoxin (Yu et al., 2004), via the product template (PT) domain which is necessary for synthesizing these ring-shaped products (Chooi and Tang, 2012); partially-reducing PKSs (PR-PKSs) that produce compounds such as 6-methylsalicylic acid (6-MSA) (Beck et al., 1990); and highly-reducing PKSs (HR-PKSs) that produce more reduced compounds such as lovastatin (Chooi and Tang, 2012; Cox, 2007). In addition, an HR-PKS can be fused to a nonribosomal peptide synthetase (NRPS) module to form a hybrid PKS-NRPS, which can lead to the biosynthesis of tetramic acid-containing products (Boettger and Hertweck, 2013). Collectively, the different combinations and programming rules of the iPKSs, along with further tailoring of the initial scaffolds by other enzymes, have led to the tremendous structural and functional diversity of polyketides isolated from fungal species.
Figure 2.
The domains of several types of iterative type I polyketide synthases (iPKSs) are shown here. The PKS domains abbreviated here are as follows: KS (Keto-synthase), AT (Acyltransferase), DH (Dehydratase), CmeT (C-methyltransferase), ER (Enoylreductase), KR (Ketoreductase), ACP (Acyl carrier protein), TE (Thioesterase), PT (Product template), MAT (malonyl-CoA:ACP acyltransferase), and CON (Condensation).
The PKS-NRPS hybrid also contains Non Ribosomal Peptide Synthetase domains as follows: C (Condensation), A (Adenylation), T (Thiolation), and R (Reduction).
The example below shows the action of a nonaketide synthase, LovB, and its partner enoylreductase, LovC, from the lovastatin pathway of Aspergillus terreus.
3. The S. cerevisiae toolbox for cloning and enzyme reconstitution
3.1 Molecular biology tools of S. cerevisiae for cloning and reconstitution of heterologous pathways
Polyketides and other secondary metabolites are biosynthesized by a series of enzymes encoded by genes that are typically clustered together in the genomes of the producing organisms (Smith et al., 1990a). The clustering of related genes has been instrumental in the discovery and engineering of natural product pathways in both bacteria and fungi (Keller et al., 2005). Due to this clustering, the cloning and expression of polyketide biosynthetic enzymes can be accomplished with only one continuous genomic DNA fragment containing the entire cluster. However, the cloning or assembly of the pathway in a suitable vector is still a challenge because the large size of most gene clusters, or even a single PKS-encoding gene (iPKSs genes are typically ~10 kB), is usually too large to be amplified by PCR efficiently and correctly.
One strategy to capture entire gene clusters is to construct a genomic DNA library with a suitable vector and chemically screen for clones that may carry a functional cluster. For example, in the first heterologous expression of penicillin, a cosmid containing the penicillin biosynthetic gene cluster from Penicillium chrysogenum was screened from a cosmid library using a DNA probe of the homologous isopenicillin N synthetase from Flavobacterium sp. SC 12,154. This cosmid was then transformed into Neurospora crassa and Aspergillus niger, which led to the production of authentic penicillin V in both transformed hosts (Smith et al., 1990b). However, this method can fail when the size of the gene cluster is larger than the capacity of a cosmid, or the library does not include a cosmid clone that contains all of the genes involved in the biosynthesis of the natural product. Recently, Bok et al. constructed an unbiased shuttle BAC library of Aspergillus terreus ATCC2054 with the vector containing both the E. coli replicon and Aspergillus autonomously replication sequence (AMA1). The average insert size was about 100 kb, which can cover all genes and regulatory elements of the biosynthetic pathway and be used successfully in the heterologous expression of secondary metabolites (Bok et al., 2015). However, the construction of high quality unbiased BAC library is time-consuming and labor-intensive, so alternative solutions have been explored using S. cerevisiae.
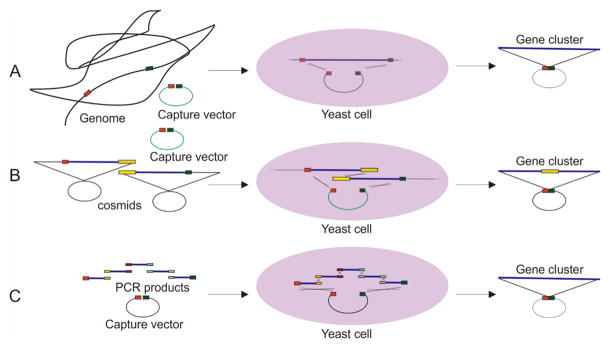
Yeast has been developed extensively as a synthetic biology tool for the cloning and capture of entire biosynthetic gene clusters (Figure 3). One significant advantage of yeast is that homologous recombination takes place far more frequently than ligation or non-homologous end joining during S. cerevisiae DNA repair (Van Leeuwen et al., 2015). This feature has been exploited to construct vectors containing large gene clusters. Overlapping DNA pieces and a linearized vector can be co-transformed into yeast spheroplasts, and the DNA fragments can then be joined by homologous recombination to form intact, selectable vectors. This method is generally known as Transformation-associated Recombination (TAR) as well as other names such as DNA assembler (Larionov et al., 1996; Shao et al., 2009). Oldenburg et al. investigated the efficiency of TAR cloning with different lengths of homologous overlaps, and confirmed that while 40 bp of overlap produced optimal results, as few as 20 bp of overlap could generate the desired product (Oldenburg et al., 1997). TAR cloning is exceptionally simple and efficient, especially in the rapid cloning of large DNA genes or gene clusters, without using restriction enzymes or being limited by PCR product sizes (Kouprina and Larionov, 2006).
Figure 3.
Cloning of large DNA fragments based on the recombination of yeast
A Capture of large DNA fragments from genomic DNA
B Assembly of interested gene cluster from overlapping cosmids
C Assembly of interested pathway from overlapping PCR products
TAR has been used to assemble two or more overlapping cosmids in one step. Feng et al used TAR in S. cerevisiae to assemble the entire fluostatins biosynthetic gene cluster from a bacterial environmental DNA (eDNA) library. Initially, when the cosmid library was screened, they found that the fluostatins cluster was located across two different cosmids. In order to assemble the entire cluster, a S. cerevisiae/Escherichia coli/Streptomyces capture vector was designed with 1 kb homology regions matched to the boundaries of the gene cluster. The linearized vector and the two cosmids were then co-transformed into yeast and homologously recombined (Feng et al., 2010). Yeast based TAR has also been used to capture large DNA fragments from the genome directly (Kouprina and Larionov, 2006). Analysis of the genome of a marine bacterium, Saccharomonospora sp. CNQ-490, by Yamanaka et al revealed a putative NRPS gene cluster similar to the gene cluster responsible for synthesis of the antibiotic daptomycin in Streptomyces roseosporus. To mine this gene cluster, 1 kb DNA fragments matching the boundaries of the targeted gene cluster were cloned by PCR and inserted into the capture vector. The linearized capture plasmid and genomic DNA were co-transformed into yeast to capture the cluster. The resultant plasmid carrying the cluster was directly used for heterologous expression, leading to the heterologous production of taromycin A (Yamanaka et al., 2014).
For accurate and efficient cloning of large fungal iPKSs genes, PCR combined with yeast homologous recombination has been widely used in recent years. A common strategy to clone large intron-free iPKSs genes from cDNA for yeast expression is to first PCR amplify several overlapping fragments from the cDNA of the iPKSs gene, followed by one-step recombination into the desired expression vector in yeast. The use of cDNA is to guarantee the correct mRNA processing in the yeast. One recent example is the cloning and reconstitution of iPKSs AurA from the aurovertin biosynthetic gene cluster in Calcarisporium arbuscula. Mao et al amplified three overlapping fragments covering the intron-free aurA cDNA. The two 5′ and 3′ pieces also contained overlapping regions with the 2μ yeast vector pXW55, which led to placement of the entire aurA gene in the vector under control of the ADH2 promoter. The resulting yeast strain containing the desired plasmid was then used directly to elucidate the product of the iPKSs through expression of the encoded enzymes and analysis of the resulting products (Mao et al., 2015). Other examples of TAR-based assembly of iPKSs include cazF and cazM from the chaetoviridin biosynthetic gene cluster in Chaetomium globosum (Winter et al., 2015; Winter et al., 2012a); bref-PKS from the biosynthetic gene cluster of brefeldin A in Eupenicillium brefeldianum (Zabala et al., 2014); and fma-PKS from the biosynthetic gene cluster of fumagillin in Aspergillus fumigatus (Lin et al., 2013). A variation of this strategy is to first assemble the entire iPKSs gene using overlap extension PCR (OE-PCR), followed by transformation into yeast along with the vector to yield the intact expression plasmid via recombination. This ExRec (overlap Extension PCR-yeast homologous recombination) method described by Ishiuchi et al was used to successfully reconstitute iPKSs from Chaetomium globosum and Coprinopsis cinerea (Ishiuchi et al., 2012).
Yeast homologous recombination has also been used in the refactoring of multi-gene fungal biosynthetic pathways for heterologous reconstitution in model hosts. Pahirulzaman et al reconstructed the four gene tenellin biosynthetic pathway from Beauveria bassiana for expression in Aspergillus oryzae. The three tailoring genes were first recombined into a single expression vector under the control of three different promoters. The PKS-NRPS gene was first cloned in a separate plasmid, followed by insertion into the three-gene plasmid via Gateway-mediated recombination in vitro (Pahirulzaman et al., 2012). Kakule et al also employed yeast homologous recombination to construct cryptic heterologous fungal PKS pathways in Fusarium heterosporum. The genes of interest were combined into a S. cerevisiae/E. coli /F. heterosporum shuttle vector via yeast homologous recombination, amplified in E. coli, and expressed in F. heterosporum. The PKS CpaS from Aspergillus flavus, the PKS LovB and enoylreductase LovC from A. terreus, and two putative PKS-NRPSs, PrlS and PrlC, from endophytic fungus NRRL 50135 were all successfully reconstituted in F. heterosporum via this strategy, leading to production of up to 1 g/L of the encoded polyketide (Kakule et al., 2014).
3.2 S. cerevisiae as a platform for PKS protein expression and purification
Purified iPKSs enzymes from fungal PKS biosynthetic pathways are needed for the functional characterization of their unique programming rules. Complete understanding of PKSs will also enable their abilities to be exploited to benefit the structural diversification, activity optimization, and generation of “unnatural” natural products (Tibrewal and Tang, 2014). As an expression host for fungal proteins, S. cerevisiae has many advantages compared to prokaryotic or more complex eukaryotic hosts. Unlike bacterial hosts, such as E.coli, S. cerevisiae has the machinery for secretory pathways and post-translational modifications (Mattanovich et al., 2012). Compared to mammalian and plant cells, S. cerevisiae has fast growth and allows easy genetic manipulation.
Nevertheless, there are some genetic modifications that were made to S. cerevisiae to ensure functional expression of iPKSs. Deletion of the two vacuolar protease genes (PEP4 and PRB1), encoding the aspartyl protease and proteinase B respectively, significantly increases the expression level of recombinant fungal iPKSs. The strain BJ5464 (MATα ura3-52 his3-Δ200 leu2-Δ1 trp1 pep4::HIS3 prb1 Δ1.6R can1 GAL) was therefore chosen for protein expression (Cardenas and Da Silva, 2014; Jones, 1991). In addition, the yeast host must ensure correct post-translational modification of the ACP domain of iPKSs. The active site serine of ACPs must be modified with a phophopantetheinyl moeity to afford the holo version. While yeast has its endogenous 4′-phosphopantetheine (pPant) transferase that transfers pPant from coenzyme A to ACP domain of fatty acid synthases, the ACP domain of most fungal iPKSs cannot be modified and hence the iPKSs is not active. Kealey et al confirmed that there were almost no functional 6-methylsalicylic acid synthases (6-MSAS) expressed in the S. cerevisiae without a heterologous pPant transferase (Kealey et al., 1998; Lee et al., 2009). To this end, npgA, a pPant transferase gene from Aspergillus nidulans (Lee et al., 2009; Wattanachaisaereekul et al., 2007) was integrated into the genome of S. cerevisiae BJ5464 to yield BJ5464-NpgA. Using a sensitive fluorescent assay, the authors showed that expressed fungal PKSs were efficiently phosphopantetheinylated in the engineered strain (Lee et al., 2009). This strain has subsequently been used widely in the expression and purification of functional iPKSs, as first demonstrated with the 335 kDa lovastatin nonaketide synthase LovB (Ma et al., 2009).
4. Yeast as a platform for studying the function of fungal iPKSs
S. cerevisiae has been used extensively as a host for the expression and characterization of iPKS pathways, and to link fungal polyketide natural products to the gene clusters that produce them. For example, while investigating the biosynthesis of the protein transport-inhibitor Brefeldin A (BFA), Zabala et al sequenced the producing organism Eupenicillium brefeldianum ATCC 58665. From the numerous gene clusters that contained iPKSs, one putative gene cluster encoding an HR-PKS and numerous P450 genes was proposed to be most likely involved in BFA biosynthesis. Genetic manipulation of the producing organism proved to be extraordinarily difficult. Therefore, the authors performed direct expression in yeast to investigate the role of the HR-PKS. The HR-PKS gene and the partnering thiohydrolase gene were cloned into two vectors from cDNA and heterologously expressed in S. cerevisiae BJ5464-NpgA. While the authors did not observe the completed core structure of BFA, an acyclic octaketide consistent with the length of BFA was recovered. The oxidation patterns of the octaketide product were consistent with those observed in BFA, thereby providing strong evidence linking this gene cluster to BFA (Zabala et al., 2014).
Yeast has also been used to connect an orphan NR-PKS gene cluster to the biosynthesis of fungal aromatic polyketide TAN-1612. Li et al identified a candidate cluster in Aspergillus niger that was presumed to be involved in the production of known compound TAN-1612. To verify the function of this cluster, the putative NR-PKS encoded by the adaA gene was cloned from cDNA into a yeast 2μ expression plasmid and the three tailoring genes adaB-D from the cluster were cloned into a separate plasmid. After two days of culturing, S. cerevisiae BJ5464-NpgA expressing these two plasmids produced TAN-1612 (Li et al., 2011). Subsequent investigations using the yeast host revealed the product of the NR-PKS alone, as well as the individual functions of the tailoring enzymes. Similarly, Zhou et al utilized yeast to confirm that two iPKSs, an HR-PKS Rdc5 and an NR-PKS Rdc1, were involved in the biosynthesis of the radicicol precursor in Pochonia chlamydosporia. Heterologous expression of these iPKSs, cloned from genomic DNA, was performed in S. cerevisiae BJ5464-NpgAand led to the production of (R)-monocillin II, an intermediate in radicicol biosynthesis. This result confirmed that the two iPKSs function collaboratively in the biosynthesis of the resorcylic acid lactone and allowed a closer study of the functions of these enzymes (Zhou et al., 2010). This led to the yeast-based reconstitution of other dual iPKSs clusters from fungi, including those of chaetoviridine, resorcylides, lasiodiplodins, and cuvularins (Xu et al., 2014).
Chooi et al. sought to identify virulence factors from Parastagonospora nodorum, a wheat pathogen affecting wheat yields globally. In the course of their investigation, they found that SN477, a PR-PKS gene, was highly upregulated during the pathogen infection. When SN477, was cloned from cDNA under the ADH2 promoter and transformed into S. cerevisiae BJ5464-NpgA, (R)-mellein was produced by the yeast host, revealing that SN477 is the only enzyme required for (R)-mellein synthesis. Though (R)-mellein showed no relevance to the virulence against wheat, it was able inhibit the germination of wheat seeds (Chooi et al., 2015).
In addition to serving as a host for linking polyketide metabolites to their corresponding iPKSs, the yeast iPKS expression platform has also proven to be useful in the mechanistic studies of these highly programmed machineries. One example is the characterization of LovB, an HR-PKS from Aspergillus terreus that is involved in the biosynthesis of the cholesterol-lowering compound lovastatin (Kennedy et al., 1999). The low yield of functional LovB (335 kDa) recovered from Aspergillus-based expression hosts significantly hindered the biochemical study of this model HR-PKS. When expressed from S. cerevisiae BJ5464-NpgA, a purified LovB yield of ~4.5 mg/L was achieved, thereby providing sufficient amounts for in vitro biochemical investigations (Ma et al., 2009). Reconstitution of LovB with its enoylreductase LovC demonstrated that the enzyme is precisely programmed to synthesize the expected product dihydromonacolin L when the needed cofactors (NADPH and SAM) are supplied. After perturbing the system through removal of one or more of the required cofactors or LovC, LovB produced a series of conjugated α–pyrones that are drastically different in structure from dihydromonacolin L. However, detailed structural characterization of these shunt products revealed that releasing pyrones is one mechanism by which LovB can clear its ACP of incorrectly tailored intermediates, thereby providing insight into how iPKSs can maintain their product fidelity. Subsequently, using the yeast expression platform and in vitro characterization, Xu et al discovered a previously hidden thioesterase, LovG, encoded in the pathway to be the enzyme responsible for both the release of dihydromonacolin L from LovB and significant increases in its turnover rate (Xu et al., 2013a).
Using the same yeast expression host, Xie et al. also reconstituted the activities of the lovastatin diketide synthase (LovF) using purified enzymes. While architecturally similar to LovB, the authors showed that this enzyme produces an enzyme-bound α-methylbutyrate diketide using Fourier Transform Mass Spectrometry (FTMS) of proteolyzed LovF fragments. Offloading of the product was demonstrated to be carried out by the acyltransferase LovD, which transfers the α-methylbutyrate to the C8-hydroxyl group of monacolin J to yield the final product, lovastatin. Kinetic analysis using LovF and smaller acyl mimics such as α-methylbutyryl-CoA or α-methylbutyryl-SNAC demonstrated protein-protein interactions between the LovF ACP domain and LovD are highly specific, as significant penalties to the acyltransfer reaction were observed when small molecules thioester carriers were used in place of the ACP domain. This was the first demonstrated acyltransferase mediated product release from an iPKSs and since then this mechanism has been found to be widely adopted by other fungal PKS pathways (Chooi and Tang, 2012; Xie et al., 2009). Together with the functional reconstitution of the lovastatin P450 LovA in yeast (Barriuso et al., 2011), the entire six-gene biosynthetic pathway of lovastatin has been functionally elucidated in S. cerevisiae, paving the way for heterologous pathway construction and engineering.
In another example, Wang et al. demonstrated the aryl-aldehyde formation in the biosynthesis of an NR-PKS through heterologous expression of a cryptic NR-PKS and an NRPS-like gene from Aspergillus terreus in yeast. When the cryptic NR-PKS ATEG03629 was expressed in S. cerevisiae BJ5464-NpgA, 5-methyl orsellinic acid (5-MOA) was produced. However, when both ATEG03629 and ATEG03630, an NRPS-like gene with a terminal reductase domain, were co-transformed into the yeast host, 2,4-dihydroxy-5,6-dimethyl benzaldehyde was produced in addition to 5-MOA. Both ATEG03629 and ATEG03630 were cloned from genomic DNA using exons predicted from bioinformatics analysis. The in vivo results indicated that the NRPS-like protein catalyzes the aryl-acid to aryl-aldehyde conversion. To further confirm the activity of ATEG03630, the enzyme was purified from BJ5464-NpgA. In vitro experiments of ATEG03630 with the substrate 5-MOA and cofactor NADPH confirmed the catalytic activity of ATEG03630. Though these compounds had been reported previously, their biosynthetic origins had never been established (Wang et al., 2014).
As the above examples demonstrate, yeast is a useful and versatile tool for production of diverse polyketides and their associated mega-enzymes in sufficient quantities for in vitro mechanistic studies of iPKSs. These applications of S. cerevisiae improve the knowledge of biosynthesis of natural products and accumulate enzyme tools for drug discovery and the synthesis of new natural products.
5. Engineering production of fungal PKSs and polyketides in yeast
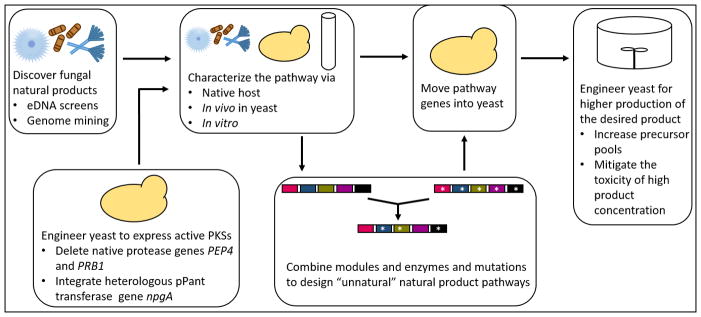
The deletion of native proteases and the integration of a heterologous pPant transferase have been discussed earlier as essential for the functional expression of fungal iPKSs in S. cerevisiae. Other methods of increasing the titer of the final polyketide product include increasing the supply of the common polyketide precursors acetyl-CoA and malonyl-CoA, and introducing self-resistance genes to mitigate the toxicity of final product. These metabolic engineering strategies can be employed in the production of both natural and engineered, unnatural polyketides in yeast. The route from the discovery of novel polyketides to high production in yeast is outlined in Figure 4.
Figure 4.
This figure outlines the route from the discovery of novel polyketides to high production in yeast, using the tools and methods discussed in this review.
5.1 Strain engineering for high titers via increased precursor production
Increasing the supply of the precursors is one common metabolic engineering strategy to increase the titer of the final product (Krivoruchko and Nielsen, 2015). Common polyketide precursors acetyl-CoA and malonyl-CoA are produced endogenously by yeast, but metabolic engineering is required to produce larger quantities of these substrates. Recent efforts to increase precursor production have focused both on enzyme overexpression and engineering, as well as on directing carbon towards the desired precursors via deletion of competing pathways.
The ACC1 gene encodes acetyl-CoA carboxylase, which catalyzes the conversion of acetyl-CoA to malonyl-CoA. Wattanachaisaereekul et al overexpressed the native yeast ACC1 through replacing the native promoter of ACC1 with a strong constitutive TEF1 promoter in a 6-MSA polyketide production S. cerevisiae strain, increasing the titer of 6-MSA by 60% (Wattanachaisaereekul et al., 2008). Choi et al. improved the activity of ACC1 by site-directed mutagenesis, leading to 3-fold increases in 6-MSA levels (Choi and Da Silva, 2014).
Metabolic pathway analysis in the well-studied S. cerevisiae has been used to determine how to direct carbon towards the desired product. Cardenas et al specifically increased the titer of precursor acetyl-CoA through the deletion of 15 genes identified during pathway analysis of glucose-6-phosphate, an acetyl-CoA and malonyl-CoA precursor (Cardenas and Da Silva, 2014). Lian et al redirected glycolytic flux to the production of acetyl-CoA via gene inactivation of ADH1 and ADH4 from the competing ethanol pathway and GPD1 and GPD2 from the competing glycerol pathway (Lian et al., 2014). Chen et al over-expressed the endogenous ADH2 and ALD6 genes as well as a mutant heterologous acetyl-CoA synthetase from Salmonella enterica to redirect carbon flux from acetaldehyde to cytosolic acetyl-CoA. They also reduced carbon loss from the pool of acetyl-CoA by inhibiting CIT2, a peroxisomal citrate synthase, and MLS1, a cytosolic malate synthase. Using this platform to produce α-santalene led to a four-fold increase in titer compared to the reference strain (Chen et al., 2013).
5.2 Strain engineering for high titers via toxicity mitigation
One challenge of using yeast as a cell factory to produce high titers of fungal polyketides is that many bioactive secondary metabolites are toxic in high concentrations (Martín et al., 2005). Some strategies for producing toxic compounds in cell factories include using inducible promoters to decouple cell growth from compound production (Nevoigt, 2008); overlaying an organic solvent like dodecane on the top of the culture to remove the toxic compound as it is produced (Rodriguez et al., 2014); and overexpressing native, broadly-specific transport proteins to remove toxic compounds from the cell (Valle Matta et al., 2001). Native producers often evolve their own solutions to protect themselves from the presence of toxic compounds, such as specific transporter proteins or other enzymes that confer self-resistance to the native host (Martín et al., 2005). These self-resistance mechanisms can take the form of product-specific transporters like the putative transporter cazK from Chaetomium globosum and the putative efflux pump lovI from A. terreus (Kennedy et al., 1999; Winter et al., 2012b). Recently, Ley et al showed that integration of the putative efflux pump mlcE, from the compactin PKS cluster of P. citrinum, into the S. cerevisiae genome conferred increased resistance to mevastatin, lovastatin, and simvastatin, as compared to the wild type strain. MlcE was shown to be a specific transporter and restored the growth rates of yeast in the presence of up to ~800 mg/L of exogenously added lovastatin (Ley et al., 2015). Past analyses of fungal PKS clusters have primarily focused on reconstitution of the enzymes necessary for the biosynthesis of the encoded natural product, but, as Ley, et al showed, future work incorporating more of the putative self-resistance genes into heterologous polyketide production hosts has the potential to significantly increase final product titers (Ley et al., 2015).
6. Combinatorial biosynthesis of fungal PKSs in S. cerevisiae
Due to the contributions of natural products to the development of pharmaceuticals, there has been significant research toward discovering new natural products and engineering the biosynthesis of novel, unnatural bioactive compounds (Butler et al., 2013; Harvey, 2008; Newman and Cragg, 2012). One future goal is to set up an algorithm which takes a molecule of interest as input and outputs a sequence of natural PKS modules to produce the desired molecule (Poust et al., 2014). Although the synthesis of new “unnatural” natural products via combinatorial biosynthesis has been pursued in bacterial polyketide antibiotics for more than 15 years (McDaniel et al., 1999), there had been little progress in the field of fungal PKSs until recently. In 2013, Xu et al. reported the reprogramming of the first-ring cyclization of two benzenediol lactones (BDLs) (Xu et al., 2013c). BDLs are a family of fungal polyketides with diverse structural features and wide-ranging bioactivities. The BDL family is composed of resorcylic acid lactones (RALs), connected at C2–C7, and dihydroxyphenylacetic acid lactones (DALs), which feature a C3–C8 bond (Shen et al., 2014). The biosynthesis of these fungal polyketides involves pairs of collaborating iPKSs: an HR-PKS which passes its product to an NR-PKS for further modifications (Zhou et al., 2008). Xu et al reconstituted heterologous HR-PKS and NR-PKS pairs from A. terreus and Chaetomium chiversii in BJ5464-NpgA and confirmed that the product template (PT) domains of the fungal NR-PKSs regiospecifically catalyzed the first-ring aldol cyclization, leading to the characteristically different polyketide folding modes of RALs and DALs. Next, rational reprogramming of the regiospecific first-ring cyclization was realized by domain replacement and site-directed mutagenesis (Xu et al., 2013c). Using the same BJ5464-NpgA system, Xu et al. expressed chimeric iPKSs enzyme pairs, resulting in the biosynthesis of unnatural BDLs, and found that the thioesterase (TE) domain acts as the decision gate for releasing the final product from a fungal NR-PKS. This indicated that in combinatorial biosynthesis, the TE domain must be able to accept altered polyketide intermediates and release unnatural natural products with the desired structure (Xu et al., 2013b).
After the characterization of different domains of HR-PKSs and NR-PKSs in the biosynthetic pathways of BDLs, Xu et al. deployed yeast as a tool to investigate whether noncognate HR-PKS and NR-PKS pairs could interact with each other efficiently (Xu et al., 2014). Through the combinatorial expression of random pairs of iPKSs subunits from four BDL biosynthetic pathways from A. terreus, C. chiversii, Lasiodiplodia theobromae, and Acremonium zeae in BJ5464-NpgA, a diverse library of BDL congeners was created. One of these unnatural polyketides had heat shock response-inducing activity that had previously been shown to block multiple cancer-causing pathways (Xu et al., 2014). This combinatorial work by Xu et al. provided more insight into PKS design rules, which will assist in future engineering of diverse natural products.
Another method for the biosynthesis of unnatural natural products is to feed artificial precursors to organisms heterologously expressing PKSs. Zhou et al and Gao et al both fed large acyl SNAC (N-acetylcysteamine thioester) substrates to fungal iPKSs expressed or purified from BJ5464-NpgA (Zhou et al., 2010; Gao et al., 2013). Both groups were able to produce unnatural analogs of the relevant natural product. Gao et al found that the efficiency of incorporation of the unnatural precursor analogs depended on the nature of the structural changes between these analogs and the natural precursors (Gao et al., 2013).
Finally, novel scaffolds can be found by combining existing heterologous natural product pathways in yeast and analyzing the resulting compounds. Klein et al took genes from known natural product pathways for alkaloids, benzoxazinoids, flavonoids, flavonols, lignans, polyunsaturated fatty acids, tetra- and diterpenoids, and type III polyketides and 14 libraries of cDNA from 17 different organisms including plants, animals, fungi, and amaurochaetes and expressed them in different combinations on yeast artificial chromosomes in S. cerevisiae. The resulting compounds were screened for useful pharmaceutical activities and for the novelty of their scaffolds (Klein et al., 2014).
The successful combinatorial biosynthesis of fungal PKSs in S. cerevisiae has not only shown the utility of yeast for engineering improved production of natural products, but also extended this paradigm from bacterial polyketides to fungal polyketides (Agarwal and Moore, 2014).
7. Discovering new natural products through genome mining and expression in S. cerevisiae
Another method of discovering new natural product scaffolds is to find novel natural product gene clusters. Top-down approaches, in which newly-discovered fungi are cultivated and their products analyzed, can lead to the discovery of new polyketides but are limited to compounds that are naturally synthesized in relative abundance in the native environment or under laboratory conditions. However, recent advances in genome sequencing and increased availability of fungal genomes can facilitate the discovery and analysis of new polyketides from putative clusters in previously studied organisms.
For over two decades, it has been known that fungal secondary metabolites are often synthesized by genes physically clustered in the genome (Smith et al., 1990a). As more sequenced fungal genomes become available, extensive genome mining efforts have been launched, leading to improved algorithms to annotate putative polyketide clusters in various fungi (Khaldi et al., 2010; Medema et al., 2011; Priebe et al., 2011). However, the metabolites encoded by clusters located this way are often unknown, and subsequently termed cryptic or orphan. Some silent clusters found via genome mining have not lead to any product in the native host under laboratory conditions (Brakhage, 2013; Yin et al., 2013).
The products of fungal iPKSs cannot be predicted solely from their sequences, so the cluster must be induced in the native host, or expressed and analyzed in a heterologous host (Anyaogu and Mortensen, 2015; Bergmann et al., 2007). Despite the many advantages S. cerevisiae has as a heterologous host, introns in the fungal gene cluster coding regions must be completely removed before the heterologous pathway is expressed in yeast due to the significant differences between the introns of yeast and filamentous fungi (Anyaogu and Mortensen, 2015; Kupfer et al., 2004). However, the accuracy of fungal intron prediction has been improved by programs like FGENESH and homology alignment of known related genes to a putative cluster (Cacho et al., 2014; Zhang, 2002). Using intron prediction programs or direct cloning from RNA, if possible, should allow entire clusters discovered through genome mining to be cloned and expressed in yeast. As discussed previously, Ishiuchi et al used their ExRec method of cloning from a pool of total RNA isolated from various fungi to express several heterologous PKSs in yeast (Ishiuchi et al., 2012). Despite this progress, and its success as a platform for the exploration of silent bacterial PKS clusters, S. cerevisiae has been underutilized for exploring new silent fungal PKS clusters (Feng et al., 2010; Kang and Brady, 2014; Montiel et al., 2015). The technology and tools are available to discover new fungal natural products with yeast if, as these authors hope, more groups begin to utilize them.
8. Conclusions
A number of useful synthetic biology strategies have been developed in S. cerevisiae that make it a versatile tool for the discovery, characterization, and production of PKSs and their products. Yeast has been used as a host for cloning or purifying protein and as a tool for ascertaining the function of enzymes in a PKS cluster as well as for individual modules of PKSs. In addition, this organism has been engineered as a host for testing biosynthesis of unnatural “natural” products, for screening bioactive compounds to find treatments for specific conditions, and for use as an industrial production host for heterologous pathways.
With such a versatile tool as yeast available, it is remarkable that more work has not been done with fungal PKSs in yeast. The literature is abundant with examples of yeast as a heterologous host for plant and bacterial PKSs, but yeast has been surprisingly underutilized for fungal PKSs. There is a rich variety of fungal natural products left to be discovered, characterized, and engineered. The future of this field will involve a greater utilization of heterologous fungal PKS expression in yeast, especially of cryptic clusters, in order to aid in the discovery and production of chemically diverse compounds that will have impacts in fields such as fuels and drug discovery.
The well-developed genetic toolbox of Saccharomyces cerevisiae has been used to circumvent the challenge of genetic manipulation of polyketide pathways in native filamentous fungi hosts.
Yeast homologous recombination has been a powerful tool for exploration of fungal polyketides and their biosynthetic pathways.
Metabolic engineering in yeast has led to high titers of functional polyketide synthases and increased polyketide titers.
Acknowledgments
Research activities in this area in our lab has been supported by funds from NIH (5 DP1 GM106413, 2 R01 GM085128 and 1 U01 GM110706) and the Packard Foundation. We thank Dr. Wei Xu and Anthony DeNicola for helpful discussions. Carly Bond is supported by the NIH T32 Biotechnology Training Grant (2 T32 GM067555), Li Li is supported by Educational Commission of Fujian Province, China (JB13014) and State Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University (MMLKF13-03).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal V, Moore BS. Fungal polyketide engineering comes of age. Proc Natl Acad Sci USA. 2014;111:12278–12279. doi: 10.1073/pnas.1412946111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anyaogu DC, Mortensen UH. Heterologous production of fungal secondary metabolites in Aspergilli. Front Microbiol. 2015;6:77. doi: 10.3389/fmicb.2015.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athlin L, Domellöf L, Norberg B. Therapeutic concentrations of griseofulvin do not affect yeast cell phagocytosis by monocytes. Dermatologica. 1987;174:122–127. doi: 10.1159/000249002. [DOI] [PubMed] [Google Scholar]
- Barrios-González J, Miranda R. Biotechnological production and applications of statins. Appl Microbiol Biotechnol. 2010;85:869–883. doi: 10.1007/s00253-009-2239-6. [DOI] [PubMed] [Google Scholar]
- Barriuso J, Nguyen DT, Li JW, Roberts JN, MacNevin G, Chaytor JL, … Ro DK. Double oxidation of the cyclic nonaketide dihydromonacolin L to monacolin J by a single cytochrome P450 monooxygenase, LovA. J Am Chem Soc. 2011;133:8078–8081. doi: 10.1021/ja201138v. [DOI] [PubMed] [Google Scholar]
- Baxter A, Fitzgerald BJ, Hutson JL, McCarthy AD, Motteram JM, Ross BC, … Williams RJ. Squalestatin 1, a potent inhibitor of squalene synthase, which lowers serum cholesterol in vivo. J Biol Chem. 1992;267:11705–11708. [PubMed] [Google Scholar]
- Beck J, RIPKA S, SIEGNER A, SCHILTZ E, SCHWEIZER E. The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum. Eur J Biochem. 1990;192:487–498. doi: 10.1111/j.1432-1033.1990.tb19252.x. [DOI] [PubMed] [Google Scholar]
- Bentley R. Mycophenolic acid: a one hundred year odyssey from antibiotic to immunosuppressant. Chem Rev. 2000;100:3801–3826. doi: 10.1021/cr990097b. [DOI] [PubMed] [Google Scholar]
- Bergmann S, Schümann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- Boettger D, Hertweck C. Molecular diversity sculpted by fungal PKS-NRPS hybrids. ChemBioChem. 2013;14:28–42. doi: 10.1002/cbic.201200624. [DOI] [PubMed] [Google Scholar]
- Bok JW, Ye R, Clevenger KD, Mead D, Wagner M, Krerowicz A, Wu CC. Fungal artificial chromosomes for mining of the fungal secondary metabolome. BMC Genomics. 2015;16:1–10. doi: 10.1186/s12864-015-1561-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakhage AA. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- Butler MS, Blaskovich MA, Cooper MA. Antibiotics in the clinical pipeline in 2013. J Antibiot (Tokyo) 2013;66:571–591. doi: 10.1038/ja.2013.86. [DOI] [PubMed] [Google Scholar]
- Cacho RA, Tang Y, Chooi YH. Next-generation sequencing approach for connecting secondary metabolites to biosynthetic gene clusters in fungi. Front Microbiol. 2014;5:774. doi: 10.3389/fmicb.2014.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas J, Da Silva NA. Metabolic engineering of Saccharomyces cerevisiae for the production of triacetic acid lactone. Metab Eng. 2014;25:194–203. doi: 10.1016/j.ymben.2014.07.008. [DOI] [PubMed] [Google Scholar]
- Chen Y, Daviet L, Schalk M, Siewers V, Nielsen J. Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism. Metab Eng. 2013;15:48–54. doi: 10.1016/j.ymben.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Choi JW, Da Silva NA. Improving polyketide and fatty acid synthesis by engineering of the yeast acetyl-CoA carboxylase. J Biotechnol. 2014;187:56–59. doi: 10.1016/j.jbiotec.2014.07.430. [DOI] [PubMed] [Google Scholar]
- Chooi YH, Krill C, Barrow RA, Chen S, Trengove R, Oliver RP, Solomon PS. An in planta-expressed polyketide synthase produces (R)-mellein in the wheat pathogen Parastagonospora nodorum. Appl Environ Microbiol. 2015;81:177–186. doi: 10.1128/AEM.02745-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi YH, Tang Y. Navigating the fungal polyketide chemical space: from genes to molecules. J Org Chem. 2012;77:9933–9953. doi: 10.1021/jo301592k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RJ. Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org Biomol Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- Dechert-Schmitt AM, Schmitt DC, Gao X, Itoh T, Krische MJ. Polyketide construction via hydrohydroxyalkylation and related alcohol C-H functionalizations: reinventing the chemistry of carbonyl addition. Nat Prod Rep. 2014;31:504–513. doi: 10.1039/c3np70076c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmoucelles C, Pinson B, Saint-Marc C, Daignan-Fornier B. Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J Biol Chem. 2002;277:27036–27044. doi: 10.1074/jbc.M111433200. [DOI] [PubMed] [Google Scholar]
- Du L, Lou L. PKS and NRPS release mechanisms. Nat Prod Rep. 2010;27:255–278. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- Fasullo M, Sun M, Egner P. Stimulation of sister chromatid exchanges and mutation by aflatoxin B1-DNA adducts in Saccharomyces cerevisiae requires MEC1 (ATR), RAD53, and DUN1. Mol Carcinog. 2008;47:608–615. doi: 10.1002/mc.20417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Kim JH, Brady SF. Fluostatins produced by the heterologous expression of a TAR reassembled environmental DNA derived type II PKS gene cluster. J Am Chem Soc. 2010;132:11902–11903. doi: 10.1021/ja104550p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Wang J, Norquay AK, Qiao K, Tang Y, Vederas JC. Investigation of fungal iterative polyketide synthase functions using partially assembled intermediates. J Am Chem Soc. 2013;135:1735–1738. doi: 10.1021/ja4001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey AL. Natural products in drug discovery. Drug Discov Today. 2008;13:894–901. doi: 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- He B, Wang Y, Zheng Y, Chen W, Zhu Q. Synthesis and cytotoxic evaluation of acylated brefeldin a derivatives as potential anticancer agents. Chem Biol Drug Des. 2013;82:307–316. doi: 10.1111/cbdd.12154. [DOI] [PubMed] [Google Scholar]
- Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- Ishiuchi Ki, Nakazawa T, Ookuma T, Sugimoto S, Sato M, Tsunematsu Y, … Moriya H. Establishing a new methodology for genome mining and biosynthesis of polyketides and peptides through yeast molecular genetics. ChemBioChem. 2012;13:846–854. doi: 10.1002/cbic.201100798. [DOI] [PubMed] [Google Scholar]
- Jones EW. Methods Enzymol. Academic Press; 1991. Tackling the protease problem in Saccharomyces cerevisiae; pp. 428–453. [DOI] [PubMed] [Google Scholar]
- Kakule TB, Lin Z, Schmidt EW. Combinatorialization of Fungal Polyketide Synthase–Peptide Synthetase Hybrid Proteins. J Am Chem Soc. 2014;136:17882–17890. doi: 10.1021/ja511087p. [DOI] [PubMed] [Google Scholar]
- Kang HS, Brady SF. Mining Soil Metagenomes to Better Understand the Evolution of Natural Product Structural Diversity: Pentangular Polyphenols as a Case Study. J Am Chem Soc. 2014;136:18111–18119. doi: 10.1021/ja510606j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kealey JT, Liu L, Santi DV, Betlach MC, Barr PJ. Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc Natl Acad Sci U S A. 1998;95:505–509. doi: 10.1073/pnas.95.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keatinge-Clay AT. The structures of type I polyketide synthases. Nat Prod Rep. 2012;29:1050–1073. doi: 10.1039/c2np20019h. [DOI] [PubMed] [Google Scholar]
- Keller NP, Turner G, Bennett JW. Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- Kelly EJ, Erickson KE, Sengstag C, Eaton DL. Expression of human microsomal epoxide hydrolase in Saccharomyces cerevisiae reveals a functional role in aflatoxin B1 detoxification. Toxicol Sci. 2002;65:35–42. doi: 10.1093/toxsci/65.1.35. [DOI] [PubMed] [Google Scholar]
- Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science. 1999;284:1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Heal JR, Hamilton WD, Boussemghoune T, Tange TO, Delegrange F, … Heim J. Yeast synthetic biology platform generates novel chemical structures as scaffolds for drug discovery. ACS Synth Biol. 2014;3:314–323. doi: 10.1021/sb400177x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Larionov V. TAR cloning: insights into gene function, long-range haplotypes and genome structure and evolution. Nat Rev Genet. 2006;7:805–812. doi: 10.1038/nrg1943. [DOI] [PubMed] [Google Scholar]
- Krivoruchko A, Nielsen J. Production of natural products through metabolic engineering of Saccharomyces cerevisiae. Curr Opin Biotechnol. 2015;35:7–15. doi: 10.1016/j.copbio.2014.12.004. [DOI] [PubMed] [Google Scholar]
- Kupfer DM, Drabenstot SD, Buchanan KL, Lai H, Zhu H, Dyer DW, … Murphy JW. Introns and splicing elements of five diverse fungi. Eukaryot Cell. 2004;3:1088–1100. doi: 10.1128/EC.3.5.1088-1100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuranda K, François J, Palamarczyk G. The isoprenoid pathway and transcriptional response to its inhibitors in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2010;10:14–27. doi: 10.1111/j.1567-1364.2009.00560.x. [DOI] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Graves J, Chen XN, Korenberg JR, Resnick MA. Specific cloning of human DNA as yeast artificial chromosomes by transformation-associated recombination. Proc Natl Acad Sci USA. 1996;93:491–496. doi: 10.1073/pnas.93.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KKM, Silva NAD, Kealey JT. Determination of the extent of phosphopantetheinylation of polyketide synthases expressed in Escherichia coli and Saccharomyces cerevisiae. Anal Biochem. 2009;394:75–80. doi: 10.1016/j.ab.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Lee SA, Kim YJ, Lee CS. Brefeldin A Induces Apoptosis by Activating the Mitochondrial and Death Receptor Pathways and Inhibits Focal Adhesion Kinase-Mediated Cell Invasion. Basic Clin Pharmacol Toxicol. 2013;113:329–338. doi: 10.1111/bcpt.12107. [DOI] [PubMed] [Google Scholar]
- Leszczynska A, Burzynska B, Plochocka D, Kaminska J, Zimnicka, … Szkopinska A. Investigating the effects of statins on cellular lipid metabolism using a yeast expression system. PloS One. 2009;4:e8499. doi: 10.1371/journal.pone.0008499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley A, Coumou HC, Frandsen RJN. Heterologous expression of MlcE in Saccharomyces cerevisiae provides resistance to natural and semi-synthetic statins. Metab Eng Commun. 2015;2:117–123. doi: 10.1016/j.meteno.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chooi YH, Sheng Y, Valentine JS, Tang Y. Comparative characterization of fungal anthracenone and naphthacenedione biosynthetic pathways reveals an alpha-hydroxylation-dependent Claisen-like cyclization catalyzed by a dimanganese thioesterase. J Am Chem Soc. 2011;133:15773–15785. doi: 10.1021/ja206906d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian J, Si T, Nair NU, Zhao H. Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains. Metab Eng. 2014;24:139–149. doi: 10.1016/j.ymben.2014.05.010. [DOI] [PubMed] [Google Scholar]
- Lin HC, Chooi YH, Dhingra S, Xu W, Calvo AM, Tang Y. The Fumagillin Biosynthetic Gene Cluster in Aspergillus fumigatus Encodes a Cryptic Terpene Cyclase Involved in the Formation of β-trans-Bergamotene. J Am Chem Soc. 2013;135:4616–4619. doi: 10.1021/ja312503y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SM, Li JW, Choi JW, Zhou H, Lee KK, Moorthie VA, … Tang Y. Complete reconstitution of a highly reducing iterative polyketide synthase. Science. 2009;326:589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejak A, Leszczynska A, Warchol I, Gora M, Kaminska J, … Sojka M. The effects of statins on the mevalonic acid pathway in recombinant yeast strains expressing human HMG-CoA reductase. BMC Biotechnol. 2013;13:68. doi: 10.1186/1472-6750-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao XM, Zhan ZJ, Grayson MN, Tang MC, Xu W, Li YQ, … Tang Y. Efficient Biosynthesis of Fungal Polyketides Containing the Dioxabicyclo-octane Ring System. J Am Chem Soc. 2015;137:11904–11907. doi: 10.1021/jacs.5b07816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín JF, Casqueiro J, Liras P. Secretion systems for secondary metabolites: how producer cells send out messages of intercellular communication. Curr Opin Microbiol. 2005;8:282–293. doi: 10.1016/j.mib.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Mattanovich D, Branduardi P, Dato L, Gasser B, Sauer M, Porro D. Recombinant gene expression. Springer; 2012. Recombinant protein production in yeasts; pp. 329–358. [DOI] [PubMed] [Google Scholar]
- McDaniel R, Thamchaipenet A, Gustafsson C, Fu H, Betlach M, Betlach M, Ashley G. Multiple genetic modifications of the erythromycin polyketide synthase to produce a library of novel “unnatural” natural products. Proc Natl Acad Sci USA. 1999;96:1846–1851. doi: 10.1073/pnas.96.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, … Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucl Acids Res. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel D, Kang HS, Chang FY, Charlop-Powers Z, Brady SF. Yeast homologous recombination-based promoter engineering for the activation of silent natural product biosynthetic gene clusters. Proc Natl Acad Sci USA. 2015;112:8953–8958. doi: 10.1073/pnas.1507606112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevoigt E. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2008;72:379–412. doi: 10.1128/MMBR.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg K, TK, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucl Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahirulzaman K, Williams K, Lazarus CM. A toolkit for heterologous expression of metabolic pathways in Aspergillus oryzae. Methods Enzymol. 2012;517:241–260. doi: 10.1016/B978-0-12-404634-4.00012-7. [DOI] [PubMed] [Google Scholar]
- Petersen AB, Ronnest MH, Larsen TO, Clausen MH. The chemistry of griseofulvin. Chem Rev. 2014;114:12088–12107. doi: 10.1021/cr400368e. [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- Poust S, Hagen A, Katz L, Keasling JD. Narrowing the gap between the promise and reality of polyketide synthases as a synthetic biology platform. Curr Opin Biotechnol. 2014;30:32–39. doi: 10.1016/j.copbio.2014.04.011. [DOI] [PubMed] [Google Scholar]
- Priebe S, Linde J, Albrecht D, Guthke R, Brakhage AA. FungiFun: a web-based application for functional categorization of fungal genes and proteins. Fungal Genet Biol. 2011;48:353–358. doi: 10.1016/j.fgb.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Rodriguez S, Kirby J, Denby CM, Keasling JD. Production and quantification of sesquiterpenes in Saccharomyces cerevisiae, including extraction, detection and quantification of terpene products and key related metabolites. Nat Protoc. 2014;9:1980–1996. doi: 10.1038/nprot.2014.132. [DOI] [PubMed] [Google Scholar]
- Roze LV, Hong SY, Linz JE. Aflatoxin biosynthesis: current frontiers. Annu Rev Food Sci Technol. 2013;4:293–311. doi: 10.1146/annurev-food-083012-123702. [DOI] [PubMed] [Google Scholar]
- Schümann J, Hertweck C. Advances in cloning, functional analysis and heterologous expression of fungal polyketide synthase genes. J Biotechnol. 2006;124:690–703. doi: 10.1016/j.jbiotec.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Shah N, Klausner RD. Brefeldin A reversibly inhibits secretion in Saccharomyces cerevisiae. J Biol Chem. 1993;268:5345–5348. [PubMed] [Google Scholar]
- Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucl Acids Res. 2009;37:e16–e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr Opin Chem Biol. 2003;7:285–295. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- Shen W, Mao H, Huang Q, Dong J. Benzenediol lactones: a class of fungal metabolites with diverse structural features and biological activities. Eur J Med Chem. 2014 doi: 10.1016/j.ejmech.2014.11.067. [DOI] [PubMed] [Google Scholar]
- Siddiqui MS, Thodey K, Trenchard I, Smolke CD. Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Res. 2012;12:144–170. doi: 10.1111/j.1567-1364.2011.00774.x. [DOI] [PubMed] [Google Scholar]
- Smith DJ, Burnham MK, Bull JH, Hodgson JE, Ward JM, Browne P, … Turner G. Beta-lactam antibiotic biosynthetic genes have been conserved in clusters in prokaryotes and eukaryotes. EMBO J. 1990a;9:741–747. doi: 10.1002/j.1460-2075.1990.tb08168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Burnham MK, Edwards J, Earl AJ, Turner G. Cloning and heterologous expression of the penicillin biosynthetic gene cluster from Penicillium chrysogenum. Nat Biotechnol. 1990b;8:39–41. doi: 10.1038/nbt0190-39. [DOI] [PubMed] [Google Scholar]
- Tibrewal N, Tang Y. Biocatalysts for natural product biosynthesis. Annu Rev Chem Biomol Eng. 2014;5:347–366. doi: 10.1146/annurev-chembioeng-060713-040008. [DOI] [PubMed] [Google Scholar]
- Tsunematsu Y, Ishiuchi Ki, Hotta K, Watanabe K. Yeast-based genome mining, production and mechanistic studies of the biosynthesis of fungal polyketide and peptide natural products. Nat Prod Rep. 2013;30:1139–1149. doi: 10.1039/c3np70037b. [DOI] [PubMed] [Google Scholar]
- Udagawa T, Yuan J, Panigrahy D, Chang YH, Shah J, D’Amato RJ. Cytochalasin E, an Epoxide ContainingAspergillus-Derived Fungal Metabolite, Inhibits Angiogenesis and Tumor Growth. J Pharmacol Exp Ther. 2000;294:421–427. [PubMed] [Google Scholar]
- Valle Matta MA, Jonniaux JL, Balzi E, Goffeau A, van den Hazel B. Novel target genes of the yeast regulator Pdr1p: a contribution of the TPO1 gene in resistance to quinidine and other drugs. Gene. 2001;272:111–119. doi: 10.1016/s0378-1119(01)00558-3. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen J, Andrews B, Boone C, Tan G. Construction of Multifragment Plasmids by Homologous Recombination in Yeast. Cold Spring Harb Protoc. 2015;2015 doi: 10.1101/pdb.top084111. pdb. top084111. [DOI] [PubMed] [Google Scholar]
- Wang M, Beissner M, Zhao H. Aryl-aldehyde formation in fungal polyketides: discovery and characterization of a distinct biosynthetic mechanism. Chem Biol. 2014;21:257–263. doi: 10.1016/j.chembiol.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattanachaisaereekul S, Lantz AE, Nielsen ML, Andresson OS, Nielsen J. Optimization of heterologous production of the polyketide 6-MSA in Saccharomyces cerevisiae. Biotechnol Bioeng. 2007;97:893–900. doi: 10.1002/bit.21286. [DOI] [PubMed] [Google Scholar]
- Wattanachaisaereekul S, Lantz AE, Nielsen ML, Nielsen J. Production of the polyketide 6-MSA in yeast engineered for increased malonyl-CoA supply. Metab Eng. 2008;10:246–254. doi: 10.1016/j.ymben.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Winter JM, Cascio D, Dietrich D, Sato M, Watanabe K, Sawaya MR, … Tang Y. Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis. J Am Chem Soc. 2015;137:9885–9893. doi: 10.1021/jacs.5b04520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter JM, Sato M, Sugimoto S, Chiou G, Garg NK, Tang Y, Watanabe K. Identification and Characterization of the Chaetoviridin and Chaetomugilin Gene Cluster in Chaetomium globosum Reveal Dual Functions of an Iterative Highly-Reducing Polyketide Synthase. J Am Chem Soc. 2012a;134:17900–17903. doi: 10.1021/ja3090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter JM, Sato M, Sugimoto S, Chiou G, Garg NK, Tang Y, Watanabe K. Identification and characterization of the chaetoviridin and chaetomugilin gene cluster in Chaetomium globosum reveal dual functions of an iterative highly-reducing polyketide synthase. J Am Chem Soc. 2012b;134:17900–17903. doi: 10.1021/ja3090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong FT, Khosla C. Combinatorial biosynthesis of polyketides--a perspective. Curr Opin Chem Biol. 2012;16:117–123. doi: 10.1016/j.cbpa.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Meehan MJ, Xu W, Dorrestein PC, Tang Y. Acyltransferase mediated polyketide release from a fungal megasynthase. J Am Chem Soc. 2009;131:8388–8389. doi: 10.1021/ja903203g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Chooi YH, Choi JW, Li S, Vederas JC, Da Silva NA, Tang Y. LovG: the thioesterase required for dihydromonacolin L release and lovastatin nonaketide synthase turnover in lovastatin biosynthesis. Angew Chem Int Ed. 2013a;52:6472–6475. doi: 10.1002/anie.201302406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhou T, Zhang S, Espinosa-Artiles P, Wang L, Zhang W, … Molnár I. Diversity-oriented combinatorial biosynthesis of benzenediol lactone scaffolds by subunit shuffling of fungal polyketide synthases. Proc Natl Acad Sci USA. 2014;111:12354–12359. doi: 10.1073/pnas.1406999111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhou T, Zhang S, Xuan LJ, Zhan J, Molnár In. Thioesterase domains of fungal nonreducing polyketide synthases act as decision gates during combinatorial biosynthesis. J Am Chem Soc. 2013b;135:10783–10791. doi: 10.1021/ja4041362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhou T, Zhou Z, Su S, Roberts SA, Montfort WR, … Lin M. Rational reprogramming of fungal polyketide first-ring cyclization. Proc Natl Acad Sci USA. 2013c;110:5398–5403. doi: 10.1073/pnas.1301201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe K, Chihaya N, Hatabayashi H, Kito M, Hoshino S, Zeng H, Cai J, Nakajima H. Production of M-/GM-group aflatoxins catalyzed by the OrdA enzyme in aflatoxin biosynthesis. Fungal Genet Biol. 2012;49:744–754. doi: 10.1016/j.fgb.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, … Moore BS. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci USA. 2014;111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin WB, Chooi YH, Smith AR, Cacho RA, Hu Y, White TC, Tang Y. Discovery of Cryptic Polyketide Metabolites from Dermatophytes Using Heterologous Expression in Aspergillus nidulans. ACS Synth Biol. 2013;2:629–634. doi: 10.1021/sb400048b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Xu F, Zeng J, Zhan J. Type III polyketide synthases in natural product biosynthesis. IUBMB life. 2012;64:285–295. doi: 10.1002/iub.1005. [DOI] [PubMed] [Google Scholar]
- Yu J, Chang PK, Ehrlich KC, Cary JW, Bhatnagar D, Cleveland TE, … Bennett JW. Clustered pathway genes in aflatoxin biosynthesis. Appl Environ Microbiol. 2004;70:1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala AO, Chooi YH, Choi MS, Lin HC, Tang Y. Fungal Polyketide Synthase Product Chain-Length Control by Partnering Thiohydrolase. ACS Chem Biol. 2014;9:1576–1586. doi: 10.1021/cb500284t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MQ. Computational prediction of eukaryotic protein-coding genes. Nat Rev Genet. 2002;3:698–709. doi: 10.1038/nrg890. [DOI] [PubMed] [Google Scholar]
- Zhou H, Qiao K, Gao Z, Vederas JC, Tang Y. Insights into radicicol biosynthesis via heterologous synthesis of intermediates and analogs. J Biol Chem. 2010;285:41412–41421. doi: 10.1074/jbc.M110.183574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zhan J, Watanabe K, Xie X, Tang Y. A polyketide macrolactone synthase from the filamentous fungus Gibberella zeae. Proc Natl Acad Sci USA. 2008;105:6249–6254. doi: 10.1073/pnas.0800657105. [DOI] [PMC free article] [PubMed] [Google Scholar]